Abstract
To date, the recently discovered SARS‐CoV‐2 virus has afflicted >6.9 million people worldwide and disrupted the global economy. Development of effective vaccines or treatments for SARS‐CoV‐2 infection will be aided by a molecular‐level understanding of SARS‐CoV‐2 proteins and their interactions with host cell proteins. The SARS‐CoV‐2 nucleocapsid (N) protein is highly homologous to the N protein of SARS‐CoV, which is essential for viral RNA replication and packaging into new virions. Emerging models indicate that nucleocapsid proteins of other viruses can form biomolecular condensates to spatiotemporally regulate N protein localization and function. Our bioinformatic analyses, in combination with pre‐existing experimental evidence, suggest that the SARS‐CoV‐2 N protein is capable of forming or regulating biomolecular condensates in vivo by interaction with RNA and key host cell proteins. We discuss multiple models, whereby the N protein of SARS‐CoV‐2 may harness this activity to regulate viral life cycle and host cell response to viral infection.
Keywords: enveloped viruses, liquid‐liquid phase separation, low‐complexity domain, stress granule, viral capsid
Abbreviations
- ERGIC
ER‐Golgi intermediate compartment
- LCD
low‐complexity domain
- LLPS
liquid‐liquid phase separation
- N
nucleocapsid
- RNP
ribonucleoprotein
- SR‐domain
serine/arginine domain
- UPR
unfolded protein response
1. INTRODUCTION
SARS‐CoV‐2 has recently emerged as the seventh coronavirus known to infect humans. 1 Since its discovery, SARS‐CoV‐2 has resulted in >6.9 million documented human infections worldwide and >400 000 deaths reported to date, according to the World Health Organization (https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019/situation‐reports; accessed on 6/9/20). Given the magnitude of this ongoing pandemic, a molecular‐level understanding of SARS‐CoV‐2 infection and host interaction is of paramount importance for rational drug development. The nucleocapsid (“N”) protein of the closely related SARS‐CoV 2 , 3 is essential for the formation of new virions and is the most common antigen of host‐produced antibodies during infection by the closely related SARS‐CoV virus, 4 and the SARS‐CoV‐2 N protein is considered a promising molecular target for effective drug treatments and vaccines. 5 , 6 We hypothesize that the N protein of SARS‐CoV‐2 is capable of forming and altering biomolecular condensates, which may fulfill multiple roles in regulating viral replication and host cell response during infection. The N protein of SARS‐CoV‐2 contains all of the hallmark features of proteins known to form biomolecular condensates in vivo, including multiple domains capable of binding RNA, multiple low‐complexity regions, an oligomerization domain that mediates N‐N homotypic interactions, a high predicted phase separation propensity, and direct physical interactions with multiple stress granule components. These features may be involved in the regulation of host cell biomolecular condensates (namely, stress granules) as well as the formation of ribonucleoprotein (RNP) condensates during viral RNA genome packaging into nascent virions. In the ensuing sections, we discuss the evidence for and implications of this hypothesis.
2. THE SARS‐CoV‐2 N PROTEIN CONTAINS A MODULAR DOMAIN ARCHITECTURE CHARACTERISTIC OF PROTEINS RECRUITED TO BIOMOLECULAR CONDENSATES
The N protein of SARS‐CoV‐2 is a putative RNA‐binding protein responsible for assembling the viral RNA genome into compact RNP complexes for encapsulation in the viral membrane. Sequence alignment of human coronavirus N proteins indicates that the SARS‐CoV‐2 N protein closely resembles the SARS‐CoV N protein (and, to a lesser extent, the MERS‐CoV N protein) but not other human coronavirus N proteins (Figure 1A). The high level of sequence alignment between the N proteins of SARS‐CoV‐2 and SARS‐CoV suggests that these proteins exhibit substantial overlap in overall domain organization, structure, and function. Therefore, many of the features governing N protein activity in SARS‐CoV may extrapolate to SARS‐CoV‐2 N protein.
FIGURE 1.
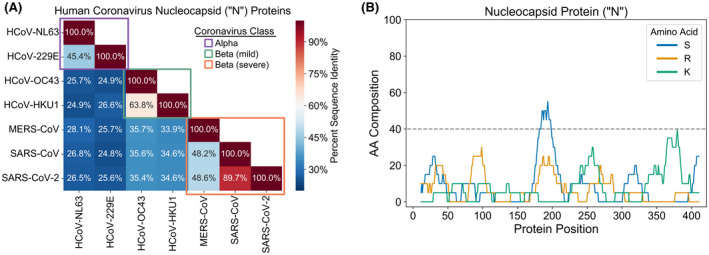
The N protein of SARS‐CoV‐2 resembles the SARS‐CoV N protein and contains multiple LCDs. A, Multiple sequence alignment of N proteins from the seven coronavirus strains known to infect humans. B, Composition scan depicting the local S, R, and K content of the N protein sequence from SARS‐CoV‐2. The SARS‐CoV‐2 N protein sequence was scanned with a 20aa window and the percent composition of each amino acid was calculated at each position, similar to previous studies. 90 , 91 A region was considered an LCD if any single amino acid constituted ≥40% of the window sequence
The SARS‐CoV N protein consists of two structured domains, referred to as NTD and CTD, separated by a disordered linker and flanked on both termini by disordered tails. 4 The NTD is primarily responsible for RNA‐binding, although the middle linker and CTD are also capable of binding RNA, and all three disordered regions enhance the affinity for RNA of both the NTD and the CTD. 4 The N protein also contains two distinct LCDs; an S‐rich domain (containing a secondary bias for R) within the middle “linker” region and a K‐rich domain within the C‐terminal disordered tail (Figure 1B).
