Abstract
Coronavirus disease 2019, caused by severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), leads to a series of clinical symptoms of respiratory and pulmonary inflammatory reactions via unknown pathologic mechanisms related to the viral infection process in tracheal or bronchial epithelial cells. Investigation of this viral infection in the human bronchial epithelial cell line (16HBE) suggests that SARS‐CoV‐2 can enter these cells through interaction between its membrane‐localized S protein with the angiotensin‐converting enzyme 2 molecule on the host cell membrane. Further observation indicates distinct viral replication with a dynamic and moderate increase, whereby viral replication does not lead to a specific cytopathic effect but maintains a continuous release of progeny virions from infected cells. Although messenger RNA expression of various innate immune signaling molecules is altered in the cells, transcription of interferons‐α (IFN‐α), IFN‐β, and IFN‐γ is unchanged. Furthermore, expression of some interleukins (IL) related to inflammatory reactions, such as IL‐6, IL‐2, and IL‐8, is maintained at low levels, whereas that of ILs involved in immune regulation is upregulated. Interestingly, IL‐22, an IL that functions mainly in tissue repair, shows very high expression. Collectively, these data suggest a distinct infection process for this virus in respiratory epithelial cells, which may be linked to its clinicopathological mechanism.
Keywords: ACE2, human bronchial epithelial cell line (16HBE), IL‐22, SARS‐CoV‐2 (COVID‐19)
Highlights
SARS‐CoV‐2 does not lead to a specific cytopathic effect in 16HBE cells, but maintains continuous release of progeny virions from infected cells. During infection, IL‐22, an interleukin that functions mainly in tissue repair, showed very high expression.
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), a new member of the beta coronavirus family named by the World Health Organization (WHO), 1 , 2 has been identified as the causative pathogen of viral coronavirus disease 2019 (COVID‐19) pneumonia. The clinical symptoms of COVID‐19 include fever, cough, fatigue, shortness of breath, and multiple patchy shadows in both lungs; this disease was reported primarily in Wuhan, China, beginning in December 2019. 3 , 4 However, SARS‐CoV‐2 has currently been a worldwide concern for some time. 5 SARS‐CoV‐2 shows approximately 70% similarity in genomic sequence with SARS‐CoV but is more highly contagious among adults, 6 which has led to more than 80 000 cases of infection, including 1000 severe cases in Wuhan city over a 2‐month period. 7 The fact that the high risk of spreading this viral infection is observed not only during the period of clinical disease but also during the 14 days of the asymptomatic latent period suggests a distinct infection process for this virus in the respiratory tract, especially in bronchial and alveolar epithelial cells. 8 , 9 Indeed, this may be logically inferred to be pathologically related to the severe lesions clinically observed in lung tissues. 10 Based on this hypothesis, integrated clinical and epidemiological data and the physiological structure of the respiratory tract, 11 , 13 we used the human bronchial epithelial cell line (16HBE) in this study to investigate the characteristics of SARS‐CoV‐2 infection in an in vitro culture system and show the distinct infection process of this virus. The cell line 16HBE is derived from a healthy individual, and according to various studies of viral infection, including SARS‐CoV and MERS infections, the cells express angiotensin‐converting enzyme 2 (ACE2) in the membrane. 14 , 16 Our findings are anticipated to provide data useful for understanding the clinicopathological characteristics of COVID‐19, even though this cell line, as one of the human airway epithelial cells available, originated from a child and might not accurately represent the epithelial cells in bronchial tissues.
2. MATERIALS AND METHODS
2.1. Cells
The Vero cells (American Type Culture Collection, Manassas, VA) used in this work were provided by the WHO and were permissible for use in viral vaccine production at passages 142 to 148. 17 The cells were cultured in dulbecco's modified eagle medium (DMEM; Corning, NY) supplemented with 5% fetal bovine serum (FCS; HyClone, Logan, UT) and grown to a monolayer after passaging at a ratio of 1:2. Human bronchial epithelial 16HBE cells purchased from Corning were grown in‐high glucose DMEM (Corning) supplemented with 5% FCS and formed a monolayer at 48 hours after passaging.
