Abstract
Lab- and pilot-scale simultaneous nitrification, denitrification and phosphorus removal-sequencing batch reactors were operated under cyclic anaerobic and micro-aerobic conditions. The use of oxygen, nitrite, and nitrate as electron acceptors by Candidatus Accumulibacter phosphatis during the micro-aerobic stage was investigated. A complete clade-level characterization of Accumulibacter in both reactors was performed using newly designed qPCR primers targeting the polyphosphate kinase gene (ppk1). In the lab-scale reactor, limited-oxygen conditions led to an alternated dominance of Clade IID and IC over the other clades. Results from batch tests when Clade IC was dominant (i.e., >92% of Accumulibacter) showed that this clade was capable of using oxygen, nitrite and nitrate as electron acceptors for P uptake. A more heterogeneous distribution of clades was found in the pilot-scale system (Clades IIA, IIB, IIC, IID, IA, and IC), and in this reactor, oxygen, nitrite and nitrate were also used as electron acceptors coupled to phosphorus uptake. However, nitrite was not an efficient electron acceptor in either reactor, and nitrate allowed only partial P removal. The results from the Clade IC dominated reactor indicated that either organisms in this clade can simultaneously use multiple electron acceptors under micro-aerobic conditions, or that the use of multiple electron acceptors by Clade IC is due to significant microdiversity within the Accumulibacter clades defined using the ppk1 gene.
Keywords: Low-DO nutrient removal, SNDPR, PAO, DPAO, Accumulibacter, ppk1 gene
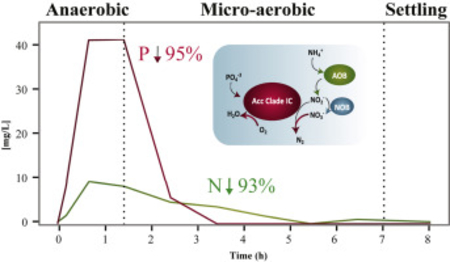
Graphical Abstract
1. Introduction
Phosphorus (P) and nitrogen (N) are the main nutrients triggering the growth of algae, which cause eutrophication in water bodies. Hence, the removal of N and P from wastewater before being discharged into the environment has become a worldwide concern. In conventional biological nutrient removal (BNR) systems, N removal is achieved by a two-stage treatment, that is, aerobic nitrification followed by anoxic denitrification. During the aerobic stage, ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB) oxidize NH4+ to NO2−, and NO2− to NO3−, respectively (Metcalf et al., 2013). These oxidized nitrogen species are used as electron acceptors during denitrification. P removal is achieved by polyphosphate-accumulating organisms (PAOs) (Comeau et al., 1986) through enhanced biological phosphorus removal (EBPR) under alternating anaerobic–aerobic conditions. Processes for BNR typically consist of anaerobic, anoxic, and aerobic zones for phosphorus release, denitrification and, nitrification and phosphorus uptake, respectively.
BNR processes are typically operated under elevated levels of dissolved oxygen (DO) in the aeration basin to ensure complete nitrification and P removal. However, aeration is an energy-intensive operation, which can account for 60% or more of the overall power consumption at a wastewater treatment plant (WWTP) (WEF, 1997). In addition, P release and denitrification reactions require a biodegradable organic carbon source to serve as an electron donor, and the availability of organic matter is often an essential limiting factor when simultaneous N and P removal is desired. When WWTPs need to meet rigorous effluent N limits, external carbon addition to support denitrification is necessary. Consequently, reducing energy requirements during aeration and optimizing the use of the organic matter available in the wastewater are current emphases in BNR research (Fitzgerald et al., 2015, Wang et al., 2015, Zhang et al., 2009, Zheng et al., 2009).
Simultaneous nitrification, denitrification, and P removal (SNDPR), achieved by alternating between anaerobic and low DO conditions, has the potential of significantly reducing both energy and carbon requirements in BNR. The SNDPR process has been previously demonstrated in lab-scale reactors (de Kreuk et al., 2005, Peng et al., 2008, Wang et al., 2015, Zeng et al., 2003, Zeng et al., 2004). It has been hypothesized that oxic and anoxic zone formation within microbial flocs facilitates the occurrence of simultaneous nitrification and denitrification (SND), as nitrite and nitrate produced by nitrifying organisms can be reduced by denitrifying PAOs (DPAOs) (Coma et al., 2010, Wang et al., 2015, Yilmaz et al., 2008).
The uncultured ‘Candidatus Accumulibacter phosphatis’ (hereafter referred to as Accumulibacter), a well-known PAO found in most full-scale EBPR systems and easily enriched in lab-scale reactors, seems to have the ability to denitrify (Flowers et al., 2009, Kim et al., 2013), and therefore, it is also considered a DPAO. Based on phylogenetic distance of the polyphosphate kinase gene (ppk1), Accumulibacter has been classified into two major divisions (Type I and II), each comprised of several distinct clades (He et al., 2007, Peterson et al., 2008). It has been proposed that environmental conditions determine the population structure of the Accumulibacter lineage (Peterson et al., 2008), and that the ability to denitrify may be associated with specific Accumulibacter clades or strains (Carvalho et al., 2007, Flowers et al., 2009, Kim et al., 2013).
Thus, the aim of this work was to investigate the clade-level population structure of Accumulibacter in reactors performing efficient P and N removal under micro-aerobic conditions. To achieve this goal, we first developed a new quantitative PCR (qPCR) primers collection targeting the ppk1 gene in each clade identified in the Accumulibacter lineage, and then applied this new primer collection to study two sequencing batch reactors (SBR) operated to achieve total nutrient removal under cyclic anaerobic and micro-aerobic conditions. A lab-scale SBR used acetate as the sole organic substrate, whereas a pilot-scale SBR reactor used primary effluent from the Nine Springs WWTP (Madison, WI). We demonstrate the improved performance of the new primer collection to differentiate Accumulibacter clades, and infer the ability of the organisms represented in clade IC to simultaneously use multiple electron acceptors under micro-aerobic conditions.
2. Material and methods
2.1. Operation of lab-scale sequencing batch reactor
A laboratory-scale SBR was originally inoculated with activated sludge obtained from the Nine Springs WWTP in Madison, WI, which uses a modified version of the University of Cape Town (UCT) process designed to achieve biological P removal (Zilles et al., 2002) and operates with high aeration rates (Park et al., 2006). Synthetic wastewater containing acetate as the sole carbon source was used for the feed. The 2-L reactor was operated under alternating anaerobic and low oxygen 8-h cycles. Each cycle consisted of 1.5 h of anaerobic reaction, 5.5 h of aerobic low-DO reaction, 50 min of settling and 10 min of decanting. During the micro-aerobic stage, an on/off control system was used to limit the amount of oxygen pumped to the reactor (0.02 L/min) and maintain low oxygen concentrations in the mixed liquor. During the first hours of aeration, the controller was not activated since the oxygen provided was close to the oxygen uptake, maintaining the DO concentration near 0.05 mg/L. When the oxygen uptake rate decreased as the substrates were consumed, the control system became active and kept the DO near the 0.2 mg/L set-point. A volume of 600 mL of synthetic wastewater was pumped into the reactor in the first 16 min of the anaerobic stage. The feed contained (per liter) 720 mg sodium acetate, 107 mg ammonium chloride, 48.4 mg potassium phosphate, 500 mg sodium bicarbonate, 37 mg potassium sulfate, 244 mg calcium chloride dehydrated, 166 g magnesium sulfate and 30 mL of trace element mixtures (Goel and Noguera, 2006). This feed corresponded to influent concentrations of 500 mg COD/L, 25.2 mg NH4+-N/L, and 9.88 mg PO43−-P/L. The reactor was operated with a hydraulic retention time (HRT) of 24 h and a sludge retention time (SRT) of 80 days. The pH in the system was controlled at 7.0–7.5 during the anaerobic and aerobic phases.
2.2. Operation of pilot-scale sequencing batch reactor
A pilot scale SBR with a working volume of 130 gallons was also operated to achieve SNDPR under cyclic anaerobic/micro-aerobic conditions (DO between 0.2 and 0.7 mg/L), similar to the operation of the lab-scale reactor. Each cycle consisted of an initial 2 h idle phase, 1.5 h of anaerobic conditions, 3.5 h of micro-aerobic conditions and 1 h of settling and decanting. This reactor received primary effluent from the Nine Springs WWTP, amended with acetate to provide an additional source of easily biodegradable organics. A summary of the chemical characteristics of the primary effluent measured throughout the course of this study is found in Supplementary Table 1. Wastewater temperatures ranged from 11 to 26 °C. The pH of the system was monitored but not controlled, and fluctuated between 7.0 and 8.0. The HRT and SRT of the reactor were 24 h and 80 days, respectively, similar to the operational conditions in the lab-scale reactor.
2.3. Sample collection and analytical tests
To monitor reactor performance, mixed liquor and effluent samples were collected, filtered through a membrane filter (0.45 μm; Whatman, Maidstone, UK) and analyzed for acetate, PO43−–P, NH4+-N, NO3−-N, and NO2−-N. Unfiltered samples were also weekly retrieved for volatile suspended solids (VSS) and total suspended solids (TSS) measurements. The concentrations of PO43−-P, VSS, and TSS were determined according to Standard Methods (APHA, 2005). Total ammonia (NH3 + NH4+) concentrations were analyzed using the salicylate method (Method 10031, Hach Company, Loveland, CO). Acetate, nitrite and nitrate were measured using high-pressure liquid chromatography on a Prevail™ Organic Acids (Discovery Sciences, Deerfield, IL) column with a mobile phase consisting of 25 mM KH2PO4, pH adjusted to 2.5 with phosphoric acid, and detection by UV at 210 nm for acetate and 214 nm for nitrite and nitrate.
Biomass samples from the reactors and from the full-scale activated sludge basin at the Nine Springs WWTP were collected weekly and stored at −80 °C until DNA extraction was performed. DNA was extracted using UltraClean® Soil DNA Isolation Kit (MoBIO Laboratories, Carlsbad, CA). Extracted DNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and stored at −80 °C.