This combination of protein features––multiple RNA‐binding domains, an oligomerization domain, and multiple LCDs––matches the prototypical architecture of proteins recruited to stress granules and other membraneless organelles. These organelles are often described as “biomolecular condensates” that form through liquid‐liquid phase separation (LLPS). Based on current models, this process is mediated by weak, multivalent interactions through a combination of (1) homotypic (“self”) interactions involving LCDs and oligomerization domains, and (2) heterotypic protein‐protein and protein‐RNA interactions involving RNA‐binding domains, LCDs, and/or specific protein‐protein interaction domains. 7 By two leading phase separation prediction methods, 8 , 9 the N protein achieves the highest overall score for the SARS‐CoV‐2 proteome (Figure 2A). In both cases, the peak phase separation score corresponds to the S/R‐rich LCD (“SR‐domain”), with a secondary (albeit lower) score corresponding to the RNA‐binding NTD (Figure 2B,C). Additionally, the SR‐domain passes the previously defined confidence threshold for the PSP method (Figure 2B), suggesting that the phase separation score is sufficiently high to be considered a likely LLPS protein. catGRANULE does not provide a prediction threshold, though the cumulative density function value for the N protein suggests that it scores well above average relative to eukaryotic proteins (Table 1). The phase separation scores for the N protein are consistent with the lower range of phase separation scores for human proteins with RNA‐binding and prion‐like domains that have characterized phase separation ability (Table 1; 10 ). Collectively, the reasonably high‐scoring SR‐domain, in combination with the RNA‐binding domains and oligomerization domain, support a role for the SARS‐CoV‐2 protein in recruitment to and/or regulation of biomolecular condensates.
FIGURE 2.
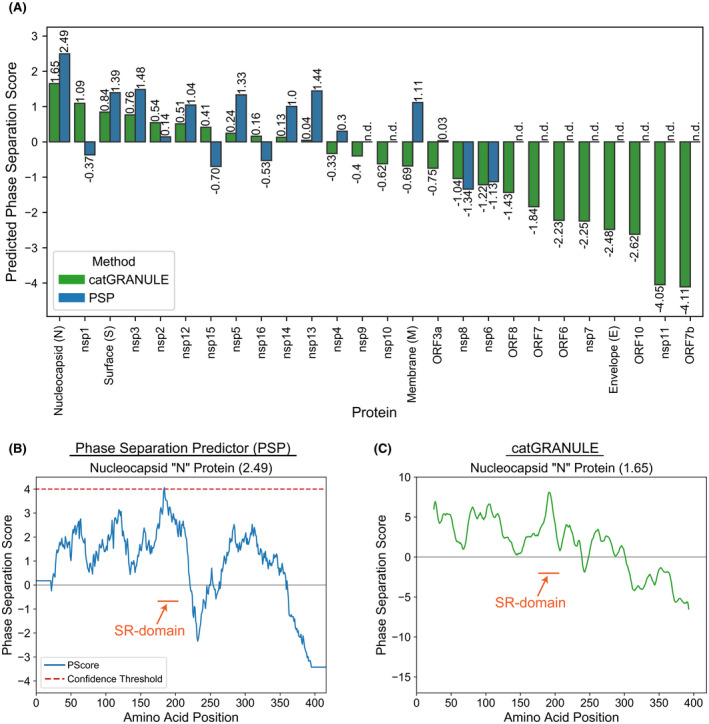
The SARS‐CoV‐2 N protein has an above‐average predicted phase separation propensity, with the SR‐domain being the highest‐scoring region. A, Phase separation scores for all SARS‐CoV‐2 proteins with both PSP 9 and catGRANULE. 8 A consensus sequence was built for each SARS‐CoV‐2 N protein by calculating the most frequent amino acid at each position from multiple sequence alignment of ~1900 sequences available on the NCBI Virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/; downloaded on 5/6/2020). ORF1ab sequences were parsed into separate sequences for the 16 nsp proteins in SARS‐CoV‐2 according to the cleavage sites in Chan et al 92 and separately aligned. B, Phase separation score profile for the N protein from SARS‐CoV‐2 using PSP. C, Phase separation score profile for the N protein from SARS‐CoV‐2 using catGRANULE
TABLE 1.
Phase separation prediction of the SARS‐CoV‐2 N protein and known human LLPS proteins
| Protein | Pscore | catGRANULE |
|---|---|---|
| FUS (NM_004960) | 13.96 | 5.75 |
| hnRNPA1 (NM_031157) | 13.89 | 4.89 |
| EWSR1 (NM_013986) | 9.41 | 3.34 |
| hnRNPA2B1 (NM_031243) | 14.06 | 4.66 |
| hnRNPDL (NM_031372) | 16.83 | 2.09 |
| TAF15 (NM_139215) | 18.48 | 6.29 |
| TDP43 (NM_007375) | 5.95 | 2.04 |
| TIA1 (NM_022173) | 5.57 | 0.97 |
| N Protein (SARS‐CoV‐2) | 2.49 a | 1.65 b |
The Pscore and catGRANULE score for RNA‐binding proteins commonly linked to stress granules and LLPS were calculated using PSP 9 and catGRANULE, 8 respectively.
In the 86th percentile for all proteins in the human proteome (Uniprot proteome UP000005640).
catGRANULE currently does not support whole‐proteome analyses. However, another human protein, TRA2A, has a score of 2.14 and ranks 188th out of 20 190 human proteins, placing it in the 99th percentile. 11
3. MULTIPLE DISTINCT LINES OF EVIDENCE CONNECT THE N PROTEIN WITH STRESS GRANULES
Many viruses can influence host cell stress granule formation by either inhibiting or enhancing stress granule formation. 12 Additionally, some viruses can induce the formation of stress granule‐like compartments containing canonical stress granule markers but with otherwise distinct molecular compositions. 13 , 14 , 15 , 16 To our knowledge, the effect of SARS‐CoV or SARS‐CoV‐2 on host cell stress granule formation has not yet been examined experimentally. However, multiple observations converge to suggest biologically meaningful connections between SARS‐CoV/SARS‐CoV‐2 and stress granules.