2.2. Virus
The KMS‐1 strain of SARS‐CoV‐2 was isolated from respiratory secretions from an adult male patient diagnosed with COVID‐19 at Yunnan Hospital of Infectious Diseases in Kunming in January 2020. Vero cells were inoculated with the sputum of this patient and further incubated at 37°C, and a typical cytopathic effect (CPE) was observed within 5 days (Figure S2). This replicated virus replicated continuously in Vero cells for two passages and was selected by plaque cloning. The cloned virus was identified via genomic sequencing and named KMS‐1 (GenBank No: MT226610.1).
2.3. Viral titration
Virus samples were serially diluted 10‐fold with serum‐free DMEM (Corning). Different dilutions of the virus were added to a 96‐well plate with a quantitative sampler. Each dilution (100 µL per well) was added to eight parallel wells, after which 100 µL of Vero cell suspension was added to each well at a concentration of 2.5 × 105 cells/mL. After the 96‐well plate was placed in an atmosphere of 5% carbon dioxide and incubated in a 37°C constant temperature incubator for 6 to 7 days, the cell lesions were assessed with an inverted microscope; more than 50% of the cells were observed to exhibit lesions. A plaque assay for viral titration was performed based on a previously described protocol. 18 Briefly, 10‐fold serial dilutions of samples were prepared in DMEM without FBS and added to monolayer Vero cells overlaid with 0.6% agarose in growth medium (DMEM containing 5% FBS) at 37°C. The cells were fixed and stained with 0.2% crystal violet at 4 days after infection.
2.4. Quantitation of the viral genome by qRT‐PCR
16HBE cells in a T25 culture flask were infected with SARS‐CoV‐2. Samples of supernatant and cells were collected at different time points. The primers used for quantitative real‐time PCR (qRT‐PCR) were specific for N and ORF1ab sequences in the SARS‐CoV‐2 genome (Table 1). Reactions were performed using a One Step PrimeScript RT‐PCR Kit (Perfect Real Time).
Table 1.
Sequences of primers for qPCR
| Primers | Sequences (5′→3′) |
|---|---|
| ORF1ab F | CCCTGTGGGTTTTACACTTAA |
| ORF1ab R | ACGATTGTGCAGCTGA |
| ORF1ab Probe | 5′‐FAM‐ CCGTCTGCGGTATGTGGAAAGGTTATGG‐BHQ1‐3′ |
| N F | GGGGAACTTCTCCTGCTAGAAT |
| N R | CAGACATTTTGCTCTCAAGCTG |
| N Probe | 5′‐FAM—TTGCTGCTGCTTGACAGATT‐TAMRA‐3′ |
| IFN‐α F | ACCCCTGCTATAACTATGACC |
| IFN‐α R | CTAACCACAGTGTAAAGGTGC |
| IFN‐β F | AACTCCACCAGCAGACAG |
| IFN‐β R | GAGAGCAGTTGAGGACATC |
| RANKL F | GGAGGAAGCACCAAGTATT |
| RANKL R | CCTCTCCAGACCGTAACT |
| IFN‐γ F | ATGAACGCTACACACTGCATC |
| IFN‐γ R | CCATCCTTTTGCCAGTTCCTC |
| TL1A F | AAGCCAGACTCCATCACT |
| TL1A R | TACCTACTTCGCATACAGAC |
| IFN‐λ F | GGACGCCTTGGAAGAGTCACT |
| IFN‐λ R | AGAAGCCTCAGGTCCCAATTC |
| TNF‐α F | GTGAGGAGGACGAACATC |
| TNF‐α R | TGAGCCAGAAGAGGTTGA |
| LIGHT F | TCTTGCTGTTGTTCATTGC |
| LIGHT R | CCTTCTTGGATGCTTCATTC |
| LTa3 F | GATGTCTGTCTGGCTGAG |
| LTa3 R | CCTGCTCTTCCTCTGTGT |
| GMCSF F | TCCTGMCCTGAGTAGAGACAC |
| GMCSF R | TGCTGCTTGTAGTGGCTGG |
| IL‐1β F | GCAACTGTTCCTGAACTCAACT |
| IL‐1β R | ATCTTTTGGGGTCCGTCAACT |
| IL‐2 F | TCCTGTCTTGCATTGCACTAAG |