2.4. Batch tests
During reactor operation, batch tests were carried out to investigate N and P removal under different electron accepting (oxygen, nitrate, and nitrite) conditions. Batch test experiments with sludge from the lab-scale reactor were performed in the same reactor vessel. Tests with sludge from the pilot-scale reactor were performed with 2L of sludge retrieved from the end of the anaerobic stage. The anoxic or aerobic phase was introduced by injecting a concentrated solution of nitrite or nitrate (25 mg N/L in the reactor), or by sparging the reactor with air, maintaining the same oxygen level as in the corresponding parent reactor. Allylthiourea (ATU) was added to oxic batch tests (5 mg/L in the reactor) with sludge from the pilot- and lab-scale reactors to prevent nitrification. Similar experiments with simultaneous presence of nitrate and oxygen were performed in the lab-scale SBR.
2.5. Ribosomal RNA gene-based tag sequencing
The bacterial composition in samples from each bioreactor and from the Nine Springs WWTP was determined via the analysis of high-throughput sequencing of 16S rRNA gene fragments. 45 samples from the lab-scale SBR retrieved during 2-years of operation, 23 samples from the pilot-scale SBR retrieved during 7 months, and 38 samples from the full-scale WWTP Nine Springs stored during 10 months were included in this analysis. The hyper-variable V3–V4 regions of the bacterial 16S rRNA gene were amplified using the primers 515f/806r (Caporaso et al., 2011). PCR products used to generate the sequencing libraries were generated in 25 μL reaction volumes using the Ex Taq kit (Takara) with 200 nM each of the forward and reverse primer and 2 μL of template nucleic acid. Cycling conditions involved an initial 5 min denaturing step at 95 °C, followed by 35 cycles at 95 °C for 45 s, 50 °C for 60 s, 72 °C for 90 s, and a final elongation step at 72 °C for 10 min. Each barcode corresponded to an eight-base sequence unique to each sample. Amplicons were visualized on an agarose gel to confirm product sizes. Purified amplicons were pooled in equimolar quantities and sequenced on an Illumina Miseq benchtop sequencer using pair-end 250 bp kits at the Cincinnati Children’s Hospital DNA Core facility.
Paired-end reads obtained were merged, aligned, filtered and binned into operational taxonomic units (OTU) with 97% identity using the QIIME pipeline (Caporaso et al., 2010). Chimeric sequences were removed using UCHIME (Edgar et al., 2011). The most representative sequences from each OTU were taxonomically classified using the MIDas-DK database (Mielczarek et al., 2013).
2.6. ppk1 gene clone library construction
PCR amplification of Accumulibacter ppk1 fragments from six sludge samples (3 lab-scale SBR samples, 2 pilot-scale SBR samples and 1 sample from the Nine Springs WWTP) was carried out on extracted genomic DNA in a 25 μL reaction volume with 400 nM of each forward and reverse primer (Acc-ppk1–254f and Acc-ppk1–1376r) (McMahon et al., 2007). The PCR program consisted of an initial 10-min denaturation step at 95 °C, followed by 30 cycles of 95 °C for 30 s, 68 °C for 1 min, and 72 °C for 2 min, and then a final extension at 72 °C for 5 min. The amplified ppk1 fragments (1137 bp) were then purified, cloned using a TOPO TA cloning kit (Invitrogen, CA) according to the manufacturer’s instructions. A total of 721 ppk1 fragments were single-pass Sanger sequenced, and these sequences were further grouped into OTUs based on 99% identity of DNA sequences within each library. ppk1 sequence classification was conducted by sequence alignment to ppk1 sequences available in the literature and phylogenetic tree reconstruction as described below.
2.7. Phylogenetic analysis
ppk1 sequences recovered from the clone library were aligned using the ‘AlignSeqs’ command in the DECIPHER “R” package (Wright, 2015) along with a dataset of Accumulibacter-ppk1 sequences (>1000 bp) retrieved from previous studies (Gonzalez-Gil and Holliger, 2011, He et al., 2007, Kim et al., 2010, Kim et al., 2013, Mao et al., 2015, McMahon et al., 2002, McMahon et al., 2007, Peterson et al., 2008, Wexler et al., 2009, Wilmes et al., 2008). Chimeric sequences were removed using the de novo chimera detection feature of UCHIME (Edgar et al., 2011). The alignment was trimmed to 1020 bp, prior to phylogenetic tree construction using neighbor-joining criterion with 1000 bootstrap tests for every node using the MEGA6 software package (Tamura et al., 2013) (Fig. S2).
2.8. Clade-specific primer design
Since existing primer sets targeting Accumulibacter clades amplify only a subset of known diversity within the lineage (current targets are Type I, Clades IIA, IIB, IIC and IID (He et al., 2007)), and the database of ppk sequences has significantly increased in the last decade, we re-evaluated the original primers and designed a new primer collection to target the ppk1 gene in each of the fourteen clades identified in the Accumulibacter lineage (clades A-E in Type I and clades A-I in Type II) (Table 1).
Table 1.
Primers designed to target 14 different Accumulibacter clades, with qPCR conditions and performance.
| Target clade | Name | Forward primer (5′–3′) | Reverse primer (5′–3′) | Amplicon size (bp) | qPCR performance |
||
|---|---|---|---|---|---|---|---|
| Std curve correlation coefficient | PCR efficiency (%) | Cross-hybridization (%) | |||||
| IA | AccIA-ppk1–978f/1058r | GCGCGACAATCTCAAATTCAA | ATGGCCTCGAAGAGGTTGC | 81 | 0.999 ± 0.001 | 105.3 ± 3.1 | 0.1 |
| TGGCCTCGAAAACGTTGC | |||||||
| IB | AccIB-ppk1–884f/1009r | TGCTTGGCCACTTCAACC | GCTTGAAGGGCTGGAACT | 126 | 0.999 ± 0.000 | 102.0 ± 3.5 | 0.12 |
| IC | AccIC-ppk1–815f/911r | GCGACACTTTGGTAATGCG | CGCTCGGTGAGGTCGAA | 96 | 0.998 ± 0.001 | 103.0 ± 4.6 | 0.15 |
| CGACACTTTGGCAATGCG | |||||||
| ID | AccID-ppk1–559f/666r | GGCCGCAGCTCTGATGC | GACAGGAAGACGAAGGTGTAT | 108 | 0.996 ± 0.005 | 107.3 ± 7.0 | 0 |
| GGTCGCAGCTCTAACGC | CGACAGGAATACGAAGGTGTAT | ||||||
| IE | AccIE-ppk1–782f/954r | AATCTGCGCACCAAGATC | GGCGGTGCAGATTGACC | 175 | 1.000 ± 0.000 | 100.0 ± 1.0 | 0 |
| ATCTGCGCGCCAAAATC | AGTCGGTGCAGATTGACC | ||||||
| IIA | AccIIA-ppk1–344f/447r | ACGACGAAATCCTGCCC | GCGGAAGAAGAAGTCGCT | 104 | 0.999 ± 0.001 | 102.3 ± 2.3 | 0.02 |
| IIB | AccIIB-ppk1–327f/654r | GCAGTACCAGTTGCTCAATG | GAAGGCGTACTCGACATC | 328 | 0.996 ± 0.004 | 98.7 ± 1.5 | 0.01 |
| IIC | AccIIC-ppk1–635f/794r | GCGACAGTGAGTACGCCT | TTGGCGCGCAGATTGGT | 160 | 0.999 ± 0.000 | 99.7 ± 6.0 | 0.04 |
| IID | AccIID-ppk1–892f/1237r | CAGTTCAACCTCACCGAGAC | GGTTGCCCAGCCGATATTG | 408 | 0.999 ± 0.001 | 97.3 ± 5.9 | 0.01 |
| IIE | AccIIE-ppk1–646f/768r | ATTCGTTCGTTTTCCTGTCTT | TCGTCCACGAAGAGGTC | 122 | 0.993 ± 0.000 | 91.7 ± 5.8 | 0.02 |
| IIF | AccIIF-ppk1–887f/1057r | TCGGCCAGTTCAATCTGAC | TGCCGTCGAAGACGCTC | 171 | 0.999 ± 0.001 | 92.0 ± 7.2 | 0.01 |
| IIG | AccIIG-ppk1–410f/945r | CCGAGCAACGCGAATGG | AGGTTGACCGGGCCAAG | 535 | 0.997 ± 0.001 | 94.7 ± 5.5 | 0 |
| IIHa | AccIIH-ppk1–642f/934r | GAATACTCCTTCGTATTCCTCTCTTC | GGCCTGCCACCCGGTAG | 292 | – | – | – |
| II-Ia | AccII-I-ppk1–820f/1164r | CTGCTCGGACAGTTCAATCTTT | TTCTGCGCGGCTCCTAT | 344 | – | – | – |
Primers targeting Clades IIH and II-I were not validated in this study.
For the design of new primers, a list of aligned ppk1 sequences retrieved from the clone library and literature was submitted to the DECIPHER’s Design Primers web tool (Wright et al., 2014) and using the following parameters: primers length ranging from 17 to 26 nucleotides with up to 2 permutations, PCR product amplicon length between 75 and 400 bp and at least 90% of sequence coverage. All primers were designed to function at the same annealing temperature (64 °C) in the PCR reaction. The program selected specific regions within the highly similar 14 Accumulibacter clades, which resulted in a set of primers with high coverage and high specificity for each clade (Table 1).
