Summary
Peripheral T‐cell lymphomas (PTCL) and natural killer (NK)/T‐cell lymphomas (NKTCL) are a heterogeneous group of aggressive malignancies with dismal outcomes and limited treatment options. While the phosphatidylinositol 3‐kinase (PIK3) pathway has been shown to be highly activated in many B‐cell lymphomas, its therapeutic relevance in PTCL and NKTCL remains unclear. The aim of this study is to investigate the expression of PIK3 and phosphatase and tensin homolog (PTEN) in these subtypes of lymphoma and to identify potential therapeutic targets for clinical testing. Therefore, the expression of PIK3α, PIK3β, PIK3γ, PIK3δ and PTEN was analyzed in 88 cases of PTCL and NKTCL samples by immunohistochemistry. All PTCL and NKTCL samples demonstrated high expression of PIK3 isoforms. In particular, high PIK3α expression was significantly associated with poor survival, even after adjustment for age, International Prognostic Index (IPI) score and anthracycline‐based chemotherapy in first line. Notably, copanlisib, a pan‐class I inhibitor with predominant activities towards PIK3α and PIK3δ isoforms, effectively inhibited phosphorylation of AKT, 4E‐BP‐1 and STAT3, causing G0/G1 cell cycle arrest and resulting in suppression of tumour cell growth in vitro and in vivo. This study provides evidence that targeting the PIK3 pathway, particularly simultaneous inhibition of PIK3α and δ, could be a promising approach for the treatment of PTCL and NKTCL.
Keywords: phosphatidylinositol 3‐kinase, peripheral T‐cell lymphoma, NK/T‐cell lymphoma, copanlisib, PI3K
Mature T‐cell lymphomas, including peripheral T‐cell lymphoma (PTCL) and natural killer (NK)/T‐cell lymphoma (NKTCL), are a heterogeneous group of aggressive non‐Hodgkin lymphomas, with poorer treatment outcomes compared to those of their B‐cell counterparts. The World Health Organization (WHO) classification recognizes a number of distinct subtypes of PTCL, including PTCL not otherwise specified (PTCL‐NOS), angioimmunoblastic T‐cell lymphoma (AITL), ALK‐positive (ALK+) and ALK‐negative (ALK−) anaplastic large‐cell lymphoma (ALCL), monomorphic epitheliotropic intestinal T‐cell lymphoma (MEITL; previously known as type II enteropathy‐associated T‐cell lymphoma) and cutaneous T‐cell lymphoma (CTCL) (Vose et al., 2008). With the exception of ALK+ ALCL, the majority of patients relapse rapidly after treatment with five‐year overall survival (OS) rates of 10–30% (Abubaker et al., 2007). In patients with relapsed/refractory (R/R) PTCL and NKTCL, few treatment options are available (Coiffier et al., 2014). The notable exception is CD30‐positive ALCL treated with brentuximab vedotin in which patients achieved an objective response rate of 80% (Horwitz et al., 2014). A better understanding of the signalling pathways regulating the growth and progression in PTCL and NKTCL is required to facilitate development of novel targeted therapies for the treatment of patients with R/R PTCL and NKTCL.
The oncogenic phosphatidylinositol 3‐kinase (PIK3)/AKT pathway plays a key role in multiple critical cellular functions, including growth, differentiation, metabolism, survival and cellular proliferation. Studies have reported that dysregulation of the PIK3/AKT pathway contributes to tumourigenesis, metastasis and resistance to chemotherapy in human cancers including Hodgkin (HL) and non‐Hodgkin lymphomas (NHL) (Engelman, 2009; Polak & Buitenhuis, 2012; Blachly & Baiocchi, 2014; Westin, 2014). There are two subclasses of PIK3 related to cancer: class IA (p110α, β and δ isoforms of the catalytic subunit) which is activated by tyrosine kinases, and class IB (p110γ isoform) which is activated by G‐protein‐coupled receptors. It is increasingly recognized that different PIK3 isoforms have distinct expression profiles and functions in oncogenic signalling and play non‐redundant roles in particular tumour types, which has prompted the development of isoform‐selective inhibitors in recent years with the aim of improving efficacy, while decreasing undesirable side effects (Rommel et al., 2007). The recent approval of the PIK3δ‐selective inhibitor idelalisib for the treatment of R/R chronic lymphocytic leukaemia (CLL), follicular lymphoma (FL), and small lymphocytic leukaemia (SLL) and the PIK3α/δ predominant inhibitor copanlisib for the treatment of relapsed FL confirmed the utility of PIK3‐selective inhibitors in cancer treatment (Gopal et al., 2014; Kahl et al., 2014; Patnaik et al., 2016).
In PTCL, preclinical and clinical studies have also shown that targeting the PIK3/AKT pathway could be a promising therapeutic approach. Activated phosphorylated AKT (pAKT) has been reported to be abnormally overexpressed and to be an indicator of poor prognosis in PTCL patients (Hong et al., 2015). Duvelisib, a PIK3δ/γ‐specific inhibitor, demonstrated promising clinical activity with an overall response rate (ORR) of 40% (14/35) and an acceptable safety profile in R/R PTCL, as well as preclinical evidence of both tumour cell‐autonomous and immune‐mediated effects (Flinn et al., 2018; Horwitz et al., 2018). In a phase II study, treatment with copanlisib, a PIK3α/δ‐predominant inhibitor, achieved an ORR of 3/14 (21·4%) in R/R PTCL patients (Dreyling et al., 2017).
However, whether the PIK3/AKT pathway is a potential target for therapeutic intervention in NKTCL is not known. The prognostic role and clinical significance of PIK3 isoforms in different lymphoma subtypes in PTCL and NKTCL are also not well‐defined. In this study, we investigated the relative expression and functional significance of class I PIK3 isoforms with the aim of identifying the specific PIK3 isoforms to be targeted for PTCL and NKTCL.
Patients and methods
Patient populations and cell lines
Eighty‐eight patients diagnosed with PTCL between 1993 and 2015 (inclusive) at the National Cancer Centre Singapore (NCCS) were selected for this study, including 34 AITL, 5 ALK+ and 4 ALK− ALCL, 19 NKTCL, 12 PTCL‐NOS, 12 MEITL and 2 CTCL. The study was approved by the SingHealth Centralised Institutional Review Board, study number 2004/407/F. The baseline characteristics of patients are summarized in Table 1. The median age at initial diagnosis was 61 years old (range = 13·8 to 83·3 years old) and 62 patients (71%) were male. Sixty patients (68%) were Eastern Cooperative Oncology Group (ECOG) status 0 to 1 and 58 patients (66%) were disease stage III–IV at initial diagnosis. Fourteen PTCL cell lines were used in this study. Detailed information on these is listed in Table SI. All cell lines were negative for mycoplasma contamination as assessed by the MycoAlertTM Mycoplasma Detection Kit (Lonza, Basel, Switzerland).
Table 1.
Patient demographics and disease characteristics at diagnosis by PIK3/AKT classifications.