First, SARS‐CoV infection activates both PKR and PERK, 17 which are cytoplasmic and ER kinases, respectively, that phosphorylate eIF2α and induce the formation of stress granules. 18 PKR senses double‐stranded RNA (dsRNA) by direct binding, 19 and SARS‐CoV infection produces an abundance of dsRNA in infected cells. 17 PERK is activated in response to the ER‐associated unfolded protein response (UPR) pathway, 19 and the UPR is also induced by SARS‐CoV infection. 20 While both PERK and PKR are activated during SARS‐CoV infection, PERK appears to be predominantly responsible for subsequent eIF2α phosphorylation, and the typical antiviral activity associated with PKR activation is suppressed during infection. 17 Importantly, activation of PKR and PERK, as well as phosphorylation of eIF2α do not impair viral replication. This indicates that SARS‐CoV possesses mechanisms to induce host cell stress but evade counteractive host cell responses by inhibiting downstream events in the PERK and PKR pathways.
Second, host cell translation is inhibited by multiple mechanisms in SARS‐CoV–infected cells, 21 , 22 , 23 , 24 and translation inhibition is commonly associated with stress granule formation. 18 The SARS‐CoV nonstructural protein 1 (nsp1) directly inhibits translation by binding to the host cell ribosomal components 21 , 24 ; Lokugamage et al speculated that this could lead to the shuttling of abortive transcripts to stress granules or processing bodies. nsp1 also induces degradation of host cell mRNA, 22 , 23 including the mRNA of the antiviral factor IFN‐β, 23 while sparing its own viral genomic RNA. 25 Additionally, stress granules were shown experimentally to form in response to translation inhibition and eIF2α phosphorylation (albeit at different stages of infection) in transmissible gastroenteritis virus (TGEV) and murine hepatitis virus (MHV), two other members of the Coronaviridae family. 26 , 27 The parallels observed between translation inhibition and eIF2α phosphorylation suggest that stress granule formation may also occur in SARS‐CoV and SARS‐CoV‐2. It is worth noting that MERS, another member of the Coronaviridae family, has been shown to inhibit stress granule formation, 28 , 29 although this occurs via inhibition of PKR, 29 which is contrary to the PKR activation observed during SARS‐CoV infection.
Third, the N protein of SARS‐CoV is recruited to stress granules via its SR‐domain and can be phosphorylated at multiple sites within the SR‐domain in vitro by SRPK1, 30 the mammalian homolog of a yeast SR‐kinase that regulates stress granules. 31 The SR‐domain is also phosphorylated by the host cell kinase GSK‐3 32 which is dependent on phospho‐serine within its recognition motif, 33 suggesting that multi‐site phosphorylation of the SR‐domain by SRPK1 may also initiate subsequent phosphorylation by GSK‐3. The N protein colocalizes with the stress granule markers PABP1 and TIA‐1 under sodium arsenite stress, but this colocalization is suppressed when SRPK1 is overexpressed, 30 which is reminiscent of the role of yeast Sky1 in stress granule disassembly. 31 The SR‐domain of the SARS‐CoV N protein has a high affinity for the prion‐like domain of human hnRNPA1, 34 which is another known component of stress granules and plays important roles in RNA metabolism and disease. 35 Finally, the N protein undergoes oligomerization and can suppress translation in vitro, yet both activities are inhibited upon SR‐domain phosphorylation. While further experiments are required to definitively resolve the relationship between SR‐domain phosphorylation and N protein oligomerization, 30 , 36 the evidence suggests that modulation of the electrostatic properties of the SR‐domain by phosphorylation can act as a switch to spatiotemporally regulate N protein activity and function during different stages of SARS‐CoV infection cycle.
Fourth, two independent studies report comprehensive sets of protein‐protein interactions between SARS‐CoV‐2 proteins and human proteins. In both cases, the core stress granule components G3BP1 and G3BP2, as well as other components, co‐precipitate with the N protein [37 and Li et al (doi: https://doi.org/10.1101/2020.03.31.019216); at the time of this writing, the study by Li et al has not formally completed peer review]. G3BP1 and G3BP2 are essential stress granule components, and G3BP1 was recently shown by three independent groups to undergo LLPS and drive the formation of biomolecular condensates (namely, stress granules) in mammalian cells. 38 , 39 , 40 Interaction between the SARS‐CoV‐2 N protein and G3BP1/G3BP2 could either enhance stress granule induction or inhibit stress granule formation by sequestering G3BP1/G3BP2, as observed for multiple flavivirus nucleocapsid proteins. 41
In summary, (1) the activation of stress granule‐inducing kinases PKR and PERK, (2) the concurrent phosphorylation of eIF2α, (3) the observed host translation shutoff, (4) the suppression of host cell responses often related to stress granule formation, (5) the demonstrated ability of N protein to join stress granules in a regulated manner, (6) the above‐average predicted phase separation propensity, (7) the multivalent domain architecture of the N protein (including domains involved in both RNA‐binding and oligomerization), and (8) physical interaction with multiple stress granule components collectively suggest that the N protein may aid in modulating stress granule formation or function.
