Abstract
Compared to their linear counterparts, cyclic peptides show better biological activities, such as antibacterial, immunosuppressive, and anti-tumor activities, and pharmaceutical properties due to their conformational rigidity. However, cyclic peptides could form numerous putative metabolites from potential hydrolytic cleavages and their fragments are very difficult to interpret. These characteristics pose a great challenge when analyzing metabolites of cyclic peptides by mass spectrometry. This study was to assess and apply a software-aided analytical workflow for the detection and structural characterization of cyclic peptide metabolites. Insulin and atrial natriuretic peptide (ANP) as model cyclic peptides were incubated with trypsin/chymotrypsin and/or rat liver S9, followed by data acquisition using TripleTOF® 5600. Resultant full-scan MS and MS/MS datasets were automatically processed through a combination of targeted and untargeted peak finding strategies. MS/MS spectra of predicted metabolites were interrogated against putative metabolite sequences, in light of a, b, y and internal fragment series. The resulting fragment assignments led to the confirmation and ranking of the metabolite sequences and identification of metabolic modification. As a result, 29 metabolites with linear or cyclic structures were detected in the insulin incubation with the hydrolytic enzymes. Sequences of twenty insulin metabolites were further determined, which were consistent with the hydrolytic sites of these enzymes. In the same manner, multiple metabolites of insulin and ANP formed in rat liver S9 incubation were detected and structurally characterized, some of which have not been previously reported. The results demonstrated the utility of software-aided data processing tool in detection and identification of cyclic peptide metabolites.
Keywords: Atrial natriuretic peptide, Metabolism of cyclic peptide, High resolution mass spectrometry, Insulin, Software-aided data processing
Graphical abstract
Highlights
-
•
A software-aided workflow enabling detection and characterization of cyclic peptide metabolites by LC/HRMS.
-
•
Automatically data processing through a combination of targeted and untargeted peak finding strategies.
-
•
MS/MS spectra of predicted metabolites interrogated against putative metabolite sequences.
-
•
Rapidly determining metabolite profiles of insulin and atrial natriuretic peptide in rat liver S9.
-
•
Potentially applicable to metabolic soft spot analysis and in vitro metabolism across species in drug discovery.
1. Introduction
Cyclic peptides are a class of peptides containing cyclic ring structure, which can be formed by folding the peptide chain with an amide bond, or other chemically stable bonds such as lactone, ether, thioether, disulfide bond [1,2]. In the past decades, several cyclic peptide drugs have been developed for clinical therapy [3], like cyclosporine A, gramicidin-S, vasopressin, oxytocin, vancomycin, and insulin [[4], [5], [6], [7], [8]]. As a feature in these therapeutic compounds, peptide cyclization can improve the potency [9,10] and proteolysis stability of peptides [11,12], as well as pharmacokinetic property and intracellular activity such as membrane permeability [13]. Apart from the advantageous conformational rigidity, the special structures of cyclic peptides also lead to great challenge for the detection and identification of cyclic peptide metabolites with mass spectrometry (MS). Firstly, the flexible starting point as well as the stochastic fragment lengths of a cyclic peptide would derive numerous possibilities of generating metabolites via peptide hydrolysis. For example, based on simulation, insulin could generate over 46000 metabolites via hydrolysis (Fig. S1) and each of these metabolites could generate multiple molecular ions of different charge states. These vast amounts of potential metabolites make it impossible to rely on manpower to search for predicted metabolites of cyclic peptides. Furthermore, for the linear peptides, the fragmentation under gas phase collision induced dissociation (CID) is well understood [14,15]. CID, electron transfer dissociation (ETD) and electron capture dissociation (ECD) are the regular ways to produce b/y, a/x, c/z fragment ions from linear peptides [16,17]. As the most commonly used activation technique in tandem mass spectrometry, CID produces a series of b/y ions, which are widely used in peptide sequencing and proteomics study [[18], [19], [20], [21]]. Many software and databases are capable of efficiently determining sequences and structures of linear peptides [[22], [23], [24]]. However, for the cyclic peptides, the C-terminus and N-terminus may not be present due to the complex cyclization types. In addition, cyclic peptides with disulfide bond [25,26] or other linkage structures [27] will resist CID fragmentation at lower collisional energy, while in high-collision energy condition they generate only nonspecific small immonium ions that are not suitable for spectral interpretation. Thus, the in-silico tools developed for the analysis of linear peptide sequences and modifications in proteomics studies are not useful for the assignment of sequence and modification sites of cyclic peptide metabolites [[28], [29], [30], [31], [32]].
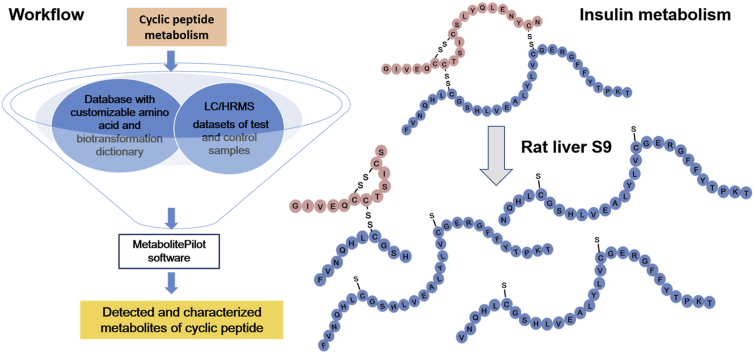
In this study, we evaluated and applied a recently implemented MetabolitePilot Software for the automatic detection and structure characterization of cyclic peptide metabolites. Insulin and atrial natriuretic peptide (ANP), which are biologically active cyclic peptides formed with three and one disulfide bonds, respectively (Fig. 1) [33,34], were selected as model cyclic peptides. Like LC/MS analysis of cyclic peptides with a variety of linkage structures, studying metabolism of both insulin and ANP faced the same challenges: enormous potential metabolites could be formed via peptide bond hydrolysis and product ion spectra are very difficult to interpret. The first experiment was to detect and structurally characterize metabolites formed in the incubation of insulin with a combination of trypsin and chymotrypsin [35,36]. Since peptide hydrolytic sites by these enzymes are known and metabolites from the incubations are predicable, results from this experiment can allow us to evaluate the effectiveness of the data processing workflow (Fig. 2) in studying metabolism of cyclic peptides in vitro. The second experiment was to investigate unknown metabolites of insulin and ANP formed in incubations with rat liver S9 that have a variety of peptide hydrolytic enzymes. Results from this study demonstrated that the novel data processing workflow was able to rapidly detect and characterize metabolites of cyclic peptides formed in biological matrix.
