Abstract
A reduced peripheral blood absolute lymphocyte count with an elevated neutrophil count has been a consistent observation in hospitalized coronavirus disease 2019 (COVID‐19) patients. In this brief meta‐analysis, the reduction of lymphocyte subset counts in COVID‐19 patients was investigated across 20 peer‐reviewed studies meeting criteria for reporting lymphocyte subset counts and COVID‐19 disease severity. CD4+ T cell, CD8+ T cell, B cell, NK cell, and total lymphocyte cell counts all showed statistically significant reduction in patients with severe/critical COVID‐19 disease compared to mild/moderate disease. T‐cell subsets showed the largest standardized magnitude of change. In some studies, multivariate analysis has shown that CD4 and/or CD8 T‐cells counts are independently predictive of patient outcomes. © 2020 International Society for Advancement of Cytometry
Keywords: COVID‐19, immunophenotyping, lymphocyte subset, T‐cell subset, flow cytometry
In December 2019, a series of patients with pneumonia of unknown etiology was reported in Wuhan, Hubei Province, China (1, 2). Within weeks, a novel coronavirus, now named Severe Acute Respiratory Syndrome Corona Virus 2 (SARS‐CoV‐2), was identified as the cause of this disease. SARS‐CoV‐2, a member of the genus beta coronavirus, has spread globally, leading to a pandemic that has infected over 7 million people and caused over 400,000 deaths (as of June 7, 2020) in over 180 countries/regions. This new pandemic has disrupted the global economy and put an enormous strain on global health care systems. The disease has been designated as coronavirus disease 2019 (COVID‐19) by the World Health Organization and often presents clinically as fever, fatigue, muscle pain, diarrhea, and pneumonia and can cause death in severe cases. The severity of the disease has been shown to be related to age and the presence of co‐morbidities, such as diabetes, obesity, and heart disease. Several reports coming out of China have revealed that patients with the most severe cases of COVID‐19 have abnormalities in many laboratory parameters, such as elevated procalcitonin, lactate dehydrogenase, d‐dimer, C‐reactive protein, neutrophil counts, and pro‐inflammatory cytokines, such as interleukin‐6 (3). Lymphopenia and thrombocytopenia are also associated with severe COVID‐19 disease. There is a growing list of publications indicating that the assessment of lymphocyte subset counts, such as CD4 and CD8 T cells, B cells, and NK cells, may provide prognostic information for COVID‐19 disease severity and convalescence when considered in conjunction with other clinical information (4, 5). To obtain a clearer picture of this emerging data, we performed a meta‐analysis of studies that included measurement of lymphocyte subset counts and disease severity in patients hospitalized with COVID‐19.
methods
A PubMed search (https://pubmed.ncbi.nlm.nih.gov/) on May 23 for “COVID‐19 Lymphocyte” produced a list of 258 publications. These publications were examined to exclude those that did not report patient clinical characterization data and lymphocyte subset evaluations. There were 16 publications that evaluated lymphocyte subset counts in COVID‐19 patients with well‐characterized degrees of disease severity. An additional four publications meeting these criteria were found using a Google search. CD4+ and/or CD8+ T‐cell counts from COVID‐19 patients with different disease severity status were reported in all 20 publications, and 10 of them also included CD19+ B cell and CD16 + CD56+ NK cell counts. These 20 peer‐reviewed publications were selected for meta‐analysis in this brief report (6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25).
