Abstract
Clinical evidence indicates that the fatal outcome observed with severe acute respiratory syndrome-coronavirus-2 infection often results from alveolar injury that impedes airway capacity and multi-organ failure—both of which are associated with the hyperproduction of cytokines, also known as a cytokine storm or cytokine release syndrome. Clinical reports show that both mild and severe forms of disease result in changes in circulating leukocyte subsets and cytokine secretion, particularly IL-6, IL-1β, IL-10, TNF, GM-CSF, IP-10 (IFN-induced protein 10), IL-17, MCP-3, and IL-1ra. Not surprising, therapies that target the immune response and curtail the cytokine storm in coronavirus 2019 (COVID-19) patients have become a focus of recent clinical trials. Here we review reports on leukocyte and cytokine data associated with COVID-19 disease in 3939 patients in China and describe emerging data on immunopathology. With an emphasis on immune modulation, we also look at ongoing clinical studies aimed at blocking proinflammatory cytokines; transfer of immunosuppressive mesenchymal stem cells; use of convalescent plasma transfusion; as well as immunoregulatory therapy and traditional Chinese medicine regimes. In examining leukocyte and cytokine activity in COVID-19, we focus in particular on how these levels are altered as the disease progresses (neutrophil NETosis, macrophage, T cell response, etc.) and proposed consequences to organ pathology (coagulopathy, etc.). Viral and host interactions are described to gain further insight into leukocyte biology and how dysregulated cytokine responses lead to disease and/or organ damage. By better understanding the mechanisms that drive the intensity of a cytokine storm, we can tailor treatment strategies at specific disease stages and improve our response to this worldwide public health threat.
Keywords: COVID-19, cytokine storm, immunotherapy, leukocyte, SARS-CoV-2
Graphical Abstract
Reviews clinical data on leukocyte and cytokine changes during mild-to-severe infection, including a detailed overview of current therapy strategies targeting the cytokine storm.
Introduction
Since December 2019, the frequent incidence of pneumonia has become a distinctive feature of the infection caused by a novel coronavirus (severe acute respiratory syndrome-coronavirus-2, SARS-CoV-2) in Wuhan, Hubei province, China.1 On February 11, 2020, the World Health Organization (WHO) officially named this pneumonia coronavirus disease 2019 (COVID-19).2 As of May 7, 2020, a total of 3,672,238 confirmed cases and 254,045 deaths have been reported in more than 214 countries (86,095 confirmed cases and 4643 deaths in China).3 Considering its worldwide reach and severity, WHO declared the COVID-19 outbreak a global pandemic—the first pandemic sparked by a coronavirus.4 In contrast to the global infection rate reached by SARS-CoV-2, severe acute respiratory syndrome-coronavirus (SARS-CoV, 2002–2003) and Middle East respiratory syndrome coronavirus (MERS-CoV, 2012) were largely localized to China and Saudi Arabia, respectively.5 Like SARS-CoV-2, the outbreak of SARS-CoV and MERS-CoV were also caused by the zoonotic coronavirus crossing the species barrier. The diseases caused by these three coronaviruses share similarities in clinical presentations, such as progression toward severe acute respiratory syndrome6; yet COVID-19′s clinical features remain distinct, including unique pulmonary presentation on CT.7 Specifically, with the SARS epidemic, dysregulation of the immune system resulted in acute fevers and decreased CD4+ and CD8+ T cell counts, yet lacked any evidence of pulmonary changes upon imaging.8
The most common symptoms observed in COVID-19 patients are malaise, dry cough, and high fever. Symptoms of diarrhea, hemoptysis, and headache are not uncommon.9 Recently the Centers for Disease Control and Prevention (USA) expanded target symptoms of COVID-19 to include chills, repeated shaking with chills, muscle pain, headache, sore throat, and loss of taste or smell. Disease can be mild or progress toward dyspnea and/or hypoxemia, or even acute respiratory distress syndrome (ARDS) and septic shock, which then can lead to multiple organ dysfunction syndrome (MODS).10 The main reason for death outcomes following SARS-CoV-2 infection is respiratory failure, with changes in heart and liver function as a secondary or related consequence of disease.11–13 Epidemiologic data does suggest certain groups are at particular risk for severe disease outcomes (i.e., elderly, select preexisting conditions). Although we know there is a link between disease severity, viral production, and a cytokine storm or cytokine release syndrome (CRS), it is still unclear which molecular triggers fuel the onset of the cytokine storm and why it can quickly advance to ARDS or MODS, with a fatal outcome in a subset of patients.14,15 It remains unknown if the observed cytokine storm and resulting leukocyte changes impact high- and low-risk individuals in the same way, and how these factors can turn an otherwise natural protective cytokine response against infection into a lethal pathogenic process.
In this article, we summarize reports on blood cytokine levels and leukocyte activation to examine for differences in immunopathogenesis between mild and severe cases of COVID-19 in patients from China. Data were obtained from reports from April 2019 to April 2020 (Supporting Information Fig. S1). We also discuss emerging concepts on the interphase between leukocyte biology and disease pathogenesis. Last, we summarize findings from ongoing clinical trials in China and United States that seek to control the onset or damage caused by the cytokine storm to reduce both morbidity and mortality.
Cytokine Release with Emerging Corona Viral Infections: Protective Versus Pathogenic Cytokine Storm Responses
The release of cytokines in response to infection can lead to mild or severe clinical manifestations. The hallmarks of a mild/nonlethal cytokine release response to infection include increased local temperature (heat), myalgia, arthralgia, nausea, rash, depression, and other mild flu-like symptoms. Concurrent to immune activation, the body launches compensatory-repair processes to restore tissue and organ function. The term “cytokine storm” was first coined in 1993 to describe a graft- vs.-host disease.16 The term has since been extended to describe the similar sudden cytokine releases associated with autoimmune, hemophagocytic lymphohistiocytosis, sepsis, cancers, acute immunotherapy responses, and infectious diseases17–20 Cytokine storm occurs when an immune system is overactivated by infection, drug, and/or some other stimuli, leading to high levels of cytokines (IFN, IL, chemokines, CSF, TNF, etc.) being released into circulation with a widespread and detrimental impact on multiple organs.21 The severe inflammatory responses induced by a cytokine storm start locally and spread systemically, causing collateral damage in tissues.22,23 To date, it is unclear what determinants of the host response to infection are responsible for triggering the inflammatory sequence leading to the clinical syndrome associated with high cytokine release. In general, it is believed to be caused by an imbalance in immune-system regulation (i.e., increase in immune cell activation via TLR or other mechanism, decrease in anti-inflammatory response, etc.), although the specific dysregulated molecular causes are still unknown. A cytokine storm is nearly always pathogenic because of its detrimental effects on the host. On the other hand, local and systemic cytokine responses to infection are essential parts of the host’s initial response to infection. Release of cytokines by natural killer (NK) cells and macrophages, along with activated T cells and humoral responses can help resolve infection, accompanied by effector mechanisms such as antibody-dependent cell-mediated cytotoxicity (ADCC).24 These responses are triggered to keep the pathogen in check. For example, the local cytokines, such as IFN-α/β and IL-1β, produced by epithelial cells can protect nearby cells by stimulating IFN-stimulated gene expression and concurrently activating immune competent cells such as NK cells. This increases the NK cell’s lytic potential and fuels the secretion of IFN-γ.25 In addition to NK cells, once myeloid cells such as resident macrophages are activated by IFN-γ, it amplifies subsequent TLR-mediated stimulation. This includes the release of high levels of TNF, IL-12, and IL-6, which, in turn, can further modulate NK cells.26 Although IL-12 acts to increase NK IFN-γ secretion, high IL-6 levels also may limit the immune response by its effects on the cytotoxic activity of NK cells via the down-regulation of intracellular perforin and granzyme B levels.27 As disease progresses, T cell and antibody responses give rise to additional cytokine responses, leading to greater or sustained antigen release and added TLR ligands from viral-induced cytotoxicity.28 Once these responses are in motion, other host- or pathogen-related factors (i.e., decreases in pathogen load, anti-inflammatory responses, genetics) work together to prevent a dysregulated response or a CRS that, if allowed to develop, could itself cause tissue damage and organ failure. For example, a lack of a negative feedback mechanism by IL-10 and IL-4 would be expected to increase the severity of cytokine responses toward a pathogenic CRS or cytokine storm.21 On the other hand, targeting treatment to disrupt the formation of cytokine storm by using pharmacologic agents, such as tocilizumab (anti-IL-6), may stabilize the advanced cases from transitioning to a more critical state.27,29
At the onset of SARS-CoV-2 infection, there typically is a preferential infection of the respiratory track as a consequence of droplet-based viral transfer. However, a recent study30 supports the theory that SARS-CoV-2 also could potentially infect intestine enterocytes directly through angiotensin-converting enzyme 2 (ACE2). ACE2 is highly expressed on differentiated enterocytes and may help to explain why diarrhea occurs with acute infection as well as the observed fecal shedding. Consequently, having a broader infection footprint may impact the source of inflammatory cascades to include tissues other than the lung.
In COVID-19 disease, a cytokine storm is common in patients with severe-to-critical symptoms; at the same time, lymphocytes and NK cell counts are sharply reduced with elevations in levels of D-dimer, C-reactive protein (CRP), ferritin, and procalcitonin.31 Consequences of a lethal cytokine storm exhibit diffuse alveolar damage characterized by hyaline membrane formation and infiltration of interstitial lymphocytes.32,33 The collateral tissue damage, organ failure, and poor outcomes of people with COVID-19 and its accompanying uncontrolled inflammatory responses share similarities with SARS and MERS. In severe SARS patients, the serum levels of IFN-γ, IL-1, IL-6, IL-12, TGF-β, MCP-1, and IL-8 were higher than patients with mild-to-moderate symptoms.34,35 IL-1β, IL-6, and IL-8 levels also were increased in the patients severely infected by MERS-CoV.36 Leukocyte and cytokine changes with severe-to-critical SARS-CoV-2 infection are further detailed below.
Characteristics of Leukocyte Changes and Cytokine Release During Mild and Severe Covid-19 Infection
Developing criteria to predict and diagnose a cytokine storm in COVID-19 patients with surrogate biomarkers is key because the peak levels of circulating cytokines are not routinely monitored for a change in kinetics. In addition to CRP and ferritin, increases in D-dimer and procalcitonin have also been associated with a higher likelihood of developing or continuing a cytokine storm in COVID-19 patients.37 Of interest, elevated CRP and ferritin levels are associated with the onset of a cytokine storm in patients receiving chimeric antigen receptor T cell therapy.38 Further study of the changes that occur in leukocyte and cytokine parameters, as described later, may help identify biomarkers for a cytokine storm in COVID-19 patients. In the following sections, we review 28 recently published studies and examine the changes in circulating leukocytes and cytokine profiles of 3939 COVID patients (see schematic for information on the process used for selecting literature in Supporting Information Fig. S1).
Aberrant activation of innate immune cells in COVID-19 infection
Monocyte and macrophage
In humans, both monocytes and macrophages express ACE2 and consequently can be infected by SARS-CoV and SARS-CoV-2,39 which results in the activation and transcription of proinflammatory genes.40 Intriguingly, infection caused by SARS-CoV-2 appears to markedly down-regulate the expression of ACE2 on peripheral blood (PB) monocytes on a per cell basis, which may be a secondary outcome to viral binding.41 Whether this down-regulation of ACE2 receptors is a surrogate to viremia remains to be determined. In addition, the expression of ACE2 was found on CD68+ and CD169+ macrophages in spleen and lymph nodes of COVID-19 patients, providing further evidence that SARS-CoV-2 infection may target ACE2 positive myeloid cells throughout the body, including the spleen and lymph nodes.42 Infected CD169+ macrophages are mainly found in the red pulp section of spleens. Moreover, macrophage-rich areas at marginal sites of lymph nodes were more likely to test positive for viral nucleocaspid protein antigens. Of interest, previous studies have demonstrated that CD169+ macrophages are responsible for controlled levels of viral replication in support of developing immunity as a result of a refractory state to type I IFN-dependent activation.43 This indicates that infection of CD169+ macrophages might be a conduit for translocation of SARS-CoV-2 to spleens and lymph nodes. This could be responsible for added systemic viral replication, and may contribute to lower immunity (see antigen-specific T cell section below). Human monocytic cell lines THP-1 and U939 express ACE2,41 and thus may be useful in the in vitro study of myeloid modulation and role of TLR interactions in triggering activation before and after infection. Likewise, hACE2 transgenic mice should be harnessed as an animal model to study the biophysiology as well as the pharmacology of SARS-CoV-2 infection.44
Monocytes from COVID-19 patients, although normal in number, show an activated phenotype, as evidenced by their morphology (FSC-high) and their capacity to produce IL-6, IL-10, and TNF.41 Activated monocytes present in the PB of COVID-19 patients were characterized by surface expression of CD11b, CD14, CD16, CD68, CD80, CD163, and CD206. Activation of the PB monocytes was particularly associated with disease severity and a poor prognosis.41 Expression of CD163 and CD206 by PB monocytes from COVID-19 patients suggest a bias toward a M2 or regulatory phenotype, which could impact adaptive antiviral effector T cell responses. Apart from immune effects, expression of CD163 in activated monocyte/macrophage has also been associated with hemophagocytic lymphohistiocytosis syndrome,45 which could potentially contribute to the hyperproduction of cytokine and immunologic pathogenesis in COVID-19 patients.46
Wen et al.47 also observed that an abundance of inflammatory CD14++IL1β+ and IFN-activated monocytes existed in the PB of COVID-19 patients. In addition, SARS-CoV-2 was found to trigger macrophages through ACE2, generating IL-6 expression in the spleen and lymph nodes and IL-6, TNF, IL-10, and PD-1 expression from the alveolar macrophages.47,48 This added mechanism might promote lymphocytopenia and contribute to a cytokine storm, initiating in the lung as viral levels rise.42,48
Autopsy reports indicated that inflammatory macrophages accumulated in the lungs of COVID-19 patients.49 Single cell RNA sequencing confirmed that monocyte-derived FCN1+ macrophages were the predominant subset found in the bronchoalveolar lavage fluid (BALF) of COVID-19 patients with ARDS, which might be indicative of cells with high inflammatory and chemokine production.50 The transcriptional analysis of BALF and PB mononuclear cells from COVID-19 patients revealed high levels of IFN-induced protein 10 (IP-10) and MCP-1, which likely attracted the trafficking of macrophages to the site of infection, a finding that is consistent with autopsy reports.51
Orchestrated by delayed Type I IFN signaling, the activation and accumulation of inflammatory monocyte/macrophage can result in a dysregulated inflammatory response and apoptosis of T cells.52 The results of one ex vivo experiment in lung tissue53 showed that SARS-CoV-2 produced three times more infectious virus particles than did SARS-CoV after 48 h. However, SARS-CoV-2 triggered fewer IFNs and proinflammatory mediators than did SARS-CoV. This may contribute to the virus’s ability to evade intrinsic innate responses at initial infection. Early type I IFN’s response may be particularly important. Preliminary reports showed IFN-α2b significantly reduced the length of time the virus could be detected in the upper respiratory tract; this, in turn, reduced the duration of elevated blood levels for the inflammatory markers IL-6 and CRP.54 However, the mechanism underlying SARS-CoV-2′s ability to dampen initial innate responses at acute infection and suppress Type-I IFNs to maximize viral release still needs additional study.