Experimental validation of primer design was carried out for 12 of the 14 primer sets. Templates containing the ppk1 sequence amplified with primers Acc-ppk1–254f and Acc-ppk1–1376r were obtained from either the clone library constructed in this study (clades IA-IC and IIA-IIG) or by gene synthesis (clade ID and IE) (IDT, Inc., Coralville, IA). Templates for clades IIH and II-I were not available in our collection, and commercial gene synthesis of these ppk1 fragments was not successful. Therefore, validation of the primers targeting these two clades was not carried out. The presence and sizes of the amplification products were determined by agarose (2%) gel electrophoresis of the reaction product, using as template ppk1 from clones and DNA samples from the lab- and pilot-scale reactors (Fig. S3). The new set of primers consistently amplified the expected DNA fragment (Table 1) in all samples with positive results and no other band was obtained from PCR. For the validated primers, measured PCR efficiencies ranged from 91.7% to 107.3%, and the standard curve correlation coefficients were always greater than 0.993, verifying the quality of the qPCR amplifications (Table 1). In addition, specificity was evaluated by having each primer undergo combinatorial testing in reactions that contained 107 copies of template from each of the sub-clades. The high specificity observed in this rigorous test (Fig. S4), where qPCR was performed in the absence of perfectly matched template and the presence of the highest concentrations of mismatched templates used in the calibration curves, demonstrates the effectiveness of the design approach. The highest potential for cross-hybridization occurred with the IC primer set and the IE template, and with the primer set IB and IC template. In both cases, amplification was observed after 30 qPCR cycles, resulting in quantifications that were at least 3 orders of magnitude lower than the template concentration (i.e., 0.15% and 0.11% cross-hybridization potential, respectively) and indicating that such level of cross-hybridization would have negligible effects on quantifications from samples that are expected to have much lower concentrations of the ppk1 gene (i.e., highest quantification was ∼104 copies in this study).
For the re-evaluation of the original primer set (He et al., 2007), similar cross-hybridization experiments as described above were performed in reactions containing ppk1 template from each of the sub-clades (Fig. S5). These experiments revealed amplification of clade IID with the IIA primer set and clade IIE with the IID primer set (4.27% and 33.85% of cross-hybridization potential). Complete cross-hybridization (∼100% amplification) of IIC primers with ppk1 sequences from clade IA and IC and a lower cross-hybridization extent with clades IB, IIB, IID, IIE, IIF and IIG (79%, 61%, 42%, 88%, 77% and 90% of amplification, respectively) was also identified.
In addition, quantification of clades IIA, IIB, IIC and IID using these set of primers was performed in 16 samples from both the lab- and pilot-scale reactors and the results were compared to the number of ppk1 copies/ng of DNA estimated using the primers designed in this study (Fig. S6). Results from this analysis indicated over-estimation in the abundance of clades IIA and IIC in most samples when using the previously designed primers. Overall, these results revealed the lack of specificity from the previous Accumulibacter clade-specific primer set due to cross-hybridization with non-target clades, and therefore, confirmed the need to re-design clade-specific ppk1-targeting primers.
2.9. Quantitative polymerase chain reaction (qPCR)
Quantification of total 16S rRNA genes, Accumulibacter 16S rRNA genes, and ppk1 in each DNA sample was carried out by qPCR. All qPCR reactions were run in a LightCycler 480 system (Roche applied Science, Indianapolis, IN). Each reaction volume was 20 μL and contained 10 μL iQ™ SYBR® Green Supermix (BioRAD Laboratories, Hercules, CA), 0.8 μL each of 10 μM forward and reverse primer, 2.4 μL nuclease free water and 4 μL of sample. Templates for calibration curves of 16S rRNA-targeted PCR were obtained from the Accumulibacter 16S gene clone library generated in (He et al., 2007). Templates for ppk1-based qPCR were obtained from clone collections or gene synthesis, as described above. In all cases, ten-fold serial dilutions of each template (ranging from 101 to 107 copies per reaction) were used to generate qPCR calibration curves. All samples were processed in triplicates and each reaction plate contained non-template controls. Amplification efficiency of PCR was estimated from the slope of the standard curve by the formula 10−1/slope − 1. Standard deviations were calculated from the average of the triplicate runs.
The 16S rRNA-targeted primer pair 341F/534R was used to quantify total cell abundance, according to Yoshida et al. (2005). To quantify total Accumulibacter 16S rRNA genes, universal primer 518f and PAO-846r were used according the procedure described by He et al. (2007). Clade-specific quantification with the formerly designed ppk1 primer set targeting Clade IIA (Acc-ppk1–893f and Acc-ppk1–997r), Clade IIB (Acc-ppk1–870f and Acc-ppk1–1002r), Clade IIC (Acc-ppk1–254f and Acc-ppk1–460r) and Clade IID (Acc-ppk1–375f and Acc-ppk1–522r) was carried out according to the conditions described in He et al. (2007). The thermal cycling protocol for clade-specific quantification with the ppk1 primers (Table 1) was as follows: initial denaturation at 95 °C for 30 s, followed by 45 cycles of denaturation at 95 °C for 30 s, annealing at 64 °C for 30 s, and extension at 72 °C for 30 s.
The fractional relative abundance of Accumulibacter (%Acc) within the bacterial community was calculated by two different methods: (1) based on the Accumulibacter ppk1 (Accppk1) abundance with respect to the total bacterial 16S rRNA (Bact16SrRNA) and (2) based on the quantifications of Accumulibacter 16S rRNA (Acc16SrRNA) with respect to the total bacterial 16S rRNA (Bact16SrRNA) (Eqs. (1), (2))). It was assumed that the Accumulibacter genome has 1 copy of the ppk1 gene and 2 copies of the rrn operon while bacterial genomes on average carried 4.1 copies of rrn operon, as has been assumed previously (He et al., 2007, Kaetzke et al., 2005, Mao et al., 2015).
| (1) |
| (2) |
A comparison of the results of these two methods, along with the sum of individual Accumulibacter clade abundance quantified with the primers designed in this study is presented in Table S2. Although there are some discrepancies in the quantifications by different methods, the trends of abundance over time were highly correlated, and thus, any of the methods can be used to compare relative abundances across samples.
2.10. Validation of the accumulibacter population structure by metagenomic analysis
Metagenomic data from lab-scale (days 218, 478 and 671) and pilot-scale SBR (day 305) samples (NCBI Short Read Archive, accession SRP075714) was used to confirm the Accumulibacter population structure predicted by qPCR. Paired-end DNA reads were filtered and quality trimmed with Sickle (https://github.com/ucdavis-bioinformatics/sickle.git). The filtered reads were then mapped to the Accumulibacter-ppk1 database generated in this study using the software package BBMap version 35.85 (https://sourceforge.net/projects/bbmap/). For each clade, the number of reads mapping to ppk1 sequences with a minimum alignment identity of 95% were quantified. The relative abundance of each Accumulibacter-clade in each sample was calculated as the number of reads mapping to ppk1 from the clade divided by the total number of reads mapping to the database (Fig. S7).
2.11. Nucleotide sequence accession numbers
The GenBank accession numbers for the ppk1 nucleotide sequences determined in this study are KU529869-KU529939.
3. Results
3.1. Electron acceptors used for phosphorus removal in lab-scale reactor
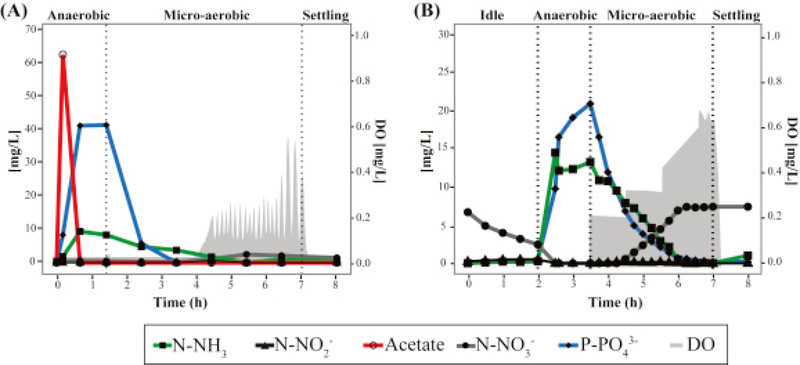
Results from a typical cycle of the lab-scale SBR at steady-state operation are shown in Fig. 1A. Acetate added at the beginning of the anaerobic stage is completely consumed within an hour. Simultaneously, P release to the mixed liquor is observed. In the subsequent micro-aerobic stage, DO is kept at 0.05 ± 0.01 mg/L during active P uptake by providing aeration at a rate equivalent to the oxygen uptake rate. The oxygen uptake rate decreases after all of the P is removed (2 h into the micro-aerobic stage), and consequently, the DO concentration begins to rise. To control DO after this point, the aeration rate is decreased by turning off aeration when DO gets over a set point of 0.2 mg/L, and turning aeration back on when DO goes below this set point. Nitrification occurs without nitrite or nitrate accumulation during the portion of the micro-aerobic stage when P is actively taken up, and then nitrate begins accumulating only after all of the P has been removed (Fig. 1A). This mode of operation results in efficient total N and P removal with minimal aeration. During the time this reactor operated, nitrate accumulation averaged 11% of the ammonium present at the beginning of the micro-aerobic stage, indicating a significant level of simultaneous nitrification and denitrification. During two years of stable operation, N and P removal in the reactor were 93 ± 9% and 95 ± 10%, respectively.
Fig. 1.
Nutrients profile in a regular cycle during day 909 and day 555 of operation of the (A) lab-scale and (B) pilot-scale SBRs, respectively. Grey areas correspond to the oxygen concentration during the micro-aerobic stage.
In order to characterize nitrogen transformations under low-DO conditions a batch test where acetate was removed from the influent was carried (Fig. S1). In this case, 67% of the ammonium in the influent was transformed to nitrate, while no nitrite accumulation was observed in the system, consistent with the presence of ammonia-oxidizing and nitrite-oxidizing organisms. Additionally, when ATU was added to the sludge to inhibit nitrification (Fig. 2C), only 27% of N-NH3 was removed from the reactor during the micro-aerobic stage, confirming that the majority of nitrogen removed in this system was due to simultaneous nitrification and denitrification.