These publications compared the results of peripheral blood lymphocyte subset counts in patients with mild/moderate disease to those with severe/critical disease hospitalized in China with a diagnosis of COVID‐19 pneumonia. For the majority of the publications, the lymphocyte subset count data were gleaned from patient data registry, and reagents and flow cytometry instruments used in the measurement were not disclosed. A few publications reported the use of commercial in vitro diagnostic (IVD) products and lab developed tests (Supporting Information Table S1). The disease status assignments varied across studies, including survival vs. nonsurvival, moderate vs. severe/critical, aggravation vs. nonaggravation, and critical vs. noncritical. For the meta‐analysis, COVID‐19 patients were categorized into two groups: mild/moderate and severe/critical. The criteria used in the selected publications were consolidated as follows: mild, survival, noncritical, and patients with nonaggravation disease were all classified into the “mild/moderate” group; deceased, nonsurvival, critical and patients with disease aggravation were all classified into the “severe/critical” group.
results
The 20 publications selected for meta‐analysis included a total of 3,017 subjects with CD4+ cell counts where 2,311 were classified as “mild/moderate” (76.6%) and 706 were classified as “severe/critical” (23.4%). The sample sizes of subjects with CD4+ and CD8+ T‐cell counts per publication varied from 17 to 499, with 10 to 479 in the mild/moderate group and 5 to 105 in the severe/critical group. A smaller subset of patients in the data set had absolute lymphocyte subset counts for B cells and NK cells reported. For publications with median and IQR or range reported, the mean and standard deviation were extrapolated according to Wan et al. (26) There is an apparent outlier in reported lymphocyte counts in one publication with increased total lymphocyte counts but decreased CD4+ and CD8+ T‐cells counts in critical vs. noncritical patients, and this outlier in lymphocyte count was removed from the data analysis (16). The mean cell counts in “mild/moderate” and “severe/critical” COVID‐19 groups reported in each publication are shown in Figure 1. The cell counts were consistently decreased in the severe/critical group, and the differences in the weighted mean value of cell counts between the two patient groups are statistically significant for all cell types. Of note, the fold changes between mild/moderate and severe/critical groups for mean CD4 and CD8 T‐cell counts are larger than for mean B cell, NK cell, and total lymphocyte cell counts.
Figure 1.
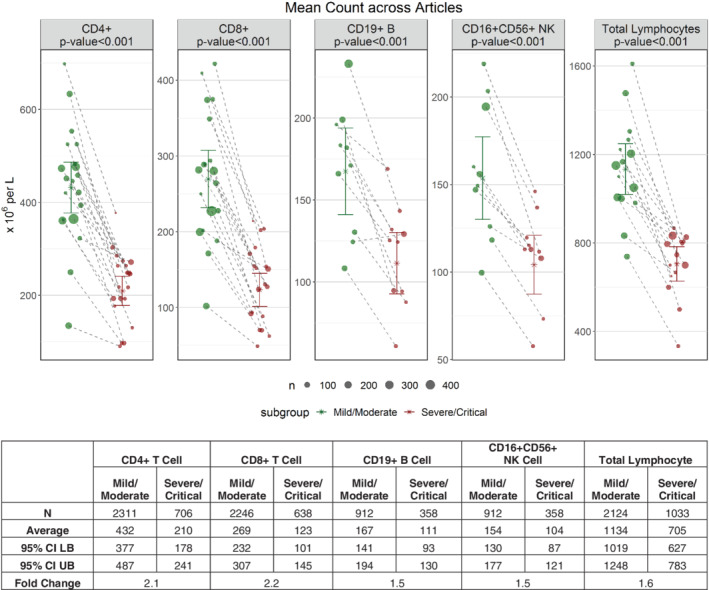
Mean cell counts for each individual lymphocyte subset in mild/moderate and severe/critical COVID‐19 patients across articles. Each solid circle represents the mean cell count in one article and the size of the circle represents the relative sample size for the mean. Paired data points from the same article are connected. The weighted mean value of cell counts across all articles is also shown with 95% confidence interval (CI) based on the random variability across articles. [Color figure can be viewed at wileyonlinelibrary.com]
Meta‐analysis was performed to calculate the standardized mean difference (SMD) and the 95% confidence interval (95% CI) between the mild/moderate and severe/critical groups for total lymphocytes, CD4+ T cell, CD8+ T‐cell, CD19+ B‐cell, and CD16 + CD56+ NK‐cell counts. R version 3.6.1 (July 05, 2019) (27) with metafor (28) package was used for the analysis. A random effects model was used to account for heterogeneity between publications. The SMD for each parameter is summarized in Figure 2. For all parameters, the lymphocyte subset absolute counts were found to be significantly lower, on average, in subjects in the severe/critical group vs. the absolute counts in the mild/moderate group.
