Highlights
-
•
Myoglobin exhibited low digestibility during pepsin and pancreatin digestion.
-
•
More changes occurred in structure and digestibility for pancreatin digestion.
-
•
Only four hydrogen bonds existed in the myoglobin-pepsin complex.
Keywords: Myoglobin, Digestibility, Molecular dynamics, Molecular docking, Liquid chromatography-tandem mass spectrometry
Abstract
Myoglobin, a critical protein responsible for meat color, has been shown insusceptible to digestion. The underlying mechanism is not clear. The present study aimed to evaluate whether the structural properties of myoglobin are associated with its insusceptibility to digestion using spectroscopic and computational techniques. Myoglobin was degraded by only 7.03% by pepsin and 33.00% by pancreatin. The structure of myoglobin still maintained α-helix after the two-step digestion, with the exposure of some aromatic residues. In addition, molecular dynamics modeling suggested that hydrophobic amino acid residues (Phe 111, Leu 10, Ala 115, Pro 116) in pepsin and polar amino acid residues (Tyr 146, Thr 95) in myoglobin were found in the proximity of binding sites, which could result in the low digestibility of myoglobin. Our findings provide a new insight into the underlying mechanisms on the difficulty in digestion of myoglobin.
1. Introduction
Myoglobin is a critical component in meat and its chemical status determines meat color (Suman & Joseph, 2013). Previous studies have paid much attention to the associations of myoglobin status with lipid oxidation and meat color (Faustman et al., 2010, Nguyen et al., 2019, Suman and Joseph, 2013). However, several studies for different purposes have shown that myoglobin has a relatively low digestibility. In an analytically chemical study, Park and Russell (2000) observed that myoglobin was more difficult to digest than cytochrome c, but thermal denaturation (90 °C, 20 min), to some extent, increased the rate of enzymatic digestion and the diversity of digest fragments of myoglobin, which could be due to the conformational changes during heating (Zhu, Zhang, Zhou, Xu, & Li, 2018). In a meat authentication study in which myoglobin peptides were detected by multiple reaction monitoring mass spectrometry, Watson, Gunning, Rigby, Philo, and Kemsley (2015) also observed that myoglobin was resistant to digest under several digestion protocols that were based on either ammonium bicarbonate, RapiGest, urea or heat. Ideally, myoglobin may be cleaved by trypsin into many different fragments, but Marchetti and Guiochon (2005) only isolated 14 peptides from the digested products of myoglobin using reverse phase liquid chromatography, which may also reflect a relatively low digestibility of myoglobin. In practice, meat proteins undergo chemical changes during processing that may modify the protein conformations and the protein digestibility (Du et al., 2018a, Du et al., 2018b, He et al., 2018, Li et al., 2017). The secondary and tertiary structures of myoglobin was found to change greatly during heating above 70 °C by ultraviolet–visible absorption, fluorescence and circular dichroism measurements (Zhu et al., 2018).
In the digestive tract, the majority of dietary proteins are firstly digested by pepsin in the stomach, and further degraded by proteases and peptidases, and absorbed in the small intestine. Bauchart et al. (2007) identified reproducible peptides derived from myoglobin and other muscle proteins in duodenal and jejunal contents of pigs that were fed cooked beef and fish. Those authors stated that such peptides could be the most resistant to digestion (Bauchart et al., 2007). In a recent study, Zhang, Zou, Zhao, and Li (2020) identified many peptides derived from diet sources in the colonic contents of mice fed different meat proteins, including a peptide from myoglobin, VEADVAGHGQEVLIR. The abundances of this peptide ranged from 0.14% to 1.74 in mice fed cooked pork protein diet and below 0.42% in mice fed dry-cured pork protein diet (Zhang, Zou et al., 2020)). This finding further indicates that myoglobin could not be well digested and absorbed in the small intestine. Zhang, Zou et al. (2020) also observed some alterations in gene expression in colon tissue involving cellular transition metal ion homeostasis and oxidative stress, in particular to heme-dependent hmox1, and they associated the insusceptibility of myoglobin with the changes in gut microbiota composition and host physiological responses. Thus, more attention should be paid to the digestibility of meat proteins and its potential impact on health.
Taken together, the existing studies indicate that myoglobin may not be easily digested. This could be related to the binding capacity of myoglobin to the enzymes that is affected by the structure of myoglobin. However, the underlying mechanism is unclear. The objectives of this study were to determine the digestibility of myoglobin in an in vitro simulation condition and examine its structural changes during pepsin and pancreatin digestion using multispectral technologies. Moreover, the mode of myoglobin binding to pepsin was evaluated by molecular dynamics modeling. These results may help understand the digestion behavior of myoglobin, and develop techniques to improve the digestibility and nutritional values of myoglobin.
2. Materials and methods
2.1. Reagents
Myoglobin from equine skeletal muscle (CAS No. 100684-32-0), pepsin from porcine gastric mucosa (CAS No. 9001-75-6), pancreatin from porcine pancreas (CAS No. 8049-47-6, a combination of trypsin, chymotrypsin, lipase and amylase) and fluorescamine (CAS No. 38183-12-9) were purchased from Sigma Aldrich (St. Louis, MO, USA). All chemicals were of analytical grade.
2.2. In vitro static digestion
An in vitro static simulated gastrointestinal digestion system was applied according to the procedures of Brodkorb et al. (2019) with minor modifications. In brief, myoglobin (6.5 mg) was dissolved in 1.3 mL simulated gastric fluid. Before being mixed with pepsin, myoglobin was pre-heated at 37 °C for 5 min. Then, pepsin (5 mg/mL) was added to obtain a final activity of 500 U/mL. Myoglobin and pepsin were mixed at ratio of 25: 1 (w: w). The mixtures were incubated at 37 °C for 120 min (pH 2.5) with interval shaking at 250 rpm to ensure enough mixing of myoglobin with pepsin. At different time points of incubation (0, 3, 60 and 120 min), 140 μL samples were taken and immediately mixed with 10 μL simulated intestinal fluid to stop the reaction by elevating the pH to 7.0.
For the intestinal phase, 30 μL pancreatin (5 mg/mL) was added and incubated at 37 °C for 120 min. At 1, 3, 15, 60 and 120 min of incubation, 140 μL of digested samples were withdrawn for further analysis. Heat treatment was not applied to deactivate enzyme because heat may induce irreversible protein denaturation and aggregation.