Based on this evidence, we hypothesize that the multiple known interactions between N protein and stress granule components (including N protein homotypic interactions) will have biologically relevant effects on stress granule formation, regulation, or function during SARS‐CoV‐2 infection (Figure 3A). We propose three possible models for SARS‐CoV‐2 N protein participation in stress granules (Figure 3B). In the first model, N protein may be recruited to canonical host cell stress granules and either become a “passive observer” (exerting little to no effect on stress granule nucleation, morphology, or function) or play an active role in either nucleating stress granules or regulating stress granule function. In this model, N protein could be contributing to the translation suppression observed in SARS‐CoV–infected cells, 30 while apparently maintaining translation of its own proteins and sustaining viral replication. 17 In the second model, SARS‐CoV‐2 proteins may induce the formation of unique types of granules, either independently of stress granules or via initial recruitment to but subsequent separation from stress granules. Support for this type of model exists for unrelated viruses, whereby viral proteins actually induce stress granules that differ from canonical stress granules in their molecular composition or co‐opt stress granule components for viral replication or viral mRNA translation. 13 , 14 , 15 , 16 , 41 , 42 In the third model, N protein may inhibit the formation of stress granules by physical interaction and sequestration of key stress granule components, including G3BP1, G3BP2, and hnRNPA1. While phosphorylation of eIF2α and translation suppression typically coincide with stress granule formation, stress granules do not form during Zika virus infection despite observed eIF2α phosphorylation, PKR activation, translation suppression, and induction of the UPR. 41 Additionally, the N protein of influenza A virus is also apparently capable of inhibiting stress granules in spite of elevated eIF2α phosphorylation, 43 although the N protein of influenza A exhibits virtually no sequence similarity with SARS‐CoV‐2 N protein (~11% sequence identity in a pairwise alignment with UniprotID P03466 using the EMBOSS Needle server; https://www.ebi.ac.uk/Tools/psa/emboss_needle/). Therefore, although this mode of action on stress granules is unusual, it is not without precedent.
FIGURE 3.
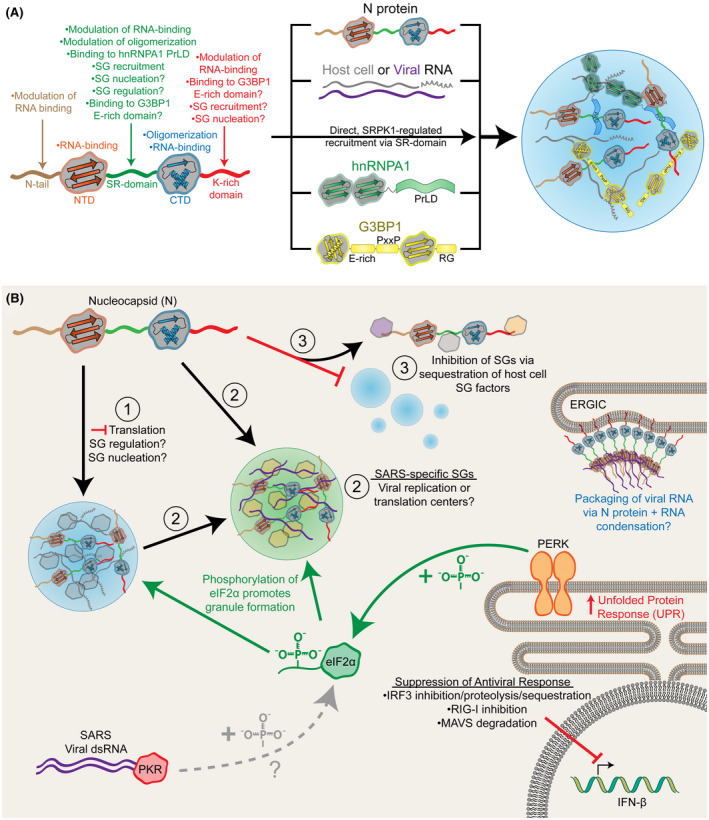
Proposed models for the influence of the SARS‐CoV‐2 N protein on the formation and regulation of biomolecular condensates. A, Putative or directly observed interactions between the SARS‐CoV‐2 N protein and stress granule (SG) components. B, Three possible models for how the N protein of SARS‐CoV‐2 could affect stress granules in host cells: (1) N protein could be recruited to canonical host cell stress granules, which could have subtle or no effect on stress granules (“passive observer”), or could alter stress granule function by contributing to translation suppression, altering stress granule interactions, or remodeling stress granules; (2) N protein could recruit specific stress granule components to form unique, SARS‐specific stress granules that could serve as sites of viral translation or replication; or (3) N protein could inhibit the formation of canonical host cell stress granules by sequestering critical stress granule components. SARS‐CoV‐2 may inhibit the typical stress granule‐associated antiviral responses in host cells by suppressing downstream events that activate IFN‐β expression. Additionally, the features of the SARS‐CoV‐2 N protein that facilitate interaction with stress granules may also facilitate biomolecular condensation of N protein and genomic RNA during nascent virion formation at the ER‐Golgi intermediate compartment (ERGIC)
4. POTENTIAL MODULATION OF STRESS GRANULE‐ASSOCIATED INNATE IMMUNE RESPONSES BY SARS‐CoV‐2
Both PKR and RIG‐I have been identified in virus‐induced stress granules, 44 , 45 , 46 and stress granules can exert antiviral effects by influencing or directly mediating host cell innate immune responses. 44 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 While it is possible that SARS‐CoV‐2 blocks stress granule formation despite hallmark stress granule indicators (model three above), SARS‐CoV suppresses host cell antiviral responses by multiple mechanisms, some of which occur downstream of stress granule formation. In addition to inducing IFN‐β mRNA degradation, 23 nsp1 also suppresses IFN‐β expression via cytosolic sequestration, 55 inhibition, 55 , 56 ubiquitin‐dependent degradation, 57 and cleavage 58 of its transcription factor, IRF3. The N protein of both SARS‐CoV and SARS‐CoV‐2 can act as suppressors of the host cell RNAi response to infection. 59 , 60 Independently of PKR, the host cell proteins RIG‐I, MDA5, and OAS‐RNase L are also capable of sensing dsRNA resulting from viral infection, 61 but these antiviral innate immunity pathways are all inhibited at various stages by SARS‐CoV proteins 62 , 63 or other coronavirus proteins. 64 , 65 , 66 Finally, two recent independent studies indicate that SARS‐CoV‐2 upregulates the expression of pro‐inflammatory cytokines and interferon‐stimulated genes (ISGs), but fails to induce IFN‐I and IFN‐III interferons, including IFN‐β. 67 , 68 Therefore, like SARS‐CoV, SARS‐CoV‐2 is capable of suppressing the type‐I IFN innate immune pathway. Collectively, blockage of the innate immune pathways downstream of stress granules by SARS‐CoV‐2 proteins may enable the formation of stress granules, while suppressing specific stress granule‐associated host cell responses.
