Abstract
Background
Respiratory syncytial virus (RSV) typically causes winter outbreaks in temperate climates. During summer 2017, the Minnesota Department of Health received a report of increased cases of severe RSV-B infection.
Methods
We compared characteristics of summer 2017 cases with those of 2014–2018 summers. To understand the genetic relatedness among viruses, we performed high-throughput sequencing of RSV from patients with a spectrum of illness from sites in Minnesota and Wisconsin.
Results
From May to September 2017, 58 RSV cases (43 RSV-B) were reported compared to 20–29 cases (3–7 RSV-B) during these months in other years. Median age and frequency of comorbidities were similar, but 55% (24/43) were admitted to the ICU in 2017 compared to 12% in preceding 3 years (odds ratio, 4.84, P < .01). Sequencing was performed on 137 specimens from March 2016 to March 2018. Outbreak cases formed a unique clade sharing a single conserved nonsynonymous change in the SH gene. We observed increased cases during the following winter season, when the new lineage was the predominant strain.
Conclusions
We identified an outbreak of severe RSV-B disease associated with a new genetic lineage among urban Minnesota children during a time of expected low RSV circulation.
Keywords: respiratory syncytial virus, viral next-generation sequencing, respiratory infections, molecular epidemiology, viral pathogenesis
We describe use of whole-genome sequencing to identify viral genetic features associated with an increased number and severity of respiratory syncytial virus cases among children and adults in Minnesota.
Respiratory syncytial virus (RSV) is one of the most common viral pathogens of the respiratory tract and has global circulation. In areas with warmer climates, including parts of the southern United States [1], circulation can occur year round. However, in regions with temperate climates, such as the majority of the United States including Minnesota and Wisconsin, RSV causes seasonal outbreaks in the cooler winter months; these typically occur October through April, although there is substantial year-to-year variation in the exact timing of outbreaks [2]. The burden of disease has been most extensively studied in infants and young children in whom it is the most common cause of bronchiolitis [3] and results in >50 000 hospitalizations per year among children younger than 5 years in the United States [4]. While it is traditionally thought of as a disease of young children, multiple studies have demonstrated its pathogenic potential in adults, particularly among the elderly and adults with underlying conditions [5, 6].
RSV causes a spectrum of disease severity ranging from mild upper respiratory infections to respiratory failure and even death. Disease outcome can be influenced by many factors, including both host and viral determinants. Host age is a strong predictor of serious disease, with highest rates of hospitalization among infants younger than 6 months [4, 7–9]. Subsequent infections are typically less severe, suggesting a role for preexisting immunity in mitigating disease. Consistent with this, some studies show correlation between serum neutralizing antibodies and protection from infection [10, 11], and there is demonstrated protection offered by RSV hyperimmune globulin [12] and RSV monoclonal antibody [13]. Another factor influencing disease severity is the presence of underlying medical comorbidities, including a history of prematurity in children [14, 15] and chronic cardiopulmonary disease and immunosuppressing conditions in both children and adults. While host factors certainly influence disease severity, viral factors may also contribute.
Among the viral factors most well studied with respect to pathogenesis are the F and G glycoproteins, which mediate viral entry and are the primary targets of neutralizing antibody responses [16]. Based on differences in antibody reactivity against these surface glycoproteins, early studies of RSV identified the presence of 2 subtypes A and B [17]. The determinants of these differences in antibody reactivity were found to localize primarily to the hypervariable region of the G glycoprotein, and thus sequencing of this region forms the primary basis for subtyping. In one study, the A subtype was found to be more common and associated with increased severity of disease [18], supporting the hypothesis that viral determinants may influence disease severity. Different immune responses based on specific viral determinants have been observed in mouse models. For example, among A subtypes, the A2 and Long strains evoke a T helper (Th1)-type antiviral response in BALB/c mice [19], whereas the line 19 strain, derived via serial passage through mouse brains, elicits a Th2-type response and airway hyperresponsiveness [20].
Previous studies have sought to investigate the role that specific viral genetic determinants play in the outcome of the virus-host interaction. The RSV G protein is a likely important factor in such interactions because sequence analysis of this gene shows high rates of nonsynonymous to synonymous mutations, suggesting that these regions are under positive selection. This is frequently observed in viral proteins that interact with host defenses [21, 22]. Based on diversity in the RSV G gene among viruses circulating in New York, Peret et al [23] have identified genetic clades of RSV-A and RSV-B subtypes GA1–GA5 and GB1–GB5, respectively [24], while other studies have identified additional clades. Phylogenetic studies have shown cocirculation of multiple genetically distinct lineages in the same location as well as circulation of genetically similar lineages in widely separated geographic locations around the same time [22, 25, 26]. Despite the high level of genetic diversity within the G gene, studies that have attempted to correlate genotype with disease severity have been inconsistent [27, 28]. An important caveat is that most of these studies evaluated only a limited region of the genome, potentially missing other pathogenicity determinants in other regions. Other investigations have demonstrated a correlation between viral load associated with severe disease and longer hospitalization [24, 29]. Multiple viral genes may affect viral load, potentially implicating a role for other genes in disease outcome.
