Abstract
Selective pericyte loss, the histological hallmark of early diabetic retinopathy (DR), enhances the breakdown of the blood-retinal barrier (BRB) in diabetes. However, the role of pericytes on BRB alteration in diabetes and the signaling pathways involved in their effects are currently unknown. To understand the role of diabetes-induced molecular alteration of pericytes, we performed transcriptomic analysis of sorted retinal pericytes from mice model of diabetes. Retinal tissue from non-diabetic and diabetic (duration 3 months) mouse eyes (n=10 in each group) were used to isolate pericytes through fluorescent activated cell sorting (FACS) using pericyte specific fluorescent antibodies, PDGFRb-APC. For RNA sequencing and qPCR analysis, a cDNA library was generated using template switching oligo and the resulting libraries were sequenced using paired-end Illumina sequencing. Molecular functional pathways were analyzed using differentially expressed genes (DEGs). Differential expression analysis revealed 217 genes significantly upregulated and 495 genes downregulated, in pericytes isolated from diabetic animals. These analyses revealed a core set of differentially expressed genes that could potentially contribute to the pericyte dysfunction in diabetes and highlighted the pattern of functional connectivity between key candidate genes and blood retinal barrier alteration mechanisms. The top up-regulated gene list included: Ext2, B3gat3, Gpc6, Pip5k1c and Pten and down-regulated genes included: Notch3, Xbp1, Gpc4, Atp1a2 and AKT3. Out of these genes, we further validated one of the down regulated genes, Notch 3 and its role in BRB alteration in diabetic retinopathy. We confirmed the downregulation of Notch3 expression in human retinal pericytes exposed to Advanced Glycation End-products (AGEs) treatment mimicking the chronic hyperglycemia effect. Exploration of pericyte-conditioned media demonstrated that loss of NOTCH3 in pericyte led to increased permeability of endothelial cell monolayers. Collectively, we identify a role for NOTCH3 in pericyte dysfunction in diabetes. Further validation of other DEGs to identify cell specific molecular change through whole transcriptomic approach in diabetic retina will provide novel insight into the pathogenesis of DR and novel therapeutic targets.
Keywords: Diabetic Retinopathy, pericytes, RNA Sequencing, Notch3, blood retinal barrier
1. Introduction
Diabetic macular edema (DME) is the foremost cause of visual loss in diabetes (Das et al., 2015a; Das et al., 2015b). Alteration of the blood-retinal barrier (BRB) characterized by increased vascular permeability is the hallmark of DME (Das et al., 2015a). The pathophysiology of macular edema is still not clear, and its understanding is further limited by the cellular and molecular complexity involved in the process. An extensive body of research has shown that pericytes play a critical role in the development and healthy maintenance of the neurovascular unit through direct interaction with endothelial cells, microglial and neurons (Cogan et al., 1961).
Pericyte loss is the classic histological mark of early diabetic retinopathy in the human retina. In diabetic experimental animals, pericyte “dropout” appears to be preceded by permeability changes in the retinal microcirculation (Qaum et al., 2001; Zhang et al., 2005). These changes may include the novel mechanism of pericyte detachment and migration that may be a precursor to the pericyte loss seen in the diabetic retina (Pfister et al., 2008). The “pericyte dysfunction” in the earliest stages of diabetic retinopathy results in modification of the physical interaction of endothelial cells and pericytes and the ability of pericytes to regulate the endothelial permeability barrier. In addition to the defined roles of PDGF, TGF-β and the angiopoietins in supporting pericyte-endothelial interactions, additional factors may play important roles that could be altered in diseases affecting the retina (Bergers and Song, 2005; Gerhardt and Betsholtz, 2003; Hellstrom et al., 1999; Hirschi et al., 1998; Uemura et al., 2002). Recently, our lab has also demonstrated that pericytes increase the integrity of the endothelial cell monolayer through a sphingosine-1-phosphate (S1P) mediated mechanism (McGuire et al., 2011).
Several studies have taken various approaches to understand the molecular alterations of BRB, including hypothesis driven candidate target approach, microarray analysis, and gene expression profile approach. These studies have provided information about the gene expression alterations in diabetic retinal vasculature but are limited significantly by the cellular heterogeneity.
Whole tissue gene expression profiling is severely hampered by noise and nonspecific amplification of signals (Porter et al., 2017). Further, in vitro cell line-based approaches are severely limited by non-physiological and short time exposure of diabetic milieu and does not recapitulate the molecular signature due to complex retinal neurovascular interaction. Given the importance of pericyte in DR, it is paramount to examine the RNA expression profile and properties of individual purified in vivo retinal pericyte cell population to understanding the molecular alteration of BRB. In recent years, the advent of high-throughput genomics technology has made possible the analysis of specific cell type-high resolution expression profiling using RNAs sequencing technology. Such technologies have stirred the cataloging of RNA expression patterns to reveal novel molecular mechanism and biological insights for drug discovery (Doostparast Torshizi and Wang, 2018).
In this study, we combined functional assays with next generation transcriptomic profiling using flow cytometric sorted pericyte cell population to gain deeper insight into the pericyte factors that may alter vascular permeability in DR. Here we describe diverse pericyte transcriptional signatures, composed of key gene set that have been proposed to involve in the molecular mechanism of pericyte dysfunction. Through unbiased analysis and comprehensive overview, we also identified NOTCH3 as a novel molecular marker of pericyte dysfunction and demonstrate that defective NOTCH3 signaling in pericytes leads to alteration of endothelial barrier function.
2. Materials and Methods:
2.1. Animal Model
Male C57BL/6J mice (jax.org) were used for these studies. Mice received five daily consecutive intraperitoneal injections of streptozotocin (50 mg/kg/day) in 10 mM citrate buffer (pH 4.5). Animals with plasma glucose concentrations greater than 250 mg/dL 24 to 48 hours after streptozotocin injection were considered diabetic and were used in the study at 12 weeks after the induction of diabetes. Blood glucose levels and body weight were monitored regularly. Animals received insulin (0.5 IU) as needed to maintain body weight. Age-matched nondiabetic animals were used as comparison controls. All animal studies were consistent with and adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
2.2. Fluorescence activated cell sorting (FACS) of retinal pericytes
Retinas from non-diabetic and diabetic mouse eyes (n =10 animals in each group) were digested with 0.5 mg/mL collagenase D (Roche/Sigma-Aldrich, MO, USA) for 30 minutes at 37°C and 100 mg/mL DNase I and 1mg/ml RNAse inhibitor in enzyme free Cell Dissociation Buffer (Life technologies, CA, USA). The tissue digest was then filtered through 70 μm cell strainer, and washed with DMEM with 5% FBS for 5 mins at 400 g at 4°C. The supernatant was carefully removed, and the digested tissue pellet was resuspended in DMEM with 5% FBS to form a single cell suspension. For sorting pericytes, six retinas from each group (diabetes or control) were pooled as a sample to obtain sufficient number of single cell suspension from retinas from each group. Cells were surface stained using fluorescent conjugated monoclonal antibodies 0.25μg of PDGFRb-APC (ebioscience, CA, USA) for 30 minutes at 4°C, for sorting pericyte from each group (n=3). The cell suspension was washed with DMEM with 5% FBS for 5 mins at 400 g at 4°C, followed by resuspension in DMEM with 5% FBS. The cells were sorted using BD FACS Aria, the cells were sorted directly into 1.5ml eppendorf tubes containing 50ul of RNA lysis buffer (Figure 1).
Figure 1.