| IL‐2 R | CATCCTGGTGAGTTTGGGATTC |
| IL‐6 F | ACTCACCTCTTCAGAACGAATTG |
| IL‐6 R | CCATCTTTGGAAGGTTCAGGTTG |
| IL‐8 F | ACTGAGAGTGATTGAGAGTGGAC |
| IL‐8 R | AACCCTCTGCACCCAGTTTTC |
| IL‐12 F | ACCAGGTGGAGTTCAAGA |
| IL‐12 R | GCTCATCACTCTATCAATAGTC |
| IL‐13 F | CCTCATGGCGCTTTTGTTGAC |
| IL‐13 R | TCTGGTTCTGGGTGATGTTGA |
| IL‐33 F | GTGACGGTGTTGATGGTAAGAT |
| IL‐33 R | AGCTCCACAGAGTGTTCCTTG |
| IL‐4 F | GGTCTCAACCCCCAGCTAGT |
| IL‐4 R | GCCGATGATCTCTCTCAAGTGAT |
| IL‐5 F | CTCTGTTGACAAGCAATGAGACG |
| IL‐5 R | TCTTCAGTATGTCTAGCCCCTG |
| IL‐10 F | TACGGCGCTGTCATCGATTT |
| IL‐10 R | AAGGTTTCTCAAGGGGCTGG |
| IL‐17 F | AGATTACTACAACCGATCCACCT |
| IL‐17 R | GGGGACAGAGTTCATGTGGTA |
| IL‐22 F | GCTTGACAAGTCCAACTTCCA |
| IL‐22 R | GCTCACTCATACTGACTCCGT |
| GAPDH F | GCGAGATCCCTCCAAAATCAA |
| GAPDH R | GTTCACACCCATGACGAACAT |
Abbreviations: F, forward; GADPH, glyceraldehyde 3‐phosphate dehydrogenase; IL, interleukin; IFN, interferon; qPCR, quantitative PCR; R, reverse; TNF, tumor necrosis factor.
2.5. Infection of 16HBE cells with SARS‐CoV‐2
As 16HBE cells grew to a monolayer in DMEM‐high glucose supplemented with 5% FCS, the SARS‐CoV‐2 KMS‐1 strain was inoculated at an multiple of infection (MOI) of 0.2. After virus attachment to the receptor for 10 minutes, DMEM‐high glucose supplemented with 0.2% FCS was added. Samples, including supernatant and cell samples, were collected every 24 hours for various assessments, including viral titration and transcriptional profile analysis of certain cellular messenger RNAs (mRNAs). The samples used for electron microscope observation were collected from cells infected with the virus at an MOI of 0.5.
2.6. Immunofluorescence detection of virus in 16HBE cells
16HBE cells grown in glass plates were infected with SARS‐CoV‐2 at an MOI of 0.2, incubated at 37°C, and collected every 24 hours. After discarding the supernatant and washing with 20 mM phosphate‐buffered saline (PBS), the cells were fixed with paraformaldehyde for 24 hours. The fixed cells were first stained with antibodies against viral N protein (Snio Biological, Beijing, China), viral S1 protein (Snio Biological) and cellular ACE2 (Genetex, CA) and then with secondary antibodies labeled with different fluorophores (Genetex) according to standard protocols for further observation under a fluorescence microscope. 19
2.7. Immunoprecipitation–Western blot analysis
16HBE cells cultured in a culture flask were infected with SARS‐CoV‐2 at an MOI of 0.2, incubated at 37°C, and collected every 24 hours. After discarding the supernatant and washing with 20 mM PBS, the cells were lysed with RIPA buffer (Beyotime, China) for 24 hours. Immunoprecipitation and Western blotting were performed with specific antibodies against S (Snio Biological) and ACE2 (Genetex).
2.8. Electronic microscope observation
16HBE cells in a T25 culture flask were infected with SARS‐CoV‐2. Samples were collected at different time points, the supernatant was discarded, and the cells were centrifuged at 3000 rpm for 2 minutes. The cell precipitate was collected, resuspended in 1 mL of glutaraldehyde and fixed for 48 hours for electron microscopy.