2.3. Degree of hydrolysis
The degree of hydrolysis (DH) was determined according to the method of Zhao et al. (2018) with minor modifications. Briefly, standard (l-leucine) or sample (75 μL) was mixed with isometric 24% trifluoroacetic acid (TCA), and centrifuged at 13,000 rpm at 4 °C for 25 min. The supernatants were used for determining the fluorescamine primary amines. TCA treated standard (l-leucine) or samples (30 μL) were mixed with 900 μL of 0.1 M sodium tetraborate buffer (pH 8.0), and then 300 μL of fluorescamine (0.2 mg/mL) in acetone was added. Fluorescence was measured at excitation and emission wavelengths of 390 and 480 nm, respectively. The DH was calculated as follows:
where [–NH2] is the concentration of primary amines in the hydrolyzed samples (h) or unhydrolyzed samples (0). [–NH2]∞ is the theoretical maximal primary amine concentration assuming total digestion to free amino acids (AAs) and [–NH2]∞ is calculated as follows:
where flys is the fraction of lysine residues in the protein, CMb is the myoglobin concentration and MWAA is the mean molecular weight of the AAs in the protein.
2.4. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
SDS-PAGE was performed to separate proteins during digestion according to Wen et al. (2015) with some modifications. Briefly, samples were mixed with 5× sample buffer at a ratio of 4 to 1 and heated at 95 °C for 10 min. After cooling, 15 μL of samples (1 μg/μL) were loaded on 10% Precast gels (GenScript, Nanjing, China). A broad pre-stained protein standard (MW 5–270 kDa; GenScript, Nanjing, China) was applied to estimate molecular weights of samples. The electrophoresis was conducted at 120 V for 80 min at 4 °C in 1000 mL running buffer (25 mM Tris-HCI, pH 8.6; 192 mM glycine; 0.1% SDS). The gels were stained with Coomassie Blue R-250, and images were captured by a Molecular Imager GelDoc™ XR + system (Bio-Rad Laboratories, Hercules, CA, USA).
2.5. UV–visible spectroscopy
The UV–Vis spectra of digested samples were obtained in the range from 250 to 650 nm in a spectrophotometer (Thermo Scientific, Fremont, CA, USA). The concentration of protein used was 0.2 mg/mL. The measurements were repeated for five times.
2.6. Far-UV circular dichroism spectroscopy
Circular dichroism (CD) spectra were recorded according to Khan et al. (2019) with some modifications. Digested myoglobin samples (0.05 mg/mL) were measured on a spectropolarimeter (Chirascan, Applied Photophysics Ltd., Leatherhead, Surrey, UK) in the range from 185 to 260 nm. All of these were collected in a cell of 0.5 mm path length at room temperature (21 °C). A wavelength step of 1.0 nm and band width of 1.0 nm was applied. Each spectrum was the average of 2 scans to improve the signal-to-noise ratio.
2.7. Fluorescence spectroscopy
Fluorescence of digested samples was measured according to the procedures of Khan et al. (2019) with some modifications. Briefly, the measurements were done on a spectrofluorometer (RF-5301, Jasco, Tokyo, Japan) using fluorescence-free quartz cuvettes (1 cm path width). Myoglobin or its digested samples were diluted to a final concentration of 0.1 mg/mL. For intrinsic fluorescence assay, the excitation wavelength was 280 nm and the emission spectra were scanned from 300 to 400 nm. Excitation and emission slit widths were set at 3.0 and 5.0 nm, respectively. Synchronous fluorescence spectra were recorded from 280 to 400 nm in which the intervals of excitation and emission wavelengths were set at 20 and 60 nm, characterizing the tyrosine and tryptophan residues, respectively. The excitation and emission slit widths for Δλ20 were both set at 5.0 nm, while the slit widths for Δλ60 were set at 3.0 and 5.0 nm, respectively.
2.8. Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Digested peptides were identified by LC-MS/MS according to the method of Wen et al. (2015) with some modifications. Digested samples taken at 120 min of pepsin and pancreatin digestion were transferred to a 10 kDa ultrafiltration centrifuge tube, and centrifuged at 14,000×g for 15 min. Peptide mixtures were fractionated on a LC system (Thermo Scientific™ Q Exactive™, USA) combined with a BioBasic, 0.18 mm × 100 mm C18 sampling column and a RP-C18 separation column (0.15 mm × 150 mm). A step-gradient elution was used at a flow rate of 2.5 μL/min with solvent A (formic acid/water, 1:1000 v/v) and solvent B (formic acid/acetonitrile, 1:1000 v/v) as follows: 0–50 min, 4% to 50% solvent B; 51–54 min, 50% to 100%, 55–60 min, 100% solvent B.
The hybrid quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to identify the peptide mixture in a positive ion mode. The mass scan was set from 50 to 6000 m/z and the time was set at 60 min. The obtained MS/MS spectra were interpreted with the software MaxQuant 1.5.5.1 (Max-Planck-Institute of Biochemistry, Germany) using Uniprot database (https://www.uniprot.org/uniprot/P68082) with a false discovery rate (FDR) of 1%. Data matching was performed with a fragment ion mass tolerance of 0.1 Da and a parent ion tolerance of 20 ppm. An unspecific enzyme was used in peptic and pancreatic peptides database search. Oxidation of methionine and carbamidomethyl formation of cystine was selected as dynamic and fixed modifications, respectively, and two missing cleavages were allowed.
2.9. Molecular docking and molecular dynamics simulation
Molecular docking was done according to the procedure of Pierce et al. (2014). Briefly, the structure of myoglobin (PDB ID: 1mnh) and pepsin (PDB ID: 3UTL) were downloaded from the RSCB protein data bank (http://www.rscb.org/pdb). Then the substrate 1mnh was docked with the 3UTL enzyme active site using the ZDOCK (http://zdock.umassmed.edu). The binding complexes with top scores were applied as a starting point for subsequent molecular dynamics simulation (MDS).
MDS was performed using the GROMACS software (package version 5.1.2, BioExcel HPC Center of Excellence, the European Union). The FF99SB force field was used to simulate the changes of amino acid residues of proteins (Lindorff-Larsen et al., 2010). In the MDS system, sodium ion would be added to reach the electrical neutrality. The system would be immersed in a water box of TIP3P water model using AmberTools (University of California, San Francisco, USA) under periodic boundary conditions. During the whole MDS, energy was minimized using the steepest descent method and gradual heating was performed to 300 K with 20,000 minimization steps. After that, the system was equilibrated at 300 K for 100 ps. Then, a 10000 ps MDS was conducted under constant volume and temperature.
2.10. Statistical analysis
The effect of digestion time on measured variables was evaluated by one-way analysis of variance and least square means were compared by Tukey’s t-test. A difference was considered significant when the p value was smaller than 0.05. Statistical analysis was conducted using the SAS 8.1 program (SAS Institute Inc., Cary, NC, USA).