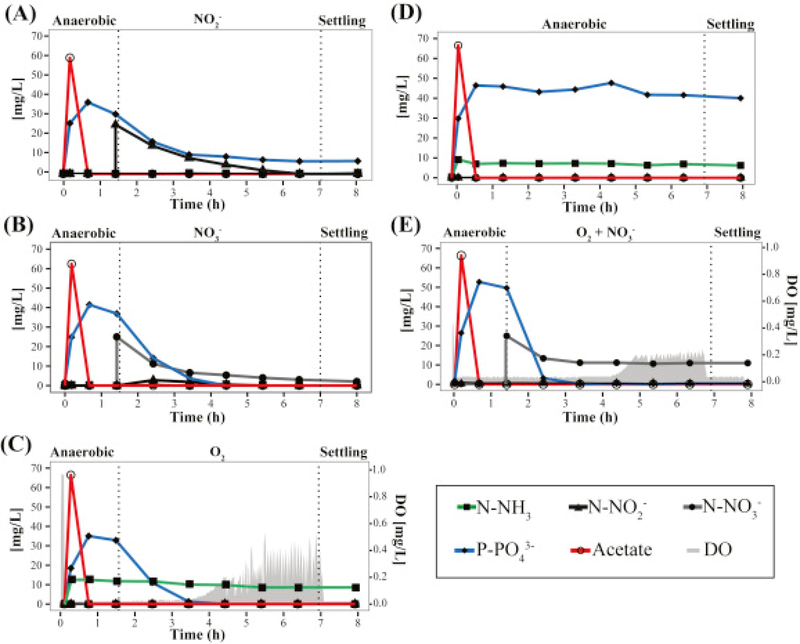
Fig. 2.
Evaluation of (A) nitrite, (B) nitrate and (C) oxygen as electron acceptors for P uptake in the lab-scale SBR. (D) Batch tests with no electron acceptor added for P uptake (anaerobic cycle). (E) Co-respiration of oxygen and nitrate during P uptake. Experiments were performed when Clade IC represented >92% of Accumulibacter in the reactor.
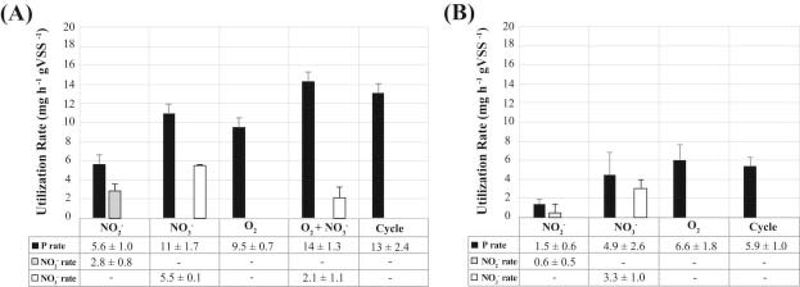
Electron acceptors available during normal operation at micro-aerobic conditions include oxygen, nitrite, and nitrate, with the latter two produced by ammonium oxidation at the low-DO conditions employed. Thus, in order to measure the contribution of each electron acceptor to P uptake, we conducted batch experiments by modifying the conditions of one of the reactor cycles (Fig. 2A–E). In all cases, the reactor was operated without the addition of ammonium to the medium. In one batch experiment (Fig. 2A) aeration was not provided and nitrite was added at the end of the anaerobic stage to create anoxic conditions with nitrite as the only electron acceptor. A similar batch experiment was performed, but with nitrate as the only electron acceptor (Fig. 2B). The observed P-uptake rates during the first hour of these tests were 5.7 ± 1.0 and 11 ± 1.7 mg P h−1 gVSS−1 for nitrite and nitrate, respectively (see Fig. 3 for rate comparisons; standard deviations represent variability from duplicate experiments). These experiments demonstrate that although nitrite and nitrate can both be used as electron acceptors by DPAO within the reactor, nitrate resulted in higher P uptake rates than nitrite. In an additional experiment with aeration and no ammonium in the medium (Fig. 2C), oxygen was the only electron acceptor present, since nitrification was not occurring. These experiments showed a phosphate uptake rate of 9.5 ± 0.7 mg P h−1 gVSS−1, which is statistically similar (P > 0.05) to the rate observed when nitrate was the sole electron acceptor (Fig. 3A). A batch experiment in which the anaerobic stage was extended to the end of the cycle, served as control of P uptake in the absence of oxygen, nitrite, or nitrate as electron acceptors. No P uptake was observed in this experiment (Fig. 2D), as expected.
Fig. 3.
P, NO2− and NO3− utilization rates in the (A) lab-scale and (B) pilot-scale SBRs during micro-aerobic (O2), anoxic (NO2− and NO3−), multiple electron acceptors (O2+NO3−) and a conventional cycle.
To test the effect of having two electron acceptors simultaneously available, we performed aerated experiments in which nitrate was also added (Fig. 2E). In this case, phosphorus was removed from the system at a much faster rate (14 ± 1.3 mg P h−1 gVSS−1) than with the individual electron acceptors (Fig. 3A). After all P was removed, the concentration of nitrate remained constant during the rest of the cycle, indicating that a fraction of the PAO population used this electron acceptor for P uptake, and that no other significant denitrification mechanism, e.g. endogenous denitrification (Choi et al., 2002, Vocks et al., 2005), was responsible for nitrate removal in the micro-aerobic condition (Fig. 2E). For comparison, the measured P uptake rate during normal operation was 13.1 ± 2.5 mg P h−1 gVSS−1, which is not significantly different to the rate when both oxygen and nitrate were present (P > 0.05) (Fig. 3A).
3.2. Phosphorus removal in pilot-scale reactor
The pilot-scale SBR has been in operation under a variety of organic loadings created by amending primary effluent with acetate. During this part of the study, the acetate addition accounted for 15–30% of the biodegradable organic matter entering the reactor. A typical cycle (Fig. 1B) consists of an initial idle phase to reduce nitrate concentrations, an anaerobic phase where the reactor receives the new batch of amended primary effluent and P is released, and a micro-aerobic phase for nitrification and P uptake. Similar to the lab-scale SBR reactor, aeration rates are matched to the oxygen uptake rate so that DO remains low and constant (0.23 ± 0.03 mg/L) during active P uptake, and are controlled with a DO set point (0.6 mg/L) at the end of the aerated phase, when all the P is taken up. In this reactor, the extent of simultaneous nitrification and denitrification is lower than in the lab-scale reactor, with nitrate accumulation effectively taking place during most of the micro-aerobic stage (Fig. 1B). During a typical cycle, about 60% of the ammonium measured at the beginning of the micro-aerobic stage is recovered as nitrate. On average, during this operational period, the N and P removal in the reactor were 42± 15% and 91± 15%, respectively.
Batch experiments with sludge from this reactor were also performed to evaluate the use of different electron acceptors coupled to P uptake (Fig. 4A–D). In the presence of nitrite, nitrate and oxygen as sole electron acceptors, the observed P uptake rates during the first hour of these tests were 1.5 ± 0.6, 4.9 ± 2.6 and 6.6 ± 1.8 mg P h−1 gVSS−1, respectively (standard deviation represent variability in duplicate experiments). As expected, no P uptake was observed in the absence of oxygen, nitrite, or nitrate as electron acceptors (Fig. 4D). Moreover, the P uptake rate during normal operation was measured to be 5.9 ± 1.0 mg P h−1 gVSS−1 (Fig. 3B).
Fig. 4.
Evaluation of (A) nitrite, (B) nitrate and (C) oxygen as electron acceptors for P uptake in the pilot-scale SBR. (D) Batch tests with no electron acceptor added for P uptake (anaerobic cycle). Experiments were performed during days 468 and 498 of operation.
All the specific P uptake rates were lower in the pilot-scale reactor than in the lab-scale reactor. Most notably, when nitrate was used as the sole electron acceptor (Fig. 4B), P uptake activity significantly decreased after one hour of anoxic conditions and nitrate accumulation was observed, suggesting that the active fraction of DPAO exhausted their denitrifying ability even when the high concentration of P in the reactor indicates that intracellular PHA was still available for further P removal. Therefore, the factor limiting denitrification in this system corresponds to the low fraction of PAOs capable of nitrate respiration and not to the amount of PHA accumulated within PAOs. Overall, these results are consistent with the lower extent of simultaneous nitrification and denitrification observed in this reactor.
3.3. Accumulibacter quantification in SBR reactors
It is well established that lab-scale SBR reactors operated to achieve EBPR under the typical anaerobic/high DO cycles and fed with acetate as the sole electron donor are enriched in Accumulibacter (Gonzalez-Gil and Holliger, 2011, He et al., 2007, He et al., 2006, He et al., 2010, Kim et al., 2010, Kim et al., 2013, Oehmen et al., 2005). Under these conditions, Accumulibacter can account for more than 70% of the microbial community (He et al., 2007, He et al., 2010). Pilot-scale and full-scale reactors treating wastewater are also most commonly enriched in Accumulibacter as the main PAO (He et al., 2007, Zilles et al., 2002), although recent reports also indicate the possibility of Tetrasphaera sp. being the prevalent PAO in some full scale plants (Nguyen et al., 2011). Because of the larger diversity of organic substrates present in wastewater, PAOs account for 5–25% of the total microbial community in pilot- and full-scale reactors treating wastewater (He et al., 2007). Accumulibacter has also been shown to be the dominant PAO in lab-scale EBPR reactors operating under cyclic anaerobic/anoxic conditions (Carvalho et al., 2007, Guisasola et al., 2009, Jiang et al., 2006, Lanham et al., 2011, Wang et al., 2014), where it can account for 20–90% of the community. In contrast, little is known about the PAO that are enriched under the anaerobic/micro-aerobic conditions used in this study.
