Abstract
The ongoing outbreak of the recently emerged 2019 novel coronavirus (nCoV), which has seriously threatened global health security, is caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) with high morbidity and mortality. Despite the burden of the disease worldwide, still, no licensed vaccine or any specific drug against 2019‐nCoV is available. Data from several countries show that few repurposed drugs using existing antiviral drugs have not (so far) been satisfactory and more recently were proven to be even highly toxic. These findings underline an urgent need for preventative and therapeutic interventions designed to target specific aspects of 2019‐nCoV. Again the major factor in this urgency is that the process of data acquisition by physical experiment is time‐consuming and expensive to obtain. Scientific simulations and more in‐depth data analysis permit to validate or refute drug repurposing opportunities predicted via target similarity profiling to speed up the development of a new more effective anti‐2019‐nCoV therapy especially where in vitro and/or in vivo data are not yet available. In addition, several research programs are being developed, aiming at the exploration of vaccines to prevent and treat the 2019‐nCoV. Computational‐based technology has given us the tools to explore and identify potentially effective drug and/or vaccine candidates which can effectively shorten the time and reduce the operating cost. The aim of the present review is to address the available information on molecular determinants in disease pathobiology modules and define the computational approaches employed in systematic drug repositioning and vaccine development settings for SARS‐CoV‐2.
Keywords: bioinformatics, COVID‐19, drug target, SARS‐CoV‐2, treatment, vaccine
Multiepitope vaccines that are designed by immunoinformatics methods are favorable for outbreaks of great proportions.
The enhancing antibody problem appears to be a contributing factor in coronavirus disease‐2019 (COVID‐19) vaccine design.
The broad global access for the pandemic COVID‐19 vaccine, if available, should be ensured to avoid the potential of disease outbreak in the future.
1. INTRODUCTION
On December 31, 2019, 27 cases of pneumonia of unknown etiology/cause in Wuhan, Hubei Province of China was reported to the World Health Organization (WHO) China Country Office (Sohrabi et al., 2020). On 12 January 2020, the World Health Organization temporarily named the causative virus as 2019 novel coronavirus (2019‐nCoV). The recent outbreak of 2019‐nCoV pandemic has so far caused more than 2,000,000 reported human infections and nearly 160,000 deaths worldwide, surpassing even the number of cases caused by severe acute respiratory syndrome coronavirus (SARS‐CoV) and Middle East respiratory syndrome coronavirus (MERS‐CoV) combined (Huang et al., 2020; Hui et al., 2020). Coronaviruses (CoVs) are a family of RNA viruses important in humans and numerous animal hosts of which six are known to infect humans: 229E, OC43, NL63, HKU1, SARS‐CoV, and MERS‐CoV. They constitute important infectious pathogens that cause severe respiratory and enteric diseases in humans and animals (Fehr & Perlman, 2015). The seventh human coronavirus also known as the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is identified to be a novel deadly and highly contagious virus that quickly spread to over 150 countries. On March 11, the WHO declared the outbreak of a pandemic (Djalante, Shaw, & DeWit, 2020). It is well recognized that the virus has an unusually high speed of transmission and can survive in the environment, on different surfaces for long hours, which poses a serious risk (Negahdaripour, 2020). Moreover, the illness causes flu‐like symptoms, and the reports show that one person infected can easily infect others (J. Xu et al., 2020).
Even though coronavirus infection of humans was identified for the first time in 1960; however, scientific research in this area grew only following the outbreak of SARS coronavirus in the year 2003 (Drosten et al., 2003; Ksiazek et al., 2003). In spite of the reaction from the scientific community that was instantaneous and nearly uniform, at the current state of our knowledge, vaccines for COVID‐19 seem months or even years away (Nabel, 2013). While there is still much to be understood about the SARS‐CoV‐2, considerable research efforts have been made to yield answers to the basic questions of the SARS‐CoV‐2 behavior. There is currently off‐label use of antiviral drugs for the treatment of this novel virus of coronaviruses that are not totally effective but only partially satisfactory treatment available. There is an urgent need for effective and specific interventions to help tackle the growing global health burden of SARS‐CoV‐2. To develop an effective care plan for SARS‐CoV‐2 outbreak, it is vital to find new targets for novel therapeutic anti‐SARS‐CoV‐2 approach. In de novo drug discovery, it is fundamental to understand the molecular entities and specific pathways driving the virus replication during infection. In this paper, we review the available information on molecular determinants in disease pathobiology modules. In addition, the computational approaches employed in systematic drug repositioning and vaccine development settings for SARS‐CoV‐2 are described. In this procedure, for example, instead of traditional epitope identification upon experimental techniques, being costly and time‐consuming (Sanchez‐Trincado, Gomez‐Perosanz, & Reche, 2017), predictive computational methods can be the key way for screening the large‐scale peptide candidates. It is hoped that the proposed methodology will open up new avenues for the rapid identification of candidate drugs for the potential prevention and treatment of SARS‐CoV‐2.
2. PATHOGENIC HUMAN CORONAVIRUSES
Coronaviruses are enveloped single‐stranded RNA viruses with one of the largest viral genomes found in the RNA viruses that the majority are disease‐causing agents (Pyrc, Berkhout, & Van Der Hoek, 2007). Several phylogenetic studies subdivided the coronaviruses into four genera known as alpha‐CoVs, beta‐CoVs, gamma‐CoVs, and delta‐CoVs, of which alpha‐ and beta‐CoVs are pathogenic viruses for mammals and gamma‐ and delta‐CoVs are known to infect mainly birds (Paules, Marston, & Fauci, 2020). Most coronaviruses are addressed as common cold viruses in humans that are of zoonotic origin and cause a more serious disease in animals (Lake, 2020). In general, the gene pools of Groups 1 and 2 coronaviruses are thought to have originated in bats which are the reservoir of diverse CoVs and further spread to humans through an intermediate host or sometimes directly (Cui, Li, & Shi, 2019). Human coronaviruses are typically associated with respiratory tract diseases such as those observed in the common cold including HCoV‐229E, HCoV‐NL63, HCoV‐OC43, and HCoV‐HKU1 and sometimes enteric illnesses. However, the emergence of highly pathogenic human coronavirus infections in the last two decades has greatly illuminated their potential to cause high morbidity and mortality in humans (Cockrell, Leist, Douglas, & Baric, 2018). The recent coronaviruses SARS‐CoV and MERS‐CoV that are two representative members of the genus Betacoronavirus emerged into the human population as zoonotic infections and were the most serious endemic illnesses associated with high mortality in the years 2003 and 2012, respectively (Peck, Burch, Heise, & Baric, 2015). While no case of SARS‐CoV infection has been around since 2004, multiple sporadic cases of MERS‐CoV have been reported in different countries. Despite the low interhuman transmissibility, MERS‐CoV has been identified as an infectious disease associated with high mortality rates which is about ~35% and much higher than that of SARS (~10%; Donnelly, Malik, Elkholy, Cauchemez, & Van Kerkhove, 2019; Fong, 2017). It is likely that discrepancy in the outcome of MERS‐CoV‐ and SARS‐CoV‐infections be explained by differential virus pathogenicity and virulence. The recently identified SARS‐CoV‐2 also belongs to the same Betacoronavirus genus. The International Committee on Taxonomy of Viruses has named the novel strain coronavirus SARS‐CoV‐2, as it is very similar to the known SARS‐CoV in terms of the whole genome sequence (R. Lu et al., 2020). SARS‐CoV‐2 has a genomic size of ~30 kilobases encoding several structural and nonstructural proteins. The major structural proteins, which form the viral particle, include spike (S), envelope (E), membrane (M), and the nucleocapsid (N) protein. Preliminary studies show that the SARS‐CoV‐2 full‐length genome is quite similar to SARS‐CoV (P. Zhou et al., 2020). However, as more data become available, the SARS‐CoV‐2 is undoubtedly distinct from SARS‐CoV and from other bat origin coronaviruses and thus it is imperative to understand the novel coronavirus demanding immediate treatment to develop effective interventions targeting SARS‐CoV‐2. In the next phase, researchers and vaccine companies will further develop new vaccines for controlling and preventing this highly pathogenic virus.
