Abstract
Purpose:
To develop a quantitative DCE MRI technique enabling entire-abdomen coverage, free-breathing acquisition, 1-second temporal resolution, and T1-based quantification of contrast agent concentration and kinetic modeling for the characterization of pancreatic ductal adenocarcinoma (PDAC).
Methods:
Segmented FLASH readouts following saturation-recovery preparation with randomized 3D Cartesian undersampling was used for incoherent data acquisition. MR Multitasking was used to reconstruct 6-dimensional images with 3 spatial dimensions, 1 T1 recovery dimension for dynamic T1 quantification, 1 respiratory dimension to resolve respiratory motion, and 1 DCE time dimension to capture the contrast kinetics. Sixteen healthy subjects and 14 patients with pathologically confirmed PDAC were recruited for the in vivo studies, and kinetic parameters vp, Ktrans, ve, and Kep were evaluated for each subject. Intersession repeatability of Multitasking DCE was assessed in 8 repeat healthy subjects. One-way unbalanced analysis of variance (ANOVA) was performed between control and patient groups.
Results:
In vivo studies demonstrated that vp, Ktrans, and Kep of PDAC were significantly lower compared with nontumoral regions in the patient group (P = .002,.003, .004, respectively) and normal pancreas in the control group (P = .011, <.001, <.001, respectively), while ve was significantly higher than nontumoral regions (P < .001) and healthy pancreas (P < .001). The kinetic parameters showed good in vivo repeatability (interclass correlation coefficient: vp, 0.95; Ktrans, 0.98; ve, 0.96; Kep, 0.99).
Conclusion:
The proposed Multitasking DCE is promising for the quantification of vascular properties of PDAC. Quantitative DCE parameters were repeatable in vivo and showed significant differences between normal pancreas and both tumor and nontumoral regions in patients with PDAC.
Keywords: DCE MRI, MR Multitasking, pancreatic ductal adenocarcinoma (PDAC), quantitative imaging, respiratory motion resolved imaging
1 |. INTRODUCTION
Pancreatic cancer is the third leading cause of cancer-related deaths in the United States.1,2 Pancreatic ductal adenocarcinoma (PDAC) represents 95% of malignant pancreatic cancers.3 It has an unusual microenvironment characterized by a highly desmoplastic stroma encompassing up to 90% of the tumor; regions with hypoxic tissue; interstitial hypertension; and heterogeneous vascularization, including low microvascular density (MVD) and reduced blood flow in the lesion area and high MVD in the surrounding area, which is inherently resistant to treatment.4 Due to technical challenges for early detection,5–8 approximately 60% of patients at the first diagnosis are deemed unable to receive macroscopic complete tumor resection, which remains the only curative treatment option for PDAC.9 Those patients who are ineligible for surgery will receive neoadjuvant chemotherapy or radiotherapy. However, the lack of effective approaches for early assessment of response to these therapies prevents optimization of the treatment design for each individual. These reasons contribute to a dismal 5-year survival rate of 8%.3
Because of its superior soft-tissue contrast, MRI plays a key role in the detection, staging, and treatment management of PDAC.10–12 The clinical MRI protocol for PDAC characterization consists of T1-weighted (T1W) gradient-echo (GRE) imaging, T2-weighted (T2W) turbo spin-echo (TSE) imaging, diffusion-weighted imaging (DWI), and gadolinium (Gd)-enhanced multiphasic T1W GRE imaging at pre-contrast phase, arterial phase (20–40 seconds after the start of injection), venous phase (45–65 seconds), and equilibrium phase (3–5 minutes).5,10 Gd-enhanced multiphasic T1W imaging has shown great tumor delineation and high sensitivity in the detection of PDAC. However, it is a qualitative technique that is subject to coil positioning, sequence parameters, and interobserver and interscanner variability.13 In addition, 4 phases are not sufficient to assess the vascular features of PDAC lesions.14
Quantitative DCE MRI has been proposed to evaluate the enhancement pattern and vascular properties of tissues. DCE MRI involves the fast acquisition of a series of T1W images at high temporal resolution during Gd-based contrast agent (CA) administration. Dynamic signal enhancement curves are then extracted per pixel or from regions of interest (ROIs) and analyzed with an appropriate pharmacokinetic model to estimate several kinetic parameters, including fractional plasma volume (vp), transfer constant (Ktrans), fractional extravascular extracellular volume (ve), and rate constant (Kep). Several investigations have shown that Ktrans and ve are positively correlated with MVD and fibrosis in PDAC, respectively.4,15 Other studies have shown that quantitative DCE MRI can characterize PDAC and monitor treatment response.16,17
Despite these encouraging results, DCE MRI of PDAC continues to face demanding technical challenges.15–17 First, imaging of the pancreas is subject to respiratory motion artifacts, which can significantly degrade image quality and affect kinetic quantification. To alleviate this problem, respiratory-triggered acquisition, sophisticated motion registration schemes,18,19 and/or patient cooperation such as prone positioning, shallow breathing, or breath-holding are usually required. Second, there is typically a trade-off among spatial resolution, temporal resolution, and spatial coverage. Third, most existing DCE techniques linearly transform the dynamic signal intensity from T1W MRI to estimate CA concentration for kinetic modeling. However, the nonlinearity between T1W MRI signal and CA concentration can introduce errors in the quantification of kinetic parameters, especially in tissues with high-contrast uptake such as blood.20 Another approach is to nonlinearly calculate the CA concentration based on a separately acquired pre-contrast T1 map and dynamic T1W signal, but this requires an additional scan and is subject to misregistration between separate imaging series.21,22
In this work, we propose a novel DCE technique based on our newly developed MR Multitasking framework23 to address the aforementioned limitations. This Multitasking DCE technique performs 6-dimensional (6D) quantitative imaging with 3 spatial dimensions, a saturation recovery dimension, a respiration dimension, and a DCE time dimension. It achieves respiratory motion–resolved, high-temporal-resolution T1 quantification of the entire abdomen in a 10-minute free-breathing scan, followed by the derivation of CA concentration and kinetic modeling. To our knowledge, this is the first abdominal DCE MRI method that simultaneously allows for 3D true free-breathing acquisition, 1-second temporal resolution, and dynamic T1 mapping.
2 |. METHODS
2.1 |. Sequence design
Three-dimensional segmented FLASH readouts following periodic nonselective saturation-recovery (SR) preparation were used to acquire data at various saturation recovery times for dynamic T1 quantification.24 Fat signal was suppressed using water excitation for the FLASH readouts. Images were acquired in a transversal orientation, covering the abdomen from the liver dome to the iliac crest. A 3D Cartesian trajectory with randomized Gaussian reordering in both phase (ky) and partition (kz) encoding was implemented to incoherently undersample k-space.24 In the Multitasking framework, training data are usually acquired frequently at the center k-space line to capture the multidimensional dynamics and estimate the temporal basis functions.23 In this work, the collection of 1 center k-space line was interleaved with the collection of 7 imaging lines, yielding a sampling period of 45 ms for the training data. The orientation of the center k-space line was modified to the superior–inferior direction (partition encoding direction, kx = ky = 0) for the improved capture of respiratory motion.
