Abstract
MicroRNAs (miRNAs) are small RNAs that do not encode for proteins and play key roles in the regulation of gene expression. miRNAs are involved in a comprehensive range of biological processes such as cell cycle control, apoptosis, and several developmental and physiological processes. Oxidative stress can affect the expression levels of multiple miRNAs and, conversely, miRNAs may regulate the expression of redox sensors, alter critical components of the cellular antioxidants, interact with the proteasome, and affect DNA repair systems. The number of publications identifying redox-sensitive miRNAs has increased significantly over the last few years, and some miRNA targets such as Nrf2, SIRT1 and NF-κB have been identified. The complex interplay between miRNAs and ROS is discussed together with their role in myocardial ischemia-reperfusion injury and the potential use of circulating miRNAs as biomarkers of myocardial infarction. Detailed knowledge of redox-sensitive miRNAs is needed to be able to effectively use individual compounds or sets of miRNA-modulating compounds to improve the health-related outcomes associated with different diseases.
Keywords: Antioxidants, Biomarkers, microRNA, Myocardial ischemia-reperfusion injury, Oxidative stress, Proteasome, Redox signaling
Abbreviations
- ADPKD
autosomal dominant polycystic kidney disease
- AGO2
Argonaute
- AIFMB
mitochondrion-associated 3
- AKT
protein kinase B
- AREs
antioxidant response elements
- ASK
apoptosis-regulating kinase
- BMSCs
bone marrow mesenchymal stromal cells
- CAMKII
Ca2+/calmodulin dependent protein kinase II
- CAT
catalase
- CBD
cAMP response element binding (CREB) binding protein
- DRGC8
Di George critical Region 8
- GPX
glutathione peroxidase
- GR
glutathione reductase
- GSH
glutathione
- H2O2
hydrogen peroxide
- HDACs
histone deacetylases
- HIF-1α
Hypoxia-inducible factor-1α
- HMOX1
Heme Oxygenase 1
- HUVECs
Human Umbilical Vein Endothelial Cells
- KEAP1
Kelch Like ECH Associated Protein
- MAPKG
mitogen-activated protein kinase 6
- MDA
malondialdehyde
- MEFs
mouse embryonic fibroblasts
- MI
myocardial infarction
- MM
multiple myeloma
- NAFT
factor of activated T cell
- NF-κB
nuclear factor κB
- NOX
NADPH oxidase
- Nrf2
nuclear factor-erythroid 2 related factor 2
- OGD/R
oxygen-glucose deprivation and reoxygenation
- POMP
Proteasome maturation protein
- PSME4
Proteasome Activator Subunit 4
- RISC
RNA-induced silencing complex
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- TGFβ1
Transforming growth factor beta 1
- UPS
Ubiquitin Proteasome System
- XPO5
Exportin 5
1. Introduction: miRNAs and oxidative stress
Oxidative stress can affect the expression levels of multiple microRNAs (miRNAs) and, conversely, miRNAs can regulate the expression of redox sensors and alter key components of the cellular antioxidant machinery [1]. This complex network between miRNAs and oxidative stress act by modulating cell homeostasis. miRNAs are short RNAs that do not encode for proteins and play key roles in the regulation of gene expression acting at the post-transcriptional level [1]. miRNAs were firstly identified in C elegans [2,3]. miRNAs are expressed in nearly all eukaryotic cells and induce gene silencing by binding to target sites found within the 3′UTR (untranslated region) of the targeted mRNA [1,4]. This interaction prevents protein production by suppressing protein synthesis and/or by initiating mRNA degradation.
miRNAs are involved in a wide range of physiological processes and as such miRNA biogenesis and function are tightly regulated at multiple levels [1,5]. Canonical biogenesis results in miRNAs being encoded as individual monocistronic genes, as a cluster containing a few to several hundreds of different miRNAs, transcribed together as polycistronic transcripts, or in introns of host genes (intronic). Primary pri-miRNAs transcripts are generated by RNA polymerase II. In the next step, pri-miRNAs are processed to pre-miRNAs by a nuclear complex, which includes the Di George critical Region 8 (DGCR8) dimer and the RNASE III Drosha [6,7]. Pri-miRNAs contained hairpins and 5′ and 3′ flanking sequences. The microprocessor complex cleaves at the stem of the hairpin and liberates a pre-miRNAs with a 5'phosphate and a 3′ hydroxyl group. The pre-miRNAs are exported to the cytoplasm by binding to the export receptor Exportin 5 (XPO5) [[8], [9], [10]], where the RNASE III enzyme Dicer (in humans Dicer functions with a trans-activation-responsive RNA-binding protein TRBP) cleaves them and generates a miRNA duplex intermediate. In the last step, one strand of mi-RNA duplex assembles with a member of the Argonaute (AGO) to form the RNA-induced silencing complex (RISC). Upon loading of miRNAs into the RISC complex, the miRNA guides RISC to complementary sequences mainly located at the 3'untranslated regions of its target mRNAs. RISC regulates gene expression through translational repression (at the early stages) or mRNA degradation (at the later stages) [11]. Besides the well-established mi-RNA biogenesis pathway, some non-canonical Drosha-independent or Dicer-independent miRNAs processor have been recently described [[12], [13], [14]]. Moreover, several cell regulatory factors such as phosphorylation [15,16], deacetylation [17], ubiquitylation [18] and SUMOylating [19], connect miRNAs biogenesis to different cell signaling pathways. Diverse studies have also revealed that some miRNAs-transcription factors are redox-sensitive [[20], [21], [22], [23]].
Alterations in miRNAs expression profiles occur during organ development, aging [24], and cell death [25]. miRNA expression is also changed during the pathophysiology of complex diseases such as inflammation [25], cardiac [26,27], and neurodegenerative diseases [28], and almost all kinds of cancer [22,29,30]. In addition to their role in regulating gene expression, miRNAs released into body fluids have emerged as potential serum biomarkers. Circulating miRNAs can be either bound to serum proteins and lipoproteins or be encircled in extracellular vesicles including exosomes, microvesicles or apoptotic bodies [25]. Currently, miRNAs are intensely studied as candidates for diagnostic and prognostic biomarkers in liver diseases [25,31], myocardial infarction [27,32] and Alzheimer disease [33].
The link between regulatory factors and miRNAs provide an interesting tool to modulate miRNAs biogenesis in certain pathologies [18,19]. Emerging evidence now suggests that reactive oxygen species (ROS) modulate some specific miRNAs biogenesis, also called “redoximiRs” [34] and that miRNAs target antioxidant responsive elements and ROS related genes, thus affecting cellular redox status. During normal oxidative metabolism, there is a constant formation of oxidative reactive species in the cell, such as ROS and reactive nitrogen species (RNS). Reactive oxidants include many ROS (such as H2O2, O2•−, •OH, 1O2, O3, RO2•, RO•) and RNS (such as •NO, •NO2, ONOO−) [35,36]. Environmental factors, as well as different physiological and pathophysiological conditions, influence the formation of radical species inside the cell. The generation of ROS and RNS elicits oxidative stress, which includes the oxidation and subsequent functional impairment of lipids, proteins and nucleic acids [37]. Increase in intracellular ROS/RNS has also been associated with cell death, having a critical role in the induction of apoptosis [38,39].
A significant disruption of redox homeostasis by ROS/RNS and antioxidant defense can induce oxidative stress. Under physiological conditions, there is a balance between factors promoting ROS/RNS formation and the cellular antioxidant pool [40,41]. The antioxidant defense system includes molecular antioxidants and enzymes. Antioxidant enzymes are proteins that convert highly reactive species into less reactive particles: superoxide dismutase (SOD) scavenges radical superoxide, while catalase (CAT) and glutathione peroxidase (GPX) detoxify hydroperoxides [42]. A broader look at the ROS/RNS defense system would include enzymes that are able to repair cellular damage [43], including enzymes involved in DNA repair and the proteasome, a system able to recognize and degrade oxidatively-modified proteins [44]. However, while at relatively high concentrations, ROS/RNS become harmful, at low levels can promote cell proliferation and survival [41]. Low ROS levels are also needed because ROS are involved in several signaling pathways [45,46].
A regulatory interplay between miRNAs and redox signaling have been increasingly reported in the literature over the last decade. While some redox-sensitive miRNAs [34] are regulated by oxidative stress, miRNAs can also regulate cellular redox status. Gene silencing by miRNAs can result in changes in ROS activators and ROS scavengers, leading to a complex interplay between oxidative stress and miRNA to modulate cellular redox homeostasis. Research published to date on the relationship between miRNAs and oxidative stress suggests that:
miRNAs biogenesis can be regulated by cellular redox status.
miRNAs can target ROS generation and modulate antioxidant signaling
miRNAs can interact with the proteasome and act as endogenous proteasome inhibitors
miRNAs cooperate with ROS to regulate cell fate, as oxidative stress-induced apoptosis and autophagy and redox regulation of DNA repair systems
miRNAs regulate ROS in many diseases such as myocardial and cerebral ischemia-reperfusion injury, as well as cancer.
miRNAs involved in cellular redox status as potential targets for therapeutics and biomarkers
Current concepts about these aspects of miRNA biology are described below.
2. miRNAs biogenesis can be regulated by cellular redox status
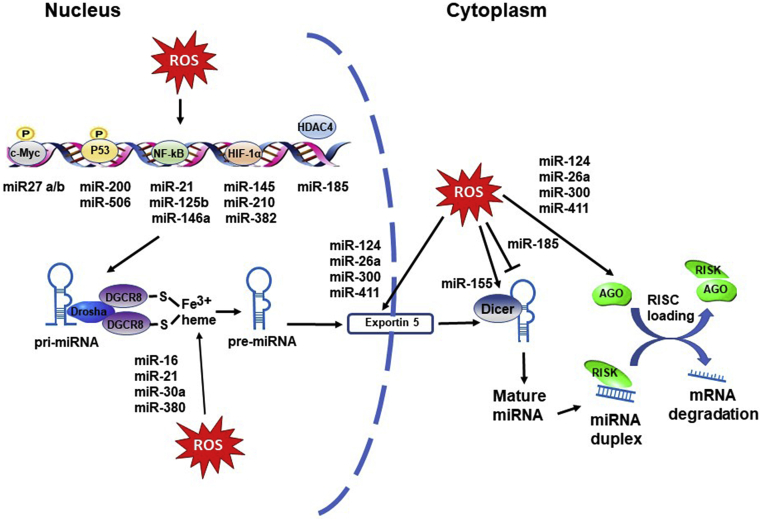
The biogenesis and regulation mechanisms of miRNAs are displayed in Fig. 1. Many scientific reports indicate that several miRNAs are regulated by redox status [34,47,48]. ROS acts by activating related transcription factors such as c-Myc, p53, Nuclear factor κB (NF-κB) and hypoxia-inducible factor 1 (HIF-1α) in response to oxidative stress. Table 1 shows a summary of studies that indicate ROS can regulate miRNAs through these transcription factors. c-Myc is a transcription factor involved in oncogenesis, and whose effects have been correlated with exposure to ROS [49]. c-Myc induces the expression of miR27a/b, which, in turn, inhibits the expression of nuclear factor-erythroid 2 related factor 2 (Nrf2) worsening the progression of cholestatic liver injury [50]. P53, a stress-related transcription factor, can be induced by ROS and activate target genes for protecting the genome stability [49]. The members of the miR-200 family are markedly enhanced in hepatic cells by hydrogen peroxide (H2O2) treatment. In liver cells, miRNA-200s are upregulated by oxidative stress-induced p38a-mediated phosphorylation of Ser33 on p53 [51]. Upregulation of miR-200-3p under these conditions promotes H2O2-induced cell death [51]. miR-506 was regulated by p53 and lead to apoptosis in lung tumor cells [52].
Fig. 1.
ROS affecting nuclear and cytoplasmic processing of miRNAs. miRNA transcription can be induced by ROS activating transcription factors (c-Myc, p53, NF-κB, HIF-1α). HDAC4 does not directly bind to DNA but suppresses transcription when bound to a promoter. ROS can affect miRNA biogenesis, translocation and maturation through regulating the activity and expression of DGCR8, drosha, exportin 5 and the processing enzyme dicer. (Arrow indicates activation, T arrow indicates inhibition).
Table 1.
