Abstract
The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in human populations sparked a global pandemic of the coronavirus disease 2019 (COVID‐19). According to preliminary data, about 14% of cases are considered severe and 5% of cases result in critical illness and, reported case fatality rates vary from 1% to more than 7%. However, the symptoms of the disease and the clinical outcome are very different in infected people. In view of these differences, it is clearly apparent that to gain insight into the biology of the SARS‐CoV‐2, it is important to study not just the infectious particle in itself but also to investigate the virus‐host cell interactions that occur during infection. This review seeks to consider the various aspects of genetic factors in determining the susceptibility and host resistance to SARS‐CoV‐2 throughout the recently published literature.
Keywords: COVID‐19, genetics, resistance, SARS‐CoV‐2, susceptibility
This review seeks to consider the various aspects of genetic factors in determining the susceptibility and host resistance to severe acute respiratory syndrome coronavirus 2 throughout the recently published literature.

1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19) caused by the acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is now in the early phases of a global pandemic (Bian et al., 2020; Sheikhshahrokh et al., 2020). The mortality rates of COVID‐19 is estimated to be 5.6% in China and 15.2% outside(Baud et al., 2020). The age gradient is the most key indication of the fatality ratio (Verity et al., 2020). People with chronic disease (e.g., lung, kidney, liver, cardiovascular disease, and diabetes) and low immune system are at high risk for severe illness from COVID‐19 (N. Chen et al., 2020). Along with clinical features, molecular markers can be very effective in identifying susceptible and resistant individuals to the SARS‐CoV‐2. Susceptibility of the COVID‐19 is suggested to have a genetic component. Obviously, the host response is as vital to defining disease outcome as the virus itself. There is, therefore, a need to understand thoroughly the host factors interacting with viral components to realize the complex processes governing SARS‐CoV‐2 virology and pathogenesis for facilitating replication, modulating immune responses, and finally determining disease severity. Due to the emergence of the SARS‐CoV‐2, few studies have yet been conducted on the genetic characteristics of the host cell in causing susceptibility of and resistance to the COVID‐19 disease. In this review, we are going to examine and compare the host genetic factors such as SARS‐CoV‐2 host cell receptors, human leukocyte antigen (HLA), ABO blood group, and female gender, which suggested from recent studies to play a role in sensitization and resistance to the SARS‐CoV‐2.
2. MAIN TEXT
2.1. Phylogeny, structural, and genomic organization
A detailed study of the SARS‐CoV‐2 genome reveals that it belongs to the genus betacoronavirus (Wu et al., 2020). The virus's original host is suggested by phylogenetic analysis to be bats (Lu et al., 2020). The betacoronavirus genus contains the three known coronaviruses that infect humans: human coronavirus‐OC43, human coronavirus‐HKU1, and SARS‐CoV (Marra et al., 2003). The genomic RNA of these coronaviruses is single‐stranded and positive‐sense (29,903 nt) associated with the Nucleoprotein (N), and the structural transmembrane proteins are the Membrane (M), Spike (S), and Envelope (E; Figure 1b; Wu et al., 2020). Two overlapping open reading frames (ORFs 1a and 1b) in the downstream regions of the genome translate the precursor polyproteins (pp1a and pp1ab), which makes the coronavirus transcription‐replication complex (Figure 1a). Moreover, ORF codes 16 nonstructural proteins (NSP) comprised of two viral cysteine proteases, including NSP3 (papain‐like) and NSP5 (primary), NSP12 (RNA‐dependent RNA polymerase), NSP13 (helicase), as well as other NSPs that are expected to engage with the virus transcription‐replication. Mutations can be seen in NSP2, NSP3, as well as the spike protein, which has a major part in the SARS‐CoV‐2 infectious ability and differentiation mechanism (Krichel, Falke, Hilgenfeld, Redecke, & Uetrecht, 2020). Two strains of coronaviruses, L‐type and S‐type, are also found. The L‐type, derived from the S‐type, is observed to be more contagious and aggressive (Tang et al., 2020). SARS‐CoV‐2 genome has shown a very high overall sequence identity with SARS‐CoV (∼80%; Zhou et al., 2020).
Figure 1.