It is important to note that a recent study reported inhibition of SARS‐CoV‐2 replication in cell culture models treated with multiple translation inhibitors, including the eIF4A inhibitor zotatifin. 37 Inhibition of eIF4A is typically associated with increased stress granule formation 69 , 70 and can block replication of certain viruses. 71 However, the mechanism by which translation inhibitors suppress SARS‐CoV‐2 replication was not examined, so it is unclear whether stress granule induction or global suppression of translation (presumably including the production of SARS‐CoV‐2 proteins) is predominantly responsible for the observed antiviral effect.
5. A POSSIBLE ROLE FOR BIOMOLECULAR CONDENSATION ACTIVITY OF N PROTEIN IN VIRAL RNP NUCLEOCAPSID ASSEMBLY AND VIRAL REPLICATION CENTERS
Although the SARS‐CoV N protein is capable of fulfilling a variety of roles, one of the best‐characterized functions of the N protein for many viruses is the packaging of the viral RNA proteome into nascent virions. 4 Interestingly, the capsid protein of human cytomegalovirus, pAP, which performs the same function as N protein in SARS viruses, has a high predicted phase separation score (3.8), and was shown to phase separate at high temperature in vitro. 9 The N protein of the measles virus undergoes LLPS with its partner P protein in vitro, which mediates RNP condensation and the assembly of capsid‐like particles. 72 Liquid‐like molecular assemblies containing the N protein were also observed specifically at sites along the ER during influenza A infection. 73 Although the function of these compartments was not determined, SARS‐CoV forms new viral nucleocapsids at the ER‐Golgi intermediate compartment (ERGIC) membrane, so it is possible that the formation of similar liquid‐like assemblies by the N protein mediates nucleocapsid formation during SARS‐CoV and SARS‐CoV‐2 infections (Figure 3B). Quite recently, purified nucleocapsid proteins from numerous retroviruses were shown to phase separate in vitro, and the HIV‐1 nucleocapsid protein forms Zn+2‐dependent, reversible foci in mammalian cells consistent with biomolecular condensation. 74 Finally, a variety of unrelated viruses are capable of forming liquid‐like compartments that specifically contain (or are even dependent upon) the N protein homolog from each of the viruses in host cells as sites of viral replication. 75 , 76 , 77 In addition to its function in nucleocapsid assembly, the SARS‐CoV N protein is also thought to play a role in SARS‐CoV replication, 4 so it is possible that the N protein joins viral replication centers during SARS infections. It is worth noting that some evidence suggests that not all viral replication centers with liquid‐like properties are entirely consistent with the LLPS model, 78 so the precise nature of these membraneless organelles requires additional study.
Cryo‐EM 79 and crystallographic 80 models have suggested a filamentous packing of SARS‐CoV genomic RNA on the inner membrane of new virions, mediated by ordered polymerization of the N protein. While a highly ordered, filamentous arrangement of N protein may appear to be at odds with an LLPS model of RNP formation, LLPS has actually been shown to enhance the nucleation of actin 81 and microtubule 82 filaments by concentrating monomeric components and assembly factors in biomolecular condensates. Additionally, filamentous actin bundles can also form dense assemblies with liquid‐like properties, 83 suggesting a role for LLPS in the macroscopic behavior and local concentration of ordered filaments. Therefore, the proposed LLPS activity of the N protein may actually be critical for the initiation and arrangement of RNPs during virion formation regardless of the degree of order in the final N protein‐RNA assemblies.
More broadly, these observations suggest that the formation of liquid‐like biomolecular condensates may be a common activity for viral nucleocapsid proteins (Figure 3B) in spite of considerable sequence differences between nucleocapsid proteins. While this is currently a speculative model, the high predicted phase separation propensity and demonstrated phase separation ability of N proteins from other viruses suggest that for SARS‐CoV‐2, as well as other viruses, RNP formation and packaging could occur via biomolecular condensation involving the multivalent N protein.
6. INDIRECT TARGETING OF N PROTEIN REGULATION VIA INHIBITION OF HOST CELL KINASES AS AN ANTIVIRAL TREATMENT STRATEGY?
The participation of the N protein in multiple processes vital for SARS‐CoV replication raises an intriguing question: how can the N protein fulfill so many distinct roles throughout the viral infection cycle, and how are these roles balanced and regulated? The conservation of an SR‐domain within the N protein across viruses of the Coronaviridae family suggests that the SR‐domain is of functional importance to the virus. Site‐specific phosphorylation of the SR‐domain by defined host cell kinases, along with observed functional consequences of these phosphorylation events, seem to suggest that the N protein hijacks host cell kinases for its spatio‐temporal regulation during the viral lifecycle. While rational drug or vaccine design targeting viral proteins is an important strategy, targeting host cell factors that appear to be advantageous to the virus may be equally effective.