3. Results
3.1. Changes of in vitro digestibility of myoglobin
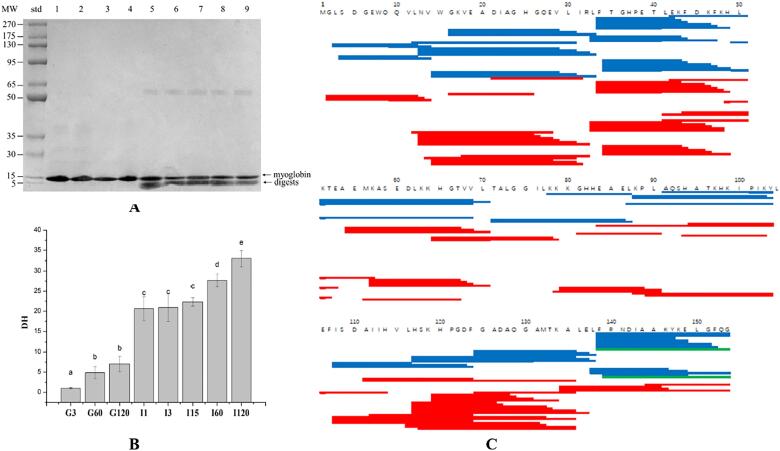
SDS-PAGE profile showed that a band close to the 15 kDa band on the calibration lane corresponded to the native myoglobin (Fig. 1A). The band did not change significantly in intensity across pepsin digestion. However, the pancreatin-treated samples had a new band close to 5 kDa band on the calibration lane (Fig. 1A), and meanwhile the band intensities showed a certain reduction during digestion. However, the band intensities were still very strong at 120 min of digestion. These observations indicate that myoglobin could be not susceptible to pepsin digestion and partially difficult to pancreatin digestion. Correspondingly, the DH values were very low across the 120 min pepsin digestion (Fig. 1B), while the pancreatin treatment increased the DH values, which reached 33.00% at 120 min of pancreatin digestion (Fig. 1B). However, the DH of 33.00% is much lower than those of other reported food proteins (Zhao et al., 2018).
Fig. 1.
Patterns reflecting the digestibility of myoglobin under in vitro digestion. (A) SDS-PAGE profiles of myoglobin and its digested products: Lane 0 shows the calibration markers (5–270 kDa). Lanes 1, 2, 3 and 4 show pepsin-treated samples for 0, 3, 60 and 120 min; and lanes 5, 6, 7, 8 and 9 show pancreatin-treated samples 1, 3, 15, 60 and 120 min after treatment by pepsin for 120 min. (B) The DH values of myoglobin in pepsin and pancreatin digestion. G0, G3, G60 and G120 refer to pepsin-treated samples for 0, 3, 60 and 120 min, respectively; I1, I3, I15, I60 and I120 refer to pancreatin-treated samples 1, 3, 15, 60 and 120 min after treatment by pepsin for 120 min, respectively. Different letters indicate a significant difference (P < 0.05). (C) Peptide matching for pepsin and pancreatin treated samples. Peptides in blue represent the pepsin-treated samples; peptides in red represent the pancreatin-treated samples; and peptides in green represent those existing in pepsin and pancreatin treated samples.
LC-MS/MS data showed that myoglobin was cleaved into a great number of fragments by pepsin and pancreatin (Fig. 1C). A total of 62 and 84 peptides were identified in the pepsin-treated and pancreatin-treated samples, respectively. The majority of identified peptides had 10 to 20 amino acid residues with a size of 1 to 3 kDa. Two peptides, RNDIAAKYKELGFQG and FRNDIAAKYKELGFQG, were found in both pepsin-treated and pancreatin-treated samples. This indicates that these two peptides were rather difficult to be digested and may have some physiological effect such as anti-digestion.
3.2. Changes of myoglobin structure during in vitro digestion
3.2.1. UV–Vis absorption spectroscopy
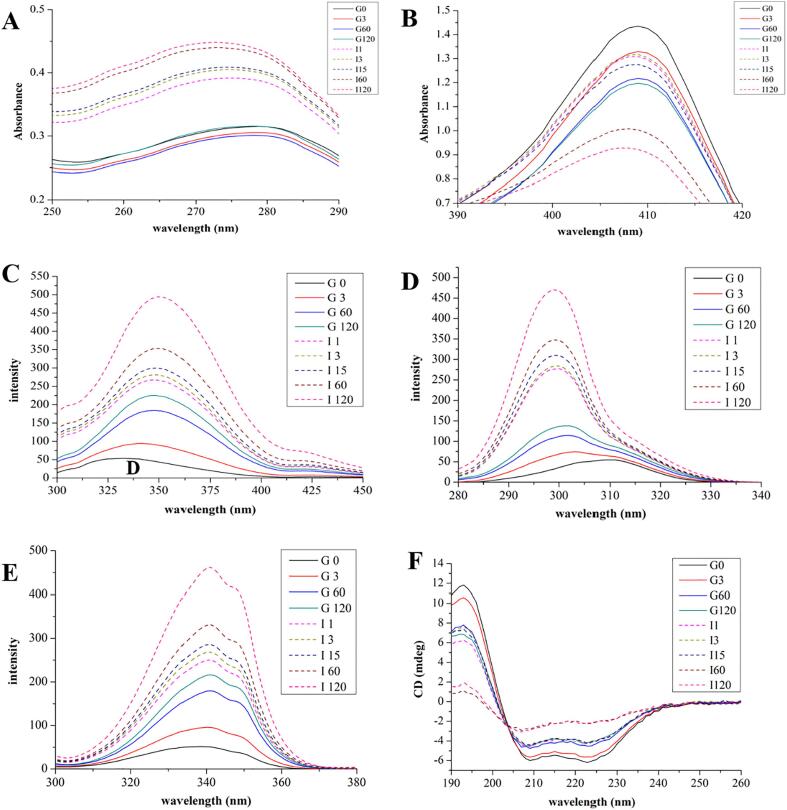
Changes in the microenvironment of myoglobin were evaluated by the UV–Vis spectra (Fig. 2A & B). Native myoglobin had two major bands at 279 nm and 409 nm. The absorbance at 279 nm increased from 0.315 to 0.436 across pepsin and pancreatin digestion (Fig. 2A). The absorbance of Soret bands due to the absorption of UV light by the heme group decreased from 1.435 at 0 min to 1.197 at 120 min in pepsin digested samples, with a significant reduction from 0 to 60 min (P < 0.05, Fig. 2B). In the pancreatin-treated samples, an obvious decline in the Soret bands was observed from 15 to 120 min of digestion.