2.2 |. Image reconstruction
2.2.1 |. Multitasking image model
Our 6D quantitative DCE technique is based on the recently developed MR Multitasking framework,23 which uses a low-rank tensor (LRT) image model to exploit the high correlation between images along multiple time dimensions,25,26 vastly accelerating the acquisition. The LRT model is a multidimensional extension of low-rank matrix models,27–30 which rely on the partial separability between spatial and temporal image dimensions to decrease the degrees of freedom and accelerate acquisition.
The abdominal DCE image is expressed as a 6D image a(x,τ,tr,td), a function of 3 spatial dimensions (x=[x,y,z]T) and 3 time dimensions (saturation recovery time τ, respiration motion tr, and DCE time course td). The strong correlation between images along and across time dimensions induces a to be partially separable in the combination of space x=[x,y,z]T, τ, tr, and td:
| (1) |
where u𝓁(x) is the 𝓁th of L spatial coefficient maps; vm(τ) is the mth of M saturation recovery basis functions; wn(tr) is the nth of N respiratory motion basis functions; qp(td) is the pth of P DCE basis functions; g𝓁mnp are the elements of the core tensor. This implies that the 4-way tensor which has modes corresponding to voxel location x and each time dimension, τ, tr and td, is low-rank. This low-rank tensor model can be expressed in matrix notation as
| (2) |
where the columns of U, V, W, and Q contain the basis functions for each dimension; ⊗ denotes the Kronecker product; the subscript (i) denotes the mode-i flattening of the tensor.31 The separation of the spatial and temporal factors decouples the tradeoff between spatial and temporal resolution, providing an avenue for accelerated acquisition. The reconstruction strategy for recovering the undersampled low-rank tensor used in this work is a factored, explicit low-rank strategy, which reconstructs the image tensor by sequentially determining each of its factors.23
2.2.2 |. Single-time-dimension reconstruction
To allow respiratory motion identification, dynamic images with a single time dimension were first generated using an explicit low-rank matrix imaging strategy, with 1 temporal dimension t (elapsed time) representing a mixture of saturation recovery times, respiratory states, and dynamic contrast changes:
| (3) |
where the single-time-dimension temporal basis function was estimated from the singular value decomposition (SVD) of the training data. Following this, the spatial coefficients were recovered by fitting the the collected imaging data using a conjugate gradient least-squares algorithm.27
2.2.3 |. Respiratory-motion compensation
The respiratory motion was binned into 6 states, as specified in the Supporting Information Section A and Supporting Information Figure S1. To enhance the correlation between images across different respiratory bins with minimal increase in the computational complexity, interbin translational registration was applied to the k-space data. To estimate translational motion, a template image of each bin was first obtained by averaging the single-time-dimension images within each respiratory state. The template image from end-expiration was set to be the reference, denoted as aref; the template images from the other 5 bins are denoted as ai, i∈1, 2,…, 5. The translation vector bi between the moving bin i and the reference was estimated as
| (4) |
with spatial Fourier transform F, a weighting window W that emphasizes the signal from moving organs of interest and reduces the signal from surrounding static muscle, linear phase modulator P(bi) (the k-space operation equivalent to image-domain translation), and mutual information metric Mt(⋅,⋅). The k-space data were then directly compensated by applying P(bi) to all k-space lines corresponding to the ith respiratory bin.
We note that the purpose of respiratory-motion compensation was not to erase the difference of images between respiratory bins, but to increase the image correlation by moving them to a similar location, thereby improving image quality.32 The superiority of respiratory-motion compensation was tested by comparing the SNR in the pancreas and sharpness of the pancreas boundary with and without compensation using a one-tailed t-test. The sharpness of the boundary was estimated using the rise-distance method.33
2.2.4 |. Tensor subspace estimation
Following respiratory-motion identification and compensation, the next stage is to recover the multidimensional temporal factor from the training data. This was divided into 2 steps: (1) predetermine the T1 recovery basis functions in V from a dictionary of SR signal curves; and (2) estimate the respiratory basis functions in W, DCE basis functions in Q, and the core tensor from the training data.
In the first step, a dictionary of feasible SR signal curves was generated ahead of time for a range of T1 values and B1 inhomogeneities based on the Bloch equations.24 The dictionary consisted of 101 T1 values logarithmically spaced from 100 ms to 3000 ms, 17 flip angles from 6° to 14° in half-degree increments, and 21 saturation pulse angles linearly spaced from 60° to 120°. The T1 recovery factor V was directly extracted from the SVD of this dictionary.
In the second step, the training data were reshaped into a 4-way tensor Dtr with 1 k-space readout mode and 3 temporal modes: τ, tr, and td. The training data were not available from every combination of respiratory state, saturation recovery time, and DCE phase, so the training data tensor was highly undersampled. In addition, a motion-removal scheme described in our previous work24 was implemented to further remove outlier readouts corresponding to abrupt motion. The undersampled training data tensor was completed by solving
| (5) |
where dtr is the collected training data; M(⋅) applies the training data sampling pattern; ‖⋅‖∗ denotes the nuclear norm; Dtr,(i) denotes the mode-i matricization of the tensor is an optional additional regularization functional, which in this work was chosen as temporal total variation (TV) along the respiratory and DCE dimensions: where ∇ is the finite-difference operator. With the completed Dtr, the core-tensor respiratory basis functions W, and DCE time-course basis functions Q can be extracted using the higher-order SVD (HOSVD),34 which provides
2.2.5 |. Spatial coefficient recovery
With a known temporal subspace spanned by the rows of the spatial factor U can be recovered by solving the following optimization problem:
| (6) |
with acquired imaging data d, undersampling operator Ω, spatial Fourier transform F, coil sensitivity operator S, and an optional regularization functional R (⋅). In this work, we used an anisotropic spatial TV regularizer to integrate compressed sensing into the low-rank framework where, for example, Δx is the voxel width in x and ∇x {⋅} is the finite-difference operator along. The final result of the reconstruction produces the 6D image with full T1 recovery curves for every combination of the respiratory states and multiple DCE phases. The MATLAB (R2018a, MathWorks, Natick, MA) p-code for the reconstruction is available upon request.
