Abstract
Purpose of Review:
Exposomics studies can measure health-relevant chemical exposures during a lifetime and estimate the “internal” environment. However, sampling limitations make these features difficult to capture directly during the critical neonatal time period.
Recent Findings:
We review the use of newborn dried bloodspots (DBS) archived from newborn screening programs for exposomic analysis in epidemiological children’s health studies. Emerging ‘omics technologies such as adductomics and metabolomics have been adapted for DBS analysis, and these technologies can now provide valuable etiological information on the complex interplay between exposures, biological response, and population phenotypes.
Summary:
Adductomics and metabolomics of DBS can provide robust measurements for retrospective epidemiological investigations. With extensive bioarchiving programs in the US and other countries, DBS are poised to substantially aid epidemiological studies, particularly for rare and low-frequency childhood diseases and disorders.
Keywords: adductomics, metabolomics, archived dried bloodspot, exposome, biomarker, exposure
Introduction
Prenatal/perinatal development is particularly vulnerable to perturbations induced by various exposures that may produce later health effects. This susceptibility may result from greater absorption, underdeveloped detoxification mechanisms, and subtle disruptions incorporated during periods of rapid organ and tissue development. Epidemiological studies link prenatal and early-life exposures from nutrition, stress, and environment with adverse child health outcomes including allergies, atypical neurodevelopment, impaired lung development and asthma, reproductive developmental toxicity, and pediatric cancers(1–6). However, many such studies lacked direct exposure measurements, instead relying on questionnaire data, maternal biomarker measurements, or external measurements that may not accurately capture fetal exposures. Obtaining direct measurements of biological samples during periods of prenatal and perinatal development is thus critically important.
Blood provides a critical matrix for investigating exposures and resulting biological responses across the lifecourse. However, collecting cord blood can be logistically challenging, and parents prefer to avoid venous blood collection of newborns and infants. In contrast, a simple heel- or finger-prick blood sample, is easier to collect, less invasive, and provides adequate sample volume for many assays. Samples placed on filter paper [e.g., Guthrie cards(7,8)] and allowed to dry, or ‘dried blood spots’ (DBS), provide a rich resource for identifying biomarkers of disease in various physiological pathways as well as exposure measurements(7–9). Further, such samples collected during newborn screening can be archived, representing a largely untapped resource of fetal/neonatal blood samples for epidemiological research(10).
Here, we review the state of the science using residual DBS for ‘exposomics’ research in children’s health. Exposomics refers to all health-relevant chemical exposures that an individual experiences, as well as the estimated ‘internal” enviroment detected using sensitive analytical platforms. Exposomic studies capture both exogenous and endogenous exposures, and downstream endogenous products along the exposure-disease continuum. We particularly focus on two complementary approaches, metabolomics and adductomics, detecting global sets of circulating small molecules (< 2 kDa) and their reactions with abundant blood proteins, respectively. Together, these approaches afford opportunities for uncovering etiologic mechanisms linking environmental exposures with adverse health outcomes. We discuss recent studies applying these approaches in children’s health epidemiology, and the advantages and challenges of using archived DBS in such research. Finally, we present software platforms currently available for merging information from metabolomics and adductomics to produce a more complete picture of the exposome.
The Exposome
In 2005, Christopher Wild proposed that the key to understanding disease etiology may hinge on measuring an individual’s environmental exposures across the lifecourse, from prenatal development onwards(11). This ‘exposome’ concept has since evolved to include not only exogenous exposures, but also those from internal sources such as gut microbiota, pre-existing disease, stress(12), and responses to environmental stressors mediated through epigenetic and metabolic pathways(13). Indeed, biological response is a critical consideration in determining relationships between exposures and health, because many exposures are transient, while biological responses persist(14). As a result, the current integrated hypothesis of disease etiology portends that most pediatric diseases reflect complex interactions between exposures (chemical, biological, psycho-social, and/or viral) during critical developmental windows and genetic factors, ultimately leading to disease resilience, initiation, and progression.
To analyze the exposome, there are two general approaches (see Table 1). “Bottom-up” approaches estimate exposures using external measurements from air, water, or diet; “top-down” approaches estimate exposures by profiling biofluids in various ‘omic layers(12). The ‘omic layers represent the system-level cascade from genomics (genome), to epigenomics (DNA methylation), to transcriptomics (RNA transcription), to proteomics (proteome/peptides), to adductomics (adducts on peptides/proteins), and finally, to metabolomics (metabolites), and the microbiome (see Figure 1)(12,15). Each ‘omic layer is a plausible point of interaction with exposures that can precipitate adverse health outcomes in children(16). Combining both bottom-up and top-down approaches can provide a comprehensive evaluation of exposures. However, top-down approaches are used more often in epidemiological research(17), because they provide a more accurate description of exposure and allow for untargeted analysis to identify unknown exposures.