Figure 2.
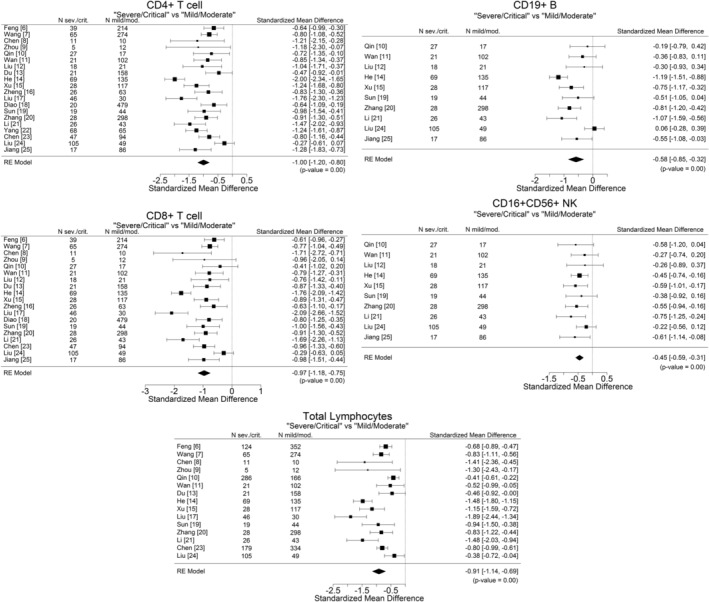
Standardized mean differences (SMDs) between mild/moderate and severe/critical disease groups, with 95% confidence interval (95%CI), in COVID‐19 patients. There is one panel for total lymphocytes, as well as panels for each subset. SMDs with CI results on the left of the 0 vertical line indicate a negative difference, i.e., the average mean count in the severe/critical group was significantly lower than in the mild/moderate group. Note that the width of the individual CIs is a function of sample size and reported variability within each publication.
These results suggest that lymphocyte subset absolute counts are linked to patient outcomes. While some of the meta‐analysis publications specified that cell counts were measured at or near hospital admission and tracked outcomes such as ICU admission or death, there is a need to assess this more thoroughly. A literature search was performed on outcome‐based studies that included multivariate analyses of laboratory measurements with at least one lymphocyte subset. Across these studies, CD4 or CD8 T cell counts were independently linked to key patient outcomes including mortality (13), ICU admission (29), viral clearance (30), and recovery (31) (see summary Table 1). In a prospective study of 179 patients with COVID‐19 pneumonia, CD8 T cells ≤75 cell/μl were independently linked to and predictive of mortality (13); other independent predictors included age > 65, cardiovascular or cerebrovascular disease, and cardiac troponin I > 0.05 ng/ml. In a retrospective study of 249 patients, CD4 T‐cell counts measured at the time of hospital admission were inversely correlated with subsequent ICU admission (29); age was the only other independent predictor. In a study of 60 patients, decreased CD8 T cells 1 week post‐treatment (as well as decreased B cells and increased CD4/CD8 ratios) were associated with poor treatment efficacy (30); this study controlled for age, sex, oxygen inhalation, antiviral treatment, disease severity on admission, and use of corticosteroid and immune enhancers. In a study of 292 patients in which 66 recovered after treatment, the CD4 T‐cell count measured before treatment was the only parameter measured that predicted the length of time before viral RNA clearance (31). These findings suggest that CD4 and CD8 T‐cell absolute counts may be valuable biomarkers in the prognosis of disease severity and recovery in COVID‐19 patients.
Table 1.