Fig. 1.
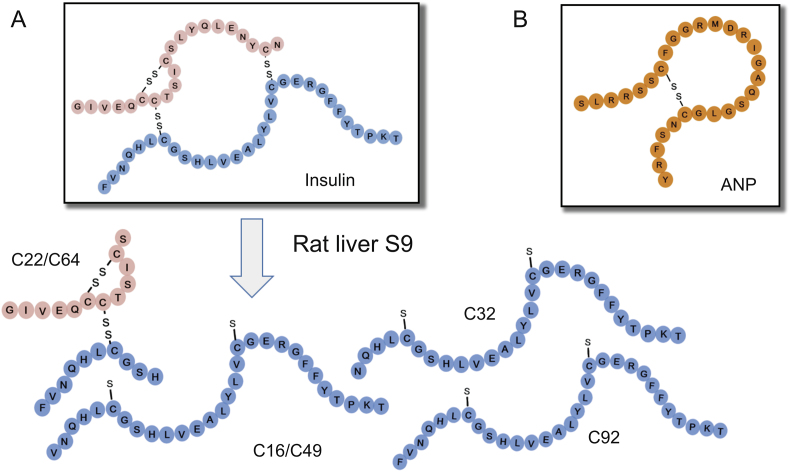
(A) Structures of insulin and its metabolites formed in liver S9 incubation. (B) Structures of ANP.
Fig. 2.
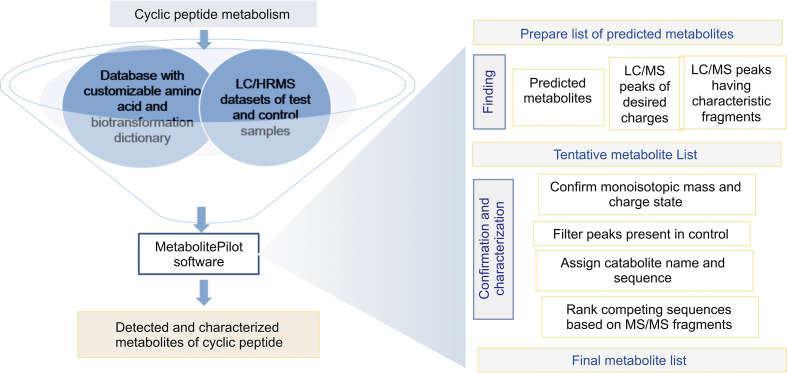
Workflow for detection, confirmation and identification of cyclic peptides metabolites using a newly developed software-aided data processing tool.
2. Experimental section
2.1. Chemicals and reagents
Human insulin and ANP (Fig. 1) were purchased from Sigma-Aldrich (Burlington, MA). Pooled rat liver S9 was obtained from Sekisui XenoTech, LLC (Kansas City, KS, USA). Trypsin and chymotrypsin were purchased from Sigma-Aldrich (Burlington, MA, USA). Ammonium bicarbonate and 0.1 M HCl were from Sigma-Aldrich (Burlington, MA, USA). Acetonitrile (ACN) methanol and water of LC-MS grade were from Merck (Kenilworth, NJ, USA). Ultrapure water was freshly prepared with Millipore purification system (Massachusetts, USA).
2.2. Enzymatic digestion of insulin
The enzymatic digestion of insulin was carried out in 200 μL of 50 mM ammonium bicarbonate (pH 7.4). Insulin was dissolved in 50 mM ammonium bicarbonate with droplet adding 0.1 M HCl until completely dissolved. In the final system, 20 μM of insulin was incubated with trypsin and chymotrypsin (5 μg/mL) under 37 °C for 0, 1, 2 and 3 h. After incubation, 500 μL of ACN was added to quench the reaction and centrifuged at 21,000 g for 10 min. The supernatant was collected and dried down under a gentle stream of N2 gas. The samples were reconstituted in LC/MS grade water (100 μL) for further liquid chromatography high resolution mass spectrometry (LC-HRMS) analysis.
2.3. Metabolism of insulin and ANP in liver S9
Insulin and ANP were incubated with rat liver S9 respectively in 200 μL of 50 mM ammonium bicarbonate (pH 7.4) for 0 and 3 h. Rat liver S9 was added prior to the addition of insulin or ANP, and pre-incubated on ice for 5 min. The final enzymatic system contained 1 mg/mL of rat liver S9 and 20 mM of insulin or ANP. After incubation, 500 μL of ACN was added to quench the reaction and centrifuged at 21,000 g for 10 min. The supernatant was collected and dried down under a gentle stream of N2 gas. The samples were reconstituted in LC/MS grade water (100 μL) for further LC-HRMS analysis.
2.4. Data acquisition for metabolites of insulin and ANP
An Agilent 1290 Infinity II LC system (Agilent Technologies, Santa Clara, US) was connected to a TripleTOF® 5600 mass spectrometer (SCIEX, Framingham, MA) for all LC-MS analysis. Mobile phase A was H2O with 0.1% formic acid and mobile phase B was acetonitrile with 0.1% formic acid. 10 μL of sample was injected onto a C18 column (Waters Acquity UPLC, BEH C18; 2.1 mm × 100 mm, 1.7 μm) for each run at a flow rate of 400 μL/min. The chromatography commenced at a solvent composition of 2% B and 98% A for 2 min, and then increased to 45% B at 45 min, and reached 90% B at 45.1 min and held until 47 min. Thereafter, the column was re-equilibrated back to the starting solvent conditions of 2% B and 98% A at 47.1 min, and held to the end of the gradient (54 min).