To better understand the role of viral determinants in disease severity, we performed RSV whole-genome sequencing (WGS) and phylogenetic analyses in the setting of a naturally occurring outbreak. We identified a cluster of severe RSV-B cases in Minnesota occurring with unusual seasonality and at a frequency above the baseline rate in the previous summers. Given the change in disease severity in a stable surveillance population, we hypothesized that a change in circulating virus would most likely account for the observed case increase. Therefore, we employed a WGS approach to characterize more comprehensively viral factors associated with disease severity and identify potential genetic markers in areas of the genome distinct from those used in traditional genotyping.
METHODS
Inpatient and Unexplained Death Surveillance Systems
The Minnesota Department of Health (MDH) has identified inpatient RSV cases via the Minnesota Severe Acute Respiratory Illness (SARI) sentinel surveillance program at 3 hospitals in Minneapolis and St Paul, including 2 general hospitals and a large pediatric hospital system, since May 2013 (described in [30]). Residual clinical specimens are also submitted to MDH for all SARI cases. All clinical testing was performed at the discretion of the treating physician or practitioner. In addition to SARI, MDH has performed prospective surveillance for RSV cases among children <2 years old since 2014. Prospective surveillance of all laboratory-confirmed cases of RSV residing in the 7-county Twin Cities metropolitan area has been established since September 2016. Laboratory-confirmed RSV cases among adults (≥18 years old) were identified from 2014 to 2016 by retrospective review. Medical records for all RSV cases were reviewed by trained MDH epidemiologists (E. B., H. F., K. M., K. C.-S., and R. L.) for demographic data, intensive care unit (ICU) admission, need for mechanical ventilation, and presence of multiple comorbidities (Supplementary Table 1) using a standardized case report form. Cases of unexplained critical illness or death with hallmarks of an infectious etiology are reported to the MDH Unexplained Critical Illnesses and Death (UNEX) program at the clinician’s or medical examiner’s discretion, with submission of residual clinical specimens including nasopharyngeal swabs and/or lung swabs. Inpatient cases in Wisconsin included in this study were identified by the Wisconsin Department of Health. The data presented are public health surveillance data and not subject to institutional review board approval for human research protections.
Outpatient Surveillance
MDH maintains year round, enhanced outpatient surveillance for influenza-like illness from 4 primary care clinics that serve patients of all ages around the state with sites in Hennepin, Kandiyohi, Kittson, and Rock counties. Clinical staff collected a deidentified upper respiratory specimen for testing in the MDH public health laboratory and a limited case report form from the first 10 patients presenting each week, as described previously [30]. Outpatient cases in Wisconsin were identified by the Wisconsin Department of Health.
RSV-B Viral Nucleic Acid Sequencing
For viral nucleic acid sequencing, both nontargeted RNA amplification and RSV-specific overlapping amplicon-based approaches were employed. For the nontargeted approach, we adapted the protocol from Ng et al [31]. Nasopharyngeal swabs were removed from viral transport media and media was subjected to centrifugation at 15 000g for 15 minutes to remove human and bacterial cells. The supernatant was subjected to nucleic acid extraction using the QIAquick viral RNA column purification system (Qiagen) followed by treatment with Baseline-ZERO DNase (Lucigen) according to the manufacturer’s instructions. The purification and concentration of viral RNA was performed using Ampure XP beads (Beckman Coulter). Reverse transcription (RT) was performed using a 28-base oligonucleotide whose 3′ end consisted of 8 Ns (that is all 4 nucleotides at each of the 8 3′ positions) and whose 5′ end 20 bases consisted of an arbitrarily designed sequence (primer N1, CCTTGAAGGCGGACTGTGAGNNNNNNNN). Second-strand synthesis was achieved in a following polymerase chain reaction (PCR) step using the RT primers. The resulting double-strand RNA was PCR amplified using AmpliTaq Gold DNA polymerase (Applied Biosystems) and a 20-base primer (the same as that described above but without the 8 Ns). The randomly amplified nucleic acid was then subjected to the NexteraXT library preparation protocol (Illumina) according to the manufacturer’s instructions and sequenced using an Illumina MiSeq platform. For RSV-specific amplification, we performed amplification of the RSV genome in 4 overlapping approximately 4-kb amplicons as previously described [32]. This method has the advantage of increased sensitivity but may result in poorer coverage of viruses substantially divergent from those used for primer design. Sequencing libraries were constructed using an identical methodology as a nontargeted approach.
Epidemiologic Data Analysis
Summer seasons were defined as 1 May through 30 September in 2014–2018. Descriptive statistics were used to summarize case characteristics for each summer. Odds ratios (OR) between RSV-B cases of the outbreak summer to prior summers were used to compare case characteristics and outcomes. Statistical significance was determined at P < .05. Data were analyzed using SAS version 9.4.
Bioinformatics Data Analysis
Sequence analysis, including trimming of raw sequencing reads, quality filtering, reference-based mapping, assembly, and consensus sequence extraction, was performed using CLC Genomics Workbench 11.0 (https://www.qiagenbioinformatics.com/). In brief, de novo assembly was used initially to assemble the RSV whole-genome sequences from high-quality sequence reads of RSV. If a complete genome was not assembled, a reference RSV sequence (GenBank accession No. KM517573) was used to guide the whole-genome assembly. Maximum likelihood phylogenetic analysis using the Tamura-Nei model with bootstrap value of 1000 was constructed using MEGA 7.0.14 [33]. Sequences were deposited in GenBank (accession numbers for SH genes MN031604–MN031740, whole genomes pending).