2.9. mRNA transcription profile of genes encoding innate immune signaling molecules
16HBE cells in a T25 culture flask were infected with SARS‐CoV‐2. Samples were collected at different time points, and the cells were suspended in TRIzol to extract total mRNA. Reactions were performed using a One Step TB Green PrimeScript PLUS RT‐PCR Kit (Perfect Real Time). The primers used for qRT‐PCR are shown in Table 1.
2.10. Statistical analysis
Data are shown as the mean and standard deviation. GraphPad Prism software (San Diego, CA) was used for statistical analyses.
3. RESULTS
3.1. The S protein of SARS‐CoV‐2 interacts with ACE2 on 16HBE cell
Recent data suggest that similar to SARS‐CoV, SARS‐CoV‐2 infects cells via the binding of its S protein to the ACE2 molecule on the host cell surface. 20 The fact that COVID‐19 caused by SARS‐CoV‐2 begins in the respiratory tract logically suggests that the virus can infect epithelial cells in the respiratory tract, probably through S‐ACE2 interaction. If so, infection of 16HBE cells with this virus should provide clues for understanding the clinicopathological mechanism of this viral infection. First, we investigated the interaction of the viral S protein with ACE2, which has been identified to be expressed on the 16HBE cell membrane. 21 Confocal fluorescence microscopy observation suggested that explicit interactions occur between the S protein and ACE2, as evidenced by the colocalization of green (S protein) and red (ACE2) fluorescence on the surface of infected 16HBE cell surfaces at 2 hours postvirus inoculation, demonstrating spatial linking of both proteins in the membrane (Figure 1A). In addition, the virus particles aggregated at the circular periphery of the cells, as evidenced by green fluorescence surrounding the cells (Figure 1B). To further confirm this interaction of the S protein with ACE2, immunoprecipitation and Western blotting were performed with specific antibodies. The antibody against ACE2 precipitated the S protein, which was recognized by the antibody against the S protein in Western blot analysis; similarly, the antibody against S precipitated ACE2 molecules (Figure 1C). These results indicate that SARS‐CoV‐2 can attach to the 16HBE cell surface through binding of the S protein to the ACE2 molecule.
Figure 1.
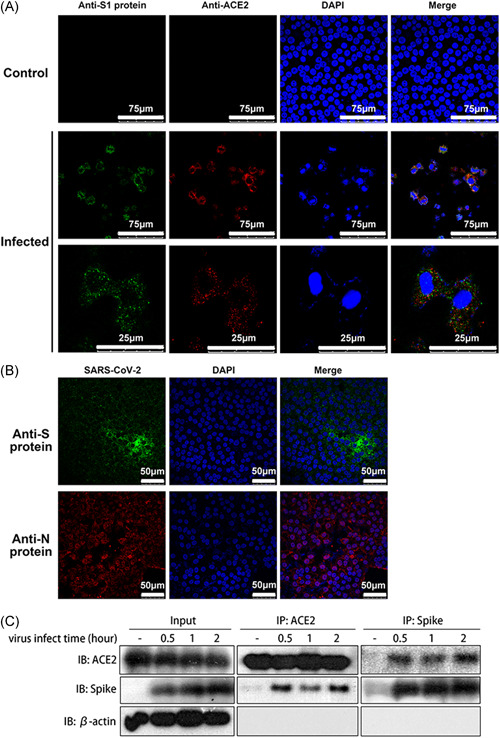
The S protein binds to the ACE2 molecule and mediates viral attachment to 16HBE cells. A, Immunofluorescence colocalization indicating the interaction between the S protein (spike protein, green) and the ACE2 molecule (red). The secondary antibodies used were goat anti‐rabbit IgG antibody (DyLight 594) and goat anti‐mouse IgG antibody (DyLight 488). Samples were obtained at 2 hpi. B, Immunofluorescence results indicating that virus particles aggregated around the 16HBE cells. The virus particles are shown by the anti‐S protein (green) antibodies and the anti‐N protein (nucleocapsid protein, red). The secondary antibodies used were goat anti‐rabbit IgG antibody (DyLight 594) and goat anti‐mouse IgG antibody (DyLight 488). Samples were obtained at 72 hpi. C, Immunoprecipitation‐Western blot analysis of the interaction between the S protein and ACE2. β‐actin as internal reference. ACE2, angiotensin‐converting enzyme 2; 16HBE, human bronchial epithelial cell line; IgG, immunoglobulin G
3.2. Replication of SARS‐CoV‐2 can be maintained in 16HBE cells for longer than 7 days
In general, most respiratory viruses, such as influenza virus, rhinovirus, and metapneumovirus, infect epithelial cells of the respiratory tract via cell lysis. 22 , 24 This leads not only to rapid viral replication for further spread in tissues of the trachea and lung but also to pathological lesions in these tissues, followed by local innate inflammatory reactions. 25 Evaluation of 16HBE cells infected with SARS‐CoV‐2 suggested that viral genome copy numbers and viral infectious titers in the supernatant of infected cells tended to increase for 7 days (Figure 2A,B). Moreover, a notable increase on the 3rd day was found after maintaining low viral genome copy numbers detected by qRT‐PCR and viral titers identified by plaque assay (Figure 2A,B), though the virus continually maintained high and stable replication in cells (Figure 2). This process is different from that observed in Vero cells (Figure S1).