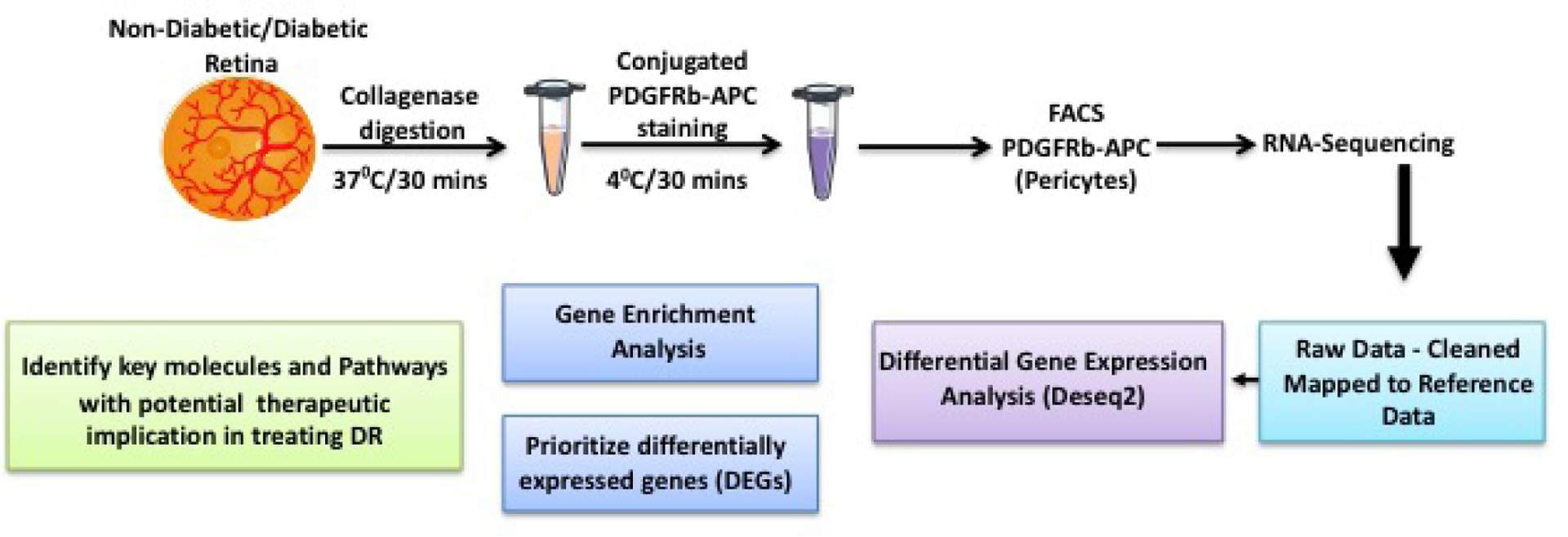
Detailed illustration of the study design.
2.3. RNA Sequencing
To enrich the number of pericytes for transcriptomic analysis we pooled at least 6 retinas per sample from age and sex matched diabetic and non-diabetic mice. RNA was extracted using Direct-zol kit (Zymo Research, CA, USA) from the sorted pericytes collected in the RNA lysis buffer and approximately 500 ng of RNA was used for cDNA cDNA conversion. Sequencing library was prepared with the TruSeq RNA Library Preparation Kit v2 (non-stranded; Illumina). Barcoded libraries were pooled (8-plex) and run on two lanes of a flow cell on an Illumina NovaSeq 6000. Sequencing was performed using 2 × 83 bp reads. Reads were aligned to the mouse genome (mm10) using STAR v2.4(Dobin et al., 2013), and quality control was performed using Picard RnaSeqMetrics (v1.128). Gene and exon read counts were generated using HTSeq(Anders et al., 2015). The purity of pericytes was confirmed by real time PCR for the expression of PDGFRb, and VE-Cad expression. The sorted cells showed expression for PDGFRb and not for VE-Cad. Normalization, differential expression analysis and correction for multiple testing were performed with DESeq2. Heatmaps were made using Euclidean distances and the complete linkage clustering method using the heatmap R-package. we conducted pairwise comparison of the expression level of diabetic vs non-diabetic pericytes and P values were adjusted for multiple comparisons using the Bonferroni method. All the analyses were conducted using R software. Raw RNA-Seq data will be deposited on Gene Expression Omnibus repository.
2.4. Cell Culture
Human retinal pericytes (HRP) and Human retinal endothelial cells (HRECs) were obtained from Cell Systems (Kirkland, WA). The cells were grown on fibronectin-coated cell culture dishes in MCDB-131 supplemented with 10% FBS, 10 ng/mL EGF, 1 μg/mL hydrocortisone, 0.2 mg/mL endothelial culture medium (EndoGro; Millipore, MA, USA), and 0.09 mg/mL heparin (pH 7.3; VEC Technologies, NY, USA). Passages 3 to 8 were used for all experiments. Both these cell types were validated for von Willebrand factor, an endothelial cell marker, Desmin, and neural/glial antigen 2 markers for pericytes (Monickaraj et al., 2018).
2.5. Advanced glycation end-product treatment
HRPs were treated with 250 and 500μg advanced glycation end-product-BSA (AGE-BSA) (Sigma-Aldrich, St. Louis, MO) for 96 hrs.
2.6. In vitro siRNA Knockdown
Notch3 gene was knockdown in HRP using Lipofectamine 3000 according to the manufacturer’s protocol (Thermofisher, CA, USA). In brief the cells were treated with Notch3 siRNA and scramble siRNA as a control, followed by lipofectamine 3000 transfection the cells were incubated for 24–48 hours followed by validation using western blotting.
2.7. Western Blot
Cells were lysed using RIPA buffer, followed by centrifugation at 12000 rpm for 10 min at 4°C. The protein concentration of each sample was estimated using Pierce BCA Protein Assay method. Protein samples were separated in precast TGX gels (Biorad, CA, USA) and transferred to blotting membranes and probed with specific primary antibodies (Abcam, Eugene, OR), followed by incubation with fluorescently-labeled secondary antibody (LI-COR, NE, USA) and images collected on the LI-COR Odyssey. Protein levels were normalized to β-tubulin (Abcam, Eugene, OR).
2.8. Real-Time PCR
Total mRNA was extracted from the HRPs and first-strand cDNA was synthesized from 1 μg total RNA (High-Capacity cDNA Reverse Transcription Kit; (Life Technologies, CA, USA) and probed with gene specific primers. Amplification and detection were then performed on a fast real-time PCR system (7500 Fast; Life Technologies). Data were derived by using the comparative Ct method for duplicate reactions with normalization to the housekeeping gene 18s RNA.
2.9. Electrical Cell-Substrate Impedance Sensing (ECIS)
Monolayer permeability was determined using the electrical cell substrate impedance sensing (ECIS) system from Applied Biophysics (NY, USA). HRECs (1×105) were plated into fibronectin-coated multiwell chambers (8W10E+) and grown for 16 hours until maximum resistance was attained (~1200 Ω). HRECs were treated with the conditioned media (CM) obtained from siRNA treated HRPs (Notch3 and Scramble) for 24–48 hrs followed by replacing of the media with fresh growth media, the cells were allowed to grow for another 48hrs then the media was centrifuged to remove the cell debris. 1:1 dilution of CM from pericytes with growth media was used. After the treatment the resistance were monitored for up to 18 hours. Resistance values for multiple wells were normalized to an identical starting resistance values and averaged and presented as normalized resistance over time.
2.10. Statistical Methods
For all quantitative experiments, statistical analyses of data were performed with either an unpaired t-test or a one-way ANOVA (Prism8 software; GraphPad). Differences among means were tested by ANOVA and corrected using the Bonferroni multiple comparison post test. Values for P < 0.05 were considered significant.
3. RESULTS:
3.1. RNA sequencing of Retinal Pericytes
In order to understand the molecular mechanism of pericyte dysfunction in DR, we set out to isolate the retinal pericytes from mice model of diabetes and perform RNA sequencing. Pdgfrb is the most commonly used markers for pericytes and we sorted Pdgfrb positive pericytes (PDGFRb-APC stained cells) from the retina of diabetic and non -diabetic mice through FACS (Figure 2). We obtained a high quality pericyte transcriptomics with >70M reads per sample (>Q30).
Figure 2.