To maximize the information acquired on the mass spectrometer for each sample, a full MS scan (m/z 300–2000) was acquired followed by top 20 information dependent acquisition (IDA) MS/MS scans (m/z 100–1600) in positive ion mode. The parameters for curtain plate (CUR), declustering potential (DP), collision energy (CE); ionspray voltage IS, Gas1, Gas2 in full MS scan mode was 30psi, 80V, 10V, 5500V, 55 psi, and 55 psi. Source temperature was set to 450 °C and tray temperature was set to 22 °C. The criteria for the IDA precursor selection were as follows: top 20 most intense peaks with charge states from 2 to 5 and intensities greater than 50 were selected. Previous candidates within the mass tolerance of 50 mDa were excluded for the duration of 3 s after 1 occurrence. Dynamic background subtraction was activated. Rolling collision energy for multiply charged peptides was enabled. Divert Valco valve was used to switch LC flow to MS between 2 and 50 min.
2.5. Data processing with MetabolitePilot™ software
The liquid chromatography/high resolution mass spectrometry (LC/HRMS) data were processed with MetabolitePilot Software 2.0; this tool facilitates automated LC/MS data processing for the characterization of therapeutic peptides, including non-linear, crosslinked and cyclic structures. This software could also deal with non-natural amino acids and modifications, targeted searching of predicted hydrolytic cleavages, calculating and assigning a-, b-, y- and internal fragments for linear and non-linear peptides. The strategies used for finding the peptide-related material were as follows: peak finding in accurate extracted ion chromatograms of hypothetical catabolites, generic LC/MS peak finding followed with charge filter that removed singly-charged peaks, and finding peaks that yielded characteristic accurate mass fragments in MS/MS data. In order to remove false positive measurements, the peak finding was followed by comparison of the data against that of a control sample; only peaks that were either absent or significantly smaller (0.5 or less) in the control sample were kept. The LC/MS peaks were matched with putative peptide catabolite names based on mass tolerance of 10 ppm and TOF isotope pattern agreement within 20%. MS and MS/MS spectra as well as metabolite chromatographic traces were saved with the peak finding results. The sequences of putative catabolites were confirmed by MS/MS annotation using theoretical a,b,y, y|a and y|b fragments and an mass tolerance of 5 ppm.
3. Results and discussion
3.1. Workflow for detection and characterization of cyclic peptides metabolites
The high-level workflow for detection and characterization of metabolites of cyclic peptides using LC/HRMS data processing software tool is shown in Fig. 2. The input for the software processing comprises the LC/HRMS data, preferentially a test sample and a control sample, and the processing instructions. The processing method combines the information regarding the peak finding strategies and settings, the studied cyclic peptide sequence in combination with the amino acid and biotransformation dictionary, and the details on potential metabolites to be considered in the target search. Since for larger cyclic peptides, the monoisotopic mass is not the most intense peak in the isotope cluster, the target extracted ion chromatography (XIC)-based search in the MetabolitePilot™ Software uses the mass to charge of the most intense peak in the cluster for the accurate ion chromatogram extraction. If the MS/MS spectrum of the studied peptide is available, it can be loaded to the method and used in untargeted peak finding strategies, such as a search for characteristic fragments.
The actual data processing has a few parts: first, all LC/MS peaks found by any chosen strategy are merged. Then the unique peaks are confirmed; peaks outside processing settings and isotope peaks leading to duplicate entries are removed. Confirmed peaks are then potentially assigned names and putative sequences, based on accurate mass. If MS/MS data are available, the sequence assignments are confirmed or ranked. In case of peptide biotransformation that can be located on multiple amino-acid residues, the interpretation considers all of these possibilities, and ranks the putative metabolite sequences based on the completeness for MS/MS peptide fragment annotation.
3.2. Metabolite identification of insulin incubated with hydrolytic enzymes
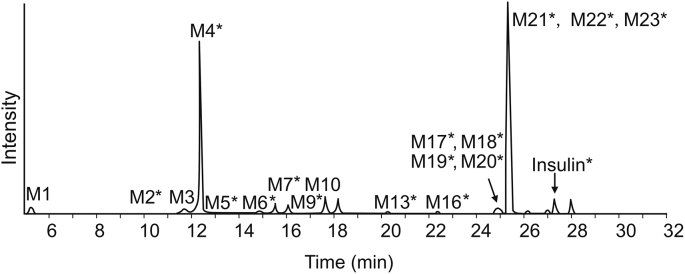
The software-aided workflow was applied to the detection and characterization of the insulin metabolites formed in the incubation with a combination of trypsin and chymotrypsin, which targeted at peptide bond between lysine and arginine, and peptide bonds with aromatic amino acids, such as tyrosine, phenylalanine, and tryptophan, respectively. As a result, 29 insulin metabolites with cyclic or linear structures were directly detected and characterized without reducing disulfide bond (Table 1). The structures of these insulin metabolites are consistent with hydrolytic sites of trypsin and chymotrypsin, which validated the effectiveness of this software-aided approach in studying biotransformation of cyclic peptides in vitro. These metabolites were initially found using multiple detection mechanisms described and further confirmed based on their MS/MS spectral data (Fig. 2). In addition, scoring and ranking of putative amino-acid sequences pointed to predicted insulin digest products. The extracted ion chromatograms of these metabolites shown in Fig. 3 indicated the relative intensities of the insulin metabolites. The accurate full-scan MS and MS/MS spectra of M4, a representative metabolite of insulin, are shown in Fig. 4. The charge state was assigned based on the isotope cluster in TOF MS, and the structure of metabolite was confirmed based on the exact masses of protonated molecule ions (Fig. 4A) product ions (Fig. 4B).
Table 1.