Figure 2.
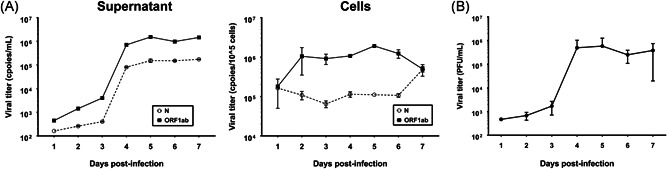
SARS‐CoV‐2 replication is maintained in human bronchial cells. A, Measurement of the viral genome copy number in the supernatant and 16HBE cells infected with SARS‐CoV‐2 by qRT‐PCR. N: Primers and probes specific for the N sequence were used to detect viral copies. ORF1ab: Primers and probes specific for the ORF1ab sequence were used to detect viral copies. B, Measurement of the virus infectious titers in the supernatant of 16HBE cells infected with SARS‐CoV‐2 by plaque assay. SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.3. SARS‐CoV‐2 infection in 16HBE cells does not lead to distinct CPEs, but the virus is released from infected cells
In epithelial cells, infection by most viruses leads to the typical outcome of the CPE, namely, cytoclasis of cells and a single release of progeny virions for rapid further infection of cells in the surrounding tissues. 26 However, in 16HBE cells, SARS‐CoV‐2 infection did not elicit the typical CPE observed with other respiratory viruses (Figure 3A), and the fluorescent assay using antibody anainst N protein showing almost 100% of cells infected by virus (Figure 3B). While electronic microscopy observation suggested that many virions were released from infected cells via membrane penetration, even during the late phase of infection (Figure 3C). These data suggest a distinct replication mode of the virus in bronchial epithelial cells that does not produce obvious damage to the cells and allows continuous virus release, as SARS‐Cov‐2 produces a typical CPE in Vero cells (Figure S2A) and 16HBE cells exhibit the typical CPEs after infection by other viruses, including enterovirus and herpes simplex virus type I (Figure S2B,C). This strategy may delay clearance and control of the immune response to viral invasion.
Figure 3.