RESULTS
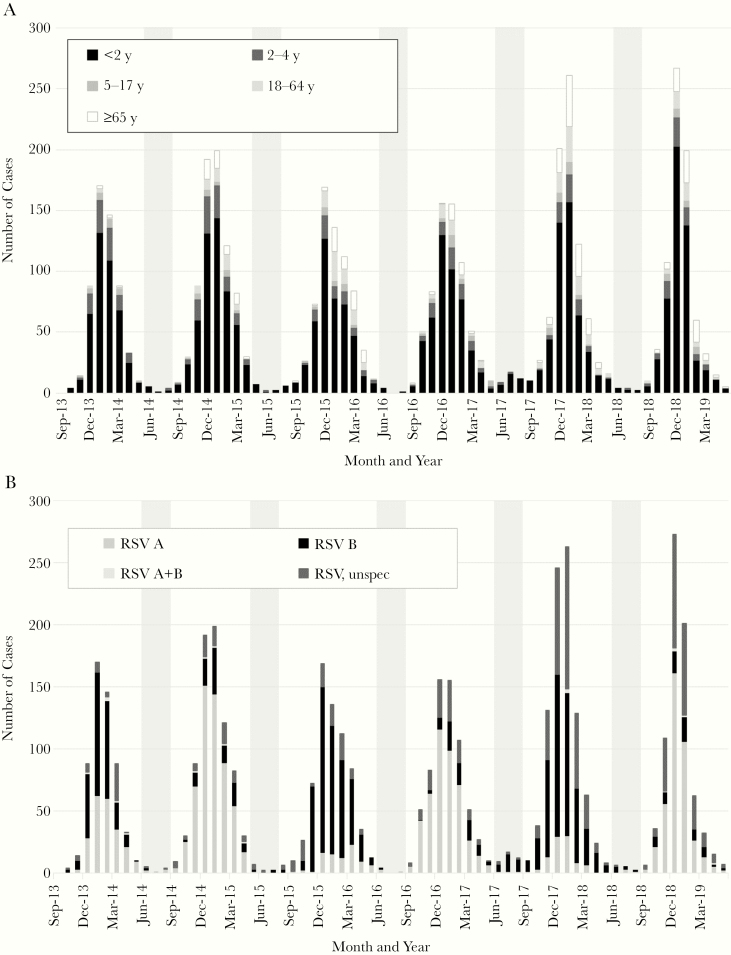
In August 2017, MDH received a report from a pediatric infectious disease physician of an increased number of severe, hospitalized cases of RSV-B among pediatric patients at an urban county hospital. Because laboratory-confirmed cases of RSV are reportable within the Minneapolis-St Paul metropolitan area, we confirmed an increased number of cases occurring in summer 2017 as compared to prior summers (Figure 1A and Table 1). This increase in reported cases continued through the following winter season and then returned to baseline in summer 2018. While the number of RSV-A cases was similar in summer 2017 as compared to the same period in prior years, the increase in case frequency was attributed to increased RSV-B cases (OR, 10.79; P < .0001) (Figure 1B and Table 1). The median age, hospital length of stay, and occurrence of comorbidities were not significantly different from prior summers. However, the rate of ICU admission was significantly higher for RSV-B cases in summer 2017 (55%) as compared to prior summers (12%; OR, 4.84; P < .01).
Figure 1.
Number of laboratory-confirmed cases of respiratory syncytial virus infection stratified by age (A) and type (B) in Minnesota, by month and year of detection. Shading indicates the period from June to August.
Table 1.