ROS-sensitive transcription factors, ROS induced epigenetic modifications of miRNAs, and ROS induced nuclear and cytoplasmatic processing of miRNAs.
| ROS inducing model | Transcription factor | Targeted miRNAs | Model and outcomes of miRNAs upregulation | References |
|---|---|---|---|---|
| ROS-sensitive transcription factors affecting miRNAs biogenesis | ||||
| MELAS syndrome | NFκ-B activation | mIR-9 | Negative regulator of GTPBP3, MTO1 and TRMU leading to aggravated mitochondrial dysfunction in platelet derived cybrids from two patients with MELAS syndrome | [169] |
| Leptin-NADPH oxidases | NFκ-B activation | miR-21 | Fibrogenesis in experimental and human Nonalcoholic Steatohepatitis (NASH) | [56] |
| High glucose | NFκ-B activation | miR-21 | Fibroblasts cells. Promotes migration | [55] |
| Inhibited GSH synthesis | C Myc overexpression | miR-27a/b | Liver cancer progression | [50] |
| Aluminium-sulfate | NFκ-B activation | miR-146a miR-125b |
Human neural cells. Promotes inflammation | [57,58] |
| H2O2 | P38/P53 phosphorylation | miR-200 | Liver cell death by inhibiting p38/p53 feed-back loop | [51] |
| Hypoxia | HIF 1-α | miR-210 miR-382 miR-145 |
Regulation of mitochondrial metabolism in human cancer cells and tumors Proliferation of pulmonary artery smooth muscle Promotes angiogenesis in human gastric cancer cells Protective effects against apoptosis in cardiomyocytes |
[63] [64] [65] [66] |
| ROS generation | P53 activation | miR-506 | Induced cell apoptosis and decreases viability in lung tumor cells | [52] |
| ROS affecting epigenetic modifications of miRNAs | ||||
| Hypoxia | HDAC4 supression | miR-185 | Lung epithelial cell death | [70] |
| Glucose deprivation | HDAC2 inhibition | miR-466h-5p | Increased apoptosis mouse cell line | [69] |
| ROS affecting nuclear and cytoplasmatic processing of miRNAs | ||||
| Hypoxia | DICER | miR-155 | Upregulation of miRNA induce glycolysis in lung alveolar epithelial cells | [73] |
| Chronic Hypoxia | DICER | miR-185 | Downregulation of Dicer-dependent miRNA maintains the induction of HIF-1α and hypoxia-responsive genes | [74] |
| Nutritional stress | XPO5 and AGO2 | miR-124 miR-26a miR-300 miR-411 |
Xpo5 and Ago2 mRNA levels are altered in malnourished mice The levels of 70% and 50% of the miRNAs, were increased in the hippocampus of LP mice |
[78] |
| Redox state favoring conversion of Fe(II) heme to Fe(III) heme |
Drosha-DGCR8 complex | miR-21 miR-30a miR-380 miR-16 |
Increase the efficiency of pri-miRNA processing | [71] [72] |
NF-κB transcriptionally regulates several miRNAs. NF-κB is a critical regulator of pro-inflammatory/stress-like responses that play important roles in DNA damage response and apoptosis in different cell type [53,54]. ROS-induced NF-κB upregulates miR-21, promoting cancer progression [55] and fibrogenesis [56]. Likewise, miR-146a and miR-125b were overexpressed after ROS-NF-κB activation in human neural cells [57,58]. The HIF-1α transcription factor is a critical oxygen sensor and a major regulator of the hypoxic adaptive response [59]. ROS directly regulate HIF-1α by oxidizing a cysteine amino residue on HIF-1α, resulting in a stabilization of the protein. HIF-1α regulates the expression of genes involved in the adaptation to hypoxia, like EPO (erythropoietin) or VEGF (Vascular endothelial growth factor) [60,61]. Likewise, HIF-1α overexpression can regulate the expression of a broad range of miRNAs, the so called hypoxamiRs [62], which constitute key regulators of cellular adaptation to hypoxia. Among them: miR-210, which regulates mitochondrial metabolism [63] and proliferation of pulmonary smooth muscle [64]; miR-382 which promotes angiogenesis [65]; and miR-145 which leads protective effects in cardiomyocytes [66].
ROS can also regulate miRNAs expression through epigenetic modifications (Table 1). Like protein coding DNA sequences, miRNAs genes may undergo DNA methylation and histone modifications. Altered epigenetic miRNA expression has been described in cancer cells [18,67,68]. Reduced activities of histone deacetylases (HDACs) under oxidative stress can alter miRNAs expression levels. Accumulation of ROS due to glucose deprivation inhibited HDAC2 in cultured mouse cells, which increased acetylation and induction of miR-466h-5p, leading to increased apoptosis [69]. Likewise, hyperoxia suppresses histone deacetylase 4 (HDAC4) and subsequently affects histone deacetylation, resulting in an elevated miR-185 transcription [70]. Functionally, miR-185 promotes lung epithelial cell death through inducing DNA damage.
ROS can modulate miRNAs biogenesis at many levels, and several enzymes and components of miRNA processing machinery can be affected by oxidative stress (Table 1). In the Drosha-DGCR8 complex, DGCR8 forms a highly stable and active complex with the ferric heme using two endogenous cysteines as axial ligands [71]. The reduction of the heme iron to the ferrous state in DGCR8 abolishes the pri-miRNA processing activity [71,72]. Dicer is a principal component of miRNA processing machinery that processes precursor miRNAs (pre-miRNAs) into mature miRNAs. There is evidence that chronic hypoxia promotes cell glucose metabolism through Dicer regulation of miR-143 and miR-155 [73] and the induction of hypoxia-responsive genes through Dicer regulation of miR-185 [74]. The relationship between the redox state and the complexes processing miRNA may be more widespread. For example, a mutant p53 inhibited the processing of pri-miRNAs by Drosha, decreasing the levels of certain mature miRNAs in cells involved in cell cycle and cell proliferation regulation [75]. In addition, mutant p53 was also reported to suppress DICER1 expression [76]. Deregulation of the miRNA biogenesis pathway is an emerging mechanism in neurodegenerative diseases. Loss of Dicer [77], as well as downregulation of Drosha and DGRC8 were potentially involved in several neurodegenerative disorders. Moreover, nutritional stress alters the expression of the Xpo5 (Exportin 5) and Ago2 (Argonaute RISC Catalytic Component 2) genes in the hippocampus of restricted protein offspring mice [78].
3. miRNAs can target ROS generation and can modulate antioxidant signaling
Oxidative stress is typically the result of an imbalance between the rate of ROS generation, and the ability to detoxify these reactive species. Strong experimental evidence exists to support that miRNAs can regulate ROS generation and alter key components of the cellular antioxidant machinery. The miRNAs affecting ROS production, antioxidants and repair systems are shown in Fig. 2.
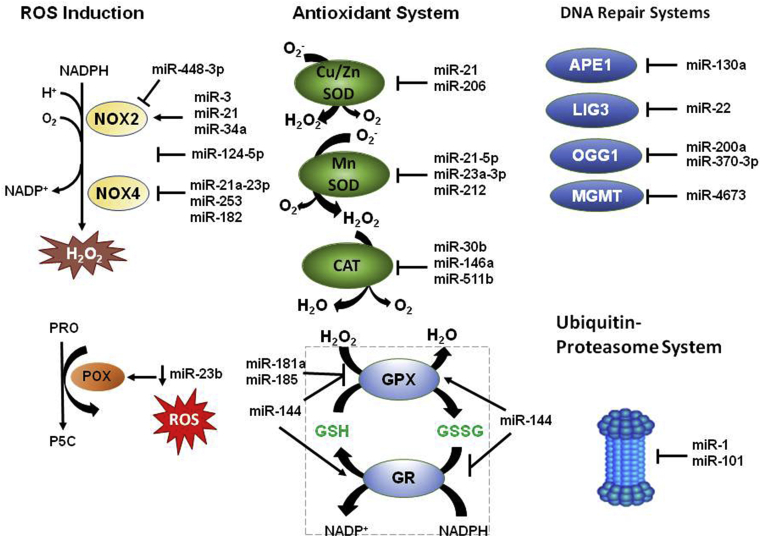
Fig. 2.
miRNAs affecting ROS production, antioxidants and repair systems. miRNAs regulate ROS levels by activating or inhibiting NOX2 (NADPH oxidase 2), NOX4 (NADPH oxidase 4) and POX (Proline oxidase) enzymes. miRNAs can affect ROS levels through activation or inhibition of antioxidants including catalase (CAT), superoxide dismutases (Cu/Zn SOD and MnSOD), glutathione peroxidase (GPX) and glutathione reductase (GR). miRNAs can interact with DNA repair systems and ubiquitin proteasome system, which both affect the cellular redox status of the cell. APE1, AP-endonuclease; LIG3, DNA ligase 3, OGG1, 8-oxoguanine glycosylase; MGMT, O-6 -methylguanine DNA methyltransferase; PRO, proline; P5C, pyrroline-5-carboxylate. (Arrow indicates activation, T arrow indicates inhibition).
NADPH oxidases (NOXs) are a family of membrane-bound enzymes that oxidize NADPH to produce ROS (either superoxide or hydrogen peroxide) as the primary species during the catalytic metabolism of oxygen for a range of host defense and signaling functions [79] and a target of several miRNAs [80] (Table 2). Enhanced expression of NOX2 isoform has been reported due to miR-34a overexpression in glioma cells [20] and after exosomes enriched miR-3 in a mouse model of cardiac injury [27]. However, other miRNAs inducing inhibition of NOX2 [81] or attenuated NOX4 activity [68] have also been reported. miR-124-5p is selectively expressed within the central nervous system (CNS) and is predicted to bind to NOX2 directly. In vivo, miR-124 overexpression improved, whereas miR-124 inhibition aggravated the injury in a cerebral I/R injury model in rats through middle cerebral artery occlusion (MCAO) surgery [81] and increased activity of miR-21a-3p targets and inhibits NOX4 to inhibit tumor formation [68] in endothelial cells. Proline oxidase (POX) is a mitochondrial inner-membrane enzyme that mediates the proline cycle to shuttle redox equivalents between mitochondria and the cytosol. POX is a miR-23b target, and in human renal carcinoma tissues, a negative correlation between miR-23b and POX protein expression has been reported [29].
Table 2.
miRNAs that modulate ROS production.
| miRNAs | Affected ROS producers | Effects | Model | References |
|---|---|---|---|---|
| Exosomes enriched miR-3 | Enhanced NOX2 expression | ROS production Angiogenesis |
Mouse model of cardiac injury | [27] |
| Inhibition of miR-21 | Decreased NOX2 | Lower ROS production | Renal inflammation in NAFLD | [170] |
| Up-regulated miR-21a-3p | Attenuated NOX4 activity | Decreased H2O2 | EOMA cells | [68] |
| Decreased miR-23b | Enhanced Proline Oxidase | ROS production Apoptosis |
Renal tumors | [29] |
| Overexpression of miR-34a | Enhanced NOX2 expression | ROS production Apoptosis |
Glioma cells | [20] |
| Overexpression of miR-124-5p | Inhibits NOX2 | Decreased ROS, MDA | Improved I/R injury in MCAO rats Decreased OS in OC-12 cells |
[81] |
| Overexpression of miR-155 | Decreased Nfe2l2, Sod1, and Hmox1 | ROS production | Induced ROS production in mesenchymal stem cells (MSCs) from aged mice | [171] |
| Decreased miR-451 | Not determined | Lower ROS production | Bone marrow derived macrophages (BMDM) from wild type and p47phox−/− mice | [172] |
The list of miRNAs that targets antioxidant enzymes continues to expand in different experimental models (Table 3). Intracellular and extracellular Cu/Zn SOD are downregulated by miRNAs in human bronchial cells [82] and in a model of atrial fibrillation [83], respectively. miR-212 suppressed MnSOD expression in human colorectal tumor [67]. miR-30b [84], miR-146a [85] and miR-551b [86] decreases CAT expression. Glutathione (GSH) is a ubiquitous low molecular antioxidant [[87], [88], [89]]. In a detoxification reaction, two molecules of GSH react with H2O2 in a GPX catalyzed reaction, giving GSSG, the disulfide-oxidized form of glutathione, and water. On the other hand, GSSG is a substrate of the glutathione reductase (GR) enzyme, which regenerates GSH. miRNAs have been regarded as potential regulators of glutathione peroxidases expression [90]. Upregulation of miR-181a [91], miR-185 [92,93] and miR-144 [94] decrease GPX expression, while miR-214 increase GPX activity [95] and decrease the GR activity [96]. GSH levels in the CNS show a circadian rhythm; therefore, a role for miRNA in the generation of the GSH rhythm has recently been suggested [97].
Table 3.
miRNAs that modulate antioxidant expression or activity.
| miRNAs | Targeted Antioxidant | Model | References |
|---|---|---|---|
| miR-7 | Inhibit Keap1/activates Nrf2pathway | Human neuroblastoma cell line | [101] |
| miR-21 | Extracellular Cu/ZnSOD inhibition MnSOD downregulation |
Human bronchial epithelial cells | [82] |
| miR 27ab | Inhibit Nrf2 expression | Chronic cholestatic liver injury | [50] |
| miR-30b | Inhibit Catalase expression | Retinal pigment epithelial cell line | [84] |
| miR-93 | Decrease Nrf2 level | Breast carcinogenesis | [99] |
| miR-101 | Inhibits Cul3/stabilizes Nrf2 | HUVEC cells and aortic rings | [173] |
| miR-144 | GR (modulated?) Decrease GPx expression Decrease Nrf2 level |
Primary erythroid progenitor cells Human neuroblastoma SH-SY5Y cells Primary erythroid progenitor cells Alveolar epithelial dysfunction in HIV-1 transgenic rats |
[94] [174] [94] [175] |
| miR-146a | Decrease Catalase expression | Human lung cancer cells | [85] |
| miR-181a | Decrease GPx expression | Rat cardiomyocyte cell line | [91] |
| miR-185 | Decrease GPx expression | Alcoholic liver disease Human endothelial cells |
[92] [93] |
| miR-200a | Inhibit Keap1/activates Nrf2 pathway | Breast carcinogenesis Liver inflammation |
[100] [176] |
| miR-212 | Supress MnSOD expression | Colorectal tumor | [67] |
| miR-214 | Increase GPx activity Decreased GR activity |
Diabetic nephropathy Alcohol induced liver injury |
[95] [96] |
| miR-206 | Intracellular Cu/ZnSOD inhibition | Canine model of atrial fibrillation | [83] |
| miR-200c | Supress Nrf2 expression | Lung cancer cells | [98] |
| miR-455 | Inhibits Cul3/activates Nrf2 | Human osteoblasts | [102] |
| miR-551b | Decrease Catalase expression | Lung Cancer model | [86] |
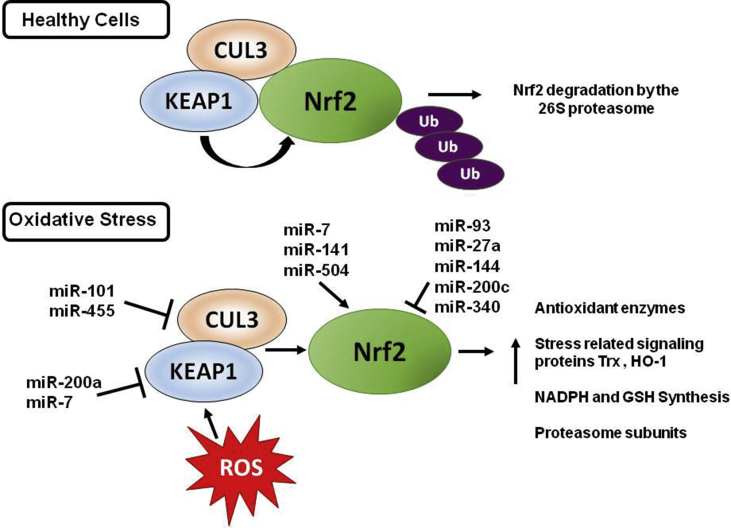
miRNAs can also target genes that indirectly modulate the antioxidant effect. Inhibition or suppression of nuclear Nrf2 expression by miR-27 a/b has been reported in chronic cholestatic liver injury [50], and by miR-200c [98] in lung cancer cells. Nrf2 is a transcriptional factor that controls cellular redox homeostasis. Nrf2 activates the transcription of genes that encode antioxidant enzymes, among others. Diverse miRNAs can decrease (miR-93) [99] or activate (miR-200a, miR-7, miR-455) [[100], [101], [102]] Nrf2 pathway in different cancer models, highlighting the importance of modulating the levels of antioxidant enzymes under tumor processes.