Insulin metabolites detected and characterized in enzymatic incubation.
| ID | Name | Neutral mass | m/z | Charge | RT (min) | Peak area | MS/MS assigned | Sequence type |
|---|---|---|---|---|---|---|---|---|
| P | Parent [M+5H]5+ | 5803.62 | 1161.7320 | 5 | 27.3 | 5.96E+05 | ✓ | C |
| M1 | YTPKT | 608.32 | 305.1688 | 2 | 5.4 | 4.26E+05 | ✓ | L |
| M2 | QLENYC [∗3]N/VC [∗3]GER | 1442.60 | 722.3065 | 2 | 9.8 | 1.71E+04 | LL | |
| M3 | FYTPKT | 755.39 | 378.7036 | 3 | 11.8 | 3.21E+05 | ✓ | L |
| M4 | QLENYC [∗3]N/LVC [∗3]GER | 1555.69 | 519.5699 | 3 | 12.4 | 6.45E+06 | ✓ | LL |
| M5 | GIVEQC [∗1]CT/IC [∗1]SLYQ + Loss of Water | 1556.66 | 779.3393 | 2 | 12.9 | 5.19E+04 | OR | |
| M6 | QLENYC [∗3]N/YLVC [∗3]GER | 1718.75 | 573.9243 | 3 | 14.9 | 1.62E+05 | ✓ | LL |
| M7 | QLENYC [∗3]N/LVC [∗3]GER + Loss of Water | 1537.67 | 769.8426 | 2 | 15.5 | 5.29E+05 | ✓ | LL |
| M8 | NYC [∗3]N/LVC [∗3]GERGFF + Hydrogenation | 1538.66 | 770.3349 | 2 | 16.1 | 4.92E+05 | ✓ | LL |
| M9 | QLENYC [∗3]N/LVC [∗3]GERGF | 1759.77 | 880.8916 | 2 | 16.4 | 5.28E+04 | LL | |
| M10 | GFFYTPK | 858.43 | 430.2247 | 2 | 17.7 | 9.40E+05 | ✓ | L |
| M11 | GFFYTPKT | 959.48 | 480.7492 | 2 | 18.2 | 8.15E+05 | ✓ | L |
| M12 | SLYQLENYC [∗3]N/LYLVC [∗3]G | 1909.86 | 637.6257 | 3 | 19.4 | 2.54E+04 | ✓ | LL |
| M13 | QLENYC [∗3]N/LVC [∗3]GERGFF | 1906.84 | 954.4265 | 2 | 20.3 | 8.16E+04 | ✓ | LL |
| M14 | GIVEQC [∗1]C [∗2]TSIC [∗1]SL/LC [∗2]GSHLVE + Loss of Water | 2188.97 | 730.6647 | 3 | 21.0 | 2.27E+04 | ✓ | RLL |
| M15 | SLYQLENYC [∗3]N/LYLVC [∗3]GERGFFY | 2709.24 | 678.3180 | 4 | 21.2 | 1.46E+04 | ✓ | LL |
| M16 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLY/FVNQHLC [∗2]GSHL | 2767.23 | 692.8140 | 4 | 22.4 | 5.88E+04 | ✓ | RLL |
| M17 | GIVEQC [∗1]C [∗2]TSIC [∗1]SL/FVNQHLC [∗2]GSHL | 2604.14 | 869.0542 | 3 | 24.9 | 2.47E+05 | ✓ | RLL |
| M18 | GIVEQC [∗1]C [∗2]TSI/C [∗1]SLYQLENYC [∗3]N/FVNQHLC [∗2]GSHLVEALYLVC [∗3]GER | 4880.18 | 814.3705 | 6 | 25.0 | 2.55E+04 | ✓ | C |
| M19 | GIVEQC [∗1]C [∗2]TSIC [∗1]S/FVNQHLC [∗2]GSHLVEALYL | 3179.46 | 795.8729 | 4 | 25.0 | 1.76E+05 | ✓ | RLL |
| M20 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLY/HLC [∗2]GSHLVEALY | 2854.28 | 952.4339 | 3 | 25.1 | 2.58E+04 | RLL | |
| M21 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLY/FVNQHLC [∗2]GSHLVEALY | 3342.53 | 836.6388 | 4 | 25.4 | 1.07E+07 | ✓ | RLL |
| M22 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLY/LC [∗2]GSHLVEALY | 2717.23 | 906.7501 | 3 | 25.6 | 2.72E+04 | ✓ | RLL |
| M23 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLY/FVNQHLC [∗2]GSHLVEALY | 3342.52 | 836.6369 | 4 | 26.0 | 3.79E+04 | ✓ | RLL |
| M24 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLYQLENYC [∗3]N/FVNQHLC [∗2]GSHLVEALYLVC [∗3]GER | 4862.17 | 973.4418 | 5 | 26.2 | 7.62E+04 | C | |
| M25 | CSLYQLENYC [∗3]N/GSHLVEALYLVC [∗3]GERGFFY | 3505.58 | 1169.5354 | 3 | 26.2 | 4.44E+04 | LL | |
| M26 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLY/FVNQHLC [∗2]GSHLVEALY | 3342.52 | 836.6381 | 4 | 27.0 | 1.82E+05 | RLL | |
| M27 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLY/FVNQHLC [∗2]GSHLVEALYL | 3455.60 | 864.9078 | 4 | 27.4 | 1.18E+05 | RLL | |
| M28 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLYQLENYC [∗3]N/FVNQHLC [∗2]GSHLVEALYLVC [∗3]GERGFF | 5213.33 | 1043.6723 | 5 | 28.0 | 2.32E+05 | C | |
| M29 | GIVEQC [∗1]C [∗2]TSIC [∗1]SLYQLENYC [∗3]N/FVNQHLC [∗2]GSHLVEALYLVC [∗3]GERGFFY | 5376.40 | 1076.2866 | 5 | 28.0 | 2.75E+05 | C |
Insulin metabolite amino-acid sequences are outlined in column Name. In the sequence representation, the chains are separated by “/” and the cysteine di-sulfide bonds are represented by shared indices in cysteine modification suffix “[∗ ]”. Sequence type column indicates type of metabolite peptide sequence: cyclic (C), linear (L), linked linear (LL), open ring (OR), linked linear with ring (RLL).