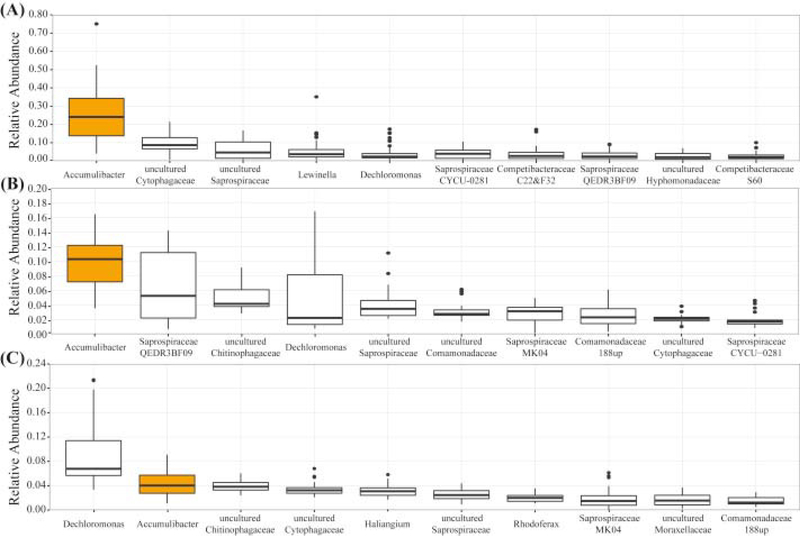
In order to estimate the contribution of Accumulibacter and Tetrasphaera to the observed P removal under cyclic anaerobic/micro-aerobic conditions, we characterized the microbial community composition within each reactor using 16S rRNA gene tag sequencing (Fig. 5). The relative abundance of each genus was estimated by the proportion of Accumulibacter reads to the total number of 16S rRNA reads per sample. In the lab-scale reactor, Accumulibacter was found to range between 4 and 75%, with a median of 24%. Tetrasphaera were detected in some samples, but not abundant, resulting in a median of 0.002% in the sample set. In the pilot-scale reactor, the Accumulibacter median was 10%, while Tetrasphaera was found in all samples with a median abundance of 0.1%, which is two orders of magnitude lower than Accumulibacter. For comparison, 16S rRNA gene tag sequencing was also performed on samples from the full-scale Nine Springs WWTP (Fig. 5C). In these samples Accumulibacter was represented with a median of 4.0%, whereas Tetrasphaera was not abundant (median of 0.6%). Likewise, the low relative abundance of Competibacter-related GAO in both pilot- and lab-scale reactors, coincides with the results previously obtained by Lemaire et al. (2006) and Carvalheira et al. (2014) in reactors operating under limited-oxygen conditions. Taken together, these results suggest that EBPR operation under cyclic anaerobic/micro-aerobic conditions did not change the dominance of Accumulibacter as the main PAO in the acetate fed reactor, or in the reactor treating primary effluent.
Fig. 5.
Phylogenetic abundance variation box plot at the genus level of the most abundant taxonomic units retrieved (A) lab-scale, (B) pilot-scale SBR and (C) Nine Springs, based on 16S rRNA gene tag sequencing data presented as the relative abundance. Results correspond to the taxonomic distribution of 45 samples from the lab-scale SBR, 23 samples from the pilot-scale SBR and 38 samples from the full-scale WWTP Nine Springs. The name of each genus-level taxon in this figure correspond to the one proposed in the MIDAS database (Mielczarek et al., 2013).
3.4. Evaluating accumulibacter diversity in the SBR reactors
Diversity within the Accumulibacter lineage has been typically described as a function of taxonomic units defined with respect to the phylogeny of the ppk1 gene (He et al., 2007, McMahon et al., 2007). With this functional gene, Accumulibacter is separated into two distinct types (Type I and II), and each Type is subdivided into several clades. There are five clades within Type I and nine clades within Type II, for a total of 14 recognized Accumulibacter sub-clades (He et al., 2007, Mao et al., 2015).
PCR primers for quantification of a number of sub-clades have been published (He et al., 2007), and have been used to characterize Accumulibacter’s diversity in different EBPR systems (Flowers et al., 2013, He et al., 2007, Mao et al., 2015). However, primers are not available for all of the clades, and therefore, a comprehensive characterization of Accumulibacter’s diversity using qPCR has not been possible. To overcome this limitation, we designed a complete set of primers to quantify 12 Accumulibacter clades. Primers targeting clades IIH and II-I were also designed but issues with the synthesis of ppk1 sequences prevented quantification of these clades. In a practical improvement over existing primer sets, the new primers were all designed to use the same annealing temperature and qPCR conditions so that they can be simultaneously used in a single thermocycler run (See Methods).
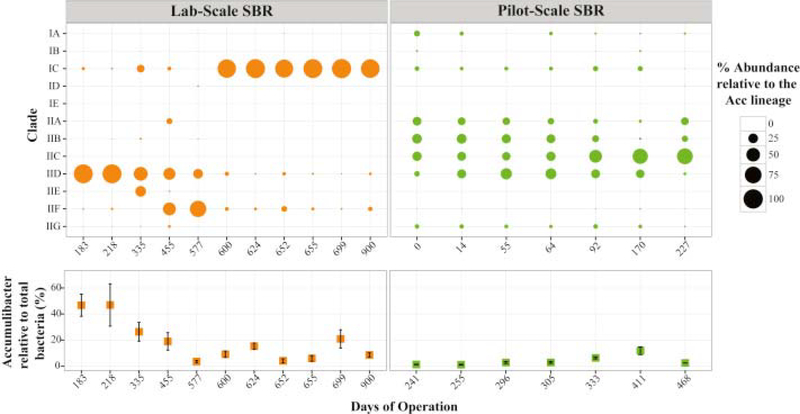
Using the newly designed primer sets, we estimated the abundance of each Accumulibacter clade in samples retrieved from the lab and pilot-scale reactors (Fig. 6). Although quantification with primers designed for clade IIH and II-I was not possible, PCR results indicated absence of these two clades in the samples analyzed in this study. In the lab-scale SBR, Accumulibacter clades IC and IID dominated the Accumulibacter population in 10 out of the 12 samples with relative abundances between 25 and 99% of the total Accumulibacter lineage (Fig. 6). A switch in the distribution from absolute dominance of Clade IID to Clade IC was observed between days 455 and 600, although no significant changes in the reactor performance were detected during this transition. Until day 218, Clade IID accounted for more than 96% of the Accumulibacter lineage, while Clade IC predominated during the last 300 days with abundances greater than 87% of the Accumulibacter population.
Fig. 6.
Estimated abundance of each clade relative to the total Accumulibacter lineage (bubble size) and the percentage of the total population of Accumulibacter relative to the bacterial community (bottom plot), in samples from the lab scale and pilot scale reactors. Values were determined using qPCR targeting the ppk1 gene and total bacterial 16S rRNA genes, as described in Methods.
In the pilot-scale reactor, the distribution of Accumulibacter clades was more heterogeneous and even than in the lab-scale reactor. Though Clade IIC was abundant in most samples (relative abundance ranging from 19 to 67%) (Fig. 6), Clades IIA, IIB and IID, and Clades IA and IC to a lesser extent, contributed to the Accumulibacter lineage too. As observed in results from 16S rRNA gene sequencing, the Accumulibacter percentage in the bacterial community was lower in this reactor, as compared to the lab-scale SBR (Fig. 6), which also reflects lower rates of P uptake (Fig. 3). Differences in the relative abundance of Accumulibacter can be attributed to the concentration of volatile fatty acids (Cao, 2011) and to the complex composition of the primary effluent, which favors microbial diversity in the pilot-scale system.
We further confirmed the dominance of each clade during different time periods using shotgun metagenomes (Fig. S7). Reference ppk1 sequences from each clade were used to map and count metagenomic reads that originated from each clade. Noticeably, ppk1 sequences from dominant clades of Accumulibacter in samples from the lab-scale reactor (IID in day 218 and IC in days 671 and 305) retrieved a greater number of metagenomic reads than other clades. Consistent with the qPCR results, the metagenomic analysis of the pilot-scale reactor reveals a more heterogeneous population of Accumulibacter, since both clade IA, IB, IIA, IIB, IIC and IID ppk1 sequences recruited a significant number of metagenomic reads. Overall, the analysis of the metagenomic reads described here validates the results obtained by quantitative PCR with the new set of primers.
4. Discussion
In this study, the population structure and metabolic behavior of Accumulibacter were evaluated in lab- and pilot-scale SNDPR reactors, treating synthetic and real wastewater, respectively. Quantifications of the relative distribution of Accumulibacter clades in these systems revealed that, in the lab-scale acetate-fed reactor, the Accumulibacter population was generally dominated by a single clade (IID or IC). A shift in the community from IID to IC dominance was observed during the time frame of the experiments. This change in the community could be due to external or internal perturbations in the reactor that shifted the PAO community away from equilibrium, as it is demonstrated by the intermediate enrichment of Clade IIF, prior to the enrichment of Clade IC. Environmental conditions that cause physiological stress have been proposed to trigger lytic-bacteriophage events that cause shifts in Accumulibacter abundance in EBPR reactors (Barr et al., 2010, Motlagh et al., 2015). However, a specific external perturbation, as related to pumping rates, substrate concentrations, temperature, and other external factors was not identified.
The pilot-scale reactor had a more diverse Accumulibacter population, consistent with a more diverse organic carbon source, external variability such as diurnal variations in primary effluent strength and seasonal variations in temperature. In addition, the DO concentration during the active P uptake was different in the lab- and pilot-scale reactors (0.05 versus 0.2 mg/L), which might have also influenced the Accumulibacter population structure. For instance, a stronger selective pressure exerted by lowering the DO in the pilot-scale reactor may shift the population towards Clades IID or IC. Further experiments are underway to test this hypothesis.
Regardless of the clade distribution, both reactors demonstrated Accumulibacter establishment as the dominant PAO under the anaerobic/micro-aerobic cycles used in reactor operation. Combined with existing demonstrations that nitrification can be accomplished with very low oxygen conditions (Fitzgerald et al., 2015, Park and Noguera, 2004, Park et al., 2006), the results of efficient EBPR in this study support the possibility of establishing efficient BNR with minimal aeration, a desired goal in the search for ways to achieve energy neutrality during WWTP operation (Stinson et al., 2013).