2.3 |. Dynamic T1 quantification and kinetic modeling
Pixelwise dynamic T1 quantification was performed following image reconstruction. Here, signal intensity is a function of R1 td = 1∕T1 td (the relaxation rate at a specific DCE time point td), amplitude A, SR pulse efficiency parameter B, FLASH readout interval TR, flip angle, and recovery time point n = 1, 2, …, N (N = 84 per SR period) such that τ = nTR. The signal intensity at a given pixel is
| (7) |
the signal intensity vector s for an entire SR period can be expressed as
| (8) |
and the 2D dynamic signal matrix S representing all DCE time bins td from 0 to Td is
| (9) |
Where r1 is the vector of R1 values containing R1(td), td = 0, 1, …, Td, and where td = 0 represents the pre-contrast time point. For each pixel and respiratory state, the dynamic relaxation curve R1(td), td = 0, 1, …, Td was fitted from the reconstructed (τ, td)-space signal using lsqnonlin (nonlinear least-square solver) in MATLAB. According to the relaxivity, the dynamic contrast agent concentration curve Ct for each pixel was directly derived as
| (10) |
with the relaxivity rate being γ = 4.0 L · mmol−1 ·s−1 in this work.35,36 Subsequently, the vascular properties were evaluated with the two-compartment extended Tofts model.37 Details of the kinetic modeling are available in Supporting Information Section B. A computational simulation in Supporting Information Section C demonstrates the improved accuracy in kinetic parameter quantification resulting from the Multitasking T1-mapping–based approach over a conventional linear approach.
2.4 |. Imaging experiments
2.4.1 |. Imaging protocol
All studies were performed on a 3T clinical MR scanner (Biograph mMR, Siemens Medical Solutions, Erlangen, Germany) with a 12-channel phase array surface coil. In the imaging session, a routine protocol was first acquired for pancreas delineation and tumor definition. It included 3D T1W volumetric interpolated breath-hold examination (VIBE) with Dixon fat suppression in axial orientation, multi-slice T2W half-Fourier acquisition single-shot turbo spin echo (HASTE) in axial and coronal orientations, and multi-slice single-shot (SS) EPI DWI. A 2D MOLLI38 sequence in oblique planes covering as much PDAC tumor mass as possible was obtained as the reference for in vivo T1 mapping. Following these sequences, the Multitasking DCE sequence was performed. The protocol was designed to cover the entire abdomen from the dome of the liver to the iliac crest. Each SR period was 500 ms, with saturation times from 5.6 ms to 470.4 ms with an increment of 5.6 ms for T1 quantification. The SR period was repeated 1200 times in a scan of 10 minutes. The DCE bin duration was selected to be 1 second by combining 2 SR periods, striking a balance between temporal resolution and SNR.24 Gd-based contrast agent (Gadavist, 0.1 mmol/kg; Bayer Schering Pharma, Berlin, Germany) was administrated intravenously 2 minutes into the scan at a rate of 2 mL/s, followed by a 20-mL saline flush at the same rate. Immediately at the end of this sequence, post-contrast MOLLI at the same slice location as pre-contrast MOLLI was collected. Detailed imaging parameters for the protocols are summarized in Table 1.
TABLE 1.
List of imaging parameters
| Parameters | T1W VIBE | T2W HASTE | MOLLI | SS-EPI DWI | Multitasking DCE |
|---|---|---|---|---|---|
| Slice thickness (mm) | 3 | 5 | 5 | 6 | 3 |
| Slice resolution | 50% | — | — | — | 50% |
| Number of slices | 72 | 86 | 1 | 60 | 120 |
| Gap | — | 1 | — | 1 | — |
| TR (ms) | 4.15 | 1000 | 280.56 | 4500 | 5.60 |
| TE (ms) | 1.39/2.65 | 99 | 1.12 | 47 | 2.45 |
| Number of averages | 1 | 1 | 1 | 6 | 1 |
| FOV (mm2) | 350 × 247 | 330 × 226 | 390 × 390 | 339 × 306 | 380 × 268 |
| In-plane resolution (mm2) | 1.2 × 1.2 | 1.3 × 1.3 | 2.0 × 2.0 | 2.0 × 2.3 | 1.2 × 1.2 |
| Flip angle (º) | 9 | 105 | 35 | 90 | 10 |
Abbreviations: HASTE, half-Fourier acquisition single-shot turbo spin echo; MOLLI, modified Look-Locker inversion recovery; SS-EPI, single-shot EPI; VIBE, volumetric interpolated breath-hold examination.
2.4.2 |. Phantom study
The phantom study was performed on the ISMRM/NIST MRI system phantom (model 130; High Precision Devices, Boulder, CO)39 to validate the T1 mapping accuracy of the proposed Multitasking DCE technique. The T1 layer with T1 varying from 50 ms to 2000 ms was selected for data analysis. As a reference, a standard 2D inversion-recovery spin-echo (IR-SE) sequence with TR = 10 000 ms and 7 different inversion times (23, 100, 400, 900, 1600, 2200, and 3000 ms) was also acquired at the center of the T1 layer with parameters: FOV = 220 × 220 mm2, in-plane spatial resolution = 1.2 mm, and slice thickness = 6 mm. Two experiments with the same protocols were conducted on 2 separate days to assess the intersession repeatability of T1 measurements in Multitasking DCE.
2.4.3 |. In vivo study
The in vivo study was approved by the local institutional review board. Written informed consent was obtained from all participating subjects before scanning. The study population included both healthy subjects (as the control group) and patients with PDAC. Sixteen volunteers (8 females, age range, 23 to 60 years) without a history of pancreatic disease were recruited as the control group, 8 of whom were able to come back for a second scan with the same protocol on a different day (with at least 1 week apart from the first scan) for the assessment of intersession repeatability.
For the patient study, the inclusion criteria were the presence of pathologically confirmed PDAC and tolerance to MRI and Gd-based contrast agent. Patients who had previously undergone pancreatic surgery were excluded from the study. The final patient group included 14 patients (7 females, age range, 51 to 77 years). The mean size of the tumors, defined as the largest diameter in axial CT images according to RECIST 1.1 criteria, is 3.9 cm, ranging from 1.6 cm to 6.7 cm. The detailed size and location information of the tumors in the patient group are provided in Supporting Information Table S4. The previous clinical contrast-enhanced CT scans and diagnosis reports for all patients were also obtained for tumor definition.
2.5 |. Image analysis
Post-processing was performed off-line in MATLAB. The pancreas boundary of each subject was defined based on the T1W VIBE images from the same session. Dynamic T1 fitting and kinetic modeling were performed on all slices involving the pancreas, for all subjects.
In the control group, the kinetic parameters vp, Ktrans, ve, and Kep obtained for each subject are the mean values of all voxels within the pancreas throughout multiple slices. For the patient group, the tumor boundary was identified by a radiologist (L.W.) with 11 years of experience in reading MR images for PDAC, by cross-referencing T1W VIBE, T2W HASTE, SS-EPI, and clinical contrast-enhanced CT images. The kinetic parameters for PDAC were the mean from all voxels within the tumor. The region of interest for the nontumoral area included all voxels in the pancreas but outside the tumor.