Fig. 3.
Metabolite profile of insulin incubated with hydrolytic enzymes (1 h); ∗represents the metabolite containing disulfide bond.
Fig. 4.
Full-scan MS (A) and MS/MS (B) spectra of insulin metabolite M4 from enzymatic incubation.
3.3. Metabolite identification of insulin and ANP formed in incubations with rat liver S9
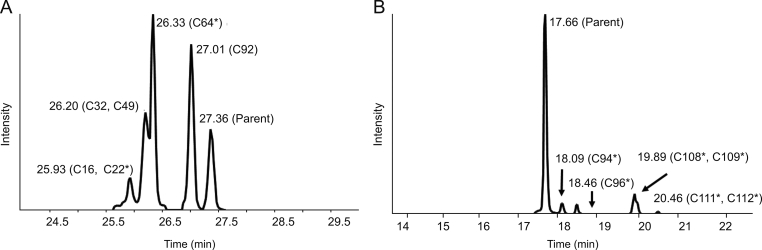
Insulin and ANP were further incubated with rat liver S9, which contained a variety of peptide hydrolases, followed by direct generation of accurate mass full-scan MS and MS/MS datasets. Major metabolites of insulin and ANP are characterized and listed in Table 2 and Table 3, some of which have not been reported in the literature. The MS responses of the metabolites relative to the parent drug increased with the incubation time. The extracted ion chromatograms of insulin and ANP metabolites are illustrated in Fig. 5. The structures of the six major insulin metabolites formed in rat liver S9 are displayed in Fig. 1. The mass spectra and proposed structures of C92 and C108, the most abundant metabolites of ANP in liver S9 incubation, are depicted in Fig. S2 and Fig. S3.
Table 2.
Time dependent metabolism of insulin in rat liver S9.
| ID | Name | m/z | RT(min) | MS area |
|||
|---|---|---|---|---|---|---|---|
| T0 | T0.5h | T1.5h | T3h | ||||
| Insulin | Parent [M+5H]5+ | 1161.737 | 27.24 | 3.50E06 | 2.08E06 | 1.20E06 | 6.83E05 |
| C16 | VNQHLCGSHLVEALYLVCGERGFFYTPKT + Desaturation [M+4H]4+ | 820.6582 | 25.9 | ND | 1.03E05 | 2.77E05 | 2.38E05 |
| C22 | GIVEQC [∗1]C [∗2]TSIC [∗1]S/FVNQHLC [∗2]GSH [M+2H]2+ | 1189.9946 | 25.92 | 4.02E04 | 4.45E+05 | 5.49E05 | 4.57E05 |
| C32 | NQHLCGSHLVEALYLVCGERGFFYTPKT + Desaturation [M+4H]4+ | 795.8898 | 26.17 | ND | 2.68E04 | 1.66E05 | 3.10E05 |
| C49 | VNQHLCGSHLVEALYLVCGERGFFYTPKT + Desaturation [M+3H]3+ | 1093.8723 | 26.22 | ND | 2.03E05 | 5.67E05 | 6.60E05 |
| C64 | GIVEQC [∗1]C [∗2]TSIC [∗1]S/FVNQHLC [∗2]GSH [M+2H]2+ | 1189.9919 | 26.33 | 8.67E04 | 9.42E05 | 1.31E06 | 1.27E06 |
| C92 | FVNQHLCGSHLVEALYLVCGERGFFYTPKT + Desaturation [M+4H]4+ | 857.4257 | 26.96 | 4.00E05 | 2.73E06 | 2.29E06 | 1.35E06 |
As shown in Fig. 1, C22 and C64 were a pair of isomers. C16 and C49 were another pair of isomes.
ND: Not detected.
Table 3.
Time dependent metabolism of ANP in rat liver S9.
| ID | Name | m/z | RT (min) | MS area |
|
|---|---|---|---|---|---|
| T0 | T3h | ||||
| ANP | Parent [M+5H]5+ | 616.6964 | 17.54 | 7.59E06 | 1.70E06 |
| C94 | SLRRSSC [∗1]FGGRMDRIGAQSGLGC [∗1]NSF [M+4H]4+ | 690.8263 | 18.09 | 7.80E04 | 8.13E04 |
| C96 | RSSC [∗1]FGGRMDRIGAQSGLGC [∗1]NSFRY [M+4H]4+ | 681.5643 | 18.41 | 3.52E04 | 7.44E04 |
| C108 | SSC [∗1]FGGRMDRIGAQSGLGC [∗1]NSFRY [M+4H]4+ | 642.5375 | 19.8 | ND | 1.44E05 |
| C109 | SC [∗1]FGGRMDRIGAQSGLGC [∗1]NSFRY [M+4H]4+ | 620.7803 | 19.97 | ND | 7.32E04 |
| C111 | SSC [∗1]FGGRMDRIGAQSGLGC [∗1]NSF [M+3H]3+ | 749.9938 | 20.4 | ND | 9.74E03 |
| C112 | SC [∗1]FGGRMDRIGAQSGLGC [∗1]NSF [M+3H]3+ | 720.9815 | 20.46 | ND | 9.96E03 |
ND: Not detected.
Fig. 5.
Metabolite profile of cyclic peptides incubated in rat liver S9. (A) Insulin; (B) ANP. ∗represents the metabolite containing disulfide bond.