4. miRNAs can interact with the proteasome and act as endogenous proteasome inhibitors
A recent exciting development in the protein homeostasis field was the discovery that miR-101 targets and inhibits the protein POMP (proteasome maturation protein), a protein that is needed for the assembly of constitutive proteasomes and immunoproteasomes [103]. By inhibiting POMP miR-101 causes impaired proteasome assembly and reduced activity. It was previously known that miR-101 is reduced in several cancers [104,105] and restoration of miR-101 inhibits cancer cell proliferation.
The proteasome is part of the ubiquitin-proteasome system (UPS) that is responsible for the degradation of more than 60% of intracellular proteins. The proteasome itself is a large complex that could be dived up into two parts, the 20S and the 19S, which together form the 26S proteasome [106,107]. The 20S proteasome is composed of 28 subunits in a barrel shape structure with four rings (two α and two β rings). The 20S proteasome has three independent proteolytic β subunits (each occurring in duplicate) with caspase-like (β1), trypsin-like (β2), and chymotrypsin-like activities (β5) that are responsible for the cleavage of proteins that enters the proteasome. The 19S complex is the regulatory complex that is responsible for recognizing poly-ubiquitinated proteins, removing the ubiquitins attached to proteins targeted for degradation, and unfolding the proteins for entry into the 20S proteolytic core. Inhibition of the proteasome is associated with many diseases, including cardiac diseases, Alzheimer's, Parkinson's, diabetes and others, and significant inhibition of the proteasome leads to cell death [107].
The proteasome is also very important in reduced intracellular oxidative stress by degrading oxidized proteins. Another form of the proteasome, call the immunoproteasome, seems to be optimized for degrading oxidized proteins [108]. Although not measured immunoproteasome activity would also likely decrease if POMP is impaired since POMP is also important in the assembly of immunoproteasomes. The activity of the proteasome can also be regulated by oxidative stress. The apoptosis-regulating kinase ASK1 is activated by oxidative stress (such as H2O2), and can interact with and phosphorylate the RPT5 subunit of the 19S complex [109]. Phosphorylation of the proteasome by ASK1 results in all three activities of the proteasome being reduced and may play a role in apoptosis [109]. Formation of a functional proteasome 26S complex is a multifaceted process involving the synthesis of the individual subunits, partial assembly, full assembly, and then maturation. This latter process in humans requires POMP, a chaperone that forms the 20S half-structures by (referred to as 16S proteasome precursors) [[110], [111], [112], [113]]. POMP is degraded after proteasome maturation. The cancer cell proliferation that is inhibited by miR-101 can be rescued by overexpression of POMP. The ability of miR-101 to inhibit proteasome activity is interesting because the proteasome inhibitor, bortezomib (which inhibits the β5 proteasome subunit) has been used as an anti-cancer drug for over a decade [114]. Other reports also suggest that cancer cells are more vulnerable to proteasome inhibition than control cells [115].
While miR-101 is important in redox cellular biology because of its direct link to regulating proteasome activity and hence the levels of oxidized protein in a cell, many other miRNAs act by regulating the Nrf2 pathway. The Nrf2 pathway is one of the most important pathways for intracellular protection during oxidative stress as Nrf2 regulates the expression of several cytoprotective and stress-related genes including thioredoxin (Trx) and HO-1 [[116], [117], [118]]. In healthy cells, Nrf2 is readily polyubiquitinated by the BCR(KEAP1) ubiquitin ligase complex, and then becomes a substrate for the 26S proteasome, resulting in low levels of Nrf2. KEAP1 (Kelch Like ECH Associated Protein 1) is a substrate adaptor that interacts with Nrf2 and the BCR complex, which under conditions of oxidative stress, undergoes oxidation of its reactive cysteine residues, resulting in Nrf2 dissociating from KEAP1 (Fig. 3) [119]. Free Nrf2 can translocate to the nucleus where it can interact with antioxidant response elements (AREs) [120]. In mammalian cells, most of the genes encoding proteasome subunits, POMP, and other assembly partners contain AREs [121]. Nrf1 and Nrf2 can upregulate the expression of proteasome subunits and POMP by binding to the ARE in response to proteasome inhibition [121,122].
Fig. 3.
miRNAs regulating the Nrf2 pathway. In healthy cells, Nrf2 is polyubiquitinated by the KEAP1 ubiquitin ligase complex, becoming a substrate for the 26S proteasome. Under conditions of oxidative stress, KEAP1 undergoes oxidation of its reactive cysteine residues, resulting in Nrf2 dissociating from KEAP1. Free Nrf2 can translocate to the nucleus where it can interact with antioxidant response elements (AREs). CUL3, cullin-3, KEAP1, Kelch-like ECH-associated protein 1, Nrf2, nuclear factor-erythroid 2-related factor 2, Ub, Ubiquitin. (Arrow indicates activation, T arrow indicates inhibition).
Nrf2 has also been shown to promote biogenesis [123] and regulate ROS levels in mitochondria via many mechanisms including increasing the synthesis of NADPH and GSH, regenerating Trx2 and GSH and increasing the detoxification of peroxides by GPx and Prx3 [124]. Pickering et al., 2012 found that the addition of H2O2 induced binding of Nrf2 to the ARE of the proteasome β5 gene, resulting in increased β5 mRNA levels. Additionally, inducers of Nrf2 also upregulate proteasome subunits and activity in different cell types [125,126]. As such, induction of proteasome subunits by the Nrf2-pathway is likely to be an important way for a cell undergoing oxidative stress to increase its capacity to remove damaged and oxidized proteins. miR-155 levels were lower in plasma cells from multiple myeloma (MM) patients when compared to control patients [127]. Addition of synthetic miR-155 mimics into MM cell lines resulted in increased pro-apoptotic effects and decreased cell viability. In MM cells resistant to the proteasome inhibitor, bortezomib, when the miR-155 mimics were added, they enhanced bortezomib anti-tumor activity. These results suggest that miR-155, like miR-101 exerts its effect by proteasome inhibition.
Although bortezomib has helped with the management of MM patients, resistance to bortezomib occurs over time [128]. Using genome-wide profiling of bortezomib-resistant myeloma cells, MiR-29b was significantly reduced in bortezomib-resistant cells [128]. Further investigation showed that miR-29b targeted the proteasome subunit PSME4 (Proteasome Activator Subunit 4). PSME4 encodes one of the subunits of the PA200 complex which can replace the 19S complex and interact with the 20S proteasome. The PA200 has been shown to be involved in degrading histones following DNA double-strand breaks. Synthetic miR-29b mimics diminished the growth of myeloma cells, xenotransplants, and patient tumor cells [128]. These miR-29b mimics also reduced proteasome activity and has the potential to synergistically enhance the anti-tumour effects of proteasome inhibitors.
Using a combination of approaches, miR-200c was identified as a negative regulator of Noxa expression [129]. Noxa is a pro-apoptotic protein that contributes to p53-mediated apoptosis under certain conditions such as radiation exposure. miR-200c overexpression resulted in increased susceptibility to bortezomib in several cell lines. When cells lacking Noxa were used to overexpress miR-200c these cells had greater apoptosis induced by proteasomal inhibition compared to cells treated with proteasome inhibitors but without overexpression of miR-200c, suggesting that miR-200c is an enhancer of bortezomib-induced cell death. The results also suggest that multiple miRNAs may be working together acting on different aspects of the proteasome (proteasome assembly, proteasome subunits, etc.) to intensify the proteasome inhibition. Overall, these results suggest that the miRNAs that act by inhibiting proteasome activity are good targets for potential anti-cancer therapeutic strategies.
Recently, in silico tools were used to determine which miRNAs are involved in oxidative stress regulation [130]. Literature with information on miRNAs that changed expression levels in the presence of oxidative stress damage was reviewed and miRNA data extracted and utilized with several databases and prediction software to gene targets and pathways of oxidative stress-modulated miRNAs. This approach allowed the identification of potential miRNAs that will target the oxidative stress-related miRNA gene targets and pathways. One of the major pathways identified was the ubiquitination pathway. One example of a miRNA affecting the UPS would be miR-501-5p. Autosomal dominant polycystic kidney disease (ADPKD) cells and tissues show upregulated miR501-5p, which induces mTOR kinase activation [131]. The mTOR kinase increases the expression of the E3 ubiquitin ligase MDM2, which ubiquitinates p53, increasing its rate of degradation by the proteasome. miR501-5p overexpression inhibits mTOR activity and increases cell proliferation in kidney cells. Reduced expression of miR501-5p as well as the proteasome and mTOR inhibitors activate apoptosis and lessens cell growth in autosomal dominant polycystic kidney disease (ADPKD) cells [131]. Several miRNAs have also been shown to be associated with both ubiquitination and autophagy, including miR-9, miR-16, miR-17, miR-93, miR-101, miR-124, miR-128, miR-200, miR-429, and miR-497 [130].
5. miRNAs cooperate with ROS to regulate cell fate, as oxidative stress-induced apoptosis and autophagy and redox regulation of DNA repair systems
Several miRNAs are involved in redox regulation of DNA damage and DNA repair pathways (Table 4) [132]. miRNAs play an important role in regulating mitochondrial signaling pathways, including the apoptotic pathway [133]. Ionizing radiation (IR) induces the generation and accumulation of mitochondrial ROS and causes DNA damage, which ultimately results in apoptosis in bone marrow mesenchymal stromal cells (BMSCs). The mitochondrial ROS can damage the mitochondria resulting in a dysfunction mitochondrial antioxidant system, resulting in further ROS buildup. IR is used experimentally to investigate the effects of radiotherapy, which is commonly performed as part of the treatment for many malignancies such as cancer. IR induces miR-22 in many cell types, including bone marrow mesenchymal stromal cells (BMSCs) [134]. Overexpression of miR-22 increases mitochondrial ROS and cellular apoptosis. Redd1 was found to be a target for miR-22 and overexpression of Redd1 diminished the role of miR-22 on mitochondrial ROS generation protecting cells from miR-22 induced cell injury due to IR.
Table 4.
miRNAs mediated redox regulation of DNA damage and DNA repair pathways.
| miRNAs | Target Expression | Effects | References |
|---|---|---|---|
| miR-22 overexpression | Reduces Redd1 | Promotes mitochondrial ROS, DNA damage, and apoptosis in bone marrow mesenchymal stromal cells | [134] |
| Decreased miR-130a | Increase APE1 | Confers resistence to temozolomide in glioma cells | [177] |
| miR-135b overexpression | Not determined | May play a role in Alzheimer's disease | [132] |
| miR-185 overexpression | Increased the relative apurinic/apyrimidinic (AP) sites in genomic DNA Downregulated 14-3-3δ signaling pathway |
DNA damage in lung epithelial cells Induced apoptosis |
[70] |
| miR-200a overexpression | Down-modulates OGG1, APE1, LIG3 and XRCC1 | Required for repairing 8-OH-dG in senescent primary human keratinocytes | [178] |
| Overexpression of miR-370-3p | Downregulates MGMT | Stimulate sensitization to temozolomide in glioblastoma cells | [135] |
| miR-4673 overexpression | Down-modulates OGG1 | Required for repairing 8-OH-dG in human lung cancer cell line | [136] |
In mouse hippocampal neurons, H2O2 upregulated miR-135b and miR-708, and their targets were predicted to be involved in DNA recombination and protein ubiquitination [132]. miRNA-370-3p was the most downregulated miRNA in tissues of glioblastoma multiforme (GBM) chemotherapy cells and temozolomide resistance cells [135]. A miRNA-370-3p mimic repressed the self-reparative ability of GBM cell DNA and increased the sensitivity of these cells to temozolomide. The target gene of miR-370-3p was O(6)-methylguanine-DNA methyltransferase (MGMT). toxicity In A549 and H1299 cells, increased miR-4673 expression due to paclitaxel (PTX) resulted in increased ROS and apoptosis and reduced cell viability [136]. miR-4673 was found to target and reduce 8-Oxoguanine-DNA Glycosylase-1 (OGG1). Increased levels of OGG1 reduced PTX induced ROS, apoptosis and cell death. Although beyond the scope of this review, many miRNAs are pro-apoptotic, including miR-34a, miR-144, miR-155, and miR-200, while others are anti-apoptotic, including miR-210, miR-21, and miR-146a [137]. Experimental data also suggest that miRNAs can initiate apoptotic pathways in mitochondria during myocardial ischemia-reperfusion-injury [133].