Patient Demographic and Viral Characteristics of Cases of Respiratory Syncytial Virus (RSV) in Minnesota From May 1 Through September 30 by Year
| Characteristic | 2014 | 2015 | 2016 | 2017 | 2018 |
|---|---|---|---|---|---|
| Subtype, No. | |||||
| RSV-A | 19 | 2 | 15 | 10 | 7 |
| RSV-B | 3 | 7 | 7 | 42 | 10 |
| Unspecified | 7 | 18 | 3 | 6 | 13 |
| Total | 29 | 27 | 26 | 58* | 30 |
| All cases | |||||
| Median age, mo | 7 | 8 | 7 | 7 | 9 |
| Sex, No. (%) | |||||
| Male | 13 (45) | 18 (67) | 8 (31) | 38 (66) | 17 (57) |
| Female | 16 (55) | 9 (33) | 18 (69) | 20 (34) | 13 (43) |
| Race, No. (%) | |||||
| White | 6 (21) | 2 (7) | 6 (23) | 17 (29) | 17 (57) |
| Black | 14 (48) | 21 (78) | 11 (42) | 31 (53) | 10 (33) |
| Asian/Pacific Islander | 2 (7) | 1 (4) | 3 (12) | 2 (3) | 0 (0) |
| American Indian/Alaska Native | 3 (10) | 1 (4) | 0 (0) | 1 (2) | 1 (3) |
| Multiracial | 0 (0) | 0 (0) | 1 (4) | 3 (5) | 0 (0) |
| Unknown | 4 (14) | 2 (7) | 5 (19) | 4 (7) | 2 (7) |
| Median length of stay, d | 3 | 3 | 3 | 4 | 4 |
| Admitted to ICU, No. (%) | 5 (17) | 6 (22) | 2 (8) | 27 (47) | 13 (43) |
| Comorbidities, No. (%) | 11 (38) | 9 (33) | 9 (32) | 22 (38) | 15 (50) |
| RSV-B | |||||
| Median age, mo | 2.5 | 7 | 7 | 6 | 19 |
| Sex, No. (%) | |||||
| Male | 2 (67) | 4 (57) | 2 (29) | 31 (74) | 7 (70) |
| Female | 1(33) | 3 (43) | 5 (71) | 11 (26) | 3 (30) |
| Race, No. (%) | |||||
| White | 0 (0) | 0 (0) | 1 (14) | 10 (24) | 6 (60) |
| Black | 1 (33) | 6 (86) | 5 (71) | 26 (62) | 3 (30) |
| Asian/Pacific Islander | 0 (0) | 1 (14) | 0 (0) | 2 (5) | 0 (0) |
| American Indian/Alaska Native | 1 (33) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Multiracial | 0 (0) | 0 (0) | 1 (14) | 2 (5) | 0 (0) |
| Unknown | 1 (33) | 0 (0) | 0 (0) | 2 (5) | 1 (10) |
| Median length of stay, d | 3 | 2 | 3 | 4.5 | 3 |
| Admitted to ICU, No. (%) | 0 (0) | 1 (14) | 1 (14) | 23 (55)** | 5 (50) |
| Comorbidities, No. (%) | 1 (33) | 2 (29) | 2 (29) | 14 (33) | 7 (70) |
*P < .0001, **P < .01 for 2017 compared to prior 2014–2016 summers.
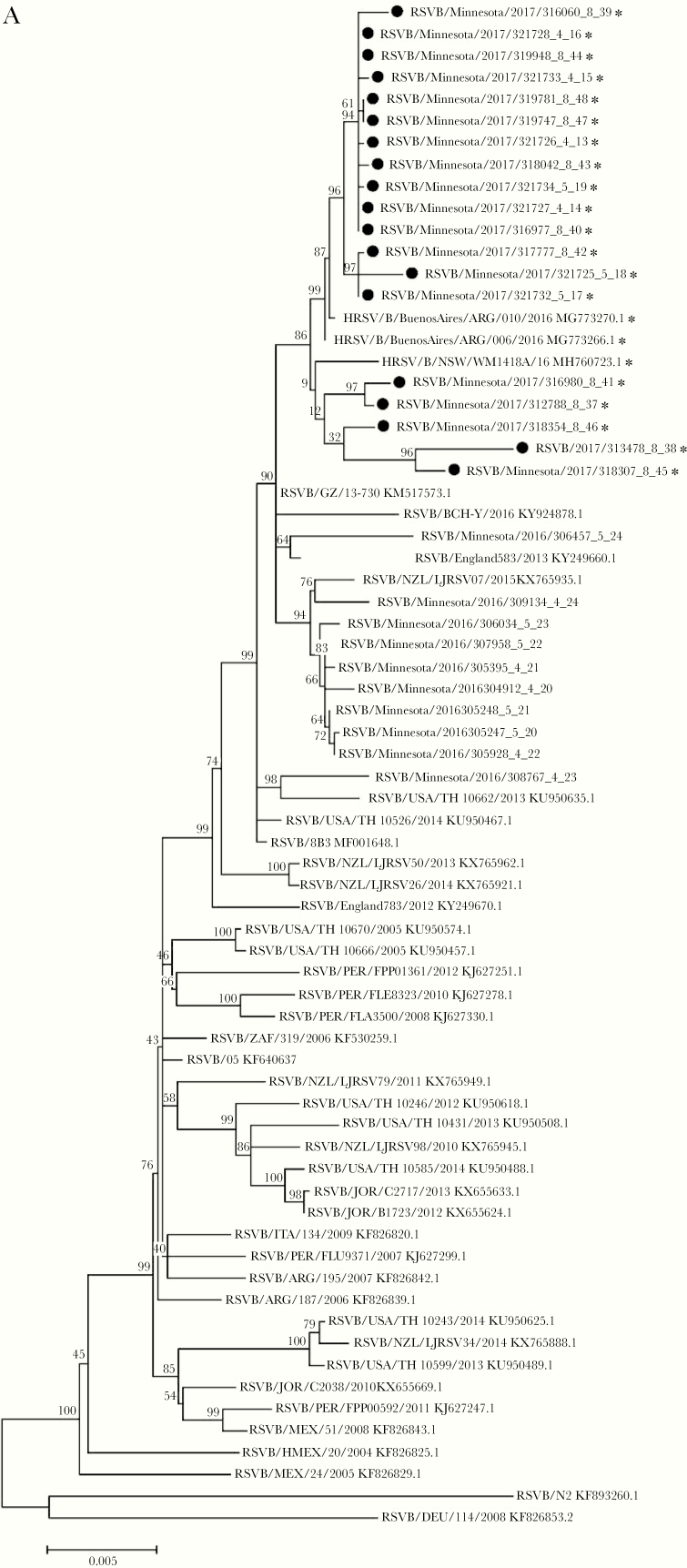
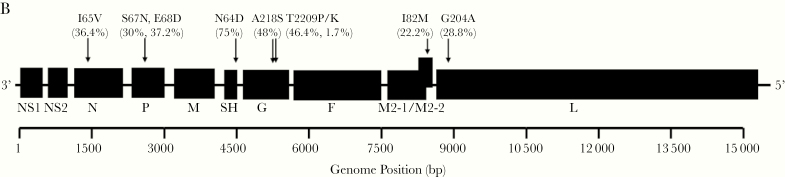
To correlate viral genetic characteristics with clinical outcomes, we attempted WGS in 11 cases that presented to care between June and August 2017 and had submitted residual clinical material to MDH through the established SARI surveillance program. We obtained adequate genomic coverage from 7 of these 11 specimens. To obtain an increased number of specimens for analysis and more broadly sample the metropolitan area, we attempted WGS on additional specimens submitted through SARI from inpatients at a large pediatric hospital serving the same region. For comparison, we performed WGS on control SARI cases from Minnesota in 2016 prior to the outbreak during a similar time period, though notably the control cases occurred earlier in spring as there were very few summer cases in 2016. Phylogenetic analysis of the 19 assembled whole-genome viral sequences from summer 2017 and 10 from 2016 demonstrated clustering of the outbreak cases into a monophyletic clade (Figure 2A). While the 2017 outbreak cases clustered separately from the majority of reference sequences from GenBank, the outbreak strain was closely related to sequences found in Argentina and Australia in 2016. We performed phylogenetic analysis of G nucleotide sequences, which are used for traditional genotyping, but this tree did not distinguish the 2017 lineage from prior isolates (Supplementary Figure 1). To further understand the specific genetic changes that separated the 2017 lineage from other strains, we performed a detailed gene-by-gene analysis of sequence to identify specific polymorphisms that accounted for the structure of the phylogenetic tree. We