4.1. Co-respiration of different electron acceptors during P uptake under micro-aerobic conditions
Oxygen, nitrite, and nitrate are possible electron acceptors under micro-aerobic conditions. Accordingly, P uptake rates were independently estimated when these substrates were the sole electron acceptor in the reactors (Fig. 3). This analysis revealed that although the three substrates could serve as electron acceptors during P uptake, nitrite by itself was an inefficient acceptor. Nitrate was a poor electron acceptor in the pilot-scale reactor, whereas the rates of P uptake with oxygen or nitrate in the lab-scale reactor were comparable. Moreover, the simultaneous presence of oxygen and nitrate in the lab-scale reactor resulted in a significantly higher rate of P uptake (Fig. 3A), demonstrating that both electron acceptors could be used at the same time. The simultaneous respiration of oxygen and nitrate would be prone to occur during a normal reactor cycle, in which ammonia is consumed without accumulation of nitrite and nitrate during active P uptake, and nitrate accumulates only when P removal ceases (Fig. 1A).
Previous studies have also reported PAO enrichments capable of using more than one electron acceptor for P uptake (Ahn et al., 2001, Hu et al., 2003, Kerrnjespersen and Henze, 1993, Wang et al., 2007, Wang et al., 2014, Zeng et al., 2011), but the relative concentration of specific clades was not established in those studies. Since the batch experiments in the lab-scale reactor were performed when Clade IC dominated the PAO population (92–98% of the Accumulibacter lineage), we postulate here that this clade is capable of simultaneous respiration with multiple electron acceptors under micro-aerobic conditions. Precedent for the possibility of co-respiration has been established with pure cultures of Thiosphaera pantotropha, Pseudomonas aeruginosa and Sulfurospirillum barnesii (He et al., 2015, Oremland et al., 1999, Robertson and Kuenen, 1990). Additionally, previous kinetic studies have documented simultaneous respiration of oxygen and nitrate by several species of denitrifying Pseudomonas species (Bonin and Gilewicz, 1991, Chen et al., 2003, Takaya et al., 2003), with greater denitrification rates when the oxygen concentration in the environment is limited. This metabolic strategy can play an important role in the competitive success of Clade IC under micro-aerobic conditions.
An alternative explanation to the co-respiration premise is that the Accumulibacter clade definition based on the ppk1 gene does not offer sufficient separation of metabolic traits, resulting in further diversity within Clade IC that includes organisms that can denitrify and organisms that cannot denitrify. This is plausible since the ppk1 gene is not directly related to respiratory pathways and within-clade maximum ppk1 sequence divergence is 12%, substantially higher than would be expected for coherent ecotypes or species (Caro-Quintero and Konstantinidis, 2012, Varghese et al., 2015). Clearly, much remains to be learned about ecophysiological differentiation within and across Accumulibacter clades.
4.2. Denitrifying capabilities revealed by the genome content
Draft genomes have been published for several Accumulibacter clades, including the principal clades discussed in this study: Clade IC (lab-scale) and IIC (pilot-scale) (Skennerton et al., 2015). Genes related to nitrogen metabolism have been annotated and compared among genomes (Skennerton et al., 2015). The three draft genomes of Clade IIC (74.41, 94.26 and 97.17% of completeness and <2.23% of contamination) did not encode a full denitrification pathway, lacking nitric oxide reductase (nor) and nitrous oxide reductase (nos) genes. Since the genomes are incomplete, it is not possible to draw strong conclusions on the end products of denitrification by this clade. Nevertheless, an incomplete denitrification pathway would suggest the inability to efficiently use nitrite and nitrate as electron acceptors (Skennerton et al., 2015), in agreement with the low DPAO activity observed in the pilot-scale reactor. In contrast, Accumulibacter Clade IIC was previously enriched in an anaerobic-anoxic-oxic SBR fed with nitrate during the anoxic stage, with efficient P uptake during this stage (Kim et al., 2013), suggesting a role of Clade IIC in DPAO activity. In our experiments Clade IIC was only abundant in the pilot-scale SBR, which exhibited some P removal with nitrate, but since there were several other clades in the reactor, it is not possible to completely attribute denitrification capability to this clade. Regardless, based on the efficient P removal rates with minimal aeration in the pilot-scale reactor, our results suggest that Clade IIC and the other clades enriched in the pilot-scale reactor have high affinity for oxygen as an electron acceptor.
Only one draft genome of Clade IC has been published (92.74% completeness and 1.74% of contamination) (Skennerton et al., 2015). This genome encoded a more complete denitrification pathway, including a periplasmic nitrate reductase (nap), nitrite reductase (nir) and nitric oxide reductase (nor). Based on the P uptake rates observed in the reactor when Clade IC was dominating the Accumulibacter lineage (>99%), we conclude that this clade is able to use oxygen, nitrite and nitrate as electron acceptors for P removal, albeit the use of nitrite was less efficient than nitrate or oxygen. Furthermore, since the P uptake rates were significantly higher when oxygen and nitrate were both added (Fig. 3A), this clade potentially has a selective advantage under micro-aerobic conditions by being able to simultaneously use multiple electron acceptors.
At the moment, no genome is available for Accumulibacter Clade IID and just one previous study has reported enrichment of this clade, revealing it as the dominant denitrifying P removing bacteria in a municipal treatment system (Zeng et al., 2015). Since our batch tests performed in the lab-scale reactor were conducted after Clade IC became dominant, we do not have any kinetic information to associate clade IID with specific electron acceptors other than oxygen. However, as denitrification linked to P removal was also achieved in the lab-scale reactor when clade IID was dominant, we speculate that clade IID is also able to use multiple electron acceptors during micro-aerobic conditions.
4.3. Differences in P and N removal rates under anoxic conditions
Differences in the P uptake and N reduction rates were observed when nitrite and nitrate were used as electron acceptors to drive anoxic P uptake (Fig. 3). Both the lab-scale and pilot-scale reactors presented greater anoxic P uptake and denitrification rates when NO3− was added to the anoxic stage instead of NO2−. This indicates that the DPAO community enriched under these conditions has a greater denitrifying P removal capacity when complete nitrification occurs during the micro-aerobic stage, which could be potentially overlapping with the nitrite shunt (Verstraete and Philips, 1998) pathway in these reactors. Previous studies have reported dissimilar results for N and P uptake rates on anoxic P uptake (Table 2). Wang et al. (2014) and Guisasola et al. (2009) estimated faster P uptake and N reduction rates with NO2− as electron acceptor, which they attributed to the location of nitrite reduction enzymes within the cell and the contribution of other organisms capable of NO2− reduction. In contrast Jiang et al. (2006) and Hu et al. (2003) reported low anoxic P uptake per N-NO2− denitrified compared to NO3−, suggesting that nitrate is preferentially used over nitrite for anoxic P uptake. Differences in P removal rates when using nitrite or nitrate as electron acceptors have been attributed to the higher amount of energy obtained per mole of NO3− in comparison with NO2− (Flickinger and Drew, 1999). However, discrepancies in the performance of denitrifying EBPR systems could be also related to the Accumulibacter clade structure and the presence/absence of enzymes involved in denitrification pathways. Unfortunately, none of these studies analyzed the type of Accumulibacter present in the reactors. Alternatively, inhibitory effects of nitrite on EBPR have been previously described, particularly when DPAOs biomass are not pre-cultivated with high nitrite concentrations (Saito et al., 2004, Zeng et al., 2011, Zhou et al., 2007, Zhou et al., 2010), which might limit P and N removal rates in these systems. Greater denitrification rates of nitrate over nitrite have also been reported in other denitrifying bacteria (Almeida et al., 1995, Carlson and Ingraham, 1983).
Table 2.
Comparison of anoxic P and N utilization rates found in this study with values reported in the literature.
| System | N utilization rate (mg N/h g VSS) | P utilization rate (mg P/h g VSS) | ||
|---|---|---|---|---|
| NO2− | NO3− | NO2− | NO3− | |
| This study LS-SBR | 2.8 | 5.5 | 5.7 | 11 |
| This study PS-SBR | 0.6 | 3.4 | 1.5 | 4.9 |
| (Wang et al., 2014) | 6.8 | 3.6 | 9.3 | 4.3 |
| (Guisasola et al., 2009) | 5.8 | 0.7 | 3.8 | 0.2 |
| (Jiang et al., 2006) | 9.0 | 12 | 20 | 30 |
| (Hu et al., 2003) | NA | NA | 25 | 29 |
Both P and N utilization rates when nitrite was used as electron acceptor were considerably lower in the pilot-scale SBR. The limited capability of nitrite reduction for P uptake potentially led to NOB outcompeting PAOs for the available nitrite and therefore, producing greater nitrate concentrations in the effluent. Interestingly, though batch tests with sludge from this reactor revealed nitrate-denitrifying capabilities for the PAO community, nitrate produced from nitrification was used only poorly for P uptake under micro-aerobic conditions. This difference can reflect a stronger affinity for oxygen compared to nitrate, thus PAOs would preferentially use oxygen when it becomes available. A different behavior was observed in the lab-scale SBR, where nitrate production only occurred after complete P uptake, indicating the use of nitrite and nitrate even when oxygen was present in the system.
5. Conclusions
Efficient nitrogen and phosphorus removal was demonstrated in lab- and pilot- scale reactors that used cyclic anaerobic/micro-aerobic conditions.
Simultaneous nitrification, denitrification, and phosphorus removal (SNDPR) was effectively maintained in a lab-scale SBR in which DO concentrations were maintained at 0.05 mg/L during active phosphorus uptake. Less efficient SNDPR was obtained in the pilot-scale reactor, where the DO was maintained at 0.2 mg/L during active phosphorus uptake.
Real-time PCR based-quantification, using a new set of Accumulibacter-ppk1 primers, revealed Clade IID and IC were the dominant PAO in the lab-scale reactor, but at different times during the operational period.
Kinetic experiments when Clade IC represented >92% of the Accumulibacter population in the reactor suggest that this organism was capable of simultaneously using oxygen, nitrite and nitrate as electron acceptors for P uptake, but with inefficient use of nitrite compared to nitrate and oxygen.