2.6 |. Statistical analysis
Statistical analysis was conducted in SPSS version 24 (IBM, Armonk, NY). The T1 measurement agreement between the Multitasking DCE and reference methods for both phantom and in vivo studies was evaluated by the intraclass correlation coefficient (ICC) and paired t-tests. The intersession repeatability of the proposed technique was assessed on the repeated data from the 8 healthy volunteers who were scanned twice on separate days. Bland-Altman analysis, ICC, and coefficient of variation (CoV) of the 4 kinetic parameters vp, Ktrans, ve, and Kep were obtained.
A significance test using one-way unbalanced analysis of variance (ANOVA) was applied to identify the significant difference of each kinetic parameter obtained between (1) PDAC tumor mass and nontumoral pancreatic tissue in the patient group, (2) PDAC tumor mass in the patient group and normal pancreatic tissue in control group, and (3) nontumoral pancreatic tissue in the patient group and normal pancreatic tissue in control group. Holm-Bonferroni correction was also implemented to correct the familywise error in this multi-group test.
3 |. RESULTS
3.1 |. Phantom measurements
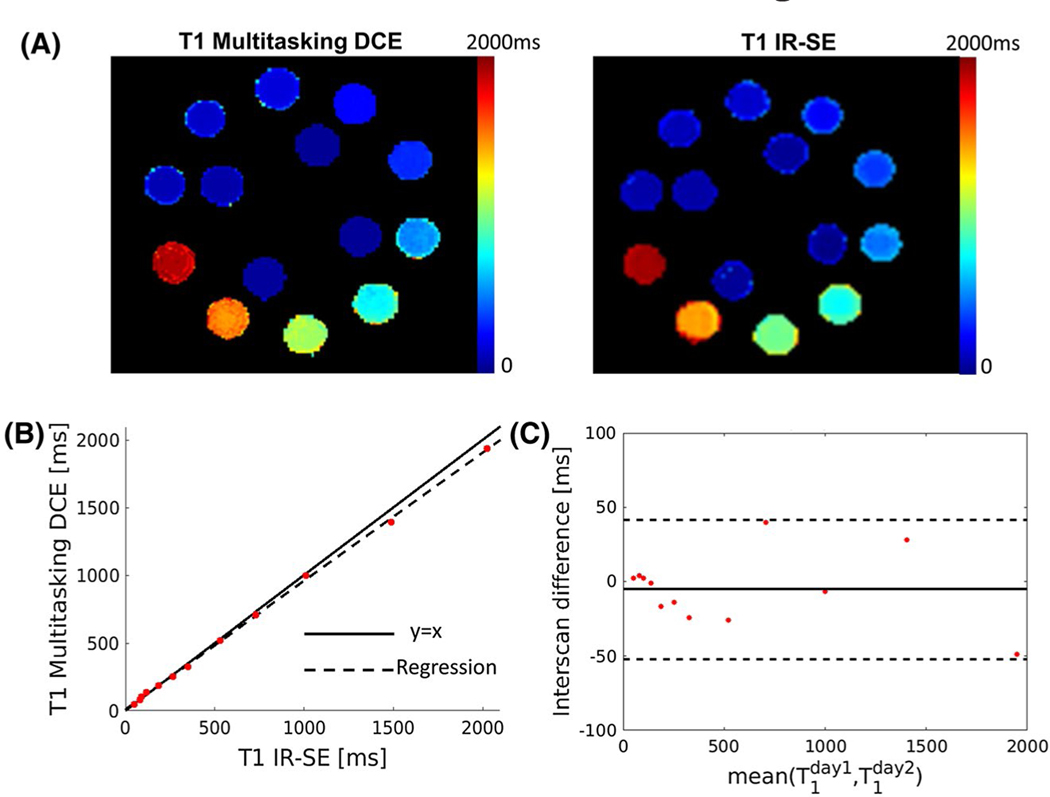
The color maps in Figure 1A display the T1 maps from the Multitasking DCE and reference IR-SE sequence. Figure 1B shows the linear regression of the T1 measurements between the 2 methods, indicating that the T1 values from Multitasking DCE were in good agreement with reference values (linear regression slope = 0.972, R2 = 0.970, ICC = 0.999). Across all spheres in the phantom, the T1 measured using Multitasking DCE shows no significant difference with the T1 measured by IR-SE (P = .073). The intersession repeatability of the T1 mapping in Multitasking DCE was illustrated in Figure 1C. The mean absolute interscan differences were 2.55% with a relatively low CoV (9%), indicating that the T1 mapping of Multitasking DCE is repeatable.
FIGURE 1.
Phantom measurements. (A) T1 maps from the Multitasking DCE and reference inversion-recovery spin-echo (IR-SE) sequence of the T1 layer of ISMRM/NIST MRI system phantom. (B) Regression of T1 of the proposed Multitasking DCE versus IR-SE. The solid line represents y = x, whereas the dashed lines represent the regression of the T1 from 2 methods (R2 = 0.970). (C) The Bland–Altman plot shows intersession repeatability of the T1 measurement of the Multitasking DCE measured on 2 different days. The solid line and dashed lines indicate the mean value and 1.96 of the SD of the T1 between different measurements, respectively
3.2 |. Numerical simulation
Supporting Information Figure S2 shows true CA concentration curves as well as estimated concentration curves for blood, normal pancreas, and PDAC using both the T1-based Multitasking approach and the conventional linear approach in a numerical simulation. The linear approach has a fundamental bias in the estimation of kinetic parameters, as it does not account for the nonlinearity of T1W signal intensity as a function of CA concentration, whereas the T1-based Multitasking approach provides estimated parameters consistent with the true parameters, as listed in Supporting Information Table S2. Supporting Information Figure S3 and Supporting Information Table S3 compare the kinetic parameter estimates from a Multitasking patient case using Multitasking and linear approaches, showing the same trends as the numerical simulation.