Table 1.
| Bottom-up Exposomics | Top-down Exposomics | |
|---|---|---|
| Measurements | “External” environment | “Internal” environment |
| Analysis performed | Direct measurement of external exposures to determine associations between exposure(s) and health outcome(s) | Direct biological measurement of exogenous and endogenous analytes to determine intermediate biomarkers associated with health outcome(s) |
| Follow-up studies | Targeted toxicological and metabolism studies | Targeted search for associations between biomarkers and retrospective external exposures |
| Major advantages | Enables direct link between exposure and health outcome for faster implementation of abatement strategies | Includes: exogenous measurements; endogenous measurements that reflect biological response of genetic susceptibility or of transient exposure from chemicals with short biological half-lives; and includes measurement of reactive compounds |
Figure 1. Theoretical framework for newborn internal exposome.
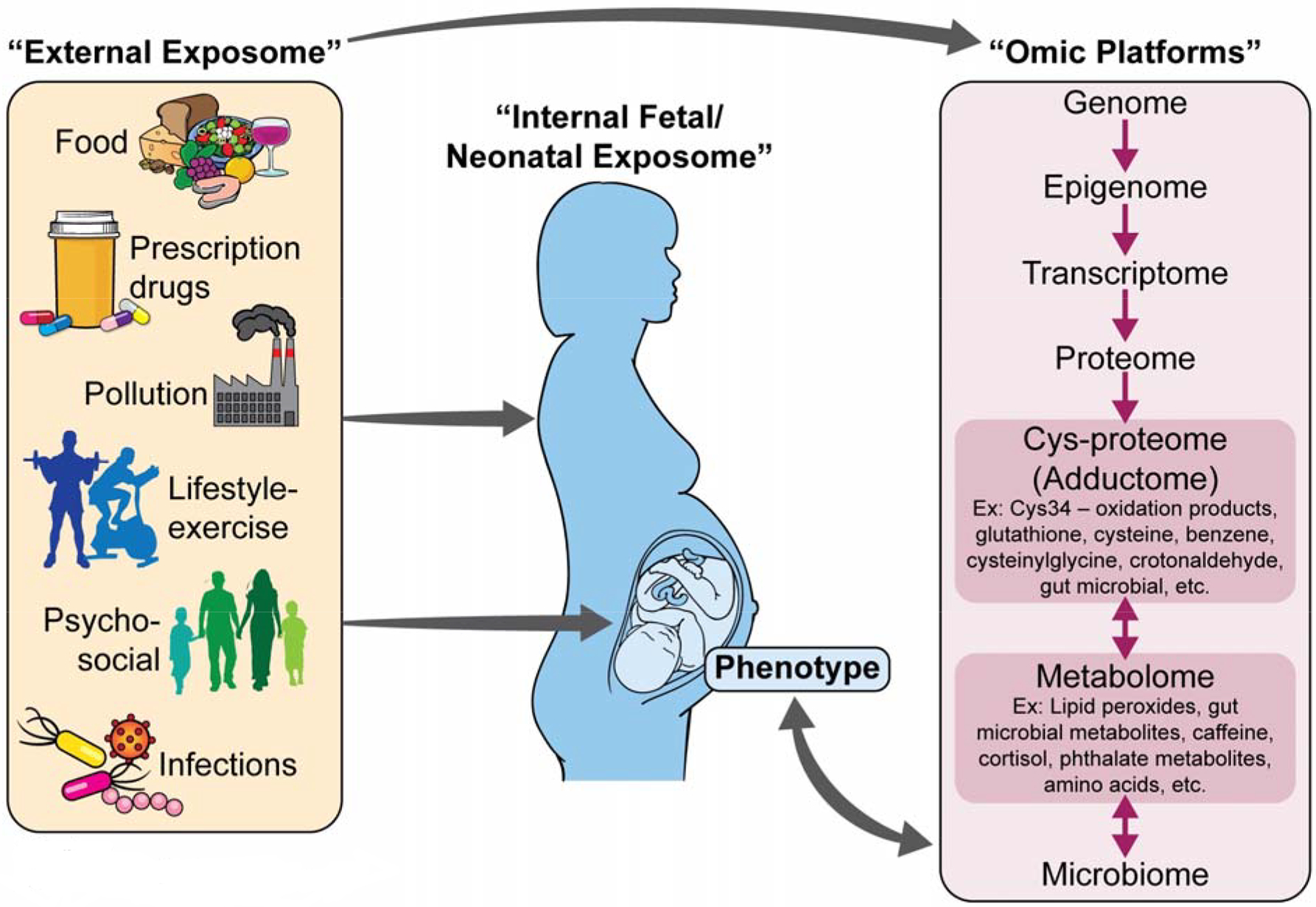
External exposures from food, prescription drugs, pollution, lifestyle-exercise, psychosocial factors, and infections mediated by the mother during fetal development or directly received by the newborn at birth can be directly measured in biological samples such as DBS. The cys-proteome and metabolome describe measurable readouts of reactive small molecules binding with proteins (adducts) and endogenous and exogenous small molecules (metabolites) due to interaction between external exposures (maternal or newborn), ‘omic layers and microbiome (maternal, fetal, or newborn) and small molecules produced from adverse health itself (maternal, fetal, or newborn)(12,15).