| Frequency (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PIK3‐⍺ | PIK3‐β | PIK3‐δ | PIK3‐ | ||||||
| Total | Low | High | Negative | Positive | Low | High | Negative | Positive | |
| Characteristics | N = 88 | n = 74 | n = 14 | n = 9 | n = 79 | n = 30 | n = 58 | n = 38 | n = 50 |
| Age | |||||||||
| Median | 61 | 61 | 56 | 61 | 61 | 56 | 61 | 63 | 55 |
| IQR | 47·3, 71·1 | 47·3, 71·1 | 48·7, 70·3 | 54·7, 71·9 | 47·3, 70·9 | 43·2, 71·3 | 48·5, 69·5 | 54·0, 71·9 | 41·4, 70·1 |
| Range | 13·8–83·3 | 13·8–83·3 | 32·9–81·5 | 25·7–81·9 | 13·8–83·3 | 24·8–81·5 | 13·8–83·3 | 21·3–83·0 | 13·8–83·3 |
| P = 1·0* | P = 0·7* | P = 0·7* | P = 0·046* | ||||||
| Age group | |||||||||
| <60 | 41 (47·1) | 33 (45·2) | 8 (57·1) | 3 (33·3) | 38 (48·7) | 16 (53·3) | 25 (43·9) | 14 (36·8) | 27 (55·1) |
| ≥60 | 46 (52·9) | 40 (54·8) | 6 (42·9) | 6 (66·7) | 40 (51·3) | 14 (46·7) | 32 (56·1) | 24 (63·2) | 22 (44·9) |
| P = 0·4** | P = 0·5 | P = 0·4** | P = 0·09** | ||||||
| Gender | |||||||||
| Female | 26 (29·6) | 20 (27·0) | 6 (42·9) | 2 (22·2) | 24 (30·4) | 10 (33·3) | 16 (27·6) | 9 (23·7) | 17 (34·0) |
| Male | 62 (70·5) | 54 (73·0) | 8 (57·1) | 7 (77·8) | 55 (69·6) | 20 (66·7) | 42 (72·4) | 29 (76·3) | 33 (66·0) |
| P = 0·3 | P = 1·0 | P = 0·6** | P = 0·3** | ||||||
| IPI score | |||||||||
| Low (0–2) | 35 (39·8) | 30 (40·5) | 5 (35·7) | 4 (44·4) | 31 (39·2) | 11 (36·7) | 24 (41·4) | 14 (36·8) | 21 (42·0) |
| High (3–5) | 35 (39·8) | 30 (40·5) | 5 (35·7) | 3 (33·3) | 32 (40·5) | 11 (36·7) | 24 (41·4) | 20 (52·6) | 15 (30·0) |
| Not evaluated | 18 (20·5) | 14 (18·9) | 4 (28·6) | 2 (22·2) | 16 (20·3) | 8 (26·7) | 10 (17·2) | 4 (10·5) | 14 (28·0) |
| P = 0·7 (P = 1·0**) | P = 1·0 (P = 1·0) | P = 0·6** (P = 1·0**) | P = 0·05** (P = 0·2**) | ||||||
| ECOG | |||||||||
| 0–1 | 60 (68·2) | 53 (71·6) | 7 (50·0) | 7 (77·8) | 53 (67·1) | 16 (53·3) | 44 (75·9) | 28 (73·7) | 32 (64·0) |
| 2–4 | 11 (12·5) | 9 (12·2) | 2 (14·3) | 0 (0·0) | 11 (13·9) | 5 (16·7) | 6 (10·3) | 6 (15·8) | 5 (10·0) |
| Unknown | 17 (19·3) | 12 (16·2) | 5 (35·7) | 2 (22·2) | 15 (19·0) | 9 (30·0) | 8 (13·8) | 4 (10·5) | 13 (26·0) |
| P = 0·2 (P = 0·6) | P = 0·7 (P = 0·6) | P = 0·1 (P = 0·3) | P = 0·2 (P = 0·6**) | ||||||
| Stage | |||||||||
| I–II | 30 (34·1) | 25 (33·8) | 5 (35·7) | 4 (44·4) | 26 (32·9) | 12 (40·0) | 18 (31·0) | 9 (23·7) | 21 (42·0) |
| III–IV | 58 (65·9) | 49 (66·2) | 9 (64·3) | 5 (55·6) | 53 (67·1) | 18 (60·0) | 40 (69·0) | 29 (76·3) | 29 (58·0) |
| P = 1·0 | P = 0·5 | P = 0·4** | P = 0·07** | ||||||
| Elevated LDH | |||||||||
| No | 14 (15·9) | 11 (14·9) | 3 (21·4) | 1 (11·1) | 13 (16·5) | 4 (13·3) | 10 (17·2) | 7 (18·4) | 7 (14·0) |
| Yes | 67 (76·1) | 57 (77·0) | 10 (71·4) | 7 (77·8) | 60 (76·0) | 23 (76·7) | 44 (75·9) | 29 (76·3) | 38 (76·0) |
| Unknown | 7 (8·0) | 6 (8·1) | 1 (7·1) | 1 (11·1) | 6 (7·6) | 3 (10·0) | 4 (6·9) | 2 (5·3) | 5 (10·0) |
| P = 0·9 (P = 0·7) | P = 0·8 (P = 1·0) | P = 0·8 (P = 0·8) | P = 0·7 (P = 0·6**) | ||||||
| Anthracycline‐based chemo in first line | |||||||||
| No | 35 (39·8) | 29 (39·2) | 6 (42·9) | 6 (66·7) | 29 (36·7) | 12 (40·0) | 23 (39·7) | 16 (42·1) | 19 (38·0) |
| Yes | 44 (50·0) | 37 (50·0) | 7 (50·0) | 2 (22·2) | 42 (53·2) | 14 (46·7) | 30 (51·7) | 19 (50·0) | 25 (50·0) |
| Unknown | 9 (10·2) | 8 (10·8) | 1 (7·1) | 1 (11·1) | 8 (10·1) | 4 (13·3) | 5 (8·6) | 3 (7·9) | 6 (12·0) |
| P = 1·0 (P = 0·9**) | P = 0·2 (P = 0·1) | P = 0·8 (P = 0·8**) | P = 0·9 (P = 0·8**) | ||||||
| Disease type | |||||||||
| AITL | 34 (38·6) | 33 (44·6) | 1 (7·1) | 3 (33·3) | 31 (39·2) | 4 (13·3) | 30 (51·7) | 26 (68·4) | 8 (16·0) |
| NKTCL | 19 (21·6) | 13 (17·6) | 6 (42·9) | 4 (44·4) | 15 (19·0) | 12 (40·0) | 7 (12·1) | 3 (7·9) | 16 (32·0) |
| ALCL, ALK+ve | 5 (5·7) | 5 (6·8) | 0 (0·0) | 0 (0·0) | 5 (6·3) | 2 (6·7) | 3 (5·2) | 0 (0·0) | 5 (10·0) |
| ALCL, ALK−ve | 4 (4·6) | 4 (5·4) | 0 (0·0) | 0 (0·0) | 4 (5·1) | 1 (3·3) | 3 (5·2) | 3 (7·9) | 1 (2·0) |
| MEITL | 12 (13·6) | 10 (13·5) | 2 (14·3) | 2 (22·2) | 10 (12·7) | 6 (20·0) | 6 (10·3) | 3 (7·9) | 9 (18·0) |
| PTCL,NOS | 12 (13·6) | 7 (9·5) | 5 (35·7) | 0 (0·0) | 12 (15·2) | 5 (16·7) | 7 (12·1) | 2 (5·3) | 10 (20·0) |
| CTCL | 2 (2·3) | 2 (2·7) | 0 (0·0) | 0 (0·0) | 2 (2·5) | 0 (0·0) | 2 (3·5) | 1 (2·6) | 1 (2·0) |
| P = 0·01 | P = 0·6 | P = 0·003 | P < 0·001 | ||||||
IQR, interquartile range; IPI, International Prognostic Index; ECOG, Eastern Cooperative Oncology Group; LCH, lactate dehydrogenase; AITL, angioimmunoblastic T‐cell lymphoma; NKTCL, natural killer/T‐cell lymphoma; ALCL, anaplastic large‐cell lymphoma; MEITL, monomorphic epitheliotropic intestinal T‐cell lymphoma; PTCL‐NOS, peripheral T‐cell lymphoma not otherwise specified; CTCL, cutaneous T‐cell lymphoma.
P value in parentheses calculated excluding the categories ‘Not evaluated’ and ‘Unknown’.
P value calculated using Fisher’s exact test unless otherwise stated.
P value calculated using the Mann–Whitney U test.
P value calculated using the chi‐squared test.
Time to event measurements
Duration of follow‐up was measured from the date of diagnosis to date of death or date of last follow‐up for surviving patients. Duration of OS was measured from the date of diagnosis to date of death or date of last follow‐up for surviving patients. Surviving patients were censored at date of last follow‐up. Duration of relapse‐free survival (RFS) was measured from the date of diagnosis to date of first relapse or death (whichever occurs first) or date of last follow‐up. Surviving patients who did not experience a relapse were censored at date of last follow‐up. Patients with date of diagnosis where both the day and month were missing were excluded from the survival analysis.
Immunohistochemistry
Expression of PIK3 p110 (p110α, p110β, p110δ, p110ɤ), phosphatase and tensin homolog (PTEN), phosphorylated Stat3 (pSTAT3) and CD30 was analysed by immunostaining on tissue microarray (TMA) sections (4 µmol/l). Immunohistochemistry staining was performed using a 1:200 dilution for PIK3 p110α and PIK3 p110δ, 1:400 dilution for PIK3 p110β, PIK3 p110ɤ and PTEN, and 1:200 dilution for pSTAT3. Scoring was performed as follows: 0, negative staining; 1+, mild expression; 2+, moderate expression and 3+, strong expression. The slides were all evaluated by a pathology associate. The details of tissue evaluation and immunohistochemistry (IHC) scoring are elaborated in Data S1. For statistical analysis, each of the stains has been classified as low (0, 1+) or high (2+, 3+). The antibodies are listed in Table SII.
Chemicals
Alpelisib (BYL‐719) and idelalisib (CAL‐101) were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Copanlisib was supplied by Bayer AG (Leverkusen, Germany).