Neutrophils
As COVID-19 progresses, the number of neutrophils in circulation gradually increase; thus, elevated neutrophil levels may be useful for predicting the severity of disease.55 Zhang et al.56 reported that the neutrophil-to-lymphocyte ratio (NLR) combined with IgG might be a better predictor than neutrophil count alone in predicting the severity of COVID-19. Neutrophil extracellular traps (NETs), which are extracellular webs of DNA/histones released by neutrophils to control infections, also are known to exacerbate inflammation.57,58 Previous studies have revealed that aberrant NETs might contribute to ARDS, cystic fibrosis, excessive thrombosis, and cytokine storm (IL-1β).59–62 In severe cases of COVID-19, elevated levels of NETosis with cell-free DNA and myeloperoxidase (MPO)-DNA have been noted frequently.63 Indeed, when neutrophils from uninfected persons were exposed to serum from COVID-19 patients in vitro, it triggered NETosis, indicating that patient sera may have the capacity to promote NETosis in neutrophils.63,64 Apart from contributing to a cytokine storm, increased NETosis likely also impacts the onset of venous and arterial thrombosis in COVID-19 patients.65 This finding raises the hypothesis that NETs may be a central dysregulated pathologic mechanism driving cytokine release and multi-organ damage, ultimately leading to respiratory failure and coagulopathy. Moreover, the transcriptional analysis51 of BALF and PB mononuclear cells from COVID-19 patients revealed that high levels of CXCL-2 and CXCL-8 may contribute to the recruitment of neutrophils to the site of infection, further aggravating the pulmonary inflammatory response. Given these findings, NETosis may represent a viable therapeutic target for inhibiting CRS and organ damage. Other important areas in neutrophil response such as interactions with humoral (the role of IgA and CD89, IgG, and CD16, etc.) or myeloid subsets remain unstudied.
NK cells
Data on the NK cell response in COVID-19 is limited. In SARS, NK cells were found to be useful in predicting disease severity and CD158b+ NK cells were associated with the presence of anti-SARS-CoV-specific antibodies.66,67 Similarly, a recent study showed that the number of NK cells in PB was decreased in patients infected with SARS-CoV-2, especially in severe cases.68 But a separate report showed no difference in the number of CD16+CD56+ NK cells in mild vs. severe cases.69 Therefore, whether NK cells could be a predictor of COVID-19 remains to be determined. Additional studies are needed to determine whether NK cells can impact viral control (i.e., directly or through ADCC-mediated mechanisms) or contribute to cytokine release during the disease course of COVID-19.
Dendritic cells and intraepithelial lymphocytes
Despite the role of dendritic cells and γδ T cells in respiratory infections,70,71 there is currently no available evidence to show a link between SARS-CoV2 infection and the modulation of myeloid or plasmacytoid dendritic cells nor reports of how γδ T cells may impact the disease.72
Aberrant activation and changes in adaptive immune cells during COVID-19 infection
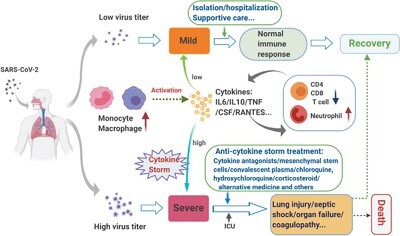
Compelling evidence exists that COVID-19 is characterized by a marked decrease in circulating T and B lymphocytes, particularly in severe and critical stages of COVID-19 infection.47,68,73 Of note, such lymphopenia in patients with severe COVID-19 frequently occurs along with aberrant activation of monocytes/macrophages and an increase of neutrophils.74 Changes in leukocyte subsets associated with mild vs. severe infection outcomes are described in the following sections and summarized in Figure 1.
FIGURE 1.

Major blood leukocyte, cytokine changes, and therapy strategies in mild vs. severe SARS-CoV-2 infection. Conceptual model of the interplay between immune activation and clinical pathology from patients with mild vs. severe infection, as well as current therapeutic strategies and possible outcome. Figure is made with BioRender (https://app.biorender.com/)
Dysregulation of T cells after SARS-CoV-2 infection
T cell subsets change as COVID-19 infection progresses from mild to severe. A single-cell sequencing study found target inflammatory genes were highly expressed by CD4 T cells, and CD8 CTLs underwent clonal expansion in the recovery stage of COVID-19 patients.47 However, in patients advancing to severe disease, the number of T lymphocytes remained low as compared with mild-stage patients, with the levels of helper T cell subsets, including Th1, Th2, and Th17, at below-normal levels.68 Indeed, numerous clinical reports show that the decreased frequency of circulating lymphocytes, including CD4+ and CD8+ T cells, is closely related to disease severity of COVID-19.75,76 Wang et al.77 found that elevated IL-6 levels occurred 1–2 d prior to decreases in CD8+ and CD4+ T cells. Liu et al.78 observed that CD4+ and CD8+ T cell levels dropped to their lowest levels after 4 to 6 d of illness, whereas IL-10, IL-2, TNF, and other cytokines reached peak levels. These studies suggest a negative relationship between high levels of cytokines (e.g., IL-6, IL-10, IL-2, TNF) and lower circulating T cells in severe patients infected by SARS-CoV-2, likely because of redistribution of cells in tissue and/or activation of induced cell death.79 SARS-CoV-2 infection might induce lymphocyte apoptosis by enhancing the P53 signaling pathway or the Fas signaling pathway.42,51 A decrease in circulating lymphocytes as a result of tissue redistribution is consistent with the development of interstitial pneumonitis, which is caused by mononuclear cell infiltration in severe cases.
Weiskopf et al.80 reported that SARS-CoV-2-specific CD4+ and CD8+ T cells were detected in the PB of COVID-19 patients with ARDS at 14 d after the onset of symptoms. Furthermore, antigen-specific central and effector memory responses were detected in virus-specific CD4+ T cells and CD8+ T cells of ARDS patients, respectively. With regard to cytokine secretion, they proposed that in cases of severe disease, the CD8+ cytotoxic T cells predominantly secrete IFN-γ, whereas virus-specific CD4+ T cells secrete expected levels of Th1 (IFN-γ, TNF, and IL-2) and Th2 (IL-5, IL-9, and IL-10) cytokines. In addition, Zheng et al.81 reported that the proportion of multifunctional CD4+ T cells (IFN-γ, TNF, and IL-2, with more than two positive cytokines among them) was reduced in severe COVID-19 patients when compared to those with mild or no infection. As multifunctional T cells are associated with better outcomes after vaccination,82 a decrease in multifunctional CD4+ T cells might infer a poor clinical outcome in patients with severe COVID-19. In COVID-19 patients with ARDS, a decrease in CD4+ and CD8+ T cells was associated with an increase in CD38+ (CD8 39.4%) HLA-DR+ (CD4 3.47%) T cells.32 Zheng et al.81 and Chen et al.83 further reported that CD8+ T cells in the PB of COVID-19 patients exhibited an overactivated phenotype, which is indicative of a sustained adaptive immune response (whether effective or not) in addition to an innate immune response. Of interest, activated CD8+ T cell responses appear to be ineffective as patients advance to severe stages of disease, as evidenced by the exhaustive phenotype of CD8 T cells (HLADR+TIGIT+CD8+ T cells).81 Consistent with an onset of T cell exhaustion, Chen et al.83 found that T cells in the PB of severe COVID-19 patients expressed high levels of PD-1 and Tim-3. The expression levels of PD-1 and Tim-3 on T cells also was positively correlated with disease severity.
Activated T cells could also add fuel to the cytokine storm by further stimulating inflammatory responses from innate immune cells. For example, it was shown that the expression of IL-1β, CSF1, and CSF2 on T cells may bind to the IL-1R and colony stimulating factor receptor (CSFR) expressed on monocytes, and further stimulate the activation of monocytes.47 Th1 cells in patients with severe COVID-19 were reported to stimulate the production of IL-6 by inflammatory monocytes,39 which, together with Th17 cells,84 may directly join innate immune cells in sparking the release of proinflammatory cytokines further contributing to the cytokine storm and subsequent organ damage.47,84
Regulatory T cells (Tregs) play a critical role in dampening an excessive inflammatory response as well as in antiviral immune responses.85 Therefore, Tregs may be central to maintaining a balance between antiviral immunity and the harmful cytokine storm. To date, the reports regarding the Treg cells in COVID-19 patients remain inconsistent. Some studies found up-regulated Treg cells in severe illness, whereas others reported that the number of Tregs was reduced or unchanged in COVID-19 patients.68,77,86 Therefore, the role of Treg cells in the pathogenesis of COVID-19 needs to be further clarified.
B cells and antibody production
Circulating B cells appear to be restored to normal levels in patients upon recovery from COVID-19 and convalescent plasma (CP) infusion (further discussed later) has been shown to be a potentially effective treatment as reported in 6 severe COVID-19 patients.87 These findings support the hypothesis that humoral immune responses that boost SARS-CoV-2-specific antibodies are important in the host’s resolution of SARS-CoV-2 infection and likely would help protect against reinfection.51,88,89 Of interest, this ability to develop neutralizing antibody production (and memory) after infection may not be the same in all recovered patients. Recent data suggests that as many as 30% of recovered patients who had a milder disease course may develop low titers of neutralizing antibodies, which may convey a higher risk for reinfection.90 Indeed, several studies have detected higher antibody titers after recovery from more severe disease when compared with milder cases (i.e., higher antibody titer in recovered elderly patients as compared with younger patients that often have mild or asymptomatic disease). This suggests a disease course with a stronger immune response may also lead to greater protection against reinfection.90,91 It is important to note that binding antibodies can also play a role in antibody-mediated phagocytosis and antibody-dependent cytotoxicity. It remains to be determined if patients with low total or neutralizing antibody titers will have higher reinfection rates.
Similar to CD4+ T, CD8+ T cells, and NK cells, B cells were also markedly decreased in severe COVID-19 patients, as compared with mild patients, and the counts of B lymphocytes were negatively associated with viral burden.51 Aside from protection, it also remains undetermined if prior exposure may prime the body’s response, leading to antibody-dependent enhancement (ADE) effects for greater infection or an amplification of inflammation cascades. Previously, ADE had been reported in other viral infections, such as SARS-CoV; thus, ADE, if present, may hinder the host’s ability to manage inflammation in lung and other tissues.52 An analysis of 173 COVID-19 patients, analyzed for SARS-CoV-2-specific IgM and IgGs, found that those with severe or critical disease had a high titer of total antibodies as well as a high IgG response, which were associated with a poor outcome and prognosis.92 Whether the increase in antibody titers during disease represents a secondary outcome of the host’s response to high viral titers or whether an ADE mechanism could contribute to a sudden rise in viral load and onset of a cytokine storm remains undetermined.
Aberrant cytokines associated with cytokine storm in COVID-19 patients
Many proinflammation cytokines, such as IL-6, TNF, IL-1, IL-2, IL-17, IFN-γ, G-CSF, MCP-1 (macrophage inflammatory protein 1), IP-10 (IFN-γ-induced protein 10), and others, were found to be significantly elevated in severe COVID-19 patients (Table 1 and Fig. 1), a profile that is similar to that found in patients with SARS and MERS.36,93,94
TABLE 1.