compared the consensus sequences of the 2016 and 2017 clades and detected single nucleotide polymorphisms (SNPs) in nearly every gene, including 8 that were predicted to produce change in the amino acid code (Figure 2B). Among these, an SNP in the SH gene (A194G) predicted to cause an N64D amino acid substitution, best correlated with the whole-genome phylogenetic trees, occurring in 18 of 19 summer 2017 cases and none of the cases from 2016. We searched GenBank for RSV sequences containing this point mutation and identified only 2 from Vietnam containing this variant prior to 2016 (KJ939919.1 and KJ939922.1) but numerous sequences from both Argentina and Australia beginning in 2016 [34, 35].
Figure 2.
A, Phylogenetic tree constructed from respiratory syncytial virus (RSV) whole-genome nucleotide sequences of 19 clinical isolates from inpatients and outpatients in Minnesota from March to May 2016 and June to August 2017. Clinical isolates collected from the summer of 2017 are marked with (•) and sequences encoding aspartic acid (D) at position 64 of SH protein are marked with (*). A subset of whole genomes from diverse geographic sites from GenBank are included for reference. B, RSV genome denoting nonsynonymous sequence polymorphisms between 2016 and 2017 cases. The frequency of these point mutations among viruses collected in Minnesota since 2017 is included in parentheses.
In light of the increased number of cases the following autumn and winter, we asked whether this new lineage persisted through the following months. We performed sequencing of virus from an additional 32 cases occurring between September 2017 and January 2018. Interestingly, these sequences matched the 2016 nucleotide variants at many of the previously identified polymorphic sites (Supplementary Table 2). However, the SH polymorphism remained in the majority of cases (27 of 32).
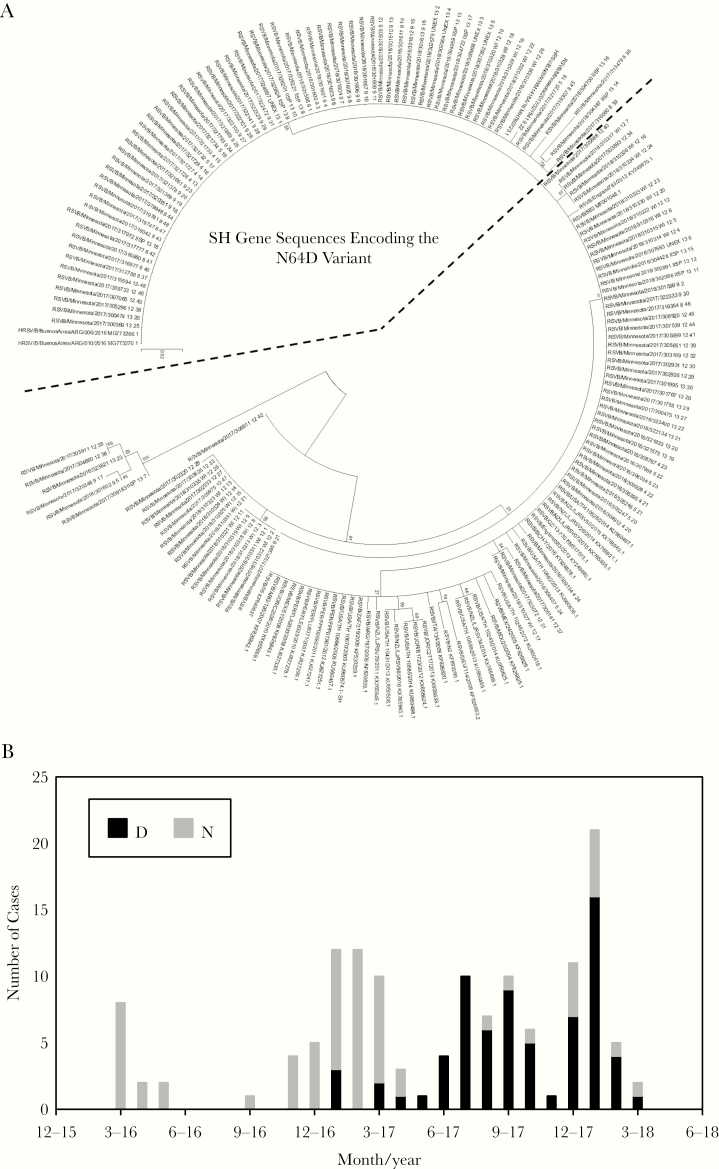
Given the rarity of this polymorphism in published sequences, we wondered when this strain entered the Minnesota population and whether this circulated in neighboring geographic regions. To address this, we performed dedicated sequencing of the 4-kb region containing the SH gene [32] to allow higher throughput and more sensitive analysis of additional specimens, including cases of RSV from an outpatient surveillance population, cases reported to MDH through UNEX, and outpatient and inpatient cases reported to the Wisconsin Department of Health. Phylogenetic analysis of the SH gene demonstrated a distinct clade that included sequences from all the additional surveillance populations (Figure 3A). We first identified this polymorphism among cases as early as January 2017 (Figure 3B). Beginning with the summer 2017 outbreak, the lineage containing this change became the dominant circulating strain among those sequenced, occurring in both inpatients (Supplementary Figure 1A), outpatients (Supplementary Figure 1B), and 5 of 6 UNEX cases. This lineage was also detected in 5 of 26 Wisconsin specimens, although it was not the dominant lineage as it was among the Minnesota cases.
Figure 3.
A, Phylogenetic tree constructed from nucleotide sequence of the SH gene from 137 clinical specimens collected between March 2016 and March 2018 and from indicated reference sequences in GenBank. The dashed line indicates a cluster SH gene sequences encoding the N64D variant. B, Number of sequences encoding either the asparagine (N) or aspartic acid (D) variant at amino acid 64 of SH protein by month of collection among all patients (n = 137).
Discussion