Lymphocyte subsets associated with COVID‐19 outcomes in multivariate analyses
| Lymph subset | Outcome | Study type | N | Statistical analysis (Odds Ratio, 95% CI, P value) | Ref |
|---|---|---|---|---|---|
| CD8 T | Mortality | Prospective | 179 (21 died) |
CD8 T cell <75/μl (3.982, 1.132–14.006, <0.001) |
(13) |
| CD4 T | ICU Admission | Retrospective | 249 (22 admitted ICU) | CD4 T cell at hospital admission (0.55 per 100 cells/μl increase, 0.33–0.92, 0.02) | (29) |
| CD8 T CD19 B | Treatment efficacy | Prospective | 60 (37 responders) |
Post‐treatment decrease CD8 T (0.0056, 0.006–0.516, 0.011) CD19 B (0.033, 0.002–0.439, 0.010) |
(30) |
| CD4 T | Viral clearance (stool) | Retrospective | 292, 66 recovered, 55 viral clearance (stool) | CD4 T cell (P = 0.010) | (31) |
discussion
Since the outbreak of COVID‐19 in December 2019, a remarkable amount of observational clinical data has been published within a relatively short period of time. Although our understanding of viral replication and clinical manifestation is still in the early stages, some consistent clinical characteristics of the disease are emerging. Systematic review and meta‐analysis studies of several clinical parameters have revealed that the severity of COVID‐19 disease correlates with low blood albumin (32), hypertension (33), thrombocytopenia (34), and increased blood levels of IL‐6 or Procalcitonin (35, 36). A recent meta‐analysis reported by Huang et al. showed that lymphopenia correlates with several poor patient outcomes, including mortality, Acute Respiratory Distress Syndrome, ICU care, and severe diseases (37). COVID‐19 disease severity has also been linked to the lymphocyte‐to‐neutrophil cell count ratio and lymphocyte‐to‐CRP (C‐reactive protein) ratio (38).
This meta‐analysis shows that absolute counts of major lymphocyte subsets are significantly and substantially decreased in severe COVID‐19 disease. The results remain consistent despite the differences in the definition of disease severity across the studies, the variations in blood specimen acquisition times, laboratory practices, and clinical care. Multivariate analyses reviewed here establish immune cell subset counts, particularly CD4+ and CD8+ T‐cell counts, as independent predictors of COVID‐19 outcomes. A limitation of this analysis is that all the studies were performed in China, and COVID‐19 is now a global pandemic.
The pathogenesis of COVID‐19 is still under investigation, and the precise mechanism(s) of the observed reduction in lymphocyte subset counts in the peripheral blood of patients with severe disease remain to be fully elucidated. Similar immune cell depletion has been reported in SAR‐CoV‐1 and MERS patients (39). It has been suggested that the reduction of immune cell counts in the peripheral blood during viral infection may be caused by the mobilization of immune cells to sites of infection, such as the lungs, and potentially by virus‐induced destruction of T cells (40). Future prospective studies designed to investigate the utility of lymphocyte subset measurements as prognostic biomarkers of disease severity, mortality, and response to treatment in patients infected with SARS‐CoV‐2 are strongly encouraged.
AUTHOR CONTRIBUTIONS
Wei Huang: Conceptualization; data curation; formal analysis; writing‐original draft. Julie Berube: Data curation; formal analysis; writing‐original draft. Michelle McNamara: Data curation; investigation; writing‐review and editing. Suraj Saksena: Data curation; methodology. Marsha Hartman: Data curation; investigation. Tariq Arshad: Investigation; resources; supervision; writing‐review and editing. Scott Bornheimer: Data curation; formal analysis; investigation; methodology; writing‐original draft; writing‐review and editing. Maurice O'Gorman: Data curation; writing‐original draft; writing‐review and editing.