Summary of clinical cohort data on cytokines and aberrant leukocyte changes in mild-to-severe stages of COVID-19 infection
| Leukocytes | Cytokines | |||||
|---|---|---|---|---|---|---|
| Hospital | Cases | Changes with Infection (n = cases whose leukocyte or cytokines changed/total cases) | Severe cases vs mild cases,P < 0.05 | Changes with Infection (n = cases whose leukocyte or cytokines changed/total cases) | Severe cases vs mild cases, P < 0.05 | References |
| Renmin Hospital, Wuhan University |
N = 82 (Deaths) |
Increase: Neutrophil count (55/74, but 74/74 in the last 24h); Neutrophil-to-lymphocyte ratio (NLR; 69/73) Decrease: Lymphocyte count (66/74, but 74/74 in the last 24h); CD8+ T cell count (34/58); NK cell count (87/58) |
No comparison | Increase: IL-6 (11/11) | No comparison | 11 |
| Jin Yintan Hospital |
N = 41 (28 Non-ICU cases and 13 ICU cases) |
Decrease: Lymphocyte count (26/41) |
Increase: Neutrophil count Decrease: WBC count; Lymphocyte count |
Increase:IL-1B, IL-1RA, IL-7, IL-8, IL-9, IL-10, G-CSF, GM-CSF, IFN-γ, IP-10, MCP-1, MIP-1A, MIP-1B, and TNF |
Increase: IL-2, IL-7, IL-10, G-CSF, IP-10, MCP-1, MIP-1A, and TNF |
14 |
| Xi’an No.8 Hospital and the First Affiliated Hospital of Xi’an Jiaotong University | N = 28 |
Increase: Monocytes with CD11b+, CD14+, CD16+, CD68+, CD80+, CD163+, CD206+ (FSC-high monocytes) was consistent with inflammatory phenotype |
No comparison |
Increase: IL-6, IL-10, TNF (generated by FSC-high monocytes) |
Increase: IL-6, IL-10, TNF (generated by FSC-high monocytes) |
41 |
|
Tongji hospital |
N = 452 (286 severe and 166 nonsevere cases) |
Increase: B cells; Decrease: Lymphocytes; NK cells (N = 44); Th cells (N = 44); Ts cells (N = 44); Treg cells (mainly naïve Treg) (N = 44) |
Increase: Leukocyte count; neutrophils; NLR; Decrease: Lymphocytes; Monocytes, Eosinophils, NK cells; Basophils; Th cells; Treg cells |
Increase: TNF-α; IL-2R; IL-6 |
Increase: IL-6, IL-2R, TNF, IL-8, IL-10 |
68 |
| Guangzhou Eighth People’s Hospital |
N = 56 (31 mild and 25 severe cases) |
Increase: Neutrophils; NLR; Treg cells Decrease: Lymphocytes; CD45+ lymphocytes; CD4+ T cells; CD8+ T cells; B cells; NK cells |
Increase: Neutrophils; WBC; Decrease: Lymphocyte counts |
Increase: IL-2, IL-4, IL-6, IL-10, TNF, IFN-γ |
Increase: IL-2, IL-6, IL-10, TNF |
86 |
| Wuhan Tongji hospital |
N = 21 (11 severe cases and 10 moderate cases) |
Increase: Total B lymphocytes (7/14) Decrease: Lymphocyte count (9/21); total T lymphocytes count (13/14); CD4+ T cells count (14/14); CD8+ T cells count (12/14); NK cells (8/14) |
Increase: WBC count; Neutrophil count Decrease: Lymphocyte count; total T lymphocytes; total T lymphocytes count; total B lymphocytes; CD4+T cells count; CD8 +T cells count |
Increase: IL-6 (13/16); IL-2R (9/16), TNF (11/16), and IL-10 (9/16) |
Increase: IL-6; IL-2R; IL-10; TNF |
74 |
| Chongqing Three Gorges Central Hospital |
N = 123 (102 mild and 21 severe cases) |
Decrease: CD4+ T cells (74/123); CD8+ T cells (42/123); B cells (32/123); NK cells (45/123) |
Decrease: CD4+ (54/102 in mild cases, while 20/21 in severe cases) and CD8+ T cells (29/102 in mild cases, while 13/21 in severe cases) |
Increase: IL-6 (47/123); IFN-γ (6/123) |
Increase: IL-6, IL-10 |
76 |
| The First Affiliated Hospital of Guangzhou Medical University |
N = 11 (Patients with ARDS) |
Increase: WBC count, Neutrophils; Tregs (2/11) Decrease: Lymphocyte count (11/11); NK cells (11/11); CD4 and CD8 lymphocytes (11/11); B lymphocytes (3/11) |
No comparison |
Increase: IL-6 (11/11), IL-10 (5/11), IL-4 (3/11) and IFN-γ (2/11) |
No comparison | 77 |
| Wuhan Union Hospital |
N = 40 (13 severe and 27 mild cases) |
Decrease: Lymphocytes |
Increase: WBC; Neutrophils Decrease: Lymphocyte; CD3+ T cells; CD8+ T cells |
Increase: IL-6 |
Increase: IL-6 (0–16 d); IL-10 (0–13 d); IL-2 and IFN-γ (4–6 d) |
78 |
|
Yunnan Provincial Hospital of Infectious Diseases |
N = 16 (10 mild and 6 severe cases) |
Decrease: T cells |
Increase: HLA-DR+TIGIT+CD8+ T cells increased Decrease: Granulocytes; Multifunction CD4+ T cells |
Increase: IL-6, TNF-α Decrease: IFN-γ and IL-2 (from CD4+ T cells) |
Decrease: IFN-γ (from CD4+ T cells) |
81 |
|
General hospital of central theatre command and Hanyang Hospital |
N = 262 (151 mild cases, 40 severe cases, 13 critical cases, 8 perished cases and 40 healthy control) |
Increase: PD1+ CD4+ T cells; PD1+ CD8+ T cells Decrease: Total T cells (166/222); CD4+ T cells (166/222); CD8+ T cells (156/222) |
Increase: PD1+ CD4+ T cells; PD1+ CD8+ T cells; CD8+ T cell (high expression of PD-1 and Tim-3) Decrease: Total T cells; CD4+ T cells; CD8+ T cells |
Increase: TNF, IL-10, IL-6, and IFN-γ |
Increase: TNF, IL-10, and IL-6 |
83 |
| Shenzhen Third People’s Hospital |
N = 53 (34 severe cases and 19 mild cases) |
Decrease: CD4 and CD8 counts |
Increase: Neutrophil Decrease: CD4 and CD8 counts |
Increase: IL-6, IL-2Ra, IFN-γ, IL-18, IL-10, IL-1ra, HGF, MIG, M-CSF, G-CSF, MIG-1a, CTACK, and IP-10 |
Increase: IL-1ra, IP-10, and MIG |
93 |
| Union Hospital, Tongji Medical College |
N = 80 (69 severe and 11 nonsevere cases) |
Decrease: Lymphocytes (60/80) |
Increase: Neutrophils and NK cells Decrease: Lymphocytes |
Increase: IL-6, IL-10 |
Increase: IL-6 |
95 |
| Fifth Hospital of Wuhan |
N = 36 (Nonsurvivors) |
Increase: Leukocytes (11/36); Neutrophils (17/36) Decrease: Lymphocytes (25/36) |
No comparison |
Increase: IL-6 (11/36) |
No comparison | 102 |
| From 552 hospitals |
N = 1099 (173 severe and 926 nonsevere cases) |
Decrease: Leukocyte count (731/890); Lymphocyte count (330/978) |
Decrease: Leukocyte count (106/173 in severe cases and 228/811 of mild cases); Lymphocyte count |
No test | No test | 162 |
| (147/154 in severe cases and 584/736 in mild cases) | ||||||
| The First Affiliated Hospital of University of Science and Technology |
N = 43 (10 healthy control, 21 No-ICU cases, and 12 ICU cases) |
Increase: Tim+PD-1+ CD4+ T cells; Tim+PD-1+ CD8+ T cells; GM-CSF CD4+ T cells; IL-6+CD4+ T cells; CD14+CD16+ monocytes; GM-CSF+CD14+ monocytes Decrease: T cells number; Monocytes; CD4+ T cells; CD8+ T cells |
Increase: Tim+PD-1+ CD4+ T cells; IFN-γ+GM-CSF+CD4+ T cells; GM-CSF CD4+ T cells; IL-6+CD4+ T cells; D14+CD16+ monocytes; GM-CSF+CD14+ monocytes IL-6+CD14+ monocytes Decrease: CD8+ T cells |
Increase: IL-6; IFN-γ | Increase: IL-6; IFN-γ | 39 |
|
Tongji hospital affiliated to Hua Zhong University of Science and Technology |
N = 29 (15 mild cases, 9 severe cases, and 5 critical cases) |
Decrease: Lymphocyte count (20/29) |
No significant difference |
Increase: IL-2R; IL-6; IL-8; IL-10; TNF |
Increase: IL-2R; IL-6 |
163 |
| Tongji Hospital |
N = 100 (34 mild cases, 34 severe cases and 32 critical cases) |
Decrease: Eosinophils |
Increase: Neutrophils Decrease: Eosinophils; lymphocytes |
Increase: IL-6, IL-2R, IL-8, IL-10, TNF, and IL-1β |
Increase: IL-6, IL-2R, IL-8, IL-10, and TNF |
164 |
| Union hospital in Wuhan |
N = 69 (the SpO2 ≥ 90% group (n = 55) and the SpO2 < 90% group [n = 14]) |
Increase: Neutrophils (41/67) Decrease: WBC count (36/67); Lymphocyte count (29/69); Eosinophil count (49/69) |
Increase: Neutrophil count; WBC count Decrease: Lymphocytes count |
Increase: IL-6 (11/43); IL-10 (16/43) |
Increase: IL-6, IL-10 |
165 |
|
Three designed-hospitals in Chongqing municipality |
N = 267 (217 Nonsevere cases and 50 severe cases) |
Decrease: Blood leukocyte count (118/267); Lymphocyte count (231/267); CD3 T cells (51/96); CD4 T cells (74/96) |
Increase: Neutrophils count Decrease: Blood leukocyte count (87/217 in nonsevere cases, 31/50 in severe cases); CD3 T cells (20/51 in nonsevere cases, 31/45 in severe cases); CD4 T cells (29/51 in nonsevere cases, 45/45 in severe cases) |
Increase: IL-6 (47/67); IL-17A (35/67); TNF (22/67); IL-10 (12/67); IFN-γ (11/67); IL-2 (5/67); IL-4 (4/67) |
Increase: IL-6; TNF; IL-17A |
166 |
| Zhongnan Hospital of Wuhan University |
N = 138 (36 ICU patients and 102 non-ICU patients) |
Decrease: Lymphocyte count |
Increase: WBC; Neutrophils (continued in 5 cases); Decrease: Lymphocyte count (continued in 5 cases) |
No test | No test | 167 |
| General Hospital of Central Theater Command |
N = 48 (21 mild cases, 10 severe cases, and 17 critical cases) |
Decrease: Lymphocytes |
Increase: WBC; neutrophil; Decrease: Lymphocytes |
Increase: IL-6 |
Increase: IL-6 |
96 |
| Wuhan Children’s Hospital |
N = 8 (severe or critical pediatric patients) |
Decrease: CD16+CD56+ lymphocytes (5/8); CD3+ lymphocytes (2/8), CD4+ lymphocytes (4/8) and CD8+ (1/8); One patient co-infected with influenza A virus showed significant decrease of leukocytes, neutrophils, and lymphocytes, while other patients did not show significant change |
No comparison |
Increase: IL-6 (2/8); IFN-γ (2/8); IL-10 (5/8) |
No comparison | 168 |
| Beijing Youan Hospital |
N = 11 (5 mild cases and 6 severe cases) |
Increase: Leukocytes; neutrophils Decrease: Lymphocytes |
Increase: Neutrophils |
Increase: IL-2; IL-4; IL-6; IL-10; IL-17; TNF; IFN-γ |
Increase: IL-10 |
169 |
|
Second People’s Hospital of Fuyang City |
N = 155 (30 severe cases and 125 moderate cases) |
Decrease: CD4+ T cells; CD8+ T cells |
Decrease: CD4+ T cells; CD8+ T cells |
Increase: IL-6 |
Increase: IL-6 |
170 |
| Central Theater General Hospital |
N = 13 (Death) |
Decrease: CD3+ T cells;CD4+ T cells; CD8+ T cells; (decreased more substantially during the disease course) |
No comparison |
Increase: IL-6; (Gradually increased and peaked before death) |
No comparison | 37 |
| Hubei Provincial Hospital of Integrated Chinese and Western Medicine |
N = 110 (57 moderate cases, 39 severe cases and 14 critical cases) |
Decrease: CD4+ T cells; CD8+ T cells |
Decrease: CD4+ T cells; CD8+ T cells |
Increase: IL-6; IL-10; IL-8 |
Increase: IL-6; IL-10 |
171 |
| Sino-French New City Branch of Tongji Hospital |
N = 548 (279 nonsevere cases and 269 severe cases) |
Increase: WBC (63/542); Neutrophils (118/542) Decrease: Lymphocytes (118/542) |
Increase: WBC (8/275 in nonsevere cases and 55/267 in severe cases); Neutrophils (22/275 in nonsevere cases and 96/267 in severe cases) Decrease: Lymphocytes (234/275 in nonsevere cases and 255/267 in severe cases) |
Increase: IL-1β (51/306); IL-2R (164/309); IL-6 (221/312); IL-10 (93/307); IL-8 (24/309); TNF (182/309) |
Increase: IL-2R (73/171 in nonsevere cases and 91/138 in severe cases); IL-6 (107/175 in nonsevere cases and 114/137 in severe cases); IL-10 (34/170 in nonsevere cases and 49/170 in severe cases); TNF (89/171 in nonsevere cases and 93/138 in severe cases) |
172 |
Abbrevations: COVID-19, coronavirus disease 2019; CTACK, cutaneous T cell-attracting chemokine; HFG, hepatocyte growth factor; ICU, intensive care unit; IP-10, IFN-induced protein 10; M-CSF, macrophage CSF; MIG, monokine induced by gamma IFN; MIP-1A, macrophage inflammatory protein-1 alpha; NK, natural killer; Treg, regulatory T cells; SpO2: blood oxygen saturation; and WBC, white blood cells.
Note: In column 3, N is the number of cases with available data. In addition, in columns 3 and 5, “n” is the number of cases in which leukocyte or cytokines changed overall, according to the corresponding report.
As shown in Table 1, the elevation of IL-6 was most frequently measured and detected in severe cases of SARS-CoV-2 infection. Zhou et al.39 reported that the pathogenic GM-CSF-producing Th1 cells in severe COVID-19 patients induced IL-6 production from CD14+CD16+ monocytes and, thus, accelerated the inflammation associated with a cytokine storm. Elevated levels of IL-6 also were found in patients with exacerbating disease progression as evidenced by chest CT.95 Moreover, Chen et al.96 reported that the serum SARS-CoV-2 viral load was closely associated with IL-6 levels in critical patients (R = 0.902). High levels of IL-6 may also contribute to an increase in neutrophil cells and decrease in lymphocytes. Clearly, IL-6 may impact the development of ARDS in COVID-19 patients97 and a rise in IL-6 may be a useful marker for severe disease onset. Furthermore, as lung-centric coagulopathy can also play an important role in the pathophysiology in the severe COVID-19 patients,98 IL-6 may contribute to this pathology by inducing coagulation cascades.99 However, hypercoagulability, together with high levels of D-dimers, fibrinogen, and CRP, in COVID-19 patients is distinct from the disseminated intravascular coagulation described in more severe inflammatory conditions.100,101 Further, the levels of IL-6 can vary in COVID-19 patients relative to severity of disease.11,102 It was recently shown that treatment with tocilizumab, an antibody that blocks the IL-6 receptor, resulted in poor outcomes in COVID-19 patients and did not prevent progression to secondary hemophagocytic lymphohistiocytosis.103 This suggests that IL-6 may be a major indicator but not the sole driver in the pathology of disease. It is possible too that the stage of disease may impact the benefit of IL-6 blockage therapy.
TNF is a master proinflammatory cytokine that is involved in the pathogenesis of autoimmune diseases such as rheumatoid arthritis (RA); consequently, anti-TNF biologics have become a first-line treatment for such diseases.104 High levels of TNF often are produced in the initial response to infectious disease.105 A recent study shows that the level of TNF (60-130 pg/mL) was higher than that of IL-6 (10-50 pg/mL) in the plasma of severe COVID-19 patients.14 Therefore, further investigation is warranted to define how TNF impacts immunopathology as well as the effectiveness of anti-TNF therapy in severe COVID-19 patients.
In addition to examining IL-6 and TNF, Yang et al.93 studied 48 cytokines in the PB from COVID-19 patients, 14 of which were markedly increased. Among these 14 cytokines, IP-10, MCP-3, and IL-1ra were identified as biomarkers for severity and fatal outcomes of disease. They also found that the level of IP-10 was markedly higher in patients with severe compared with mild disease. A report from 10 COVID-19 patients with severe disease showed a marked elevation of CCL5 (CC chemokine ligand 5, RANTES); importantly, treatment with CCR5-blocking antibody resulted in decreased IL-6 blood levels, lower IFN-related gene expression, reduced SARS-CoV-2 viral load, and a restoration of the immune homeostasis.106 Liu et al.107 reported that 38 cytokines in the PB of COVID-19 patients were significantly increased, and 15 cytokines (IL-12, IL-1ra, IP-10, PDGF-BB [platelet-derived growth factor-BB], TNF, IFN-γ, M-CSF [macrophage CSF], IL-17, HGF, G-CSF, IL-2, IL-4, IL-10, IL-1α, and IL-7) were associated with severity of disease. In addition, some inflammatory markers, such as CRP and D-dimer, were also markedly elevated.102 However, unlike findings in SARS patients,108 COVID-19 patients showed increases in anti-inflammatory cytokines such as IL-10 and IL-4,74 indicative of an increased Th2 response and subsequent pulmonary interstitial fibrosis. Last, the results of single-cell RNA sequence of early recovery patients infected with SARS-CoV-2, showed that IL-1β and M-CSF may be novel mediators in the inflammatory response associated with a cytokine storm.47,77
A Th17-type cytokine storm caused by a mobilization of Th17 responses has also been observed in both SARS and MERS patients.109,110 It was reported that a high number of CCR4+ CCR6+ Th17 cells, which at least partially attributed to this immunopathology, was also present in COVID-19 patient with ARDS.84 Markedly elevated cytokines (i.e., IL-1, IL-17, TNF, and GM-CSF) in COVID-19 patients have been associated with Th17 responses.107 These findings suggest that the Th17-type cytokine storm may lead or be associated with the onset of organ damage commonly observed in severe COVID-19 patients.84
Taken together, staging of SARS-CoV-2 infection are commonly divided into four stages (mild/common/severe/critical), all of which may have different magnitudes of cytokine release. It is also possible that the qualitative features of the cytokine storm between disease stages may point to distinct host or pathogen triggers (e.g., cytokine storm with or without concomitant bacterial pneumonia). Figure 2 summarizes the combined contribution of blood and lung tissue infiltrates, and shows how added factors such as an accompanying bacterial pneumonia may exacerbate cytokine responses.