Cell viability assays
For each assay, 2000 cells were seeded on a 96‐well plate and treated with indicated concentrations of alpelisib, idelalisib or copanlisib for 72 h as previously described (Nairismagi et al., 2018; Song et al., 2018). Cell viability was measured using the CellTiter‐Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) following manufacturer’s instructions. All experiments were performed in triplicate. Half maximal inhibitory concentration (IC50) values were calculated using graphpad prism (GraphPad Software, San Diego, CA, USA).
Western blot
Western blot was performed as previously described (Song et al., 2018). Briefly, total proteins were extracted, resolved and blotted onto a polyvinylidene difluoride membrane. After blocking, membranes were probed with the primary antibodies listed in Table SII, followed by horseradish peroxidase‐conjugated anti‐mouse or anti‐rabbit secondary antibodies. Signals were visualized using ChemiDoc MP System (Bio‐RAD, Hercules, CA, USA).
Cell cycle assays
Two million cells were seeded on a T‐25 flask and treated with 0·5, 1·0 and 5·0 μmol/l alpelisib, idelalisib or copanlisib for 72 h. The cells were fixed with 70% ethanol and stained with 50 μg/ml propidium iodide (Sigma‐Aldrich, St. Louis, MO, USA). The stained cells were analysed by LSRII (BD Biosciences, San Jose, CA, USA) and quantified using flowjo (v7.2.2) (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
In vivo xenograft studies
All experiments were approved by the SingHealth Institutional Animal Care and Use Committee. Male NOD/SCID/IL‐2rγnull (NSG) mice (The Jackson Laboratory, Bar Harbor, ME, USA) were inoculated subcutaneously with 5 × 106 NKS1 cells. Tumour size and body weight were monitored two to three times per week. When the tumour volume reached 200 mm3, mice were randomised according to tumour size and therapy was started. Copanlisib was dosed intravenously at 25 mg/kg on a Q2D schedule.
Statistical analysis
Patient demographics and clinical characteristics (categorical variables) were summarised as frequency and percentage, and continuous variables were summarised as median with interquartile range (IQR). Comparisons of patient demographics and clinical characteristics by PIK3/AKT strains were performed using Fisher’s exact test or the chi‐squared test for categorical variables (where appropriate) and the Mann–Whitney U test for continuous variables. Overall survival and RFS were estimated by the Kaplan–Meier method and median survival reported with 95% confidence interval (95% CI). The log‐rank test was used to determine if there was a difference in survival between different groups of patients. Univariable Cox proportional‐hazards regression analyses were performed to estimate the hazard ratio (HR) between groups of patients. Patient demographics and clinical characteristics that were associated with survival in a univariable Cox regression analysis with a significance level of P < 0·1 and were clinically meaningful were included in a multivariable Cox regression analysis for backward stepwise model selection. Model selection was performed using the likelihood ratio test with a significance threshold of P < 0·05 for inclusion in the final model. The proportional hazard assumption for using the Cox regression model was assessed using the Schoenfeld residuals test. A two‐sided P value < 0·05 was considered statistically significant. All analyses were performed in stata version 14.2 (StataCorp LLC, College Station, TX, USA).
Results
Prevalence of high PIK3 expression in PTCL and NKTCL
PIK3δhigh was found to be predominant in PTCL and NKTCL (58/88, 66%), followed by PIK3αhigh (14/88, 16%) and PIK3βhigh (8/88, 9%) with PIK3γhigh (3/88, 3%) being the lowest (Fig 1A). No PTEN staining was observed in nine (10%) cases and the expression was weak in 70 (80%) samples. Interestingly, significant differences in expression of PIK3α, PIK3β and PIK3δ isoforms were observed between lymphoma subtypes where the incidence of PIK3αhigh was significantly higher in NKTCL and PTCL‐NOS (P = 0·011), while the incidence of PIK3βhigh was significantly more prevalent in ALCL and PTCL‐NOS (P = 0·015) compared to other subtypes (Fig 1B). PIK3δ was consistently expressed at high levels across all subtypes; however, PIK3δhigh was significantly less prevalent in NKTCL compared to the other subtypes (P = 0·003). These observations suggest that PIK3 isoforms have specific role in subtypes of lymphoma. The representative expression features of PTCL and NKTCL patients are depicted in Fig 2A and Figure S1.
Figure 1.
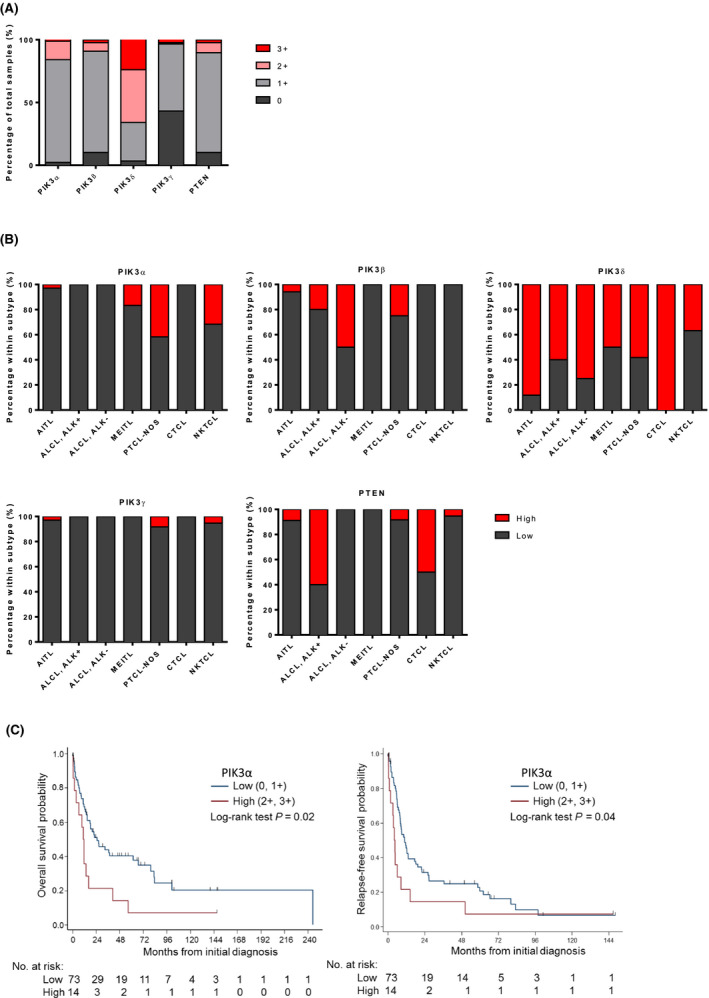
PIK3δhigh was more prevalent in PTCL and NKTCL while PIK3αhigh was more prevalent in NKTCL and PTCL‐NOS compared to other subtypes. (A) PIK3 isoforms and PTEN immunohistochemistry (IHC) staining scores in PTCL and NKTCL tumours. (B) PIK3 isoforms and PTEN IHC staining scores in seven individual PTCL and NKTCL subtypes. (C) Kaplan–Meier curves for (left panel) overall survival and (right panel) relapse‐free survival according to PIK3α status. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 2.