This study describes an outbreak of RSV-B infections in Minnesota during the summer of 2017. Initially identified by clinician report, this outbreak demonstrated an increase in frequency and disease severity with abnormal seasonality. This high frequency continued into the subsequent respiratory season but did not persist into the following summer. We utilized established statewide surveillance programs and next-generation sequencing to investigate the epidemiologic and molecular changes of circulating RSV-B viruses. While molecular epidemiology has been employed to investigate hospital-based outbreaks of RSV before [36, 37], our study shows that these techniques can be used on a larger scale to aid in understanding disease epidemiology at the state level.
By pairing epidemiologic data with specimen collection and next-generation sequencing techniques, we were able to demonstrate the emergence of a new lineage of RSV that coincided with the onset of the outbreak. This lineage was found in both inpatients and outpatients in Minnesota and Wisconsin. We identified an SNP that serves as a genetic marker for this new lineage. Based on sequence at this genetic locus, we observed 2 distinct populations of RSV-B cocirculating. This lineage was not found in Minnesota prior to 2017 and rapidly became the dominant circulating strain over the course of a single season while simultaneously appearing in other geographically distant locations. Prior studies of RSV sequences have shown temporal but not geographic clusters. Our findings, along with the published sequences of others, suggest a similar pattern of this particular SH lineage emerging in Australia, South America, and North America within a period of years [26]. Our findings suggest that the new strain emerged and rapidly became the dominant strain circulating in the community. Interestingly, this has been previously described in 1999 with the emergence of a novel B genotype containing a 60-nucleotide duplication in the C-terminal third of G. Consistent with our observation of a rapid displacement of the prior lineage, this clade (designated BA clade) rapidly disseminated worldwide and became the dominant circulating strain [38].
While our study identified a new variant in the SH surface transmembrane protein, the functional significance of this is unclear because little is known about the role of SH in RSV infection and pathogenesis in natural infections. RSV encodes 11 viral proteins, 3 of which (F or fusion, G or glycoprotein, and SH or short hydrophobic) are expressed in its lipid outer membrane [39]. SH is second only to the G protein in variability at the amino acid level with only 76% identity between RSV-A and B [39], a feature that is commonly associated with proteins that interact with the host immune system. However, compared to the other surface-expressed proteins, less is known about the function SH plays in RSV infection. SH is only found in a subset of Paramyxoviridae, including human metapneumovirus and parainfluenza virus type 5 but not in other parainfluenza viruses or measles [40]. It can be deleted without loss of replication ability in cell culture but Whitehead et al showed SH-deleted virus is attenuated in mice and chimpanzees [41]. With respect to function, studies suggest that SH forms a pentameric ion channel [42, 43]. Fuentes et al showed SH may inhibit tumor necrosis factor-α production and apoptosis [40]. Additional studies have demonstrated a role in modulating the inflammatory response to viral infection but have reported both pro- and anti-inflammatory activity [44, 45]. While our study cannot establish a causative role for this or other polymorphisms, future in vitro studies may clarify whether this change has functional significance. Furthermore, additional large-scale studies pairing clinical and viral sequencing data may help to better define the role that virologic factors play in clinical outcome.