3.4. The software features for the peak finding and confirmation of minor peptide metabolites
As a majority of therapeutic peptides entering clinical development have twenty or more amino acid residues [37], the monoisotopic peak in the theoretical isotope pattern of a typical therapeutic peptide is not the most intense peak; its relative abundance decreases with the increase of size of studied peptides. To support data mining for such larger molecules, one of unique features in MetabolitePilot™ software is to consider the isotopic distribution of the predicted metabolites and use the most intense isotope for LC/MS peak finding in XIC. Once a peak is found in an XIC trace, TOF MS confirmation includes review of the anticipated isotope pattern. In Table 4, peak index column outlines the index of the peptide isotope peak which was used for XIC extraction when finding metabolites. The monoisotopic peak index is 0, and the respective isotope indices are 1, 2, 3, etc. Once a peak is found with the base peak other than 0, the XIC trace of the base peak is provided in the result workspace. Moreover, since series of multiply-charged isotope peaks are selected in the 1 Da isolation window in the first quadrupole of mass spectrometer (Q1) and fragmented in parallel, peptide fragments exhibit isotope patterns and these patterns aid in confident MS/MS annotation. The isotopic signal of fragments improves signal to noise (S/N) of minor multiply charged peptide fragments and enables their contribution to sequence confirmation. For example, the contribution of doubly charged fragments of insulin to the overall assignment would be raised from 3.6% to 6.9% of total MS/MS ion count.
Table 4.
The representative identification of ANP metabolites in rat liver S9 with isotopic MS1 ions.
| Peak ID | Name | m/z | Charge | Peak index | % Score |
|---|---|---|---|---|---|
| 1 | FGGRMD [M+2H]2+ | 341.6519 | 2 | 0 | 57.1 |
| 2 | Loss of 1611.8223 [M+3H]3+ | 489.8823 | 3 | 0 | 0.8 |
| 3 | Loss of 2594.2036 [M+H]+ | 485.2511 | 1 | 0 | 0 |
| 4 | RIGAQSGLGCNS [M+2H]2+ | 581.7858 | 2 | 0 | 52.1 |
| 5 | Loss of 2691.2006 [M+H]+ | 388.2541 | 1 | 0 | 0 |
| 6 | Loss of 2647.1743 [M+H]+ | 432.2804 | 1 | 0 | 0 |
| 7 | Loss of 2603.1491 [M+H]+ | 476.3056 | 1 | 0 | 0 |
| 8 | Loss of 2767.2596 [M+H]+ | 312.1951 | 1 | 0 | 0 |
| 9 | Loss of 2559.1217 [M+H]+ | 520.333 | 1 | 0 | 0 |
| 10 | SLRRSSC [∗1]FGGRMDRIGAQSGLGC [∗1]NS [M+4H]4+ | 654.0593 | 4 | 1 | 54.4 |
| 11 | SLRRSSC [∗1]FGGRMDRIGAQSGLGC [∗1]NS + Loss of Water [M+5H]5+ | 519.8473 | 5 | 1 | 50 |
| 12 | SLRRSSC [∗1]FGGRMDRIGAQSGLGC [∗1]NS [M+5H]5+ | 523.4488 | 5 | 1 | 50.9 |
| 13 | SLRRSSC [∗1]FGGRMDRIGAQSGLGC [∗1]NS [M+3H]3+ | 871.7447 | 3 | 1 | 53 |
| 14 | RSSC [∗1]FGGRMDRIGAQSGLGC [∗1]NS [M+4H]4+ | 565.0045 | 4 | 1 | 48.4 |
| 15 | Loss of 2626.1117 [M+H]+ | 453.343 | 1 | 0 | 0 |
| 16 | SLRRSSC [∗1]FGGRMDRIGAQSGLGC [∗1]NSFR [M+5H]5+ | 584.0816 | 5 | 1 | 45.1 |
| 17 | Loss of 2515.0960 [M+H]+ | 564.3587 | 1 | 0 | 0 |
| 18 | RIGAQSGLGCNSF [M+2H]2+ | 655.3193 | 2 | 0 | 53.4 |
| 19 | Parent [M+5H]5+ | 616.6947 | 5 | 1 | 51 |
3.5. The software features for the structure confirmation of isobaric metabolites and modification site
For metabolite identification, one of challenges is to determine isobaric and isomeric metabolites that pose ambiguity even with high resolution mass spectrometry [38]. For large peptides, hydrolytic cleavages may have identical molecular weight, but their sequences could be different. In Table 5, for an ANP metabolite eluted by 17.01 min, two possible isobaric metabolite sequences (RIGAQSGLGCNSF or IGAQSGLGCNSFR) were proposed based on TOF MS data interpretation. For further sequence identification, MetabolitePilot™ software enabled the MS/MS information to match characterized fragments in the spectrum with predicted theoretical ones. Therefore, based on more assigned fragments (provided in Table S1), RIGAQSGLGCNSF was claimed as a “Winner” metabolite sequence, with 23.8% of total ion count that could be directly assigned to sequence fragments.
Table 5.
The sequence identification of an ANP metabolite M4 based on fragments assignment.
| Auto-generated | Rank | MSMS peak area assigned (%) | Proposed sequences | AA index | Apply to results | Fragments assigned |
|---|---|---|---|---|---|---|
| TRUE | 1 | 23.80 | RIGAQSGLGCNSF | AA (10–22) | TRUE | 9 |
| TRUE | 2 | 4.80 | IGAQSGLGCNSFR | AA (11–23) | FALSE | 3 |
The MS/MS data also aid in the characterization of cyclic peptide modifications. One of common approaches to enhancing the stability of therapeutic peptides is the structural modification; considering potential modifications and their sites will increase the number of potential metabolites to be searched for. For instance, MetabolitePilot™ Software proposed five potential metabolite sequences listed in Table 6, and four of them were linear peptides with serine amino acid residue replaced with oxoalanine at various locations. By utilizing the MS/MS spectrum information, metabolite sequence SSC [∗1]FGGRMDRIGAQSGLGC [∗1]NSFRY, having a disulfide bond between cysteines at positions 3 and 19, was selected with 37 fragments matched.
Table 6.