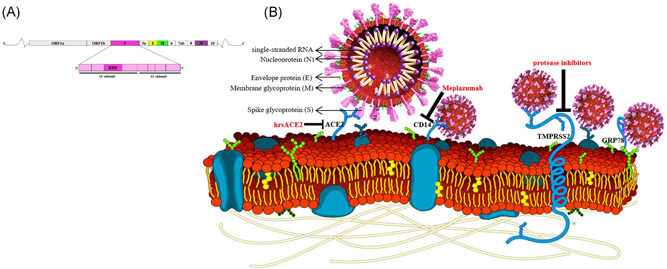
Schematic representation of the overall genome and protein of SARS‐CoV‐2 and host cell receptors and inhibitors. (a) Linear representation of the SARS‐CoV‐2 genome and the Spike protein. (b) Spike proteins on the surface of the SARS‐CoV‐2 bind to ACE‐2, TMPRSS2, GRP78, and CD147. The suggested inhibitor for each receptor is indicated. ACE2, angiotensin‐converting enzyme 2; CD147, cluster of differentiation 147; GRP78, glucose‐regulated protein 78 kDa; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane protease serine‐type 2
2.2. SARS‐CoV‐2 host cell receptors
The first interaction of SARS‐CoV‐2 with the host initiates with the attachment to a specific virus receptor on the host cell. The S protein mediates this process. The S domain is divided into two subunits: S1 and S2. S1 forms a globular structure and contains the receptor‐binding domain that facilitates the attachment of the virus to the target cell. S2 includes the stalk region of the Spike and contains a putative fusion peptide. The S protein of the SARS‐CoV‐2 requires cleavage of the S domain during the entry process (Figure 1a; Simmons et al., 2005). Receptor binding is a critical pace in the life cycle of the virus since the interaction of the S protein with the receptor defines the host range and tropism of the virus. Several receptors have been suggested for the SARS‐CoV‐2 thus far. The epithelial cells of the respiratory and intestinal tracts are prime targets of coronaviruses (Lai & Holmes, 2001). Given the genetic similarity of SARS‐CoV‐2 to SARS‐CoV and the studies to date, it can be predicted whether the viral host cell receptors can be used as molecular markers to determine the susceptibility of or resistance to COVID‐19.
2.3. Angiotensin‐converting enzyme 2
After isolation and characterization of the SARS‐CoV‐2, angiotensin‐converting enzyme 2 (ACE2) is found to be the primary receptor of the virus and facilitate viral entry into target cells (Ou et al., 2020). ACE2 has zinc metalloproteinase activity (Tipnis et al., 2000) and preferentially is expressed on the apical side of epithelial cells (Ito et al., 2005); interestingly, SARS‐CoV‐2 is shown to enter cells through the apical side of polarized epithelial cells (Milewska et al., 2020). ACE2 is an important component of the renin‐angiotensin pathway (RAAS) and also vital in regulating cardiac function (Crackower et al., 2002). It is expressed in a number of tissues, including heart, lungs, kidneys, and intestines (Utsumi, Yokota, Ishikawa, Ohnishi, & Fujiwara, 1990) which may explain the multiorgan dysfunction detected in patients with COVID ‐19 (Zhang, Penninger, Li, Zhong, & Slutsky, 2020).
Remarkably, the decreasing ACE2 expression level is shown to be a fundamental protective factor against lung failure during severe acute lung injury (Imai et al., 2005). On the other hand, ACE2 overexpression increases disease harshness in mice infected with SARS‐CoV, indicating that ACE2‐dependent viral passage in cells is a critical pace (Yang et al., 2007). ACE2, therefore, functions both as the entry receptor of SARS‐CoV and as a protection against lung injury (Zhang et al., 2020).
In alveolar epithelial type II cells expressing ACE2, SARS‐CoV‐2 can be entered, replicated, and assembled (Y. Zhao et al., 2020). Due to these shreds of evidence, scientists suggest that ACE2 can be a therapeutic target for patients with COVID‐19 using RAAS inhibitors (M. Sun, Yang, Sun, & Su, 2020; Vaduganathan et al., 2020). In a recent study, human recombinant soluble ACE2 (hrsACE2), by a factor of 1,000–5,000, prevents the coronavirus load (Figure 1b). The outcome is dose‐dependent, indicating that it varies based on the total amount of virus with compared to the total amount of hrsACE2. Furthermore, the authors confirm these data from regular cell cultures in engineered miniature replicas of blood vessels and kidneys; human stem cells trigger these organoids (Monteil et al., 2020).