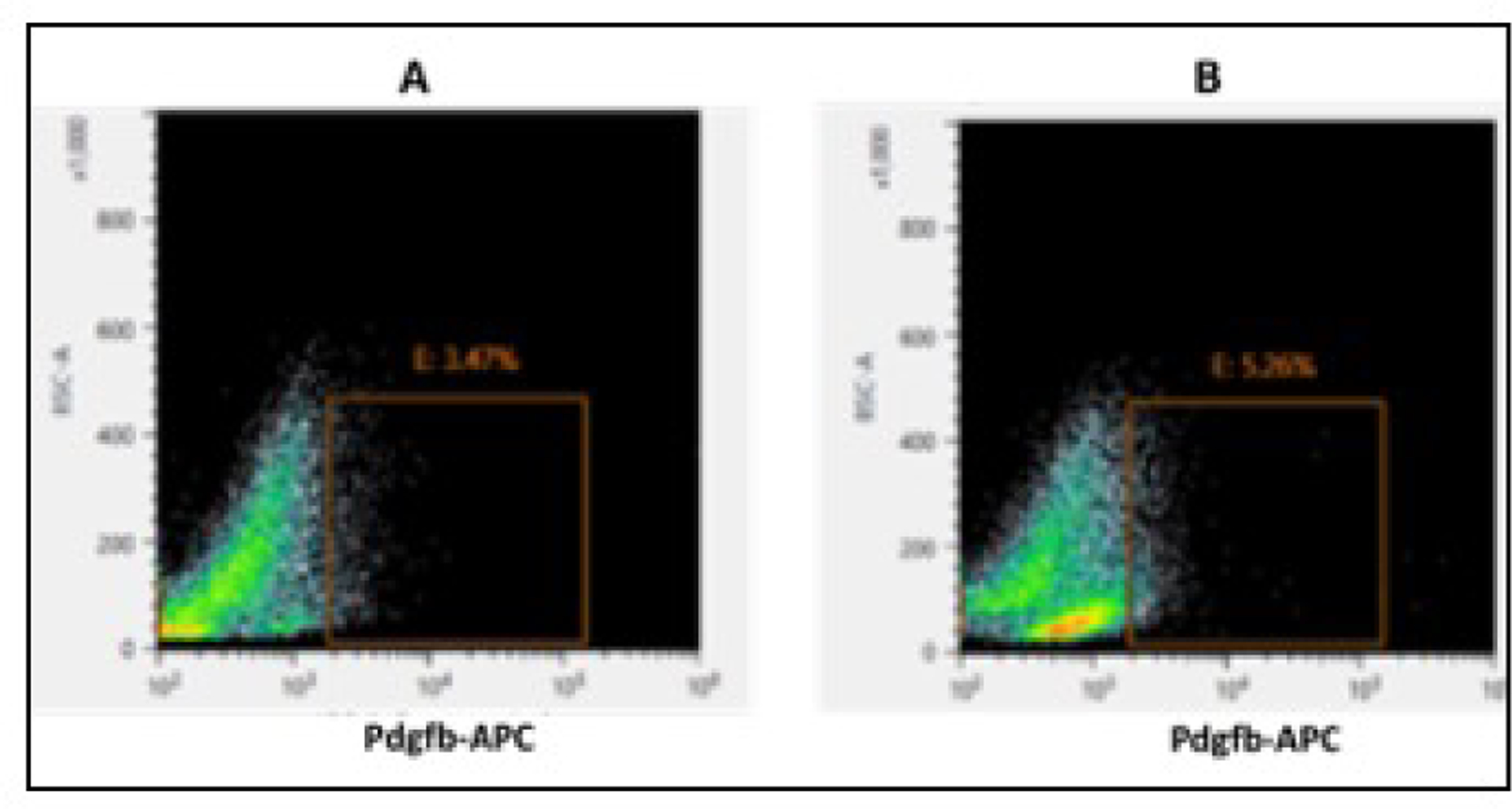
FACS purification of retinal pericytes. The representative Dot blots display the intensity of channel of analyzed retinal dissociated cells for Pdgfrb-APC staining from (A) non-diabetic mice retina and (B) diabetic mice retina (12 weeks old). The sorted, Pdgfrb-APC positive cell population for pericytes is specified by the inset.
In our initial experiment, we validated the sorted retinal pericyte cells using qRT-PCR. We confirmed the expression of pericyte marker PDGFRb (0.89-fold) relative to GAPDH mRNA from approximately 500 sorted cells. We further assessed the nature of the FACS sorted population of cells by measuring the mRNA count for pericyte specific cell markers from the RNA sequence data (Chasseigneaux et al., 2018). Expression of these markers such as PDGFRb, Rgs5, Abcc9, or Vtn as marker of pericyte cells has been previously documented in the transcriptomic studies (He et al., 2016; Zeisel et al., 2015; Zhang et al., 2014), and confirm that the purified cells are pericytes, however our study does not account for the heterogeneity of the analyzed cells (Fig. S1).
3.2. Identification of Gene Transcripts from Pericytes of Diabetic mice
To identify the pathway mediating hyperglycemia induced pericyte dysfunction, we compared the mRNA expression profile of the sorted pericyte population from diabetic and non-diabetic mice. Differential expression analysis of RNA-seq data through DESeq2 revealed a total of 712 genes were differentially regulated with a fold change in expression > 2.00 (adjusted p < 0.05) in diabetic pericytes compared to non-diabetic controls cells. Out of these, 495 mRNAs were significantly downregulated, and 217 mRNAs were upregulated in pericytes cell from diabetes condition as shown in heat map and in the volcano plot (Figure 3a). To investigate the relatedness among the samples between diabetic and non-diabetic control, differentially expressed genes (log 2fold changes, padj <0.05) were subjected to unsupervised hierarchical clustering analysis and the results are shown in heat map format (Figure 3b).
Figure 3.
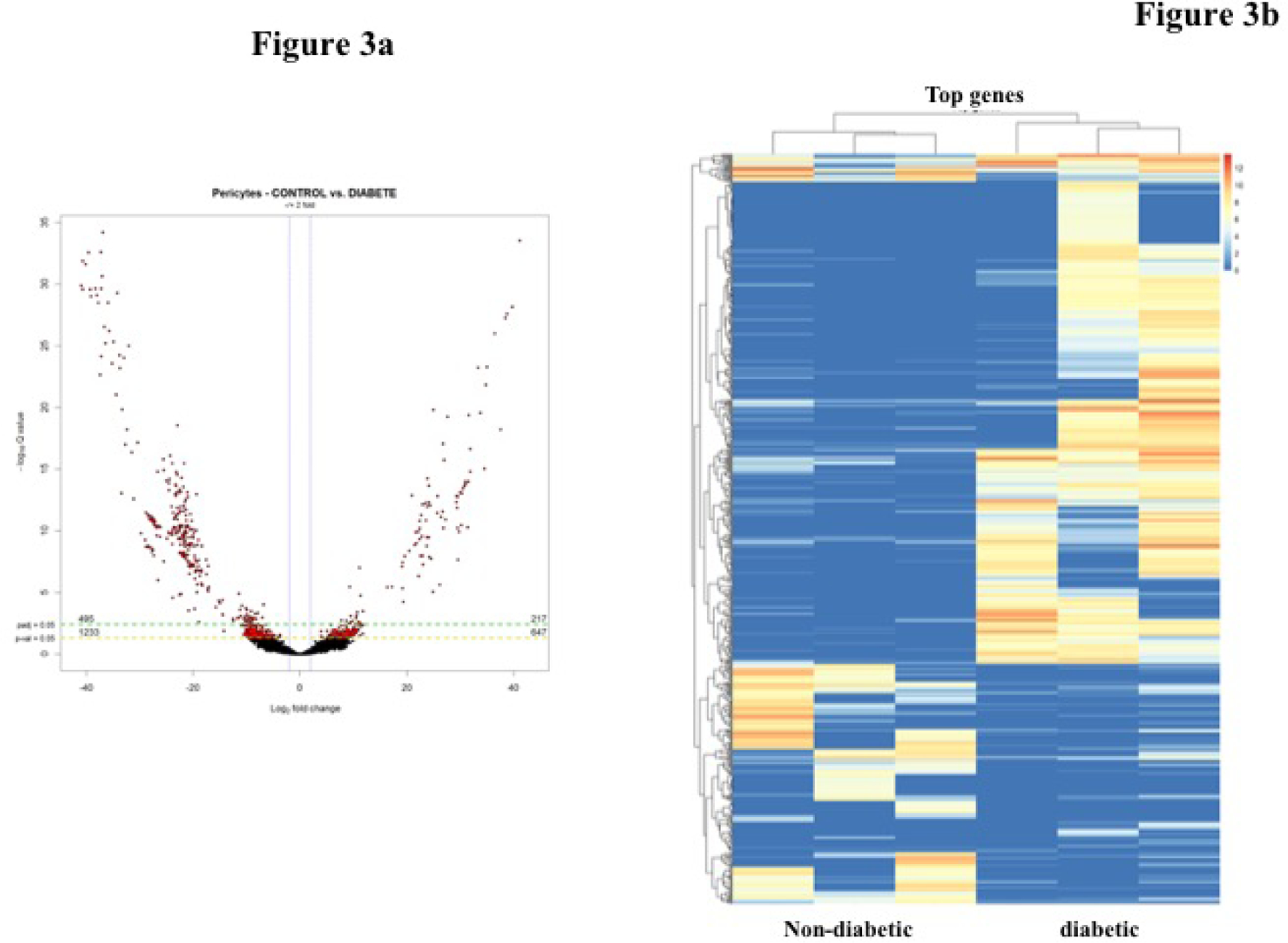
a) Volcano plot of fold change (FC) of transcripts derived using edgeR. Diabetic pericytes compared to control pericytes. Significantly upregulated genes (217) are in the right side and down regulated genes (495) in the left indicated in red circles. b) Hierarchically clustered heatmap showing transcripts of pericytes from non-diabetic and diabetic mice (n=3).