Fig. 2.
Spectra of myoglobin during in vitro digestion. (A) 279 nm bands of UV–Vis spectra; (B) 409 nm bands of UV–Vis spectra; (C) Intrinsic fluorescence spectra of Mb; (D) Synchronous fluorescence spectra for Tyr residues (Δλ = 20 nm); (E) Synchronous fluorescence spectra for Trp residues (Δλ = 60 nm); (F) CD spectra of myoglobin. G0, G3, G60 and G120 refer to pepsin-treated samples for 0, 3, 60 and 120 min, respectively; I1, I3, I15, I60 and I120 refer to pancreatin-treated samples for 1, 3, 15, 60 and 120 min after treatment by pepsin, respectively.
3.2.2. Intrinsic fluorescence spectroscopy
The intrinsic fluorescence spectra of myoglobin are shown in Fig. 2C and Table 1. When excited at 280 nm, myoglobin exhibited the maximum emission at 333.13 nm but the fluorescence intensity was low. In pepsin-treated samples, the fluorescence intensity of samples increased drastically from 54.22 at 0 min to 225.42 at 120 min along with a red shift of the maximum emission from 333.13 to 347.50 nm. However, the change in fluorescence intensity within the first 60 min was greater than that between 60 and 120 min (130.74 vs. 40.46, Table 1). Normally, in a strong acid environment, protein may be denatured and the distance between aromatic amino acid residues will be increased, which will result in the reduction in the resonance energy transfer and the fluorescence intensities of aromatic amino acid residues. However, in the present study, the fluorescence intensity was observed to increase, which indicates that the tertiary structure of myoglobin could be changed upon mixing with pepsin. The intrinsic fluorescence intensities further increased during pancreatin digestion from 271.64 at 1 min to 494.62 at 120 min (P < 0.05, Fig. 2C, Table 1). Thus, myoglobin exhibited a higher susceptibility to pancreatin digestion than to pepsin digestion, which was in line with the UV–Vis results.
Table 1.
The effect of in vitro digestion on the characteristic peaks of fluorescence spectra (Means ± SD, n = 5 each).
| Sample ID | Synchronous fluorescence |
|||||
|---|---|---|---|---|---|---|
| Intrinsic fluorescence |
Δλ = 20 nm |
Δλ = 60 nm |
||||
| wavelength at peak position | Intensity of the peak | wavelength at peak position | Intensity of the peak | wavelength at peak position | Intensity of the peak | |
| Pepsin digestion | ||||||
| G0 | 333.13 ± 2.75c | 54.22 ± 2.75d | 310.11 ± 0.93a | 54.85 ± 3.88d | 338.25 ± 1.49b | 51.55 ± 2.62d |
| G3 | 340.00 ± 1.90b | 94.52 ± 4.13c | 303.33 ± 0.52b | 74.54 ± 1.83c | 340.50 ± 0.55a | 95.88 ± 4.71c |
| G60 | 347.80 ± 1.10a | 184.96 ± 12.22b | 301.20 ± 0.45c | 118.07 ± 6.32b | 341.00 ± 0.00a | 179.51 ± 9.16b |
| G120 | 347.50 ± 1.05a | 225.42 ± 20.57a | 301.43 ± 1.13c | 138.96 ± 9.69a | 340.83 ± 0.41a | 216.48 ± 21.61a |
| Pancreatin digestion | ||||||
| I1 | 340.83 ± 0.41C | 271.64 ± 26.38C | 300.00 ± 0.00A | 279.36 ± 18.21C | 340.83 ± 0.41 | 250.33 ± 18.93D |
| I3 | 348.67 ± 1.03AB | 281.31 ± 15.02C | 299.86 ± 0.38AB | 283.97 ± 22.42C | 340.83 ± 0.41 | 268.61 ± 12.33CD |
| I15 | 348.33 ± 1.21bB | 300.26 ± 14.85C | 299.50 ± 0.55B | 310.13 ± 17.14C | 340.67 ± 0.52 | 286.08 ± 12.62C |
| I60 | 349.50 ± 1.22bAB | 353.68 ± 28.13B | 299.00 ± 0.00C | 347.42 ± 27.76B | 340.83 ± 0.41 | 331.38 ± 26.15B |
| I120 | 350.00 ± 1.26A | 494.62 ± 33.33A | 299.00 ± 0.00C | 470.27 ± 38.12A | 340.83 ± 0.41 | 462.03 ± 29.90A |
Note: G0, G3, G60, G120 refer to pepsin-treated samples for 0, 3, 60, 120 min, respectively; I1, I3, I15, I60, I120 refer to pancreatin-treated samples for 1, 3, 15, 60, 120 min after treatment by pepsin, respectively. a,b,c,d, different lower letters in the same column indicate significant differences among different time points during pepsin digestion (P < 0.05). A,B,C,D, different upper letters in the same column indicate significant differences among different time points during pepsin digestion (P < 0.05).
3.2.3. Synchronous fluorescence spectroscopy
The synchronous fluorescence spectra of tyrosine (Tyr) and tryptophan (Trp) residues in myoglobin increased during digestion (P < 0.05, Fig. 2D & E, Table 1). The fluorescence intensities of the Tyr and Trp residues increased during pepsin and pancreatin digestion, but a blue shift was observed from 310.11 to 299.00 nm for the Tyr residue and a red shift was seen from 338.25 to 340.83 nm for the Trp residue. Notably, during pepsin digestion, a 6.78 nm blue shift occurred from 0 to 3 min at Δ λ = 20 nm, with an increase in fluorescence intensity (P < 0.05, Table 1). The binding capacity of myoglobin or its fragments to pepsin did not change from 60 to 120 min because the maximum emission wavelength changed little (P > 0.05, Table 1). During pancreatin digestion, the maximum emission wavelength did not change too much, but the fluorescence intensity increased greatly, which indicates that the binding capacity of myoglobin fragments to pancreatin enzymes is relatively stable. The fluorescence intensity of the Tyr residue increased from 138.96 at 120 min of pepsin digestion to 279.36 at 1 min of pancreatin digestion (Table 1), which could be due to the pH change from 2.5 to 7.0. This is because when pH changes from acidic to neutral conditions, some hydrophobic residues will be exposed. Overall, the change in fluorescence intensity during pancreatin digestion was higher than that for pepsin digestion (190.91 vs. 84.11), indicating that pancreatin contributed more to myoglobin digestion.