The characterization of ANP metabolite C108 (Table 3) based on fragment assignments to isomeric linear and non-linear sequences.
| Auto-generated | Rank | TIC intensity assigned | MSMS peak area assigned (%) | Proposed sequences | Fragments assigned |
|---|---|---|---|---|---|
| TRUE | 1 | 483.6 | 7.20 | SSC [∗1]FGGRMDRIGAQSGLGC [∗1]NSFRY | 37 |
| TRUE | 2 | 403.6 | 6.00 | SSCFGGRMDRIGAQS [SOa]GLGCNSFRY | 19 |
| TRUE | 2 | 403.6 | 6.00 | SSCFGGRMDRIGAQSGLGCNS [SOa]FRY | 19 |
| TRUE | 3 | 356 | 5.30 | S [SOa]SCFGGRMDRIGAQSGLGCNSFRY | 18 |
| TRUE | 3 | 356 | 5.30 | SS [SOa]CFGGRMDRIGAQSGLGCNSFRY | 18 |
4. Conclusions
Detection and structural characterization of cyclic peptide metabolites in biological matrices represent great analytical challenges. The expanded MetabolitePilot™ Software 2.0 offers multiple mechanisms for targeted and non-targeted searching for intact cyclic peptide metabolites with cyclic or linear structures through processing LC/HRMS data sets, followed by automated sequence confirmation and structural identification of these metabolites (Fig. 2). Results from analyzing predicted metabolites of insulin formed by the hydrolysis of trypsin and chymotrypsin demonstrated that the approach is capable of rapidly finding and identifying metabolic products of cyclic peptides. Additionally, the features and characterization tools integrated in the software allowed for the confirmation of metabolite sequences and ranking of competing assignments. The example of applying the HRMS-based data processing tool for the direct detection and identification of unknown metabolites of insulin and ANP in rat liver S9 without reducing their disulfide bond or enzymatic hydrolysis, further indicated that it is useful for studying in vitro biotransformation of cyclic peptides with a variety of linkage structures. Potential applications of the analytical approach at the stage of drug discovery include metabolic soft spot analysis of cyclic peptides in lead optimization and in vitro metabolism comparison across species in clinical candidate characterization. The effectiveness of this workflow for analyzing in vivo metabolites of cyclic peptides remains to be evaluated, which will face additional challenges due to the interference by a large number of endogenous peptides.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.05.012.
Contributor Information
Tingting Cai, Email: cai_tingting0101@wuxiapptec.com.
Mingshe Zhu, Email: mingshe.zhu@yahoo.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Joo S.H. Cyclic Peptides as therapeutic agents and biochemical tools. Biomol. Ther. 2012;20:19–26. doi: 10.4062/biomolther.2012.20.1.019. http://doi:10.4062/biomolther.2012.20.1.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrew T.B., Cayla M.M., Lokey R.S. Form and function in cyclic peptide natural products: a pharmacokinetic perspective. Curr. Top. Med. Chem. 2013;13:821–836. doi: 10.2174/1568026611313070005. [DOI] [PubMed] [Google Scholar]
- 3.Anthi T., Minos-Timotheos M., Carmen S. Review cyclic peptides on a merry-go-round: towards drug design. Peptide Sci. 2015;104:453–461. doi: 10.1002/bip.22669. http://doi:10.1002/bip.22669 [DOI] [PubMed] [Google Scholar]
- 4.Fahr A. Cyclosporin clinical pharmacokinetics. Clin. Pharmacokinet. 1993;24:472–495. doi: 10.2165/00003088-199324060-00004. http://10.2165/00003088-199324060-00004 [DOI] [PubMed] [Google Scholar]
- 5.Kates S.A., Solé N.A., Johnson C.R. A novel, convenient, three-dimensional orthogonal strategy for solid-phase synthesis of cyclic peptides. Tetrahedron Lett. 1993;34:1549–1552. doi: 10.1016/0040-4039(93)85003-F. [DOI] [Google Scholar]
- 6.Hall R.G., Payne K.D., Bain A.M. Multicenter evaluation of vancomycin dosing: emphasis on obesity. Am. J. Med. 2008;121:515–518. doi: 10.1016/j.amjmed.2008.01.046. http://doi:10.1016/j.amjmed.2008.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emanuele E., Arra M., Pesenti S. Vasopressin and oxytocin as neurohormonal mediators of MDMA (ecstasy) sociosexual behavioural effects. Med. Hypotheses. 2006;67:1250–1251. doi: 10.1016/j.mehy.2006.05.021. http://doi:10.1016/j.mehy.2006.05.021 [DOI] [PubMed] [Google Scholar]
- 8.Bellary S., Barnett A.H. Inhaled insulin: new technology, new possibilities. Int. J. Clin. Pract. 2006;60:728–734. doi: 10.1111/j.1742-1241.2006.00976.x. http://doi:10.1111/j.1742-1241.2006.00976.x [DOI] [PubMed] [Google Scholar]
- 9.Sia S.K., Carr P.A., Cochran A.G. Short constrained peptides that inhibit HIV-1 entry. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14664–14669. doi: 10.1073/pnas.232566599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y., Kodadek T. Direct comparison of linear and macrocyclic compound libraries as a source of protein ligands. ACS Comb. Sci. 2005;17:190–195. doi: 10.1021/co500161c. http://doi:10.1021/co500161c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luca G., De M.R., Lucia C. Chemical modifications designed to improve peptide stability: incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization. Curr. Pharmaceut. Des. 2010;16:3185–3203. doi: 10.2174/138161210793292555. [DOI] [PubMed] [Google Scholar]
- 12.Di L. Strategic approaches to optimizing peptide ADME properties. AAPS J. 2015;17:134–143. doi: 10.1208/s12248-014-9687-3. http://doi:10.1208/s12248-014-9687-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rezai T., Yu B., Millhauser G.L. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J. Am. Chem. Soc. 2006;128:2510–2511. doi: 10.1021/ja0563455. http://doi:10.1021/ja0563455 [DOI] [PubMed] [Google Scholar]