3. THERAPEUTIC OPTIONS FOR 2019‐nCoV
3.1. Perspectives on targeting “structural” proteins of COVID‐19
There is currently no approved treatment for COVID‐19 in humans, and up to now, a number of antiviral treatments such as ribavirin, primarily have been used based on the experiences from SARS and MERS medication (Singhal, 2020). In addition, using chloroquine phosphate, an old antimalarial drug, is found with limited success in patients with COVID‐19 (Gao, Tian, & Yang, 2020). 2019‐nCoV is genetically closely related to SARS coronavirus (about 79% identity) and MERS‐CoV (about 50%) but more similar (with 88%) to two bat‐derived SARS‐like coronaviruses (R. Lu et al., 2020). This virus is found to be a bat CoVs of the genus Betacoronavirus, however, has displayed large genetic distances to its close relatives to be considered a new human coronavirus. However, the modeling study did indicate a similar structure in receptor‐binding domain (RBD) of 2019‐nCoV to that of SARS‐CoV with variations at some key amino acid residues. Emerging evidence suggest that 2019‐nCoV might be bound to the SARS‐CoV receptor angiotensin‐converting enzyme 2 (ACE2) as a functional cell receptor in human (R. Lu et al., 2020). Clinical features of 2019‐nCoV‐infected patients indicate multiple organ failure in addition to pneumonia. ACE2 acting in a first key step of 2019‐nCoV infection may help explain the clinical features of other systems observed in these patients. ACE2 is expressed extensively in the endothelium of the heart, liver, kidney, testis, and intestine (M. Yang, Zhao, & Zhang, 2020) and 2019‐nCoV may probably infect any tissues where ACE2 is available. The ongoing works dissecting genetic molecular interactions highlight that SARS‐CoV‐2 differ in several critical residues at RBD (particularly Gln493) from other SARS strains leading to high‐affinity interactions with ACE2 receptors, being significantly more contagious than those other viruses as a result (Wan, Shang, Graham, Baric, & Li, 2020). One prophylactic and therapeutic strategy can be through the competitive inhibition of the ACE2 receptor binding of 2019‐nCoV by using an agent that can target the receptor (M. Yang et al., 2020). The molecular docking results revealed the binding of the five natural compounds including Scutellarin, Nicotianamine, Hesperetin, Baicalin, and Glycyrrhizin to the ACE2 receptor confirming antivirus effects of these agents for preventing 2019‐nCoV disease (H. Chen & Du, 2020). Alternatively, the high identity between 2019‐nCoV and SARS‐CoV raised the possibility of cross‐reactivity of SARS‐CoV‐specific antibodies with 2019‐nCoV spike protein (S‐protein). The modeling results validate the potential interaction between the RBD in S‐protein of 2019‐nCoV and certain anti‐SARS‐CoV antibodies and suggest that SARS‐CoV‐specific human monoclonal antibody, CR3022, but not the other SARS‐CoV antibodies (e.g., m396, CR3014), may be effective in neutralizing 2019‐nCoV and blocking its cell infection (Tian et al., 2020). At present, numerous attempts are being made to develop monoclonal antibodies that are specific and effective against 2019‐nCoV. There may be more similarities in the mechanisms of cell entry for 2019‐nCoV and SARS‐CoV which employ ACE2 as entry receptor and the cellular serine protease called transmembrane protease, serine 2 for activating S‐protein for host cell entry (Hoffmann et al., 2020) which might hopefully work as candidate therapeutic target for anti‐2019‐nCoV intervention (Hoffmann et al., 2020). Thus, in this context, serine protease inhibitor camostat mesylate that is approved for human use in a different indication may provide an effective therapeutic option for the treatment of 2019‐nCoV‐infected patients (Kawase, Shirato, van der Hoek, Taguchi, & Matsuyama, 2012; Y. Zhou et al., 2015). In another effort to limit 2019‐nCoV infection, furin inhibitors have been reported to develop as a specific antiviral treatment (Coutard et al., 2020). As the priming of viral envelope glycoproteins (such spike [S] protein) by the proteases of the target cells is essential for cell tropism and pathogenicity of virus, the efficacy and extent of this cleavage event may have implication in the development of antiviral agents. Human furin is a protease highly expressed in lungs, which cleaves the 2019‐nCoV S‐protein at a specific conserved site absent in SARS‐CoV sequences (Bassi, Zhang, Renner, & Klein‐Szanto, 2017) and thereby generates mature infectious virus particles. Overall, because of the fact that S‐protein relates to degrees of pathogenicity, transmissibility, and pandemic potential, S‐protein‐based antiviral therapies offer an attractive and seemingly efficient modality into the area of anti‐nCoV treatment (Jiang, He, & Liu, 2005; X. Li, Geng, Peng, Meng, & Lu, 2020). Treatment regimens based on the use of S‐protein‐based therapeutics include antagonists of RBD–ACE2 interaction, inhibitors of S‐protein cleavage proteases, neutralizing antibodies, S‐protein inhibitors, and small interfering RNAs (Malik et al., 2020). Nevertheless, a potential concern for the treatment of COVID‐19 infection is the emergence of drug resistance by the rapid genomic evolution of virus during therapy. The higher risk of drug resistance has most often been noted to occur when targeting single virus proteins (Zumla, Chan, Azhar, Hui, & Yuen, 2016). Even though these therapeutic strategies have demonstrated in vitro efficacy, they have not undergone animal or clinical testing so far and may, therefore, be of limited use in our present COVID‐19 problem.
3.2. Perspectives on targeting “nonstructural” proteins of COVID‐19
It is indicated that the key to the development of new drugs able to control the cause of the recent outbreak of the deadly viral pneumonia in China requires the identification of a conserved target region within the whole Coronavirus genus (H. Yang et al., 2005). Despite all structural proteins including S, E, M, and N proteins, nonstructural proteins (NSPs) withstand considerable sequence variation among different CoVs (Woo et al., 2005). NSPs responsible for viral replication and transcription may be adopted to target CoV. The SARS‐CoV encodes 16 NSPs as potential virulence factors. Among these NSPs, an RNA‐dependent RNA polymerase (RdRp), the NSP12, which is the central subunit of the CoV replicative machinery and its essential cofactors NSP7 and NSP8 play a crucial role in the replication of the SARS‐CoV genome (Kirchdoerfer & Ward, 2019; Subissi et al., 2014). Accordingly, there should be possibly interference with the binding of NSP7 or NSP8 to NSP12 to inhibit the RdRp activity of NSP12 for the development of novel antiviral agents. As the comparative analyses of their amino acid sequences show high similarity with NSP7 (98.8%) and NSP8 (97.5%) of SARS‐CoV (Ruan et al., 2020), screening for compounds disrupting NSP12‐NSP7‐NSP8 complex may evolve as novel therapies of SARS‐CoV‐2. Based on virtual screening and docking methods, seven compounds (Saquinavir, Tipranavir, Lonafarnib, Tegobuvir, Olysio, Filibuvir, and Cepharanthine) have been suggested to have potential as efficacious new inhibitors of virus replication process (Ruan et al., 2020) offering more opportunities for clinical assessment of anti‐SARS‐CoV‐2 drugs.
Usually, beta‐coronaviruses produce a ~800 kDa polypeptide that is posttranscriptionally cleaved at certain processing sites into separate subunits. The papain‐like protease and 3‐chymotrypsin‐like protease (3CLpro) are main proteases usually responsible for this proteolytic processing to generate various NSPs and are thus important for viral replication (Anand, Ziebuhr, Wadhwani, Mesters, & Hilgenfeld, 2003). Recent studies revealed that 3CLpro is conserved in SARS‐CoV‐2 having 12‐point mutations identified in the receptor‐binding site that affects its binding ability to some medicinal plant compounds already used to treat viral diseases like inhibitor of SARS‐CoV 3CLpro named ML188. Therefore, further optimization and drug development efforts were made to develop new compounds to inhibit SARS‐CoV‐2 3CLpro and the results identified nine novel natural compounds predicted to bind with the receptor‐binding site of SARS‐CoV‐2 3CLpro which possess therapeutic activities (ul Qamar, Alqahtani, Alamri, & Chen, 2020). These investigations lack experimental validation, and to further address this anti‐COVID‐19 drug discovery with putative clinical application, more in vitro and in vivo studies are required. The enzyme Nsp13 helicase another potential target in the viral replication apparatus has been investigated in SARS‐CoV‐2 and could be structurally characterized by computational methods. Its nucleotide sequence predicted a protein of 596 amino acids. Similar to previous reports, molecular dynamics simulations together with virtual screening efforts were applied to identify the interaction profile of important binding site residues of SARS‐CoV‐2 Nsp13 helicase and potential therapeutic small molecules to find potent and selective inhibitors against SARS‐CoV‐2 (Mirza & Froeyen, 2020). In silico predictions were used for determining ADME parameters of the identified potential hits against SARS‐CoV‐2 Nsp13 target, such as absorption, distribution, metabolism, elimination, toxicity (ADME‐Tox) and resulting top hits were selected following removal of hits that exhibited poor ADMET‐Tox found to contain high‐risk chemical groups (Mirza & Froeyen, 2020).