Supplementary Material
Acknowledgements
This work was partially supported by funding from the National Science Foundation (CBET-1435661 and MCB-0738232) and the Madison Metropolitan Sewerage District. Additional funding from the Chilean National Commission for Scientific and Technological Research (CONICYT) as a fellowship to Pamela Camejo is also acknowledged. Vikram Kapoor was supported by U.S. Environmental Protection Agency (EPA) via a post-doctoral appointment administered by the Oak Ridge Institute for Science and Education. We thank Jaime Yañez, Jacqueline German, Jia Hui Khoo, Diana Barrera, Elizabeth Erb, Madeleine Haut, and Chloe Olson for support with laboratory analysis and reactor operation and Michael Elk for technical support during cloning experiments. The U.S. Environmental Protection Agency, through its Office of Research and Development, partially funded and collaborated in the research described herein. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
References
- Ahn J, Daidou T, Tsuneda S, Hirata A Metabolic behavior of denitrifying phosphate-accumulating organisms under nitrate and nitrite electron acceptor conditions J. Biosci. Bioeng, 92 (5) (2001), pp. 442–446 [DOI] [PubMed] [Google Scholar]
- Almeida JS, Reis MAM, Carrondo MJT Competition between nitrate and nitrite reduction in denitrification by pseudomonas-fluorescens Biotechnol. Bioeng, 46 (5) (1995), pp. 476–484 [DOI] [PubMed] [Google Scholar]
- APHA Standard Methods for the Examination of Water and Wastewater (2005) Washington, DC [Google Scholar]
- Barr JJ, Slater FR, Fukushima T, Bond PL Evidence for bacteriophage activity causing community and performance changes in a phosphorus-removal activated sludge FEMS Microbiol. Ecol, 74 (3) (2010), pp. 631–642 [DOI] [PubMed] [Google Scholar]
- Bonin P, Gilewicz M A direct demonstration of Co-Respiration of oxygen and nitrogen-oxides by Pseudomonas-Nautica – some spectral and kinetic-properties of the respiratory components FEMS Microbiol. Lett, 80 (2–3) (1991), pp. 183–188 [Google Scholar]
- Cao YS Biological Phosphorus Removal Activated Sludge Process in Warm Climates IWA Publishing; (2011) [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R QIIME allows analysis of high-throughput community sequencing data Nat. Methods, 7 (5) (2010), pp. 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample Proc. Natl. Acad. Sci. U. S. A, 108 (2011), pp. 4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Ingraham JL Comparison of denitrification by Pseudomonas-Stutzeri, Pseudomonas-Aeruginosa, and paracoccus-denitrificans Appl. Environ. Microbiol, 45 (4) (1983), pp. 1247–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Quintero A, Konstantinidis KT Bacterial species may exist, metagenomics reveal Environ. Microbiol, 14 (2) (2012), pp. 347–355 [DOI] [PubMed] [Google Scholar]
- Carvalheira M, Oehmen A, Carvalho G, Eusebio M, Reis MAM The impact of aeration on the competition between polyphosphate accumulating organisms and glycogen accumulating organisms Water Res, 66 (2014), pp. 296–307 [DOI] [PubMed] [Google Scholar]
- Carvalho G, Lemos PC, Oehmen A, Reis MAM Denitrifying phosphorus removal: linking the process performance with the microbial community structure Water Res, 41 (19) (2007), pp. 4383–4396 [DOI] [PubMed] [Google Scholar]
- Chen F, Xia Q, Ju LK Aerobic denitrification of Pseudomonas aeruginosa monitored by online NAD(P)H fluorescence Appl. Environ. Microbiol, 69 (11) (2003), pp. 6715–6722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Hong SW, Kim SJ, Chung IH Development of a biological process for livestock wastewater treatment using a technique for predominant outgrowth of Bacillus species Water Sci. Technol, 45 (12) (2002), pp. 71–78 [PubMed] [Google Scholar]
- Coma M, Puig S, Balaguer MD, Colprim J The role of nitrate and nitrite in a granular sludge process treating low-strength wastewater Chem. Eng. J, 164 (1) (2010), pp. 208–213 [Google Scholar]
- Comeau Y, Hall KJ, Hancock REW, Oldham WK Biochemical-model for enhanced biological phosphorus removal Water Res, 20 (12) (1986), pp. 1511–1521 [Google Scholar]
- de Kreuk M, Heijnen JJ, van MCM Loosdrecht Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge Biotechnol. Bioeng, 90 (6) (2005), pp. 761–769 [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R UCHIME improves sensitivity and speed of chimera detection Bioinformatics, 27 (16) (2011), pp. 2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald CM, Camejo P, Oshlag JZ, Noguera DR Ammonia-oxidizing microbial communities in reactors with efficient nitrification at low-dissolved oxygen Water Res, 70 (2015), pp. 38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger MC, Drew SW The Encyclopedia of Bioprocess Technology: Fermentation, Biocatalysis, and Bioseparation Wiley, New York: (1999) [Google Scholar]
- Flowers JJ, Cadkin TA, McMahon KD Seasonal bacterial community dynamics in a full-scale enhanced biological phosphorus removal plant Water Res, 47 (19) (2013), pp. 7019–7031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JJ, He S, Yilmaz S, Noguera DR, McMahon KD Denitrification capabilities of two biological phosphorus removal sludges dominated by different “Candidatus Accumulibacter” clades Environ. Microbiol. Rep, 1 (6) (2009), pp. 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel RK, Noguera DR Evaluation of sludge yield and phosphorus removal in a cannibal solids reduction process J. Environ. Eng. ASCE, 132 (10) (2006), pp. 1331–1337 [Google Scholar]
- Gonzalez-Gil G, Holliger C Dynamics of microbial community structure of and enhanced biological phosphorus removal by aerobic granules cultivated on propionate or acetate Appl. Environ. Microbiol, 77 (22) (2011), pp. 8041–8051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisasola A, Qurie M, Vargas MD, Casas C, Baeza JA Failure of an enriched nitrite-DPAO population to use nitrate as an electron acceptor Process Biochem, 44 (7) (2009), pp. 689–695 [Google Scholar]
- He D, Zheng MS, Ma T, Li C, Ni JR Interaction of Cr(VI) reduction and denitrification by strain Pseudomonas aeruginosa PCN-2 under aerobic conditions Bioresour. Technol, 185 (2015), pp. 346–352 [DOI] [PubMed] [Google Scholar]
- He S, Gall DL, McMahon KD “Candidatus Accumulibacter” population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes Appl. Environ. Microbiol, 73 (18) (2007), pp. 5865–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Gu AZ, McMahon KD Fine-scale differences between Accumulibacter-like bacteria in enhanced biological phosphorus removal activated sludge Water Sci. Technol, 54 (1) (2006), pp. 111–117 [DOI] [PubMed] [Google Scholar]
- He SM, Bishop FI, McMahon KD Bacterial community and “Candidatus Accumulibacter” population dynamics in laboratory-scale enhanced biological phosphorus removal reactors Appl. Environ. Microbiol, 76 (16) (2010), pp. 5479–5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JY, Ong SL, Ng WJ, Lu F, Fan XJ A new method for characterizing denitrifying phosphorus removal bacteria by using three different types of electron acceptors Water Res, 37 (14) (2003), pp. 3463–3471 [DOI] [PubMed] [Google Scholar]
- Jiang YF, Wang BZ, Wang L, Chen JM, He SB Dynamic response of denitrifying poly-P accumulating organisms batch culture to increased nitrite concentration as electron acceptor J. Environ. Sci. Health Part A Toxic Hazard. Subst. Environ. Eng, 41 (11) (2006), pp. 2557–2570 [DOI] [PubMed] [Google Scholar]
- Kaetzke A, Jentzsch D, Eschrich K Quantification of Microthrix parvicella in activated sludge bacterial communities by real-time PCR Lett. Appl. Microbiol, 40 (3) (2005), pp. 207–211 [DOI] [PubMed] [Google Scholar]
- Kerrnjespersen JP, Henze M Biological phosphorus uptake under anoxic and aerobic conditions Water Res, 27 (4) (1993), pp. 617–624 [Google Scholar]
- Kim JM, Lee HJ, Kim SY, Song JJ, Park W, Jeon CO Analysis of the fine-scale population structure of “Candidatus Accumulibacter phosphatis” in enhanced biological phosphorus removal sludge, using fluorescence in situ hybridization and flow cytometric sorting Appl. Environ. Microbiol, 76 (12) (2010), pp. 3825–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Lee HJ, Lee DS, Jeon CO Characterization of the denitrification-associated phosphorus uptake properties of “Candidatus Accumulibacter phosphatis” clades in sludge subjected to enhanced biological phosphorus removal Appl. Environ. Microbiol, 79 (6) (2013), pp. 1969–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanham AB, Moita R, Lemos PC, Reis MA Long-term operation of a reactor enriched in Accumulibacter clade I DPAOs: performance with nitrate, nitrite and oxygen Water Sci. Technol, 63 (2) (2011), pp. 352–359 [DOI] [PubMed] [Google Scholar]
- Lemaire R, Meyer R, Taske A, Crocetti GR, Keller J, Yuan ZG Identifying causes for N2O accumulation in a lab-scale sequencing batch reactor performing simultaneous nitrification, denitrification and phosphorus removal J. Biotechnol, 122 (1) (2006), pp. 62–72 [DOI] [PubMed] [Google Scholar]