6. miRNAs and ROS in cancer and myocardial ischemia-reperfusion injury
Numerous reports provide strong evidence of a reciprocal link between miRNAs and ROS in cancer [138,139]. However, the role of miRNAs and ROS in cancer will not be discussed in detail here as several great recent reviews on that topic are available [[140], [141], [142], [143], [144]]. ROS generation leads to oxidative DNA damage which has been suggested to be one of the first steps in the development of tumors [37,40]. ROS accumulation may activate oncogenic signaling and also control the expression of various tumor suppressor genes that results in tumor progression [145]. Paradoxically, radiation and various chemotherapeutic agents used to treat cancer mediate their effects through the production of ROS [39,145]. Several miRNAs behave as regulators of gene expression by interacting with oncogenic and tumor suppressor genes that contribute to tumorigenesis. Consistent with this hypothesis, recurrent genetic and epigenetic alterations of individual miRNA have been found in several tumors [20,29,135,136].
ROS-sensitive transcription factors affect the biogenesis of miRNAs with oncogenic roles. For example, ROS generated by p53 dependent mechanisms can induce the expression of miR-506, resulting in decreased viability of lung tumor cells due to apoptosis [52]. It has also been shown that miRNAs can modify ROS homeostasis during the process of cancer. In human renal carcinoma tissue a decrease in miR-23b enhanced proline oxidase protein expression leading to ROS production and apoptosis [29]. Silencing miR-517a promotes oxidative stress in melanoma cells and decreases cell proliferation [146]. Several miRNA modulations can affect the Nrf2 pathway and modify antioxidant enzymes expression in different cancer models. Enhanced expression of miR27a/b inhibits Nrf2, thereby worsening the progression of cholestatic liver injury [50]. In breast cancer cells miR-93 decreases Nrf2 level [99] and miR-28 reduces the stability of Nrf2, increasing colony formation [147]. Attempts to modify ROS production in cancer using mi-RNA as a therapeutic tool have also been reported. The miRNA-mediated targeting of the NOX family of enzymes decreases ROS production and reduces cancer aggressiveness [148]. Although the molecular mechanisms underlying the role of miRNA and ROS in tumorigenesis and chemoresistance are better understood than in other diseases, they are still currently the subject of significant research.
Several reports support a role for miRNAs in myocardial ischemia-reperfusion (I/R) injury (Table 5) [149]. When a patient is admitted to the hospital with myocardial ischemia, that patient usually undergoes re-canalization of the coronary artery to help restore blood supply to the heart. However, this restoration of the blood supply (re-oxygenation), or reperfusion, can cause oxidative stress and inflammation, which leads to tissue damage, apoptosis and necrosis of cardiac cells. Reperfusion injury results in changes in intracellular free Ca2+ concentrations and a key regulator of Ca2+ signaling is Ca2+/calmodulin dependent protein kinase II (CaMKII). CaMKII mediates several signaling pathways in the heart, including hypertrophy, apoptosis, and heart disease. In H2O2 treated cardiomyocytes, miR-145 targets and reduced CaMKIIδ protein expression resulting in suppressed ROS-induced Ca2+ increases, which will prevent apoptosis [150].
Table 5.
miRNAs involved in oxidative stress induced Cardiac Ischemia/Reperfusion Injury.
| miRNAs | Expression | Target | Effects | Model | References |
|---|---|---|---|---|---|
| miR-1 | Upregulated | E3 enzyme/19S and 20S proteasome subunits | LV end-diastolic diameter and LV mass | Mice I/R | [152] |
| miR-19a | Downregulated | PTEN/PI3K/pAKT | Cell injury/apoptosis | H9c2 | [179] |
| miR-21 | Upregulated | PI3K/AKT | ROS | H9c2 | [180] |
| miR-22 | Upregulated | SIRT-1 PI3K/AKT/βCatenin SIRT-1/PGC1α CBP |
Apoptosis Altered mitochondrial function Inhibits Apoptosis |
Cardiomyocytes H9c2 Rats I/R Cardiomyocytes |
[181] [182] [151] |
| miR-34a | Upregulated | SIRT-1 | Apoptosis/infarct size | Cardiomyocytes | [183] |
| miR-93 | Downregulated | PTEN/PI3K/pAKT | ROS/Cell injury/apoptosis | H9c2 | [184] |
| miR-126a-5p | Upregulated | Hspb8 | Cell injury/apoptosis | H9c2 Mice I/R |
[185] |
| miR-129-5p | Downregulated | PI3K/AKT/mTOR | Cell injury/autophagy | H9c2 | [186] |
| miR-141-3p | Upregulated | PI3K/AKT | Apoptosis | H9c2 | [187] |
| miR-142-3p | Downregulated | TLR4/NFk-B | Apoptosis | Mice I/R | [188] |
| miR-144 | Downregulated | FoxO1 | Apoptosis | H9c2 Rats I/R |
[189] |
| miR-145 | Downregulated | CaMKIIδ | Apoptosis | Cardiomyocytes | [150] |
| miR-153 | Upregulated | Nrf2/HO-1 | ROS/apoptosis | Cardiomyocytes | [160] |
| miR-181b-5p | Upregulated | PI3K/AKT | Cell injury/apoptosis | H9c2 Rats I/R |
[190] |
| miR-181c-5p | Upregulated | PTPN4 | Cell injury/apoptosis | H9c2 | [191] |
| miR-208 | Downregulated | P21 | ROS/apoptosis | Cardiomyocytes | [192] |
| miR-210 | Downregulated | AIFM3 | ROS/apoptosis | Cardiomyocytes | [153] |
| miR-223 | Downregulated | NLRP3 | Inflammation | H9c2 | [193] |
| miR-374a-5p | Downregulated | MAPK6 | Cell injury | H9c2 Mice I/R |
[149] |
| miR-486 | Downregulated | JNK/C-Jun NF-kB |
Cell injury/apoptosis | H9c2 | [194] |
| miR-711 | Upregulated | HIF-1α/NF-kB | apoptosis | H9c2 | [195] |
Oxidative stress following a period of hypoxia causes lipid and protein oxidations, and DNA damage which could eventually lead to cell death. In rat heart, using a myocardial I/R injury model of 30 min ischemia followed by 12h reperfusion, the infarct size, cardiomyocyte apoptosis, and levels of creatine kinase and lactate dehydrogenase released were all decreased when miR-22 was overexpressed [151]. One of the targets of miR-22, CBP (cAMP response element binding (CREB) binding protein), was inhibited by miR-22 overexpression. Downregulation of CBP resulted in decreased Bax and p21 (pro-apoptotic related genes) and reduced p53 acetylation activity [151]. This data suggest that miR-22 can inhibit cardiomyocyte apoptosis that occurs due to I/R injury by inhibiting CBP. miR-374a-5p expression was decreased in a myocardial hypoxia/reoxygenation (H/R) H9C2 cell model and a mouse I/R model [149]. miR-374a-5p over-expression diminished cardiac cell damage in both in vivo the cell H/R model and the mouse I/R models of ischemia. miR-374a-5p was found to regulate mitogen-activated protein kinase 6 (MAPK6) negatively. Increased MAPK6 activity inhibited the protective effect of miR-374a-5p in the H9C2 H/R model [149]. Hence, miR-374a-5p seems to be protective against in vitro H/R injury and in vivo cardiac I/R injury.
A mouse model of myocardial infarction (MI) showed increased expression of miRNA-1 [152]. To investigate the role of miRNA-1 on MI, mouse hearts that underwent MI, as well as sham hearts, were treated with a miRNA-1 antagomir that inhibited miRNA-1 expression, miRNA-1 lentiviral vectors that increased miRNA levels or bortezomib that decrease proteasome activity [152]. miRNA-1 upregulated components of the UPS, such as an E3 enzyme and 19S and 20S subunits. Reduced miRNA-1 levels or inhibiting proteasome activity both lessened the left ventricular (LV) end-diastolic diameter and LV mass increases that occur due to MI. Together with other experiments performed, these results suggest that UPS component are mediators of the effects of miRNA-1 on the cardiac remodeling that occurs after MI. It was disappointing that the latter study did not measure the proteasome activity after reducing miRNA levels since an increase in proteasome expression does not always result in increased proteasome activity.
Overexpression of miR-210 in cardiomyocytes reduces ROS production and cell death, while lower miR-210 levels increase ROS production after hypoxia-reoxygenation [153]. miR-210 targets mitochondrion-associated 3 (AIFM3), an apoptosis-inducing factor, but miR-210 cardioprotective effects do not seem to be via AIFM3. The upregulation of miR-210 may be via protein kinase B (Akt) and p53-dependent pathways since Akt inhibition results in lower miR-210 induction during hypoxia, and p53 overexpression in mouse embryonic fibroblasts (MEFs) induce miR-210. The miR-210 cardioprotective effects in cardiomyocytes seem to be through reducing mitochondrial ROS production and although not investigated may be occurring via Nrf2. Tingle SJ et al. [154] investigated if dual blockade of miR-24-3p and miR-145-5p will synergistically upregulate shared target genes during Human Umbilical Vein Endothelial Cells (HUVECs) I/R injury. Under hypoxic conditions miR-24-3p, miR-145-5p and ROS production are upregulated, and heme oxygenase 1 (HMOX1) and SOD1 are downregulated. miR-24-3p and miR-145-5p were highly expressed in human kidneys following extended cold ischemia. Inhibition of miR-24-3p and miR-145-5p before hypoxia-reoxygenation increased HMOX1 and SOD2, and decreased cellular ROS to lower levels than when either miR-24-3p or miR-145-5p were blocked individually.
An ingredient from the traditional Chinese medicinal plant, Rhodiola rosea, Salidroside, was found to be protective (increases antioxidant enzymes, SOD and GSH-Px, reduces ROS and malondialdehyde (MDA) levels and increased cell viability) against myocardial I/R injury in vitro and in vivo. This protective effect in H9C2 cells was found to be mediated by miR-21 as a miR-21 inhibitor reversed the effects of Salidroside [155].
These changes in miRNA expression occur mainly via modulation Nrf2, sirtuins, calcineurin/nuclear factor of activated T cell (NFAT), or NF-κB pathways. Several circulating miRNAs have been reported to be potential biomarkers of ROS-related cardiac diseases, including myocardial infarction, hypertrophy, ischemia/reperfusion, and heart failure, such as miRNA-499, miRNA-199, miRNA-21, miRNA-144, miRNA-208a, miRNA-34a, and others. While a lot of research publications and reviews suggest that circulating miRNAs are potential biomarkers for ROS-related cardiac disease and are likely to be good therapeutic targets because of the number of miRNAs and opposing actions of some miRNAs, significantly more experimental research is needed before we will know if miRNAs will be good biomarkers or therapeutic targets [156].
7. miRNAs and ROS in cerebral ischemia-reperfusion injury
Cerebral I/R injury happens when an ischemic stroke occurs, and is characterized by swelling of cells, apoptosis and necrosis [157]. The primary source of ROS in brain tissue comes from NOX2. Data from a rat I/R injury model and an SH-SY5Y cell hypoxia/reoxygenation (H/R) model showed that NOX2 is significantly increased [158]. The miR-652 was decreased in both models (I/R and H/R), and the use of a miRNA-652 agomir (which reduces miR-652 levels) decreased NOX2 expression and ROS production in brain tissue of the rat cerebral I/R model. miR-652 overexpression reduced NOX2 expression and ROS generation in the H/R treated SH-SY5Y cells [158]. These results suggest that miR-652 is protective against cerebral I/R injury by targeting NOX2.
A cellular model to mimic cerebral I/R injury, called oxygen-glucose deprivation/reoxygenation (OGD/R), has been used in several studies. Hippocampal neurons exposed to OGD/R had significantly less miR-148b-3p expression levels [159]. When miR-148b-3p was overexpressed in neurons, ROS levels and apoptosis was increased, and cell viability decreased after OGD/R. Conversely, inhibition of miR-148b-3p decreased ROS production and apoptosis and improved cell viability [159]. The miR-148b-3p target was a cytoprotective gene, Sestrin2 and Nrf2. Inhibition of miR-148b-3p upregulated both Sestrin2 and Nrf2. Consistent with these results, reducing Sestrin2 or Nrf2 considerably reversed the protective effect of miR-148-3p-inhibition in OGD/R-injured neurons. Treatment of cardiomyocytes using an oxygen-glucose deprivation and reoxygenation (OGD/R) cellular model which should be similar to an I/R model significantly upregulated miR-153 which resulted in increased ROS production and apoptosis [160]. Cardiomyocytes were protected from OGD/R treatment injury when miR-153 levels were reduced. Like many other miRNAs, miR-153 targeted Nrf2, and acts via the inhibiting the Nrf2/HO-1 pathway.
miRNAs are not just regulators of I/R injury in cardiomyocytes but many other cell types and tissues including neuronal tissue. Various miRNAs have also been found to be altered in neuronal injury during cerebral ischemia/reperfusion injury [159]. MiR-153 has also been shown to be important in regulating neuron survival during cerebral ischemia/reperfusion (I/R) injury. miR-199a-5p overexpression in HT22 neurons exposed to OGD/R treatment increased ROS production and induced apoptosis, while inhibition of miR-199a-5p prevented OGD/R-induced ROS production and apoptosis [161]. The target gene for miR-199a-5p was Brahma-related gene 1 (Brg1), which activated Nrf2/HO-1 signaling. Knockdown of Brg1 levels prevented the miR-199a-5p inhibition-mediated neuroprotective effect on neurons [161]. Hence the results suggest that lower levels of miR-199a-5p are protective for neurons exposed to OGD/R-induced injury.
More recent studies suggest that miR-224-3p and miR-10a are also involved in modulating ROS levels [162,163]. Using the OGD/R model in N2a cells, miR-224-3p overexpression reduced ROS and apoptosis due to its interaction with the FAK family‐interacting protein (FIP200) [162]. The effect of miR‐224‐3p on apoptosis was partially blocked when FIP200 was overexpressed [162]. A triterpenoid isolated from the trametes lactinea (Berk.) Pat (a type of mushroom), Trametenolic acid B (TAB) significantly reduced serum ROS levels, neuronal cell loss and apoptosis in cerebral I/R injury rats [163]. The neuroprotective effect of TAB against ODG/R and I/R injury seems to occur through miR-10a. TAB downregulates miR-10a resulting in increased activation of the PI3K/Akt/mTOR signaling pathway, which reduces mitochondrial-mediated apoptosis [163]. Li et al. found that 115 circulating miRNAs were differentially expressed in acute ischemic stroke, a form of cerebral I/R [164]. As such, several other miRNAs involved in cerebral I/R will likely be identified in the next few years that alter the cellular redox status of cells.