3.2.4. Circular dichroism spectroscopy
Far-UV CD spectroscopy is a powerful technique to shed light on the changes in secondary structures of protein. The CD spectra of myoglobin and its digestion products in the range from 190 to 260 nm are shown in Fig. 2F. There are two negative bands at 209 and 223 nm, which are characteristic of α-helix structures in proteins. The 209 nm band corresponds to π–π* transition of the α-helix and the 223 nm band corresponds to π–π* transition for both the α-helix and random coil (Xie et al., 2014). In the first 60 min of pepsin digestion, the intensities of the 209 nm and 223 nm bands decreased drastically. This reflects changes in secondary structure in myoglobin, corresponding to an obliteration of native structure and the partial unfolding of the protein, signifying that pepsin altered the helical structure of protein, and the β-sheet and random coil appeared. At 120 min, the absolute value of these two bands increased by 1.45 mdeg and 2.04 mdeg, respectively. The CD spectra of myoglobin exhibited the similar changes in the pancreatin digestion to those in the pepsin digestion (Fig. 2F). However, the α-helical structure was still predominant in pepsin- and pancreatin-treated samples of myoglobin.
3.3. Molecular dynamics simulation analysis
The above results showed that myoglobin was not well digested by pepsin. This could be associated with the “rigid structure” of myoglobin. We further explore the possible binding sites and mode between myoglobin and pepsin using molecular docking and MDS.
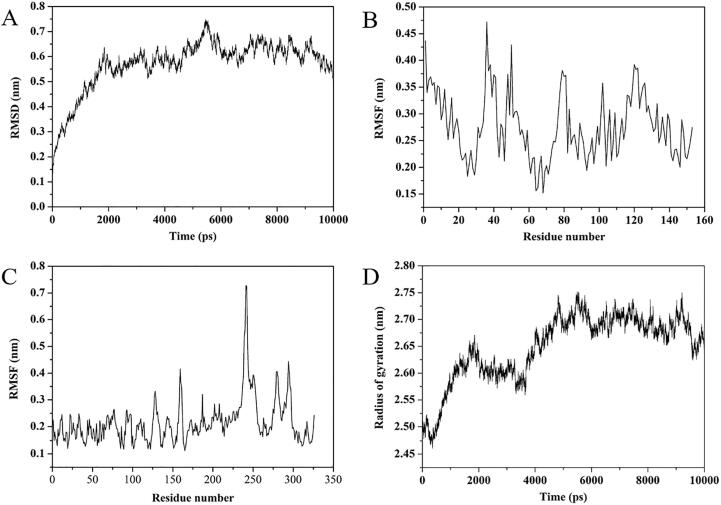
The root-mean-square deviation (RMSD) values of the backbone atoms of the system were obtained to evaluate the stability of the myoglobin-pepsin complex in the 10000 ps MDS. The RMSD values increased dramatically in the first 2000 ps, which could be attributed to the structure optimization of the system, and then the RMSD values kept constant (Fig. 3A). These results suggest that the myoglobin-pepsin system achieved an equilibrium status after 2000 ps of MDS.
Fig. 3.
Patterns related to the protein stability over 10 ns MD simulation. (A) The RMSD plot of myoglobin-pepsin complex. (B-C) The RMSF values of myoglobin and pepsin over the amino acid residue numbers, respectively. (D) Radius of gyration (Rg) values.
The root mean square fluctuation (RMSF) values of each residue of myoglobin and pepsin were also calculated to reflect the mobility of specific residues around their mean position (Fig. 3B & C). The RMSF values of myoglobin showed a great fluctuation during simulation, indicating a dynamic shift of residues from their initial position. During the whole MDS process, the residues 20–30 and 60–70 had lower RMSF values than other residues, which could be attributed to the existence of α helices that were far away from binding sites and the active sites of pepsin. The residues 34–40, 45–50, 77–80 and 120–126 located in the loop had higher RMSF values, of which the first two domains were located in the loop close to the binding sites and the other two domains were far away from the binding sites. These results suggest that these loops could be altered when myoglobin interacts with pepsin. The RMSF values of pepsin exhibited minor fluctuation during MDS (Fig. 3C). The RMSF values of most residues were less than 2 Å. There are some fluctuations around residues 239–253 which are located in the loop domains far away from active sites of pepsin, indicating that these loops could be flexible and less stable than the other residues. Thus, these loops may play a critical role in interacting with myoglobin.
In addition, the radius of gyration (Rg) of Cα was evaluated on the 10000 ps MD trajectories and plotted against the simulation time to monitor the conformational changes of the complex (Fig. 3D). The Rg values increased drastically in the first 2000 ps, and reduced in the second 2000 ps with a slight fluctuation in the subsequent simulation, indicating variations in compactness of the myoglobin-pepsin structure. Concerning the binding process, myoglobin and pepsin might unfold at the beginning of binding and then fold again when they interact. The Rg values range from 2.5 to 2.75 nm. This indicates that the myoglobin-pepsin complex could be compact, and myoglobin could not bind well with pepsin. Therefore, the rigid structure of myoglobin-pepsin system may explain the relatively low in vitro digestibility of myoglobin.
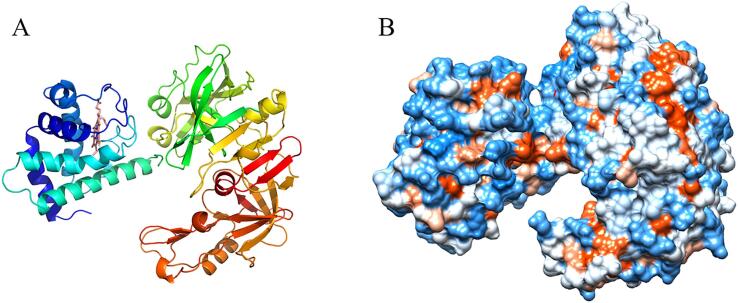
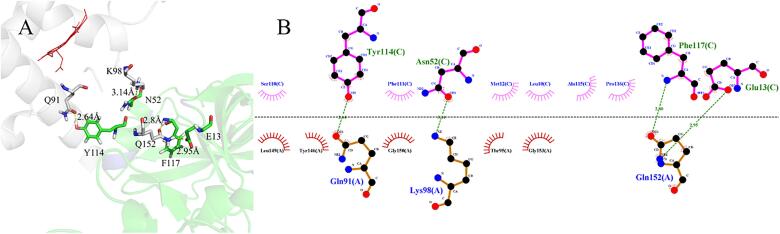
To get more information on the molecular interaction between myoglobin and pepsin, the binding modes of myoglobin-pepsin were constructed by the Pymol software based on the MDS data (Fig. 4, Fig. 5). Myoglobin may not immerse into the active cavity of pepsin, which could be associated with the size and the globular shape of myoglobin. This result is in agreement with the low digestibility observed in DH and SDS-PAGE. Furthermore, the 2D protein–protein interaction diagram revealed four hydrogen bonds existing between myoglobin and pepsin, with distances of 2.64, 3.14, 2.95 and 2.80 Å, respectively (Fig. 5B). The hydrogen bonds could play an important role in stabilizing the myoglobin-pepsin complex. Hydrogen bonds are considered strong if the distance between donor and acceptor ranges from 1.8 to 2.3 Å, while they are considered weak if the distance is 2.5–3.2 Å (Dannenberg, 1998). Thus, the hydrogen bonds between myoglobin and pepsin are weak. In addition, both hydrophobic amino acid residues (Phe 111, Leu 10, Ala 115, Pro 116) in pepsin and polar residues (Tyr 146, Thr 95) in myoglobin were found around the binding sites (Fig. 5A & B). Therefore, hydrogen bonds and hydrophobic interaction could be the main driving forces for the binding of myoglobin to pepsin.
