The global pandemic of COVID‐19, caused by severe acute respiratory syndrome (SARS) coronavirus (CoV)‐2, has seen the world's physical connectedness and economy come to a standstill. Whilst simple public health interventions—handwashing and cough etiquette, social distancing, testing and isolation—have been able to suppress SARS‐CoV‐2 spread when applied in a timely, coordinated manner, there remain many gaps in our knowledge of the disease, its origin and transmission, treatment and prevention. In this issue of Transboundary and Emerging Diseases, several articles fill some of these gaps.
To reiterate a statement made in our previous issue, Transboundary and Emerging Diseases is committed to publishing high‐quality research on SARS‐CoV‐2 (Ward, Li, & Tian, 2020).
Yu et al. (2020) in this issue describe the epidemiological and clinical characteristics of 333 confirmed COVID‐19 cases in Shanghai, during the early phase of the pandemic. Estimates of some key epidemiologic parameters—such as incubation period—are presented, and risk factors are described. Importantly, the role of travel to Wuhan features prominently. It is also an example of the successful control of COVID‐19 within a megacity.
Bauer et al. (2020) present a novel approach to choosing an animal model for SARS‐CoV‐2 studies. In the case of a newly discovered virus such as SARS‐CoV‐2, the laborious process of matching the available virus strain with an animal model can be substantially improved by assessing genome‐wide functionalities. This has some profound implications for the development of diagnostics, vaccines and treatments, all of which are dependent on effective animal models.
A key gap in our knowledge of SARS‐CoV‐2 transmission is the role of animals, and despite a consensus that this virus arose from an animal reservoir, a fundamental and intriguing question is which species? Deng et al. (2020) present the results of a SARS‐CoV‐2 serological survey in 35 animal species in China, including the dog of a COVID‐19 patient and an additional two in‐contact dogs. Despite testing 1,914 sera using a double antigen sandwich ELISA, SARS‐CoV‐2‐specific antibodies were not detected. As stressed by Li (2020) in a Letter to the Editors, pets such as cats can be infected when the infection pressure is sufficiently high (such as in Wuhan, China during the epidemic); however, accumulating evidence suggests that pets such as cats are unlikely to be a reservoir of SARS‐CoV‐2. In addition, diagnostic methods used on animals require validation. Tests available for the detection of SARS‐CoV‐2 are comprehensively described in this issue of Transboundary and Emerging Diseases (Li & Ren, 2020).
In addition to the publication of new knowledge about SARS‐CoV‐2 in this issue of Transboundary and Emerging Diseases, new ideas are also presented. Roe (2020a) has suggested an explanation for the neurological symptoms (acute cerebrovascular disease) noted in COVID‐19 patients, as well as apparent virus reactivation in patients: by analogy with the known pathogenesis of SARS and Nipah viruses, selective infection of neurons enables a virus to evade attacks from the host's immune system. If demonstrated to be the case, this has public health implications that will remain well after the current pandemic of acute COVID‐19 is over. In addition, in this issue the mechanism of thrombosis in COVID‐19 patients is explored (Roe, 2020b).
In recent times, we have faced many global health crises—such as COVID‐19, African swine fever and avian influenza—to the extent that overlapping emergencies have occurred. Stoffel et al. (2020) argue in a Letter to the Editors in the previous issue of Transboundary and Emerging Diseases that to address such a situation we need a health network of global scope with rapid and open exchange of information and that a One Health approach might have the potential to provide such a network. In the case of COVID‐19, a cluster of 27 cases of 'pneumonia of unknown cause' emerged in Wuhan, Hubei province, China and was reported by local health officials at the end of December 2019 (Wang, Horby, Hayden, & Gao, 2020)—although there was evidence of human‐to‐human transmission as early as at least mid‐December (Li et al., 2020). In January, the aetiology was identified as a new coronavirus (subsequently named SARS‐CoV‐2). Thus, disease emergence was followed by discovery of a cause. The general consensus is that SARS‐CoV‐2 is of animal origin—a zoonosis—but despite various hypotheses, the species has not been identified. The process of disease emergence and then pathogen discovery (illustrated in the following Figure, upper panel) is characteristic of separate human and animal health systems, a lack of coordination between these sectors, and a response which is reactive to clinical disease occurrence. Rather than disease emergence (especially in humans in the case of a zoonosis) leading to pathogen discovery, the opposite is what we need to strive for. Discovery of zoonotic agents in animal populations, a thorough risk assessment driven by knowledge of the hazard and the likelihood of spillover, and then integrated monitoring and surveillance of animal and human populations can shift the world into a paradigm of discovery that prevents emergence and spread (as illustrated in the following Figure, lower panel). A key enabler of such a shift in our thinking and approach to disease emergence and spread is a One Health workforce capable of undertaking integrated monitoring, surveillance, risk assessment and response activities.