8. miRNAs involved in cellular redox status as potential targets for therapeutics and biomarkers
The number of publications that suggest miRNAs would be quality biomarkers for health conditions has significantly increased over the last few years. As such, the promise of miRNAs being used in the treatment of diseases is high. However, an FDA-approved miRNA for the treatment of any illness is still a few years away. Most advanced candidate miRNAs are now in phase 1 or phase 2 clinical trials. The road to approval is long, and most miRNA candidates are typically withdrawn during different stages of the clinical trials (clinicaltrials.gov). Another type of RNA, single (siRNA), had clinical trials started in 2004 and the first siRNA drug was only approved in 2018 [165]. As of April 2020, nearly 900 clinical research studies on miRNAs as biomarkers or as interventional drugs has been conducted or started (clinicaltrials.gov), with some (28 as of April 20th, 2020) of these trials already being discontinued and some (17 as of April 20th, 2020) withdrawn. miR-21, which when inhibited reduces ROS production, is in phase 1 trial for Alport syndrome (https://clinicaltrials.gov/ct2/show/NCT03373786) [166].
It is likely that a miRNA clinical biomarker will be approved before a miRNA is approved for treating disease since more than half of the current studies are screening for biomarkers or secondary studies focusing on particular miRNAs for specific diseases such as preeclampsia. While in some cases, one miRNA might be able to identify a specific health outcome, it is more likely that health conditions will require several for increased specificity of detection [167].
9. Conclusion
Oxidative stress is a key contributing factor to many diseases, including cancer and cardiovascular disease. The number of publications identifying new redox-sensitive miRNAs, as well as the roles of these miRNAs is increasing at a dramatic pace. Our understanding of the major targets of these redox-sensitive miRNAs has increased dramatically, and a few targets such as Nrf2, SIRT1, and NF-κB have been identified that are targets for multiple miRNAs. However, more experimental work is needed since miRNAs could have numerous targets and understanding the effect of the miRNA on those targets, as well as the function of these targets, is essential. Research is also needed to further determine the crosstalk between miRNAs, ROS and diseases, as well as to discover redox-sensitive miRNAs that are vital in many diseases but are not currently being investigated. Discovering new redox-sensitive miRNAs is likely to get easier as in silico-based approaches show promise to identify new miRNA targets as well as miRNAs binding to specific targets. miRNA disease association prediction will also continue to improve [168].
While the rush is on to identify miRNA biomarkers of diseases and to develop miRNA based therapeutic targets, the number of redox-sensitive miRNAs in clinical trials is limited. Hence, the use of redox-sensitive miRNAs for biomarkers of disease or as clinical therapeutics is likely several years off as numerous studies are required to validate and elaborate on current findings. Overall, redox-sensitive miRNAs have the potential to allow us to regulate oxidative stress. Once our understanding of redox-sensitive miRNAs is detailed enough to allow us to use individual or pools of miRNA modulating compounds safely, targeting miRNAs will enable us to improve health-related outcomes associated with different diseases.
Declaration of competing interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Acknowledgements
This work was supported by grants to AVG from the National Institutes of Health Superfund Research Program (P42 ES004699) and the American Heart Association (16GRNT31350040).
References
- 1.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. (80) [DOI] [PubMed] [Google Scholar]
- 3.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. (80) [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. (80) [DOI] [PubMed] [Google Scholar]
- 5.Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 7.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 8.Kim V.N. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Mt B., K C., D G. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604.Most. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu K., He J., Pu W., Peng Y. The role of exportin-5 in MicroRNA biogenesis and cancer. Genom. Proteomics Bioinf. 2018 doi: 10.1016/j.gpb.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djuranovic S., Nahvi A., Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336 doi: 10.1126/science.1215691. (80) 237 LP – 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheloufi S., Dos Santos C.O., Chong M.M.W., Hannon G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro J.S., Langlois R.A., Pham A.M., Tenoever B.R. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18:1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne). 2018;9:1–12. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paroo Z., Ye X., Chen S., Liu Q. Phosphorylation of the human MicroRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su C., Li Z., Cheng J., Li L., Zhong S., Liu L., Zheng Y., Zheng B. The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in arabidopsis. Dev. Cell. 2017;41:527–539. doi: 10.1016/j.devcel.2017.05.008. e5. [DOI] [PubMed] [Google Scholar]
- 17.Wada T., Kikuchi J., Furukawa Y. Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep. 2012;13:142–149. doi: 10.1038/embor.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao S.H., Cheng W.C., Wang Y.T., Wu H.T., Yeh H.Y., Chen Y.J., Tsai M.H., Wu K.J. Regulation of miRNA biogenesis and histone modification by K63-Polyubiquitinated DDX17 controls cancer stem-like features. Canc. Res. 2019;79:2549–2563. doi: 10.1158/0008-5472.CAN-18-2376. [DOI] [PubMed] [Google Scholar]
- 19.Yuan H., Deng R., Zhao X., Chen R., Hou G., Zhang H., Wang Y., Xu M., Jiang B., Yu J. SUMO1 modification of KHSRP regulates tumorigenesis by preventing the TL-G-Rich miRNA biogenesis. Mol. Canc. 2017;16:1–18. doi: 10.1186/s12943-017-0724-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S.-Z., Hu Y.-Y., Zhao J., Zhao Y.-B., Sun J.-D., Yang Y., Ji C.-C., Liu Z.-B., Cao W.-D., Qu Y., Liu W.-P., Cheng G., Fei Z. MicroRNA-34a induces apoptosis in the human glioma cell line, A172, through enhanced ROS production and NOX2 expression. Biochem. Biophys. Res. Commun. 2014;444:6–12. doi: 10.1016/J.BBRC.2013.12.136. [DOI] [PubMed] [Google Scholar]
- 21.Simone N.L., Soule B.P., Ly D., Saleh A.D., Savage J.E., DeGraff W., Cook J., Harris C.C., Gius D., Mitchell J.B. Ionizing radiation-induced oxidative stress alters miRNA expression. PloS One. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pradhan A.K., Bhoopathi P., Talukdar S., Scheunemann D., Sarkar D., Cavenee W.K., Das S.K., Emdad L., Fisher P.B. MDA-7/IL-24 regulates the miRNA processing enzyme DICER through downregulation of MITF. Proc. Natl. Acad. Sci. U.S.A. 2019;116:5687–5692. doi: 10.1073/pnas.1819869116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston H., Dickinson P., Ivens A., Buck A.H., Levine R.D., Remacle F., Campbell C.J. Intracellular redox potential is correlated with miRNA expression in MCF7 cells under hypoxic conditions. Proc. Natl. Acad. Sci. 2019;116:19753–19759. doi: 10.1073/PNAS.1909455116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalton S., Smith K., Singh K., Kaiser H., Kolhe R., Mondal A.K., Khayrullin A., Isales C.M., Hamrick M.W., Hill W.D., Fulzele S. Accumulation of kynurenine elevates oxidative stress and alters microRNA profile in human bone marrow stromal cells. Exp. Gerontol. 2020;130 doi: 10.1016/j.exger.2019.110800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loosen S.H., Schueller F., Trautwein C., Roy S., Roderburg C. Role of circulating microRNAs in liver diseases. World J. Hepatol. 2017;9:586–594. doi: 10.4254/wjh.v9.i12.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q.M., Maltagliati A.J. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genom. 2018;50:77–97. doi: 10.1152/physiolgenomics.00041.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Youn S.W., Li Y., Kim Y.M., Sudhahar V., Abdelsaid K., Kim H.W., Liu Y., Fulton D.J.R., Ashraf M., Tang Y., Fukai T., Ushio-Fukai M. Modification of cardiac progenitor cell-derived exosomes by miR-322 provides protection against myocardial infarction through nox2-dependent angiogenesis. Antioxidants. 2019;8 doi: 10.3390/antiox8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konovalova J., Gerasymchuk D., Parkkinen I., Chmielarz P., Domanskyi A. Interplay between MicroRNAs and oxidative stress in neurodegenerative diseases. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20236055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W., Zabirnyk O., Wang H., Shiao Y.H., Nickerson M.L., Khalil S., Anderson L.M., Perantoni A.O., Phang J.M. MiR-23b targets proline oxidase, a novel tumor suppressor protein in renal cancer. Oncogene. 2010;29:4914–4924. doi: 10.1038/onc.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang T., Wang-Johanning F., Zhou F., Kallon H., Wei Y. MicroRNAs serve as a bridge between oxidative stress and gastric cancer (Review) Int. J. Oncol. 2016;49:1791–1800. doi: 10.3892/ijo.2016.3686. [DOI] [PubMed] [Google Scholar]
- 31.Motawi T.K., Mohamed M.R., Shahin N.N., Ali M.A.M., Azzam M.A. Time-course expression profile and diagnostic potential of a miRNA panel in exosomes and total serum in acute liver injury. Int. J. Biochem. Cell Biol. 2018;100:11–21. doi: 10.1016/j.biocel.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Ong S., Katwadi K., Kwek X., Ismail N.I., Chinda K., Ong S., Hausenloy D.J. Expert Opinion on Therapeutic Targets Non-coding RNAs as therapeutic targets for preventing myocardial ischemia-reperfusion injury. Expert Opin. Ther. Targets. 2018;22:247–261. doi: 10.1080/14728222.2018.1439015. [DOI] [PubMed] [Google Scholar]
- 33.Cao F., Liu Z., Sun G. Diagnostic value of miR-193a-3p in Alzheimer's disease and miR-193a-3p attenuates amyloid-β induced neurotoxicity by targeting PTEN. Exp. Gerontol. 2020;130:1–7. doi: 10.1016/j.exger.2019.110814. [DOI] [PubMed] [Google Scholar]
- 34.Fierro-Fernández M., Miguel V., Lamas S. Role of redoximiRs in fibrogenesis. Redox Biol. 2016;7:58–67. doi: 10.1016/j.redox.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finkel T. Signal transduction by mitochondrial oxidants. J. Biol. Chem. 2012;287:4434–4440. doi: 10.1074/jbc.R111.271999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Q. Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacol. Ther. 2010;125:376–393. doi: 10.1016/j.pharmthera.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Gutteridge J.M.C., Halliwell B. NEW YORK ACAD SCIENCES; 2000. Free Radicals and Antioxidants in the Year 2000 - A Historical Look to the Future. ISI:000088609100011. [DOI] [PubMed] [Google Scholar]
- 38.Chen Q., Chai Y.C., Mazumder S., Jiang C., Macklis R.M., Chisolm G.M., Almasan A. The late increase in intracellular free radical oxygen species during apoptosis is associated with cytochrome c release, caspase activation, and mitochondrial dysfunction. Cell Death Differ. 2003;10:323–334. doi: 10.1038/sj.cdd.4401148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raj L., Ide T., Gurkar A.U., Foley M., Schenone M., Li X., Tolliday N.J., Golub T.R., Carr S.A., Shamji A.F., Stern A.M., Mandinova A., Schreiber S.L., Lee S.W. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–234. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Halliwell B., Gutteridge J.M.C. Oxford University Press; Oxford: 1999. Free Radicals in Biology and Medicin. [Google Scholar]
- 41.Park S.G., Kim J.H., Xia Y., Sung J.H. Generation of reactive oxygen species in adipose-derived stem cells: friend or foe? Expert Opin. Ther. Targets. 2011;15:1297–1306. doi: 10.1517/14728222.2011.628315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu B.P. Cellular defenses against damage from reactive oxygen species. Physiol. Rev. 1994;74:139–162. doi: 10.1152/physrev.1994.74.1.139. [DOI] [PubMed] [Google Scholar]
- 43.Jung T., Catalgol B., Grune T. The proteasomal system. Mol. Aspect. Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Höhn T.J.A., Grune T. The proteasome and the degradation of oxidized proteins: part III-Redox regulation of the proteasomal system. Redox Biol. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C., Hao X., Chang J., Geng Z., Wang Z. Mn-TAT PTD-Ngb attenuates oxidative injury by an enhanced ROS scavenging ability and the regulation of redox signaling pathway. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-56595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu S., Liu J., Lee K., Tang F., Fang K. Vol. 65. 2020. pp. 1–16. (Toxicology in Vitro Cr ( VI ) induces ROS-mediated mitochondrial-dependent apoptosis in neuronal cells via the activation of Akt/ERK/AMPK signaling pathway). [DOI] [PubMed] [Google Scholar]
- 47.He J., Jiang B.H. Interplay between reactive oxygen species and MicroRNAs in cancer. Curr. Pharmacol. Rep. 2016;2:82–90. doi: 10.1007/s40495-016-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ebrahimi S., Hashemy S.I. MicroRNA-mediated redox regulation modulates therapy resistance in cancer cells: clinical perspectives. Cell. Oncol. 2019;42:131–141. doi: 10.1007/s13402-018-00421-z. [DOI] [PubMed] [Google Scholar]
- 49.Vafa O., Wade M., Kern S., Beeche M., Pandita T.K., Hampton G.M., Wahl G.M. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell. 2002;9:1031–1044. doi: 10.1016/S1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 50.Yang H., Li T.W.H., Zhou Y., Peng H., Liu T., Zandi E., Martínez-Chantar M.L., Mato J.M., Lu S.C. Activation of a novel c-Myc-miR27-prohibitin 1 circuitry in cholestatic liver injury inhibits glutathione synthesis in mice. Antioxidants Redox Signal. 2015;22:259–274. doi: 10.1089/ars.2014.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao Y., Yan W., Lu L., Wang Y., Lu W., Cao Y., Cai W. p38/p53/miR-200a-3p feedback loop promotes oxidative stress-mediated liver cell death. Oxidative Stress. Liver Cell Death. 2015;14:1548–1558. doi: 10.1080/15384101.2015.1026491. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Yin M., Ren X., Zhang X., Luo Y., Wang G., Huang K., Feng S., Bao X., Huang K., He X., Liang P., Wang Z., Tang H., He J., Zhang B. Selective killing of lung cancer cells by miRNA-506 molecule through inhibiting NF-κB p65 to evoke reactive oxygen species generation and p53 activation. Oncogene. 2015;34:691–703. doi: 10.1038/onc.2013.597. [DOI] [PubMed] [Google Scholar]