Fig. 4.
The final structure of myoglobin-pepsin complex after MD simulation. (A) three-dimensional structure of the complex; (B) hydrophobic surface map of the complex.
Fig. 5.
Interaction mode of myoglobin-pepsin complex. (A) Three-dimensional structure of hydrogen bonds network. The residues of myoglobin and pepsin are shown in white and green sticks, respectively. The yellow dashed lines represent hydrogen bonds. (B) Two-dimensional representation of myoglobin and pepsin. The residues upon the dotted line refer to pepsin, the residues below the dotted line refer to myoglobin. The green dashed lines indicate hydrogen bonds.
4. Discussion
Myoglobin is a special sarcoplasmic protein responsible for the attractive color for the fresh meat, and the protein comprises of 153 amino acid residues and eight alpha helices (Suman & Joseph, 2013). Myoglobin has a “rigid” flattened ball, with a hydrophilic surface and hydrophobic pocket to embed heme (Kaur, Banipal, & Banipal, 2017). The heme prosthetic group comprises of four porphyrin rings and an iron atom and is enwrapped in the hydrophobic pocket (Thiansilakul, Soottawat, & Richards, 2011). Such a special structure may determine the capacities of carrying oxygen in living muscles and maintaining color stability in meat. However, the “rigid” structure of myoglobin may also affect the protein digestibility.
In the in vivo conditions, dietary proteins undergo pepsin digestion under low pH conditions, which results in the alteration of polypeptide chains and the exposure of hydrophobic amino acid residues. When the gastric fluid enters into the duodenum, the exposed hydrophobic amino acid residues of dietary proteins or their fragments may bind to trypsin, chymotrypsin and peptidases to be further degraded into smaller pieces. In the present study, myoglobin showed quite low degree of hydrolysis in pepsin digestion. Pancreatin digestion increased the digestibility of myoglobin but the degree of hydrolysis was much lower than other food proteins, which may reach 83.1% for β-casein (Zhao et al., 2018). This could be attributed to the afore-mentioned “rigid” structure of myoglobin.
It is well known that enzymes prefer to cleave the proteins at specific peptide bonds. Pepsin preferentially cleaves the bonds between hydrophobic and aromatic amino acids, e.g., tyrosine, tryptophan and phenylalanine (Mohseni-Shahri, Moeinpour, & Nosrati, 2018). Pepsin is composed of two homologous domains, including an N terminal and a C terminal, both of which are composed of β-sheets (Shen et al., 2015). The catalytic sites of pepsin are located in the cavity between these two domains with active Asp32 and Asp215 (Yue et al., 2019). As the main enzyme in pancreatin, trypsin tends to cleave proteins at the sites of Arg and Lys residues (Schuchert-Shi & Hauser, 2009). There are 2 Tyr residues, 2 Trp residues and 7 Phe residues in myoglobin. However, these amino acids are located in α-helices and buried in the hydrophobic core, which is not accessible for pepsin. This could to some extent explain the low pepsin digestibility of myoglobin. In addition, Arg and Lys residues in myoglobin are also located in α-helices, but they are exposed to the hydrophilic surface. This may interpret the relatively higher pancreatin digestibility of myoglobin. Thus, the unique structure of myoglobin determines its low digestibility. Noticeably, the number of identified peptides in the present study is much greater than previous studies (Marchetti and Guiochon, 2005, Watson et al., 2015). This could be attributed to three reasons. Firstly, pancreatin is a mixture of proteases, mainly including trypsin and chymotrypsin that have more cleavage sites. Secondly, the LC-MS/MS system could have higher sensitivity. Thirdly, in previous analytical studies, trypsin digestion time was quite long and some specific pretreatments (e.g., heat and urea) were used.
In myoglobin, porphyrin ring of heme moiety acts as a chromophore and gives rise to characteristic peak at 409 nm, which is attributed to the interaction between heme in hydrophobic environment and globin (Nasreen, Ahamad, Ahmad, Hassan, & Islam, 2018). Thus, the changes of the peak at 409 nm may reflect the environment and dissociation of heme. The 409 nm peak of Soret bands decreased during digestion, indicating the weakened interaction between polypeptide chain and heme, resulting in the unfolding of the protein. As aforementioned, tryptophan, tyrosine and phenylalanine residues are located in the hydrophobic core of the native myoglobin. Pepsin and pancreatin digestion promote the exposure of these residues, which can be reflected by the increase in intrinsic fluorescence of myoglobin. Actually, tryptophan contributes primarily to intrinsic fluorescence because its sensitivity and quantum efficiency is much higher than tyrosine and phenylalanine. Two tryptophan residues in myoglobin are located at positions 7 and 14 (designated as W7 and W14). In native myoglobin, W7 is exposed toward the polar solvent while W14 appears to be buried in the helices (Nasreen et al., 2018). Because of the proximity of two tryptophan residues to heme, fluorescence of tryptophan is largely quenched. In pepsin- and pancreatin-treated samples, the tertiary structure of myoglobin was altered. The rapid increase in fluorescence intensity and the significant red shift signify that the tryptophan residues may move away from the heme and be exposed to a more polar environment (Kohn et al., 2018, Mondal et al., 2019). In the present study, the blue shift of tyrosine residues and the red shift of tryptophan residues may result from their microenvironment alterations. In addition, the CD spectra indicate transition of α-helix structure to β-sheet or random coil structure in myoglobin.