Printed with permission from ©Mount Sinai Health System
Metabolomics is either targeted, the quantitative analysis of 10s to 100s of metabolites, or untargeted, the unbiased semi-quantitative analysis of 100s to 1000s of metabolites and unknown signals that represent the downstream readout of biological activity that relates with phenotype(18). Metabolites measured include endogenous analytes involved in various biological pathways such as purine catabolism or bile acid biosynthesis, exogenous exposures such as nicotine and its biological metabolite, cotinine, and for untargeted analysis, includes chemicals of unknown identification for discovery. Therefore, metabolomics is used to establish associations between exposures, biological response, and health effects. Adductomics measures addition products between reactive small molecules and abundant blood proteins: this approach enables assessing exposures to exogenous and endogenous chemicals over extended time periods(19). For example, stable adducts with human serum albumin (HSA) and hemoglobin persist in the blood for one month and three months, respectively(20).
Combined application of metabolomics and adductomics provides unique opportunities for characterizing the exposome, particularly for exposures that would go undetected through bottom-up strategies. While protein adducts have been used as biomarkers for estimating exposures to environmental toxicants(21–24), a large class of endogenously-produced adducts have recently emerged as important biomarkers, reflecting unique signatures of cellular by-products that can link environmental exposures with adverse health outcomes(25). Because of its important biological role binding electrophiles and other small reactive molecules in circulating blood, the HSA-Cys34 adductome, a subset of the proteome (see Figure 1), provides particularly unique opportunities for investigating environmental risk factors for chronic diseases and other adverse health outcomes. Applications of metabolomics and adductomics technologies to DBS samples provide a further advantage; achieving direct measurements for a retrospective, top-down, exposomic analysis of disease etiology using samples collected at birth, before the onset of clinical symptoms.
DBS are widely archived
DBS offer a valuable research resource because of their simplicity of collection, absence of a required cold chain before archiving, and minimal space requirements in bioarchives. With the overall success of DBS collection as part of standard newborn screening, there is long-term investment in their use for primary and secondary research. The result is substantial bioarchives holding millions of DBS. While storage temperatures and lengths of time vary, according to the NewSTEPs data repository(26), 12 US States and territories archive DBS for more than 1 year at temperatures ≤ 5°C, seven of them store samples at least 5 years and 3 that store them indefinitely (Table 2). This equates to a potential biological specimen collected at birth (i.e., archived DBS) for every child diagnosed with a disease or disorder across seven states in the last 5 years; for California and Minnesota, this can be for every child and young adult diagnosed in the last 30 years. Other countries with extensive archiving programs include Denmark, which has been storing their excess DBS at −20°C since 1982 (over 2 million currently in archive)(27,28), and Sweden which has been storing DBS at 5 °C since 1981. The potential for DBS use in etiological studies, particularly for rare childhood diseases and disorders in which it is economically and ethically impossible to collect prospective blood samples, is unprecedented.
Table 2.
US States and territories that archive excess DBS collected during newborn screening for greater than 1 year(26)
| US State/Territory | Storage Conditions (°C) | Time (yrs) |
|---|---|---|
| Alaska | RTa | 3 |
| Arkansas | −20 | 2 |
| California | −20 | Indefinitely |
| Connecticut | −80 | 3 |
| Deleware | −20 | 3 |
| Idaho | ≤ RT | 1.5 |
| Iowa | ≤ RT | 5 |
| Maine | −20 | Indefinitely |
| Maryland | 4 | 25 |
| Massachusetts | −20 | 15 |
| Michigan | ≤ RT | ≤ 100 |
| Minnesota | 4 | Indefinitely |
| Missouri | ≤ −20 | 5 |
| New Jersey | RT | 23 |
| New York | 4 | ≤ 27 |
| North Carolina | RT | 5 |
| North Dakota | N/Ab | ≤ 18 |
| Ohio | −20 | 2 |
| Puerto Rico | 4 | 2 |
| Rhode Island | N/A | 23 |
| Texas | RT | ≤ 25 |
| Washington | RT | 21 |
RT means room temperature
N/A means the temperature is not available
Moreover, retrieval of DBS for use in research studies is largely supported. Where systems are in place for protection of personal information, parental support for use of DBS is high(29,30). For example, in a study of 5,024 kids in New York State, 62% of parents provided consent for use of their child’s residual DBS for immunologic and environmental chemical analysis(31). As of 2019, the Danish Newborn Screening Biobank that archives Denmark’s remainder DBS has received less than 0.1% of citizen requests to remove samples from biorepositories, and received no reports of abuse through their 36 years of function(32). This support, combined with DBS availability through biorepositories, demonstrates the feasibility of using this resource for population-based research. In fact, a systematic review on residual DBS use following newborn screening included 654 articles published from 1973 to 2017, with over 50% published in the later decade, 30% of which originated in the US. Of US studies, 61.6% used a targeted population in which specific individuals’ DBS were requested(33). Thus, residual DBS appear well-utilized worldwide for public health research, and expansion is expected.