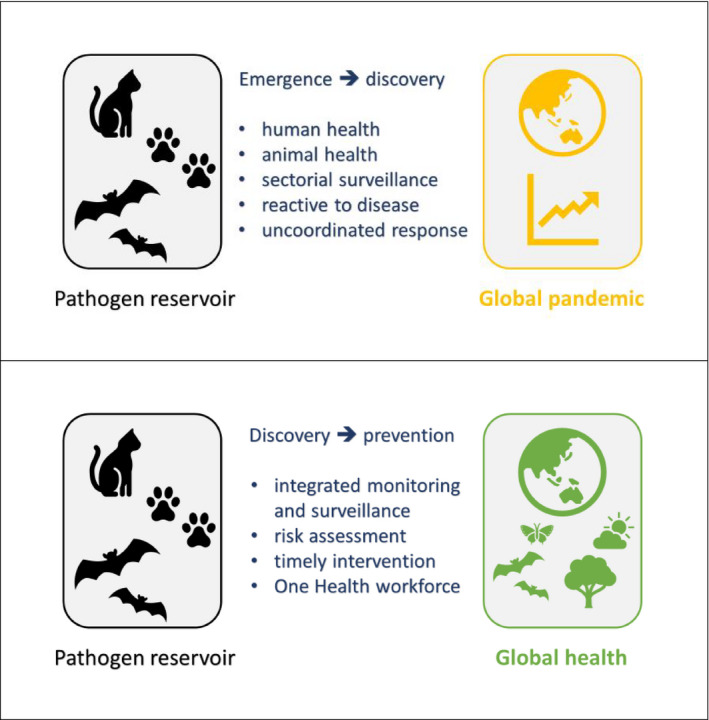
Specifically in terms of One Health surveillance, barriers to operationalization include legal issues, hurdles to data sharing, unclear responsibilities and structural barriers between human and animal authorities that prevent integrated action (Stärk et al., 2015). In addition, as identified by Stärk et al. (2015), ‘policy makers in the health sector often perceive One Health as a veterinary‐driven initiative that is not particularly relevant to their priority problems’. There is a lack of funding for the development of a sustainable One Health workforce, perhaps because of the scarcity of a strong business case (costs versus benefits) for One Health surveillance. Integration (versus specialization) in heath training at an undergraduate level, to promote the inter‐relatedness of medical, veterinary and environmental sciences, is also a need (Stärk et al., 2015). The COVID‐19 pandemic could be a catalyst for such a seismic shift in how we approach emerging infectious diseases and One Health.
The emergence of infectious diseases can be driven by interconnected economic, social and environmental changes. We have many tools available for identification, prioritization and investigation of emerging infectious diseases impacting human and animal health—including environmental and horizon scanning, surveillance, disease prioritization, risk assessment and disease modelling (Brookes, Hernandez‐Jover, Black, & Ward, 2015). EIDs are complex, multifactorial health problems for which a single tool is not sufficient; however, integration of the tools already available into a systematic approach for the development of tactical and strategic plans for emerging risk preparedness is lacking (Brookes et al., 2015).
We can be sure, even when the current COVID‐19 pandemic is resolved, that the need for surveillance, response and prevention of transboundary and emerging diseases will remain. An integrated approach to health—regardless of specific pathogens, domains or nations—is a critical need. Issues of politics, trade and commerce must be recognized, but can no longer be used as excuses for inaction.
Ethics Statement
The author confirms that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required for this editorial.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Bauer, D. C. , Tay, A. P. , Wilson, L. O. W. , Reti, D. , Hosking, C. , McAuley, A. J. , … Vasan, S. S. (2020). Supporting pandemic response using genomics and bioinformatics: A case study on the emergent SARS‐CoV‐2 outbreak. Transboundary and Emerging Diseases, 67(4), 1453–1462. 10.1111/tbed.13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes, V. J. , Hernandez‐Jover, M. , Black, P. F. , & Ward, M. P. (2015). Preparedness for emerging infectious diseases: Pathways from anticipation to action. Epidemiology and Infection, 143, 2043–2058. 10.1017/S095026881400315X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, J. , Jin, Y. , Liu, Y. , Sun, J. , Hao, L. , Bai, J. , … Tian, K. (2020). Serological survey of SARS‐CoV‐2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transboundary and Emerging Diseases, 67(4), 1745–1749. 10.1111/tbed.13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , & Ren, L. (2020). Recent progress on the diagnosis of 2019 Novel Coronavirus. Transboundary and Emerging Diseases, 67(4), 1485–1491. 10.1111/tbed.13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. (2020). Cats under the shadow of the SARS‐CoV‐2 pandemic. Transboundary and Emerging Diseases, 67(4), 1416–1417. 10.1111/tbed.13599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, K. (2020a). Explanation for COVID‐19 infection neurological damage and reactivations. Transboundary and Emerging Diseases, 67(4), 1414–1415. 10.1111/tbed.13594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, K. (2020b). High COVID‐19 virus replication rates, the creation of antigen‐antibody immune complexes, and indirect hemagglutination resulting in thrombosis. Transboundary and Emerging Diseases, 67(4), 1418–1421. 10.1111/tbed.13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stärk, K. D. C. , Kuribreña, M. A. , Dauphin, G. , Vokaty, S. , Ward, M. P. , Wieland, B. , & Lindberg, A. (2015). One health surveillance: More than a buzz word? Preventive Veterinary Medicine, 120(1), 124–130. 10.1016/j.prevetmed.2015.02.020 [DOI] [PubMed] [Google Scholar]
- Stoffel, C. , Schuppers, M. , Buholzer, P. , Muñoz, V. , Lechner, I. , Sperling, U. , … de Nardi, M. (2020). The ongoing crises in China illustrate that the assessment of epidemics in isolation is no longer sufficient. Transboundary and Emerging Diseases, 67(3), 1043–1044. 10.1111/tbed.13536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Horby, P. W. , Hayden, F. G. , & Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, M. P. , Li, X. , & Tian, K. (2020). Novel coronavirus 2019, an emerging public health emergency. Transboundary and Emerging Diseases, 67(2), 469–470. 10.1111/tbed.13509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Sun, X. , Cui, P. , Pan, H. , Lin, S. , Han, R. , … Fu, C. (2020). Epidemiological and clinical characteristics of 333 confirmed cases with coronavirus disease 2019 in Shanghai, China. Transboundary and Emerging Diseases, 67(4), 1697–1707. 10.1111/tbed.13604 [DOI] [PMC free article] [PubMed] [Google Scholar]