To improve control strategies, the research studies have attempted to develop effective treatments and prophylactic strategies including vaccination. Considering the cost per cured subject, vaccination appears more favorable. Alternatively, with rising morbidity and death rates, the benefit of the vaccine in reducing the disease burden will be higher. For this, the rest of the review will focus on what is known about the development of vaccine against COVID‐19.
4. VACCINE DEVELOPMENT AGAINST COVID‐19 CORONAVIRUS
4.1. Multiple vaccine platforms against COVID‐19 coronavirus challenge
Vaccine efficacy depends largely upon the vaccine target and platform. Among all platform technologies, whole‐virus such as live‐attenuated viral vaccines, and killed whole virus vaccines, subunit vaccines, plasmid‐based DNA vaccines, RNA replicons, and virus‐like particle have been developed to induce protective responses to viral infections (Anthony & Fauci, 2018). Choosing a suitable vaccine platform is especially challenging and important to ensure both efficacy and safety. Vaccine strategy that uses inactive viruses is a classic formula for viral vaccination (W.‐H. Chen, Strych, Hotez, & Bottazzi, 2020). In some cases, vaccination with live virus cause complications including lung destruction and pulmonary immunopathology in mice such as infiltration with eosinophils (Tseng et al., 2012). Multiple platforms are under development and/or investigation. DNA‐ and RNA‐based platforms have great potential for speeding up the research related to this field. The genetic vaccines can be made quickly requiring no culture and facilitate rapid testing allowing for more rapid development of vaccine (Lurie, Saville, Hatchett, & Halton, 2020). This is particularly important because a major problem is the time it takes to produce a vaccine. Since the genetic sequence of the 2019‐nCoV became available in early January, some companies have made significant steps forward in the early development of a vaccine for COVID‐19. Inovio Pharmaceuticals in San Diego funded by the Coalition for Epidemic Preparedness Innovations (CEPI) began animal testing of a COVID‐19 plasmid DNA vaccine called “INO‐4800” in February 2020 and this vaccine candidate is currently under early‐stage phase I clinical evaluation (Challener, 2020). Already, a messenger RNA vaccine expressing the stabilized S‐protein of COVID‐19 has been made and is being tested in a Phase I dose escalation study by Kaiser Permanente Washington Health Research Institute (KPWHRI; Cohen, 2020). Within a short period of time, at least 62 projects (list of April 4, 2020) have begun vaccine research against COVID‐19. Although each vaccine product must pass through several stages before application in the clinical setting, new technology and previous experience with vaccine against related viruses make an enormous acceleration of the process possible. This is apparently shown by the results reported by some leading companies and research groups that have made the most progress in their projects. These include the projects that have already entered the clinical testing phase for vaccine against COVID‐19 infection (see Table 1) or plan to do so within a few months (Table 2).
Table 1.
Ongoing clinical trials of vaccines against SARS‐CoV‐2
| Co. or Inst. | Vaccine platform | Technology | Study | Target antigen | No. of participants | Study start date | Clinical trial No. |
|---|---|---|---|---|---|---|---|
| VRC and Moderna | mRNA‐1273 | A lipid nanoparticle‐encapsulated mRNA‐based vaccine | Phase I | A portion of spike protein | 45 healthy participants | March 3, 2020 | NCT04283461 |
| Inovio Pharmaceuticals | INO‐4800 | A DNA‐based vaccine | Phase I | 40 healthy participants | April 6, 2020 | NCT04336410 | |
| University of Oxford | ChAdOx1 nCoV‐19 | Attenuated adenovirus capable of producing the spike protein of SARS‐CoV‐2 | Phase I/II single‐blinded | Spike protein | 510 healthy participants | March 27, 2020 | NCT04324606 |
| CanSino | Ad5‐nCoV | Replication‐defective adenovirus Type 5 as the vector to express SARS‐CoV‐2 spike protein | Phase I | Spike protein | 108 healthy participants | March 17, 2020 | NCT04313127 |
| Shenzhen Geno‐Immune Medical Institute | LV‐SMENP‐DC vaccine and antigen‐specific cytotoxic T cell vaccine | Lentiviral SMENP minigenes to expressing COVID‐19 antigens | Phase I | 100 healthy and Covid‐19‐positive volunteers | February 19, 2020 | NCT04276896 | |
| Phase II | |||||||
| Shenzhen Geno‐Immune Medical Institute | COVID‐19/aAPC | Lentivirus modification including immune‐modulatory genes and the viral minigenes to the pathogen‐specific aAPC cells | Phase I | Conserved structural and protease protein domains | 100 healthy and COVID‐19‐positive volunteers | March 9, 2020 | NCT04299724 |
Abbreviations: aAPCs, artificial antigen‐presenting cells; Co., company; COVID, coronavirus disease; Inst., institute; mRNA, messenger RNA; SARS ‐CoV‐2, severe acute respiratory syndrome coronavirus 2; VRC, NIAID's Vaccine Research Center.
Table 2.
Top COVID‐19 vaccine candidates undergoing preclinical testing
| Co. or Inst. | Vaccine platform | Target antigen | Expected time to enter clinical trial |
|---|---|---|---|
| Sanofi and GlaxoSmithKline (GSK) | A recombinant DNA vaccine expressed in baculovirus system | Spike protein COVID‐19 antigen | In the second half of 2020 |
| Novavax | Protein nanoparticle technology platform | Antigens derived from the spike protein+ saponin‐based Matrix‐M™ adjuvant | In late spring of 2020 |
| NuGenerex Immuno‐Oncology (NGIO) | Synthetic peptide | Spike protein | – |
| Johnson & Johnson and Biomedical Advanced Research and Development Authority (BARDA) | Recombinant adenovirus‐based vaccine | – | In September 2020 |
| Serum Institute and Codagenix | Live‐attenuated vaccine | – | Be market ready by 2022 |
| Clover (Sichuan) Biopharmaceuticals | COVID‐19 S‐Trimer | Combined with CpG 1018, a proprietary TLR9 agonist adjuvant | – |
| Altimmune and University of Alabama at Birmingham (UAB) | AdCOVID | – | In the third quarter of 2020 |
| MIGAL Galilee Research Institute | Avian coronavirus Infectious Bronchitis Virus (IBV) vaccine | – | – |
| Tonix Pharmaceuticals | TNX‐1800 | Live horsepox virus vaccine expressing a COVID‐19 protein | – |
| Vaxart | Vaxart's COVID‐19 oral vaccine | Different SARS‐CoV‐2 antigens combination | In the second half of 2020 |
| Applied DNA and Takis Biotech | Linear DNA synthetic gene | – | – |
Abbreviations: Co., company; CoV, coronavirus; Inst., institute; SARS ‐CoV‐2, severe acute respiratory syndrome coronavirus 2; TLR9, toll‐like receptor 9.
Source: (Hodgson, 2020).