Metabolomics of DBS
Metabolomics is an essential part of exposomics analysis that can be robustly applied to DBS. Comparisons of metabolomics applied to experimental DBS versus experimental venous plasma support that the breadth of metabolites measured and the variability (coefficient of variation, CV) are comparably low in both targeted (34) and untargeted analysis(35). In a targeted metabolomics panel of 350 analytes, only 15% of metabolites detected in blood were lost in experimental DBS(36), and untargeted metabolomics profiling of 103 archived DBS collected between 1988 and 2005 measured > 1000 metabolites over a broad range of chemical classes expected in blood(37). This breadth of measurements supported the first case-control DBSmetabolomics discovery studies in autism spectrum disorder, colorectal cancer, and hypertension(38–40), and prompted emerging applications of targeted and untargeted metabolomics to epidemiological samples.
Traditional metabolomics studies in archived DBS sought to discover biomarkers for second-tier tests of genetic disorders and inborn errors in metabolism(41,42), and for newborn nutrition. Heel-prick DBS from 3-month-old infants (stored ~2–9 years at −20°C) detected distinct lipid profiles for exclusively breastfed and exclusively formula-fed infants, and intermediate profiles observed for mixed-fed infants(43). In a follow-up validation study, three key lipids — phosphatidylcholine PC(35:2) and sphingomyelins SM(36:2) and SM(39:1) — enabled distinguishing breastfed from formula-fed infants and suggested that nutrition can significantly alter lipid metabolism early during child development(44). Furthermore, a nested case-control study of archived DBS revealed early-life metabolite profiles linked with later onset of acute lymphoblastic leukemia (ALL)(45). In a stratified analysis of 118 diagnosed ALL cases ages 6–14 years old and 117 matched controls, 18 metabolites were positively associated with case status. Additionally, linoleic acid and other 18:2 fatty acid chain metabolites were detected at higher levels in formula-fed compared to breast-fed infants and increased with the mother’s pre-pregnancy body mass index, providing support for early-life nutrition as a risk factor for ALL. Together, these studies demonstrate the feasibility of archived DBS metabolomics to provide valuable etiological information on the complex interplay between exposures, biological responses, and population phenotypes.
Adductomics of DBS
Chemical toxicants are often reactive electrophiles with short life-spans in vivo (e.g., alkylating and acylating agents, aldehydes, alkylnitrosamines, dialkylsulfates, oxiranes, quinones, reactive oxygen and nitrogen species)(46); measuring these toxicants in target tissues is rarely possible. However, their adducts formed with abundant blood proteins, particularly hemoglobin and HSA, can serve as exposure biomarkers. Targeted hemoglobin and HSA adducts have been measured in human blood for many environmental toxicants that are either electrophiles or their precursors, i.e., ethylene oxide, benzene, 1,3-butadiene, acrylamide, aflatoxin B1, a variety of aromatic amines, and polycyclic aromatic hydrocarbons [reviewed in(47)]. Still, as with metabolomics, the need to obtain venous blood samples has limited the utility of adductomics for large exposure studies. DBS provide the opportunity to leverage adductomics for epidemiologic research.
The first targeted DBS assay quantified benzene oxide adducts on hemoglobin as a biomarker of prenatal benzene exposure, a known risk factor for leukemia(21). In this study, newborn DBS samples were extracted in an ethanol solution to selectively precipitate hemoglobin, and quantified benzene-oxide using gas chromatography-mass spectrometry (GC-MS). More recently, methanol was used to selectively remove interfering proteins from DBS samples and performed untargeted adductomics with HSA using the unprecipitated protein fraction(48). Liquid chromatography - high resolution MS (LC-HRMS) yielded measures of HSA-Cys34 adducts in 49 archived DBS from newborns whose mothers either actively smoked during pregnancy or that were nonsmokers. In total, twenty-six HSA-Cys34 adducts were detected, including cyanide adducts of HSA-Cys34 which enabled discriminating between infants with smoking and nonsmoking mothers. While other studies have utilized untargeted adductomics for measuring HSA-Cys34 adducts in plasma or serum(49–53), this study provided proof-of-concept that newborn DBS can be used in discovery-based experiments for prenatal/perinatal exposures.
Challenges of DBS
Validation of methodologies and improvements in measurement quantification are now accelerating the use of DBS in population-based studies. DBS sampling through the neonatal screening program operates under stringent quality assurance/quality control, using only certified, high quality filter paper that meets performance standards for lot-to-lot variation and sample volume absorbency(54). These standards enable precise and reproducibly measurements from DBS, on par with standard blood collection methods (e.g. vacuum tubes and capillary pipettes)(55). Nonetheless, there remain some challenges regarding the quantification and semi-quantification of metabolites and protein adducts in archived DBS for children’s etiological research. For example, initial evaluations of long-term DBS storage conditions suggest analyte stability at low temperatures(34,36,43,56), and strategies such as potassium, hemoglobin, or mass spectral ion suppression have been proposed to overcome concerns regarding unknown absolute volumes of blood on the card or in a uniform punch(57–60). A robust determination of how these factors affect metabolite and protein adduct measurements will enhance the breadth, precision, and accuracy of measurements, allowing for the detection of smaller effect sizes and improving analysis and interpretation in children’s health studies.