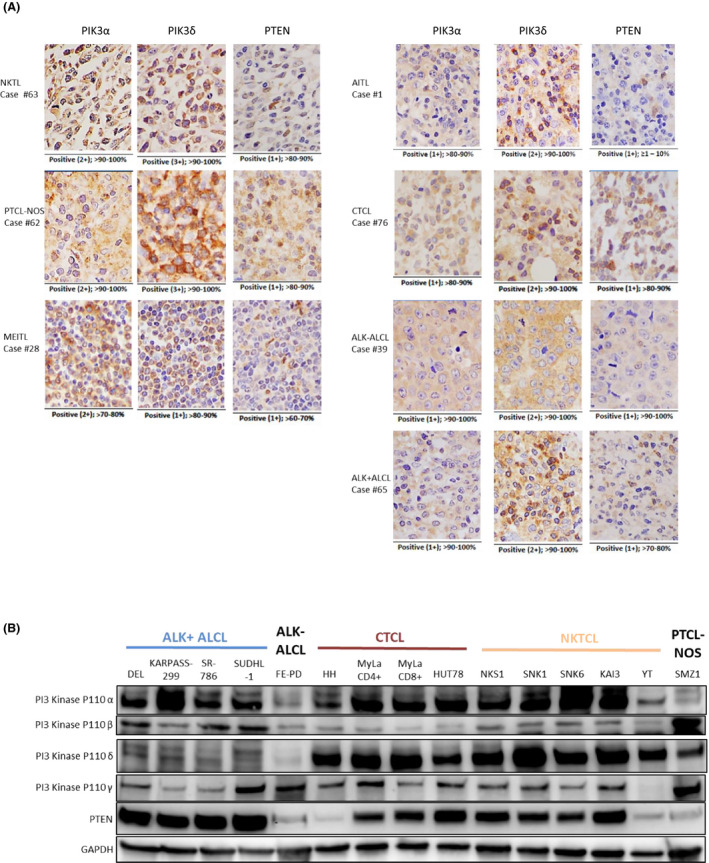
Four PIK3 isoforms and PTEN were expressed in PTCL patients and cell lines. (A) Immunohistochemistry staining of PIK3α, PIK3δ, and PTEN in representative PTCL and NKTCL samples. (B) PIK3 and PTEN protein levels in 15 PTCL and NKTCL cell lines. [Colour figure can be viewed at wileyonlinelibrary.com]
PIK3α expression is associated with overall survival and relapse‐free survival in PTCL and NKTCL patients
No significant associations were observed between PIK3 catalytic subunits (p110α, p110β, p110δ, p110ɤ) with patient characteristics; patient demographics and disease characteristics are presented in Table 1. Loss and low expression of PTEN were associated with older age at diagnosis (P = 0·02). One patient was excluded from the survival analysis as the date of diagnosis was not recorded. With a median follow‐up of 15 months (range: 0·03 to 245·2 months), thirty patients (34%) were alive. In the univariable analysis, PIK3αhigh, older age, high IPI, high ECOG score and absence of anthracycline‐based chemotherapy in first line were significantly associated with inferior OS. In the multivariable analysis, PIK3α, age, IPI, disease type and first‐line anthracycline‐based chemotherapy remained significantly associated with OS. After adjusting for age at diagnosis, IPI score, disease type and first‐line anthracycline‐based chemotherapy, patients with PIK3αhigh expression had an OS hazard ratio of 2·77 (95% CI, 1·41 to 5·46; likelihood ratio P value = 0·006) as compared to patients with PIK3αlow expression. Multivariable OS estimates are presented in Table 2 and Fig 1C, left panel. The median OS of patients with PIK3αlow was 25 months (95% CI: 14·8‐ 67·0 months) compared to 11 months (95% CI: 1·8–16·3 months) in patients with PIK3αhigh.
Table 2.
Univariable and multivariable overall survival analyses.
| OS (Events/Pts = 57/87) | Univariable | Multivariable | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | Adjusted HR (95% CI) | P value | P value (LR test) | |
| PIK3α | |||||
| Low | 1 | 1 | 0·006 | ||
| High | 2·09 (1·12–3·91) | 0·02 | 2·77 (1·41–5·46) | 0·003 | |
| Age at diagnosis | |||||
| ≤60 | 1 | 1 | 0·002 | ||
| >60 | 2·91 (1·64–5·17) | <0·001 | 3·35 (1·53–7·31) | 0·002 | |
| IPI score | |||||
| Low (0–2) | 1 | 1 | 0·006 | ||
| High (3–5) | 3·02 (1·62–5·63) | 0·001 | 2·58 (1·31–5·09) | 0·006 | |
| Not evaluated | 1·81 (0·85–3·83) | 0·1 | 2·85 (1·26–6·45) | 0·01 | |
| Disease type | |||||
| Non‐NKTCL | 1 | 1 | 0·002 | ||
| NKTCL | 2·04 (1·12–3·71) | 0·02 | 3·23 (1·57–6·64) | 0·001 | |
| Anthracycline‐based chemo in first line | |||||
| No | 1 | 1 | 0·01 | ||
| Yes | 0·27 (0·15–0·49) | <0·001 | 0·38 (0·20–0·71) | 0·003 | |
| Unknown | 0·31 (0·07–1·32) | 0·1 | 0·53 (0·11–2·55) | 0·4 | |
| ECOG | |||||
| 0–1 | 1 | ||||
| 2–4 | 4·55 (1·94–10·66) | <0·001 | |||
| Unknown | 1·18 (0·61–2·27) | 0·6 | |||
| Elevated LDH | |||||
| No | 1 | ||||
| Yes | 1·86 (0·87–3·96) | 0·1 | |||
| Unknown | 1·49 (0·39–5·65) | 0·6 | |||
| PTEN | |||||
| Low | 1 | ||||
| High | 0·30 (0·07–1·21) | 0·09 | |||
| Gender | |||||
| Female | 1 | ||||
| Male | 1·67 (0·90–3·11) | 0·1 | |||
| Stage | |||||
| I–II | 1 | ||||
| III–IV | 1·61 (0·88–2·94) | 0·1 | |||
| PIK3‐β | |||||
| Negative | 1 | ||||
| Positive | 0·59 (0·27–1·33) | 0·2 | |||
| PIK3δ | |||||
| Low | 1 | ||||
| High | 0·66 (0·38–1·14) | 0·1 | |||
| PIK3γ | |||||
| Negative | 1 | ||||
| Positive | 1·15 (0·67–1·96) | 0·6 | |||
P value calculated using a Wald test or likelihood ratio test (indicated by LR test). IPI, International Prognostic Index; NKTCL, natural killer/T‐cell lymphoma; ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase.
PIK3δlow and PIK3αhigh, male gender, high IPI high ECOG score and absence of anthracycline‐based chemotherapy in first line were associated with worse RFS in the univariable analysis. In the multivariable analysis, PIK3α, gender, ECOG score, stage and first‐line anthracycline‐based chemotherapy remained significantly associated with RFS. After adjusting for gender, ECOG score, stage and first‐line anthracycline‐based chemotherapy, patients with PIK3αhigh expression had a HR of relapse of 3·07 (95% CI, 1·53 to 6·17; likelihood ratio P value = 0·003) as compared to patients with PIK3αlow expression. Multivariable RFS estimates are presented in Table 3 and Fig 1C, right panel. For RFS, the median survival was 11 months (95% CI: 8·0–17·6 months) for patients with PIK3αlow and four months (95% CI: 1·1–8·6 months) for patients with PIK3αhigh.
Table 3.
Univariable and multivariable relapse‐free survival analyses.
| Relapse‐free survival (Events/Pts = 68/87) | Univariable | Multivariable | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | Adjusted HR (95% CI) | P value | P value (LR test) | |
| PIK3α | |||||
| Low | 1 | 1 | 0·003 | ||
| High | 1·87 (1·02–3·45) | 0·04 | 3·07 (1·53–6·17) | 0·002 | |
| Gender | |||||
| Female | 1 | 1 | 0·01 | ||
| Male | 1·86 (1·06–3·29) | 0·03 | 2·16 (1·16–4·05) | 0·02 | |
| ECOG | |||||
| 0–1 | 1 | 1 | <0·001 | ||
| 2–4 | 8·81 (3·45–22·48) | <0·001 | 11·33 (3·96–32·42) | <0·001 | |
| Unknown | 1·44 (0·81–2·57) | 0·2 | 2·16 (1·06–4·38)* | 0·03 | |
| Stage | |||||
| I–II | 1 | 1 | 0·005 | ||
| III–IV | 1·57 (0·93–2·67) | 0·09 | 2·16 (1·23–3·78) | 0·007 | |
| Anthracycline‐based chemo in first line | |||||
| No | 1 | 1 | <0·001 | ||
| Yes | 0·41 (0·24–0·68) | 0·001 | 0·22 (0·12–0·41) | <0·001 | |
| Unknown | 0·40 (0·12–1·36)* | 0·1 | 0·42 (0·11–1·55)* | 0·2 | |
| Age at diagnosis | |||||
| ≤60 | 1 | ||||
| >60 | 1·57 (0·96–2·56) | 0·07 | |||
| IPI score | |||||
| Low (0–2) | 1 | ||||
| High (3–5) | 2·28 (1·31–3·97) | 0·004 | |||
| Not evaluated | 1·75 (0·92–3·32) | 0·09 | |||
| Disease type | |||||
| Non‐NKTCL | 1 | ||||
| NKTCL | 1·69 (0·96–2·97) | 0·07 | |||
| Elevated LDH | |||||
| No | 1 | ||||
| Yes | 1·94 (0·98–3·83) | 0·06 | |||
| Unknown | 1·41 (0·44–4·54) | 0·6 | |||
| PIK3δ | |||||
| Low | 1 | ||||
| High | 0·59 (0·36–0·97) | 0·04 | |||
| PIK3β | |||||
| Negative | 1 | ||||
| Positive | 0·73 (0·35–1·54) | 0·4 | |||
| PIK3γ | |||||
| Negative | 1 | ||||
| Positive | 1·09 (0·67–1·77) | 0·7 | |||
| PTEN | |||||
| Low | 1 | ||||
| High | 0·67 (0·27–1·68) | 0·4 | |||
P value calculated using Wald test or likelihood ratio test (indicated by LR test). ECOG, Eastern Cooperative Oncology Group; IPI, International Prognostic Index; NKTCL, natural killer/T‐cell lymphoma; LDH, lactate dehydrogenase.