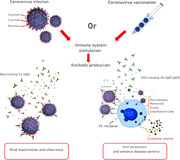
Although the virus's S‐protein is considered a promising immunogen for inclusion in a nucleic acid COVID‐19 vaccine, there continues to be debate over the best approach to ensure optimal immune response. For example, in the current debate, target antigen design in a full‐length protein or the receptor‐binding domain is challenging (Lurie et al., 2020). However, several areas of consensus that are herein disclosed have emerged in this area. Beginning from the basics, it has been demonstrated that immune response to coronavirus infection or vaccination involves early neutralizing immunoglobulin G (IgG) antibody response against viral surface proteins (Zand & Wang, 2020). There is also some evidence that coronaviruses make use of certain antibodies for entry to the cell as an alternative mechanism of target cell binding (Wan et al., 2020). The phenomenon called antibody‐dependent enhancement (ADE) of virus infection is thought to occur when antivirus antibodies attach surface viral antigens and subsequently cause uptake of the virus‐IgG complex via the Fc family of receptors leading to viral infection of cells expressing Fc receptor including macrophages, monocytes, B cells, and vascular endothelium which may contribute to disease severity (Wan et al., 2020). Binding of IgG subclasses IgG1 and IgG3 targeting the receptor‐binding domain of coronavirus S‐protein function as a mechanism for viral inactivation and clearance, at the same time provides a route for viral persistence via ADE and can cause severe disease by induction of cytokine storm through a mechanism that depends on these enhancing antibodies (as illustrated in Figure 1; Jaume et al., 2011; X. Li et al., 2020). Importantly, specific protein sequences responsible for ADE have been recognized on the S‐proteins of SARS and COVID‐19 coronaviruses (Wang et al., 2016). It is also clinically demonstrated that sera from SARS patients apparently contain both neutralizing IgG antibodies and those that cause ADE (Wang et al., 2016). This is particularly important when modeling optimal vaccine strategy for reducing severe disease. Of note, within the different vaccine types (except for live‐attenuated virus vaccines), the general goal is to induce adaptive immune response against S or N viral proteins and also the increase in high‐affinity IgG production is of the utmost importance for planning vaccine actions. These data collectively suggest that one approach may be to manipulate and modify the protein sequences to disable the potential for ADE if possible (Zand & Wang, 2020). In addition, given that such an adverse effect may be associated with T helper type 2 (Th2) immune responses, extended human safety testing will be critical. To this point, using a suitable adjuvant with a capacity to trigger a type 1T helper (Th1) response and high neutralizing antibody titers, theoretically, is likely to be protective to avoid immunopathology (Lurie et al., 2020). It worth noting that the use of an adjuvant in the vaccine is a reasonable approach in a pandemic that may substantially reduce the amount of vaccine material required for immunization. Owing to the limited worldwide production capacity for vaccine, this strategy will allow more vaccine doses to be produced for widespread vaccination of populations and therefore contributes to protection of more people (Hotez & Bottazzi, 2020).
Figure 1.
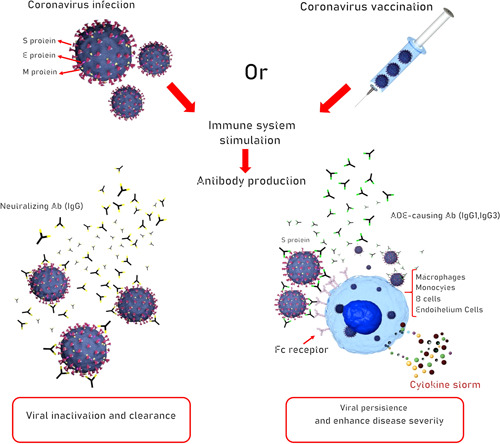
The putative neutralizing and infection‐enhancing antibody response to human SARS‐CoV‐2 virus infection. The host's immune response elicited by either natural infection or vaccination targeting a surface antigen influences the proliferative activity of live virus through antibody‐mediated mechanisms. Infected individuals develop specific antibodies capable of neutralizing the ability of virus to enter certain cell types. These neutralizing antibodies to the viral envelope can protect against reinfection with SARS‐CoV‐2. In addition, nonneutralizing antibodies to the viral surface proteins provide no protection against viral challenge and potentiate the uptake of virus particles by Fc receptor‐bearing cells including macrophages, monocytes, B cells, and endothelium cells leading to viral persistence. The so‐called enhancing antibodies have substantial effects through antibody Fc‐mediated effector function which amplify the overwhelming cytokine storm and, potentially, exacerbate disease pathologies. This phenomenon is called antibody‐dependent enhancement (ADE) of infection. The role of infectivity‐enhancing antibodies raises issues about the development of SARS‐CoV‐2 virus vaccines and the use of passive antibody therapy. This is perhaps an important challenge in SARS‐CoV‐2 vaccine research. The goal of the current vaccine research programs should be therefore to develop antibodies for the virus eradication. Ab, antibody; IgG, immunoglobulin G; SARS ‐CoV‐2, severe acute respiratory syndrome coronavirus 2
The E protein has also been proposed as an attractive vaccine target for developing live‐attenuated vaccines, which is based on prior studies of related coronaviruses that cause MERS and SARS‐CoV (Almazán et al., 2013). For both SARS and MERS‐CoV, the construction of an attenuated virus deleted in E protein could result in a replication‐competent but propagation‐defective virus that supports the efficacy of the attenuated viruses as vaccine candidates (Almazán et al., 2013). Deletion of E protein can be accomplished to 2019‐nCoV and it is expected to lead to a similar breakthrough given its phylogenetic clustering with human SARS‐CoV E protein (Ralph et al., 2020).
As reported, the long‐term immunologic outcomes of SARS survivors have been correlated with antiviral antibody responses that persisted for up to 6 years after infection and T cell responses detectable for as long as 11 years (Tang et al., 2011). In a research published in 2016, a novel subunit vaccine based on a native conformational epitope from a MERS‐CoV envelope glycoprotein was first developed (Tai et al., 2016) and gave encouraging results that trimeric form of MERS‐CoV spike receptor‐binding domain is capable of inducing protective immunity in a human DPP4 transgenic (hDPP4‐Tg) mouse model. This vaccine candidate containing residues 377–588 of MERS‐CoV S‐protein was shown to adhere the cell‐associated DPP4 receptor and block MERS pseudovirus entry into target cells displaying high cross‐neutralizing activity of the epitope that give rise to the therapeutic efficacy of the vaccine. Therefore, learning from the study of SARS‐ and MERS‐CoVs, it should be noted that a protective immune response from a vaccine must lead to antibody‐dependent or –independent virus neutralization and potent cytotoxicity.