- 53.Lingappan K. NF-κB in oxidative stress. Curr. Opin. Toxicol. 2018;7:81–86. doi: 10.1016/j.cotox.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markopoulos G.S., Roupakia E., Tokamani M., Alabasi G., Sandaltzopoulos R., Marcu K.B., Kolettas E. Roles of NF-κB signaling in the regulation of miRNAs impacting on inflammation in cancer. Biomedicines. 2018;6:1–19. doi: 10.3390/biomedicines6020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madhyastha R., Madhyastha H., Pengjam Y., Nakajima Y., Omura S., Maruyama M. NFkappaB activation is essential for miR-21 induction by TGFβ1 in high glucose conditions. Biochem. Biophys. Res. Commun. 2014;451:615–621. doi: 10.1016/J.BBRC.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 56.Dattaroy D., Pourhoseini S., Das S., Alhasson F., Seth R.K., Nagarkatti M., Michelotti G.A., Diehl A.M., Chatterjee S. Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via leptin-mediated NADPH oxidase in experimental and human nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308:G298–G312. doi: 10.1152/ajpgi.00346.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pogue A.I., Li Y.Y., Cui J.-G., Zhao Y., Kruck T.P.A., Percy M.E., Tarr M.A., Lukiw W.J. Characterization of an NF-κB-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J. Inorg. Biochem. 2009;103:1591–1595. doi: 10.1016/J.JINORGBIO.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Pogue A.I., Percy M.E., Cui J.G., Li Y.Y., Bhattacharjee S., Hill J.M., Kruck T.P.A., Zhao Y., Lukiw W.J. Up-regulation of NF-kB-sensitive miRNA-125b and miRNA-146a in metal sulfate-stressed human astroglial (HAG) primary cell cultures. J. Inorg. Biochem. 2011;105:1434–1437. doi: 10.1016/j.jinorgbio.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Semenza G.L. Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim. Biophys. Acta Mol. Cell Res. 2011;1813:1263–1268. doi: 10.1016/j.bbamcr.2010.08.006. http://www.sciencedirect.com/science/article/pii/S0167488910002223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coimbra-Costa D., Alva N., Duran M., Carbonell T., Rama R. Oxidative stress and apoptosis after acute respiratory hypoxia and reoxygenation in rat brain. Redox Biol. 2017;12:216–225. doi: 10.1016/j.redox.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.chao Yang M., li You F., Wang Z., nan Liu X., feng Wang Y. Salvianolic acid B improves the disruption of high glucose-mediated brain microvascular endothelial cells via the ROS/HIF-1α/VEGF and miR-200b/VEGF signaling pathways. Neurosci. Lett. 2016;630:233–240. doi: 10.1016/j.neulet.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Greco S., Gaetano C., Martelli F. HypoxamiR regulation and function in ischemic cardiovascular diseases. Antioxidants Redox Signal. 2014;21:1202–1219. doi: 10.1089/ars.2013.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Favaro E., Ramachandran A., McCormick R., Gee H., Blancher C., Crosby M., Devlin C., Blick C., Buffa F., Li J.L., Vojnovic B., das Neves R.P., Glazer P., Iborra F., Ivan M., Ragoussis J., Harris A.L. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PloS One. 2010;5 doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu H., Ding Y., Wang Y., Geng S., Liu J., He J., Lu Y., Li X., Yuan M., Zhu S., Zhao S. MitoKATP channels promote the proliferation of hypoxic human pulmonary artery smooth muscle cells via the ROS/HIF/miR-210/ISCU signaling pathway. Exp. Ther. Med. 2017;14:6105–6112. doi: 10.3892/etm.2017.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seok J.K., Lee S.H., Kim M.J., Lee Y.M. MicroRNA-382 induced by HIF-1α is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014;42:8062–8072. doi: 10.1093/nar/gku515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun N., Meng F., Xue N., Pang G., Wang Q., Ma H. Inducible miR-145 expression by HIF-1α protects cardiomyocytes against apoptosis via regulating SGK1 in simulated myocardial infarction hypoxic microenvironment. Cardiol. J. 2018;25:268–278. doi: 10.5603/CJ.a2017.0105. [DOI] [PubMed] [Google Scholar]
- 67.Meng X., Wu J., Pan C., Wang H., Ying X., Zhou Y., Yu H., Zuo Y., Pan Z., Liu R., Huang W. Genetic and epigenetic down-regulation of MicroRNA-212 promotes colorectal tumor metastasis via dysregulation of MnSOD. Gastroenterology. 2013;145:426–436. doi: 10.1053/J.GASTRO.2013.04.004. e6. [DOI] [PubMed] [Google Scholar]
- 68.Gordillo G.M., Biswas A., Khanna S., Pan X., Sinha M., Roy S., Sen C.K. Dicer knockdown inhibits Endothelial cell tumor growth via microRNA 21a-3p targeting of nox-4. J. Biol. Chem. 2014;289:9027–9038. doi: 10.1074/jbc.M113.519264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Druz A., Betenbaugh M., Shiloach J. Glucose depletion activates mmu-miR-466h-5p expression through oxidative stress and inhibition of histone deacetylation. Nucleic Acids Res. 2012;40:7291–7302. doi: 10.1093/nar/gks452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang D., Lee H., Cao Y., Dela Cruz C.S., Jin Y. miR-185 mediates lung epithelial cell death after oxidative stress. Am. J. Physiol. Lung Cell Mol. Physiol. 2016;310:L700–L710. doi: 10.1152/ajplung.00392.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barr I., Smith A.T., Chen Y., Senturia R., Burstyn J.N., Guo F. Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc. Natl. Acad. Sci. U.S.A. 2012;109:1919–1924. doi: 10.1073/pnas.1114514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen T.A., Park J., Dang T.L., Choi Y.G., Kim V.N. Microprocessor depends on hemin to recognize the apical loop of primary microRNA. Nucleic Acids Res. 2018;46:5726–5736. doi: 10.1093/nar/gky248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yao M., Wang X., Tang Y., Zhang W., Cui B., Liu Q., Xing L. Dicer mediating the expression of miR-143 and miR-155 regulates hexokinase II associated cellular response to hypoxia. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;307:L829–L837. doi: 10.1152/ajplung.00081.2014. [DOI] [PubMed] [Google Scholar]
- 74.Ho J.J.D., Metcalf J.L., Yan M.S., Turgeon P.J., Wang J.J., Chalsev M., Petruzziello-Pellegrini T.N., Tsui A.K.Y., He J.Z., Dhamko H., Man H.S.J., Robb G.B., Teh B.T., Ohh M., Marsden P.A. Functional importance of dicer protein in the adaptive cellular response to hypoxia. J. Biol. Chem. 2012;287:29003–29020. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 76.Su X., Chakravarti D., Cho M.S., Liu L., Gi Y.J., Lin Y.L., Leung M.L., El-Naggar A., Creighton C.J., Suraokar M.B., Wistuba I., Flores E.R. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haramati S., Chapnik E., Sztainberg Y., Eilam R., Zwang R., Gershoni N., McGlinn E., Heiser P.W., Wills A.M., Wirguin I., Rubin L.L., Misawa H., Tabin C.J., Brown R., Chen A., Hornstein E. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berardino B.G., Fesser E.A., Cánepa E.T. Perinatal protein malnutrition alters expression of miRNA biogenesis genes Xpo5 and Ago2 in mice brain. Neurosci. Lett. 2017;647:38–44. doi: 10.1016/j.neulet.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 79.Crosas-Molist E., Fabregat I. Role of NADPH oxidases in the redox biology of liver fibrosis. Redox Biol. 2015;6:106–111. doi: 10.1016/j.redox.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Z., Tuo Y.H., Chen J.W., Wang Q.Y., Li S., Li M.C., Dai G., Wang J.S., Zhang Y.L., Feng L., Shi Z.S. NADPH oxidase inhibitor regulates microRNAs with improved outcome after mechanical reperfusion. J. Neurointerventional Surg. 2017;9:702–706. doi: 10.1136/neurintsurg-2016-012463. [DOI] [PubMed] [Google Scholar]
- 81.Wu Y., Yao J., Feng K. miR-124-5p/NOX2 Axis modulates the ROS production and the inflammatory microenvironment to protect against the cerebral I/R injury. Neurochem. Res. 2020;45:404–417. doi: 10.1007/s11064-019-02931-0. [DOI] [PubMed] [Google Scholar]
- 82.Zhang X., Ng W.L., Wang P., Tian L.L., Werner E., Wang H., Doetsch P., Wang Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFα. Canc. Res. 2012;72:4707–4713. doi: 10.1158/0008-5472.CAN-12-0639. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 83.Zhang Y., Zheng S., Geng Y., Xue J., Wang Z., Xie X., Wang J., Zhang S., Hou Y. MicroRNA profiling of atrial fibrillation in canines: MiR-206 modulates intrinsic cardiac autonomic nerve remodeling by regulating SOD1. PloS One. 2015;10:1–16. doi: 10.1371/journal.pone.0122674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haque R., Chun E., Howell J.C., Sengupta T., Chen D., Kim H. MicroRNA-30b-mediated regulation of catalase expression in human ARPE-19 cells. PloS One. 2012;7 doi: 10.1371/journal.pone.0042542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Q., Chen W., Bai L., Chen W., Padilla M.T., Lin A.S., Shi S., Wang X., Lin Y. Receptor-interacting protein 1 increases chemoresistance by maintaining inhibitor of apoptosis protein levels and reducing reactive oxygen species through a microRNA-146a-mediated catalase pathway. J. Biol. Chem. 2014;289:5654–5663. doi: 10.1074/jbc.M113.526152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu X., Wells A., Padilla M.T., Kato K., Kim K.C., Lin Y. A signaling pathway consisting of miR-551b, catalase and MUC1 contributes to acquired apoptosis resistance and chemoresistance. Carcinogenesis. 2014;35:2457–2466. doi: 10.1093/carcin/bgu159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carretero J., Obrador E., Pellicer J.A., Pascual A., Estrela J.M. Mitochondrial glutathione depletion by glutamine in growing tumor cells. Free Radic. Biol. Med. 2000;29:913–923. doi: 10.1016/S0891-5849(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 88.Vairetti M., Griffini P., Pietrocola G., Richelmi P., Freitas I. Cold-induced apoptosis in isolated rat hepatocytes: protective role of glutathione. Free Radic. Biol. Med. 2001;31:954–961. doi: 10.1016/S0891-5849(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 89.Carbonell T., Alva N., Sanchez-Nunõ S., Dewey S., Gomes A.V.A.V. Subnormothermic perfusion in the isolated rat liver preserves the antioxidant glutathione and enhances the function of the ubiquitin proteasome system. Oxid. Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/9324692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matoušková P., Hanousková B., Skálová L. Micrornas as potential regulators of glutathione peroxidases expression and their role in obesity and related pathologies. Int. J. Mol. Sci. 2018;19:1–23. doi: 10.3390/ijms19041199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang L., Huang H., Fan Y., Kong B., Hu H., Hu K., Guo J., Mei Y., Liu W.L. Effects of downregulation of microrna-181a on h2O 2-induced H9c2 Cell apoptosis via the mitochondrial apoptotic pathway. Oxid. Med. Cell. Longev. 2014;2014 doi: 10.1155/2014/960362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y., Chen S.H., Jin X., Li Y.M. Analysis of differentially expressed genes and microRNAs in alcoholic liver disease. Int. J. Mol. Med. 2013;31:547–554. doi: 10.3892/ijmm.2013.1243. [DOI] [PubMed] [Google Scholar]
- 93.La Sala L., Cattaneo M., De Nigris V., Pujadas G., Testa R., Bonfigli A.R., Genovese S., Ceriello A. Oscillating glucose induces microRNA-185 and impairs an efficient antioxidant response in human endothelial cells, Cardiovasc. Diabetol. 2016;15:1–9. doi: 10.1186/s12933-016-0390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sangokoya C., Telen M., Chi J. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116(20):4338–4348. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang S., Fei X., Lu Y., Xu B., Ma Y., Wan H. miRNA-214 suppresses oxidative stress in diabetic nephropathy via the ROS/Akt/mTOR signaling pathway and uncoupling protein 2. Exp. Ther. Med. 2019:3530–3538. doi: 10.3892/etm.2019.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dong X., Liu H., Chen F., Li D., Zhao Y. MiR-214 promotes the alcohol-induced oxidative stress via down-regulation of glutathione reductase and cytochrome P450 oxidoreductase in liver cells, alcohol. Clin. Exp. Res. 2014;38:68–77. doi: 10.1111/acer.12209. [DOI] [PubMed] [Google Scholar]
- 97.Kinoshita C., Aoyama K., Nakaki T. Neuroprotection afforded by circadian regulation of intracellular glutathione levels: a key role for miRNAs. Free Radic. Biol. Med. 2018;119:17–33. doi: 10.1016/j.freeradbiomed.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 98.Cortez M.A., Valdecanas D., Zhang X., Zhan Y., Bhardwaj V., Calin G.A., Komaki R., Giri D.K., Quini C.C., Wolfe T., Peltier H.J., Bader A.G., Heymach J.V., Meyn R.E., Welsh J.W. Therapeutic delivery of mir-200c enhances radiosensitivity in lung cancer. Mol. Ther. 2014;22:1494–1503. doi: 10.1038/mt.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Singh B., Ronghe A.M., Chatterjee A., Bhat N.K., Bhat H.K. MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis. 2013;34:1165–1172. doi: 10.1093/carcin/bgt026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eades G., Yang M., Yao Y., Zhang Y., Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J. Biol. Chem. 2011;286:40725–40733. doi: 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kabaria S., Choi D.C., Chaudhuri A.D., Jain M.R., Li H., Junn E. MicroRNA-7 activates Nrf2 pathway by targeting Keap1 expression. Free Radic. Biol. Med. 2015;89:548–556. doi: 10.1016/J.FREERADBIOMED.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu D., Zhu H., Wang C., Zhu X., Liu G., Chen C., Cui Z. microRNA-455 targets cullin 3 to activate Nrf2 signaling and protect human osteoblasts from hydrogen peroxide. Oncotarget. 2017;8:59225–59234. doi: 10.18632/oncotarget.19486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang X., Schulz R., Edmunds S., Krüger E., Markert E., Gaedcke J., Cormet-Boyaka E., Ghadimi M., Beissbarth T., Levine A.J., Moll U.M., Dobbelstein M. MicroRNA-101 suppresses tumor cell proliferation by acting as an endogenous proteasome inhibitor via targeting the proteasome assembly factor POMP. Mol. Cell. 2015;59:243–257. doi: 10.1016/j.molcel.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 104.Kottakis F., Polytarchou C., Foltopoulou P., Sanidas I., Kampranis S.C., Tsichlis P.N. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol. Cell. 2011;43(2):285–298. doi: 10.1016/j.molcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Varambally S., Cao Q., Mani R., Shankar S., Wang X., Ateeq B., Laxman B., Cao X., Jing X., Ramnarayanan K., Brenner J.C., Yu J., Kim J.H., Han B., Tan P., Kumar-sinha C., Lonigro R.J., Maher C.A., Chinnaiyan A.M. vol. 322. 2009. pp. 1695–1699. (Genomic Loss of micro-RNA-101 Leads to Overexpression of Histone). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gilda J.E., Gomes A.V. Proteasome dysfunction in cardiomyopathies. J. Physiol. 2017;595:4051–4071. doi: 10.1113/JP273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.V Gomes A. Genetics of proteasome diseases. Sci. (Cairo) 2013 doi: 10.1155/2013/637629. (n.d.) 637629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cui Z., Hwang S.M., V Gomes A. Identification of the immunoproteasome as a novel regulator of skeletal muscle differentiation. Mol. Cell Biol. 2014;34:96–109. doi: 10.1128/MCB.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ji W.U., Eunju I., Joongkyu P., Yohan O., Boram M., Hyun J.L., Jong B.Y., Chung K.C. ASK1 negatively regulates the 26 S proteasome. J. Biol. Chem. 2010;285:36434–36446. doi: 10.1074/jbc.M110.133777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hirano Y., Hendil K.B., Yashiroda H., Iemura S.I., Nagane R., Hioki Y., Natsume T., Tanaka K., Murata S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- 111.Hirano Y., Kaneko T., Okamoto K., Bai M., Yashiroda H., Furuyama K., Kato K., Tanaka K., Murata S. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008;27(16):2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heink S., Ludwig D., Kloetzel P.M., Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fricke B., Heink S., Steffen J., Kloetzel P.M., Kruger E. The proteasome maturation protein POMP facilitates major steps of 20S proteasome formation at the endoplasmic reticulum. EMBO Rep. 2007;8:1170–1175. doi: 10.1038/sj.embor.7401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ruschak A.M., Slassi M., Kay L.E., Schimmer A.D. 6). Novel proteasome inhibitors to overcome bortezomib resistance. Journal of the National Cancer Institute. 2011, July doi: 10.1093/jnci/djr160. [DOI] [PubMed] [Google Scholar]