Proportional hazard assumption violated.
Note that PIK3α was preferentially expressed in subsets of PTCL and NKTCL with poor prognosis instead of ALK+ ALCL with superior outcomes. We further evaluated the prognosis value by excluding ALK+ ALCL. After adjusting for the key prognostic factor, PIK3α remained an independent prognostic factor for OS and RFS in the multivariable analysis, indicating that PIK3α might play an important role in disease progression and portend a poor prognosis (Tables SIII, SIV). In this group of patients (excluding ALK+ ALCL cases), the adjusted OS and RFS HR for PIK3αhigh versus PIK3αlow was 2·68 (95% CI, 1·35 to 5·30; likelihood ratio P value = 0·008) and 3·38 (95% CI, 1·65 to 6·91; likelihood ratio P value = 0·002) respectively.
Simultaneous inhibition of PIK3α/δ using copanlisib is essential for potent and broad anti‐tumour activity in PTCL and NKTCL independent of subtype
Since PIK3α was prognostic of OS and RFS and PIK3δ was highly expressed in PTCL and NKTCL (Fig 2A), we proposed the rationale of testing PIK3α and/or PIK3δ inhibitors in PTCL and NKTCL cell lines. Before evaluating the efficacy of PIK3 inhibitors, we first screened the expression levels of four PIK3 isoforms and PTEN in 15 NKTCL and PTCL cell lines representing ALCL, PTCL‐NOS and CTCL subtypes, using Western blot analysis. The results showed that PIK3α protein level was high in CTCL and NKTCL cell lines, low in ALK+ ALCL and absent in a PTCL‐NOS cell line (SMZ1). PIK3β and γ isoforms were evenly expressed in all PTCL cell lines. Interestingly, PIK3δ protein was much lower in ALK+ ALCL cell lines (DEL, KAPRAS‐229, SUDHL‐1 and SUDHL) than in the remaining PTCL cell lines. PTEN expression level was heterogeneous even in the same subtype of cell lines (Fig 2B). In summary, the Western blot result showed that all of the PTCL cell lines examined expressed PIK3α or/and PIK3δ suggesting they might respond to PIK3 inhibitor treatment.
Therefore, we proceeded to evaluate IC50 of PIK3α‐selective inhibitor alpelisib (BYL‐719), PIK3δ‐specific inhibitor idelalisib (CAL‐101) and PIK3α/δ‐predominant inhibitor copanlisib (BAY 80‐6946) in nine selected PTCL cell lines. The PTCL cell lines were treated with increasing concentrations of the respective PIK3 inhibitors for 72 h, the cell viability was measured and the IC50 was calculated by non‐linear regression. These three PIK3 inhibitors revealed differential activity profiles: copanlisib demonstrated the most potent activity (IC50 in the range 0·14–1·68 μmol/l) while alpelisib (IC50 in the range 0–111 μmol/l) and idelalisib (IC50 in the range 6–49 μmol/l) revealed only moderate to no cytotoxic effect (Fig 3A and Figure S2). Since PIK3α and PIK3δ are known to activate the AKT‐mTOR pathway (Heavican et al., 2019), we analysed the phosphorylation status of AKT S473 (pAKTS473) and EIF4EBP1 (pEIF4EBP1) in response to 0·5, 1·0 and 5·0 µmol/l of PIK3 inhibitors in three NKTCL cell lines, SNK6, NKS1 and YT. The results showed that copanlisib had a greater effect on inhibiting pAKTS473 and pEIF4EBP1 than alpelisib and idelalisib. Interestingly, a dose‐dependent decrease in pSTAT3 was also observed, which was greater in copanlisib than in alpelisib and idelalisib (Fig 3B). These results indicate that the pharmacological inhibition of both PIK3α and PIK3δ isoforms using pan‐PIK3 inhibitor was more cytostatic and effective in inhibiting the AKT‐mTOR pathway than single isoform inhibition in PTCL across all subtypes.
Figure 3.
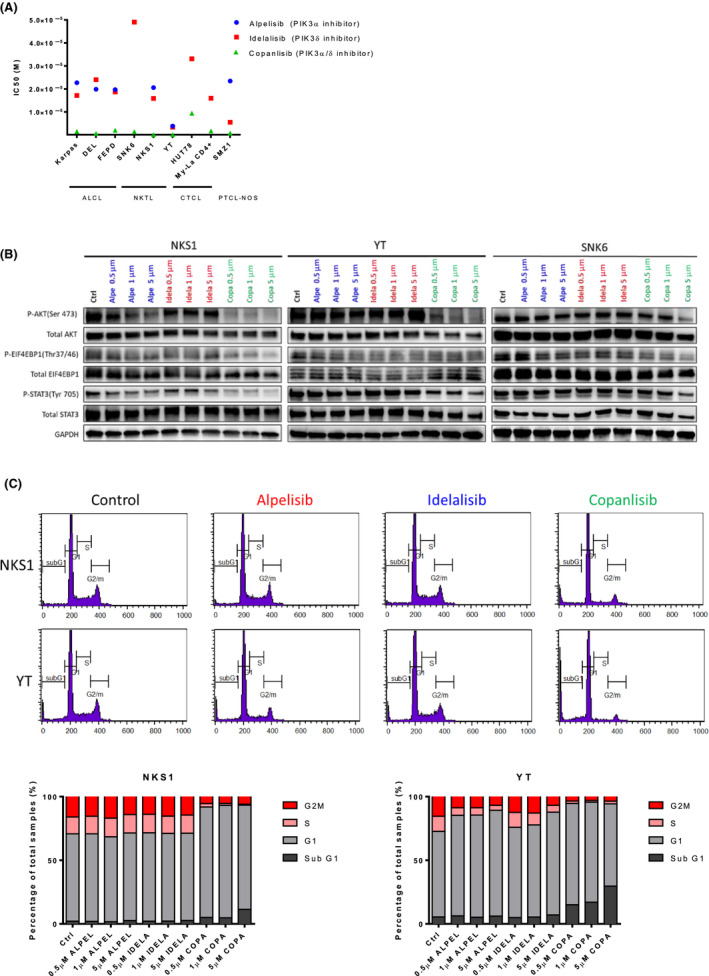
The efficacy of PIK3 inhibitors on PTCL and NKTCL cell lines. (A) The IC50 of Alpelisib, Idelalisib and Copanlisib in nine PCTL and NKTCL cell lines. (B) Inhibition of the PIK3 pathway was assessed by p‐AKT (S473), p‐EIF4EBP1 (S65) and p‐STAT3 in NKS1 and YT cells. (C) The effect of alpelisib, idelalisib and copanlisib on cell cycle arrest in NKS1 and YT cells. [Colour figure can be viewed at wileyonlinelibrary.com]
We further analysed the anti‐proliferative activity of PIK3 inhibitors on NKS1 and YT cells. The cells were treated with 0·5, 1·0 and 5·0 µmol/l of PIK3 inhibitors for 72 h followed by ethanol fixation and propidium iodide staining. The cell cycles were then determined by flow cytometry. The results showed that copanlisib had a greater effect on cell cycle arrest at the G0/G1 phase than alpelisib and idelalisib (Fig 3C). There was no observable increase in apoptosis after PIK3 inhibitor treatment. Indeed, a marginal reduction in cell viability was observed only at 5·0 µmol/l of copanlisib treatment. Consistent with cell cycle analysis results, 0·5 µmol/l of copanlisib treatment but not alpelisib and idelalisib abolished cell proliferation in NKS1 and YT cells (Fig 4A). These observations suggested that copanlisib could effectively inhibit mTOR and STAT3 signalling pathways leading to cell cycle arrest at G0/G1, resulting in proliferation suppression in NKTCL.
Figure 4.