4.2. Vaccine design against 2019‐nCoV; the current efforts of immunoinformatics approach
It is best to use vaccines to protect healthy individuals from infection as well as create herd immunity and to reduce the burden on health services. The rapid global spread of COVID‐19 prompts the urgent action to control the public health risk of this novel coronavirus. For this purpose, applying analysis tools and databases, several modeling approaches have been carried out to elucidate the genetic and antigenic properties of the virus to accelerate novel vaccine design. Peptide‐based vaccines have received more attention due to their ease of production by the chemical synthesis of dominant protective epitopes (Dudek, Perlmutter, Aguilar, Croft, & Purcell, 2010). Just as neutralizing the free virus particles before cell entry is required to prevent cells from being infected by COVID‐19, rational vaccine design strategies based on the characterization of epitopes on the most exposed protein of the virus that can be recognized by B cell antibodies and T lymphocytes may be able to overcome the ongoing outbreak. Using immunoinformatics approach to find protein epitopes for vaccine development against COVID‐19 infection, several putative immune epitopes (including YLQPRTFLL, GVYFASTEK, EPVLKGVKL, VVNQNAQAL, WTAGAAAYY as potential CTL epitopes, and CVNLTTRTQLPPAYTN, NVTWFHAIHVSGTNGT, and SFSTFKCYGVSPTKLNDL peptides as B cell/antibody epitopes) from surface glycoprotein of 2019‐nCoV were identified (Baruah & Bose, 2020). These assessments need to be followed to ensure work with related antigens in vaccine development attempts for preventive strategy of the novel virus. Interestingly, modeling of COVID‐19 glycoprotein could describe multiple epitope specificity of cytotoxic T cells (CTL) and B cells generating immune responses to COVID‐19 and when compared with CTL epitopes predicted in the S‐protein of MERS‐CoV, revealed one overlapping peptide epitope, but no comparable epitopes with SARS‐CoV were detected (Baruah & Bose, 2020). This is an important issue because even though the genomes of COVID‐19 share better sequence homology with SARS‐CoV and despite greater genotype distance from MERS‐CoV (X. Xu et al., 2020), it may exhibit similar immunogenicity with MERS‐CoV which may contribute to the immunopathological outcomes. Nevertheless, delineating peptide‐binding properties of MHC/HLA molecules, to which peptide antigens are attached, is also crucial to examine subunit vaccine efficacy. Antigen presentation by MHC molecules is essential to immune responses mediated by CD4+ and CD8+ T cells. In essence, the characterization of the corresponding HLA alleles can take a step toward designing T‐cell‐based vaccines. For that, common epitopes of four structural proteins of 2019‐nCoV and the dominant HLA‐DR alleles prevalent in the five ethnic populations were mapped using molecular modeling techniques with the Immune Epitope Database and Analysis Resource consensus tool and the results illustrated eight high‐affinity epitopes in the functional regions of the S and membrane proteins of the 2019‐nCoV strain Wuhan‐Hu‐1 recognized by HLA‐DR. Furthermore, the authors suggested that the administration of these immunodominant epitopes from 2019‐nCoV, as a multiepitopic peptide vaccine, able to provide protective antiviral antibody and T‐cell responses may represent a universally potent subunit vaccine effective in different ethnic populations (Ramaiah & Arumugaswami, 2020). In another study, bioinformatic analysis recognized various B‐cell and T‐cell epitopes within the 2019‐nCoV surface glycoprotein. A wide variety of sequential linear B‐cell epitopes and candidate T‐cell epitopes with MHC‐I and MHC‐II binding affinity were initially obtained. However, the antigenicity analysis pointed out that only 13 MHC‐I epitopes and 3 MHC‐II epitopes had the antigenic propensity (Bhattacharya et al., 2020). Furthermore, of the S‐protein oligopeptides, the most immunogenic and uniquely viral epitopes were identified through comparative Homo sapiens‐coronavirus proteome analysis. Using the Peptide Match program, the 2109‐nCoV and the human proteomes were compared and searched for viral peptides that are absent in the human proteome to increase antiviral specificity and minimizing the risk for cross‐reactions (Lucchese, 2020). A total of 66 non‐self sequences in the virus genome were obtained as a result that represents the best targets for vaccine development due to their potential for immunogenicity (Richman, Vonderheide, & Rech, 2019). In addition, the identification of the potential T cell epitopes of the E protein of SARS‐CoV‐2 proposed three peptides of YVYSRVKNL, SLVKPSFYV, and LAILTALRL with high binding affinity to MHC‐I molecules as promising vaccine candidates for COVID‐19 (Abdelmageed et al., 2020).
Previous experience with developing anti‐SARS coronavirus vaccines, since the last pandemic nearly 17 years ago, have opened the doors to rationally develop vaccine approaches designed to counter the COVID‐19 pandemic. Preliminary studies suggest that in convalescent SARS‐CoV patients, immune protection is mainly associated with T cell responses against structural (e.g., the S and N proteins) rather than nonstructural CoV proteins, indicating that T‐cell epitopes in SARS‐CoV structural proteins have been found to be the most immunogenic as compared with the nonstructural proteins (C. k.‐f. Li et al., 2008). The authors claim that immune targeting of SARS‐CoV‐derived B cell and T cell epitopes that map identically to SARS‐CoV‐2 proteins may offer substantial protection against the new virus (Ahmed, Quadeer, & McKay, 2020). What strengthens the idea is that the determined epitopes comprise no mutations in the available SARS‐CoV‐2 sequences (Ahmed et al., 2020).
Since genomic variations of the pathogen have significant clinical consequences, the genetic diversity of SARS‐CoV‐2 and strategies to develop vaccine candidates are described in detail in the next section.
4.3. The genetic diversity and rapid evolution of SARS‐CoV‐2: Impact on treatment and vaccine development
One hypothesis is that the rapid spread of SARS‐CoV‐2 may be due to its evolution driven by viral mutations. Understandably, the evolution of RNA viruses continues to occupy a great deal of attention (Dolan, Whitfield, & Andino, 2018). The rapid evolution of RNA viruses that utilize a highly divergent RNA‐dependent RNA polymerase is often more evident than those of DNA‐based viruses. In fact, enormous genetic diversity is a critical determinant of their adaptative capacity (Lauring, Frydman, & Andino, 2013). Nucleotide substitution in DNA is often involved as one of the most important mechanisms provoking viral evolution in nature (Lauring & Andino, 2010). In the last few months, genome‐sequencing projects have been conducted by the worldwide scientific community to understand the evolutionary dynamics and characteristics of this virus. A comprehensive assessment using whole‐genome sequencing discovered 93 mutations comprising 42 missense mutations across the entire genomes of SARS‐CoV‐2 (Phan, 2020). Among the viral proteins, all the major nonstructural and structural proteins showed missense mutations and most variations occur toward the ORF1ab polyprotein (29 missense mutations) and in the spike surface glycoprotein (eight missense mutations). Specifically, three of these mutations were positioned in the receptor‐binding domain of spike surface glycoprotein (in residues: D354, Y364, and F367) and were expected to affect its conformational changes that probably lead to antigenic variation and the loss of binding to antibodies against S‐protein. Of note, no mutations were detected in the envelope (E) protein of SARS‐CoV‐2 (Phan, 2020). A comparison of 10 genomic information obtained from 2019‐nCoV samples suggest that the 2019‐nCoV genomes would likely have low heterogeneity (R. Lu et al., 2020). Data from 56 genomic sequences of distinct patient infected with 2019‐nCoV (showing >99% sequence identity) have reported two hypervariable genomic hotspots and confirmed previous results of a high level of genomic conservation within 2019‐nCoV (Ceraolo & Giorgi, 2020). With potential implications in epitope definition, the proteome analysis also reported the polymorphic variants corresponding to the two variable regions. The viral ORF8‐encoded protein‐bearing Serine/Leucine variation and a large gene encoding for a polyprotein (ORF1ab) translation of which results an ∼800 kDa polyprotein were described as the two most variable locations in the core genome (Ceraolo & Giorgi, 2020). Another important aspect of virus replication may be at the functional contact between virus and host. Several viruses, including coronaviruses, require host cellular factors to replicate (Zumla et al., 2016). Furthermore, repeated observations in other families of RNA viruses indicate that many, if not most, mutations are surface‐located and that the conserved residues are probably core residues of viral proteins (Cheng & Brooks III, 2013; Patel & Kukol, 2017; Warren, Wan, Conant, & Korkin, 2013). A more in‐depth analysis of the intraviral and virus‐host protein–protein interactions of SARS‐CoV‐2 has revealed several findings (Srinivasan et al., 2020): (a) the spatial patterns of mutations that affect protein functional surfaces are bidentified in a substantial number of SARS‐CoV‐2 proteins and were also analyzed in terms of ligand‐binding abilities of human SARS‐CoV‐2, and (b) all protein‐binding sites of nonstructural proteins revealed fully conserved residues supporting the notion that groups of mutations unlikely disrupt the protein–protein interaction of viral proteins (Srinivasan et al., 2020). The SARS‐CoV‐2 sequenced genome is predicted to have 16 nonstructural proteins: NSP1‐NSP16 involving in the intraviral heteromeric complexes such as NSP7‐NSP8‐NSP12, NSP10‐NSP16, and NSP10‐NSP14 (Srinivasan et al., 2020), targeting of which may be critical for developing effective antiviral SARS‐CoV‐2 therapies and vaccine. Taking advantage of all the currently available information, we postulated that the low variability within the new pandemic virus 2019‐nCoV contributes to mount the possibility of developing a practical vaccine that would confer protection against the new virus strain.