Even though Asian populations are not different from the others in ACE2 expression level (Y. Chen, Shan, & Qian, 2020); however, having much higher allele frequency in the expression quantitative trait loci variants related to higher ACE2 can suggest different susceptibility or response to SARS‐CoV‐2 from different populations under the similar conditions (Cao et al., 2020). No polymorphisms or mutation in ACE2 related to S‐protein binding‐resistance have been reported yet in different populations (Cao et al., 2020). Recently, Stawiski et al. (2020) using analysis of several large genomic datasets including over 400 population group distinguished variants of ACE2 that alter the interaction between host cell and SARS‐CoV‐2. Variants such as K31R, N33I, H34R, E35K, E37K, D38V, Y50F, N51S, M62V, K68E, F72V, Y83H, G326E, G352V, D355N, Q388L, and D509Y have les affinity to bind to SARS‐CoV‐2 and can play a role in resistance to COVID ‐19 (Stawiski et al., 2020). However, in general, these variants are very rare However, this study could be the basis for further studies to examine the clinical outcomes of patients with different genetic backgrounds during the course of the disease.
2.4. Transmembrane protease serine‐type 2
Transmembrane protease serine‐type 2 (TMPRSS2) is a protease from the type II transmembrane serine protease family; the spike protein is activated by it to induce virus‐cell membrane fusion at the cell surface, and also coronaviruses entry into the host cell is facilitated by it (Gierer et al., 2013; Matsuyama et al., 2010; Shirato, Kawase, & Matsuyama, 2018). TMPRSS2 cleaves the S protein in the SARS‐CoV‐infected people lung, causing the promotion of viral spread and pathogenesis via viral recognition decrease by activating S protein and neutralizing antibodies for virus‐cell and cell‐cell fusion (Glowacka et al., 2011). In mice model, TMPRSS2 has a part in the coronaviruses spread and immunopathology in the airways (Iwata‐Yoshikawa et al., 2019). It has been established that SARS‐CoV‐2 uses the cellular protease TMPRSS2 for S protein priming (Hoffmann, Kleine‐Weber, Krüger et al., 2020; Lukassen et al., 2020). Furthermore, a cell line expressing TMPRSS2 is highly susceptible to SARS‐CoV‐2 infection (Matsuyama et al., 2020). Therefore, protease inhibitors, like Nafamostat, may block SARS‐CoV‐2 entry into host cells and is expected to prevent COVID‐19 transmission (Figure 1b; Hoffmann, Kleine‐Weber, Schroeder et al., 2020).
The TMPRSS2 Isoform 1 contains 37 amino acids in its tail that are not available in isoform 2; it may activate the S proteins of SARS‐CoV and promote viral spread in the infected host (Zmora, Moldenhauer, Hofmann‐Winkler, & Pöhlmann, 2015). TMPRSS2 Isoform 1 activates respiratory (Z. Cheng et al., 2015). Two single‐nucleotide polymorphisms (SNPs; rs2070788 and rs383510) are related to increased TMPRSS2 gene expression and are connected significantly to the susceptibility to H7N9 influenza (Z. Cheng et al., 2015). Perhaps TMPRSS2 SNPs in host cells modulate COVID‐19. Therefore, studies on the connection of TMPRSS2 SNPs with the severity of the disease will be very useful in determining the susceptibility and resistance of individuals to the SARS‐CoV‐2.
2.5. Glucose‐regulated protein 78 kDa
Glucose‐regulated protein 78 kDa (GRP78) or immunoglobulin heavy chain binding protein (BiP) belongs to the Heat Shock Protein 70 family. It is present in eukaryotes all on the endoplasmic reticulum membrane (Ibrahim, Abdelmalek, & Elfiky, 2019). The latest findings propose that GRP78 is cell surface‐localized, where it performs physiological roles to adjust signaling and cellular homeostasis (Lee, 2014). It is, using the structural alignments method, determined that Region IV of the S protein is the main driving force for GRP78 binding (Ibrahim, Abdelmalek, Elshahat, & Elfiky, 2020). This bond may represent one of the ways the virus takes to enter the host cell by increasing the percentage of GRP78 protein linked to the elderly and people with chronic diseases and tumors (Manevski et al., 2020). Chu et al. (2018) have revealed that GRP78 is capable of facilitating surface attachment of Middle East respiratory syndrome coronavirus (MERS‐CoV) and bat coronavirus HKU9 (bCoV‐HKU9) to host cell (Chu et al., 2018). The authors have stated that GRP78 remark the significance of monitoring the bCoV‐HKU9 development that can jump the interspecies fence into humans, resulting in another critical epidemic in the future (Chu et al., 2018). In SARS‐CoV‐infected cells, GRP78 is induced (Chan et al., 2006). Studies on the expression of this GRP78 and its polymorphisms and their role in determining the susceptibility and resistance to coronaviruses are very limited; however, it may be responsible for increasing the rate of infection and death. Therefore, it may be applied to produce an anticorrelation drug against COVID‐19. Also, antagonism of GR78 may lead to a therapeutic for COVID‐19.