Interestingly, SRPK1 appears to play an important role in viral replication for numerous distinct viruses, as either inhibition or activation of SRPK1 can be advantageous to viruses. 84 , 85 , 86 , 87 , 88 In the case of Hepatitis B virus, this effect is exerted through modulation of RNA packaging in the nucleocapsid, despite a radically different SR‐domain compared to the SR‐domain of the SARS‐CoV‐2 N protein. 83 The substrate‐kinase balance and full phosphorylation/dephosphorylation cycles involving SRPK1 have been proposed to be important for at least a subset of these viruses, 85 , 86 suggesting that both inhibition and overactivation of SRPK1 may independently be effective antiviral strategies. Furthermore, inhibition of SRPK1/2 has been shown to dampen viral replication in cell culture models. 86 , 87 , 88 Similarly, inhibition of GSK‐3 has been shown to reduce viral replication of SARS‐CoV. 32 SRPK1/2 inhibitors continue to be developed as potential cancer therapeutics, and one FDA‐approved kinase inhibitor, Alectinib, potently cross‐reacts with and inhibits SRPK1. 89 Re‐purposing of SRPK1/2 or GSK‐3 inhibitors, or development of new inhibitors, could be viable strategies for disrupting SARS‐CoV‐2 viral replication and spread, although it must be emphasized that this is currently speculative and requires extensive testing to determine the safety and efficacy in humans.
7. CONCLUSION
Collectively, sequence analyses in combination with emerging and pre‐existing experimental evidence suggest that the formation and regulation of biomolecular condensates could be vital activities of the essential N protein of SARS‐CoV‐2. We propose that the N protein of SARS‐CoV‐2 may harness the ability to form or join biomolecular condensates to dysregulate stress granules, enhance viral replication or translation of viral proteins, and package the viral RNA genome into new virions. Given the vital role of the N protein in multiple stages of the viral lifecycle, modulation of N protein regulation by treatments targeting host cell kinases or membraneless organelles may be viable strategies for combating existing SARS‐CoV‐2 infections.
CONFLICT OF INTEREST
The authors declare that they have no conflicting interests.
AUTHOR CONTRIBUTIONS
S.M. Cascarina conceived of initial hypotheses, designed software, analyzed data, and wrote the initial manuscript; S.M. Cascarina and E.D. Ross contributed to hypothesis refinement, analytical approach, and manuscript composition/editing.
ACKNOWLEDGMENTS
We thank Lindsey Bush for editing suggestions during final manuscript preparation. This work was supported by the National Institute of General Medical Sciences (https://www.nigms.nih.gov/), grant number R35GM130352 awarded to EDR.
Cascarina SM, Ross ED. A proposed role for the SARS‐CoV‐2 nucleocapsid protein in the formation and regulation of biomolecular condensates. The FASEB Journal. 2020;34:9832–9842. 10.1096/fj.202001351
This article was fast‐tracked under a recently instituted interim policy in which the editors may, at their discretion, accept coronavirus‐related manuscripts submitted for the Review, Perspectives and Hypotheses categories without additional review.
[Correction added on July 2, 2020, after first online publication: The SARS‐CoV 9192 under introduction has been corrected to “The nucleocapsid (“N”) protein of the closely related SARS‐CoV”.]
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300;1394‐1399. 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 3. Marra MA, Jones SJM, Astell CR, et al. The genome sequence of the SARS‐associated coronavirus, Science. 2003;300:1399‐1404. 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 4. McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991‐3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS‐CoV‐2. Cell Host Microbe. 2020;27:671‐680.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang S, Yang M, Hong Z, et al. Crystal structure of SARS‐CoV‐2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites, Acta Pharmaceutica Sinica B. 2020. 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alberti S, Dormann D. Liquid‐liquid phase separation in disease. Annu Rev Genet. 2019;53:171‐194. [DOI] [PubMed] [Google Scholar]
- 8. Bolognesi B, Gotor NL, Dhar R, et al. A concentration‐dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 2016;16:222‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vernon RMC, Chong PA, Tsang B, et al. Pi‐Pi contacts are an overlooked protein feature relevant to phase separation. eLife. 2018;7:e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J, Choi JM, Holehouse AS, et al. A molecular grammar governing the driving forces for phase separation of prion‐like RNA binding proteins. Cell. 2018;174:688‐699.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cid‐Samper F, Gelabert‐Baldrich M, Lang B, et al. An integrative study of protein‐RNA condensates identifies scaffolding RNAs and reveals players in fragile X‐associated tremor/ataxia syndrome. Cell Rep. 2018;25:3422‐3434.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaete‐Argel A, Márquez CL, Barriga GP, Soto‐Rifo R, Valiente‐Echeverría F. Strategies for success. viral infections and membraneless organelles. Front Cell Infect Microbiol. 2019;9:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Piotrowska J, Hansen SJ, Park N, Jamka K, Sarnow P, Gustin KE. Stable formation of compositionally unique stress granules in virus‐infected cells. J Virol. 2010;84:3654‐3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scholte FEM, Tas A, Albulescu IC, et al. Stress granule components G3BP1 and G3BP2 play a proviral role early in chikungunya virus replication. J Virol. 2015;89:4457‐4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brocard M, Iadevaia V, Klein P, et al. Norovirus infection results in eIF2α independent host translation shut‐off and remodels the G3BP1 interactome evading stress granule formation. PLoS Pathog. 2020;16:e1008250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burke JM, Lester ET, Tauber D, Parker R. RNase L promotes the formation of unique ribonucleoprotein granules distinct from stress granules. J Biol Chem. 2020;295:1426‐1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krahling V, Stein DA, Spiegel M, Weber F, Muhlberger E. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J Virol. 2009;83:2298‐2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141‐150. [DOI] [PubMed] [Google Scholar]