3.3. Differentially Expressed Genes (DEGs) and Pathway Analysis
Next, we explored the molecular functions and the pathways from differentially expressed genes (DEGs) through gene set enrichment analysis (GSEA) and identified highly significant pathways associated with top genes from the overall profile (Figure 4). We further used hierarchical approach determined by the gradual convergence of biologically relevant function to pick the top gene pertinent to DR pathogenesis based on four -tier approach through: a) pericyte cell-enriched gene catalogue b) modifiers of pro-inflammatory reaction, c) molecular factor involved in cell growth and apoptosis, d) cell signaling pathway regulating cell adhesion and vascular barrier permeability. Through this approach, we identified top five up-regulated genes (Table 1) and down-regulated genes (Table 2). Further, mapping the mRNA for pericyte specificity and abundance, we identified Notch3 expression as a top-ranking down regulated candidate gene that may play a role in the molecular mechanism of pericyte dysfunction in DR. The expression of Notch 3 in vascular mural cells and the evidence for Notch signaling pathway in the regulation of vascular integrity strongly suggest a role in the diabetic pericytes.
Figure 4.
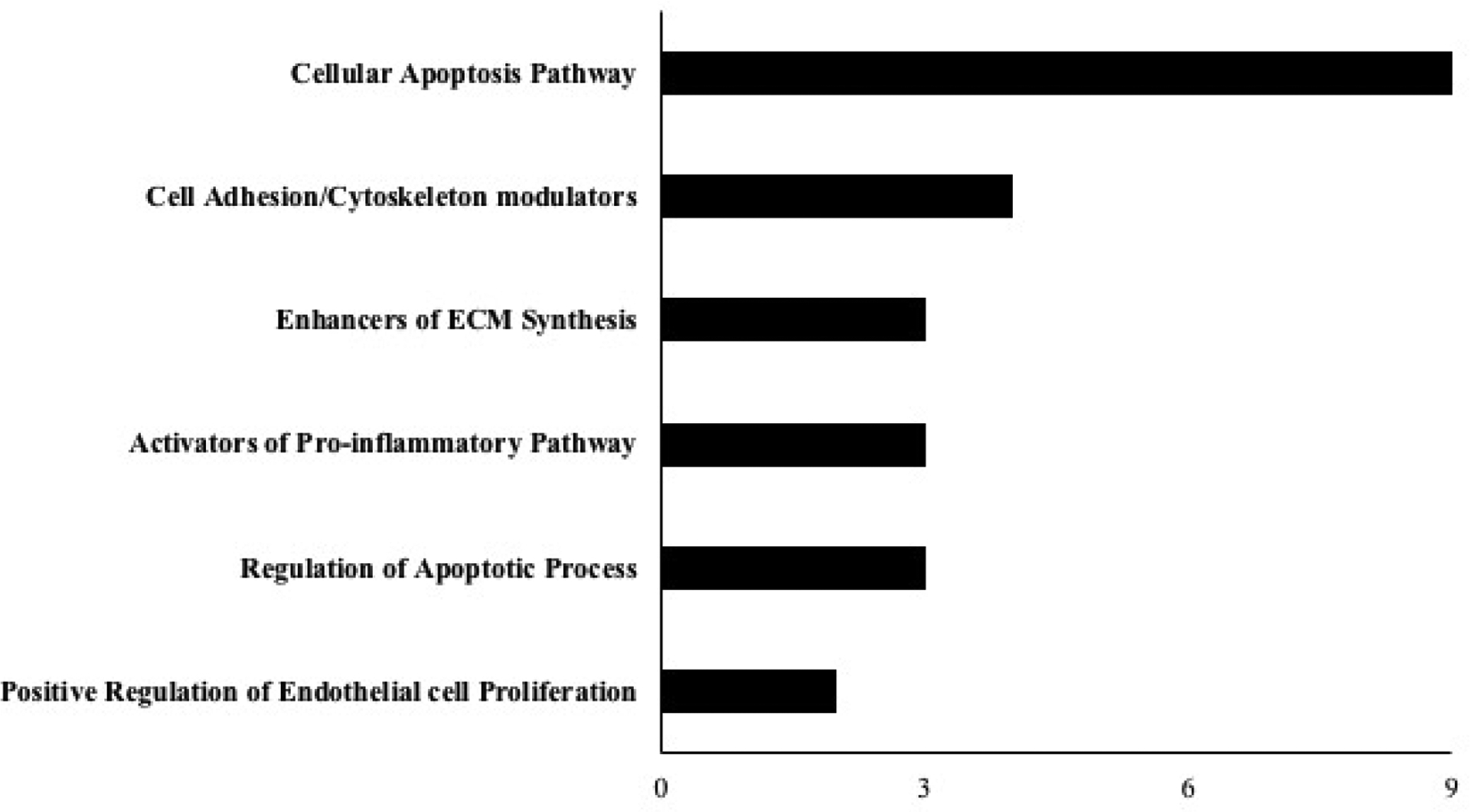
Gene set enrichment analysis (GSEA) was done for genes that were regulated in the diabetic pericytes with two-fold regulation (p<0.05).
Table 1.
List of pericyte specific top 5 up-regulated genes.
| Gene | Log2 FC | P-Value | Functions |
|---|---|---|---|
| EXT2- Exostosin-2 |
24.02597 | 1.20E-14 | Enhanced Protein Glycosylation; increased proteoglycan biosynthesis and ECM synthesis; PG plays an important role in the formation of neovascularization; Potential Factor for initiating Pericyte migration |
| B3GAT3- Beta-1,3-Glucuronyltransferase 3 |
21.58965 | 1.48E-10 | |
| GPC6- Glypican 6 |
19.36 | 9.11E-10 | Highly expressed in pericytes; Promotes cell migration and immune cell infiltration |
| PIP5K1C- Phosphatidylinositol-4- Phosphate 5-Kinase Type 1 Gamma |
5.47144 | 0.00233 | Pro-inflammatory Factor that promotes immune cell infiltration |
| PTEN- Phosphatase And Tensin Homolog |
5.14992 | 0.00277 | Regulates angiogenesis and a modulator of actin cytoskeletal dynamics resulting in increased paracellular permeability. Promotes Cell Apoptosis |
Table 2.
List of pericyte specific top 5 down-regulated genes.
| Gene | Log2 FC | P-Value | Functions |
|---|---|---|---|
| NOTCH3- Notch Receptor 3 |
−29.9702 | 6.18E-16 | Pericyte survival Factor and regulator of vascular integrity |
| XBP1- X-Box Binding Protein 1 |
−22.6406 | 6.83E-14 | Inhibition potentiates cytokine and ER stress-induced apoptosis |
| GPC4- Glypican 4 |
−11.77120 | 0.00017 | Critical for endostatin binding and regulating endothelial barrier function |
| ATP1A2- ATPase Na+/K+ Transporting Subunit Alpha 2 |
−9.2817 | 1.16E-07 | Maintain contractibility of Perivascular cells and barrier function |
| AKT3- AKT Serine/Threonine Kinase 3 |
−6.0302 | 0.00059 | Critical prosurvival factor |
3.4. Notch3 expression decreased in Pericytes treated with AGE
To mimic glycated hemoglobin in diabetes, human retinal pericytes (HRPs) were treated with 250 and 500μg AGE (advanced glycation end-product) for 96 hrs. There was a significant decrease in Notch3 mRNA expression in cells treated with 500μg AGE compared with untreated cells (Ctrl) (p<0.027) (Figure 5).
Figure 5.