2.6. Cluster of differentiation 147
Extracellular matrix metalloproteinase inducer or cluster of differentiation 147 (CD147), well known also as Basigin, is expressed by several cell types, such as endothelial cells, epithelial cells, and leukocytes (Yurchenko, Constant, & Bukrinsky, 2006). A recent study has revealed that CD147 is a novel receptor of the SARS‐CoV‐2 S protein, and the interaction of CD147 and S protein assists the viral invasion for host cells; Meplazumab, a humanized anti‐CD147 antibody, may prevent the viruses from attacking host cells through blocking CD147 efficiently (Figure 1b; Wang et al., 2020). Meplazumab has effectively recovered patients with SARS‐CoV‐2 pneumonia with a satisfactory safety profile in a recent clinical trial (Bian et al., 2020). It has been reported that cyclophilin A (CyPA) bound to N protein of SARS‐CoV interacts with CD147. In cells with highly expressed CD147, CyPA is more integrated with SARS‐CoV. These findings indicate that CD147, mediated by CyPA bound to SARS‐CoV N protein, has a practical part in assisting the invasion of host cells by SARS‐CoV (Z. Chen et al., 2005).
3. HUMAN LEUKOCYTE ANTIGEN
Human leukocyte antigen (HLA) is one of the most polymorphic antigen systems. Silico analysis revealed that among all known HLA ‐A, ‐B, and ‐C genotypes with affinity to binding to all SARS‐CoV‐2 peptides, HLA‐B*46:01 has the least number of peptides binding site for SARS‐CoV‐2, signifying that people with this allele can mainly be susceptible to COVID‐19. On the contrary, it is found that HLA‐B*15:03 can present the SARS‐CoV‐2 peptides that are very conserved and common among human coronaviruses with greatest capability, signifying that it may allow cross‐protective T‐cell‐based immunity (Nguyen et al., 2020). Although such data need clinical test; however, the results can improve the ongoing work of immune landscape profiling. Nonetheless, the previous study of SARS‐CoV has revealed that individuals with the HLA‐B*46:01 genotype show increased disease severity (Lin et al., 2003). Another research has shown that HLA‐Cw*1502 and DRB1*0301 genes can be the resistance factors of SARS‐CoV infection. HLA‐B*0703, on the other hand, has displayed a significant rise in the SARS‐CoV patient group (Y. Sun & Xi, 2014). Given that SARS‐CoV‐2‐related antigen epitopes are all more or less associated with HLA antigen recognition, as inferred by known SARS‐CoV‐2 gene and protein sequences, a close association between a COVID‐19 pandemic and the HLA system is hypothesized here.
4. ABO BLOOD GROUP
Based on an epidemiologic study in Wuhan and Shenzhen, China, Group O people are more resistant to SARS‐CoV because of ABO antibodies, whereas blood group A is related to a higher risk of getting COVID‐19 as compared to non‐A blood groups (J. Zhao et al., 2020). These results are in line with earlier reported investigations in the Hong Kong outbreak. The authors find that Group O people are quite resistant to infection and can decline the infection rate in the population (Guillon et al., 2008). The ABO histo‐blood group comprises two antigens (A and B antigens), which are the ABO gene product. Group O people express the H antigen, the biosynthetic precursor to A and B antigens. Besides red cells, numerous tissues and secretions, such as endothelium, intestinal mucosa, heart, kidney, as well as other organs, express ABH antigens. ABH is a carbohydrate antigen expressed on glycoproteins and glycosphingolipids (GSLs; Cooling, 2015). Glycan analysis of SARS‐CoV S protein reveals a broad range of structures, such as complex N‐glycans with 2–4 antennae able to support ABH epitopes (Y. Cheng et al., 2005). It is highly probable that on the S protein and host envelope GSLs, most human isolates express ABH antigens since the virus targets respiratory and gastrointestinal mucosa. Monoclonal anti‐A and human anti‐A may block S protein‐expressing A antigen (Guillon et al., 2008).