5. THE CHALLENGES FOR SARS‐2 VACCINE DEVELOPMENT
Generally, the clinical development of a new vaccine requires many years of efforts in this area. However, with the milestone experience of a successful vaccine for the eradication of severe foot‐and‐mouth disease caused by EV71 virus, it has been possible to develop vaccines against a modern highly pathogenic emerging virus (S. Lu, 2014; J. Xu et al., 2010). Moreover, having the results of previous efforts related to SARS‐CoV and MERS vaccine research available, first steps have been taken toward achieving an effective vaccine to respond to the threat of this emerging virus. This is readily apparent from a recent study begun at KPWHRI in Seattle, a part of the National Institutes of Health that was allowed to quickly proceed to Phase I trial to determine human safety and efficacy of an investigational vaccine designed to protect against COVID‐19 which was related to their experience as an NIH clinical trials center since 2007 (Challener, 2020). The question then at issue became factors affecting the choice of vaccines. For vaccine developers, high‐level attention is focused on the safety evaluation of a new candidate vaccine against SARS‐2 in humans. As the immune response to virus plays a major role in the pathogenesis of SARS‐2 infection, it is important to ensure that vaccine should not induce immunopathological injury of host cells. In this regard, the foremost issue to be considered is the type of vaccines and the immunogens selected. Indeed, the promise of future vaccine development would be most applicative by rational design of new vaccine candidates. Subunit vaccines are generally safe to use. This vaccine type is most often composed of one or multiple recombinant protein‐ or synthetic peptide‐based formulations (Deng, Hu, Wang, & Deng, 2012). Subunit vaccines offer many advantages over live‐attenuated or inactivated organisms. Unlike inactivated or live‐attenuated virus vaccines, subunit vaccines are known to provide better efficacy as they are unable to revert to the virulent form of the pathogen and eliminate the risk of incomplete inactivation, thus making them biologically safe; however, they typically require coadministration with adjuvants (Zhang et al., 2012). Based on this information, the subunit candidates such as full‐length S‐protein and/or the RBD element of SARS‐CoV‐2 prove valuable for prevention against COVID‐19 (Shang, Yang, Rao, & Rao, 2020), though they are expensive to manufacture and need for repeated injections (Phillpotts, Venugopal, & Brooks, 1996). Moreover, subunit vaccines can be designed to contain well‐characterized neutralizing epitopes defined as those epitopes which elicit a neutralizing antibody response but avoiding epitopes with pathological ADE effects to improve immunogenicity (Naz & Dabir, 2007). Another scientific challenge for vaccine development may be dependent to the strategies of vaccine delivery. Evidence of respiratory tract infection and findings of SARS‐2 in stool (Holshue et al., 2020; Q. Li et al., 2020) suggest that mucosal delivery routes by oral or aerosol administration of vaccine appear to be the possible modes of SARS‐CoV‐2 immunization.
Epidemiological studies have shown that the disease caused by SARS‐CoV‐2 is significantly associated with a higher age in which individuals above 50 years of age often exhibit more severe pathology following SARS‐CoV‐2 infection and are more likely to die from infection. In many viral infections, the naive younger individuals experience milder manifestations of the disease. Because it affects the health of older people more strongly, vaccine as a public health measure, must protect this vulnerable population (Amanat & Krammer, 2020). Unfortunately, it is precisely in this population that vaccines are the least efficacious because of immune senescence (Sambhara & McElhaney, 2009). As of current influenza vaccines, specific formulations including high‐dose antigen or using an adjuvant may be particularly useful for this segment of the population (Amanat & Krammer, 2020; DiazGranados et al., 2013). Interestingly, even if vaccination does not result in protection in all recipients, it is still able to stop the transmission of the virus and thereby benefits older individuals (Amanat & Krammer, 2020).
Finally, once a successful SARS‐2 vaccine is developed and available, the responsibility to let the global access to it should be a part of planning. Nonetheless, apart from the availability of the vaccine, still social, clinical, and economic hurdles face SARS‐2 vaccine and vaccination programmes that include, for example, willingness of the public to get a new vaccination, the potential variation in efficiency for quite different populations, as well as severe adverse reactions arising from the new vaccine (Pang et al., 2020).
6. CONCLUDING REMARKS
Our review of the available relevant literature summarized the published information regarding COVID‐19 vaccine research and development. In summary, extensive bioinformatics analysis, in the past few months, has helped provide a unique opportunity to a better understanding of determinants of immunogenicity, immunodominance, and structure‐function relationships that can to a great extent reduce the experimental cost in epitope identification for vaccine design as well as limit the pool of peptides available for analysis. While research into such efforts is in its early stages, the prospect of a preventative or therapeutic vaccine seems realistic as a result of a more intense global cooperation and also by taking advantage of the gathered data on SARS and MERS. As data collection continues, we are getting closer to finding better ways to conquer this disease. First and foremost, accurate and up to date data on the status and timing of vaccine production and release must be accessible. Meanwhile, the collaboration of countries from all over the world is clearly needed in producing a preventive or therapeutically desired result.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
M. G. and M. A. were involved in drafting the manuscript. M. A. has critically reviewed the manuscript content.
Ghaebi M, Osali A, Valizadeh H, Roshangar L, Ahmadi M. Vaccine development and therapeutic design for 2019‐nCoV/SARS‐CoV‐2: Challenges and chances. J Cell Physiol. 2020;235:9098–9109. 10.1002/jcp.29771
REFERENCES
- Abdelmageed, M. I. , Abdelmoneim, A. H. , Mustafa, M. I. , Elfadol, N. M. , Murshed, N. S. , Shantier, S. W. , & Makhawi, A. M. (2020). Design of multi epitope‐based peptide vaccine against E protein of human 2019‐nCoV: An immunoinformatics approach. BioRxiv. 10.1101/2020.02.04.934232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S. F. , Quadeer, A. A. , & McKay, M. R. (2020). Preliminary identification of potential vaccine targets for the COVID‐19 coronavirus (SARS‐CoV‐2) based on SARS‐CoV immunological studies. Viruses, 12(3),E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán, F. , DeDiego, M. L. , Sola, I. , Zuñiga, S. , Nieto‐Torres, J. L. , Marquez‐Jurado, S. , … Enjuanes, L. (2013). Engineering a replication‐competent, propagation‐defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio, 4(5),e00650‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat, F. , & Krammer, F. (2020). SARS‐CoV‐2 vaccines: Status report, immunity. Immunity, 52, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, K. , Ziebuhr, J. , Wadhwani, P. , Mesters, J. R. , & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti‐SARS drugs. Science, 300(5626), 1763–1767. [DOI] [PubMed] [Google Scholar]
- Baruah, V. , & Bose, S. (2020). Immunoinformatics‐aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019‐nCoV. Journal of Medical Virology, 92, 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi, D. E. , Zhang, J. , Renner, C. , & Klein‐Szanto, A. J. (2017). Targeting proprotein convertases in furin‐rich lung cancer cells results in decreased in vitro and in vivo growth. Molecular Carcinogenesis, 56(3), 1182–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, M. , Sharma, A. R. , Patra, P. , Ghosh, P. , Sharma, G. , Patra, B. C. , … Chakraborty, C. (2020). Development of epitope‐based peptide vaccine against novel coronavirus 2019 (SARS‐COV‐2): Immunoinformatics approach. Journal of Medical Virology, 92, 618–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceraolo, C. , & Giorgi, F. M. (2020). Genomic variance of the 2019‐nCoV coronavirus. Journal of Medical Virology, 92, 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challener, C. A. (2020). Can vaccine development be safely accelerated?
- Chen, H. , & Du, Q. (2020). Potential natural compounds for preventing 2019‐nCoV infection.