Supporting information
Table S1 Study characteristic and demographics of patients in publications included in the meta‐analysis
LITERATURE CITED
- 1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med 2020;382:1199–1207. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cao X. COVID‐19: Immunopathology and its implications for therapy. Nat Rev Immunol 2020;20(5):269–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID‐19): A meta‐analysis. Clin Chem Lab Med 2020;58(7):1021–1028. 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 5. Kermali M, Khalsa RK, Pillai K, Ismail Z, Harky A. The role of biomarkers in diagnosis of COVID‐19 ‐ A systematic review. Life Sci 2020;254:117788. 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feng Y, Ling Y, Bai T, Xie Y, Huang J, Li J, Xiong W, Yang D, Chen R, Lu F, et al. COVID‐19 with different severity: A multi‐center study of clinical features. Am J Respir Crit Care Med 2020;201(11):1380–1388. 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang L, He W, Yu X, Hu D, Bao M, Liu H, Zhou J, Jiang H. Coronavirus disease 2019 in elderly patients: Characteristics and prognostic factors based on 4‐week follow‐up. J Infect 2020;80(6):639–645. 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020;130(5):2620–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Y, Zhang Z, Tian J, Xiong S. Risk factors associated with disease progression in a cohort of patients infected with the 2019 novel coronavirus. Ann Palliat Med 2020;9(2):428–436. [DOI] [PubMed] [Google Scholar]
- 10. Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, et al. Dysregulation of Immune Response in Patients with COVID‐19 in Wuhan, China. Clin Infect Dis 2020. ciaa248. 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wan S, Yi Q, Fan S, Lv J, Zhang X, Guo L, Lang C, Xiao Q, Xiao K, Yi Z, et al. Relationships among lymphocyte subsets, cytokines, and the pulmonary inflammation index in coronavirus (COVID‐19) infected patients. Br J Haematol 2020;189(3):428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Z, Long W, Tu M, Chen S, Huang Y, Wang S, Zhou W, Chen D, Zhou L, Wang M, et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID‐19. J Infect 2020. 10.1016/j.jinf.2020.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Du RH, Liang LR, Yang CQ, Wang W, Cao TZ, Li M, Guo GY, Du J, Zheng CL, Zhu Q, et al. Predictors of mortality for patients with COVID‐19 pneumonia caused by SARS‐CoV‐2: A prospective cohort study. Eur Respir J 2020;55(5):2000524. 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He R, Lu Z, Zhang L, Fan T, Xiong R, Shen X, Feng H, Meng H, Lin W, Jiang W, et al. The clinical course and its correlated immune status in COVID‐19 pneumonia. J Clin Virol 2020;127:104361. 10.1016/j.jcv.2020.104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu B, Fan CY, Wang AL, Zou YL, Yu YH, He C, Xia WG, Zhang JX, Miao Q. Suppressed T cell‐mediated immunity in patients with COVID‐19: A clinical retrospective study in Wuhan, China. J Infect. 2020;81(1):e51–e60. 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng Y, Xu H, Yang M, Zeng Y, Chen H, Liu R, Li Q, Zhang N, Wang D. Epidemiological characteristics and clinical features of 32 critical and 67 noncritical cases of COVID‐19 in Chengdu. J Clin Virol 2020;127:104366. 10.1016/j.jcv.2020.104366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation between relative nasopharyngeal virus RNA load and lymphocyte count disease severity in patients with COVID‐19. Viral Immunol 2020. 10.1089/vim.2020.0062. [DOI] [PubMed] [Google Scholar]
- 18. Bo D, Wang C, Tan Y, Xiewan C, Ying L, Lifen N, Li C, Min L, Yueping L, Wang G, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID‐19). Front Immunol 2020;11:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun Y, Dong Y, Wang L, Xie H, Li B, Chang C, Wang FS. Characteristics and prognostic factors of disease severity in patients with COVID‐19: The Beijing experience. J Autoimmun 2020;2020:102473. 10.1016/j.jaut.2020.102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X, Tan Y, Ling Y, Lu G, Liu F, Yi Z, Jia X, Wu M, Shi B, Xu S, et al. Viral and host factors related to the clinical outcome of COVID‐19. Nature 2020. 