Our study provides insights into the molecular epidemiology of RSV in the upper Midwest; however, it does have certain limitations. Because WGS is labor and resource intensive, we were only able to sequence a sample of the cases during any time period and thus these sequences may not fully reflect the circulating strains. However, aside from the outbreak cases, all other cases were selected at random from reported cases so are likely to be representative of the circulating population. If the novel lineage is more pathogenic, patients infected with this lineage may be more likely to seek care and have specimens collected. However, the same pattern of strain replacement was seen both in inpatients and outpatients, suggesting it also became more common among those with milder disease. We also considered the possibility that the increase in cases seen in winter 2017 could be an artifact of our reporting system because RSV mandatory reporting in the Minneapolis-St Paul metropolitan area was implemented in September 2016. However, we do not think this accounts for the increased number of cases in summer 2017 as we did not see a similarly increased number of cases in summer 2018, which would have been expected had the increase in numbers been solely due to increased reporting. We did not see a similar increase in the number of subtype A cases, also arguing that the detected outbreak is not an artifact of a change in reporting method.
Thus, in summary we have combined traditional epidemiology and emerging WGS techniques to characterize a summer outbreak of severe RSV-B disease in Minnesota. The virus associated with this outbreak contains unique genetic polymorphisms, suggesting these changes may be associated with increased disease severity. The new tools of WGS allow for broader investigation of the role of viral determinants in understanding viral pathogenesis and identifying points of host-pathogen interaction.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Stacy Holzbauer for critical review of data and support of Unexplained Critical Illnesses and Death (UNEX) project, Richard Danila for support of this project, and Larry Anderson and Suman Das for their helpful discussion.
Financial support. This work was supported by the Centers for Disease Control and Prevention (grant number CDC-RFA-CK17-170102CONT18 cooperative agreement to the Emerging Infections Program); and the National Institutes of Health (grant number T32 5T32AI055433-14 to B. K. T.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: IDWeek 2018, San Francisco, CA, 3–7 October 2018.
References
- 1. Light M, Bauman J, Mavunda K, Malinoski F, Eggleston M. Correlation between respiratory syncytial virus (RSV) test data and hospitalization of children for RSV lower respiratory tract illness in Florida. Pediatr Infect Dis J 2008; 27:512–8. [DOI] [PubMed] [Google Scholar]
- 2. Mullins JA, Lamonte AC, Bresee JS, Anderson LJ. Substantial variability in community respiratory syncytial virus season timing. Pediatr Infect Dis J 2003; 22:857–62. [DOI] [PubMed] [Google Scholar]
- 3. Meissner HC. Viral bronchiolitis in children. N Engl J Med 2016; 374:62–72. [DOI] [PubMed] [Google Scholar]
- 4. Hall CB, Weinberg GA, Iwane MK, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med 2009; 360:588–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 2005; 352:1749–59. [DOI] [PubMed] [Google Scholar]
- 6. Widmer K, Zhu Y, Williams JV, Griffin MR, Edwards KM, Talbot HK. Rates of hospitalizations for respiratory syncytial virus, human metapneumovirus, and influenza virus in older adults. J Infect Dis 2012; 206:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fjaerli HO, Farstad T, Bratlid D. Hospitalisations for respiratory syncytial virus bronchiolitis in Akershus, Norway, 1993–2000: a population-based retrospective study. BMC Pediatr 2004; 4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forster J, Ihorst G, Rieger CH, et al. Prospective population-based study of viral lower respiratory tract infections in children under 3 years of age (the PRI.DE study). Eur J Pediatr 2004; 163:709–16. [DOI] [PubMed] [Google Scholar]
- 9. Eriksson M, Bennet R, Rotzén-Ostlund M, von Sydow M, Wirgart BZ. Population-based rates of severe respiratory syncytial virus infection in children with and without risk factors, and outcome in a tertiary care setting. Acta Paediatr 2002; 91:593–8. [DOI] [PubMed] [Google Scholar]
- 10. Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 1986; 140:543–6. [DOI] [PubMed] [Google Scholar]
- 11. Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis 1991; 163:693–8. [DOI] [PubMed] [Google Scholar]
- 12. Reduction of respiratory syncytial virus hospitalization among premature infants and infants with bronchopulmonary dysplasia using respiratory syncytial virus immune globulin prophylaxis. The PREVENT Study Group. Pediatrics 1997; 99:93–9. [DOI] [PubMed] [Google Scholar]
- 13. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. The IMpact-RSV Study Group. Pediatrics 1998; 102:531–7. [PubMed] [Google Scholar]
- 14. Langley GF, Anderson LJ. Epidemiology and prevention of respiratory syncytial virus infections among infants and young children. Pediatr Infect Dis J 2011; 30:510–7. [DOI] [PubMed] [Google Scholar]
- 15. Sommer C, Resch B, Simões EA. Risk factors for severe respiratory syncytial virus lower respiratory tract infection. Open Microbiol J 2011; 5:144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murphy BR, Alling DW, Snyder MH, et al. Effect of age and preexisting antibody on serum antibody response of infants and children to the F and G glycoproteins during respiratory syncytial virus infection. J Clin Microbiol 1986; 24:894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anderson LJ, Hierholzer JC, Tsou C, et al. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis 1985; 151:626–33. [DOI] [PubMed] [Google Scholar]