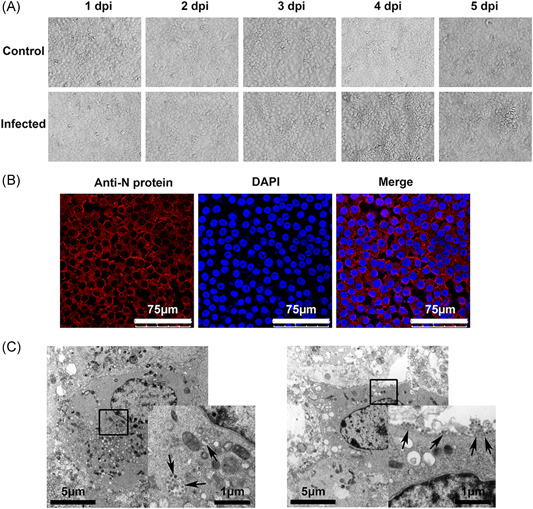
16HBE cells infected with SARS‐CoV‐2 do not show CPEs as the virus is released. A, CPEs were observed in infected 16HBE cells by SARS‐CoV‐2. Magnification, ×200. B, The percentage of cells are getting infected at the 3rd day after infection by Immunofluorescence. The virus particles are shown by the anti‐N protein (nucleocapsid protein, red). The secondary antibodies used were goat anti‐rabbit IgG antibody (DyLight 594). C, Electron micrograph of SARS‐CoV‐2‐infected 16HBE cells. SARS‐CoV‐2 virus particles are indicated by the arrows. CPE, cytopathic effect; 16HBE, human bronchial epithelial cell line; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.4. SARS‐CoV‐2 infection in 16HBE cells leads to a modified transcription profile of various innate immune signaling molecules
During the process of respiratory viral infection, epithelial cells of the trachea and bronchial tissues play important roles in the innate immune response via intracellular pattern recognition receptors, which recognize viral pathogen‐associated molecular patterns (PAMPs). This recognition can initiate specific transcriptional responses based on the NF‐kB pathway to upregulate various immune signaling molecules, including members of the interferon (IFN), tumor necrosis factor (TNF), and interleukin (IL) families. 27 These signaling molecules play important roles in activating and/or recruiting innate immune cells, such as dendritic cells, innate lymphoid cells, and macrophages. 28 , 29 In addition, these events lead to local inflammatory reactions and activation of the innate immune response, in turn, eliciting an adaptive immune response via antigen presentation to T cells. 30 , 31 In this study, we logically hypothesized that the transcription profile of some immune signaling molecules in 16HBE cells infected with SARS‐CoV‐2 is modified by viral PAMPs, which might be important for viral pathological mechanisms. The observation of the mRNA profile of genes encoding various immune signaling molecules suggested the following. First, the levels of kinase molecules involved in the NF‐kB signaling pathway, including NIK, TAK, and IKKα, were not apparently changed during 7 days postinfection—except for IKKβ, which showed a trend of upregulation via an unknown mechanism (Figure 4A). Second, the mRNA transcripts of some molecules mediating the antiviral response to viral infection, including IFNα, IFNβ, IFNγ, RANKL, and TL1A, among others, did not change, but those of IFNλ, TNF‐α, LIGHT, and LTa3 GMCSF were upregulated with an increasing trend (Figure 4B). Some ILs related to inflammatory reactions, including IL‐1β, IL‐2, IL‐6, IL‐8, IL‐12, IL‐13, and IL‐33, were downregulated, but those involved in immune regulation, such as IL‐4, IL‐5, IL‐10, and IL‐17, were upregulated (Figure 4C). Interestingly, expression of IL‐22, the IL reported to be related to tissue repair, 32 was much higher in infected cells than in control cells, revealing not only the mRNA transcriptional level (300‐fold) but also the expression level (4‐fold) (Figure 4D,E). These data suggest that SARS‐CoV‐2 interferes with the innate immune response and/or stress response in infected bronchial epithelial cells through an unknown mechanism, which probably leads to the distinct chronic infection process of this virus.
Figure 4.