- 14.Christopher K.B., Richard A.J.O. Gas-phase peptide fragmentation: how understanding the fundamentals provides a springboard to developing new chemistry and novel proteomic tools. J. Mass Spectrom. 2008;43:1301–1319. doi: 10.1002/jms.1469. http://doi:10.1002/jms.1469 [DOI] [PubMed] [Google Scholar]
- 15.Alex G.H. To b or not to b: the ongoing saga of peptide b ions. Mass Spectrom. Rev. 2009;28:640–654. doi: 10.1002/mas.20228. http://doi:10.1002/mas.20228 [DOI] [PubMed] [Google Scholar]
- 16.Chi H., Chen H., He K. pNovo+: de novo peptide sequencing using complementary HCD and ETD tandem mass spectra. J. Proteome Res. 2013;12:615–625. doi: 10.1021/pr3006843. [DOI] [PubMed] [Google Scholar]
- 17.Quan L.D., Liu M. CID, ETD and HCD fragmentation to study protein post-translational modifications. Mod. Chem. Appl. 2013;1:e102. http://doi:10.4172/2329-6798.1000e102 [Google Scholar]
- 18.Medzihradszky K.F., Campbell J.M., Baldwin M.A. The characteristics of peptide collision-induced dissociation using a high-performance MALDI-TOF/TOF tandem mass spectrometer. Anal. Chem. 2000;72:552–558. doi: 10.1021/ac990809y. http://doi:10.1021/ac990809y [DOI] [PubMed] [Google Scholar]
- 19.Wysocki V.H., Resing K.A., Zhang Q. Mass spectrometry of peptides and proteins. Methods. 2005;35:211–222. doi: 10.1016/j.ymeth.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Ioannis A.P. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom. Rev. 1995;14:49–73. http://doi:10.1002/mas.1280140104 [Google Scholar]
- 21.Béla P., Sándor S. Fragmentation pathways of protonated peptides, Mass Spectrom. Rev. 2005;24:508–548. doi: 10.1002/mas.20024. http://doi:10.1002/mas.20024 [DOI] [PubMed] [Google Scholar]
- 22.Strohalm M., Kavan D., Novák P. mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal. Chem. 2010;82:4648–4651. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- 23.Rusconi F. massXpert 2: a cross-platform software environment for polymer chemistry modelling and simulation/analysis of mass spectrometric data. Bioinformatics. 2009;25:2741–2742. doi: 10.1093/bioinformatics/btp504. http://doi:10.1093/bioinformatics/btp504 [DOI] [PubMed] [Google Scholar]
- 24.Swaminathan J., Varatharajan S. Peptide fragment ion analyzer (PFIA): a simple and versatile tool for the interpretation of tandem mass spectrometric data and de novo sequencing of peptides. Rapid Commun. Mass Spectrom. 2007;21:3033–3038. doi: 10.1002/rcm.3179. http://doi:10.1002/rcm.3179 [DOI] [PubMed] [Google Scholar]
- 25.Ciccimaro E., Ranasinghe A., D’Arienzo C. Strategy to improve the quantitative LC-MS analysis of molecular ions resistant to gas-phase collision induced dissociation: application to disulfide-rich cyclic peptides. Anal. Chem. 2014;86:11523–11527. doi: 10.1021/ac502678y. [DOI] [PubMed] [Google Scholar]
- 26.Xia Y.Q., Ciccimaro E., Zheng N. Differential mobility spectrometry combined with multiple ion monitoring for bioanalysis of disulfide-bonded peptides with inefficient collision-induced dissociation fragmentation. Bioanalysis. 2017;9:183–192. doi: 10.4155/bio-2016-0190. http://doi:10.4155/bio-2016-0190 [DOI] [PubMed] [Google Scholar]
- 27.Fu Y., Xia Y., Flarakos J. Differential mobility spectrometry coupled with multiple ion monitoring in regulated LC-MS/MS bioanalysis of a therapeutic cyclic peptide in human plasma. Anal. Chem. 2016;88:3655–3661. doi: 10.1021/acs.analchem.5b04408. [DOI] [PubMed] [Google Scholar]
- 28.Arnold R.J., Jayasankar N., Aggarwal D. A machine learning approach to predicting peptide fragmentation spectra. P. Pac. Symp. Biocomput. 2006;11:219–230. doi: 10.1142/9789812701626_0021. [DOI] [PubMed] [Google Scholar]
- 29.Frank A.M. Predicting intensity ranks of peptide fragment ions. J. Proteome Res. 2009;8:2226–2240. doi: 10.1021/pr800677f. http://doi:10.1021/pr800677f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb-Robertson B.J.M., Cannon W.R. Current trends in computational inference from mass spectrometry-based proteomics. Briefings Bioinf. 2007;8:304–317. doi: 10.1093/bib/bbm023. http://doi:10.1093/bib/bbm023 [DOI] [PubMed] [Google Scholar]
- 31.Li S., Arnold R.J., Tang H. On the accuracy and limits of peptide fragmentation spectrum prediction. Anal. Chem. 2011;83:790–796. doi: 10.1021/ac102272r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z. Prediction of low-energy collision-induced dissociation spectra of peptides. Anal. Chem. 2004;76:3908–3922. doi: 10.1021/ac049951b. [DOI] [PubMed] [Google Scholar]
- 33.Hirsch I.B. Insulin analogues. N. Engl. J. Med. 2005;352:174–183. doi: 10.1056/NEJMra040832. http://doi: 10.1056/NEJMra040832 [DOI] [PubMed] [Google Scholar]
- 34.Philippe R.B. Structure activity in the atrial natriuretic peptide (ANP) family. Med. Res. Rev. 1990;10:115–142. doi: 10.1002/med.2610100105. http://doi:10.1002/med.26 [DOI] [PubMed] [Google Scholar]
- 35.Olsen J.V., Ong S., Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. http://doi: 10.1074/mcp.T400003-MCP200 [DOI] [PubMed] [Google Scholar]
- 36.Feher J. second ed. Academic Press; Boston: 2017. 8.5 Digestion and Absorption of the Macronutrients, Quantitative Human Physiology; pp. 821–833. [Google Scholar]
- 37.Lau J.L., Dunn M.K. Therapeutic peptides: historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018;26:2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 38.Yao M., Chen B., Zhao W. LC-MS differential analysis for fast and sensitive determination of biotransformation of therapeutic proteins. Drug Metab. Dispos. 2018;46:451–457. doi: 10.1124/dmd.117.077792. https://doi: 10.1124/dmd.117.077792 Epub 2018 Jan 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.