- 19. Pakos‐Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17:1374‐1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan C‐P, Siu K‐L, Chin K‐T, Yuen K‐Y, Zheng B, Jin D‐Y. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2006;80:9279‐9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. A two‐pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol. 2009;16:1134‐1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kamitani W, Narayanan K, Huang C, et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A. 2006;103:12885‐12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Narayanan K, Huang C, Lokugamage K, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type i interferon, in infected cells. J Virol. 2008;82:4471‐4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lokugamage KG, Narayanan K, Huang C, Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J Virol. 2012;86:13598‐13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. SARS coronavirus nsp1 protein induces template‐dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1‐induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sola I, Galan C, Mateos‐Gomez PA, et al. The polypyrimidine tract‐binding protein affects coronavirus RNA accumulation levels and relocalizes viral rnas to novel cytoplasmic domains different from replication‐transcription sites. J Virol. 2011;85:5136‐5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raaben M, Groot Koerkamp MJA, Rottier PJM, de Haan CAM. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 2007;9:2218‐2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakagawa K, Narayanan K, Wada M, Makino S. Inhibition of stress granule formation by middle east respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J Virol. 2018;92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rabouw HH, Langereis MA, Knaap RCM, et al. Middle east respiratory coronavirus accessory protein 4a inhibits PKR‐mediated antiviral stress responses. PLoS Pathog. 2016;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng TY, Lee KR, Tarn WY. Phosphorylation of the arginine/serine dipeptide‐rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152‐4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shattuck JE, Paul KR, Cascarina SM, Ross ED. The prion‐like protein kinase Sky1 is required for efficient stress granule disassembly. Nat Commun. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu CH, Yeh SH, Tsay YG, et al. Glycogen synthase kinase‐3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus mucleocapsid protein and viral replication. J Biol Chem. 2009;284:5229‐5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fiol CJ, Mahrenholzs AM, Wangg Y, et al. Formation of protein kinase recognition sites by covalent modification of the substrate. J Biol Chem. 1987;262:14042‐14048. [PubMed] [Google Scholar]
- 34. Luo H, Chen Q, Chen J, Chen K, Shen X, Jiang H. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579:2623‐2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim HJ, Kim NC, Wang YD, et al. Mutations in prion‐like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang C‐K, Chen C‐M, Chiang M‐H, Hsu Y‐L, Huang T‐H. Transient oligomerization of the SARS‐CoV N protein—implication for virus ribonucleoprotein packaging. PLoS One. 2013;8:e65045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang P, Mathieu C, Kolaitis RM, et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181:325‐345.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guillén‐Boixet J, Kopach A, Holehouse AS, et al. RNA‐induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181:346‐361.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanders DW, Kedersha N, Lee DSW, et al. Competing protein‐RNA interaction networks control multiphase intracellular organization. Cell. 2020;181:306‐324.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hou S, Kumar A, Xu Z, et al. Zika virus hijacks stress granule proteins and modulates the host stress response. J Virol. 2017;91:e00474‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hosmillo M, Lu J, McAllaster MR, et al. Noroviruses subvert the core stress granule component G3BP1 to promote viral VPg‐dependent translation. eLife. 2019;8:e46681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. Influenza a virus host shutoff disables antiviral stress‐induced translation arrest. PLoS Pathog. 2014;10:e1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Onomoto K, Jogi M, Yoo JS, et al. Critical role of an antiviral stress granule containing RIG‐I and PKR in viral detection and innate immunity. PLoS One. 2012;7:e43031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reineke LC, Kedersha N, Langereis MA, van Kuppeveld FJM, Lloyd RE. Stress granules regulate double‐stranded RNA‐dependent protein kinase activation through a complex containing G3BP1 and Caprin1. mBio. 2015;6:e02486‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reineke LC, Lloyd RE. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J Virol. 2015;89:2575‐2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim SSY, Sze L, Lam KP. The stress granule protein G3BP1 binds viral dsRNA and RIG‐I to enhance interferon‐β response. J Biol Chem. 2019;294:6430‐6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rozelle DK, Filone CM, Kedersha N, Connor JH. Activation of stress response pathways promotes formation of antiviral granules and restricts virus replication. Mol Cell Biol. 2014;34:2003‐2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoo JS, Takahasi K, Ng CS, et al. DHX36 enhances RIG‐I signaling by facilitating PKR‐mediated antiviral stress granule formation. PLoS Pathog. 2014;10:e1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simpson‐Holley M, Kedersha N, Dower K, et al. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol. 2011;85:1581‐1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manivannan P, Siddiqui MA, Malathi K. RNase L amplifies interferon signaling by inducing PKR‐mediated antiviral stress granules, J Virol. 2020. 10.1128/jvi.00205-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Law LMJ, Razooky BS, Li MMH, et al. ZAP’s stress granule localization is correlated with its antiviral activity and induced by virus replication. PLOS Pathog. 2019;15:e1007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oh SW, Onomoto K, Wakimoto M, et al. Leader‐containing uncapped viral transcript activates RIG‐I in antiviral stress granules. PLoS Pathog. 2016;12:e1005444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ng CS, Jogi M, Yoo J‐S, et al. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol. 2013;87:9511‐9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Spiegel M, Pichlmair A, Martinez‐Sobrido L, et al. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two‐step model for activation of interferon regulatory factor 3. J Virol. 2005;79:2079‐2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol. 2007;81:11620‐11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin‐dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Devaraj SG, Wang N, Chen Z, et al. Regulation of IRF‐3‐dependent innate immunity by the papain‐like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282:32208‐32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mu J, Xu J, Zhang L, et al. SARS‐CoV‐2‐encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells, Sci China Life Sci. 2020:1‐4. 