3.3 |. Effect of respiratory-motion compensation
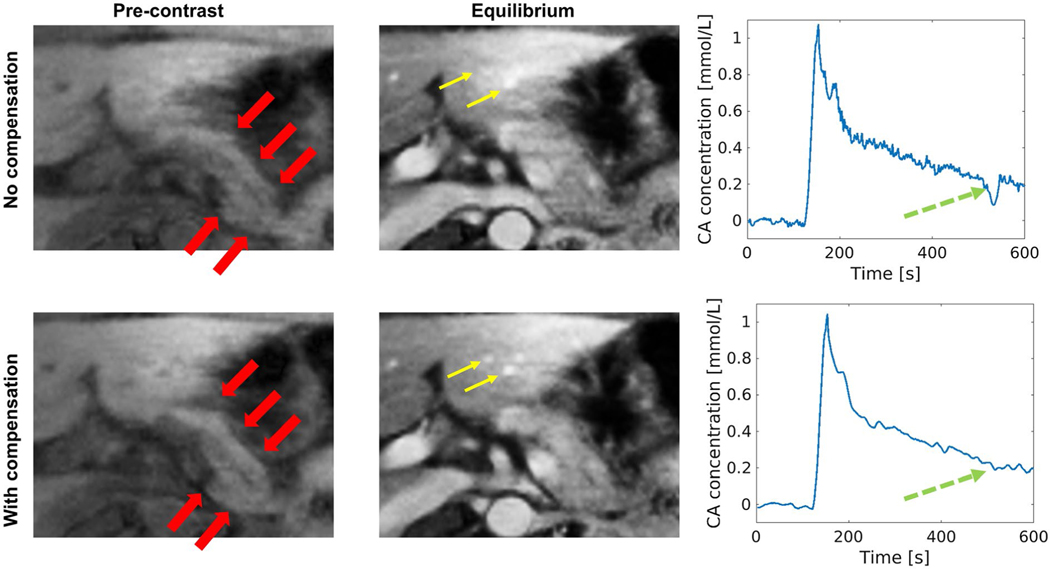
The respiratory-motion compensation scheme was applied to all subjects. Figure 2 compares the image quality and CA concentration curves in the pancreas with and without respiratory-motion compensation in a representative subject. The subject had an irregular respiratory pattern with nonu-niformly distributed bins, resulting in poor image quality at bins with fewer image lines. In this example, respiratory-motion compensation increased the interbin image correlation and therefore improved image quality. The CA concentration curve with motion compensation is much cleaner with less oscillation from respiratory motion. The SNR and sharpness measurements before and after motion compensation for both pre-contrast and equilibrium phases (td = 4 minutes) are listed in Table 2 for comparison. A one-tailed t-test shows that motion compensation produced significantly superior SNR in the pancreas and sharpness of the pancreas boundary (P = .006, .008, .013, <.001, respectively).
FIGURE 2.
Effect of respiratory-motion compensation. The images without and with respiratory compensation are shown at 2 DCE phases: 1 pre-contrast phase and 1 equilibrium phase (td = 4 minutes). The contrast agent (CA) concentration curve of the pancreas is displayed. Images with compensation show less artifacts and improved structure delineation. The red thick arrows mark the pancreas, which is much sharper with less artifacts on images with compensation. The yellow thin arrows mark small vessels, which show clearer boundary and improved contrast on images with compensation. The green dashed arrows point out that the CA concentration curve with compensation is much cleaner with far less oscillation
TABLE 2.
Comparison of SNR and sharpness without motion compensation and with motion compensation over all subjects (N = 30)
| SNR pre-contrast | SNR equilibrium | SHP pre-contrast (mm−1) | SHP equilibrium (mm−1) | |
|---|---|---|---|---|
| NMC | 29.83 ± 14.21 | 35.22 ± 13.07 | 0.39 ± 0.15 | 0.49 ± 0.16 |
| MC | 33.95 ± 17.09 | 40.50 ± 16.10 | 0.47 ± 0.15 | 0.59 ± 0.18 |
| P-value | .006 | .008 | .013 | <.001 |
Note: One-tailed t-tests were performed and the p-values are listed in the table. The equilibrium phase is td = 4 minutes, where td is the time since injection. Abbreviations: MC, motion compensation; NMC, no motion compensation; SHP, sharpness.
3.4 |. In vivo measurements
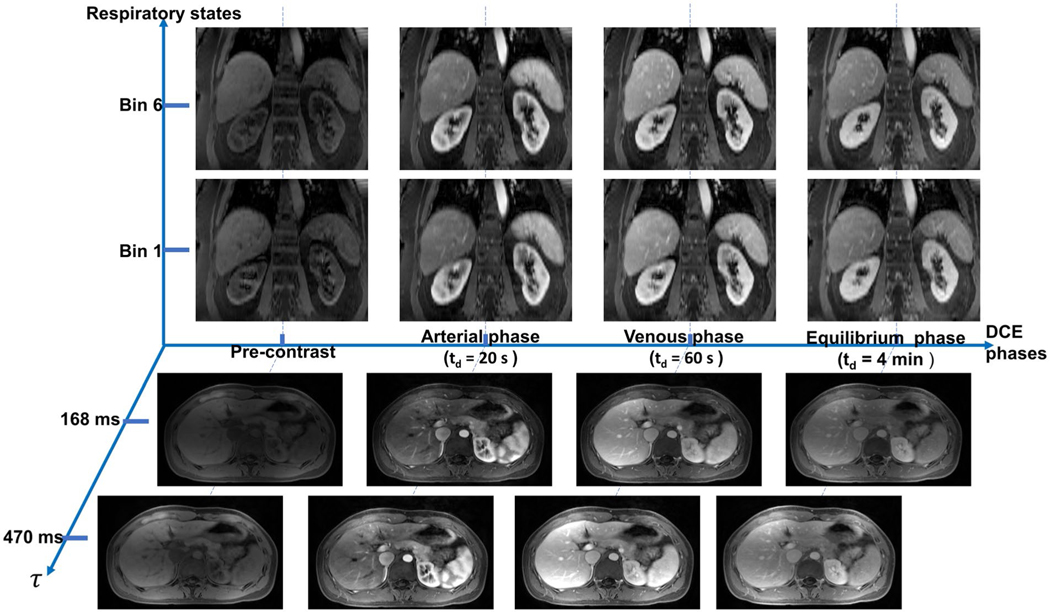
The in vivo protocol including the conventional sequences and Multitasking DCE was successfully applied to all healthy volunteers and patients. A demonstration of the 6D images from Multitasking DCE is shown in Figure 3. As described previously, the DCE bin duration in the reconstruction was chosen to be 1 second, yielding 600 DCE phases, each of which contains 84 saturation times. Combined with 6 respiratory bins, the reconstructed image tensor possesses a total of 600 × 84 × 6 = 302 400 timepoints, each corresponding to a 3D (3 spatial dimensions) image volume. Figure 3 displays the images in either coronal or axial orientation at 4 key DCE phases (pre-contrast phase, arterial phase with td = 20 seconds, venous phase with td = 60 seconds, and equilibrium phase with td = 4 minutes) at 2 saturation times and 2 respiratory states. Coronal orientation provides an appropriate view of the respiratory motion, while axial orientation shows excellent pancreas delineation. Supporting Information Video S1 further demonstrates the 6D images from Multitasking DCE.
FIGURE 3.