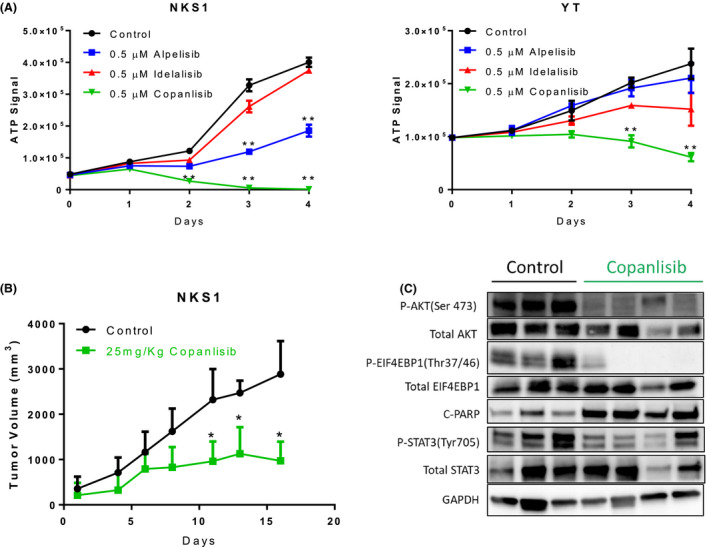
Copanlisib inhibited NKTCL tumour cell growth in vitro and in vivo. (A) 0·5 µM of copanlisib but not alpelisib, idelalisib abolished NKS1 and YT cell proliferation. (B) Tumour growth curves of the NKS1 xenograft model. treatment with 25 mg/kg copanlisib significantly decreases tumour growth compared with vehicle‐control‐only treated mice. (C) Copanlisib inhibited phosphorylation of AKT, EIF4EBP1 and STAT3, and increased cleaved‐PAPRP protein level in NKS1‐engrafted tumours. Error bars indicate the standard error of the mean. *, P < 0·05, **, P < 0·01. [Colour figure can be viewed at wileyonlinelibrary.com]
Copanlisib treatment resulted in tumour growth inhibition in the NKTCL xenograft model
The in vivo efficacy of copanlisib was determined using our established xenograft model. Treatment of the NKS1 xenograft model with 25 mg/kg of copanlisib resulted in a significant decrease in tumour growth and reduced tumour weight (P < 0·01) (Fig 4B, Figure S3). Consistent with the in vitro results, treatment of NKS1 with copanlisib also led to inhibition of AKT Ser473, EIF4EBP1 and STAT3 phosphorylation (Fig 4C). Interestingly, the cleaved PARP protein level was also significantly increased after copanlisib treatment, which was not observed in vitro. These data suggested that copanlisib might be a potent inhibitor for the treatment of NKTCL.
Discussion
By means of IHC in patient tumour samples, we demonstrated that PIK3δ is highly expressed in PTCL and NKTCL independent of lymphoma subtype. PIK3δ has been described to play a role in the differentiation, proliferation and survival of haematopoietic cells, including myeloid cells, B cells, and T cells through tumour cell‐autonomous effects (Okkenhaug et al., 2002; Ali et al., 2004). Mice with inactivated p110δ are resistant to tumourigenesis, and p110δ inactivation in Treg is necessary to confer this tumour resistance, suggesting that p110δ plays a role in mediating anti‐tumour immunity (Ali et al., 2014). High expression of PIK3δ in PTCL has been previously reported, confirming the results of our study. Sanchez et al. reported that the PIK3CD gene was overexpressed in ALCL and CTCL cell lines and primary samples, and correlated with survival pathways such as the T‐cell receptor, nuclear factor‐κB and CD40 pathways (Martin‐Sanchez et al., 2013). In addition, we observed that PIK3α might play an important role in disease progression and portend a poor prognosis in PTCL and NKTCL. Mutations in PIK3CA, the gene encoding p110α, are commonly seen in mantle cell lymphoma (MCL) and diffuse large B‐cell lymphoma, further indicating its importance in these malignancies (Abubaker et al., 2007).
In our study, we further demonstrated that simultaneous inhibition of PIK3α and PIK3δ isoforms the using PIK3δ and PIK3α dual‐inhibitor copanlisib was more efficacious than inhibiting either isoform alone for the treatment of PTCL and NKTCL. In vitro, cell lines were arrested at the G0/G1 phase and none of them underwent apoptosis. Overall, the response was mainly cytostatic, as widely reported (Foukas et al., 2010; Martin‐Sanchez et al., 2013). Similar to other studies, our results confirmed that the inhibition of PIK3α or PIK3δ alone was not sufficient to reduce PTCL and NKTCL cell survival. Genetic and pharmacological inhibition of PIK3δ using CAL‐101 had no effects on the survival of ALCL and CTCL cell lines (Martin‐Sanchez et al., 2013). In the study of Horwitz et al., copanlisib induced more growth inhibition in PTCL cell lines than duvelisib, a PIK3γδ‐specific inhibitor, while idelalisib had no impact on growth inhibition as assessed by GR50 (50% growth reduction compared with day 0) (Horwitz et al., 2018). In primary CLL cells, copanlisib and idelalisib treatment revealed IC50 values of 450 nmol/l and >10 µmol/l, respectively (Gockeritz et al., 2015). Treatment of MCL cell lines and primary samples with the PIK3110α/δ inhibitor pictilisib, was more effective at blocking activation of the downstream BCR pathway and inducing apoptosis than the use of idelalisib alone (Iyengar et al., 2013). A plausible explanation could be that PIK3δ inhibition induced feedback activation of the PIK3α isoform (Pongas et al., 2017). In contrast, Katsuya demonstrated that idelalisib promoted apoptosis in ex vivo adult T‐cell lymphoma (ATL) samples and blocked the CCL22 pathway involved in the survival of ATL cells in part through the PIK3/AKT pathway (Katsuya et al., 2018). Copanlisib, which was recently approved by the FDA for the treatment of relapsed FL, has been reported to have promising efficacy and manageable toxicity in patients with relapsed or refractory indolent or aggressive lymphoma including PTCL. Among the PTCL patients, 14/32 were eligible for response assessment: two (14·3%) patients obtained a CR and one (7·1%) patient a partial response (PR), yielding an ORR of 21·4% (given that the ORR for the whole aggressive cohort was 29·4%) (Dreyling et al., 2017).
Interestingly, we found that STAT3 phosphorylation was significantly suppressed upon PIK3 inhibitor treatment in PTCL and NKTCL cell lines. This is of relevance as our study and others have previously shown that alterations in the JAK/STAT signalling pathway are highly prevalent in PTCL and NKTCL in which STAT3 is one of the most frequently mutated genes (Song et al., 2018). Cooperation between the PIK3/AKT and the JAK/STAT pathways has been extensively described in many types of cells. In mouse fibroblasts, activation of STAT3 by PIK3 appears to be mediated by a member of the Tec kinase family (Hart et al., 2011). In non‐small cell lung cancer, pharmacological or genetic inhibition of PIK3/AKT signalling induced compensatory activation of STAT3 and upregulation of the expression of its downstream genes. (Bian et al., 2018) In B‐cell lymphomas, constitutive activation of STAT3 depends on upstream signalling through PIK3 (Han et al., 2010). Recently, we have demonstrated that activated STAT3 binds to the promoter of PD‐L1 and is a direct regulator of PD‐L1 (Song et al., 2018). Copanlisib might be a potential immune‐modulating drug that could combine with immune checkpoint inhibitors such as PD1 or PD‐L1 inhibitors.
In conclusion, our findings have clinical implications in PTCL and NKTCL and suggest that both PIK3δ and PIK3α would need to be pharmacologically targeted to achieve maximal clinical activity.
Funding
This work was supported by research funding from the National Medical Research Council of Singapore (MOHIAFCat1‐11015 and NMRC‐OFIRG16NOV090), Pharmaceuticals, Bayer AG, TANOTO Foundation (NRDUKST18101), LING Foundation (NRDUKSN18101) and New Century Foundation (NCCRF‐YR2014‐SEP‐SD2).
Author contributions
DC and TL designed the analysis and wrote the paper; ML, YL, WL, DM, EK, GC and TJ collected the data; SH, JQ and BC analyzed the data; JY, TP and NS provided the clinical samples; CL evaluated the IHC results; OP, NL, ST and CK supervised the project.
Conflicts of interest
OCK received research funding from Pharmaceuticals, Bayer AG. The remaining authors declare to have no conflicts of interest.
Ethical approval and consent to participate
The human subject study was approved by the SingHealth Centralised Institutional Review Board, study number 2004/407/F. The animal study was approved by SingHealth Instructions Animal Care and Use Committee (IACUC). The study number is 2018/SHS/1423.
Supporting information
Figure S1. Immunohistochemistry (IHC) scores for 88 cases. Scoring was performed as follows: 0, negative staining; 1+, mild expression; 2+, moderate expression and 3+, strong expression.
Figure S2 . Half maximal inhibitory concentration (IC50) curves of PIK3 inhibitors on peripheral T‐cell lymphoma (PTCL) and NK/T‐cell lymphoma (NKTCL) cell lines. Alpelisib, idelalisib and copanlisib 72‐h dose–response curve and IC50 analysis of PTCL and NKTCL cell lines. Error bars indicate the standard error of the mean.