- 115.Masdehors P., Omura S., Merle-Beral H., Mentz F., Cosset J.M., Dumont J., Magdelénat H., Delic J. Increased sensitivity of CLL-derived lymphocytes to apoptotic death activation by the proteasome-specific inhibitor lactacystin. Br. J. Haematol. 1999;105:752–757. doi: 10.1046/j.1365-2141.1999.01388.x. [DOI] [PubMed] [Google Scholar]
- 116.Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxidants Redox Signal. 2018;29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Furukawa M., Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Eggler A.L., Liu G., Pezzuto J.M., Breemen R.B., Mesecar A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sekhar K.R., Rachakonda G., Freeman M.L. Cysteine-based regulation of the CUL3 adaptor protein Keap1. Toxicol. Appl. Pharmacol. 2010;244:21–26. doi: 10.1016/j.taap.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Taguchi K., Motohashi H., Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Gene Cell. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 121.Steffen J., Seeger M., Koch A., Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 122.Radhakrishnan S.K., Lee C., Young P., Beskow A., Chan J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Piantadosi C.A., Carraway M.S., Babiker A., Suliman H.B. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ. Res. 2008;103:1232–1240. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sc L., HM PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kapeta S., Chondrogianni N., Gonos E.S. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J. Biol. Chem. 2010;285:8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kwak M.K., Wakabayashi N., Itoh K., Motohashi H., Yamamoto M., Kensler T.W. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 127.Amodio N., Gallo Cantafio M., Botta C., Agosti V., Federico C., Caracciolo D., Ronchetti D., Rossi M., Driessen C., Neri A., Tagliaferri P., Tassone P. Replacement of miR-155 elicits tumor suppressive activity and antagonizes bortezomib resistance in multiple myeloma. Cancers. 2019;11:236. doi: 10.3390/cancers11020236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jagannathan S., Vad N., Vallabhapurapu S., Vallabhapurapu S., Anderson K.C., Driscoll J.J. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia. 2015;29:727–738. doi: 10.1038/leu.2014.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lerner M., Haneklaus M., Harada M., Grander D. MiR-200c regulates Noxa expression and sensitivity to proteasomal inhibitors. PloS One. 2012;7 doi: 10.1371/journal.pone.0036490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Engedal N., Žerovnik E., Rudov A., Galli F., Olivieri F., Procopio A., Rippo M., Monsurrò V., Betti M., Albertini M. From oxidative stress damage to pathways, networks, and autophagy via MicroRNAs. Oxid. Med. Cell Longev. 2018;2018:4968321. doi: 10.1155/2018/4968321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.SL, de Stephanis L., Mangolini A., Servello M., Harris P.C., Dell'Atti L., Pinton P., Aguiari G. MicroRNA501-5p induces p53 proteasome degradation through the activation of the mTOR/MDM2 pathway in ADPKD cells. J. Cell. Physiol. 2018;233:6911–6924. doi: 10.1002/jcp.26473. [DOI] [PubMed] [Google Scholar]
- 132.Xu S., Zhang R., Niu J., Cui D., Xie B., Zhang B., Lu K., Yu W., Wang X., Zhang Q. Oxidative stress mediated-alterations of the microRNA expression profile in mouse hippocampal neurons. Int. J. Mol. Sci. 2012;13:16945–16960. doi: 10.3390/ijms131216945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Makhdoumi P., Roohbakhsh A., Karimi G. MicroRNAs regulate mitochondrial apoptotic pathway in myocardial ischemia-reperfusion-injury. Biomed. Pharmacother. 2016;84:1635–1644. doi: 10.1016/j.biopha.2016.10.073. [DOI] [PubMed] [Google Scholar]
- 134.Liu Z., Li T., Zhu F., Deng S., Li X., He Y. Regulatory roles of miR-22/Redd1-mediated mitochondrial ROS and cellular autophagy in ionizing radiation-induced BMSC injury. Cell Death Dis. 2019;10 doi: 10.1038/s41419-019-1373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gao Y.T., Chen X.B., Liu H.L. Up-regulation of miR-370-3p restores glioblastoma multiforme sensitivity to temozolomide by influencing MGMT expression. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep32972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang H.L., Shi Y.P., He H.J., Wang Y.H., Chen T., Yang L.W., Yang T., Chen J., Cao J., Yao W.M., Liu G. MiR-4673 modulates paclitaxel-induced oxidative stress and loss of mitochondrial membrane potential by targeting 8-oxoguanine-DNA glycosylase-1. Cell. Physiol. Biochem. 2017;42:889–900. doi: 10.1159/000478644. [DOI] [PubMed] [Google Scholar]
- 137.Dehaini H., Awada H., El-Yazbi A., Zouein F.A., Issa K., Eid A.A., Ibrahim M., Badran A., Baydoun E., Pintus G., Eid A.H. MicroRNAs as potential pharmaco-targets in ischemia-reperfusion injury compounded by diabetes. Cells. 2019;8:152. doi: 10.3390/cells8020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mateescu B., Batista L., Cardon M., Gruosso T., De Feraudy Y., Mariani O., Nicolas A., Meyniel J.P., Cottu P., Sastre-Garau X., Mechta-Grigoriou F. MiR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 2011;17:1627–1635. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 139.Wen J., Xiong K., Aili A., Wang H., Zhu Y., Yu Z., Yao X., Jiang P., Xue L., Wang J. PEX5, a novel target of microRNA-31-5p, increases radioresistance in hepatocellular carcinoma by activating Wnt/β-catenin signaling and homologous recombination. Theranostics. 2020;10:5322–5340. doi: 10.7150/thno.42371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lan J., Huang Z., Han J., Shao J., Huang C. Redox regulation of microRNAs in cancer. Canc. Lett. 2018;418:250–259. doi: 10.1016/J.CANLET.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 141.Zhang W.C. MicroRNAs tune oxidative stress in cancer therapeutic tolerance and resistance. Int. J. Mol. Sci. 2019;20:6090. doi: 10.3390/ijms20236094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Babu K.R., Tay Y. The Yin-Yang regulation of reactive oxygen species and microRNAs in cancer. Int. J. Mol. Sci. 2019;20:5335. doi: 10.3390/ijms20215335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin Y.H. MicroRNA networks modulate oxidative stress in cancer. Int. J. Mol. Sci. 2019;20:4497. doi: 10.3390/ijms20184497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ventura A., Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gupta S.C., Hevia D., Patchva S., Park B., Koh W., Aggarwal B.B. Upsides and downsides of reactive oxygen species for Cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxidants Redox Signal. 2012;16:1295–1322. doi: 10.1089/ars.2011.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang C., Yan Z., Hu F., Wei W., Sun Z., Xu W. Silencing of microRNA-517a induces oxidative stress injury in melanoma cells via inactivation of the JNK signaling pathway by upregulating CDKN1C. Canc. Cell Int. 2020;20:1–14. doi: 10.1186/s12935-019-1064-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang Q., Muhua, Yao Yuan, Eades Gabriel, Zhang Yongshu, Zhou MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Canc. Res. Treat. 2011;129:983–991. doi: 10.1007/s10549-011-1604-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kushwaha P.P., Gupta S., Singh A.K., Prajapati K.S., Shuaib M., Kumar S. MicroRNA targeting nicotinamide adenine dinucleotide phosphate oxidases in cancer. Antioxidants Redox Signal. 2020;32:267–284. doi: 10.1089/ars.2019.7918. [DOI] [PubMed] [Google Scholar]
- 149.Huang Z.Q., Xu W., Wu J.L., Lu X., Chen X.M. MicroRNA-374a protects against myocardial ischemia-reperfusion injury in mice by targeting the MAPK6 pathway. Life Sci. 2019;232:116619. doi: 10.1016/j.lfs.2019.116619. [DOI] [PubMed] [Google Scholar]
- 150.Cha M.-J., Jang J.-K., Ham O., Song B.-W., Lee S.-Y., Lee C.Y., Park J.-H., Lee J., Seo H.-H., Choi E., Jeon W., Hwang H.J., Shin H.-T., Choi E., Hwang K.-C. MicroRNA-145 suppresses ROS-induced Ca2+ overload of cardiomyocytes by targeting CaMKIIδ. Biochem. Biophys. Res. Commun. 2013;435:720–726. doi: 10.1016/J.BBRC.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 151.Yang J., Chen J., Yang J., Ding J., Li S., Wu H., Jing Z., Fan Z., Dong W., Xinxin L. MicroRNA-22 targeting CBP protects against myocardial ischemia-reperfusion injury through anti-apoptosis in rats. Mol. Biol. Rep. 2014;41:555–561. doi: 10.1007/s11033-013-2891-x. [DOI] [PubMed] [Google Scholar]
- 152.Wei L., Zhang Y., Qi X., Sun X., Li Y., Xu Y. Ubiquitinproteasomes are the dominant mediators of the regulatory effect of microRNA1 on cardiac remodeling after myocardial infarction. Int. J. Mol. Med. 2019;44:1899–1907. doi: 10.3892/ijmm.2019.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mutharasan R.K., Nagpal V., Ichikawa Y., Ardehali H. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1519–H1530. doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tingle S.J., Sewpaul A., Bates L., Thompson E., Shuttleworth V., Figueiredo R., Ibrahim I.K., Ali S., Wilson C., Sheerin N. Dual microRNA blockade increases expression of antioxidant protective proteins: implications for ischaemia reperfusion injury. Transplantation. 2020 doi: 10.1097/tp.0000000000003215. [DOI] [PubMed] [Google Scholar]
- 155.Liu B., Wei H., Lan M., Jia N., Liu J., Zhang M. MicroRNA-21 mediates the protective effects of salidroside against hypoxia/reoxygenation-induced myocardial oxidative stress and inflammatory response. Exp. Ther. Med. 2020;19:1655–1664. doi: 10.3892/etm.2020.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kura B., Bacova B.S., Kalocayova B., Sykora M., Slezak J. Oxidative stress-responsive microRNAs in heart injury. Int. J. Mol. Sci. 2020;21:358. doi: 10.3390/ijms21010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Spescha R.D., Klohs J., Semerano A., Giacalone G., Derungs R.S., Reiner M.F., Gutierrez D.R., Mendez-Carmona N., Glanzmann M., Savarese G., Kränkel N., Akhmedov A., Keller S., Mocharla P., Kaufmann M.R., Wenger R.H., Vogel J., Kulic L., Nitsch R.M., Beer J.H., Peruzzotti-Jametti L., Sessa M., Lüscher T.F., Camici G.G. Post-ischaemic silencing of p66Shc reduces ischaemia/reperfusion brain injury and its expression correlates to clinical outcome in stroke. Eur. Heart J. 2015;36:1590–1600. doi: 10.1093/eurheartj/ehv140. [DOI] [PubMed] [Google Scholar]
- 158.Zuo M.L., Wang A.P., Song G.L., Yang Z.B. miR-652 protects rats from cerebral ischemia/reperfusion oxidative stress injury by directly targeting NOX2. Biomed. Pharmacother. 2020;124:109860. doi: 10.1016/j.biopha.2020.109860. [DOI] [PubMed] [Google Scholar]
- 159.Du Y., Ma X., Ma L., Li S., Zheng J., Lv J., Cui L., Lv J. Inhibition of microRNA-148b-3p alleviates oxygen-glucose deprivation/reoxygenation-induced apoptosis and oxidative stress in HT22 hippocampal neuron via reinforcing Sestrin2/Nrf2 signalling. Clin. Exp. Pharmacol. Physiol. 2020;47:561–570. doi: 10.1111/1440-1681.13231. [DOI] [PubMed] [Google Scholar]
- 160.Zhu X., Zhao Y., Hou W., Guo L. MiR-153 regulates cardiomyocyte apoptosis by targeting Nrf2/HO-1 signaling. Chromosome Res. 2019;27:167–178. doi: 10.1007/s10577-019-09608-y. [DOI] [PubMed] [Google Scholar]
- 161.Li F., Liang J., Tong H., Zhu S., Tang D. Inhibition of microRNA-199a-5p ameliorates oxygen-glucose deprivation/reoxygenation-induced apoptosis and oxidative stress in HT22 neurons by targeting Brg1 to activate Nrf2/HO-1 signalling. Clin. Exp. Pharmacol. Physiol. 2020 doi: 10.1111/1440-1681.13265. [DOI] [PubMed] [Google Scholar]
- 162.Deng Y., Ma G., Dong Q., Sun X., Liu L., Miao Z., Gao F. Overexpression of miR-224-3p alleviates apoptosis from cerebral ischemia reperfusion injury by targeting FIP200. J. Cell. Biochem. 2019;120:17151–17158. doi: 10.1002/jcb.28975. [DOI] [PubMed] [Google Scholar]
- 163.Wang J., Wang A., He H., She X., He Y., Li S., Liu L., Luo T., Huang N., Luo H., Zou K. Trametenolic acid B protects against cerebral ischemia and reperfusion injury through modulation of microRNA-10a and PI3K/Akt/mTOR signaling pathways. Biomed. Pharmacother. 2019;112:108692. doi: 10.1016/j.biopha.2019.108692. [DOI] [PubMed] [Google Scholar]
- 164.Li P., Teng F., Gao F., Zhang M., Wu J., Zhang C. Identification of circulating MicroRNAs as potential biomarkers for detecting acute ischemic stroke. Cell. Mol. Neurobiol. 2015;35:433–447. doi: 10.1007/s10571-014-0139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ozcan G., Ozpolat B., Coleman R.L., Sood A.K., Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug Deliv. Rev. 2015;87:108–119. doi: 10.1016/J.ADDR.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.ClinicalTrials.gov. National Library of Medicine (U.S.). (December 14, 2017- May 24, 2019 ). A study of RG-012 in subjects with Alport syndrome. Identifier NCT03373786. Retrieved from https://clinicaltrials.gov/ct2/show/NCT03373786, (n.d.).