Representative 6D images with 3 spatial dimensions, a saturation-recovery (SR) dimension, a respiratory dimension, and a DCE time course. Images are displayed at pre-contrast phase, arterial phase (td = 20 seconds), venous phase (td = 60 seconds), and equilibrium phase (td = 4 minutes). For each DCE phase, images of end-inspiration (bin 1) and end-expiration (bin 6) phases at τ = 470 ms are shown in coronal orientation; images of different τs (τ = 168 ms and τ = 470 ms) at end-expiration phase are shown in transversal view with clear pancreas delineation
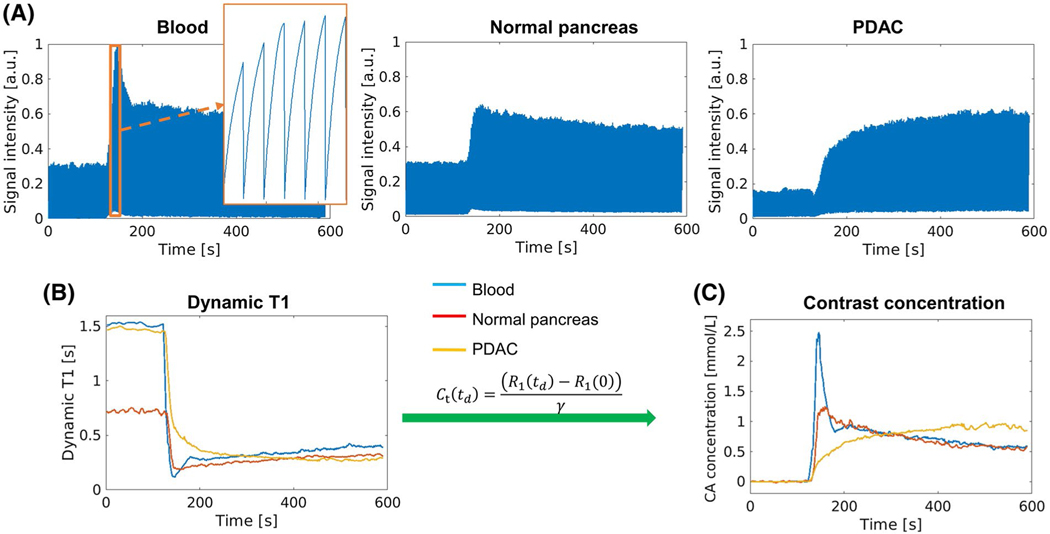
The typical process of the conversion from dynamic signal to CA concentration is illustrated in Figure 4. Figure 4A displays example signal intensity curves over all saturation times and DCE phases at end-expiration, displaying blood, normal pancreas, and PDAC mass. Figure 4B,C shows the corresponding dynamic T1 curves and CA concentration curves for each of these tissues. The shape of the concentration curves is consistent with the findings in other studies.15
FIGURE 4.
Transformation from signal intensity to CA concentration. (A) Representative signal profiles of blood, normal pancreas, and pancreatic ductal adenocarcinoma (PDAC) displayed at end-expiration. The zoomed-in area in the image of blood signal shows the SR curves during the peak enhancement. (B) Dynamic T1 mapping based on SR periods. (C) Contrast agent (CA) concentration curves derived directly from the T1 mapping according to contrast media relaxivity
The agreement of in vivo T1 measurements between Multitasking DCE and MOLLI was assessed for normal pancreas in the control group (denoted as control), PDAC mass in the patient group (denoted as tumor), and nontumoral areas (denoted as non-tumor) in the patient group (Table 3). The ICC of the 3 categories for pre-contrast T1 (ICC = 0.820, 0.904, and 0.973, respectively) and post-contrast T1 (ICC = 0.924, 0.958, and 0.922, respectively) indicate good agreement of in vivo T1 measurements between Multitasking DCE and MOLLI; the p-values given by paired t-tests for the 3 categories are 0.024, 0.180, and 0.428, respectively, for pre-contrast T1, and <.001, .013, and .043, respectively, for post-contrast T1.
TABLE 3.
Comparison of in vivo T1 measurements between Multitasking DCE and MOLLI
| Multitasking (ms) | MOLLI (ms) | ICC | Paired t-test (p-value) | ||
|---|---|---|---|---|---|
| Pre-contrast | Control | 757 ± 55 | 818 ± 46 | 0.820 | .024 |
| Tumor | 1571 ± 210 | 1621 ± 171 | 0.904 | .180 | |
| Non-tumor | 1183 ± 338 | 1245 ± 352 | 0.973 | .428 | |
| Post-contrast | Control | 419 ± 74 | 470 ± 80 | 0.924 | <.001 |
| Tumor | 386 ± 63 | 426 ± 56 | 0.958 | .013 | |
| Non-tumor | 417 ± 58 | 460 ± 49 | 0.922 | .043 |
Note: The mean values and SD of pre-contrast and post-contrast T1 from Multitasking DCE and MOLLI are listed for normal pancreas in control group, tumor, and nontumoral regions in patients. The ICC and p-value by paired t-test between T1 measured by MOLLI and Multitasking are provided. The ICC of each category indicates good agreement of in vivo T1 measurement between Multitasking DCE and MOLLI.
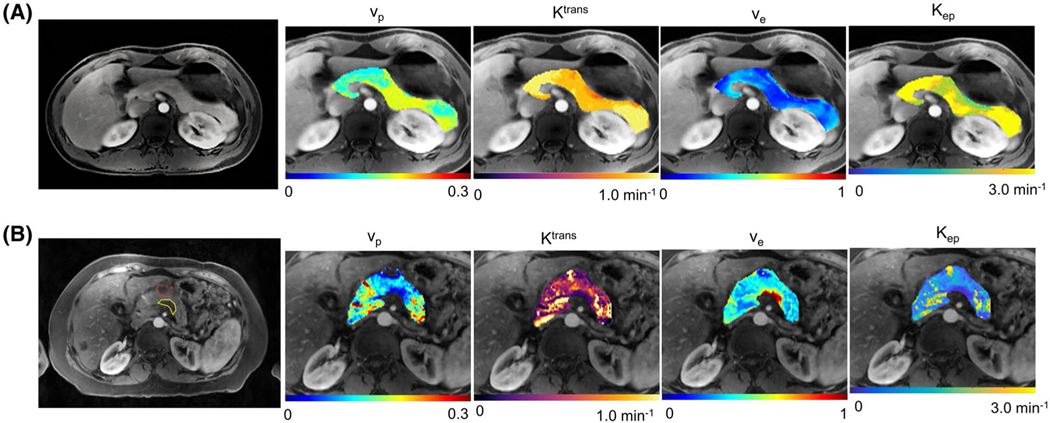
Examples of the kinetic parametric maps are shown in Figure 5. Figure 5A shows a healthy subject with a normal pancreas. The gray-scale images display the anatomical structure of a representative slice at the last saturation time of the arterial phase. The overlaid color voxels show the results of parametric mapping. Figure 5B represents a PDAC case with tumor located within the pancreatic body. The tumor mass was marked by a yellow boundary on the gray-scale images by the radiologist, and a benign cyst is marked by a dashed red boundary.
FIGURE 5.