Figure S3 . Treatment with 25 mg/kg of copanlisib significantly reduced NKS1 xenograft tumour size (left panel) and tumour weight (right panel). Bar: 1 cm, **: P < 0.01. Error bars indicate the standard error of the mean.
Table SI. List of cell lines in this study.
Table SII. List of antibodies. IHC, immunohistochemistry; WB, Western blot.
Table SIII. Univariable and multivariable overall survival analyses excluding ALK+ ALCL cases.
Table SIV. Univariable and multivariable relapse‐free survival analyses excluding ALK+ ALCL cases.
Table SV. Sensitivity analyses of survival differences between patients with known and unknown values of International Prognostic Index (IPI) score, Eastern Cooperative Oncology Group (ECOG) score, lactate dehydrogenase (LDH) and anthracycline‐based chemo in first line.
Table SVI. IC50 of alpelisib, idelalisib and copanlisib in the peripheral T‐cell lymphoma (PTCL) cell lines.
Data S1 . Supplementary methods.
Acknowledgements
We thank all the participants in the study, Jeslin Chian Hung Ha, Rebecca Kee, Khoo Lay Poh from the Division of Medical Oncology, National Cancer Centre Singapore and the SingHealth Tissue Repository for their great assistance in collecting and collating samples and clinical data from patients.
Contributor Information
Soon Thye Lim, Email: lim.soon.thye@singhealth.com.sg.
Choon Kiat Ong, Email: cmrock@nccs.com.sg.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
References
- Abubaker, J. , Bavi, P.P. , Al‐Harbi, S. , Siraj, A.K. , Al‐Dayel, F. , Uddin, S. & Al‐Kuraya, K. (2007) PIK3CA mutations are mutually exclusive with PTEN loss in diffuse large B‐cell lymphoma. Leukemia, 21, 2368–2370. [DOI] [PubMed] [Google Scholar]
- Ali, K. , Bilancio, A. , Thomas, M. , Pearce, W. , Gilfillan, A.M. , Tkaczyk, C. , Kuehn, N. , Gray, A. , Giddings, J. , Peskett, E. , Fox, R. , Bruce, I. , Walker, C. , Sawyer, C. , Okkenhaug, K. , Finan, P. & Vanhaesebroeck, B. (2004) Essential role for the p110delta phosphoinositide 3‐kinase in the allergic response. Nature, 431, 1007–1011. [DOI] [PubMed] [Google Scholar]
- Ali, K. , Soond, D.R. , Pineiro, R. , Hagemann, T. , Pearce, W. , Lim, E.L. , Bouabe, H. , Scudamore, C.L. , Hancox, T. , Maecker, H. , Friedman, L. , Turner, M. , Okkenhaug, K. & Vanhaesebroeck, B. (2014) Inactivation of PI(3)K p110delta breaks regulatory T‐cell‐mediated immune tolerance to cancer. Nature, 510, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, C. , Liu, Z. , Li, D. & Zhen, L. (2018) PI3K/AKT inhibition induces compensatory activation of the MET/STAT3 pathway in non‐small cell lung cancer. Oncology Letters, 15, 9655–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachly, J.S. & Baiocchi, R.A. (2014) Targeting PI3‐kinase (PI3K), AKT and mTOR axis in lymphoma. British Journal of Haematology, 167, 19–32. [DOI] [PubMed] [Google Scholar]
- Coiffier, B. , Federico, M. , Caballero, D. , Dearden, C. , Morschhauser, F. , Jager, U. , Trumper, L. , Zucca, E. , Gomes da Silva, M. , Pettengell, R. , Weidmann, E. , d'Amore, F. , Tilly, H. & Zinzani, P.L. (2014) Therapeutic options in relapsed or refractory peripheral T‐cell lymphoma. Cancer Treatment Reviews, 40, 1080–1088. [DOI] [PubMed] [Google Scholar]
- Dreyling, M. , Morschhauser, F. , Bouabdallah, K. , Bron, D. , Cunningham, D. , Assouline, S.E. , Verhoef, G. , Linton, K. , Thieblemont, C. , Vitolo, U. , Hiemeyer, F. , Giurescu, M. , Garcia‐Vargas, J. , Gorbatchevsky, I. , Liu, L. , Koechert, K. , Pena, C. , Neves, M. , Childs, B.H. & Zinzani, P.L. (2017) Phase II study of copanlisib, a PI3K inhibitor, in relapsed or refractory, indolent or aggressive lymphoma. Annals of Oncology, 28, 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman, J.A. (2009) Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nature Reviews Cancer, 9, 550–562. [DOI] [PubMed] [Google Scholar]
- Flinn, I.W. , O'Brien, S. , Kahl, B. , Patel, M. , Oki, Y. , Foss, F.F. , Porcu, P. , Jones, J. , Burger, J.A. , Jain, N. , Kelly, V.M. , Allen, K. , Douglas, M. , Sweeney, J. , Kelly, P. & Horwitz, S. (2018) Duvelisib, a novel oral dual inhibitor of PI3K‐delta, gamma, is clinically active in advanced hematologic malignancies. Blood, 131, 877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foukas, L.C. , Berenjeno, I.M. , Gray, A. , Khwaja, A. & Vanhaesebroeck, B. (2010) Activity of any class IA PI3K isoform can sustain cell proliferation and survival. Proceedings of the National Academy of Sciences, USA, 107, 11381–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gockeritz, E. , Kerwien, S. , Baumann, M. , Wigger, M. , Vondey, V. , Neumann, L. , Landwehr, T. , Wendtner, C.M. , Klein, C. , Liu, N. , Hallek, M. , Frenzel, L.P. & Krause, G. (2015) Efficacy of phosphatidylinositol‐3 kinase inhibitors with diverse isoform selectivity profiles for inhibiting the survival of chronic lymphocytic leukemia cells. International Journal of Cancer, 137, 2234–2242. [DOI] [PubMed] [Google Scholar]
- Gopal, A.K. , Kahl, B.S. , de Vos, S. , Wagner‐Johnston, N.D. , Schuster, S.J. , Jurczak, W.J. , Flinn, I.W. , Flowers, C.R. , Martin, P. , Viardot, A. , Blum, K.A. , Goy, A.H. , Davies, A.J. , Zinzani, P.L. , Dreyling, M. , Johnson, D. , Miller, L.L. , Holes, L. , Li, D. , Dansey, R.D. , Godfrey, W.R. & Salles, G.A. (2014) PI3Kdelta inhibition by idelalisib in patients with relapsed indolent lymphoma. New England Journal of Medicine, 370, 1008–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S.S. , Yun, H. , Son, D.J. , Tompkins, V.S. , Peng, L. , Chung, S.T. , Kim, J.S. , Park, E.S. & Janz, S. (2010) NF‐kappaB/STAT3/PI3K signaling crosstalk in iMyc E mu B lymphoma. Molecular Cancer, 9, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, J.R. , Liao, L. , Yates, J.R. 3rd & Vogt, P.K. (2011) Essential role of Stat3 in PI3K‐induced oncogenic transformation. Proceedings of the National Academy of Sciences, USA, 108, 13247–13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavican, T.B. , Bouska, A. , Yu, J. , Lone, W. , Amador, C. , Gong, Q. , Zhang, W. , Li, Y. , Dave, B.J. , Nairismagi, M.L. , Greiner, T.C. , Vose, J. , Weisenburger, D.D. , Lachel, C. , Wang, C. , Fu, K. , Stevens, J.M. , Lim, S.T. , Ong, C.K. , Gascoyne, R.D. , Missiaglia, E. , Lemonnier, F. , Haioun, C. , Hartmann, S. , Pedersen, M.B. , Laginestra, M.A. , Wilcox, R.A. , Teh, B.T. , Yoshida, N. , Ohshima, K. , Seto, M. , Rosenwald, A. , Ott, G. , Campo, E. , Rimsza, L.M. , Jaffe, E.S. , Braziel, R.M. , d'Amore, F. , Inghirami, G. , Bertoni, F. , de Leval, L. , Gaulard, P. , Staudt, L.M. , McKeithan, T.W. , Pileri, S. , Chan, W.C. & Iqbal, J. (2019) Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T‐cell lymphoma. Blood, 133, 1664–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, J.Y. , Hong, M.E. , Choi, M.K. , Chang, W. , Do, I.G. , Jo, J.S. , Jung, S.H. , Park, S. , Kim, S.J. , Ko, Y.H. & Kim, W.S. (2015) The clinical significance of activated p‐AKT expression in peripheral T‐cell lymphoma. Anticancer Research, 35, 2465–2474. [PubMed] [Google Scholar]