FIGURE 2.
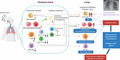
Severe SARS-CoV-2 infection: summary of aberrant activation of leukocytes and cytokine production contributing to a cytokine storm and pathology. Conceptual model of observations associated with severe SARS-CoV-2 infection. The activation of monocytes/macrophages and lymphocyte subsets in blood are likely to be major sources of cytokine release, together with the infiltration of leukocytes into lung tissue. Alveolar injury is shown to be associated with cell infiltrates, the release of neutrophil extracellular traps (NETs), hyperplasia of type II pneumocytes, among others, all of which could result in ARDS, lung insufficiency and a cytokine storm (exacerbated if combined with a superimposed bacterial infection). COVID-19/bacterial pneumonia image provided by: Dr. Ana S. Kolansky, University of Pennsylvania. Figure is made with BioRender (https://app.biorender.com/)
Treatments Under Investigation Against the Cytokine Storm in Covid-19 Infection
Because of its central role in the pathogenesis of SARS-CoV-2 infection, the cytokine storm and its accompanying excessive inflammatory responses have become a therapeutic target in the treatment of COVID-19 patients. Immunotherapy strategies aimed at curtailing the cytokine storm are under investigation in several countries (Table 2). Additional therapies to indirectly reduce viral burden and secondarily reduce the incidence of a cytokine storm are summarized in Supporting Information Tables S1–S4. Strategies to date do not address a specific type of cytokine storm but assume a “one size fits all” approach in treating severe disease in all patients.
TABLE 2.
Ongoing Clinical Trials: therapeutics against cytokine storm (up to May 6, 2020)
| Register Number | Title | Drugs/Strategies | Therapeutic Target | Study design | Samples | Phase |
|---|---|---|---|---|---|---|
| ChiCTR2000029765 | A multicenter, randomized controlled trial for the efficacy and safety of tocilizumab in the treatment of new coronavirus pneumonia (COVID-19) | Tocilizumab | Anti-IL-6 Receptor | RCT | 188 | 4 |
| ChiCTR2000030196 | A multicenter, single arm, open label trial for the efficacy and safety of CMAB806 in the treatment of cytokine release syndrome of novel coronavirus pneumonia (COVID-19) | Tocilizumab | Anti-IL-6 Receptor | Single arm | 60 | 2 |
| NCT04317092 | Tocilizumab in COVID-19 Pneumonia (TOCIVID-19) | Tocilizumab | Anti-IL-6 Receptor | Single arm | 330 | 2 |
| NCT04320615 | A Study to Evaluate the Safety and Efficacy of Tocilizumab in Patients with Severe COVID-19 Pneumonia | Tocilizumab | Anti-IL-6 Receptor | RCT | 330 | 3 |
| NCT04310228 | Favipiravir Combined with Tocilizumab in the Treatment of Corona Virus Disease 2019 | Tocilizumab | Anti-IL-6 Receptor | RCT | 150 | N/A |
| NCT04306705 | Tocilizumab vs CRRT in Management of Cytokine Release Syndrome (CRS) in COVID-19 | Tocilizumab | Anti-IL-6 Receptor | Cohort study | 120 | N/A |
| NCT04315480 | Tocilizumab for SARS-CoV2 Severe Pneumonitis | Tocilizumab | Anti-IL-6 Receptor | Single arm | 30 | 2 |
| NCT04331795 | Tocilizumab to Prevent Clinical Decompensation in Hospitalized, Non-critically Ill Patients With COVID-19 Pneumonitis | Tocilizumab | Anti-IL-6 Receptor | RCT | 50 | 2 |
| NCT04331808 | CORIMUNO-19 - Tocilizumab Trial - TOCI (CORIMUNO-TOCI) | Tocilizumab | Anti-IL-6 Receptor | RCT | 240 | 2 |
| NCT04332913 | Efficacy and Safety of Tocilizumab in the Treatment of SARS-Cov-2 Related Pneumonia | Tocilizumab | Anti-IL-6 Receptor | Cohort study | 30 | N/A |
| NCT04335071 | Tocilizumab in the Treatment of Coronavirus Induced Disease (COVID-19) | Tocilizumab | Anti-IL-6 Receptor | RCT | 100 | 2 |
| NCT04346355 | Efficacy of Early Administration of Tocilizumab in COVID-19 Patients | Tocilizumab | Anti-IL-6 Receptor | RCT | 398 | 2 |
| NCT04356937 | Efficacy of Tocilizumab on Patients With COVID-19 | Tocilizumab | Anti-IL-6 Receptor | RCT | 300 | 3 |
| NCT04359667 | Serum IL-6 and Soluble IL-6 Receptor in Severe COVID-19 Pneumonia Treated With Tocilizumab | Tocilizumab | Anti-IL-6 Receptor | Case | 30 | N/A |
| NCT04361032 | Assessment of Efficacy and Safety of Tocilizumab Compared to Deferoxamine, Associated With Standards Treatments in COVID-19 (+) Patients Hospitalized In Intensive Care in Tunisia | Tocilizumab/Deferoxamine | Anti-IL-6 Receptor | RCT | 260 | 3 |
| NCT04361552 | Tocilizumab for the Treatment of Cytokine Release Syndrome in Patients With COVID-19 (SARS-CoV-2 Infection) | Tocilizumab | Anti-IL-6 Receptor | RCT | 180 | 3 |
| NCT04363736 | A Study to Investigate Intravenous Tocilizumab in Participants With Moderate to Severe COVID-19 Pneumonia | Tocilizumab | Anti-IL-6 Receptor | RCT | 100 | 2 |
| NCT04363853 | Tocilizumab Treatment in Patients With COVID-19 | Tocilizumab | Anti-IL-6 Receptor | Single arm | 200 | 2 |
| NCT04370834 | Tocilizumab for Patients With Cancer and COVID-19 Disease | Tocilizumab | Anti-IL-6 Receptor | Single arm | 200 | 2 |
| NCT04372186 | A Study to Evaluate the Efficacy and Safety of Tocilizumab in Hospitalized Participants With COVID-19 Pneumonia | Tocilizumab | Anti-IL-6 Receptor | RCT | 379 | 3 |
| NCT04322773 | Anti-il6 Treatment of Serious COVID-19 Disease with Threatening Respiratory Failure | Tocilizumab + Sarilumab | Anti-IL-6 Receptor | RCT | 200 | 2 |
| NCT04345445 | Study to Evaluate the Efficacy and Safety of Tocilizumab Versus Corticosteroids in Hospitalised COVID-19 Patients with High Risk of Progression | Tocilizumab or Corticosteroids |
Anti-IL-6 Receptor Glucocorticoid |
RCT | 310 | 3 |
| NCT04315298 | Evaluation of the Efficacy and Safety of Sarilumab in Hospitalized Patients With COVID-19 | Sarilumab | Anti-IL-6 Receptor | RCT | 400 | 3 |
| NCT04324073 | Cohort Multiple Randomized Controlled Trials Open-label of Immune Modulatory Drugs and Other Treatments in COVID-19 Patients - Sarilumab Trial - CORIMUNO-19 - SARI | Sarilumab | Anti-IL-6 Receptor | RCT | 240 | 3 |
| NCT04357808 | Efficacy of Subcutaneous Sarilumab in Hospitalised Patients With Moderate-severe COVID-19 Infection (SARCOVID) | Sarilumab | Anti-IL-6 Receptor | RCT | 30 | 2 |
| NCT04357860 | Clinical Trial of Sarilumab in Adults With COVID-19 | Sarilumab | Anti-IL-6 Receptor | RCT | 120 | 2 |
| NCT04359901 | Sarilumab for Patients With Moderate COVID-19 Disease: A Randomized Controlled Trial With a Play-The-Winner Design | Sarilumab | Anti-IL-6 Receptor | RCT | 120 | 2 |
| NCT04322188 | An Observational Case-control Study of the Use of Siltuximab in ARDS Patients Diagnosed With COVID-19 Infection | Siltuximab | Anti-IL-6 | Case-Control | 50 | N/A |
| NCT04343989 | A Randomized Placebo-controlled Safety and Dose-finding Study for the Use of the IL-6 Inhibitor Clazakizumab in Patients with Life-threatening COVID-19 Infection | Clazakizumab | Anti-IL-6 | RCT | 30 | 2 |
| NCT04348500 | Clazakizumab (Anti-IL- 6 Monoclonal) Compared to Placebo for COVID19 Disease | Clazakizumab | Anti-IL-6 | RCT | 60 | 2 |
| NCT04363502 | Use of the Interleukin-6 Inhibitor Clazakizumab in Patients With Life-threatening COVID-19 Infection | Clazakizumab | Anti-IL-6 | RCT | 30 | 2 |
| ChiCTR2000030196 | A multicenter, single arm, open label trial for the efficacy and safety of CMAB806 in the treatment of cytokine release syndrome of novel coronavirus pneumonia (COVID-19) |
Conventional therapy+ Tocilizumab |
Anti-IL-6R | Single arm | 60 | 2 |
| NCT04362813 | Study of Efficacy and Safety of Canakinumab Treatment for CRS in Participants With COVID-19-induced Pneumonia | Canakinumab | Anti-IL-1β | RCT | 450 | 3 |
| NCT04365153 | Canakinumab to Reduce Deterioration of Cardiac and Respiratory Function Due to COVID-19 | Canakinumab | Anti-IL-1β | RCT | 45 | 2 |
| NCT04362111 | Early Identification and Treatment of Cytokine Storm Syndrome in Covid-19 | Anakinra | Anti-IL-1 Receptor | RCT | 20 | 3 |
| NCT04357366 | suPAR-guided Anakinra Treatment for Validation of the Risk and Management of Respiratory Failure by COVID-19 (SAVE) | Anakinra or trimethoprim/sulfamethoxazole | Anti-IL-1 Receptor or Anti-inflammation | Single arm | 100 | 2 |
| NCT04364009 | Anakinra for COVID-19 Respiratory Symptoms | Anakinra | Anti-IL-1 Receptor | RCT | 240 | 3 |
| NCT04366232 | Efficacy of Intravenous Anakinra and Ruxolitinib During COVID-19 Inflammation (JAKINCOV) | Anakinra+ Ruxolitinib | Anti-IL-1 Receptor+ JAK inhibitor | RCT | 50 | 2 |
| NCT04330638 | Treatment of COVID-19 Patients with Anti-interleukin Drugs |
Anakinra+ Siltuximab+ Tocilizumab |
IL-1 receptor Antagonist+ Anti-IL-6+ Anti-IL-6 Receptor |
RCT | 342 | 3 |
| ChiCTR2000030089 | A clinical study for the efficacy and safety of Adalimumab Injection in the treatment of patients with severe novel coronavirus pneumonia (COVID-19) | Adalimumab | Anti-TNF-alpha | RCT | 60 | 4 |
| ChiCTR2000030580 | Efficacy and safety of adamumab combined with tozumab in severe and critical patients with novel coronavirus pneumonia (COVID-19) | Adalimumab and Tocilizumab |
Anti-TNF-alpha Anti-IL-6 Receptor |
RCT | 60 | 4 |
| NCT04324021 | Efficacy and Safety of Emapalumab and Anakinra in Reducing Hyperinflammation and Respiratory Distress in Patients With COVID-19 Infection. | Emapalumab and Anakinra |
Anti-IFN-γ IL-1 receptor Antagonist |
RCT | 54 | 3 |
| ChiCTR2000030703 | A randomized, blinded, controlled, multicenter clinical trial to evaluate the efficacy and safety of Ixekizumab combined with conventional antiviral drugs in patients with novel coronavirus pneumonia (COVID-19) | Ixekizumab | Anti-IL-17A | RCT | 40 | 0 |
| NCT04347226 | Anti-Interleukin-8 (Anti-IL-8) for Cancer Patients With COVID-19 | BMS-986253 | Anti-IL-8 | RCT | 138 | 2 |
| NCT04275245 | Clinical Study of Anti-CD147 Humanized Meplazumab for Injection to Treat With 2019-nCoV Pneumonia | Humanized Meplazumab | Anti-CD147 | Single arm | 20 | 2 |
| NCT04337216 | Mavrilimumab to Reduce Progression of Acute Respiratory Failure in Patients with Severe COVID-19 Pneumonia and Systemic Hyper-inflammation | Mavrilimumab | Anti-GM-CSF-R | Single arm | 10 | 2 |
| NCT04341116 | Study of TJ003234 (Anti-GM-CSF Monoclonal Antibody) in Subjects With Severe Coronavirus Disease 2019 (COVID-19) | Anti-GM-CSF Monoclonal Antibody | Anti-GM-CSF | RCT | 144 | 3 |
| NCT04343651 | Study to Evaluate the Efficacy and Safety of Leronlimab for Mild to Moderate COVID-19 | Leronlimab | CCR5 blockading | RCT | 75 | 2 |
| NCT04347239 | Study to Evaluate the Efficacy and Safety of Leronlimab for Patients With Severe or Critical Coronavirus Disease 2019 (COVID-19) | Leronlimab | CCR5 blockading | RCT | 390 | 2 |
| ChiCTR2000030262 | Clinical study for combination of anti-viral drugs and type I interferon and inflammation inhibitor TFF2 in the treatment of novel coronavirus pneumonia (COVID-19) | type I interferon and TFF2 |
Boost innate resistance Anti-inflammatory peptide |
RCT | 30 | 0 |
| ChiCTR2000029572 | Safety and efficacy of umbilical cord blood mononuclear cells in the treatment of severe and critically 2019-nCoV pneumonia (novel coronavirus pneumonia, NCP): a randomized controlled clinical trial | Umbilical cord blood mononuclear cells | Anti-inflammatory, Anti-fibrotic | RCT | 30 | 0 |
| ChiCTR2000029606 | Clinical Study for Human Menstrual Blood-Derived Stem Cells in the Treatment of Acute Novel Coronavirus Pneumonia (NCP) | Human Menstrual Blood-Derived Stem Cells | Anti-inflammatory, Anti-fibrotic | RCT | 63 | 0 |
| ChiCTR2000029990 | Clinical trials of mesenchymal stem cells for the treatment of pneumonitis caused by novel coronavirus pneumonia (COVID-19) | Mesenchymal stem cells | Anti-inflammatory, Anti-fibrotic | RCT | 120 | 2 |
| ChiCTR2000030116 | Safety and effectiveness of human umbilical cord mesenchymal stem cells in the treatment of acute respiratory distress syndrome of severe novel coronavirus pneumonia (COVID-19) | Human umbilical cord mesenchymal stem cells | Anti-inflammatory, Anti-fibrotic | CCT | 16 | N/A |
| ChiCTR2000030866 | Open-label, observational study of human umbilical cord derived mesenchymal stem cells in the treatment of severe and critical patients with novel coronavirus pneumonia (COVID-19) | Human umbilical cord derived mesenchymal stem cells | Anti-inflammatory, Anti-fibrotic | Single arm | 30 | 0 |
| NCT04299152 | Stem Cell Educator Therapy Treat the Viral Inflammation Caused by Severe Acute Respiratory Syndrome Coronavirus 2 | Stem Cell | Immune suppression | RCT | 20 | 2 |
| NCT04333368 | Cell Therapy Using Umbilical Cord-derived Mesenchymal Stromal Cells in SARS-CoV-2-related ARDS | Umbilical Cord-derived Mesenchymal Stromal Cells | Anti-inflammatory, Anti-fibrotic | RCT | 60 | 2 |
| NCT04345601 | Mesenchymal Stromal Cells for the Treatment of SARS-CoV-2 Induced Acute Respiratory Failure (COVID-19 Disease) | Mesenchymal Stromal Cells | Anti-inflammatory, Anti-fibrotic | Single arm | 30 | 1 |
| NCT04361942 | Treatment of Severe COVID-19 Pneumonia With Allogeneic Mesenchymal Stromal Cells (COVID_MSV) | Allogeneic Mesenchymal Stromal Cells | Anti-inflammatory, Anti-fibrotic | RCT | 24 | 2 |
| NCT04366063 | Mesenchymal Stem Cell Therapy for SARS-CoV-2-related Acute Respiratory Distress Syndrome | Mesenchymal Stem Cell | Anti-inflammatory, Anti-fibrotic | RCT | 60 | 3 |
| NCT04366830 | Intermediate-size Expanded Access Program (EAP), Mesenchymal Stromal Cells (MSC) for Acute Respiratory Distress Syndrome (ARDS) Due to COVID-19 Infection | EAP+MSCs | Anti-inflammatory, Anti-fibrotic | N/A | N/A | N/A |
| NCT04371393 | MSCs in COVID-19 ARDS | MSCs | Anti-inflammatory, Anti-fibrotic | RCT | 300 | 3 |