- Chen, W. H. , Strych, U. , Hotez, P. J. , & Bottazzi, M. E. (2020). The SARS‐CoV‐2 vaccine pipeline: An overview. Current Tropical Medicine Reports, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S. , & Brooks, C. L., III . (2013). Viral capsid proteins are segregated in structural fold space. PLoS Computational Biology, 9(2),e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell, A. S. , Leist, S. R. , Douglas, M. G. , & Baric, R. S. (2018). Modeling pathogenesis of emergent and pre‐emergent human coronaviruses in mice. Mammalian Genome, 29(7‐8), 367–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J. (2020). Vaccine designers take first shots at COVID‐19. Science, 368, 14–16. [DOI] [PubMed] [Google Scholar]
- Coutard, B. , Valle, C. , de Lamballerie, X. , Canard, B. , Seidah, N. , & Decroly, E. (2020). The spike glycoprotein of the new coronavirus 2019‐nCoV contains a furin‐like cleavage site absent in CoV of the same clade. Antiviral Research, 176, 104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J. , Li, F. , & Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology, 17(3), 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, M. P , Hu, Z. H , Wang, H. l , & Deng, F. (2012). Developments of subunit and VLP vaccines against influenza A virus. Virologica Sinica, 27(3), 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiazGranados, C. A. , Dunning, A. J. , Jordanov, E. , Landolfi, V. , Denis, M. , & Talbot, H. K. (2013). High‐dose trivalent influenza vaccine compared to standard dose vaccine in elderly adults: Safety, immunogenicity and relative efficacy during the 2009–2010 season. Vaccine, 31(6), 861–866. [DOI] [PubMed] [Google Scholar]
- Djalante, R. , Shaw, R. , & DeWit, A. (2020). Building resilience against biological hazards and pandemics: COVID‐19 and its implications for the Sendai Framework. Progress in Disaster Science, 6, 100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan, P. T. , Whitfield, Z. J. , & Andino, R. (2018). Mapping the evolutionary potential of RNA viruses. Cell Host & Microbe, 23(4), 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly, C. A. , Malik, M. R. , Elkholy, A. , Cauchemez, S. , & Van Kerkhove, M. D. (2019). Worldwide reduction in MERS cases and deaths since 2016. Emerging Infectious Diseases, 25(9), 1758–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten, C. , Günther, S. , Preiser, W. , Van Der Werf, S. , Brodt, H.‐R. , Becker, S. , … Fouchier, R. A. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New England Journal of Medicine, 348(20), 1967–1976. [DOI] [PubMed] [Google Scholar]
- Dudek, N. L. , Perlmutter, P. , Aguilar, I. , Croft, N. P. , & Purcell, A. W. (2010). Epitope discovery and their use in peptide based vaccines. Current Pharmaceutical Design, 16(28), 3149–3157. [DOI] [PubMed] [Google Scholar]
- Fehr, A. R. , & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis In H. Maier, E. Bickerton, & P. Britton (Eds.), Coronaviruses. Methods in Molecular Biology (Vol. 1282). New York, NY: Humana Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, I. W. (2017). Emerging animal coronaviruses: First SARS and now MERS. Emerging Zoonoses. Emerging Infectious Diseases of the 21st Century, Cham: Springer. [Google Scholar]
- Gao, J. , Tian, Z. , & Yang, X. (2020). Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID‐19 associated pneumonia in clinical studies. BioScience Trends, 14, 72–73. [DOI] [PubMed] [Google Scholar]
- Graham, B. S. , Mascola, J. R. , & Fauci, A. S. (2018). Novel vaccine technologies essential components of an adequate response to emerging viral diseases. JAMA, 319, 1431–1432. [DOI] [PubMed] [Google Scholar]
- Hodgson, J. (2020). The pandemic pipeline. Nature Biotechnology, 38(5), 523–532. [DOI] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Krueger, N. , Mueller, M. A. , Drosten, C. , & Pöhlmann, S. (2020). The novel coronavirus 2019 (2019‐nCoV) uses the SARS‐coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. 10.1101/2020.01.31.929042 [DOI] [Google Scholar]
- Holshue, M. L. , DeBolt, C. , Lindquist, S. , Lofy, K. H. , Wiesman, J. , Bruce, H. , … Pillai, S. K. (2020). First case of 2019 novel coronavirus in the United States. The New England Journal of Medicine, 382, 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez, P. J. , & Bottazzi, M. E. (2020). Developing a low‐cost and accessible COVID‐19 vaccine for global health. Preprints, 2020030464. [DOI] [PMC free article] [PubMed]
- Huang, C. , Wang, Y. , Li, X. , Ren, L. , Zhao, J. , Hu, Y. , … Gu, X. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, D. S. , Azhar, E. I. , Madani, T. A. , Ntoumi, F. , Kock, R. , Dar, O. , … Drosten, C. (2020). The continuing 2019‐nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. International Journal of Infectious Diseases, 91, 264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaume, M. , Yip, M. S. , Cheung, C. Y. , Leung, H. L. , Li, P. H. , Kien, F. , … Altmeyer, R. (2011). Anti‐severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH‐and cysteine protease‐independent FcγR pathway. Journal of Virology, 85(20), 10582–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , He, Y. , & Liu, S. (2005). SARS vaccine development. Emerging Infectious Diseases, 11(7), 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase, M. , Shirato, K. , van der Hoek, L. , Taguchi, F. , & Matsuyama, S. (2012). Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. Journal of Virology, 86(12), 6537–6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer, R. N. , & Ward, A. B. (2019). Structure of the SARS‐CoV nsp12 polymerase bound to nsp7 and nsp8 co‐factors. Nature Communications, 10(1),2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek, T. G. , Erdman, D. , Goldsmith, C. S. , Zaki, S. R. , Peret, T. , Emery, S. , … Lim, W. (2003). A novel coronavirus associated with severe acute respiratory syndrome. New England Journal of Medicine, 348(20), 1953–1966. [DOI] [PubMed] [Google Scholar]
- Lake, M. A. (2020). What we know so far: COVID‐19 current clinical knowledge and research. Clinical Medicine, 20(2), 124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring, A. S. , & Andino, R. (2010). Quasispecies theory and the behavior of RNA viruses. PLoS Pathogens, 6(7),e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring, A. S. , Frydman, J. , & Andino, R. (2013). The role of mutational robustness in RNA virus evolution. Nature Reviews Microbiology, 11(5), 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. k.‐f , Wu, H. , Yan, H. , Ma, S. , Wang, L. , Zhang, M. , … Brenchley, J. M. (2008). T cell responses to whole SARS coronavirus in humans. The Journal of Immunology, 181(8), 5490–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Guan, X. , Wu, P. , Wang, X. , Zhou, L. , Tong, Y. , … Wong, J. Y. (2020). Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. New England Journal of Medicine, 382, 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Geng, M. , Peng, Y. , Meng, L. , & Lu, S. (2020). Molecular immune pathogenesis and diagnosis of COVID‐19. Journal of Pharmaceutical Analysis, 10, 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. , Wu, H. , … Zhu, N. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. (2014). EV71 vaccines: A milestone in the history of global vaccine development. Emerging Microbes & Infections, 3(4),e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchese, G. (2020). Epitopes for a 2019‐nCoV vaccine. Cellular & Molecular Immunology, 17, 539–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie, N. , Saville, M. , Hatchett, R. , & Halton, J. (2020). Developing COVID‐19 vaccines at pandemic speed. New England Journal of Medicine, 382, 1969–1973. [DOI] [PubMed] [Google Scholar]
- Malik, Y. S. , Sircar, S. , Bhat, S. , Sharun, K. , Dhama, K. , Dadar, M. , … Chaicumpa, W. (2020). Emerging novel coronavirus (2019‐nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Veterinary Quarterly, 40(1), 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza, M. U. , & Froeyen, M. (2020). Structural elucidation of SARS‐CoV‐2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 RNA‐dependent RNA polymerase and Nsp13 helicase. Journal of Pharmaceutical Analysis. 10.1016/j.jpha.2020.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel, G. J. (2013). Designing tomorrow's vaccines. New England Journal of Medicine, 368(6), 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz, R. K. , & Dabir, P. (2007). Peptide vaccines against cancer, infectious diseases, and conception. Frontiers in Bioscience, 12, 1833–1844. [DOI] [PubMed] [Google Scholar]