- 167.Hanna J., Hossain G.S., Kocerha J. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019;10 doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jiang L., Zhu* J. Review of MiRNA-disease association prediction. Curr. Protein Pept. Sci. 2020;21:1–10. doi: 10.2174/1389203721666200210102751. [DOI] [PubMed] [Google Scholar]
- 169.Meseguer S., Martínez-Zamora A., García-Arumí E., Andreu A.L., Armengod M.-E. The ROS-sensitive microRNA-9/9* controls the expression of mitochondrial tRNA-modifying enzymes and is involved in the molecular mechanism of MELAS syndrome. Hum. Mol. Genet. 2014;24:167–184. doi: 10.1093/hmg/ddu427. [DOI] [PubMed] [Google Scholar]
- 170.Alhasson F., Seth R.K., Sarkar S., Kimono D.A., Albadrani M.S., Dattaroy D., Chandrashekaran V., Scott G.I., Raychoudhury S., Nagarkatti M., Nagarkatti P., Diehl A.M., Chatterjee S. High circulatory leptin mediated NOX-2-peroxynitrite-miR21 axis activate mesangial cells and promotes renal inflammatory pathology in nonalcoholic fatty liver disease. Redox Biol. 2018;17:1–15. doi: 10.1016/J.REDOX.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Onodera Y., Teramura T., Takehara T., Obora K., Mori T., Fukuda K. miR-155 induces ROS generation through downregulation of antioxidation-related genes in mesenchymal stem cells. Aging Cell. 2017;16:1369–1380. doi: 10.1111/acel.12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ranjan R., Lee Y.G., Karpurapu M., Syed M.A., Chung S., Deng J., Jeong J.J., Zhao G., Xiao L., Sadikot R.T., Weiss M.J., Christman J.W., Park G.Y. p47phox and reactive oxygen species production modulate expression of microRNA-451 in macrophages. Free Radic. Res. 2015;49:25–34. doi: 10.3109/10715762.2014.974037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kim J.H., Lee K.S., Lee D.K., Kim J., Kwak S.N., Ha K.S., Choe J., Won M.H., Cho B.R., Jeoung D., Lee H., Kwon Y.G., Kim Y.M. Hypoxia-responsive MicroRNA-101 promotes angiogenesis via heme oxygenase-1/vascular endothelial growth factor axis by targeting cullin 3. Antioxidants Redox Signal. 2014;21:2469–2482. doi: 10.1089/ars.2014.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhou C., Zhao L., Zheng J., Wang K., Deng H., Liu P., Chen L., Mu H. MicroRNA-144 modulates oxidative stress tolerance in SH-SY5Y cells by regulating nuclear factor erythroid 2-related factor 2-glutathione axis. Neurosci. Lett. 2017;655:21–27. doi: 10.1016/j.neulet.2017.06.045. [DOI] [PubMed] [Google Scholar]
- 175.Kukoyi A.T., Fan X., Staitieh B.S., Hybertson B.M., Gao B., McCord J.M., Guidot D.M. MiR-144 mediates Nrf2 inhibition and alveolar epithelial dysfunction in HIV-1 transgenic rats. Am. J. Physiol. Cell Physiol. 2019;317:C390–C397. doi: 10.1152/ajpcell.00038.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhao X.J., Yu H.W., Yang Y.Z., Wu W.Y., Chen T.Y., Jia K.K., Kang L.L., Jiao R.Q., Kong L.D. Polydatin prevents fructose-induced liver inflammation and lipid deposition through increasing miR-200a to regulate Keap1/Nrf2 pathway. Redox Biol. 2018;18:124–137. doi: 10.1016/j.redox.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chen H., Li X., Li W., Zheng H. miR-130a can predict response to temozolomide in patients with glioblastoma multiforme, independently of O6-methylguanine-DNA methyltransferase. J. Transl. Med. 2015;13:1. doi: 10.1186/s12967-015-0435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tinaburri L., D'Errico M., Sileno S., Maurelli R., Degan P., Magenta A., Dellambra E., Karihtala P. MIR-200a modulates the expression of the DNA repair protein OGG1 playing a role in aging of primary human keratinocytes. Oxid. Med. Cell. Longev. 2018;2018 doi: 10.1155/2018/9147326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sun G., Lu Y., Li Y., Mao J., Zhang J., Jin Y., Li Y., Sun Y., Liu L., Li L. MiR-19a protects cardiomyocytes from hypoxia/reoxygenation-induced apoptosis via PTEN/PI3K/p-Akt pathway. Biosci. Rep. 2017;37:1–11. doi: 10.1042/BSR20170899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang L., Ren Y., Pan W., Yu Z., Tong L., Li N., Tang B. Fluorescent nanocomposite for visualizing cross-talk between microRNA-21 and hydrogen peroxide in ischemia-reperfusion injury in live cells and in vivo. Anal. Chem. 2016;88:11886–11891. doi: 10.1021/acs.analchem.6b03701. [DOI] [PubMed] [Google Scholar]
- 181.Zhang S., Zhao Y. Lentinan protects cardiomyocytes against hypoxia-induced injury by regulation of microRNA-22/Sirt1, Artif. Cells. Nanomed. Biotechnol. 2018;47:3938–3946. doi: 10.1080/21691401.2019.1666863. [DOI] [PubMed] [Google Scholar]
- 182.Du J.K., Cong B.H., Yu Q., Wang H., Wang L., Wang C.N., Tang X.L., Lu J.Q., Zhu X.Y., Ni X. Upregulation of microRNA-22 contributes to myocardial ischemia-reperfusion injury by interfering with the mitochondrial function. Free Radic. Biol. Med. 2016;96:406–417. doi: 10.1016/j.freeradbiomed.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 183.Fu B.C., Lang J.L., Zhang D.Y., Sun L., Chen W., Liu W., Liu K.Y., Ma C.Y., Jiang S.L., Li R.K., Tian H. Suppression of miR-34a expression in the myocardium protects against ischemia-reperfusion injury through SIRT1 protective pathway. Stem Cell. Dev. 2017;26:1270–1282. doi: 10.1089/scd.2017.0062. [DOI] [PubMed] [Google Scholar]
- 184.Ke Z.P., Xu P., Shi Y., Gao A.M. MicroRNA-93 inhibits ischemia-reperfusion induced cardiomyocyte apoptosis by targeting PTEN. Oncotarget. 2016;7:28796–28805. doi: 10.18632/oncotarget.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jiang B., Liu Y., Liang P., Li Y., Liu Z., Tong Z., Lv Q., Liu M., Xiao X. MicroRNA-126a-5p enhances myocardial ischemia-reperfusion injury through suppressing Hspb8 expression. Oncotarget. 2017;8:94172–94187. doi: 10.18632/oncotarget.21613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang H., Zhang X., Zhang J. MiR-129-5p inhibits autophagy and apoptosis of H9c2 cells induced by hydrogen peroxide via the PI3K/AKT/mTOR signaling pathway by targeting ATG14. Biochem. Biophys. Res. Commun. 2018;506:272–277. doi: 10.1016/j.bbrc.2018.10.085. [DOI] [PubMed] [Google Scholar]
- 187.Qin Q., Cui L., Zhou Z., Zhang Z., Wang Y., Zhou C. Inhibition of microRNA-141-3p reduces hypoxia-induced apoptosis in H9c2 rat cardiomyocytes by activating the RP105-Dependent PI3K/AKT signaling pathway. Med. Sci. Monit. 2019;25:7016–7025. doi: 10.12659/MSM.916361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhao Z., Qu F., Liu R., Xia Y. Differential expression of miR-142-3p protects cardiomyocytes from myocardial ischemia-reperfusion via TLR4/NFkB axis. J. Cell. Biochem. 2019:1–12. doi: 10.1002/jcb.29506. [DOI] [PubMed] [Google Scholar]
- 189.E L., Jiang H., Lu Z. MicroRNA-144 attenuates cardiac ischemia/reperfusion injury by targeting FOXO1. Exp. Ther. Med. 2019:2152–2160. doi: 10.3892/etm.2019.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yuan L., Fan L., Li Q., Cui W., Wang X., Zhang Z. Inhibition of miR-181b-5p protects cardiomyocytes against ischemia/reperfusion injury by targeting AKT3 and PI3KR3. J. Cell. Biochem. 2019;120:19647–19659. doi: 10.1002/jcb.29271. [DOI] [PubMed] [Google Scholar]
- 191.Ge L., Cai Y., Ying F., Liu H., Zhang D., He Y., Pang L., Yan D., Xu A., Ma H., Xia Z. MiR-181c-5p exacerbates hypoxia/reoxygenation-induced cardiomyocyte apoptosis via targeting PTPN4. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/1957920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu C., Zheng H., Xie L., Zhang J. Decreased miR-208 induced ischemia myocardial and reperfusion injury by targeting p21. Pharmazie. 2016;71:719–723. doi: 10.1691/ph.2016.6740. [DOI] [PubMed] [Google Scholar]
- 193.Meng Q., Huo X., Sun H., Wang H., Luan Z., Wang S. miR-223 regulates myocardial ischemia-reperfusion damage via targeting NLRP3 in vitro and in vivo. Int. J. Clin. Exp. Med. 2018;11:2004–2013. [Google Scholar]
- 194.Zhang X., Zhang C., Wang N., Li Y., Zhang D., Li Q. MicroRNA-486 alleviates hypoxia-induced damage in H9c2 cells by targeting NDRG2 to inactivate JNK/C-Jun and NF-κB signaling pathways. Cell. Physiol. Biochem. 2018;48:2483–2492. doi: 10.1159/000492686. [DOI] [PubMed] [Google Scholar]
- 195.Zhao D., Zheng H., Greasley A., Ling F., Zhou Q., Wang B., Ni T., Topiwala I., Zhu C., Mele T., Liu K., Zheng X. The role of miR-711 in cardiac cells in response to oxidative stress and its biogenesis: a study on H9C2 cells. Cell. Mol. Biol. Lett. 2020;25:26. doi: 10.1186/s11658-020-00206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]