| ChiCTR2000029898 | A Randomized, Open-label, Parallel, Controlled Trial for Evaluation of the Efficacy and Safety of Chloroquine Phosphate in the treatment of Severe Patients with Novel Coronavirus Pneumonia (COVID-19) | Chloroquine Phosphate | Immune suppression | RCT | 100 | 4 |
| NCT04323631 | Hydroxychloroquine for the Treatment of Patients with Mild to Moderate COVID-19 to Prevent Progression to Severe Infection or Death | Hydroxychloroquine | Immune suppression | RCT | 1116 | 1 |
| NCT04323527 | Chloroquine Diphosphate for the Treatment of Severe Acute Respiratory Syndrome Secondary to SARS-CoV2 | Chloroquine Diphosphate | Immune suppression | RCT | 440 | 2 |
| NCT04358068 | Evaluating the Efficacy of Hydroxychloroquine and Azithromycin to Prevent Hospitalization or Death in Persons With COVID-19 | Hydroxychloroquine +Azithromycin | Anti-inflammation | RCT | 2000 | 2 |
| NCT04358081 | Hydroxychloroquine Monotherapy and in Combination With Azithromycin in Patients With Moderate and Severe COVID-19 Disease | Hydroxychloroquine+ Monotherapy+ Azithromycin | Anti-inflammation | RCT | 444 | 3 |
| NCT04362332 | Chloroquine, Hydroxychloroquine or Only Supportive Care in Patients AdmItted With Moderate to Severe COVID-19 | Chloroquine+ Hydroxychloroquine | Anti-inflammation | RCT | 950 | 4 |
| ChiCTR2000029757 | Convalescent plasma for the treatment of severe novel coronavirus pneumonia (COVID-19): a prospective randomized controlled trial | Convalescent plasma | Convalescent plasma | RCT | 200 | 0 |
| ChiCTR2000029850 | Study for convalescent plasma treatment for severe patients with novel coronavirus pneumonia (COVID-19) | Convalescent plasma | Convalescent plasma | CCT | 20 | 0 |
| ChiCTR2000030010 | A randomized, double-blind, parallel-controlled, trial to evaluate the efficacy and safety of anti-SARS-CoV-2 virus inactivated plasma in the treatment of severe novel coronavirus pneumonia patients (COVID-19) | Anti-SARS-CoV-2 virus inactivated plasma | Convalescent plasma | RCT | 100 | N/A |
| ChiCTR2000030929 | A randomized, double-blind, parallel-controlled trial to evaluate the efficacy and safety of anti-SARS-CoV-2 virus inactivated plasma in the treatment of severe novel coronavirus pneumonia (COVID-19) | Anti-SARS-CoV-2 virus inactivated plasma | Convalescent plasma | RCT | 60 | N/A |
| NCT04346446 | Efficacy of Convalescent Plasma Therapy in Severely Sick COVID-19 Patients | Convalescent Plasma in severe COVID-19 Patients | Convalescent Plasma | RCT | 20 | 2 |
| NCT04347681 | Potential Efficacy of Convalescent Plasma to Treat Severe COVID-19 and Patients at High Risk of Developing Severe COVID-19 | Convalescent Plasma | Convalescent Plasma | CCT | 40 | 2 |
| NCT04353206 | Convalescent Plasma in ICU Patients With COVID-19-induced Respiratory Failure | Convalescent Plasma in ICU Patients | Convalescent plasma | Single arm | 90 | 1 |
| NCT04359810 | Plasma Therapy of COVID-19 in Critically Ill Patients | Convalescent Plasma | Convalescent plasma | RCT | 105 | 2 |
| ChiCTR2000030475 | Cytosorb in Treating Critically Ill Hospitalized Adult Patients with novel coronavirus pneumonia (COVID-19) | Cytosorb | Broad Cytokine/Toxin Removal | Single arm | 19 | 0 |
| NCT04324528 | Cytokine Adsorption in Severe COVID-19 Pneumonia Requiring Extracorporeal Membrane Oxygenation |
Cytokine Adsorption+ Extracorporeal Membrane Oxygenation |
Broad Cytokine/Toxin Removal | Case reports | 30 | R/A |
| NCT04344080 | Effect of CytoSorb Adsorber on Hemodynamic and Immunological Parameters in Critical Ill Patients With COVID-19 | CytoSorb | Broad Cytokine/Toxin Removal | RCT | 24 | N/A |
| NCT04358003 | Plasma Adsorption in Patients With Confirmed COVID-19 | Plasma Adsorption | Broad Cytokine/Toxin Removal | Single arm | 2000 | N/A |
| NCT04374149 | Therapeutic Plasma Exchange Alone or in Combination With Ruxolitinib in COVID-19 Associated CRS | Plasma Exchange | Broad Cytokine/Toxin Removal | CCT | 20 | 2 |
| NCT04374539 | Plasma Exchange in Patients With COVID-19 Disease and Invasive Mechanical Ventilation: a Randomized Controlled Trial | Plasma Exchange | Broad Cytokine/Toxin Removal | RCT | 116 | 2 |
| NCT04273581 | The Efficacy and Safety of Thalidomide Combined with Low-dose Hormones in the Treatment of Severe COVID-19 | Thalidomide and Interferon-alpha | Prevent lung injury Boost Innate resistance | RCT | 40 | 2 |
| NCT04293887 | Efficacy and Safety of IFN-α2β in the Treatment of Novel Coronavirus Patients | IFN-α2β | Boost Innate Resistance | RCT | 328 | 1 |
| NCT04343768 | An Investigation into Beneficial Effects of Interferon Beta 1a, Compared to Interferon Beta 1b And The Base Therapeutic Regiment in Moderate to Severe COVID-19: A Randomized Clinical Trial | Interferon Beta 1a or Interferon Beta 1b | Boost Innate Resistance | RCT | 60 | 4 |
| NCT04325061 | Efficacy of Dexamethasone Treatment for Patients with ARDS Caused by COVID-19 | Dexamethasone | Anti-inflammatory | RCT | 200 | 4 |
| NCT04347980 | Dexamethasone Treatment for Severe Acute Respiratory Distress Syndrome Induced by COVID-19 | Dexamethasone | Anti-inflammatory | RCT | 122 | 3 |
| NCT04327401 | COVID-19-associated ARDS Treated with Dexamethasone: Alliance Covid-19 Brasil III | Dexamethasone | Immune Suppression | RCT | 290 | 3 |
| NCT04358627 | Dexmedetomidine to Improve Outcomes of ARDS in Critical Care COVID-19 Patients | Dexmedetomidine | Immune Suppression | Case | 80 | N/A |
| NCT04355247 | Prophylactic Corticosteroid to Prevent COVID-19 Cytokine Storm | Corticosteroid | Immune Suppression | Single arm | 20 | 2 |
| NCT04360876 | Targeted Steroids for ARDS Due to COVID-19 Pneumonia: A Pilot Randomized Clinical Trial | Steroids | Immune Suppression | RCT | 90 | 2 |
| NCT04323592 | Efficacy of Methylprednisolone for Patients With COVID-19 Severe Acute Respiratory Syndrome | Methylprednisolone | Anti-inflammatory | CCT | 104 | 3 |
| NCT04343729 | Methylprednisolone in the Treatment of Patients with Signs of Severe Acute Respiratory Syndrome in Covid-19 | Methylprednisolone | Anti-inflammatory | RCT | 420 | 2 |
| NCT04306393 | Nitric Oxide Gas Inhalation in Severe Acute Respiratory Syndrome in COVID-19 | Nitric Oxide Gas | Pulmonary vasodilator | RCT | 200 | 2 |
| NCT04358588 | Pulsed Inhaled Nitric Oxide for the Treatment of Patients With Mild or Moderate COVID-19 | Nitric Oxide | Anti-inflammation | N/A | N/A | N/A |
| NCT04244591 | Glucocorticoid Therapy for Novel Coronavirus Critically Ill Patients With Severe Acute Respiratory Failure | Glucocorticoid | Immune suppression | RCT | 80 | 3 |
| NCT04320277 | Baricitinib in Symptomatic Patients Infected by COVID-19: an Open-label, Pilot Study. | Baricitinib | JAK inhibitor | CCT | 60 | 3 |
| NCT04340232 | Safety and Efficacy of Baricitinib for COVID-19 | Baricitinib | JAK inhibitor | Single arm | 80 | 3 |
| NCT04359290 | Ruxolitinib for Treatment of Covid-19 Induced Lung Injury ARDS | Ruxolitinib | JAK inhibitor | Single arm | 15 | 2 |
| NCT04355793 | Expanded Access Program of Ruxolitinib for the Emergency Treatment of Cytokine Storm From COVID-19 Infection | Ruxolitinib | JAK inhibitor | N/A | N/A | N/A |
| NCT04361903 | Ruxolitinib for the Treatment of Acute Respiratory Distress Syndrome in Patients With COVID-19 Infection | Ruxolitinib | JAK inhibitor | Cohort | 13 | N/A |
| NCT04362137 | Phase 3 Randomized, Double-blind, Placebo-controlled Multi-center Study to Assess the Efficacy and Safety of Ruxolitinib in Patients With COVID-19 Associated Cytokine Storm (RUXCOVID) | Ruxolitinib | JAK inhibitor | RCT | 402 | 3 |
| NCT04321993 | Treatment of Moderate to Severe Coronavirus Disease (COVID-19) in Hospitalized Patients |
Baricitinib+ Sarilumab+Lopinavir/ritonavir+Hydroxychloroquine sulfate |
JAK inhibitor+ Anti-IL-6+ Anti-virus+ Anti-inflammatory |
CCT | 1000 | 2 |
| NCT04373044 | Antiviral Therapy and Baricitinib for the Treatment of Patients With Moderate or Severe COVID-19 | Antiviral Therapy+Baricitinib | Anti-virus+JAK inhibitor | Single arm | 59 | 2 |
| NCT04348383 | Defibrotide as Prevention and Treatment of Respiratory Distress and Cytokine Release Syndrome of Covid 19. | Defibrotide | Anti-inflammatory | RCT | 120 | 2 |
| NCT04357444 | Low Dose of IL-2 In Acute Respiratory DistrEss Syndrome Related to COVID-19 | IL-2 | Anti-inflammation | RCT | 30 | 2 |
| NCT04355364 | Efficacy and Safety of Aerosolized Intra-tracheal Dornase Alpha Administration in Patients With COVID19-induced ARDS (COVIDORNASE) | Aerosolized Intra-tracheal Dornase Alpha | Anti-inflammation | RCT | 100 | 3 |
| NCT04363437 | COlchicine in Moderate Severity Hospitalized Patients Before ARDS to Treat COVID-19 | Colchicine | Anti-inflammation | RCT | 70 | 2 |
| NCT04366791 | Radiation Eliminates Storming Cytokines and Unchecked Edema as a 1-Day Treatment for COVID-19 | Radiation | Other | Single arm | 10 | 2 |
Abbrevation: ChiCTR, Chinese Clinical Trials Register; NCT, National Clinical Trails; RCT: Random clinical trial; CCT: Controlled clinical trial;
Blockade of proinflammatory cytokine
Clinical reports showing that elevated levels of IL-6 are associated with the immunopathology and disease severity of COVID-19111 have provided a strong scientific rationale for examining the effects of IL-6 or its receptor antagonists (e.g., siltuximab and clazakizumab or sarilumab and tocilizumab). In fact, tocilizumab has been recommended in China to treat COVID-19 patients with bilateral pulmonary damage and severe symptoms.112 Early clinical reports of tocilizumab showed that fevers subsided in 20 severe patients within 1 d after the treatment and 95% of these patients achieved recovery sufficient to allow them to be released from the hospital within 2 wk.113 Guo et al.114 showed that tocilizumab treatment (400 mg once through an i.v. drip115), attenuated overactivated inflammatory immune responses and boosted antivirus immune responses mediated by B cells and CD8+ T cells. Furthermore, Luo et al.116 recommended that tocilizumab be given repeatedly in low doses (80–240 mg per time) for maximal benefit. Taken together, therapy to block IL-6 remains the most prevalent anticytokine treatment under investigation. By contrast, there are no registered studies in the Chinese Clinical Trials Register (ChiCTR) to test the effect of IL-1 inhibitors for the treatment of COVID-19. Although IL-6 is an attractive therapy target, blocking IL-6 may not be beneficial in all patients. As noted above, treatment with tocilizumab can result in the onset of secondary hemophagocytic lymphohistiocytosis in COVID-19 patients.103 Similar to anti-IL-6, the effect of antagonists against other inflammatory cytokines in attenuating a cytokine storm is under investigation. As of May 6, 2020, a total of 49 clinical trials targeting cytokine inhibition were registered in the ChiCTR and the National Institutes of Health’s Clinical Research Trials (Table 2). In addition to anti-IL-6 strategies, antagonistic antibodies directed against IFN-γ, IL-1, IL-1R, TNF, IL-8, GM-CSF, GM-CSF receptor, IL-17A, and CCR-5 are also under evaluation for their effect on the cytokine storm or in mediating excessive immune activation (Table 2). Other approaches to suppress proinflammatory cytokine in COVID-19 patients have also been proposed. For example, given IL-37′s ability to inhibit IL-1β, IL-6, TNF, and CCL2, Conti and colleagues hypothesized that IL-37 may be useful in the treatment of COVID-19 patients with CRS.117 In another study, the CCL5-CCR5 axis was found to contribute to immunopathology106 in critically ill COVID-19 patients with elevated levels of CCL5 (RANTES), as compared with patients who had mild or moderate cases of disease. Blocking the CCR5 antibody with Leronlimab reduced IL-6 levels in critically ill COVID-19 patients and restored the CD4/CD8 ratio, leading to a marked reduction in SARS-CoV-2 plasma viremia.106 Deng and colleagues have also suggested that upstream targets, such as cyclic guanosine monophosphate (GMP) – adenosine monophosphate (AMP) synthase (cGAS), anaplastic lymphoma kinase (ALK), and stimulator of interferon genes (STING), may help reduce cell activation and cytokine release.118 Likewise, JAK-STAT signaling inhibitors (Baricitinib and Ruxolitinib) have also been proposed for preventing CRS119 (Table 2). Apart from anticytokine treatment, cytokine therapy with IFN-α2b was reported by Huazhong University of Science and Technology (Wuhan, China), where treated patients saw reductions in their levels of IL-6 as well as a shorter duration in viral shedding.54 IFN-beta-1b has also been noted as beneficial when added to antiviral regimen that includes lopinavir and ritonavir.120
Transplantation of mesenchymal stem cells (MSCs)
MSCs are adult stem cells that have the ability to self-replicate and which show potential for differentiation into multiple cell types.121 MSCs have the potential to impact anti-inflammatory activities by producing immunosuppressive cytokines and by directly interacting with and inhibiting the activation of immune cells.121 Leng et al88 reported that 7 COVID-19 patients in Beijing Youan Hospital (Beijing, China) were given MSCs (ChiCTR2000029990). In this report, 14 d after transplantation with MSCs, patients showed an increase in peripheral lymphocytes and CD14+CD11c+CD11bmid regulatory DC cells, accompanied by an increase in IL-10 and a decrease in TNF. Those patients also saw a decrease in cytokine-producing CXCR3+CD4+ T cells, CXCR3+CD8+ T cells, and CXCR3+ NK cells. As of May 6, 2020, a total of 40 clinical trials evaluating the efficacy of MSCs transplantation in the treatment of COVID-19 patients have been registered in ChiCTR and NIH (Table 2 and Supporting Information Table S1).