Software for multi-omics integration
‘Omics assessments produce large and diverse data; thus, data integration through computational methods represents a key component of delineating inter-relationships between multi-scale clinical, molecular, and environmental data for improved disease diagnosis, prognosis, and targeted therapies(61). Recently developed bioinformatics tools and methods promote integrative analysis of heterogeneous datasets(62). These methods fall into two main categories: knowledge-base driven and data driven. Knowledge-based methods utilize known pathway level information from databases such as KEGG(63) to aggregate multi-omics data at the pathway level; such tools include MetaboAnalyst 4.0(64), iPEAP (65), IMPaLA (66), and MetScape 2.0(67), or text mining of scientific literature to identify relationships between biomedical terms (68,69). Data driven methods use univariate and multivariate statistical approaches such as Pearson correlation and partial least squares (PLS) regression to identify correlations between variables (e.g., genes, metabolites, proteins, adducts on proteins, environmental exposures) across two or more ‘omics datasets collected from the same samples. Currently, more than 20 different dimension reduction techniques support integrative analysis of two or more ‘omics (or other biomedical) datasets(70). Algorithms have been implemented in web-based, R packages, and other open-source tools such as 3Omics(71), mixOmics(72), xMWAS(73), and omicsade4(74). xMWAS offers additional capabilities such as detection of communities (or clusters) of tightly connected nodes for discovery of novel associations between clinical, molecular, and environmental variables. Further, xMWAS performs differential centrality analysis to identify network nodes that undergo changes in their association patterns between different conditions. Applying such computational tools to identify associations across multiple ‘omics and other biomedical datasets is critical to improve our understanding of the complex inter-relationships between clinical, molecular, and environmental data for identifying disease markers, screening tools for early diagnosis, and identifying modifiable risk factors of childhood disease.
Conclusions
Archived DBS analysis provides direct measurements of early-life exposures for retrospective epidemiological research that can be conducted more rapidly and cost-effectively than what was envisioned when Christopher Wild first suggested performing population-based exposome studies in prospective cohorts to understand etiology(11). With the advancement of metabolomics and adductomics technologies and software available for multi- omic integration, exposomic analysis of archived DBS provides a powerful tool for etiological research, particularly for rare and low-frequency pediatric diseases and disorders.
Key points.
Dried blood spots (DBS) following newborn screening are routinely archived for potential secondary research
Metabolomics and adductomics are complementary approaches that together measure environmental exposures, reactive compounds, and endogenous metabolites
Exposomic analysis of archived DBS can provide direct biological measurements, retrospectively, of a critical early-life exposure window for etiological research
Acknowledgments
The authors are supported by the National Institute of Environmental Health Sciences grants 2U2CES026561-02 (LP), 1U2CES030859-01 (LP), P30ES23515 (LP), R21ES030882-01 (LP), R21ES026776-01 (WF), and U24OD023319-01 (WF); National Institute on Alcohol Abuse and Alcoholism grant R21AA026928-02 (KU); National Institute of Diabetes and Digestive and Kidney Diseases grant U24DK112341 (KU); and National Institute on Aging grant RF1AG057470 (KU).
Footnotes
Conflicts of Interest
none
References
- 1.Schmidt RJ, Kogan V, Shelton JF, et al. Combined Prenatal Pesticide Exposure and Folic Acid Intake in Relation to Autism Spectrum Disorder. Environ Health Perspect. 2017. September 22;125(9):097007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng X-W, Bloom MS, Dharmage SC, et al. Prenatal exposure to perfluoroalkyl substances is associated with lower hand, foot and mouth disease viruses antibody response in infancy: Findings from the Guangzhou Birth Cohort Study. Science of The Total Environment. 2019. May;663:60–7. [DOI] [PubMed] [Google Scholar]
- 3.Wenzel AG, Bloom MS, Butts CD, et al. Influence of race on prenatal phthalate exposure and anogenital measurements among boys and girls. Environment International. 2018. January 1;110:61–70. [DOI] [PubMed] [Google Scholar]
- 4.Gaudriault P, Mazaud-Guittot S, Lavoue V, et al. Endocrine Disruption in Human Fetal Testis Explants by Individual and Combined Exposures to Selected Pharmaceuticals, Pesticides, and Environmental Pollutants. Environ Health Perspect. 2017. August 4;125(8):087004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosa MJ, Lee AG, Wright RJ. Evidence establishing a link between prenatal and early-life stress and asthma development Current Opinion in Allergy and Clinical Immunology. 2018. April;18(2):148–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkeleit J, Riise T, Bjørge T, et al. Maternal exposure to gasoline and exhaust increases the risk of childhood leukaemia in offspring – a prospective study in the Norwegian Mother and Child Cohort Study. Br J Cancer. 2018. October;119(8):1028–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDade TW, Williams SA, Snodgrass JJosh. What a Drop Can Do: Dried Blood Spots as a Minimally Invasive Method for Integrating Biomarkers Into Population-Based Research. Demography. 2007;44(4):899–925. [DOI] [PubMed] [Google Scholar]