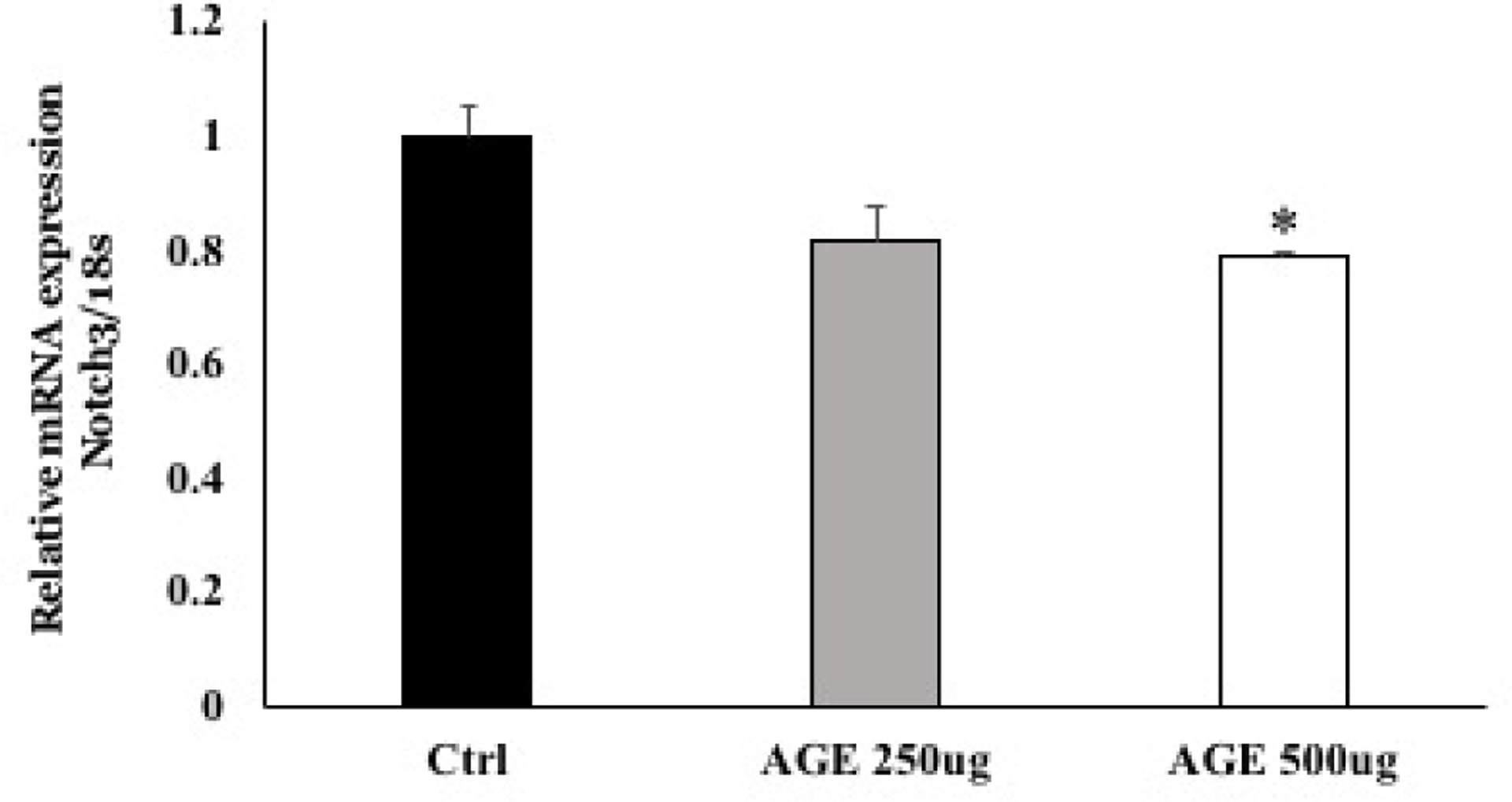
mRNA expression of Notch3 in HRPs treated with 500μg AGE was significantly decreased compared to untreated cells (*p=0.027).
3.5. Notch3 downregulation leads to increased expression of inflammatory and autophagy genes
In order to find the influence of Notch3 downregulation on inflammation and autophagy in pericytes, we knocked down Notch3 in human retinal pericytes using siRNA (Figure 6a). Knocking down Notch3 led to significant increase in inflammatory genes like angiopoietin2 (Ang2), CCL2, ICAM1, VEGF and MMP2 in comparison with cells treated with scrambled siRNA (Figure 6b). To further explain the association of notch3 downregulation and pericyte loss in diabetic retinopathy, we looked for autophagy markers (ATG5, BECN1, LC3B, p62) in pericytes knocked out for Notch3. Autophagy markers were significantly upregulated with notch3 down regulation (Figure 6c).
Figure 6. Functional study on the impact of Notch3 knockdown in HRPs.
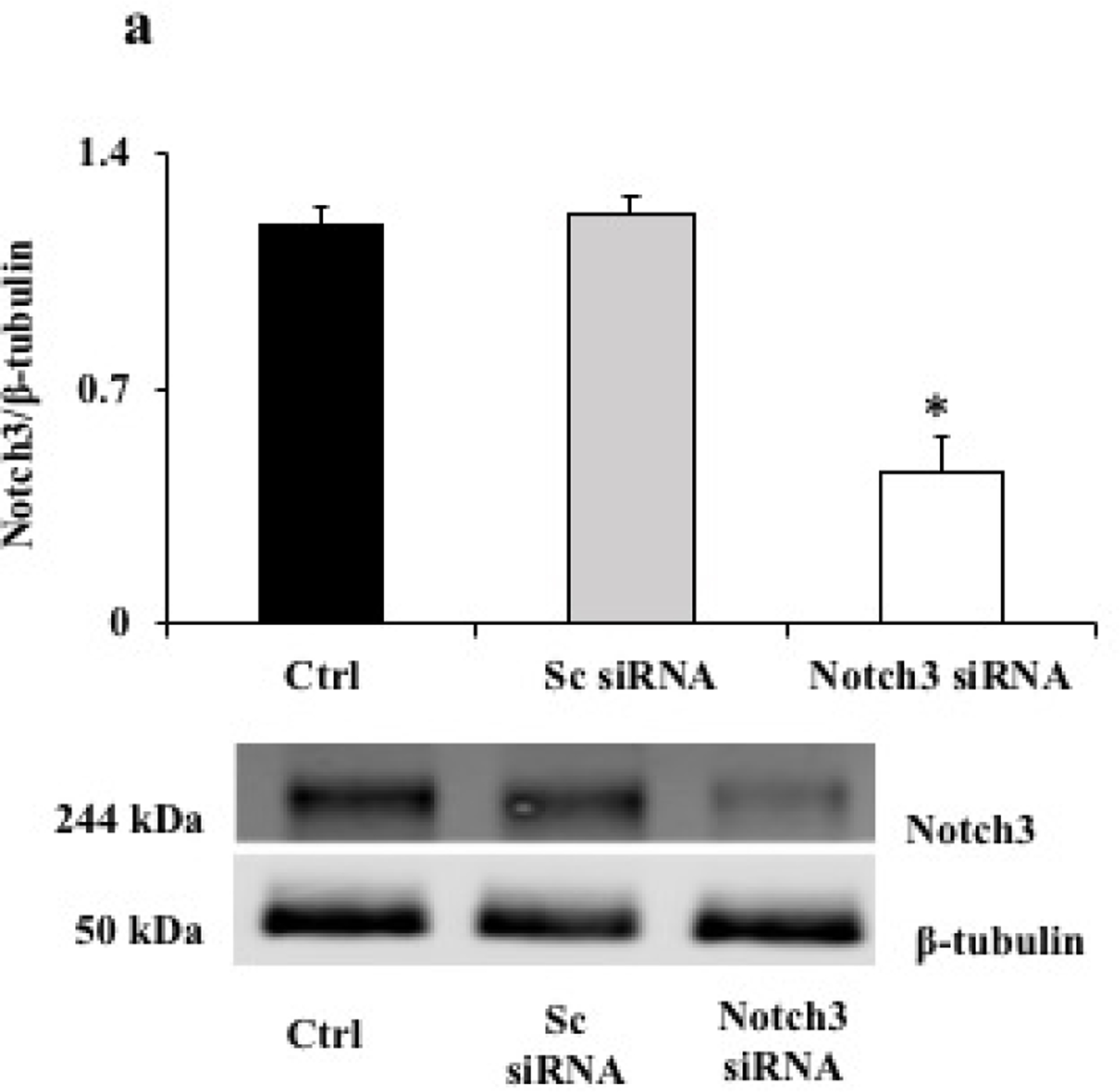
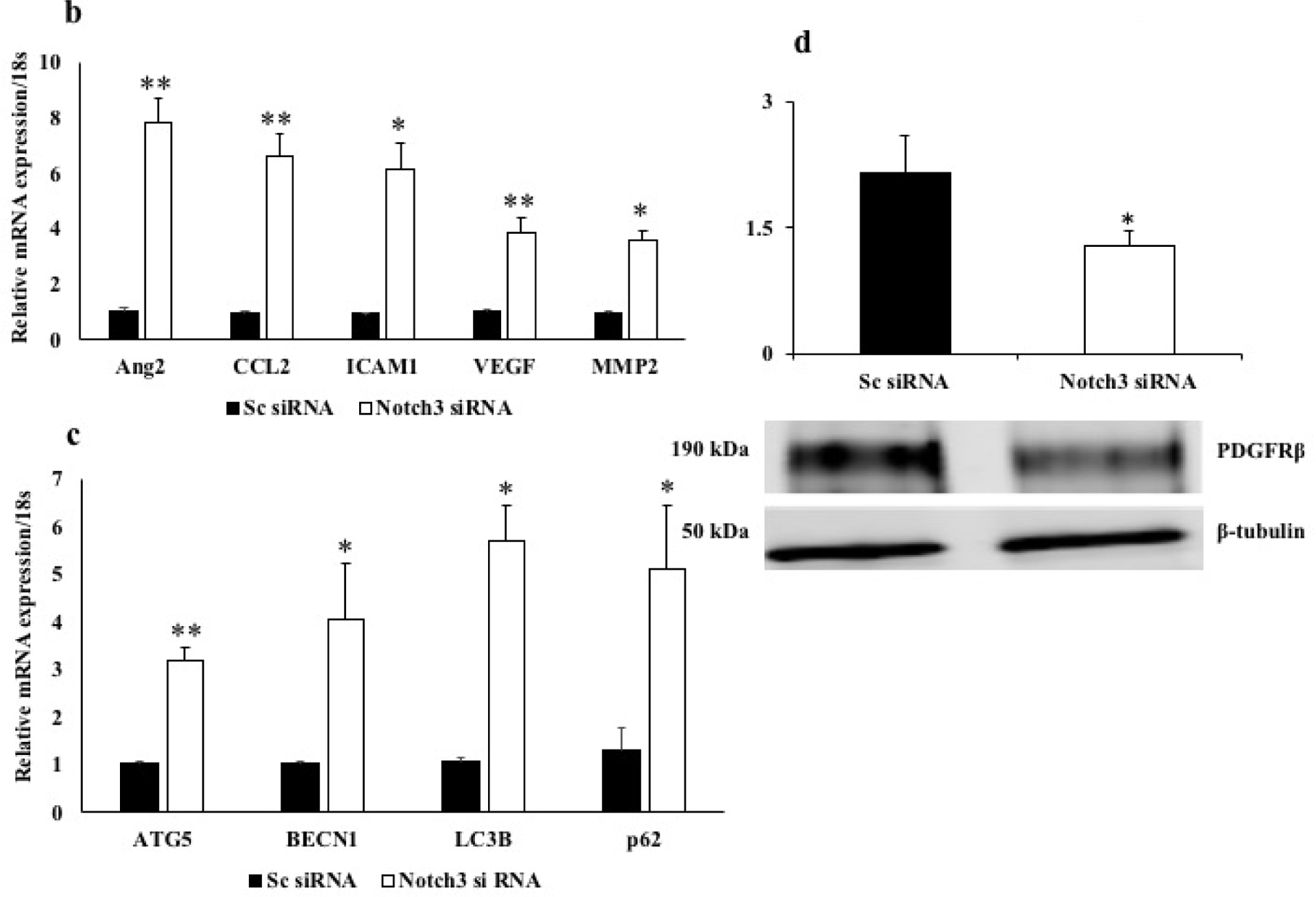
a. Representative western blot image of Notch3 and β-tubulin protein in HRPs knockdown for Notch3 using siRNA. Control (untreated), Notch3 siRNA and scrambled (Sc) siRNA. HRPs treated with Notch3 siRNA resulted in a significant reduction in the Notch3 protein expression in comparison with cells treated with Sc siRNA or untreated cells (*p≤0.002). b. mRNA expression of inflammatory genes- angiopoietin2 (Ang2), CCL2, ICAM1, VEGF and MMP2 were significantly increased in Notch3 