Representative maps of kinetic parameters of a 32-year-old subject in control group (A) and a 57-year-old subject with PDAC marked by a yellow boundary (B). The gray-scale image in each case is a representative slice with excellent pancreas delineation at the arterial phase. The overlaid color maps display vp, Ktrans, ve, and Kep of the pancreas. In (B), the tumor region has lower vp, Ktrans, and Kep, and higher ve than the surrounding nontumoral regions. A hypo-enhancing cyst in (B) is marked by a dashed red boundary
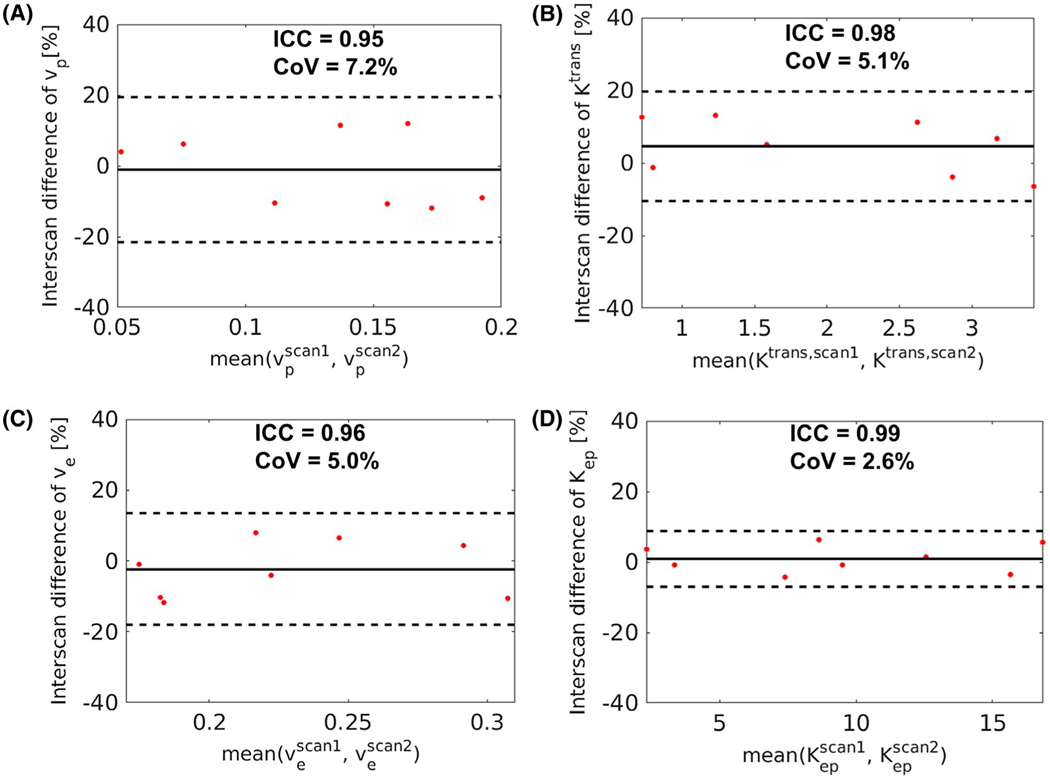
The intersession repeatability of the kinetic parameters from the proposed Multitasking DCE method was also evaluated, with Bland-Altman plots shown in Figure 6. The ICC of vp, Ktrans, ve, and Kep were 0.95, 0.98, 0.96, and 0.98, respectively. The CoV of the 4 parameters were 7.2%, 5.1%, 5.0%, and 2.6%, respectively.
FIGURE 6.
Bland-Altman plots evaluating the in vivo intersession repeatability of the kinetic parameters from Multitasking DCE on the 8 subjects in the control group who received the scan twice on separate days. The solid lines indicate the mean bias, and the dashed lines indicate the 95% limit of agreement. The interclass correlation coefficient (ICC) and coefficient of variation (CoV) of each parameter are listed on top of the corresponding plot
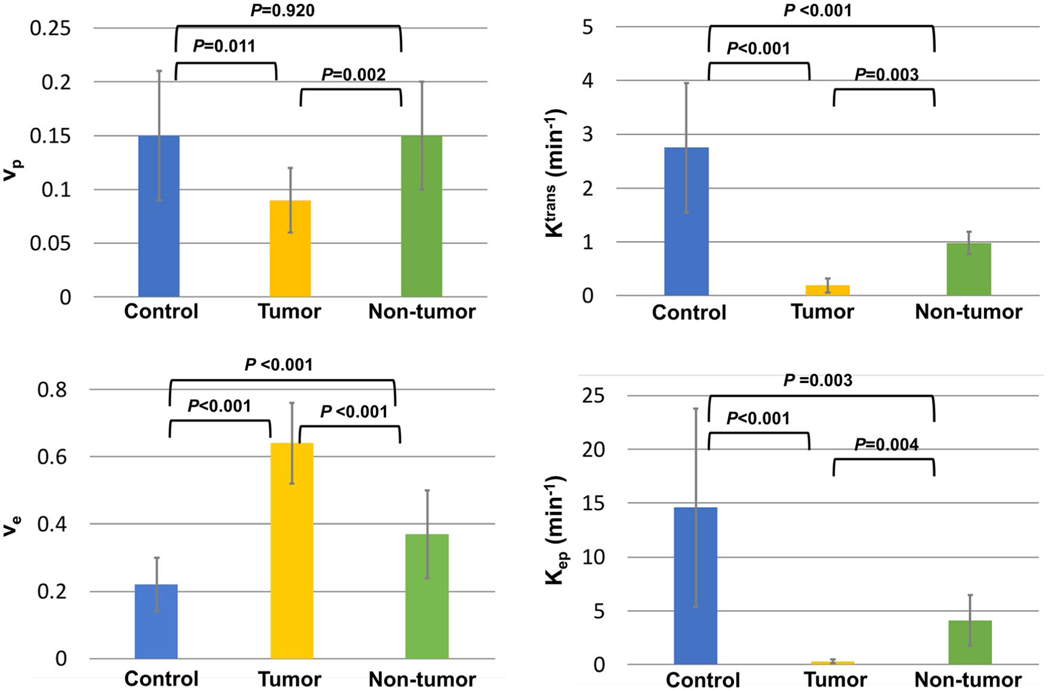
Figure 7 contains a bar graph showing the mean and SD measurements of vp, Ktrans, ve, and Kep from the control, tumor, and nontumoral regions. The results for all categories were in general agreement with published values from literature.4,15–17 An ANOVA analysis with Holm-Bonferroni correction indicated that all 4 parameters were significantly different between tumor with non-tumor (P = .002, .003, <.001, and .004, respectively) and between tumor and control (P = .0011, <.001, <.001, and <.001, respectively). The Ktrans, ve, and Kep between non-tumor and control also showed a significant difference (P < .001, <.001, and =.003, respectively). Detailed results of the statistical analyses are listed in Supporting Information Table S1. Supporting Information Figure S4 shows the correlation between the size of PDAC and each kinetic parameter estimated by Multitasking DCE.
FIGURE 7.