5. CONCLUSION
While a detailed clinical picture of the COVID‐19 pandemic continues to emerge, there remain substantial unanswered questions regarding the role of individual genetic variability in susceptibility and severity of SARS‐CoV‐2. Similarities, including genomic and immune system responses between SARS‐CoV‐2 and other coronaviruses, especially SARS‐CoV and MERS‐CoV, are topics of ongoing active research results of which may provide an understanding of the infection severity (Nguyen et al., 2020). Similar to most coronaviruses, each pace of the SARS‐CoV‐2 life cycle makes use of the host cell's different machinery and structural components. Host cell modifications induced by viral infections constitute the basis of pathogenesis at the level of the patients (Shereen, Khan, Kazmi, Bashir, & Siddique, 2020). Since the beginning of the COVID‐19 pandemic, studies have been conducted in this field. In this review, we discussed about the expression level or mutation and variation in some host genetic factors such as ACE2, TMPRSS2, GRP78, CD147, HLA, and ABO blood group.
The answer to the question of what is the basis in terms of the network of interactions established between SARS‐CoV‐2 and cellular proteins explains the pathogenesis of viruses, studying virus‐host cell relationships and the high‐throughput screening application. Such studies may shed light on paces the virus takes on to enter cells, transcribe, reproduce its RNA genome, synthesize its component proteins, assemble, and exit the infected cell. The results can provide a better understanding of the problem of host cell susceptibility and outcome of SARS‐CoV‐2 infections. Using system biology approaches in the study of SARS‐CoV‐2 can certainly help in both deciphering the molecular interactions that occur during infection, the emergence of pathogenesis, and also developing novel therapeutic strategies for coronavirus COVID‐19 pandemic.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
R. R. and M. H. designed the study. S. M., A. T., and M. L. contributed to data gathering and paper writing. All authors revised the paper.
ACKNOWLEDGMENT
The authors would like to thank the Clinical Research Development Unit of Baqiyatallah Hospital, Tehran, Iran, for guidance and advice.
Mohammadpour S, Torshizi Esfahani A, Halaji M, Lak M, Ranjbar R. An updated review of the association of host genetic factors with susceptibility and resistance to COVID‐19. J Cell Physiol. 2020;236:49–54. 10.1002/jcp.29868
DATA AVAILABILITY STATEMENT
Data will be made available upon request.
REFERENCES
- Baud, D. , Qi, X. , Nielsen‐Saines, K. , Musso, D. , Pomar, L. , & Favre, G. (2020). Real estimates of mortality following COVID‐19 infection. The Lancet Infectious Diseases, S1473‐3099, 30195‐X. 10.1016/S1473-3099(20)30195-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, H. , Zheng, Z.‐H. , Wei, D. , Zhang, Z. , Kang, W.‐Z. , Hao, C.‐Q. , & Zhu, P. (2020). Meplazumab treats COVID‐19 pneumonia: An open‐labelled, concurrent controlled add‐on clinical trial. medRxiv, 10.1101/2020.03.21.20040691 [DOI] [Google Scholar]
- Cao, Y. , Li, L. , Feng, Z. , Wan, S. , Huang, P. , Sun, X. , & Wang, W. (2020). Comparative genetic analysis of the novel coronavirus (2019‐nCoV/SARS‐CoV‐2) receptor ACE2 in different populations. Cell Discovery, 6(1), 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C.‐P. , Siu, K.‐L. , Chin, K.‐T. , Yuen, K.‐Y. , Zheng, B. , & Jin, D.‐Y. (2006). Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. Journal of Virology, 80(18), 9279–9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Shan, K. , & Qian, W. (2020). Asians and other races express similar levels of and share the same genetic polymorphisms of the SARS‐CoV‐2 cell‐entry receptor. Preprints. 10.20944/preprints202002.0258.v1 [DOI] [Google Scholar]