- Horwitz, S.M. , Advani, R.H. , Bartlett, N.L. , Jacobsen, E.D. , Sharman, J.P. , O'Connor, O.A. , Siddiqi, T. , Kennedy, D.A. & Oki, Y. (2014) Objective responses in relapsed T‐cell lymphomas with single‐agent brentuximab vedotin. Blood, 123, 3095–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz, S.M. , Koch, R. , Porcu, P. , Oki, Y. , Moskowitz, A. , Perez, M. , Myskowski, P. , Officer, A. , Jaffe, J.D. , Morrow, S.N. , Allen, K. , Douglas, M. , Stern, H. , Sweeney, J. , Kelly, P. , Kelly, V. , Aster, J.C. , Weaver, D. , Foss, F.M. & Weinstock, D.M. (2018) Activity of the PI3K‐delta, gamma inhibitor duvelisib in a phase 1 trial and preclinical models of T‐cell lymphoma. Blood, 131, 888–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar, S. , Clear, A. , Bodor, C. , Maharaj, L. , Lee, A. , Calaminici, M. , Matthews, J. , Iqbal, S. , Auer, R. , Gribben, J. & Joel, S. (2013) P110alpha‐mediated constitutive PI3K signaling limits the efficacy of p110delta‐selective inhibition in mantle cell lymphoma, particularly with multiple relapse. Blood, 121, 2274–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl, B.S. , Spurgeon, S.E. , Furman, R.R. , Flinn, I.W. , Coutre, S.E. , Brown, J.R. , Benson, D.M. , Byrd, J.C. , Peterman, S. , Cho, Y. , Yu, A. , Godfrey, W.R. & Wagner‐Johnston, N.D. (2014) A phase 1 study of the PI3Kdelta inhibitor idelalisib in patients with relapsed/refractory mantle cell lymphoma (MCL). Blood, 123, 3398–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuya, H. , Cook, L.B.M. , Rowan, A.G. , Satou, Y. , Taylor, G.P. & Bangham, C.R.M. (2018) Phosphatidylinositol 3‐kinase‐delta (PI3K‐delta) is a potential therapeutic target in adult T‐cell leukemia‐lymphoma. Biomarker Research, 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Sanchez, E. , Rodriguez‐Pinilla, S.M. , Sanchez‐Beato, M. , Lombardia, L. , Dominguez‐Gonzalez, B. , Romero, D. , Odqvist, L. , Garcia‐Sanz, P. , Wozniak, M.B. , Kurz, G. , Blanco‐Aparicio, C. , Mollejo, M. , Alves, F.J. , Menarguez, J. , Gonzalez‐Palacios, F. , Rodriguez‐Peralto, J.L. , Ortiz‐Romero, P.L. , Garcia, J.F. , Bischoff, J.R. & Piris, M.A. (2013) Simultaneous inhibition of pan‐phosphatidylinositol‐3‐kinases and MEK as a potential therapeutic strategy in peripheral T‐cell lymphomas. Haematologica, 98, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairismagi, M. , Gerritsen, M.E. , Li, Z.M. , Wijaya, G.C. , Chia, B.K.H. , Laurensia, Y. , Lim, J.Q. , Yeoh, K.W. , Yao, X.S. , Pang, W.L. , Bisconte, A. , Hill, R.J. , Bradshaw, J.M. , Huang, D. , Song, T.L.L. , Ng, C.C.Y. , Rajasegaran, V. , Tang, T. , Tang, Q.Q. , Xia, X.J. , Kang, T.B. , Teh, B.T. , Lim, S.T. , Ong, C.K. & Tan, J. (2018) Oncogenic activation of JAK3‐STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T‐cell lymphoma. Leukemia, 32, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug, K. , Bilancio, A. , Farjot, G. , Priddle, H. , Sancho, S. , Peskett, E. , Pearce, W. , Meek, S.E. , Salpekar, A. , Waterfield, M.D. , Smith, A.J. & Vanhaesebroeck, B. (2002) Impaired B and T cell antigen receptor signaling in p110delta PI 3‐kinase mutant mice. Science, 297, 1031–1034. [DOI] [PubMed] [Google Scholar]
- Patnaik, A. , Appleman, L.J. , Tolcher, A.W. , Papadopoulos, K.P. , Beeram, M. , Rasco, D.W. , Weiss, G.J. , Sachdev, J.C. , Chadha, M. , Fulk, M. , Ejadi, S. , Mountz, J.M. , Lotze, M.T. , Toledo, F.G. , Chu, E. , Jeffers, M. , Pena, C. , Xia, C. , Reif, S. , Genvresse, I. & Ramanathan, R.K. (2016) First‐in‐human phase I study of copanlisib (BAY 80–6946), an intravenous pan‐class I phosphatidylinositol 3‐kinase inhibitor, in patients with advanced solid tumors and non‐Hodgkin's lymphomas. Annals of Oncology, 27, 1928–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak, R. & Buitenhuis, M. (2012) The PI3K/PKB signaling module as key regulator of hematopoiesis: implications for therapeutic strategies in leukemia. Blood, 119, 911–923. [DOI] [PubMed] [Google Scholar]
- Pongas, G.N. , Annunziata, C.M. & Staudt, L.M. (2017) PI3Kdelta inhibition causes feedback activation of PI3Kalpha in the ABC subtype of diffuse large B‐cell lymphoma. Oncotarget, 8, 81794–81802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel, C. , Camps, M. & Ji, H. (2007) PI3K delta and PI3K gamma: partners in crime in inflammation in rheumatoid arthritis and beyond? Nature Reviews Immunology, 7, 191–201. [DOI] [PubMed] [Google Scholar]
- Song, T.L. , Nairismagi, M.L. , Laurensia, Y. , Lim, J.Q. , Tan, J. , Li, Z.M. , Pang, W.L. , Kizhakeyil, A. , Wijaya, G.C. , Huang, D.C. , Nagarajan, S. , Chia, B.K. , Cheah, D. , Liu, Y.H. , Zhang, F. , Rao, H.L. , Tang, T. , Wong, E.K. , Bei, J.X. , Iqbal, J. , Grigoropoulos, N.F. , Ng, S.B. , Chng, W.J. , Teh, B.T. , Tan, S.Y. , Verma, N.K. , Fan, H. , Lim, S.T. & Ong, C.K. (2018) Oncogenic activation of the STAT3 pathway drives PD‐L1 expression in natural killer/T‐cell lymphoma. Blood, 132, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vose, J. , Armitage, J. , Weisenburger, D. & International lymphoma T‐cell Project (2008) International peripheral T‐cell and natural killer/T‐cell lymphoma study: pathology findings and clinical outcomes. Journal of Clinical Oncology, 26, 4124–4130. [DOI] [PubMed] [Google Scholar]
- Westin, J.R. (2014) Status of PI3K/Akt/mTOR pathway inhibitors in lymphoma. Clinical Lymphoma Myeloma and Leukemia, 14, 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Immunohistochemistry (IHC) scores for 88 cases. Scoring was performed as follows: 0, negative staining; 1+, mild expression; 2+, moderate expression and 3+, strong expression.
Figure S2 . Half maximal inhibitory concentration (IC50) curves of PIK3 inhibitors on peripheral T‐cell lymphoma (PTCL) and NK/T‐cell lymphoma (NKTCL) cell lines. Alpelisib, idelalisib and copanlisib 72‐h dose–response curve and IC50 analysis of PTCL and NKTCL cell lines. Error bars indicate the standard error of the mean.
Figure S3 . Treatment with 25 mg/kg of copanlisib significantly reduced NKS1 xenograft tumour size (left panel) and tumour weight (right panel). Bar: 1 cm, **: P < 0.01. Error bars indicate the standard error of the mean.
Table SI. List of cell lines in this study.
Table SII. List of antibodies. IHC, immunohistochemistry; WB, Western blot.
Table SIII. Univariable and multivariable overall survival analyses excluding ALK+ ALCL cases.
Table SIV. Univariable and multivariable relapse‐free survival analyses excluding ALK+ ALCL cases.
Table SV. Sensitivity analyses of survival differences between patients with known and unknown values of International Prognostic Index (IPI) score, Eastern Cooperative Oncology Group (ECOG) score, lactate dehydrogenase (LDH) and anthracycline‐based chemo in first line.
Table SVI. IC50 of alpelisib, idelalisib and copanlisib in the peripheral T‐cell lymphoma (PTCL) cell lines.
Data S1 . Supplementary methods.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.