- 8. *.Freeman JD, Rosman LM, Ratcliff JD, et al. State of the Science in Dried Blood Spots. Clinical Chemistry. 2018. April 1;64(4):656–79. [DOI] [PubMed] [Google Scholar]; This article summarizes the 1001 small molecules reported to be measured in DBS from a scoping review of reviews approach
- 9.Zakaria R, Allen KJ, Koplin JJ, et al. Advantages and Challenges of Dried Blood Spot Analysis by Mass Spectrometry Across the Total Testing Process. EJIFCC. 2016. December 1;27(4):288–317. [PMC free article] [PubMed] [Google Scholar]
- 10.Brindle E, Connor KAO, Garrett DA. Applications of Dried Blood Spots in General Human Health Studies. Dried blood spots applications and techniques. Hoboken: John Wiley and Sons; 2014. 114 p. [Google Scholar]
- 11.Wild CP. Complementing the Genome with an “Exposome”: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology. Cancer Epidemiol Biomarkers Prev. 2005. August 1;14(8):1847–50. [DOI] [PubMed] [Google Scholar]
- 12.Rappaport SM, Smith MT. Environment and Disease Risks. Science. 2010. October 22;330(6003):460–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller GW, Jones DP. The Nature of Nurture: Refining the Definition of the Exposome. Toxicological Sciences. 2014. January;137(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis KK, Auerbach SS, Balshaw DM, et al. The Importance of the Biological Impact of Exposure to the Concept of the Exposome. Environ Health Perspect. 2016. October;124(10):1504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Go Y-M, Chandler JD, Jones DP. The cysteine proteome. Free Radical Biology and Medicine. 2015. July 1;84:227–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niedzwiecki MM, Walker DI, Vermeulen R, et al. The Exposome: Molecules to Populations. Annual Review of Pharmacology and Toxicology. 2019;59(1):107–27. [DOI] [PubMed] [Google Scholar]
- 17.Xue J, Lai Y, Liu C-W, Ru H. Towards Mass Spectrometry-Based Chemical Exposome: Current Approaches, Challenges, and Future Directions. Toxics. 2019. September;7(3):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patti GJ, Yanes O, Siuzdak G. Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012. April;13(4):263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rappaport SM, Li H, Grigoryan H, et al. Adductomics: Characterizing exposures to reactive electrophiles. Toxicology Letters. 2012. August 13;213(1):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart AJ, Blindauer CA, Berezenko S, et al. Role of Tyr84 in controlling the reactivity of Cys34 of human albumin. The FEBS Journal. 2005;272(2):353–62. [DOI] [PubMed] [Google Scholar]
- 21.Funk WE, Waidyanatha S, Chaing SH, Rappaport SM. Hemoglobin Adducts of Benzene Oxide in Neonatal and Adult Dried Blood Spots. Cancer Epidemiol Biomarkers Prev. 2008. August 1;17(8):1896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeowell-O’Connell K, Rothman N, Waidyanatha S, et al. Protein Adducts of 1,4-Benzoquinone and Benzene Oxide among Smokers and Nonsmokers Exposed to Benzene in China. Cancer Epidemiol Biomarkers Prev. 2001. August 1;10(8):831–8. [PubMed] [Google Scholar]
- 23.Waidyanatha S, Zheng Y, Serdar B, Rappaport SM. Albumin Adducts of Naphthalene Metabolites as Biomarkers of Exposure to Polycyclic Aromatic Hydrocarbons. Cancer Epidemiol Biomarkers Prev. 2004. January 1;13(1):117–24. [DOI] [PubMed] [Google Scholar]
- 24.Rappaport SM, Yeowell-O’Connell K. Protein adducts as dosimeters of human exposure to styrene, styrene-7,8-oxide, and benzene. Toxicology Letters. 1999. September 5;108(2):117–26. [DOI] [PubMed] [Google Scholar]
- 25.Grigoryan H, Li H, Iavarone AT, et al. Cys34 adducts of reactive oxygen species in human serum albumin. Chem Res Toxicol. 2012. August 20;25(8):1633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NewSTEPs Data Repository - DBS Retention Report [Internet]. [cited 2019 Oct 28]. Available from: https://data.newsteps.org/newsteps-web/reports/profile/dbsRetention.action
- 27.Nørgaard-Pedersen B, Hougaard DM. Storage policies and use of the Danish Newborn Screening Biobank. J Inherit Metab Dis. 2007. August;30(4):530–6. [DOI] [PubMed] [Google Scholar]
- 28.Statens Serum Institut. Our Biological Samples [Internet]. [cited 2019 Oct 28]. Available from: https://www.danishnationalbiobank.com/our-samples
- 29.Jansen ME, van den Bosch LJM, Hendriks MJ, et al. Parental perspectives on retention and secondary use of neonatal dried bloodspots: a Dutch mixed methods study. BMC Pediatrics. 2019. July 9;19(1):230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Botkin JR, Rothwell E, Anderson R, et al. Public Attitudes Regarding the Use of Residual Newborn Screening Specimens for Research. 2012;129(2):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeung EH, Louis GB, Lawrence D, et al. Eliciting parental support for the use of newborn blood spots for pediatric research. BMC Med Res Methodol. 2016. February 4;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hougaard DM, Bybjerg-Grauholm J, Christiansen M, Nørgaard-Pedersen B. Response to “Newborn dried blood spot samples in Denmark: the hidden figures of secondary use and research participation. ” Eur J Hum Genet. 2019. June 28;1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. *.Rothwell E, Johnson E, Riches N, Botkin JR. Secondary research uses of residual newborn screening dried bloodspots: a scoping review. Genet Med. 2019. July;21(7):1469–75. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article summarizes the increasing interest in the use of archived excess DBS following newborn screening in research from 1973–2017 within and outside the US. It shows that many of the 1,627,751 DBS used in non-newborn screening related research to date was focused on individual and public health needs, mostly from those bioarchives that are large and open to research collaborations such as California and New York in the US.