- Chen, Z. , Mi, L. , Xu, J. , Yu, J. , Wang, X. , Jiang, J. , & Zhu, P. (2005). Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. Journal of Infectious Diseases, 191(5), 755–760. 10.1086/427811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N. , Zhou, M. , Dong, X. , Qu, J. , Gong, F. , & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Cheng, G. , Chui, C. , Lau, F. , Chan, P. K. , & Wong, R. S. (2005). ABO blood group and susceptibility to severe acute respiratory syndrome. Journal of the American Medical Association, 293(12), 1447–1451. [DOI] [PubMed] [Google Scholar]
- Cheng, Z. , Zhou, J. , To, K. K.‐W. , Chu, H. , Li, C. , Bossé, Y. , … Yuen, K.‐Y. (2015). Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A (H1N1) influenza and A (H7N9) influenza. Journal of Infectious Diseases, 212(8), 1214–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Chan, C.‐M. , Zhang, X. , Wang, Y. , Yuan, S. , Zhou, J. , & Yuen, K.‐Y. (2018). Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. Journal of Biological Chemistry, 293, 11709–11726. 10.1074/jbc.RA118.001897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooling, L. (2015). Blood groups in infection and host susceptibility. Clinical Microbiology Reviews, 28(3), 801–870. 10.1128/cmr.00109-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower, M. A. , Sarao, R. , Oudit, G. Y. , Yagil, C. , Kozieradzki, I. , Scanga, S. E. , & Pei, Y. (2002). Angiotensin‐converting enzyme 2 is an essential regulator of heart function. Nature, 417(6891), 822–828. [DOI] [PubMed] [Google Scholar]
- Gierer, S. , Bertram, S. , Kaup, F. , Wrensch, F. , Heurich, A. , Krämer‐Kühl, A. , & Drosten, C. (2013). The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. Journal of Virology, 87(10), 5502–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka, I. , Bertram, S. , Müller, M. A. , Allen, P. , Soilleux, E. , Pfefferle, S. , & Pöhlmann, S. (2011). Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. Journal of Virology, 85(9), 4122–4134. 10.1128/jvi.02232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon, P. , Clément, M. , Sébille, V. , Rivain, J.‐G. , Chou, C.‐F. , Ruvoën‐Clouet, N. , & Le Pendu, J. (2008). Inhibition of the interaction between the SARS‐CoV spike protein and its cellular receptor by anti‐histo‐blood group antibodies. Glycobiology, 18(12), 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Krüger, N. , Mueller, M. A. , Drosten, C. , & Pöhlmann, S. (2020). The novel coronavirus 2019 (2019‐nCoV) uses the SARS‐coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. 10.1101/2020.01.31.929042 [DOI] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Krüger, N. , Herrler, T. , Erichsen, S. , … Pöhlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181, 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, I. M. , Abdelmalek, D. H. , & Elfiky, A. A. (2019). GRP78: A cell's response to stress. Life Sciences, 226, 156–163. 10.1016/j.lfs.2019.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, I. M. , Abdelmalek, D. H. , Elshahat, M. E. , & Elfiky, A. A. (2020). COVID‐19 Spike‐host cell receptor GRP78 binding site prediction. Journal of Infection, 80, 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, Y. , Kuba, K. , Rao, S. , Huan, Y. , Guo, F. , Guan, B. , & Leong‐Poi, H. (2005). Angiotensin‐converting enzyme 2 protects from severe acute lung failure. Nature, 436(7047), 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, N. , Mossel, E. C. , Narayanan, K. , Popov, V. L. , Huang, C. , Inoue, T. , & Makino, S. (2005). Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. Journal of Virology, 79(5), 3182–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata‐Yoshikawa, N. , Okamura, T. , Shimizu, Y. , Hasegawa, H. , Takeda, M. , & Nagata, N. (2019). TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. Journal of Virology, 93(6), e01815–e01818. 10.1128/jvi.01815-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichel, B. , Falke, S. , Hilgenfeld, R. , Redecke, L. , & Uetrecht, C. (2020). Processing of the SARS‐CoV pp1a/ab nsp7–10 region. Biochemical Journal, 477(5), 1009–1019. 10.1042/bcj20200029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M. , & Holmes, K. (2001). Coronaviridae: The viruses and their replication. In Knipe B. N. & Howley D. M. (Eds.), Fields virology (pp. 1163–1185). Philadephia, PA: Lippincott‐Raven. [Google Scholar]