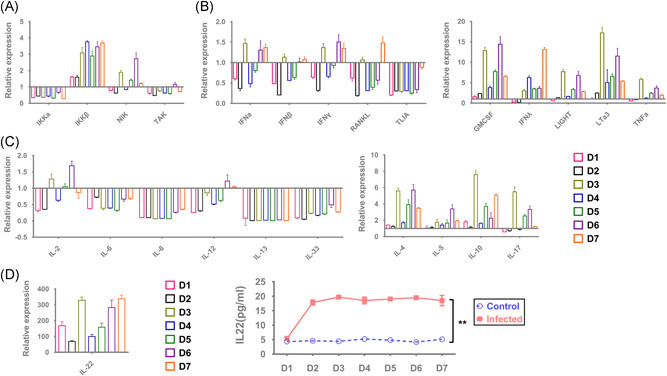
Modified mRNA expression profiles of various immune signaling molecules in infected 16HBE cells. A, mRNA expression profile of NF‐kB signaling pathway‐related molecules. B, mRNA expression profile of interferon, tumor necrosis factor, colony‐stimulating factor, etc. C, mRNA expression profile of multiple interleukins. D, mRNA and protein expression of IL‐22. Samples were obtained within 7 dpi. *P < .05; **P < .001. IL, interleukin; mRNA, messenger RNA
4. DISCUSSION
COVID‐19, an acute contagious respiratory disease caused by SARS‐CoV‐2 with high infectivity and a poor clinical outcome, is a worldwide concern. 33 Much work is needed to understand this disease and its causative agent. Since the first report of COVID‐19, clinical observations have indicated that most patients experience a latent infection continuing for 1 to 2 weeks, with a high possibility of transmission to other individuals. 34 In addition, clinical manifestations, including cough, fever, and lung inflammatory features, are observed and, in some cases, followed by respiratory failure due to lesions in lung tissue. 4 This clinicopathological process suggests a weaker innate immune response and/or stress response in respiratory tissue during the early phase of SARS‐CoV‐2 infection, even though the virus is replicating, which might lead to severe inflammation in the lung tissue and a protracted disease course. Undoubtedly, this clinical feature is related to the distinct infection process of this virus in respiratory epithelial cells. Our findings in 16HBE cells infected with SARS‐CoV‐2 suggest a critical role for its pathological infection mechanism, which allows the virus to spread in the latent period and escape monitoring and clearance by the innate immune response. Thus, the virus does not induce damage to epithelial cells and the tissue structure, as was observed in infected 16HBE cells, which maintained their structure and morphology for at least 1 week and confirmed in our other study of rhesus macaques infected with the virus, in which only metaplasia in trachea and bronchial tissues was observed. 35 The fact that the virus chronically infects 16HBE cells and is continuously released from infected cells for a long time might be why some patients with COVID‐19 are asymptomatic but contagious. Nonetheless, this distinct infection process in respiratory epithelial cells might attenuate and delay host inflammatory reactions and innate immune responses to the virus and allow it to replicate in local tissue. Indeed, downregulation of various innate immune signaling molecules, including IFN family members and some ILs involved in inflammatory reactions, in infected cells, as we identified, facilitates this process. Collectively, these data suggest that SARS‐CoV‐2 uses a clever infection strategy and that suitable preventive countermeasures should be considered. Regardless, the reason for the very high expression of IL‐22 in infected 16HBE cells remains unclear. This IL has been identified as functioning in tissue repair and immune regulation. 36 Determining whether this high level of expression might explain why virus‐infected cells maintain their structure and morphology or whether it might be part of the viral strategy to maintain its transmission ability needs further study. Although these data were obtained using 16HBE cells, a continuous cell line originating from the bronchial tissue of a 1‐year‐old child that might not accurately represent the physiologic state of epithelial cells in the human airway, they provide basic knowledge for understanding the pathological mechanism by which COVID‐19 is induced by SARS‐CoV‐2.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Supporting information
Supporting information
Liao Y, Li X, Mou T, et al. Distinct infection process of SARS‐CoV‐2 in human bronchial epithelial cell lines. J Med Virol. 2020;92:2830–2838. 10.1002/jmv.26200
Yun Liao, Xueqi Li, and Tangwei Mou are equal contributors.
Contributor Information
Longding Liu, Email: liuld@imbcams.com.cn.
Qihan Li, Email: liqihan@imbcams.com.cn.
DATA AVAILABILITY STATEMENT
The corresponding authors have full access to all the data generated in the present study and assume full responsibility for the final submission of this manuscript for publication.
REFERENCES
- 1. World_Health_Organization . Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: Interim Guidance. 2020; https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected. Accessed March 27, 2020.
- 2. Wu Y, Ho W, Huang Y, et al. SARS‐CoV‐2 is an appropriate name for the new coronavirus. Lancet. 2020;395(10228):949‐950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hui DS, Azhar EI, Madani TA, et al. The continuing 2019‐nCoV epidemic threat of novel coronaviruses to global health — the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human‐pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. National_Medical_Products_Administration . Update on the epidemic situation of new coronavirus pneumonia as of 24:00 on March 23. 2020; http://www.nmpa.gov.cn/WS04/CL2577/375996.html. Accessed March 27, 2020.