Transfusion of convalescent plasma
CP transfusion has been recommended for the treatment of patients experiencing a sudden disease progression and those with critical and severe disease, according to the Guidelines of Diagnosis and Treatment of COVID-19 (7th ed., China).112 It was previously shown that CP treatment could reduce the levels of cytokines in patients with severe influenza.122 Yang et al.73 reported that the viremia of 10 COVID-19 patients was controlled and the lymphocyte counts increased after 7 d of CP transfusion. Similar beneficial effects also were reported in 6 severe COVID-19 patients.87 However, results from patients receiving CP remain limited as questions remain related to the optimal dose and best therapeutic window for administering CP. For example, patients in the early phases of infection have not been studied. In addition, CP often is combined with other treatments making it difficult to conclusively evaluate the true benefit of CP alone.119 Importantly, the question of whether harmful ADE could be induced by CP therapy remains under study.119 Directly removing cytokines (and other toxins) through blood purification using continuous renal replacement therapy, hemoadsorption, hemoperfusion, and blood exchange have also been proposed as a way of containing the cytokine storm. Luo et al.123 reported findings from three patients treated with plasma exchange in the First Affiliated Hospital of Bengbu Medical College (Bengbu, China). Their results showed that the level of IL-6 was decreased and lymphocyte count increased after treatment. Current clinical trials testing the therapeutic effect of CP and blood purification are summarized in Table 2 and Supporting Information Table S2.
Chloroquine (CQ) or hydroxychloroquine (HCQ) plus azithromycin
CQ or HCQ have been proposed as antiviral agents in the treatment of both mild and severe cases of COVID-19.124 These compounds increase the pH of endosomes, which may reduce SARS-CoV-2 infection by inhibiting Cathepsin L, an enzyme that is vital for cleavage of the viral Spike protein,125 leading to viral entry. CQ or HCQ also have the anti-inflammatory effects, regulating myeloid activity by limiting its impact on endocytic TLR receptors (TLR 3, 7, 9) and reducing peptide binding on MHC class II proteins.126,127 Use of CQ or HCQ as treatment for COVID-19 has commonly been joined with azithromycin to prevent bacterial infections. Azithromycin also is often used clinically to dampen lung inflammation in other conditions such as cystic fibrosis and chronic obstructive pulmonary disease, providing added rationale for its use in COVID-19.128 Gautret et al.129 treated COVID-19 patients (n = 20) with 600 mg of HCQ daily, with some patients (n = 6) also receiving concurrent azithromycin, depending on their clinical manifestation. Results support the finding that HCQ treatment can reduce viral load and its effect might be reinforced by concurrent use of azithromycin, which as noted above could be the result of the antibiotic’s immunomodulatory effect as seen with other respiratory diseases. Lu et al. (n = 13)130 and Zhang et al. (n = 31)131 (registered number: NCT04261517 and ChiCTR2000029559, shown in Supporting Information Table S3) also reported that early treatment with HCQ showed better outcomes (negative COVID-19 nucleic acid of throat swabs, shorter time to clinical recovery, or shorter cough remission time). In a retrospective study from Yu et al.,132 HCQ reduced the level of IL-6 and decreased mortality of critical COVID-19 patients (n = 48), suggesting that HCQ could be an effective treatment in critical COVID-19 patients.
Of note, it is still unknown if HCQ has differing effects on cytokine and leukocyte levels when it is administered at different stages of disease; therefore, the effect of HCQ on the immunopathology of CRS in COVID-19 patients has yet to be determined. Likewise, the therapeutic benefit of HCQ has been challenged by data showing that long-term use of HCQ in systemic lupus erythematosus (SLE) patients did not prevent the onset of severe COVID-19 in a subset of SLE patients (n = 17).133 Furthermore, high CQ doses (600 mg) used in severe cases may lead to arrhythmias and cardiac complications.134 Although anecdotal reports suggest that CQ or HCQ may possess better efficacy in the treatment of early COVID-19, to date, limited clinical studies have been published. Whether the efficacy of HCQ is different in patients with varying degrees of severity and whether its clinical benefit, if confirmed, is because the drug limits immune responses (i.e., preventing a cytokine storm) or because it directly inhibits viral infection has not been fully explored. So far, a total of 83 clinical trials testing the preventative or therapeutic effects of HCQ or CQ on COVID-19 have now been registered (Table 2 and Supporting Information Table S3).
Traditional Chinese medicine (TCM), steroids, and other therapeutic approaches
According to the Guidelines of Diagnosis and Treatment of COVID-19 (7th ed., China), TCM is recommended in the management and treatment of COVID-19 patients in China. Inspired by the purported efficacy of TCM in the treatment of SARS, more than 85% of patients in different stages of COVID-19 in China were treated with a variety of TCM herbal medicines (see Yang et al.,135). TCM strategies selected for COVID-19 were selected based on their immunosuppressive and anti-inflammatory properties, as determined by their use in clinics.136–142 There also is evidence that the immunosuppressive features of TCM may convey beneficial effects in the prevention and treatment of a cytokine storm.114 A retrospective study reported that matrine and sodium chloride Injection could markedly improve lymphopenia in COVID-19 patients, possibly by inhibiting pulmonary inflammatory cytokines.143,144 In an in vitro experiment, Zhu et al.145 found that liquiritin could markedly inhibit replication of SARS-CoV-2 by mimicking type I IFN. Ma Xing Shi Gan decoction (MXSGD) is a multiple component TCM preparation recommended by the Guidelines for use in China. Findings from a network pharmacologic study and KEGG enrichment analysis indicate that MXSGD has a number of beneficial effects, including inhibiting cytokine storm.39 To identify additional TCM with the capacity to inhibit cytokine storm in COVID-19 patients, Ren and colleagues146 screened the TCM database 2009 (TCMD 2009). They searched for compounds that impacted the arachidonic acid (AA) metabolic pathway because it is essential for the synthesis of cytokines. Their study revealed magnolignan I, lonicerin, and physcion-8-O-β-D-glucopy-ranoside were the most potent AA inhibitors and, consequently, suggested that herbal medicines which included these agents might be most useful in dampening a cytokine storm in patients.146 We anticipate additional TCM strategies will be identified for the management of cytokine storm in COVID-19 patients. For example, ulinastatin, a natural anti-inflammatory substance from fresh human urine is believed to inhibit cytokine release and subsequent tissue injury147 and is currently under investigation as a treatment of COVID-19 patients.148
In addition to TCM, corticosteroid treatment has been shown to dampen the inflammatory cascade from a cytokine storm.149 A recent study found that early and short-term administration of methylprednisolone could improve the clinical outcome in moderate-to-severe COVID-19 patients.150 Although useful in critical patients as a tool for managing host inflammatory responses, corticosteroids are not recommended in high doses nor for prolonged periods of time as they may dampen normal immune responses that could help contain the viral infection.146,151,152 Nevertheless, there are a number of ongoing clinical trials (Table 2 and Supporting Information Table S4) that may show if and when safe and beneficial corticosteroid-based treatment is indicated for treating in COVID-19.
Last, several immunomodulatory therapies, including recombinant human IFN-alpha, rhG-CSF, i.v. immunoglobulin, antibody, vaccine, nutritional supplements (zinc, vitamin C, and vitamin D3), colchicine, and cellular therapy are also under investigation for inhibiting a cytokine storm (Table 2 and Supporting Information Table S4).
Closing Remarks
It is generally believed that the cytokine storm triggered by SARS-CoV-2 infection is a central mediator for the lung injury and resulting ARDS found in cases of severe or critical COVID-19 patients. We present several leukocyte and cytokine changes that may help define the progression of COVID-19 from an initial to a late stage in both mild and severe cases. However, more needs to be learned regarding how elevations in specific cytokine(s) combinations (using standardized methods and defined ranges) could be used to describe each stage of the inflammatory response as the disease progresses. Apart from commonly used surrogates of disease progression (D-dimer, CRP, ferritin, and procalcitonin levels) with emphasis on D-dimer and ferritin, cytokine-based biomarkers still lack consensus on cut-off values for assessing disease progression (other than a pattern of a rise with severity). Disease staging with an integration of cellular (neutrophilia, decreases in lymphocytes, etc.), soluble plasma markers (D-dimer, etc.) together with cytokine changes also remains to be pursued. Based on currently reported clinical data to date, priority could be placed on combining elevations of IL-6, TNF, IL-1β, IP-10, D-dimer, and ferritin with changes in circulating and activation states of myeloid, neutrophil, and T cell responses, which together could provide improved biomarkers for progression to CRS and severe COVID-19 patients. Although age, combined comorbidities (cardiovascular disease, diabetes, etc.), and bacterial infection have also emerged as risk factors for severe disease,153–155 it remains unclear how each of these conditions changes the immunoregulatory cascade, characteristics of the cytokine storm, coagulopathy, and other inflammatory outcomes. Defining shared or population-specific “triggers and amplifiers” of a cytokine storm at specific disease stages will advance novel precision medicine strategies.
Other factors known to contribute to disease pathogenesis (such as host genetics and epigenetics, microbiome, mucosal infection apart from lung, immunoregulatory networks, aging, co-occurring disease, exosomes, complement, neurologic, endocrine, and polydrug environments, etc.) have yet to be fully investigated. For example, Fogarty et al.98 reported a potential 3–4-fold higher thrombotic risk in Caucasians, which underscores the potential impact that race and ethnicity may have on disease outcomes. Although coagulopathy seems to be a severe complication in COVID-19 infection, it may not necessarily be related to disease severity or exclusively present during or after a cytokine storm.156 It remains undetermined how the onset of coagulopathy in the absence of compromised lung capacity (ventilation/perfusion) or CRS may be related to early press reports of higher stroke incidence in young and middle-aged infected patients in the United States. Indeed, it has been proposed that, in predisposed individuals, alveolar viral damage may trigger an underlying inflammatory reaction promoting a microvascular pulmonary thrombosis or endothelial thromboinflammatory syndrome affecting microvascular beds beyond the lung (e.g., brain and other vital organs).157 How immune factors identified in this review (e.g., NETosis, IL-6, macrophage activation, antibody response, etc.) may contribute to damage to microvascular beds or how this damage predisposes incidence of a CRS remains to be determined. Therefore, there is a need to identify leading molecular triggers of multi-organ failure after a cytokine storm in order to prevent added lethal outcomes.
Regarding therapy, the timing and the best ways to combine various treatments also remains to be determined (e.g., early antiviral therapy followed by anti-CRS therapy when that occurs).158 There is also a need to extend immunopathogenesis studies beyond adults. Based on studies of COVID-19 clinical features in pregnancy, it is known that neonates can be different from adults.159,160 For example, children rarely progress with a disease course requiring ICU care. Regrettably, the limited clinical trial activity focusing on children remains a missed opportunity to better address how to treat and limit transmission in this group.161
Greater advances in all of these areas will allow for selective treatments both to target key pathogenic components of the cytokine storm and further advance highly targeted precision medicine strategies.
Authorship
J.W., M.J., X.C. and L.J.M. wrote manuscript and designed figures. J.W. and M.J. contributed equally to this work.
Disclosures
The authors declare no conflicts of interest.
Supplementary Material
Supporting Information.
Acknowledgments
This work was supported by a grant to LJM by the Philadelphia Foundation (Robert I. Jacobs Fund), Ken Nimblett and The Summerhill Trust, the Kean Family Professorship, and the Wistar Institute. This work was funded by The Science and Technology Development Fund, Macau (File no. FDCT014/2015/A1, FDCT201/2017/A3 and FDCT0056/2019/AFJ) and funded by University of Macau (File no. MYRG2016-00023-ICMS-QRCM, MYRG2017-00120-ICMS and MYRG2019-00169-ICMS).
Abbreviations
- AA
Arachidonic acid
- ACE2
Angiotensin-converting enzyme 2
- ADCC
Antibody-dependent cell-mediated cytotoxicity
- ADE
Antibody-dependent enhancement
- ARDS
Acute respiratory distress syndrome
- ALK
Anaplastic Lymphomas Kinase
- BALF
Bronchoalveolar lavage fluid
- cGAS
cyclic GMP-AMP synthase
- ChiCTR
Chinese Clinical Trials Register
- COVID-19
Coronavirus disease 2019
- CP
Convalescent plasma
- CQ
Chloroquine
- CRP
C-reactive protein
- CRS
Cytokine release syndrome
- HCQ
hydroxychloroquine
- IP-10
IFN-induced protein 10
- M-CSF
Macrophage CSF
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MIG
Monokine induced by gamma IFN
- MIP-1A
Macrophage inflammatory protein-1 alpha
- MODS
Multiple organ dysfunction syndrome
- MSCs
Mesenchymal stem cells
- MXSGD
Ma Xing Shi Gan decoction
- NETs
Neutrophil extracellular traps
- NK
Natural killer cell
- NLR
Neutrophil-to-lymphocyte ratio
- PB
peripheral blood
- PDGF-BB
Platelet-derived growth factor-BB
- RA
Rheumatoid arthritis
- SARS-CoV
Severe acute respiratory syndrome-coronavirus
- SARS-CoV-2
Severe acute respiratory syndrome-coronavirus-2
- SpO2
Blood oxygen saturation
- STING
Stimulator of interferon genes
- TCM
Traditional Chinese medicine
- Treg
regulatory T cells
Contributor Information
Jin Wang, State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macau, SAR, China.