10.1007/s11427-020-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cui L, Wang H, Ji Y, et al. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J Virol. 2015;89:9029‐9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hur S. Double‐stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol. 2019;37:349‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hu Y, Li W, Gao T, et al. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25‐mediated RIG‐I ubiquitination. J Virol. 2017;91:e02143‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shi C‐S, Qi H‐Y, Boularan C, et al. SARS‐coronavirus open reading frame‐9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol. 2014;193:3080‐3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117:8094‐8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Goldstein SA, Thornbrough JM, Zhang R, et al. Lineage A betacoronavirus NS2 proteins and the homologous torovirus berne pp1a carboxy‐terminal domain are phosphodiesterases that antagonize activation of RNase L. J Virol. 2017;91:e02201‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao L, Jha BK, Wu A, et al. Antagonism of the interferon‐induced OAS‐RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607‐616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhou Z, Ren L, Zhang LI, et al. Heightened innate immune responses in the respiratory tract of COVID‐19 patients. Cell Host Microbe. 2020;27:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Blanco‐Melo D, Nilsson‐Payant BE, Liu W‐C, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181:1036‐1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dang Y, Kedersha N, Low WK, et al. Eukaryotic initiation factor 2α‐independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem. 2006;281:32870‐32878. [DOI] [PubMed] [Google Scholar]
- 70. Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, Parker R. Modulation of RNA condensation by the DEAD‐box protein eIF4A. Cell. 2020;180:411‐426.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Slaine PD, Kleer M, Smith NK, Khaperskyy DA, McCormick C. Stress granule‐inducing eukaryotic translation initiation factor 4A inhibitors block influenza A virus replication. Viruses. 2017;9:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Guseva S, Milles S, Jensen MR, et al. Measles virus nucleo‐ and phosphoproteins form liquid‐like phase‐separated compartments that promote nucleocapsid assembly. Sci Adv. 2020;6:eaaz7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Alenquer M, Vale‐Costa S, Etibor TA, Ferreira F, Sousa AL, Amorim MJ. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat Commun. 2019;10:1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Monette A, Niu M, Chen L, Rao S, Gorelick RJ, Mouland AJ. Pan‐retroviral nucleocapsid‐mediated phase separation regulates genomic RNA positioning and trafficking. Cell Rep. 2020;31:107520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Heinrich BS, Maliga Z, Stein DA, Hyman AA, Whelan SPJ. Phase transitions drive the formation of vesicular stomatitis virus replication compartments. mBio. 2018;9:e02290‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou Y, Su JM, Samuel CE, Ma D. Measles virus forms inclusion bodies with properties of liquid organelles. J Virol. 2019;93;e00948‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nikolic J, Le Bars R, Lama Z, et al. Negri bodies are viral factories with properties of liquid organelles. Nat Commun. 2017;8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. McSwiggen DT, Hansen AS, Teves SS, et al. Evidence for DNA‐mediated nuclear compartmentalization distinct from phase separation. eLife. 2019;8:e47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Neuman BW, Adair BD, Yoshioka C, et al. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80:7918‐7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chen C‐Y, Chang C‐K, Chang Y‐W, et al. Structure of the SARS coronavirus nucleocapsid protein RNA‐binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. 2007;368:1075‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Case LB, Zhang X, Ditlev JA, Rosen MK. Stoichiometry controls activity of phase‐separated clusters of actin signaling proteins. Science. 2019;363:1093‐1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hernández‐Vega A, Braun M, Scharrel L, et al. Local nucleation of microtubule bundles through Tubulin concentration into a condensed tau phase. Cell Rep. 2017;20:2304‐2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Weirich KL, Banerjee S, Dasbiswas K, Witten TA, Vaikuntanathan S, Gardel ML. Liquid behavior of cross‐linked actin bundles. Proc Natl Acad Sci U S A. 2017;114:2131‐2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Prescott EL, Brimacombe CL, Hartley M, Bell I, Graham S, Roberts S. Human papillomavirus type 1 E1^E4 protein is a potent inhibitor of the serine‐arginine (SR) protein kinase SRPK1 and inhibits phosphorylation of host SR proteins and of the viral transcription and replication regulator E2. J Virol. 2014;88:12599‐12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Heger‐Stevic J, Zimmermann P, Lecoq L, Böttcher B, Nassal M. Hepatitis B virus core protein phosphorylation: identification of the SRPK1 target sites and impact of their occupancy on RNA binding and capsid structure. PLOS Pathog. 2018;14:e1007488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Takamatsu Y, Krähling V, Kolesnikova L, et al. Serine‐arginine protein kinase 1 regulates ebola virus transcription. mBio. 2020;11:e02565‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fukuhara T, Hosoya T, Shimizu S, et al. Utilization of host SR protein kinases and RNA‐splicing machinery during viral replication. Proc Natl Acad Sci U S A. 2006;103:11329‐11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Karakama Y, Sakamoto N, Itsui Y, et al. Inhibition of hepatitis C virus replication by a specific inhibitor of serine‐arginine‐rich protein kinase. Antimicrob Agents Chemother. 2010;54:3179‐3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hatcher JM, Wu G, Zeng C, et al. SRPKIN‐1: a covalent SRPK1/2 inhibitor that potently converts VEGF from pro‐angiogenic to anti‐angiogenic isoform. Cell Chem Biol. 2018;25:460‐470.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cascarina SM, Ross ED. Proteome‐scale relationships between local amino acid composition and protein fates and functions. PLOS Comput Biol. 2018;14:e1006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cascarina SM, Elder MR, Ross ED. Atypical structural tendencies among low‐complexity domains in the Protein Data Bank proteome. PLOS Comput Biol. 2020;16:e1007487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chan JFW, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human‐pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]