- Mao YP, Graham DW, Tamaki H, Zhang T Dominant and Novel Clades of Candidatus Accumulibacter Phosphatis in 18 Globally Distributed Full-scale Wastewater Treatment Plants Scientific Reports 5 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KD, Dojka MA, Pace NR, Jenkins D, Keasling JD Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal Appl. Environ. Microbiol, 68 (10) (2002), pp. 4971–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KD, Yilmaz S, He S, Gall DL, Jenkins D, Keasling JD Polyphosphate kinase genes from full-scale activated sludge plants Appl. Microbiol. Biotechnol, 77 (1) (2007), pp. 167–173 [DOI] [PubMed] [Google Scholar]
- Metcalf E, Tchobanoglous G, Stensel HD, Tsuchihashi R, Burton F Wastewater Engineering: Treatment and Resource Recovery McGraw-Hill, New York, NY, USA: (2013) [Google Scholar]
- Mielczarek AT, Saunders AM, Larsen P, Albertsen M, Stevenson M, Nielsen JL, Nielsen PH The microbial database for Danish wastewater treatment plants with nutrient removal (MiDas-DK) – a tool for understanding activated sludge population dynamics and community stability Water Sci. Technol, 67 (11) (2013), pp. 2519–2526 [DOI] [PubMed] [Google Scholar]
- Motlagh AM, Bhattacharjee AS, Goel R Microbiological study of bacteriophage induction in the presence of chemical stress factors in enhanced biological phosphorus removal (EBPR) Water Res, 81 (2015), pp. 1–14 [DOI] [PubMed] [Google Scholar]
- Nguyen HTT, Le VQ, Hansen AA, Nielsen JL, Nielsen PH High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems FEMS Microbiol. Ecol, 76 (2) (2011), pp. 256–267 [DOI] [PubMed] [Google Scholar]
- Oehmen A, Yuan ZG, Blackall LL, Keller J Comparison of acetate and propionate uptake by polyphosphate accumulating organisms and glycogen accumulating organisms Biotechnol. Bioeng, 91 (2) (2005), pp. 162–168 [DOI] [PubMed] [Google Scholar]
- Oremland RS, Blum JS, Bindi AB, Dowdle PR, Herbel M, Stolz JF Simultaneous reduction of nitrate and selenate by cell suspensions of selenium-respiring bacteria Appl. Environ. Microbiol, 65 (10) (1999), pp. 4385–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HD, Noguera DR Evaluating the effect of dissolved oxygen on ammonia-oxidizing bacterial communities in activated sludge Water Res, 38 (14–15) (2004), pp. 3275–3286 [DOI] [PubMed] [Google Scholar]
- Park HD, Whang LM, Reusser SR, Noguera DR Taking advantage of aerated-anoxic operation in a full-scale University of Cape Town process Water Environ. Res, 78 (6) (2006), pp. 637–642 [DOI] [PubMed] [Google Scholar]
- Peng Y, Hou H, Wang S, Cui Y, Zhiguo Nitrogen Y and phosphorus removal in pilot-scale anaerobic-anoxic oxidation ditch system J. Environ. Sci. (China), 20 (4) (2008), pp. 398–403 [DOI] [PubMed] [Google Scholar]
- Peterson SB, Warnecke F, Madejska J, McMahon KD, Hugenholtz P Environmental distribution and population biology of Candidatus Accumulibacter, a primary agent of biological phosphorus removal Environ. Microbiol, 10 (10) (2008), pp. 2692–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LA, Kuenen JG Combined heterotrophic nitrification and aerobic denitrification in thiosphaera-pantotropha and other bacteria Ant. Van Leeuwenhoek Int. J. General Mol. Microbiol, 57 (3) (1990), pp. 139–152 [DOI] [PubMed] [Google Scholar]
- Saito T, Brdjanovic D, van MC Loosdrecht Effect of nitrite on phosphate uptake by phosphate accumulating organisms Water Res, 38 (17) (2004), pp. 3760–3768 [DOI] [PubMed] [Google Scholar]
- Skennerton CT, Barr JJ, Slater FR, Bond PL, Tyson GW Expanding our view of genomic diversity in Candidatus Accumulibacter clades Environ. Microbiol, 17 (5) (2015), pp. 1574–1585 [DOI] [PubMed] [Google Scholar]
- Stinson B, Murthy S, Bott C, Wett B, Al-Omari A, Bowden G, Mokhyerie Y, De H Clippeleir Roadmap toward energy neutrality & chemical optimization at enhanced nutrient removal facilities Proc. Water Environ. Fed, 2013 (4) (2013), pp. 702–731 [Google Scholar]
- Takaya N, Catalan-Sakairi MAB, Sakaguchi Y, Kato I, Zhou ZM, Shoun H Aerobic denitrifying bacteria that produce low levels of nitrous oxide Appl. Environ. Microbiol, 69 (6) (2003), pp. 3152–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S MEGA6: molecular evolutionary genetics analysis version 6.0 Mol. Biol. Evol, 30 (12) (2013), pp. 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, Kyrpides NC, Pati A Microbial species delineation using whole genome sequences Nucleic Acids Res, 43 (14) (2015), pp. 6761–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraete W, Philips S Nitrification-denitrification processes and technologies in new contexts Environ. Pollut, 102 (1998), pp. 717–726 [Google Scholar]
- Vocks M, Adam C, Lesjean B, Gnirss R, Kraume M Enhanced post-denitrification without addition of an external carbon source in membrane bioreactors Water Res, 39 (14) (2005), pp. 3360–3368 [DOI] [PubMed] [Google Scholar]
- Wang XX, Wang SY, Xue TL, Li BK, Dai X, Peng YZ Treating low carbon/nitrogen (C/N) wastewater in simultaneous nitrification-endogenous denitrification and phosphorous removal (SNDPR) systems by strengthening anaerobic intracellular carbon storage Water Res, 77 (2015), pp. 191–200 [DOI] [PubMed] [Google Scholar]
- Wang YY, Pan ML, Yan M, Peng YZ, Wang SY Characteristics of anoxic phosphors removal in sequence batch reactor J. Environ. Sci. China, 19 (7) (2007), pp. 776–782 [DOI] [PubMed] [Google Scholar]
- Wang YY, Zhou S, Ye L, Wang H, Stephenson T, Jiang XX Nitrite survival and nitrous oxide production of denitrifying phosphorus removal sludges in long-term nitrite/nitrate-fed sequencing batch reactors Water Res, 67 (2014), pp. 33–45 [DOI] [PubMed] [Google Scholar]
- WEF Energy Conservation in Wastewater Treatment Facilities Water Environment Federation, Alexandria, VA, USA: (1997) [Google Scholar]
- Wexler M, Richardson DJ, Bond PL Radiolabelled proteomics to determine differential functioning of Accumulibacter during the anaerobic and aerobic phases of a bioreactor operating for enhanced biological phosphorus removal Environ. Microbiol, 11 (12) (2009), pp. 3029–3044 [DOI] [PubMed] [Google Scholar]
- Wilmes P, Andersson AF, Lefsrud MG, Wexler M, Shah M, Zhang B, Hettich RL, Bond PL, VerBerkmoes NC, Banfield JF Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal ISME J, 2 (8) (2008), pp. 853–864 [DOI] [PubMed] [Google Scholar]
- Wright ES DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment BMC Bioinform, 16 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ES, Yilmaz LS, Ram S, Gasser JM, Harrington GW, Noguera DR Exploiting extension bias in polymerase chain reaction to improve primer specificity in ensembles of nearly identical DNA templates Environ. Microbiol, 16 (5) (2014), pp. 1354–1365 [DOI] [PubMed] [Google Scholar]
- Yilmaz G, Lemaire R, Keller J, Yuan Z Simultaneous nitrification, denitrification, and phosphorus removal from nutrient-rich industrial wastewater using granular sludge Biotechnol. Bioeng, 100 (3) (2008), pp. 529–541 [DOI] [PubMed] [Google Scholar]
- Yoshida N, Takahashi N, Hiraishi A Phylogenetic characterization of a polychlorinated-dioxin- dechlorinating microbial community by use of microcosm studies Appl. Environ. Microbiol, 71 (8) (2005), pp. 4325–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng RJ, Lemaire R, Yuan Z, Keller Simultaneous nitrification J, denitrification, and phosphorus removal in a lab-scale sequencing batch reactor Biotechnol. Bioeng, 84 (2) (2003), pp. 170–178 [DOI] [PubMed] [Google Scholar]
- Zeng RJ, Lemaire R, Yuan Z, Keller J A novel wastewater treatment process: simultaneous nitrification, denitrification and phosphorus removal Water Sci. Technol, 50 (10) (2004), pp. 163–170 [PubMed] [Google Scholar]
- Zeng W, Li B, Wang X, Bai X, Peng Y Influence of nitrite accumulation on “Candidatus Accumulibacter” population structure and enhanced biological phosphorus removal from municipal wastewater Chemosphere, 144 (2015), pp. 1018–1025 [DOI] [PubMed] [Google Scholar]
- Zeng W, Li L, Yang YY, Wang XD, Peng YZ Denitrifying phosphorus removal and impact of nitrite accumulation on phosphorus removal in a continuous anaerobic-anoxic-aerobic (A2O) process treating domestic wastewater Enzyme Microb. Technol, 48 (2) (2011), pp. 134–142 [DOI] [PubMed] [Google Scholar]
- Zhang HM, Wang XL, Xiao JN, Yang FL, Zhang J Enhanced biological nutrient removal using MUCT-MBR system Bioresour. Technol, 100 (3) (2009), pp. 1048–1054 [DOI] [PubMed] [Google Scholar]
- Zheng X, Tong J, Li HJ, Chen YG The investigation of effect of organic carbon sources addition in anaerobic-aerobic (low dissolved oxygen) sequencing batch reactor for nutrients removal from wastewaters Bioresour. Technol, 100 (9) (2009), pp. 2515–2520 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ganda L, Lim M, Yuan Z, Kjelleberg S, Ng WJ Free nitrous acid (FNA) inhibition on denitrifying poly-phosphate accumulating organisms (DPAOs) Appl. Microbiol. Biotechnol, 88 (1) (2010), pp. 359–369 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Pijuan M, Yuan Z Free nitrous acid inhibition on anoxic phosphorus uptake and denitrification by poly-phosphate accumulating organisms Biotechnol. Bioeng, 98 (4) (2007), pp. 903–912 [DOI] [PubMed] [Google Scholar]
- Zilles JL, Peccia J, Kim MW, Hung CH, Noguera DR Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants Appl. Environ. Microbiol, 68 (6) (2002), pp. 2763–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.