- Lee, A. S. (2014). Glucose‐regulated proteins in cancer: Molecular mechanisms and therapeutic potential. Nature Reviews Cancer, 14(4), 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M. , Tseng, H.‐K. , Trejaut, J. A. , Lee, H.‐L. , Loo, J.‐H. , Chu, C.‐C. , & Tsai, Z.‐U. (2003). Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Medical Genetics, 4(1), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. , Wu, H. , & Zhu, N. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen, S. , Chua, R. L. , Trefzer, T. , Kahn, N. C. , Schneider, M. A. , Muley, T. , & Boots, A. W. (2020). SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are predominantly expressed in a transient secretory cell type in subsegmental bronchial branches. bioRxiv.39(10), e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manevski, M. , Muthumalage, T. , Devadoss, D. , Sundar, I. K. , Wang, Q. , Singh, K. P. , & Rahman, I. (2020). Cellular stress responses and dysfunctional Mitochondrial–cellular senescence, and therapeutics in chronic respiratory diseases. Redox Biology, 20, 101443. 10.1016/j.redox.2020.101443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M. A. , Jones, S. J. , Astell, C. R. , Holt, R. A. , Brooks‐Wilson, A. , Butterfield, Y. S. , & Chan, S. Y. (2003). The genome sequence of the SARS‐associated coronavirus. Science, 300(5624), 1399–1404. [DOI] [PubMed] [Google Scholar]
- Matsuyama, S. , Nagata, N. , Shirato, K. , Kawase, M. , Takeda, M. , & Taguchi, F. (2010). Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. Journal of Virology, 84(24), 12658–12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama, S. , Nao, N. , Shirato, K. , Kawase, M. , Saito, S. , Takayama, I. , & Kato, F. (2020). Enhanced isolation of SARS‐CoV‐2 by TMPRSS2‐expressing cells. Proceedings of the National Academy of Sciences of the United States of America, 117(13), 7001–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska, A. , Kula‐Pacurar, A. , Wadas, J. , Suder, A. , Szczepanski, A. , Dabrowska, A. , & Rajfur, Z. (2020). Replication of SARS‐CoV‐2 in human respiratory epithelium. bioRxiv. 10.1101/2020.03.20.999029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil, V. , Kwon, H. , Prado, P. , Hagelkrüys, A. , Wimmer, R. A. , Stahl, M. , & Prosper, F. (2020). Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell, 181, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, A. , David, J. K. , Maden, S. K. , Wood, M. A. , Weeder, B. R. , Nellore, A. , & Thompson, R. F. (2020). Human leukocyte antigen susceptibility map for SARS‐CoV‐2, medRxiv, (–). 10.1128/JVI.00510-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, X. , Liu, Y. , Lei, X. , Li, P. , Mi, D. , Ren, L. , & Qian, Z. (2020). Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nature Communications, 11(1), 1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikhshahrokh, A. , Ranjbar, R. , Saeidi, E. , Dehkordi, F. S. , Heiat, M. , Ghasemi‐Dehkordi, P. , & Goodarzi, H. (2020). Frontier therapeutics and vaccine strategies for SARS‐CoV‐2 (COVID‐19): A review. Iranian Journal of Public Health, 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen, M. A. , Khan, S. , Kazmi, A. , Bashir, N. , & Siddique, R. (2020). COVID‐19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato, K. , Kawase, M. , & Matsuyama, S. (2018). Wild‐type human coronaviruses prefer cell‐surface TMPRSS2 to endosomal cathepsins for cell entry. Virology, 517, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, G. , Gosalia, D. N. , Rennekamp, A. J. , Reeves, J. D. , Diamond, S. L. , & Bates, P. (2005). Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proceedings of the National Academy of Sciences of the United States of America, 102(33), 11876–11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawiski, E. W. , Diwanji, D. , Suryamohan, K. , Gupta, R. , Fellouse, F. A. , Sathirapongsasuti, F. , & Seshagiri, S. (2020). Human ACE2 receptor polymorphisms predict SARS‐CoV‐2 susceptibility. bioRxiv. 10.1101/2020.04.07.024752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , & Xi, Y. (2014). Association between HLA gene polymorphism and the genetic susceptibility of SARS infection, HLA and associated important diseases (p. 311). London, UK: IntechOpen. [Google Scholar]