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 21. Li S, Jiang L, Li X, Lin F, Wang Y, Li B, Jiang T, An W, Liu S, Liu H, et al. Clinical and pathological investigation of severe COVID‐19 patients. JCI Insight 2020;2020:138070. 10.1172/jci.insight.138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang P, Wang P, Song Y, Zhang A, Yuan G, Cui Y. A retrospective study on the epidemiological characteristics and establishment of early warning system of severe COVID‐19 patients. J Med Virol 2020. 10.1002/jmv.26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen R, Sang L, Jiang M, Yang Z, Jia N, Fu W, Xie J, Guan W, Liang W, Ni Z, et al. Longitudinal hematologic and immunologic variations associated with the progression of COVID‐19 patients in China. J Allergy Clin Immunol 2020;S0091‐6749(20):30638‐2. 10.1016/j.jaci.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu R, Wang Y, Li J, Han H, Xia Z, Liu F, Wu K, Yang L, Liu X, Zhu C. Decreased T cell populations contribute to the increased severity of COVID‐19. Clin Chim Acta 2020;508:110–114. 10.1016/j.cca.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang M, Guo Y, Luo Q, Huang ZK, Zhao R, Liu SY, Le AP, Li JM, Wan LG. T cell subset counts in peripheral blood can be used as discriminatory biomarkers for diagnosis and severity prediction of COVID‐19. J Infect Dis 2020;222(2):198–202. 10.1093/infdis/jiaa252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol 2014;14:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Core Team R. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna: Austria, 2013. http://www.R-project.org/. [Google Scholar]
- 28. Viechtbauer W. Conducting meta‐analyses in R with the metafor package. J Stat Softw 2010;36(3):1–48. [Google Scholar]
- 29. Chen J, Qi T, Liu L, Ling Y, Qian Z, Li T, Li F, Xu Q, Zhang Y, Xu S, et al. Clinical progression of patients with COVID‐19 in Shanghai, China. J Infect 2020;80(5):e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang F, Nie J, Wang H, Zhao Q, Xiong Y, Deng L, Song S, Ma Z, Mo P, Zhang Y. Characteristics of peripheral lymphocyte subset alteration in COVID‐19 pneumonia. J Infect Dis 2020;221(11):1762–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai FH, Wu F, Song ZG, Huang W, Chen J, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl) 2020;133(9):1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aziz M, Fatima R, Lee‐Smith W, Assaly R. The association of low serum albumin level with severe COVID‐19: A systematic review and meta‐analysis. Crit Care. 2020;24(1):255. 10.1186/s13054-020-02995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pranata R, Lim MA, Huang I, Raharjo SB, Lukito AA. Hypertension is associated with increased mortality and severity of disease in COVID‐19 pneumonia: A systematic review, meta‐analysis and meta‐regression. J Renin Angiotensin Aldosterone Syst 2020;21(2):1470320320926899. 10.1177/1470320320926899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID‐19) infections: A meta‐analysis. Clin Chim Acta 2020;506:145–148. 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ulhaq ZS, Soraya GV. Interleukin‐6 as a potential biomarker of COVID‐19 progression. Med Mal Infect 2020;50(4):382–383. 10.1016/j.medmal.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lippi G, Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID‐19): A meta‐analysis. Clin Chim Acta 2020;505:190–191. 10.1016/j.cca.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang I, Pranata R. Lymphopenia in severe coronavirus disease‐2019 (COVID‐19): Systematic review and meta‐analysis. J Intensive Care 2020;8:36. 10.1186/s40560-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lagunas‐Rangel FA. Neutrophil‐to‐lymphocyte ratio and lymphocyte‐to‐C‐reactive protein ratio in patients with severe coronavirus disease 2019 (COVID‐19): A meta‐analysis. J Med Virol 2020. 10.1002/jmv.25819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Brüggen M‐C, O'Mahony L, Gao Y, Nadeau K, Akdis CA. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy 2020. 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merad M, Martin JC. Pathological inflammation in patients with COVID‐19: A key role for monocytes and macrophages. Nat Rev Immunol 2020;20(6):355–362. 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Study characteristic and demographics of patients in publications included in the meta‐analysis