According to spectroscopic observations, myoglobin seems susceptible to digestion by pepsin and pancreatin. However, the SDS-PAGE profile and the degree of hydrolysis showed the relatively low digestibility of myoglobin. We speculate that the low digestibility may result from its “rigid” structure that affects the binding capacity of myoglobin to digestive enzymes and the rate of digestion. Several classical hypotheses for enzymatic catalysis may give some indications. If myoglobin is considered as the key and the digestive enzymes are considered as the lock, the “rigid” myoglobin may not fit the active sites of enzymes well, in particular to pepsin (Benkovic & Hammes-Schiffer, 2003). The four relatively weak hydrogen bonds between myoglobin and pepsin may also reflect the undesirable fitness of the “key” to the “lock” (Dannenberg, 1998). On the other hand, enzymes, especially of pancreatin, may exhibit some flexibility to fit the “rigid” myoglobin in a transition state to form a macromolecule under hydrophobic and steric effects (Benkovic and Hammes-Schiffer, 2003, Estell et al., 1986, Hammes, 2002). However, the efficiency of such a reaction could be not very high based our results. In physiological conditions, the low digestibility and digestion efficiency of myoglobin may result in a low bioavailability of the protein and its accumulation in in the cecum and colon, where the protein or its fragments are fermented by gut microbiota. The fermentation leads to the production of toxic products and interference with cellular metabolism and DNA synthesis of the host (Corpet et al., 1995, Simonetti et al., 2016).
Finally, the nutritional value of dietary proteins is closely related to their susceptibility to digestion (Han, Chee, & Cho, 2015). In meat, collagen shows relatively low digestibility, too (Zhang, Zhao et al., 2020) although the majority of proteins are susceptible to digestion. In addition, protein oxidation during excessive processing may also impair the protein digestibility (He et al., 2018, Li et al., 2017). Therefore, new strategies are needed to make such proteins as myoglobin and collagen more liable to digestion and to alleviate protein oxidation.
5. Conclusion
Myoglobin showed relatively low degree of hydrolysis in pepsin and pancreatin digestion, although significant changes were observed in spectroscopic measurements and a number of peptides were identified by LC-MS/MS. The “rigid” nature of myoglobin and the relatively weak hydrogen bonds between myoglobin and digestive enzymes could be the main reason for the lowly efficient digestion of myoglobin. The findings provide a new insight into the underlying mechanisms on the difficulty in digestion of myoglobin.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the grants from NSFC (31530054), MOST (10000 Talent Project), Jiangsu Department of Finance (CX (18)2024), and Jiangsu Provincial Department of Education (PAPD).
References
- Bauchart C., Morzel M., Chambon C., Mirand P.P., Reynes C., Buffiere C., Remond D. Peptides reproducibly released by in vivo digestion of beef meat and trout flesh in pigs. British Journal of Nutrition. 2007;98(6):1187–1195. doi: 10.1017/S0007114507761810. [DOI] [PubMed] [Google Scholar]
- Benkovic S.J., Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301(5637):1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- Brodkorb A., Egger L., Alminger M., Alvito P., Assuncao R., Ballance S.…Recio I. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nature Protocols. 2019;14(4):991–1014. doi: 10.1038/s41596-018-0119-1. [DOI] [PubMed] [Google Scholar]
- Corpet D.E., Yin Y., Zhang X., Rémésy C., Stamp D., Medline A.…Archer M.C. Colonic protein fermentation and promotion of colon carcinogenesis by thermolyzed casein. Nutrition and Cancer. 1995;23(3):271–281. doi: 10.1080/01635589509514381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg J.J. An introduction to hydrogen bonding by George A Jeffrey. Journal of America Chemistry Society. 1998;120(22):5604. doi: 10.1021/ja9756331. [DOI] [Google Scholar]
- Du X., Sun Y., Pan D., Wang Y., Ou C., Cao J. Change of the structure and the digestibility of myofibrillar proteins in Nanjing dry-cured duck during processing. Journal of the Science of Food and Agriculture. 2018;98:3140–3147. doi: 10.1002/jsfa.8815. [DOI] [PubMed] [Google Scholar]
- Du X., Sun Y., Pan D., Wang Y., Ou C., Cao J. The effect of structural change on the digestibility of sarcoplasmic proteins in Nanjing dry-cured duck during processing. Poultry Science. 2018;97(12):4450–4457. doi: 10.3382/ps/pey316. [DOI] [PubMed] [Google Scholar]
- Estell D.A., Graycar T.P., Miller J.V., Powers D.B., Wells J.A., Burnier J.P., Ng P.G. Probing steric and hydrophobic effects on enzyme-substrate interactions by protein engineering. Science. 1986;233(4764):659–663. doi: 10.1126/science.233.4764.659. [DOI] [PubMed] [Google Scholar]
- Faustman C., Sun Q., Mancini R., Suman S. Myoglobin and lipid oxidation interactions: Mechanistic bases and control. Meat Science. 2010;86(1):86–94. doi: 10.1016/j.meatsci.2010.04.025. [DOI] [PubMed] [Google Scholar]
- Hammes G.G. Multiple conformational changes in enzyme catalysis. Biochemistry. 2002;41(26):8221–8228. doi: 10.1021/bi0260839. [DOI] [PubMed] [Google Scholar]
- Han S.W., Chee K.M., Cho S.J. Nutritional quality of rice bran protein in comparison to animal and vegetable protein. Food Chemistry. 2015;172:766–769. doi: 10.1016/j.foodchem.2014.09.127. [DOI] [PubMed] [Google Scholar]
- He J., Zhou G., Bai Y., Wang C., Zhu S., Xu X., Li C. The effect of meat processing methods on changes in disulfide bonding and alteration of protein structures: Impact on protein digestion products. RSC Advances. 2018;8:17595. doi: 10.1039/c8ra02310g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A., Banipal P.K., Banipal T.S. Study on the interactional behaviour of transition metal ions with myoglobin: A detailed calorimetric, spectroscopic and light scattering analysis. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2017;174:236–244. doi: 10.1016/j.saa.2016.11.041. [DOI] [PubMed] [Google Scholar]