Mengmeng Jiang, State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macau, SAR, China.
Xin Chen, State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macau, SAR, China.
Luis J Montaner, Vaccine and Immunotherapy Center, The Wistar Institute, Philadelphia, Pennsylvania, USA.
References
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AC, Williamson DA. An outbreak of COVID-19 caused by a new coronavirus: what we know so far. Med J Aust. 2020;212:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization WH, 2020 Coronavirus disease 2019 (COVID-19) Situation Report-108. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200507covid-19-sitrep-108.pdf?sfvrsn=44cc8ed8_2.
- Cepeda-Valdes R, Carrion-Alvarez D, Trejo-Castro A, Hernandez-Torre M, Salas-Alanis J. Cutaneous manifestations in COVID-19: family cluster of Urticarial Rash. Clin Exp Dermatol. 2020. [epub ahead of print, PMID:32407564]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zheng X, Tong Q, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol. 2020;92:491-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RS, Wu A, To KF, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ (Clinical research ed). 2003;326:1358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Qie S, Liu Z, Ren J, Jianing Xi J. Clinical characteristics of 50466 patients with 2019-nCoV infection. J Med Virol. 2020. [epub ahead of print, PMID:32108351]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhou P, Wei Y, et al. Histopathologic changes and SARS-CoV-2 immunostaining in the lung of a patient with COVID-19. Ann Intern Med. 2020;172:629-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhou X, Qiu Y, et al. Clinical characteristics of 82 death cases with COVID-19. medRxiv. 10.1101/2020.02.26.20028191. 2020.02.26.20028191; doi:print. [DOI] [Google Scholar]
- Tian S, Xiong Y, Liu H, et al. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod Pathol. 2020. [Epub ahead of print: PMID 32291399]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton LM, Duval EJ, Stroberg E, Ghosh S. COVID-19 Autopsies, Oklahoma, USA. Am J Clin Pathol. 2020;153:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CL, Wang YM, Li XW, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, Hui DS, Azhar EI, Memish ZA, Maeurer M. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet North Am Ed. 2020;395(10224):e35-e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara JL, Abhyankar S, Gilliland DG. Cytokine storm of graft-versus-host disease: a critical effector role for interleukin-1. Transplant Proc. 1993;25:1216-1267. [PubMed] [Google Scholar]
- Chousterman BG, Swirski FK, Weber GF. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol. 2017;39:517-528. [DOI] [PubMed] [Google Scholar]
- Shimabukuro-Vornhagen A, Godel P, Subklewe M, et al. Cytokine release syndrome. J Immunother Cancer. 2018;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crayne CB, Albeituni S, Nichols KE, Cron RQ. The immunology of macrophage activation syndrome. Front Immunol. 2019;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry SM, Johnson MA, Janossy G. Cytopathology or immunopathology? The puzzle of cytomegalovirus pneumonitis revisited. Bone Marrow Transplant. 2000;26:591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KK, Khan MA. Storm SSKCIC: a Major Threat to Human Health. J Interferon Cytokine Res. 2020;40:19-23. [DOI] [PubMed] [Google Scholar]
- Cron RQ, Behrens EM. Cytokine Storm Syndrome. Switzerland AG: Springer Nature; 2019. [Google Scholar]
- Borrok MJ, Luheshi NM, Beyaz N, et al. Enhancement of antibody-dependent cell-mediated cytotoxicity by endowing IgG with FcαRI (CD89) binding. mAbs. 2015;7:743-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatenoud L, Ferran C, Reuter A, et al. Systemic reaction to the anti-T-cell monoclonal antibody OKT3 in relation to serum levels of tumor necrosis factor and interferon-gamma. N Engl J Med. 1989;320:1420-1421. [DOI] [PubMed] [Google Scholar]
- Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifaldi L, Prencipe G, Caiello I, et al. Inhibition of natural killer cell cytotoxicity by interleukin-6: implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol. 2015;67:3037-3046. [DOI] [PubMed] [Google Scholar]
- Li T, Xie J, He Y, et al. Long-term persistence of robust antibody and cytotoxic T cell responses in recovered patients infected with SARS coronavirus. PLoS One. 2006;1:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quartuccio L, Sonaglia A, McGonagle D, et al. Profiling COVID-19 pneumonia progressing into the cytokine storm syndrome: results from a single Italian Centre study on tocilizumab versus standard of care. J Clin Virol. 2020. 10.1016/j.jcv.2020.104444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, Beumer J, van der Vaart J. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020. [Epub ahead of print, PMID: 32358202]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Hu W, Niu L, Liu H, Xu H, Xiao S. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020;15:700-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien JY, Hsueh PR, Cheng WC, Yu CJ, Yang PC. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11:715-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li J, Zhan Y, et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun. 2004;72:4410-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SKP, Lau CCY, Chan KH, et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679-2690. [DOI] [PubMed] [Google Scholar]
- Liu HC, Li L, Mao CZ, et al. Analyses of the clinical features and contributing factors in 13 fatal cases of corona virus disease 2019 Medical Journal of Chinese People’s Liberation Army. 2020:1-9. http://kns.cnki.net/kcms/detail/11.1056.R.20200424.1518.004.html
- Teachey DT, Lacey SF, Shaw PA, et al. Identification of predictive biomarkers for cytokine release syndrome after chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Cancer Discov. 2016;6:664-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Fu B, Zheng X, et al. Aberrant pathogenic GM-CSF+T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. BioRxiv. 2020. 10.1101/2020.02.12.945576. bioRxiv 2020.02.12.945576. [DOI] [Google Scholar]
- Yang XF, Dai TX, Zhou XB, et al. Analysis of adaptive immune cell populations and phenotypes in the patients infected by SARS-CoV-2. medRxiv. 10.1101/2020.03.23.20040675. 2020.03.23.20040675. [DOI] [Google Scholar]
- Zhang D, Guo R, Lei L, et al. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. medRxiv. 10.1101/2020.03.24.20042655. 2020.03.24.20042655. [DOI] [Google Scholar]
- Chen YW, Feng ZQ, Diao B, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv. 10.1101/2020.03.27.20045427. 2020.03.27.20045427. [DOI] [Google Scholar]
- Honke N, Shaabani N, Cadeddu G, et al. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat Immunol. 2011;13:51-57. [DOI] [PubMed] [Google Scholar]
- Bao L, Deng W, Huang B, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020. [Epub ahead of print, PMID: 32380511]. [DOI] [PubMed] [Google Scholar]
- Fisman DN. Hemophagocytic syndromes and infection. Emerg Infect Dis. 2000;6:601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. Hlh Across speciality collaboration U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Su WR, Tang H, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. [Epub ahead of print, PMID: 32377375]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CF, Xie J, Zhao L, et al. Aveolar macrophage activation and cytokine storm in the pathogenesis of severe COVID-19. Research Square. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao XH, Li TY, He ZC, et al. A pathological report of three COVID-19 cases by minimally invasive autopsies. Zhonghua Bing Li Xue Za Zhi. 2020;49:E009. [DOI] [PubMed] [Google Scholar]
- Liao M, Liu Y, Yuan J, et al. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. 2020. 10.1101/2020.02.23.20026690. 2020.02.23.20026690. [DOI] [Google Scholar]
- Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Chan JF, Wang Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. 2020. [Epub ahead of print, PMID:32270184]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wei XS, Xiang X, et al. Interferon-a2b treatment for COVID-19. medRxiv. 2020. 10.1101/2020.04.06.20042580. 2020.04.06.20042580. [DOI] [Google Scholar]
- Chen J, Fan H, Zhang L, et al. Retrospective analysis of clinical features in 101 death cases with COVID-19. medRxiv. 2020. 10.1101/2020.03.09.20033068. 2020.03.09.20033068. [DOI] [Google Scholar]
- Zhang BC, Zhou XY, Zhu CL, et al. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. medRxiv. 2020. 10.1101/2020.03.12.20035048. 2020.03.12.20035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasaraju T, Yang E, Samy RP, et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am J Pathol. 2011;179:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532-1535. [DOI] [PubMed] [Google Scholar]
- Lefrancais E, Looney MR. Neutralizing extracellular histones in acute respiratory distress syndrome. a new role for an endogenous pathway. Am J Respir Crit Care Med. 2017;196:122-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzenreiter R, Kienberger F, Marcos V, et al. Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J Cyst Fibros. 2012;11:84-92. [DOI] [PubMed] [Google Scholar]
- Fuchs TA, Brill A, Wagner DD. Neutrophil extracellular trap (NET) impact on deep vein thrombosis. Arterioscler Thromb Vasc Biol. 2012;32:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowicz-Scroggins ME, Dunican EM, Charbit AR, et al. Extracellular DNA, neutrophil extracellular traps, and inflammasome activation in severe asthma. Am J Respir Crit Care Med. 2019;199:1076-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Yalavarthi S, Shi H, et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5:138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med. 2020;217:e20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzini C, Girelli D. The role of Neutrophil Extracellular Traps in Covid-19: only an hypothesis or a potential new field of research?. Thromb Res. 2020;191:26-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong QM, He ZP, Zhuang H, et al. Dynamics of peripheral blood B lymphocytes and natural killer cells in patients with severe acute respiratory syndrome. Zhonghua liu xing bing xue za zhi. 2004;25:695-697. [PubMed] [Google Scholar]
- Xia CQ, Xu LL, Wang Z, et al. The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am J Clin Pathol. 2004;121:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Zhou LQ, Hu ZW, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan. China Clin Infect Dis. 2020. [Epub ahead of print, PMID:32161940]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YS, Huang Z, Ying GP, et al. Study of the lymphocyte change between COVID-19 and non-COVID-19 pneumonia cases suggesting other factors besides uncontrolled inflammation contributed to multi-organ injury. medRxiv. 2020. 10.1101/2020.02.19.20024885. 2020.02.19.20024885. [DOI] [Google Scholar]
- Dong P, Jun XW, Yan YW, et al. Gammadelta T cells provide protective function in highly pathogenic avian H5N1 influenza A virus infection. Front Immunol. 2018;9:2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russier M, Yang G, Briard B, et al. Hemagglutinin stability regulates H1N1 influenza virus replication and pathogenicity in mice by modulating type I interferon responses in dendritic cells. J Virol. 2020;94:e01423-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brufsky A, Lotze MT. DC/L-SIGNs of hope in the COVID-19 pandemic. J Med Virol. 2020. [Epub ahead of print, PMID:32374430]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Liu BD, Li CS, et al. The feasibility of convalescent plasma therapy in severe COVID-19 patients: a pilot study. medRxiv. 2020. 10.1101/2020.03.16.20036145. 2020.03.16.20036145. [DOI] [Google Scholar]
- Chen G, Wu D, Guo W, et al. Clinical and immunologic features in severe and moderate forms of coronavirus disease 2019. MedRxiv. 2020. 10.1101/2020.02.16.20023903. [DOI] [Google Scholar]
- Tan L, Wang Q, Zhang D, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Tranduct Target Ther. 2020;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan SX, Yi QJ, Fan SB, et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv. 10.1101/2020.02.10.20021832. 2020.02.10.20021832. [DOI] [Google Scholar]
- Wang WJ, He JX, Lie PY, et al. The definition and risks of cytokine release syndrome-like in 11 COVID-19-infected pneumonia critically ill patients: disease characteristics and retrospective analysis. medRxiv. 10.1101/2020.02.26.20026989. 2020.02.26.20026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li S, Liu J, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Bi R, Kim C, Chiplunkar S, Yel L, Gollapudi S. Role of NF-kappaB signaling pathway in increased tumor necrosis factor-alpha-induced apoptosis of lymphocytes in aged humans. Cell Death Differ. 2005;12:177-183. [DOI] [PubMed] [Google Scholar]
- Weiskopf D, Schmitz KS, Raadsen MP, et al. Phenotype of SARS-CoV-2-specific T-cells in COVID-19 patients with acute respiratory distress syndrome. medRxiv. 10.1101/2020.04.11.20062349. 2020.04.11.20062349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H-Y, Zhang M, Yang C-X, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17:541-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Braeckel E, Desombere I, Clement F, Vandekerckhove L, Verhofstede C, Vogelaers D. Leroux-Roels G. Polyfunctional CD4(+) T cell responses in HIV-1-infected viral controllers compared with those in healthy recipients of an adjuvanted polyprotein HIV-1 vaccine. Vaccine. 2013;31:3739-3746. [DOI] [PubMed] [Google Scholar]
- Diao B, Wang CH, Tan YG, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol. 2020;11:827. 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Yang XO. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect. 2020;53:368-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorer M, Lambert K, Rakebrandt N, et al. Rapid expansion of Treg cells protects from collateral colitis following a viral trigger. Nat Commun. 2020;11:1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YL, Tan MK, Chen X, et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. medRxiv. 10.1101/2020.03.12.20034736. 2020.03.12.20034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Xia P, Zhou Y, et al. Potential effect of blood purification therapy in reducing cytokine storm as a late complication of critically ill COVID-19. Clin Immunol (Orlando, Fla). 2020;214:108408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Z, Zhu R, Hou W, et al. Transplantation of ACE2 mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging and Disease. 2020;11:216-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CG, Wang ZQ, Zhao F, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 10.1101/2020.03.30.20047365. 2020.03.30.20047365. [DOI] [Google Scholar]
- Wang XL, Guo XH, Xin QQ, et al. Neutralizing antibodies responses to SARS-CoV-2 in COVID-19 inpatients and convalescent patients. medRxiv. 10.1101/2020.04.15.20065623. 2020.04.15.20065623. [DOI] [Google Scholar]
- Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020. [Epub ahead of print, PMID:32221519]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shen C, Li J, et al. Exuberant elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 infection is associated with disease severity and fatal outcome. medRxiv. 10.1101/2020.03.02.20029975. 2020.03.02.20029975. [DOI] [Google Scholar]
- Jiang Y, Xu J, Zhou C, et al. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med. 2005;171:850-857. [DOI] [PubMed] [Google Scholar]