- Sun, M. , Yang, J. , Sun, Y. , & Su, G. (2020). Inhibitors of RAS might be a good choice for the therapy of COVID‐19 pneumonia. Zhonghua jie he he hu xi za zhi= Zhonghua jiehe he huxi zazhi= Chinese journal of tuberculosis and respiratory diseases, 43, E014. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Wu, C. , Li, X. , Song, Y. , Yao, X. , Wu, X. , & Lu, J. (2020). On the origin and continuing evolution of SARS‐CoV‐2. National Science Review, 1–12. 10.1093/nsr/nwaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipnis, S. R. , Hooper, N. M. , Hyde, R. , Karran, E. , Christie, G. , & Turner, A. J. (2000). A human homolog of angiotensin‐converting enzyme cloning and functional expression as a captopril‐insensitive carboxypeptidase. Journal of Biological Chemistry, 275(43), 33238–33243. [DOI] [PubMed] [Google Scholar]
- Utsumi, K. , Yokota, Y. , Ishikawa, T. , Ohnishi, K. , & Fujiwara, K. (1990). Reproductive disorders in female SHR rats infected with sialodacryoadenitis virus, Coronaviruses and their diseases (pp. 525–532). Boston, MA: Springer. [DOI] [PubMed] [Google Scholar]
- Vaduganathan, M. , Vardeny, O. , Michel, T. , McMurray, J. J. , Pfeffer, M. A. , & Solomon, S. D. (2020). Renin–Angiotensin–Aldosterone system inhibitors in patients with COVID‐19. New England Journal of Medicine, 382, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verity, R. , Okell, L. C. , Dorigatti, I. , Winskill, P. , Whittaker, C. , Imai, N. , & Ferguson, N. M. (2020). Estimates of the severity of coronavirus disease 2019: A model‐based analysis. The Lancet Infectious Diseases, 20, 669–677. 10.1016/S1473-3099(20)30243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Chen, W. , Zhou, Y.‐S. , Lian, J.‐Q. , Zhang, Z. , Du, P. , & Geng, J.‐J. (2020). SARS‐CoV‐2 invades host cells via a novel route: CD147‐spike protein. bioRxiv. 10.1101/2020.03.14.988345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. , Yu, B. , Chen, Y.‐M. , Wang, W. , Song, Z.‐G. , & Zhang, Y.‐Z. (2020). Author correction: A new coronavirus associated with human respiratory disease in China. Nature, 580, E7. 10.1038/s41586-020-2202-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.‐H , Deng, W. , Tong, Z. , Liu, Y.‐X , Zhang, L.‐F , Zhu, H. , & Ma, C.‐M (2007). Mice transgenic for human angiotensin‐converting enzyme 2 provide a model for SARS coronavirus infection. Comparative Medicine, 57(5), 450–459. [PubMed] [Google Scholar]
- Yurchenko, V. , Constant, S. , & Bukrinsky, M. (2006). Dealing with the family: CD147 interactions with cyclophilins. Immunology, 117(3), 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Penninger, J. M. , Li, Y. , Zhong, N. , & Slutsky, A. S. (2020). Angiotensin‐converting enzyme 2 (ACE2) as a SARS‐CoV‐2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Medicine, 46(4), 586–590. 10.1007/s00134-020-05985-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Yang, Y. , Huang, H.‐P. , Li, D. , Gu, D.‐F. , & Liu, Y.‐K. (2020). Relationship between the ABO blood group and the COVID‐19 susceptibility. medRxiv. 10.1101/2020.03.11.20031096 [DOI] [Google Scholar]
- Zhao, Y. , Zhao, Z. , Wang, Y. , Zhou, Y. , Ma, Y. , & Zuo, W. (2020). Single‐cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019‐nCov. bioRxiv. 10.1101/2020.01.26.919985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X.‐L. , Wang, X.‐G. , Hu, B. , Zhang, L. , Zhang, W. , & Shi, Z. ‐L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora, P. , Moldenhauer, A.‐S. , Hofmann‐Winkler, H. , & Pöhlmann, S. (2015). TMPRSS2 Isoform 1 activates respiratory viruses and is expressed in viral target cells. PLoS One, 10(9), e0138380. 10.1371/journal.pone.0138380 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available upon request.