- 18. Hall CB, Walsh EE, Schnabel KC, et al. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis 1990; 162:1283–90. [DOI] [PubMed] [Google Scholar]
- 19. Moore ML, Peebles RS Jr. Respiratory syncytial virus disease mechanisms implicated by human, animal model, and in vitro data facilitate vaccine strategies and new therapeutics. Pharmacol Ther 2006; 112:405–24. [DOI] [PubMed] [Google Scholar]
- 20. Lukacs NW, Moore ML, Rudd BD, et al. Differential immune responses and pulmonary pathophysiology are induced by two different strains of respiratory syncytial virus. Am J Pathol 2006; 169:977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cane PA, Pringle CR. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J Virol 1995; 69:2918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. García O, Martín M, Dopazo J, et al. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J Virol 1994; 68:5448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peret TC, Hall CB, Hammond GW, et al. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J Infect Dis. 2000; 181:1891–6. [DOI] [PubMed] [Google Scholar]
- 24. El Saleeby CM, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis 2011; 204:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matheson JW, Rich FJ, Cohet C, et al. Distinct patterns of evolution between respiratory syncytial virus subgroups A and B from New Zealand isolates collected over thirty-seven years. J Med Virol 2006; 78:1354–64. [DOI] [PubMed] [Google Scholar]
- 26. Bose ME, He J, Shrivastava S, et al. Sequencing and analysis of globally obtained human respiratory syncytial virus A and B genomes. PLoS One 2015; 10:e0120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilca R, De Serres G, Tremblay M, et al. Distribution and clinical impact of human respiratory syncytial virus genotypes in hospitalized children over 2 winter seasons J Infect Dis 2006; 193:54–8. [DOI] [PubMed] [Google Scholar]
- 28. Martinello RA, Chen MD, Weibel C, Kahn JS. Correlation between respiratory syncytial virus genotype and severity of illness. J Infect Dis 2002; 186:839–42. [DOI] [PubMed] [Google Scholar]
- 29. DeVincenzo JP, El Saleeby CM, Bush AJ. Respiratory syncytial virus load predicts disease severity in previously healthy infants. J Infect Dis 2005; 191:1861–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thielen BK, Friedlander H, Bistodeau S, et al. Detection of influenza C viruses among outpatients and patients hospitalized for severe acute respiratory infection, Minnesota, 2013–2016. Clin Infect Dis 2018; 66:1092–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ng TFF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol 2015; 89:5340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schobel SA, Stucker KM, Moore ML, et al. Respiratory syncytial virus whole-genome sequencing identifies convergent evolution of sequence duplication in the C-terminus of the G gene. Sci Rep 2016; 6:26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Di Giallonardo F, Kok J, Fernandez M, et al. Evolution of human respiratory syncytial virus (RSV) over multiple seasons in New South Wales, Australia. Viruses 2018; 10:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goya S, Valinotto LE, Tittarelli E, et al. An optimized methodology for whole genome sequencing of RNA respiratory viruses from nasopharyngeal aspirates. PLoS One 2018; 13:e0199714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mazzulli T, Peret TC, McGeer A, et al. Molecular characterization of a nosocomial outbreak of human respiratory syncytial virus on an adult leukemia/lymphoma ward. J Infect Dis 1999; 180:1686–9. [DOI] [PubMed] [Google Scholar]
- 37. Taylor GS, Vipond IB, Caul EO. Molecular epidemiology of outbreak of respiratory syncytial virus within bone marrow transplantation unit. J Clin Microbiol 2001; 39:801–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trento A, Casas I, Calderón A, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol 2010; 84:7500–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collins PL, Fearns R, Graham BS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. In: Anderson LJ, Graham BS, eds. Challenges and opportunities for respiratory syncytial virus vaccines. Berlin, Heidelberg: Springer-Verlag,2013:3–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fuentes S, Tran KC, Luthra P, Teng MN, He B. Function of the respiratory syncytial virus small hydrophobic protein. J Virol 2007; 81:8361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Whitehead SS, Bukreyev A, Teng MN, et al. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol 1999; 73:3438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carter SD, Dent KC, Atkins E, et al. Direct visualization of the small hydrophobic protein of human respiratory syncytial virus reveals the structural basis for membrane permeability. FEBS Lett 2010; 584:2786–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gan SW, Tan E, Lin X, et al. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J Biol Chem 2012; 287:24671–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax 2013; 68:66–75. [DOI] [PubMed] [Google Scholar]
- 45. Russell RF, McDonald JU, Ivanova M, Zhong Z, Bukreyev A, Tregoning JS. Partial attenuation of respiratory syncytial virus with a deletion of a small hydrophobic gene is associated with elevated interleukin-1β responses. J Virol 2015; 89:8974–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.