- Liu T, Zhang JY, Yang YH, et al. The potential role of IL-6 in monitoring severe case of coronavirus disease 2019. medRxiv. 10.1101/2020.03.01.20029769. 2020.03.01.20029769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XH, Zhao BH, Qu YM, et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin Infect Dis. 2020. Apr 17: ciaa449. [Epub ahead of print, PMID:32301997]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Xie J, Yin M, et al. Clinical and laboratory profiles of 75 hospitalized patients with novel coronavirus disease 2019 in Hefei, China. medRxiv. 10.1101/2020.03.01.20029785. 2020.03.01.20029785. [DOI] [Google Scholar]
- Fogarty H, Townsend L, Ni Cheallaigh C, et al. COVID-19 coagulopathy in Caucasian patients. Br J Haematol. 2020. [Epub ahead of print, PMID:32330308]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr R, Stirling D, Ludlam CA. Interleukin 6 and haemostasis. Br J Haematol. 2001;115:3-12. [DOI] [PubMed] [Google Scholar]
- Zou Y, Guo HY, Zhang YY, et al. Analysis of coagulation parameters in patients with COVID-19 in Shanghai, China. Biosci Trends. 2020. [Epub ahead of print, PMID:32350161]. [DOI] [PubMed] [Google Scholar]
- Panigada M, Bottino N, Tagliabue P, et al. Hypercoagulability of COVID-19 patients in intensive care unit. A report of thromboelastography findings and other parameters of hemostasis. J Thromb Haemost. 2020. [Epub ahead of print, PMID:32302438]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yang R, Xu Y, Gong P. Clinical characteristics of 36 non-survivors with COVID-19 in Wuhan, China. Lancet. 2020;28(10229):1054-1062. 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbel J, Narayanan N, Bhatt PJ. Use of tocilizumab for COVID-19-induced cytokine release syndrome: a cautionary case report. Chest. 2020;S0012-3692:30764-30769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zheng Y, Chen X. Drugs for autoimmune inflammatory diseases: from small molecule compounds to Anti-TNF biologics. Front Pharmacol. 2017;8:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie-Silva I, Klein A, Gomes JMM, et al. Acute-phase proteins during inflammatory reaction by bacterial infection: fish-model. Sci Rep. 2019;9:4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson BK, Seethamraju H, Dhody K, et al. Disruption of the CCL5/RANTES-CCR5 pathway restores immune homeostasis and reduces plasma viral load in critical COVID-19. medRxiv. 10.1101/2020.05.02.20084673. 2020.05.02.20084673. [DOI] [Google Scholar]
- Liu Y, Zhang C, Huang F, et al. Elevated plasma level of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl Sci Rev. 2020. 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Travanty EA, Oko L, et al. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am J Respir Cell Mol Biol. 2013;48:742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E, Poissy J, Goffard A, et al. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside?. PLoS One. 2014;9:e88716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L, Menachery VD, Gralinski LE, et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio. 2013;4:e00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li M, Zhou Z, Guan X, Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)?. J Autoimmun. 2020;111:102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Han Ly, Hu Bj. Advance in cognition of revised and updated ‘guidelines of diagnosis and treatment of COVID-19. Chin J Nosocomiol. 2020:1-7. [Google Scholar]
- ECNS. 2020 China optimizes treatment for COVID-19. http://www.ecns.cn/news/society/2020-03-07/detail-ifzuhesu4122216.shtml
- Guo C, Li B, Ma H, et al. Tocilizumab treatment in severe COVID-19 patients attenuates the inflammatory storm incited by monocyte centric immune interactions revealed by single-cell analysis. bioRxiv. 10.1101/2020.04.08.029769. 2020.04.08.029769. [DOI] [Google Scholar]
- Xu XL, Han MF, Li TT, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA. 2020;117:10970-10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P, Liu Y, Qiu L, Liu X, Liu D, Li J. Tocilizumab treatment in COVID-19: a single center experience. J Med Virol. 2020. [Epub ahead of print, PMID:32253759]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti P, Ronconi G, Caraffa A, et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J Biol Regul Homeost Agents. 2020;34:1. [DOI] [PubMed] [Google Scholar]
- Deng XB, Yu CY, Pei JF. Regulation of interferon production as a potential strategy for COVID-19 treatment. ArXiv. 2020. rXiv:2003.00751. [Google Scholar]
- Stebbing J, Phelan A, Griffin I, et al. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis. 2020;20:400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IF, Lung KC, Tso EY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;S0140-6736:31042-31044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galipeau J, Sensebe L. Mesenchymal stromal cells: clinical challenges and therapeutic opportunities. Cell Stem Cell. 2018;22:824-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZH, Zhou JM, Huang YB, et al. The efficacy of convalescent plasma for the treatment of severe influenza. MedRxiv. 2020. 10.1101/2020.02.20.20025593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Yang L, Wang C, Liu C, Dianming L. Clinical observation of 6 severe COVID-19 patients treated with plasma exchange or tocilizumab. J Zhejiang Univ. 2020;49:227-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gbinigie K, Frie K. Should chloroquine and hydroxychloroquine be used to treat COVID-19? A rapid review. BJGP Open. 2020. [Epub ahead of print, PMID:32265182]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19?. Int J Antimicrob Agents. 2020;105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz KR, Gilman AL. The lysosomotropic amines, chloroquine and hydroxychloroquine: a potentially novel therapy for graft-versus-host disease. Leuk Lymphoma. 1997;24:201-210. [DOI] [PubMed] [Google Scholar]
- Kyburz D, Brentano F, Gay S. Mode of action of hydroxychloroquine in RA-evidence of an inhibitory effect on toll-like receptor signaling. Nat Clin Pract Rheumatol. 2006;2:458-459. [DOI] [PubMed] [Google Scholar]
- Vermeersch K, Gabrovska M, Aumann J, et al. Azithromycin during acute chronic obstructive pulmonary disease exacerbations requiring hospitalization (BACE). A multicenter, randomized, double-blind, placebo-controlled trial. Am J Respir Crit Care Med. 2019;200:857-868. [DOI] [PubMed] [Google Scholar]
- Gautret P, Lagier JC, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020. Mar 20: 105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen J, Liu Dp, Liu L, et al. A pilot study of hydroxychloroquine in treatment of patients with common coronavirus disease-19 (COVID-19). J Zhejiang Univ (Med Sci). 2020;49:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hu J, Zhang Z, et al. Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial. medRxiv. 10.1101/2020.03.22.20040758. 2020.03.22.20040758. [DOI] [Google Scholar]
- Yu B, Wang DW, Li C. Hydroxychloroquine application is associated with a decreased mortality in critically ill patients with COVID-19. medRxiv. 10.1101/2020.04.27.20073379. 2020.04.27.20073379. [DOI] [Google Scholar]
- Mathian A, Mahevas M, Rohmer J, et al. Clinical course of coronavirus disease 2019 (COVID-19) in a series of 17 patients with systemic lupus erythematosus under long-term treatment with hydroxychloroquine. Ann Rheum Dis. 2020. [Epub ahead of print, PMID:32332072]. [DOI] [PubMed] [Google Scholar]
- Chorin E, Dai M, Shulman E, et al. The QT interval in patients with SARS-CoV-2 infection treated with hydroxychloroquine/azithromycin. medRxiv. 10.1101/2020.04.02.20047050. 2020.04.02.20047050. [DOI] [Google Scholar]
- Yang Y, Islam MS, Wang J, Li Y, Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci. 2020;16:1708-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Howard OM, Yang X, Wang L, Oppenheim JJ, Krakauer T. Effects of shuanghuanglian and qingkailing, two multi-components of traditional Chinese medicinal preparations, on human leukocyte function. Life Sci. 2002;70:2897-2913. [DOI] [PubMed] [Google Scholar]
- Zou H, He T, Chen X. Tetrandrine inhibits differentiation of proinflammatory subsets of T helper cells but spares de novo differentiation of iTreg cells. Int Immunopharmacol. 2019;69:307-312. [DOI] [PubMed] [Google Scholar]
- Chen X, Murakami T, Oppenheim JJ, Howard OM. Triptolide, a constituent of immunosuppressive Chinese herbal medicine, is a potent suppressor of dendritic-cell maturation and trafficking. Blood. 2005;106:2409-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Oppenheim JJ, Howard OM. Chemokines and chemokine receptors as novel therapeutic targets in rheumatoid arthritis (RA): inhibitory effects of traditional Chinese medicinal components. Cell Mol Immunol. 2004;1:336-342. [PubMed] [Google Scholar]
- Chen X, Yang L, Zhang N, et al. Shikonin, a component of chinese herbal medicine, inhibits chemokine receptor function and suppresses human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2003;47:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Beutler JA, McCloud TG, et al. Tannic acid is an inhibitor of CXCL12 (SDF-1alpha)/CXCR4 with antiangiogenic activity. Clin Cancer Res. 2003;9:3115-3123. [PubMed] [Google Scholar]
- Chen X, Mellon RD, Yang L, Dong H, Oppenheim JJ, Howard OM. Regulatory effects of deoxycholic acid, a component of the anti-inflammatory traditional Chinese medicine Niuhuang, on human leukocyte response to chemoattractants. Biochem Pharmacol. 2002;63:533-541. [DOI] [PubMed] [Google Scholar]
- Yang MW, Chen F, Zhu DJ, et al. Clinical efficacy of matrine and sodium chloride injection in treatment of 40 cases of COVID-19. Chin J Chin Mater Med. 2020;1-12. http://new.big5.oversea.cnki.net/KCMS/detail/11.2272.R.20200323.1514.001.html?uid=WEEvREcwSlJHSldRa1FhdkJtNEYwbVRPQlNCZlo3UHRsSkxUbUR3THdUMD0=$9A4hF_YAuvQ5obgVAqNKPCYcEjKensW4IQMovwHtwkF4VYPoHbKxJw!!&v=MDY3NzdydkpJMTA9UHlyUmQ3RzRITkhNckkxR1pPc09ZdzlNem1SbjZqNTdUM2ZscVdNMENMTDdSN3FkWitacUZ5N2xW. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhao Rh, Guo Ss, et al. Effect of matrine sodium chloride injection on mouse model of combination of disease and syndrome of pulmonary syndrome of human coronavirus pneumonia virus virus infection. Acta Pharmaceutica Sinica. 2020;55:366-373. [Google Scholar]
- Zhu J, Deng Y-Q, Wang X, et al. An artificial intelligence system reveals liquiritin inhibits SARS-CoV-2 by mimicking type I interferon. bioRxiv. 10.1101/2020.05.02.074021. 2020.05.02.074021. [DOI] [Google Scholar]
- Lu X, Chen T, Wang Y, et al. Adjuvant corticosteroid therapy for critically ill patients with COVID-19. medRxiv. 10.1101/2020.04.07.20056390. 2020.04.07.20056390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Qiu X, Jiang H, Zhi Y, Fu J, Liu J. Ulinastatin inhibits the inflammation of LPS-induced acute lung injury in mice via regulation of AMPK/NF-kappaB pathway. Int Immunopharmacol. 2015;29:560-567. [DOI] [PubMed] [Google Scholar]
- Chinese Clinical Trial Register, 2020 A clinical trial for Ulinastatin Injection in the treatment of patients with severe novel coronavirus pneumonia (COVID-19). 2020. ChiCTR2000030779.
- Liu Q, Zhou YH, Yang ZQ. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol. 2016;13:3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel R, Morrison A, Vahia A, et al. Early short course corticosteroids in hospitalized patients with COVID-19. medRxiv. 10.1101/2020.05.04.20074609. 2020.05.04.20074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet North Am Ed. 2020;395:473-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 Pneumonia in Wuhan, China. JAMA Intern Med. 2020. Mar 13;e200994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar J, Chakrabarti P. A machine learning model reveals older age and delayed hospitalization as predictors of mortality in patients with COVID-19. medRxiv. 10.1101/2020.03.25.20043331. 2020.03.25.20043331. [DOI] [Google Scholar]
- Zeng Z, Sha T, Zhang Y, et al. Hypertension in patients hospitalized with COVID-19 in Wuhan, China: a single-center retrospective observational study. medRxiv. 10.1101/2020.04.06.20054825. 2020.04.06.20054825. [DOI] [Google Scholar]
- Shen Z, Xiao Y, Kang L, et al. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin Infect Dis. 2020;ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley TJ, Mocco J, Majidi S, et al. Large-vessel stroke as a presenting feature of Covid-19 in the young. N Engl J Med. 2020;382:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciceri F, Beretta L, Scandroglio AM, et al. Microvascular COVID-19 lung vessels obstructive thromboinflammatory syndrome (MicroCLOTS): an atypical acute respiratory distress syndrome working hypothesis. Crit Care Resusc. 2020. [Epub ahead of print: PMID: 32294809]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cron RQ, Chatham WW. Don’t forget the host: cOVID-19 cytokine storm. Rheumatologist. 2020. https://www.the-rheumatologist.org/article/dont-forget-the-host-covid-19-cytokine-storm/. [Google Scholar]
- Xia W, Shao J, Guo Y, Peng X, Li Z, Hu D. Clinical and CT features in pediatric patients with COVID-19 infection: different points from adults. Pediatr Pulmonol. 2020;55:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Curtis N. COVID-19 in children, pregnancy and neonates: a review of epidemiologic and clinical features. Pediatr Infect Dis J. 2020;39:469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JI, Ocwieja KE, Nakamura MM. A call for pediatric COVID-19 clinical trials. Pediatrics. 2020. [Epub ahead of print, PMID:32398330]. [DOI] [PubMed] [Google Scholar]
- Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020;382:1708-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu HG, Liu W, et al. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:E005. [DOI] [PubMed] [Google Scholar]
- Gong J, Dong H, Xia SQ, et al. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19 pneumonia. medRxiv. 10.1101/2020.02.25.20025643. 2020.02.25.20025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Yan X, Tang X, et al. Epidemiological and clinical features of 2019-nCoV acute respiratory disease cases in Chongqing municipality, China: a retrospective, descriptive, multiple-center study. medRxiv. 10.1101/2020.03.01.20029397. 2020.03.01.20029397. [DOI] [Google Scholar]
- Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan. China JAMA. 2020;323:1061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Li H, Lu XX, et al. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center’s observational study. World J Pediatr. 2020;1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Yin J, Wang W, et al. Down-regulated gene expression spectrum and immune responses changed during the disease progression in COVID-19 patients. Clin Infect Dis. 2020:1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Han MF, Zhao FD, Zhang T, Ma L. Clinical manifestations and sero-immunological characteristics of 155 patients with COVID-19. Chin J Nosocomio. 2020;30:961-965. [Google Scholar]
- Li GH, Li L, He M, et al. Value of various inflammatory markers combined with lymphocyte subsets on clinical diagnosis of different clinical types of COVID-19. J Chongqing Med Univ. 2020:1-5. http://new.big5.oversea.cnki.net/KCMS/detail/50.1046.r.20200428.0930.006.html?uid=WEEvREcwSlJHSldRa1FhdkJtNEYwbVRPQlNCZlo3UHRsSkxUbUR3THdUMD0=$9A4hF_YAuvQ5obgVAqNKPCYcEjKensW4IQMovwHtwkF4VYPoHbKxJw!!&v=MDcyMTJSN3FkWitacUZ5N2xWcnJMSVZrPVB6elNaYkc0SE5ITXE0MUNaT3NJWXc5TXptUm42ajU3VDNmbHFXTTBDTEw3. [Google Scholar]
- Li X, Xu S, Yu M, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. The J Allergy Clin Immunol. 2020. [Epub ahead of print, PMID:32294485]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.