- 34.Koulman A, Prentice P, Wong MCY, et al. The development and validation of a fast and robust dried blood spot based lipid profiling method to study infant metabolism. Metabolomics. 2014;10(5):1018–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michopoulos F, Theodoridis G, Smith CJ, Wilson ID. Metabolite Profiles from Dried Biofluid Spots for Metabonomic Studies using UPLC Combined with oaToF-MS. J Proteome Res. 2010. June 4;9(6):3328–34. [DOI] [PubMed] [Google Scholar]
- 36.Drolet J, Tolstikov V, Williams BA, et al. Integrated Metabolomics Assessment of Human Dried Blood Spots and Urine Strips. Metabolites. 2017. September;7(3):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrick L, Edmands W, Schiffman C, et al. An untargeted metabolomics method for archived newborn dried blood spots in epidemiologic studies. Metabolomics. 2017. March;13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barone R, Alaimo S, Messina M, et al. A Subset of Patients With Autism Spectrum Disorders Show a Distinctive Metabolic Profile by Dried Blood Spot Analyses. Front Psychiatry. 2018;9:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jing Y, Wu X, Gao P, et al. Rapid differentiating colorectal cancer and colorectal polyp using dried blood spot mass spectrometry metabolomic approach. IUBMB Life. 2017;69(5):347–54. [DOI] [PubMed] [Google Scholar]
- 40.Bai Q, Peng B, Wu X, et al. Metabolomic study for essential hypertension patients based on dried blood spot mass spectrometry approach. IUBMB Life. 2018;70(8):777–85. [DOI] [PubMed] [Google Scholar]
- 41.Dénes J, Szabó E, Robinette SL, et al. Metabonomics of Newborn Screening Dried Blood Spot Samples: A Novel Approach in the Screening and Diagnostics of Inborn Errors of Metabolism. Anal Chem. 2012. November 20;84(22):10113–20. [DOI] [PubMed] [Google Scholar]
- 42.Tortorelli S, Eckerman JS, Orsini JJ, et al. Moonlighting newborn screening markers: the incidental discovery of a second-tier test for Pompe disease. Genet Med. 2018. August;20(8):840–6. [DOI] [PubMed] [Google Scholar]
- 43.Prentice P, Koulman A, Matthews L, et al. Lipidomic Analyses, Breast- and Formula-Feeding, and Growth in Infants. J Pediatr. 2015. February;166(2):276–281.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Acharjee A, Prentice P, Acerini C, et al. The translation of lipid profiles to nutritional biomarkers in the study of infant metabolism. Metabolomics. 2017;13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. **.Petrick LM, Schiffman C, Edmands WMB, et al. Metabolomics of neonatal blood spots reveal distinct phenotypes of pediatric acute lymphoblastic leukemia and potential effects of early-life nutrition. Cancer Letters. 2019. June 28;452:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first study to demonstrate the use of untargeted metabolomics in archived DBS for discovery of early-life predictors of later childhood acute lymphoblastic leukemia (ALL) diagnosis. Linoleic acid, among other metabolites, was correlated with breastfeeding practice and pre-pregnancy maternal BMI suggesting early life nutrition as a risk factor for ALL.
- 46.Törnqvist M, Fred C, Haglund J, et al. Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. Journal of Chromatography B. 2002. October 5;778(1):279–308. [DOI] [PubMed] [Google Scholar]
- 47.LoPachin RM, DeCaprio AP. Protein Adduct Formation as a Molecular Mechanism in Neurotoxicity. Toxicological Sciences. 2005. August 1;86(2):214–25. [DOI] [PubMed] [Google Scholar]
- 48. **.Yano Y, Grigoryan H, Schiffman C, et al. Untargeted adductomics of Cys34 modifications to human serum albumin in newborn dried blood spots. Anal Bioanal Chem. 2019. April 1;411(11):2351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used untargeted adductomics to identify cyanide adducts of HSA-Cys34 in archived DBS as a predictor of active maternal smoking while pregnant.