- Negahdaripour, M. (2020). The battle against COVID‐19: Where do we stand now? Iranian Journal of Medical Sciences, 45(2), 81–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, J. , Wang, M. X. , Ang, I. Y. H. , Tan, S. H. X. , Lewis, R. F. , Chen, J. I. P. , … Yang, Q. (2020). Potential rapid diagnostics, vaccine and therapeutics for 2019 novel Coronavirus (2019‐ncoV): A systematic review. Journal of Clinical Medicine, 9(3),E623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, H. , & Kukol, A. (2017). Evolutionary conservation of influenza A PB2 sequences reveals potential target sites for small molecule inhibitors. Virology, 509, 112–120. [DOI] [PubMed] [Google Scholar]
- Paules, C. I. , Marston, H. D. , & Fauci, A. S. (2020). Coronavirus infections—more than just the common cold. Journal of the American Medical Association, 323(8), 707–708. [DOI] [PubMed] [Google Scholar]
- Peck, K. M. , Burch, C. L. , Heise, M. T. , & Baric, R. S. (2015). Coronavirus host range expansion and Middle East respiratory syndrome coronavirus emergence: Biochemical mechanisms and evolutionary perspectives. Annual review of virology, 2, 95–117. [DOI] [PubMed] [Google Scholar]
- Phan, T. (2020). Genetic diversity and evolution of SARS‐CoV‐2. Infection, Genetics and Evolution, 81, 104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillpotts, R. , Venugopal, K. , & Brooks, T. (1996). Immunisation with DNA polynucleotides protects mice against lethal challenge with St. Louis encephalitis virus. Archives of Virology, 141(3‐4), 743–749. [DOI] [PubMed] [Google Scholar]
- Pyrc, K. , Berkhout, B. , & Van Der Hoek, L. (2007). Identification of new human coronaviruses. Expert Review of Anti‐Infective Therapy, 5(2), 245–253. [DOI] [PubMed] [Google Scholar]
- ul Qamar, M. T. , Alqahtani, S. M. , Alamri, M. A. , & Chen, L.‐L. (2020). Structural basis of SARS‐CoV‐2 3CLpro and anti‐COVID‐19 drug discovery from medicinal plants. Journal of Pharmaceutical Analysis. 10.1016/j.jpha.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, R. , Lew, J. , Zeng, T. , Francis, M. , Xue, B. , Roux, M. , … Al‐Ahdal, M. N. (2020). 2019‐nCoV (Wuhan virus), a novel Coronavirus: Human‐to‐human transmission, travel‐related cases, and vaccine readiness. The Journal of Infection in Developing Countries, 14(1), 3–17. [DOI] [PubMed] [Google Scholar]
- Ramaiah, A. , & Arumugaswami, V. (2020). Insights into cross‐species evolution of novel human coronavirus 2019‐nCoV and defining immune determinants for vaccine development. BioRxiv. 10.1101/2020.01.29.925867 [DOI] [Google Scholar]
- Richman, L. P. , Vonderheide, R. H. , & Rech, A. J. (2019). Neoantigen dissimilarity to the self‐proteome predicts immunogenicity and response to immune checkpoint blockade. Cell systems, 9(4), 375–382. e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Z. , Liu, C. , Guo, Y. , He, Z. , Huang, X. , Jia, X. , & Yang, T. (2020). Potential inhibitors targeting RNA‐dependent RNA polymerase activity (NSP12) of SARS‐CoV‐2. Preprints, 2020030024. 10.20944/preprints202003.0024.v1 [DOI] [PMC free article] [PubMed]
- Sambhara, S. , & McElhaney, J. E. (2009). Immunosenescence and influenza vaccine efficacy vaccines for pandemic influenza. Current Topics in Microbiology and Immunology, 333, 413–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Trincado, J. L. , Gomez‐Perosanz, M. , & Reche, P. A. (2017). Fundamentals and methods for T‐and B‐cell epitope prediction. Journal of Immunology Research, 2017, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, W. , Yang, Y. , Rao, Y. , & Rao, X. (2020). The outbreak of SARS‐CoV‐2 pneumonia calls for viral vaccines. NPJ Vaccines, 5(1), 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal, T. (2020). A review of coronavirus disease‐2019 (COVID‐19). The Indian Journal of Pediatrics, 87, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi, C. , Alsafi, Z. , O'Neill, N. , Khan, M. , Kerwan, A. , Al‐Jabir, A. , … Agha, R. (2020). World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID‐19). International Journal of Surgery, 76, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, S. , Cui, H. , Gao, Z. , Liu, M. , Lu, S. , Mkandawire, W. , … Korkin, D. (2020). Structural genomics of SARS‐CoV‐2 indicates evolutionary conserved functional regions of viral proteins. Viruses, 12(4),E360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi, L. , Posthuma, C. C. , Collet, A. , Zevenhoven‐Dobbe, J. C. , Gorbalenya, A. E. , Decroly, E. , … Imbert, I. (2014). One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proceedings of the National Academy of Sciences, 111(37), E3900–E3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, W. , Zhao, G. , Sun, S. , Guo, Y. , Wang, Y. , Tao, X. , … Lanying, D. (2016). A recombinant receptor‐binding domain of MERS‐CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS‐CoV infection. Virology, 499, 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, F. , Quan, Y. , Xin, Z. T. , Wrammert, J. , Ma, M. J. , Lv, H. , … Liu, W. (2011). Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: A six‐year follow‐up study. The Journal of Immunology, 186(12), 7264–7268. [DOI] [PubMed] [Google Scholar]
- Tian, X. , Li, C. , Huang, A. , Xia, S. , Lu, S. , Shi, Z. , … Wu, Y. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerging Microbes & Infections, 9(1), 382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, C. T. , Sbrana, E. , Iwata‐Yoshikawa, N. , Newman, P. C. , Garron, T. , Atmar, R. L. , … Couch, R. B. (2012). Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One, 7(4),e35421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , Shang, J. , Graham, R. , Baric, R. S. , & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade‐long structural studies of SARS coronavirus. Journal of Virology, 94(7),e00127‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , Shang, J. , Sun, S. , Tai, W. , Chen, J. , Geng, Q. , … Shi, Z. (2020). Molecular mechanism for antibody‐dependent enhancement of coronavirus entry. Journal of Virology, 94(5),e02015‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Zhang, L. , Kuwahara, K. , Li, L. , Liu, Z. , Li, T. , … Xie, J. (2016). Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non‐human primates. ACS Infectious Diseases, 2(5), 361–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, S. , Wan, X. F. , Conant, G. , & Korkin, D. (2013). Extreme evolutionary conservation of functionally important regions in H1N1 influenza proteome. PLoS One, 8(11),e81027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P. C. , Lau, S. K. , Chu, C. M. , Chan, K. H. , Tsoi, H. W. , Huang, Y. , … Luk, W. K. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. Journal of Virology, 79(2), 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Qian, Y. , Wang, S. , Serrano, J. M. G. , Li, W. , Huang, Z. , & Lu, S. (2010). EV71: An emerging infectious disease vaccine target in the Far East? Vaccine, 28(20), 3516–3521. [DOI] [PubMed] [Google Scholar]
- Xu, J. , Zhao, S. , Teng, T. , Abdalla, A. E. , Zhu, W. , Xie, L. , … Guo, X. (2020). Systematic comparison of two animal‐to‐human transmitted human coronaviruses: SARS‐CoV‐2 and SARS‐CoV. Viruses, 12(2),E244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Chen, P. , Wang, J. , Feng, J. , Zhou, H. , Li, X. , … Hao, P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, 63(3), 457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Xie, W. , Xue, X. , Yang, K. , Ma, J. , Liang, W. , … Ziebuhr, J. (2005). Correction: Design of wide‐spectrum inhibitors targeting coronavirus main proteases. PLoS Biology, 3(11),e428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Zhao, J. , & Zhang, Z. (2020). More than pneumonia, the potential occurrence of multiple organ failure in 2019 novel coronavirus infection. 10.2139/ssrn.3532272 [DOI]
- Zand, M. , & Wang, J. (2020). Potential mechanisms of age related severity of COVID‐19 Infection: Implications for vaccine development and convalescent serum therapy. 10.31219/osf.io/f3pze [DOI]
- Zhang, T. T. , Kang, T. H. , Ma, B. , Xu, Y. , Hung, C.‐F. , & Wu, T.‐C. (2012). LAH4 enhances CD8+ T cell immunity of protein/peptide‐based vaccines. Vaccine, 30(4), 784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X.‐L. , Wang, X.‐G. , Hu, B. , Zhang, L. , Zhang, W. , … Huang, C.‐L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Vedantham, P. , Lu, K. , Agudelo, J. , Carrion Jr, R. , Nunneley, J. W. , … Renslo, A. R. (2015). Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Research, 116, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A. , Chan, J. F. , Azhar, E. I. , Hui, D. S. , & Yuen, K.‐Y. (2016). Coronaviruses—drug discovery and therapeutic options. Nature Reviews Drug Discovery, 15(5), 327–347. [DOI] [PMC free article] [PubMed] [Google Scholar]