siRNA HRPs compared with sc siRNA cells (**p≤0.009, *p=0.01). c. mRNA expression of autophagy markers- ATG5, BECN1, LC3B, p62 were significantly increased in Notch3 siRNA cells compared with sc siRNA cells (**p≤0.008, *p≤0.05). d. Representative western blot image of PDGFRb and β-tubulin protein in Notch3 siRNA and Sc siRNA. PDGFRb protein level was significantly decreased in cells treated with Notch3 siRNA cells compared with Sc siRNA cells (*p=0.03).
3.6. Notch3 downregulation leads to decreased PDGFRβ expression
PDGFRβ an essential marker of pericyte differentiation was significantly decreased in the Notch3 Knockdown pericytes compared with scramble siRNA pericytes (Figure 6d).
3.7. Notch3 downregulation leads to increased permeability in vitro
In order to check the influence of Notch3 downregulation in vascular permeability, we treated human retinal endothelial cells (HREC) with conditioned media from the HRPs treated with both scramble and Notch3 siRNA. Treatment with conditioned media from Notch3 siRNA cells led to increased permeability compared to cells treated with conditioned media from scramble siRNA cells (Figure 7).
Figure 7.
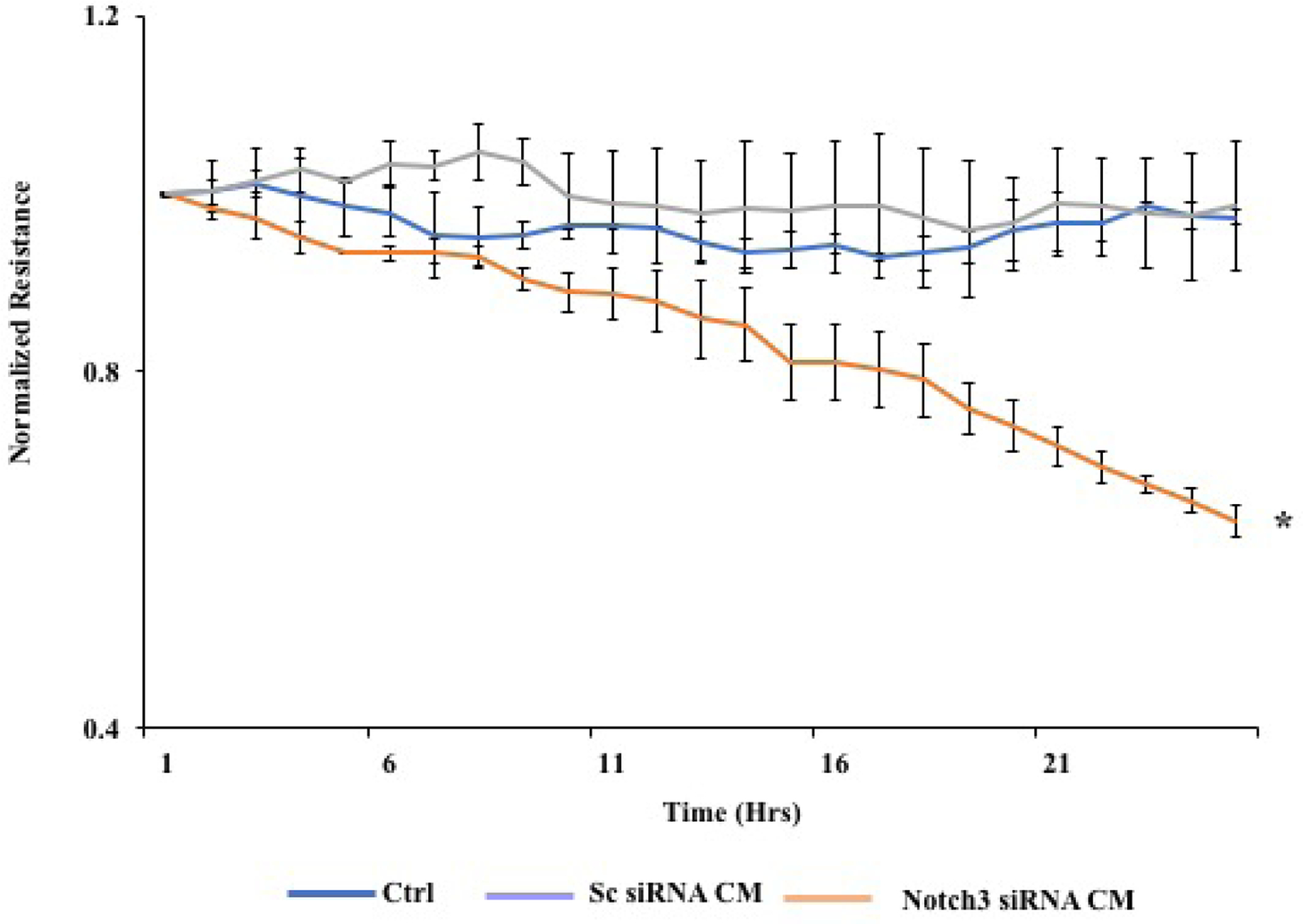
Preformed monolayers of HRECs were incubated with 50% of conditioned media from cells treated with both scrambled and Notch3 siRNA and the normalized resistance was determined. Decreased resistance which implies increased permeability was seen in cells treated with Notch3 siRNA conditioned media (CM) compared with CM from Sc siRNA cells (*p=0.01).