- 49.Grigoryan H, Edmands W, Lu SS, et al. Adductomics Pipeline for Untargeted Analysis of Modifications to Cys34 of Human Serum Albumin. Anal Chem. 2016. November 1;88(21):10504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu SS, Grigoryan H, Edmands WMB, et al. Profiling the Serum Albumin Cys34 Adductome of Solid Fuel Users in Xuanwei and Fuyuan, China. Environ Sci Technol. 2017. January 3;51(1):46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S, Grigoryan H, Edmands WMB, et al. Cys34 Adductomes Differ between Patients with Chronic Lung or Heart Disease and Healthy Controls in Central London. Environ Sci Technol. 2018. February 20;52(4):2307–13. [DOI] [PubMed] [Google Scholar]
- 52.Grigoryan H, Edmands WMB, Lan Q, et al. Adductomic signatures of benzene exposure provide insights into cancer induction. Carcinogenesis. 2018. 03;39(5):661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grigoryan H, Schiffman C, Gunter MJ, et al. Cys34 Adductomics Links Colorectal Cancer with the Gut Microbiota and Redox Biology. Cancer Res. 2019. October 22; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mei JV, Zobel SD, Hall EM, et al. Performance properties of filter paper devices for whole blood collection. Bioanalysis. 2010. August 1;2(8):1397–403. [DOI] [PubMed] [Google Scholar]
- 55.NSQAP About the Program | CDC [Internet]. 2019. [cited 2019 Oct 29]. Available from: https://www.cdc.gov/labstandards/nsqap_about.html
- 56.Zukunft S, Sorgenfrei M, Prehn C, et al. Targeted Metabolomics of Dried Blood Spot Extracts. Chromatographia. 2013. October 1;76(19):1295–305. [Google Scholar]
- 57.Richardson G, Marshall D, Keevil BG. Prediction of haematocrit in dried blood spots from the measurement of haemoglobin using commercially available sodium lauryl sulphate. Ann Clin Biochem. 2018. May;55(3):363–7. [DOI] [PubMed] [Google Scholar]
- 58.Capiau S, Wilk LS, De Kesel PMM, et al. Correction for the Hematocrit Bias in Dried Blood Spot Analysis Using a Nondestructive, Single-Wavelength Reflectance-Based Hematocrit Prediction Method. Anal Chem. 2018. 06;90(3):1795–804. [DOI] [PubMed] [Google Scholar]
- 59.Capiau S, Stove VV, Lambert WE, Stove CP. Prediction of the Hematocrit of Dried Blood Spots via Potassium Measurement on a Routine Clinical Chemistry Analyzer. Anal Chem. 2013. January 2;85(1):404–10. [DOI] [PubMed] [Google Scholar]
- 60.Chepyala D, Kuo H-C, Su K-Y, et al. Improved Dried Blood Spot-Based Metabolomics Analysis by a Postcolumn Infused-Internal Standard Assisted Liquid Chromatography-Electrospray Ionization Mass Spectrometry Method. Anal Chem. 2019. August 20;91(16):10702–12. [DOI] [PubMed] [Google Scholar]
- 61.Kramer F, Just S, Zeller T. New perspectives: systems medicine in cardiovascular disease. BMC Syst Biol. 2018. December;12(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinu FR, Beale DJ, Paten AM, et al. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites. 2019. April 18;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.KEGG: Kyoto Encyclopedia of Genes and Genomes [Internet]. [cited 2019 Nov 3]. Available from: https://www.genome.jp/kegg/
- 64.Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018. July 2;46(W1):W486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun H, Wang H, Zhu R, et al. iPEAP: integrating multiple omics and genetic data for pathway enrichment analysis. Bioinformatics. 2014. March 1;30(5):737–9. [DOI] [PubMed] [Google Scholar]
- 66.Kamburov A, Cavill R, Ebbels TMD, et al. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011. October 15;27(20):2917–8. [DOI] [PubMed] [Google Scholar]
- 67.Karnovsky A, Weymouth T, Hull T, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012. February 1;28(3):373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis AP, Grondin CJ, Johnson RJ, et al. The Comparative Toxicogenomics Database: update 2019. Nucleic Acids Res. 2019. January 8;47(D1):D948–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y, Liang Y, Wishart D. PolySearch2: a significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. 2015. July 1;43(W1):W535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meng C, Zeleznik OA, Thallinger GG, et al. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief Bioinform. 2016. July 1;17(4):628–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuo T-C, Tian T-F, Tseng YJ. 3Omics: a web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst Biol. 2013. December;7(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rohart F, Gautier B, Singh A, Cao K-AL. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLOS Computational Biology. 2017. November 3;13(11):e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. *.Uppal K, Ma C, Go Y-M, Jones DP. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics. 2018. February 15;34(4):701–2. [DOI] [PMC free article] [PubMed] [Google Scholar]; this article presents the xMWAS tool, a user-friendly, freely available software for integration of ‘omics platforms with biochemical and phenotypic data that can be used for improved understanding of pediatric disease pathophysiology
- 74.Meng C, Kuster B, Culhane AC, Gholami AM. A multivariate approach to the integration of multi-omics datasets. BMC Bioinformatics. 2014. December;15(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]