- Khan M.S., Rehman M.T., Bhat S.A., Tabrez S., Hussain A., Husain F.M.…Sumbul S. Food additive dye (quinoline yellow) promotes unfolding and aggregation of myoglobin: A spectroscopic and molecular docking analysis. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2019;214:216–226. doi: 10.1016/j.saa.2019.01.090. [DOI] [PubMed] [Google Scholar]
- Kohn E.M., Lee J.Y., Calabro A., Vaden T.D., Caputo G.A. Heme dissociation from myoglobin in the presence of the Zwitterionic detergent N, N-dimethyl-N-dodecylglycine betaine: Effects of ionic liquids. Biomolecules. 2018;8(4):126. doi: 10.3390/biom8040126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liu Y., Zou X.Y., He J., Xu X., Zhou G., Li C. In vitro protein digestibility of pork products is affected by the method of processing. Food Research International. 2017;92:88–94. doi: 10.1016/j.foodres.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins-Structure Function and Bioinformatics. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti N., Guiochon G. Separation of peptides from myoglobin enzymatic digests by RPLC. Influence of the mobile-phase composition and the pressure on the retention and separation. Analytical Chemistry. 2005;77(11):3425–3430. doi: 10.1021/ac050541c. [DOI] [PubMed] [Google Scholar]
- Mohseni-Shahri F.S., Moeinpour F., Nosrati M. Spectroscopy and molecular dynamics simulation study on the interaction of sunset yellow food additive with pepsin. International Journal of Biological Macromolecules. 2018;115:273–280. doi: 10.1016/j.ijbiomac.2018.04.080. [DOI] [PubMed] [Google Scholar]
- Mondal S., Raposo M.L., Ghosh A., Prieto G., Ghosh S. Physicochemical and conformational studies on interaction of myoglobin with an amino-acid based anionic surfactant, sodium N-dodecanoylsarcosinate (SDDS) Colloids and Surfaces A-Physicochemical and Engineering Aspects. 2019;577:167–174. doi: 10.1016/j.colsurfa.2019.05.061. [DOI] [Google Scholar]
- Nasreen K., Ahamad S., Ahmad F., Hassan M.I., Islam A. Macromolecular crowding induces molten globule state in the native myoglobin at physiological pH. International Journal of Biological Macromolecules. 2018;106:130–139. doi: 10.1016/j.ijbiomac.2017.08.014. [DOI] [PubMed] [Google Scholar]
- Nguyen T., Kim S., Kim J.G. Diffuse reflectance spectroscopy to quantify the met-myoglobin proportion and meat oxygenation inside of pork and beef. Food Chemistry. 2019;275:369–376. doi: 10.1016/j.foodchem.2018.09.121. [DOI] [PubMed] [Google Scholar]
- Park Z., Russell D. Thermal denaturation: A useful technique in peptide mass mapping. Analytical Chemistry. 2000;72:2667–2670. doi: 10.1021/ac991444k. [DOI] [PubMed] [Google Scholar]
- Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z. ZDOCK Server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30(12):1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert-Shi A., Hauser C. Peptic and tryptic digestion of peptides and proteins monitored by capillary electrophoresis with contactless conductivity detection. Analytical Biochemistry. 2009;387:202–207. doi: 10.1016/j.ab.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Shen L., Xu H., Huang F., Li Y., Xiao H., Yang Z.…Li Y. Investigation on interaction between Ligupurpuroside A and pepsin by spectroscopic and docking methods. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2015;135:256–263. doi: 10.1016/j.saa.2014.06.087. [DOI] [PubMed] [Google Scholar]
- Simonetti A., Gambacorta E., Perna A. Antioxidative and antihypertensive activities of pig meat before and after cooking and in vitro gastrointestinal digestion: Comparison between Italian autochthonous pig Suino Nero Lucano and a modern crossbred pig. Food Chemistry. 2016;212:590–595. doi: 10.1016/j.foodchem.2016.06.029. [DOI] [PubMed] [Google Scholar]
- Suman S.P., Joseph P. Myoglobin chemistry and meat color. Annual Review of Food Science and Technology. 2013;4:79–99. doi: 10.1146/annurev-food-030212-182623. [DOI] [PubMed] [Google Scholar]
- Thiansilakul Y., Soottawat B.S., Richards M.P. Isolation, characterisation and stability of myoglobin from Eastern little tuna (Euthynnusaffinis) dark muscle. Food Chemistry. 2011;124(1):254–261. doi: 10.1016/j.foodchem.2010.06.028. [DOI] [Google Scholar]
- Watson A., Gunning Y., Rigby N., Philo M., Kemsley K. Meat authentication via multiple reaction monitoring mass spectrometry of myoglobin peptides. Analytical Chemistry. 2015;87:10315–10322. doi: 10.1021/acs.analchem.5b02318. [DOI] [PubMed] [Google Scholar]
- Wen S., Zhou G., Li L., Xu X., Yu X., Bai Y., Li C. Effect of cooking on in vitro digestion of pork proteins: A peptidomic perspective. Journal of Agricultural and Food Chemistry. 2015;63:250–261. doi: 10.1021/jf505323g. [DOI] [PubMed] [Google Scholar]
- Xie W., Wei S., Liu J., Ge X., Zhou L., Zhou J., Shen J. Spectroscopic studies on the interaction of Ga3+-hypocrellin A with myoglobin. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2014;121:109–115. doi: 10.1016/j.saa.2013.10.085. [DOI] [PubMed] [Google Scholar]
- Yue Y., Wang Z., Zhang Y., Wang Z., Lv Q., Liu J. Binding of triclosan and triclocarban to pepsin: DFT, spectroscopic and dynamic simulation studies. Chemosphere. 2019;214:278–287. doi: 10.1016/j.chemosphere.2018.09.108. [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhao D., Zhu S., Nian Y., Xu X., Zhou G., Li C. Overheating induced structural changes of type I collagen and impaired the protein digestibility. Food Research International. 2020;134 doi: 10.1016/j.foodres.2020.109225. [DOI] [PubMed] [Google Scholar]
- Zhang M., Zou X., Zhao D., Li C. Pork meat proteins alter gut microbiota and lipid metabolism genes in the colon of adaptive immune-deficient mice. Molecular Nutrition and Food Research. 2020;64(9):1901105. doi: 10.1002/mnfr.201901105. [DOI] [PubMed] [Google Scholar]
- Zhao D., Li L., Le T.T., Larsen L.B., Xu D., Jiao W.…Zhang X. Digestibility of glycated milk proteins and the peptidomics of their in vitro digests. Journal of the Science of Food and Agriculture. 2018;99(6):3069–3077. doi: 10.1002/jsfa.9520. [DOI] [PubMed] [Google Scholar]
- Zhu S., Zhang M., Zhou G., Xu X., Li C. Effects of heating temperature on myoglobin structure with spectroscopic technology. Science and Technology of Food Science. 2018;39:35–39. [Google Scholar]