4. Discussion
Pericyte interaction with endothelial cells is essential for the structural organization of retinal microvasculature and BRB function(Runkle and Antonetti, 2011). Pericyte loss is considered as the histological hallmarks of early diabetic retinopathy and BRB alterations; however the underlying molecular mechanism is still poorly understood (Engerman, 1989). In this study we took advantage of RNA sequencing combined with advanced bioinformatics to decipher the molecular basis of pericyte dysfunction in diabetic mouse retina. RNA sequencing has significantly improved our ability to quantitatively understand and characterize the differentially expressed genes and pathways and decipher novel mechanism of action contributing to the disease (Costa et al., 2013).
Previous gene expression profiling studies to decipher pericyte specific molecular signature were severely limited by retinal cellular heterogeneity and lack of in vivo pericyte specific gene expression profiles for comparison. We have combined FACS purification approach to sort pericyte cell population with the next generation transcriptomic profiling of purified pericytes from the age and sex matched diabetic and non-diabetic rodent retina. Our study represents an innovative resource in the study of diabetic retinopathy for the following two reasons. We utilized PDGFRβ, a consistent and most frequently used marker to isolate the mouse retinal pericytes (Epshtein et al., 2017; Guichet et al., 2015; Lindahl et al., 1997; Lindblom et al., 2003; Winkler et al., 2010). We then performed next generation transcriptomics analysis of pericyte samples sequenced at high depth (>80 million reads per sample) that enabled the quantification of differentially expressed genes with high confidence. RNA-sequencing of enriched pericytes from both the healthy and diabetic retina has allowed us to uncover novel genes, which certainly play an important role in pericyte dysfunction and BRB alterations.
To identify potential genes that may contribute to pericyte specific dysfunction from the RNA sequencing, we further mapped the enriched genes using hierarchical approach based on known mechanisms of DR pathogenesis and mural cell abundance. Through differential expression analysis, we have generated a comprehensive list of gene transcripts significantly enriched in the retinal pericyte cell population from the diabetic mice compared to controls. Several mechanisms have been tied to retinal pericyte dysfunction and blood retinal barrier alterations in DR (Das et al., 2015a; Hammes et al., 2004; McGuire et al., 2011; Pfister et al., 2008). Compromise in microvascular integrity, altered barrier function, ischemia, and leukocyte infiltration are some of the most common mechanisms that are associated with pericyte dysfunction and DR progression (Das et al., 2015a). To identify the pericyte molecular signature linking DR pathogenesis, we performed GO enrichment analysis and detected a significant representation of genes involved in the cell apoptosis pathway (Gpc6, Bmp3, Endog, Igf2r, Prps1, Pgf, Bmp6, Ppid, Tbrg1), cell adhesion/cytoskeleton modulators pro-inflammatory pathway 9 (Ext2, Gpc6, B3gat3, Pten) and pro-inflammatory pathway (Pip5k1c, Pten, Gpc6). In addition, we observed gene expression alterations involved in endothelial cell proliferation and ECM alterations. Enrichment of potential genes linked to the biological process of DR pathogenesis may serve as molecular markers of pericyte dysfunction. However, the connection between these gene expression changes and the health of pericyte is not clear and further studies will be required to determine whether these genes directly contribute to DR phenotypes.
Pericytes play an important role in the formation, maturation, and maintenance of the microvasculature. Pericytes are contractile cells that are postulated to regulate regional flow through the microcirculation (Das et al., 1988; Kelley et al., 1987). The vascular beds found in the nervous system, in which vascular permeability is limited, have a ratio of pericytes to endothelial cells that is much higher than that found in tissues with normally higher levels of permeability (Armulik et al., 2005; McGuire et al., 2011; Ozerdem et al., 2001). Pericyte loss as seen in diabetic retinopathy results in focal endothelial cell proliferation and probably contributes to the formation of microaneurysms in the weakened wall of retinal capillaries.
Our results shown down regulation of the Notch3 gene in pericytes from diabetic animals that may play an important role in pericyte dysfunction in diabetic retinopathy. Notch signaling plays a vital role in endothelial-pericyte interactions. Notch functions as a receptor, and mammals have four Notch receptors (Notch1, Notch2, Notch3 and Notch4) and many ligands, like jagged 1 (JAG1) and JAG2 and delta-like proteins(Ables et al., 2011). Mutations in NOTCH3, causes cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), the most common monogenic cause of cerebral small vessel disease (SVD) and also associated with stroke, and vascular cognitive impairment and dementia(Machuca-Parra et al., 2017). Notch3 knockout mice revealed abnormal maturation of arterial vSCMs with reduced PDGFRb(Domenga et al., 2004; Jin et al., 2008). Using a pericyte expressed LacZ transgene (XlacZ4) to examine pericyte loss, it has been shown that Ins2Akita mice (type 1 diabetes model) in combination with the Notch3+/− deficiency increases numbers of acellular capillaries and pericyte loss (Liu et al., 2018).
Notch signaling plays a vital role in the downstream expression of canonical Notch target key transcriptional repressors (Hes and Hey), PDGFR-β, and smooth muscle α-actin (αSMA)(Guichet et al., 2015; Iso et al., 2003; Schulz et al., 2015). In our study, we found that the downregulation of NOTCH3 leads to decreased PDGFRβ expression, indicating that NOTCH3 is a potential downstream regulator of PDGFR β expression in pericytes. Animal models of diabetic retinopathy have identified that reduced PDGFRβ signaling in hyperglycemia leads to pericyte apoptosis. These results indicate that hyperglycemia-induced downregulation of NOTCH3 contributes to various signaling mechanisms leading to pericyte dysfunction.
In summary, we demonstrate that diabetes may mediate down regulation of Notch 3 expression in retinal pericytes of STZ induced diabetic mice. Further investigations on human retinal pericytes revealed loss of NOTHC3 leads to increased autophagy and pro-inflammatory response. Collectively our results identify a critical molecular signaling of DR pathogenesis and provide new insights into the molecular mechanisms of pericyte dysfunction in diabetes. Possible activation of Notch3 may prove to be a novel therapeutic strategy in management of diabetic macular edema.
Supplementary Material
Figure S1. Listed are RNA-seq expression values for the known pericyte markers. The expression quantification is given by LCF (Log Transformation). VTN-Vitronectin, PDGFRb-Platelet Derived Growth Factor Receptor Beta, RGS5-Regulator Of G Protein Signaling 5, ABCC9-ATP Binding Cassette Subfamily C Member 9.
Table S1. List of pericyte specific top 5 up-regulated genes.
Table S2. List of pericyte specific top 5 down-regulated genes.
Highlights:
Transcriptomic analysis of pericytes isolated from diabetic animals revealed a core set of differentially expressed genes.
The top up regulated and down regulated genes probably play a role in inflammation, immune cell infiltration, apoptosis, pericyte migration and barrier function alteration.
Functional validation studies show that Notch3 downregulation in pericytes in diabetes may lead to increased autophagy, inflammation and permeability changes as seen in diabetic retinopathy.
Funding:
International Retinal Research Foundation Grant #42817, NIH EY022327 and VA Merit Review Award IO 1BX001801
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicting interest statement
The authors declare that they have no competing financial interests.