- 8. Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418‐423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren LL, Wang YM, Wu ZQ, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl). 2020;133:1015‐1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greber UF, Gastaldelli M. Junctional gating: the achilles' heel of epithelial cells in pathogen infection. Cell Host Microbe. 2007;2(3):143‐146. [DOI] [PubMed] [Google Scholar]
- 12. Woodland DL, Randall TD. Anatomical features of anti‐viral immunity in the respiratory tract. Semin Immunol. 2004;16(3):163‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hiemstra PS, McCray PB Jr., Bals R. The innate immune function of airway epithelial cells in inflammatory lung disease. Eur Respir J. 2015;45(4):1150‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li W, Moore MJ, Vasilieva N, et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215‐220. [DOI] [PubMed] [Google Scholar]
- 16. Ge XY, Li JL, Yang XL, et al. Isolation and characterization of a bat SARS‐like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barrett PN, Mundt W, Kistner O, Howard MK. Vero cell platform in vaccine production: moving towards cell culture‐based viral vaccines. Expert Rev Vaccines. 2009;8(5):607‐618. [DOI] [PubMed] [Google Scholar]
- 18. Xu X, Guo Y, Fan S, et al. Attenuated phenotypes and analysis of a herpes simplex virus 1 strain with partial deletion of the UL7, UL41, and LAT genes. Virol Sin. 2017;32(5):404‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kajimura J, Ito R, Manley NR, Hale LP. Optimization of single‐ and dual‐color immunofluorescence protocols for formalin‐fixed, paraffin‐embedded archival tissues. J Histochem Cytochem. 2015;64(2):112‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade‐long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127–e00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014;14(5):315‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Makris S, Johnston S. Recent advances in understanding rhinovirus immunity. F1000Res. 2018;7. 10.12688/f1000research.15337.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soto JA, Gálvez NMS, Benavente FM, et al. Human metapneumovirus: mechanisms and molecular targets used by the virus to avoid the immune system. Front Immunol. 2018;9:2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ryu JH, Kim CH, Yoon JH. Innate immune responses of the airway epithelium. Mol Cells. 2010;30(3):173‐183. [DOI] [PubMed] [Google Scholar]
- 26. Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev. 2011;24(1):210‐229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hossain MM, Norazmi MN. Pattern recognition receptors and cytokines in Mycobacterium tuberculosis infection‐‐the double‐edged sword? Biomed Res Int . BioMed Res Int. 2013;2013:179174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang LC, Xu XH, Jin LP. Molecular characteristics and possible functions of innate lymphoid cells in the uterus and gut. Cytokine Growth Factor Rev. 2020;52:15‐24. [DOI] [PubMed] [Google Scholar]
- 29. Nakata K, Inagawa H, Nishizawa T, Kohchi C, Soma GI. Specific messenger RNA expression for signal transduction molecules by lipopolysaccharide in intestinal macrophages. Clin Exp Immunol. 2006;143(3):484‐493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Picascia A, Grimaldi V, Iannone C, Soricelli A, Napoli C. Innate and adaptive immune response in stroke: focus on epigenetic regulation. J Neuroimmunol. 2015;289:111‐120. [DOI] [PubMed] [Google Scholar]
- 31. Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perusina Lanfranca M, Lin Y, Fang J, Zou W, Frankel T. Biological and pathological activities of interleukin‐22. J Mol Med (Berl). 2016;94(5):523‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Velavan TP, Meyer CG. The COVID‐19 epidemic. Trop Med Int Health. 2020;25(3):278‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deng W, Bao L, Gao H, et al. Rhesus macaques can be effectively infected with SARS‐CoV‐2 via ocular conjunctival route. 2020. https://www.biorxiv.org/content/10.1101/2020.03.13.990036v1
- 36. Wang Q, Huang Y, Zhou R, et al. Regulation and function of IL‐22 in peritoneal adhesion formation after abdominal surgery. Wound Repair Regen. 2020;28(1):105‐117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The corresponding authors have full access to all the data generated in the present study and assume full responsibility for the final submission of this manuscript for publication.