References
- Ables JL, Breunig JJ, Eisch AJ, Rakic P, 2011. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci 12, 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W, 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A, Abramsson A, Betsholtz C, 2005. Endothelial/pericyte interactions. Circ Res 97, 512–523. [DOI] [PubMed] [Google Scholar]
- Bergers G, Song S, 2005. The role of pericytes in blood-vessel formation and maintenance. Neuro Oncol 7, 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasseigneaux S, Moraca Y, Cochois-Guegan V, Boulay AC, Gilbert A, Le Crom S, Blugeon C, Firmo C, Cisternino S, Laplanche JL, Curis E, Decleves X, Saubamea B, 2018. Isolation and differential transcriptome of vascular smooth muscle cells and mid-capillary pericytes from the rat brain. Sci Rep 8, 12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogan DG, Toussaint D, Kuwabara T, 1961. Retinal vascular patterns. IV. Diabetic retinopathy. Arch Ophthalmol 66, 366–378. [DOI] [PubMed] [Google Scholar]
- Costa V, Aprile M, Esposito R, Ciccodicola A, 2013. RNA-Seq and human complex diseases: recent accomplishments and future perspectives. Eur J Hum Genet 21, 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Frank RN, Weber ML, Kennedy A, Reidy CA, Mancini MA, 1988. ATP causes retinal pericytes to contract in vitro. Exp Eye Res 46, 349–362. [DOI] [PubMed] [Google Scholar]
- Das A, McGuire PG, Rangasamy S, 2015a. Diabetic Macular Edema: Pathophysiology and Novel Therapeutic Targets. Ophthalmology 122, 1375–1394. [DOI] [PubMed] [Google Scholar]
- Das A, Stroud S, Mehta A, Rangasamy S, 2015b. New treatments for diabetic retinopathy. Diabetes Obes Metab 17, 219–230. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z, Tournier-Lasserve E, Gridley T, Joutel A, 2004. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev 18, 2730–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doostparast Torshizi A, Wang K, 2018. Next-generation sequencing in drug development: target identification and genetically stratified clinical trials. Drug Discov Today 23, 1776–1783. [DOI] [PubMed] [Google Scholar]
- Engerman RL, 1989. Pathogenesis of diabetic retinopathy. Diabetes 38, 1203–1206. [DOI] [PubMed] [Google Scholar]
- Epshtein A, Rachi E, Sakhneny L, Mizrachi S, Baer D, Landsman L, 2017. Neonatal pancreatic pericytes support beta-cell proliferation. Mol Metab 6, 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt H, Betsholtz C, 2003. Endothelial-pericyte interactions in angiogenesis. Cell Tissue Res 314, 15–23. [DOI] [PubMed] [Google Scholar]
- Guichet PO, Guelfi S, Teigell M, Hoppe L, Bakalara N, Bauchet L, Duffau H, Lamszus K, Rothhut B, Hugnot JP, 2015. Notch1 stimulation induces a vascularization switch with pericyte-like cell differentiation of glioblastoma stem cells. Stem Cells 33, 21–34. [DOI] [PubMed] [Google Scholar]
- Hammes HP, Lin J, Wagner P, Feng Y, Vom Hagen F, Krzizok T, Renner O, Breier G, Brownlee M, Deutsch U, 2004. Angiopoietin-2 causes pericyte dropout in the normal retina: evidence for involvement in diabetic retinopathy. Diabetes 53, 1104–1110. [DOI] [PubMed] [Google Scholar]
- He L, Vanlandewijck M, Raschperger E, Andaloussi Mae M, Jung B, Lebouvier T, Ando K, Hofmann J, Keller A, Betsholtz C, 2016. Analysis of the brain mural cell transcriptome. Sci Rep 6, 35108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom M, Kalen M, Lindahl P, Abramsson A, Betsholtz C, 1999. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 126, 3047–3055. [DOI] [PubMed] [Google Scholar]
- Hirschi KK, Rohovsky SA, D’Amore PA, 1998. PDGF, TGF-beta, and heterotypic cell-cell interactions mediate endothelial cell-induced recruitment of 10T1/2 cells and their differentiation to a smooth muscle fate. J Cell Biol 141, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y, 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 194, 237–255. [DOI] [PubMed] [Google Scholar]
- Jin S, Hansson EM, Tikka S, Lanner F, Sahlgren C, Farnebo F, Baumann M, Kalimo H, Lendahl U, 2008. Notch signaling regulates platelet-derived growth factor receptor-beta expression in vascular smooth muscle cells. Circ Res 102, 1483–1491. [DOI] [PubMed] [Google Scholar]
- Kelley C, D’Amore P, Hechtman HB, Shepro D, 1987. Microvascular pericyte contractility in vitro: comparison with other cells of the vascular wall. J Cell Biol 104, 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl P, Johansson BR, Leveen P, Betsholtz C, 1997. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277, 242–245. [DOI] [PubMed] [Google Scholar]
- Lindblom P, Gerhardt H, Liebner S, Abramsson A, Enge M, Hellstrom M, Backstrom G, Fredriksson S, Landegren U, Nystrom HC, Bergstrom G, Dejana E, Ostman A, Lindahl P, Betsholtz C, 2003. Endothelial PDGF-B retention is required for proper investment of pericytes in the microvessel wall. Genes Dev 17, 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang W, Lilly B, 2018. Evaluation of Notch3 Deficiency in Diabetes-Induced Pericyte Loss in the Retina. J Vasc Res 55, 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuca-Parra AI, Bigger-Allen AA, Sanchez AV, Boutabla A, Cardona-Velez J, Amarnani D, Saint-Geniez M, Siebel CW, Kim LA, D’Amore PA, Arboleda-Velasquez JF, 2017. Therapeutic antibody targeting of Notch3 signaling prevents mural cell loss in CADASIL. J Exp Med 214, 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire PG, Rangasamy S, Maestas J, Das A, 2011. Pericyte-derived sphingosine 1-phosphate induces the expression of adhesion proteins and modulates the retinal endothelial cell barrier. Arterioscler Thromb Vasc Biol 31, e107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monickaraj F, McGuire P, Das A, 2018. Cathepsin D plays a role in endothelial-pericyte interactions during alteration of the blood-retinal barrier in diabetic retinopathy. FASEB J, fj201700781RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozerdem U, Grako KA, Dahlin-Huppe K, Monosov E, Stallcup WB, 2001. NG2 proteoglycan is expressed exclusively by mural cells during vascular morphogenesis. Dev Dyn 222, 218–227. [DOI] [PubMed] [Google Scholar]
- Pfister F, Feng Y, vom Hagen F, Hoffmann S, Molema G, Hillebrands JL, Shani M, Deutsch U, Hammes HP, 2008. Pericyte migration: a novel mechanism of pericyte loss in experimental diabetic retinopathy. Diabetes 57, 2495–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JR, Telford WG, Batchelor E, 2017. Single-cell Gene Expression Profiling Using FACS and qPCR with Internal Standards. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qaum T, Xu Q, Joussen AM, Clemens MW, Qin W, Miyamoto K, Hassessian H, Wiegand SJ, Rudge J, Yancopoulos GD, Adamis AP, 2001. VEGF-initiated blood-retinal barrier breakdown in early diabetes. Invest Ophthalmol Vis Sci 42, 2408–2413. [PubMed] [Google Scholar]
- Runkle EA, Antonetti DA, 2011. The blood-retinal barrier: structure and functional significance. Methods Mol Biol 686, 133–148. [DOI] [PubMed] [Google Scholar]
- Schulz GB, Wieland E, Wustehube-Lausch J, Boulday G, Moll I, Tournier-Lasserve E, Fischer A, 2015. Cerebral Cavernous Malformation-1 Protein Controls DLL4-Notch3 Signaling Between the Endothelium and Pericytes. Stroke 46, 1337–1343. [DOI] [PubMed] [Google Scholar]
- Uemura A, Ogawa M, Hirashima M, Fujiwara T, Koyama S, Takagi H, Honda Y, Wiegand SJ, Yancopoulos GD, Nishikawa S, 2002. Recombinant angiopoietin-1 restores higher-order architecture of growing blood vessels in mice in the absence of mural cells. J Clin Invest 110, 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler EA, Bell RD, Zlokovic BV, 2010. Pericyte-specific expression of PDGF beta receptor in mouse models with normal and deficient PDGF beta receptor signaling. Mol Neurodegener 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, Linnarsson S, 2015. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
- Zhang SX, Ma JX, Sima J, Chen Y, Hu MS, Ottlecz A, Lambrou GN, 2005. Genetic difference in susceptibility to the blood-retina barrier breakdown in diabetes and oxygen-induced retinopathy. Am J Pathol 166, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ, 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Listed are RNA-seq expression values for the known pericyte markers. The expression quantification is given by LCF (Log Transformation). VTN-Vitronectin, PDGFRb-Platelet Derived Growth Factor Receptor Beta, RGS5-Regulator Of G Protein Signaling 5, ABCC9-ATP Binding Cassette Subfamily C Member 9.
Table S1. List of pericyte specific top 5 up-regulated genes.
Table S2. List of pericyte specific top 5 down-regulated genes.