Bar graph showing the mean value and SD of vp, Ktrans, ve, and Kep for control group, tumor, and nontumoral regions in the patient group. The p-values between each pair of categories by one-way unbalanced ANOVA with Holm-Bonferroni correction are marked on top of the bar graph. All 4 parameters were significantly different in the comparison between tumor and control and between tumor and non-tumor. The Ktrans, ve, and Kep between control and non-tumor also showed a significant difference
4 |. DISCUSSION
DCE MRI has been widely applied in the study of permeability properties of lesions in oncology, especially in tumors of breast and pelvic organs.40–44 Investigations have reported that permeability parameters derived from DCE MRI are associated with the histologic criteria.15,17,40–42 However, only a limited number of studies have performed DCE MRI in the upper abdomen due to the challenges of adequate anatomical coverage, sufficient temporal resolution, as well as the challenges posed by respiratory motion. In this study, we presented a 6D quantitative DCE MRI technique based on MR Multitasking and demonstrated its feasibility of the characterization of PDAC. Multitasking DCE enables 3D free-breathing acquisition, whole-abdomen coverage, high DCE temporal resolution of 1 second, and dynamic T1 mapping in a single 10-minute scan, which overcomes all the aforementioned challenges and potentially facilitates the wide application of quantitative assessment of vascular properties in PDAC, including tumor characterization and treatment-response monitoring.
The coverage in head–foot direction of Multitasking DCE is 360 mm from the liver dome to the iliac crest. The potential utility of the large coverage includes the evaluation of vasculature involvement of PDAC as well as the detection of abdominal metastasis of PDAC,45–47 which are vital information in the diagnosis and staging of the disease.
Sufficient temporal resolution is crucial to the quantification of DCE MRI. Othman et al reported that a temporal resolution of at least 10 seconds/phase is required for the accurate diagnosis of cancer.48 Several recent investigations have also shown that the acquisition of the arterial input function, which usually changes more rapidly than the tissue of interest, requires a temporal resolution on the order of 1 second.49,50 To achieve satisfactory temporal resolution, a tradeoff usually has to be made by reducing either the coverage or spatial resolution. For example, Akisik et al used 2D-T1W acquisition for 3 axial slices and 1 sagittal slice with 4.4-second temporal resolution.17 Kim et al implemented the k-space weighted image-contrast (KWIC) technique, which allows k-space data sharing to reconstruct 3D images with high-spatial/low-temporal resolution or vice versa in one DCE session.16 In this work, the proposed technique can achieve adequate coverage and spatial resolution with a high temporal resolution that can be retrospectively set based on the choice of DCE bin duration. Here it was retrospectively set to be 1 second to balance temporal resolution and SNR, as a similar temporal resolution also yielded the highest intersession repeatability when evaluated in our previous work.24
In addition, the CA concentration in the proposed technique is directly quantified from dynamic T1 mapping to avoid the error introduced by linear approximation from T1W signal intensity. The feasibility of dynamic T1 fitting in Multitasking DCE has been assessed by numerical simulations in our previous work.24 For the in vivo study, the pre-contrast T1 shows no significant difference between the MOLLI and Multitasking measurements for tumor and non-tumoral regions in the PDAC group; a less than 10% difference (P = .024) was shown in controls, which may be due to known differences between various T1 mapping sequences.51 The post-contrast T1 measured by Multitasking is systematically 10% shorter than the T1 from the MOLLI scan collected after Multitasking for all tissue types (P < .001, P = .013, and P = .043, respectively). One possible reason is that contrast washout lengthened T1 during the time gap between the T1 measurement in the last DCE phase in Multitasking DCE and the subsequent MOLLI scan, especially in cases where subjects moved during the scan and relocalization of the pancreas was required before the post-contrast MOLLI scan.
Another feature of the proposed Multitasking DCE is the capability to resolve respiratory motion. Respiration-induced motion artifacts are a major source of degraded image quality in abdominal MRI. Acquisition with breath-holds is the most common way to reduce these artifacts, which is often unreliable and in many cases infeasible. Radial sampling patterns are more robust to motion, but take longer for image reconstruction and can still result in blurring or streaking artifacts. XD-GRASP is a technique that separates respiratory motion states and uses sparsity constraints along different time dimensions. A recent publication of DCE MRI in liver using XD-GRASP described a protocol with 3D coverage and reasonable spatial resolution but a relatively low temporal resolution of 13 seconds and no T1 quantification.52
The estimates of the kinetic parameters vp, Ktrans, ve, and Kep of control and patient groups are generally in line with the values published in the literature.15,16 Ktrans and ve have been shown to correlate with the MVD and fibrosis in PDAC lesions, respectively, and therefore can be used as imaging markers of the perfusion properties of tumor.4,15 In the TK model, Ktrans is dominated by the tissue blood flow and the capillary permeability–surface area and can have different physiological interpretations depending on the balance between the 2 factors.37 In the context of PDAC, the microvessels in the lesions are leaky and in low density, leading to high permeability and limited blood flow, in which case Ktrans is approximately equal to plasma flow per unit volume of tissue.37 This is a possible pathological reason behind the decrease of Ktrans value observed in malignant tumors compared with the nontumoral area in the patient group and normal tissue in the control group. The ve in the DCE MRI represents fractional extravascular extracellular volume. The higher ve in malignant tumor possibly results from the changes in the interstitial space due to the increased collagen content in the extracellular matrix. The nontumoral area in patients with PDAC are commonly associated with chronic obstructed pancreatitis also characterized by atrophy of the distal end toward the tail, reduced blood, and increased fibrosis,53,54 which may contribute to decreased Ktrans and increased ve in nontumoral tissues in the patient group compared with the normal tissues in the control group.55–58 Figure 5 suggests regional differences of kinetic parameters in the control group, which may be caused by the intrinsic heterogeneous tissue compositions in different parts of the pancreas.59 Previous studies have shown that some imaging biomarkers such as ADC differ significantly between the head, body, and tail of the pancreas.60
Our study had several limitations. All of the patients recruited for this pilot study were undergoing chemotherapy at the time of the study. Chemotherapy may change the microvascular structures and thus the kinetic parameters, which limits the value of the current data to represent the vascular properties of untreated PDAC tumors. In addition, this pilot study had a relatively small number of subjects for both control and patient groups. With the encouraging findings of this work, further studies to validate the clinical utility of the proposed technique in a larger patient cohort with first diagnosed PDAC are warranted. Furthermore, tumor histological validation was not available for this pilot study. Instead, the capability of Multitasking DCE in quantitatively assessing tumor vascular properties was evaluated by the intersession repeatability of the kinetic parameters. The kinetic parameters are consistent with the values in the published literatures. In future studies, the correlation between the kinetic parameters and the histological parameters including the MVD and fibrosis should be assessed.
5 |. CONCLUSIONS
A novel Multitasking abdominal DCE technique was developed, enabling free-breathing and respiratory motion–resolved image acquisition, entire-abdomen coverage, 1-second temporal resolution, dynamic T1 quantification, and kinetic modeling. Quantitative DCE parameters from the preliminary in vivo study were repeatable and showed significant differences among normal pancreas, tumor, and nontumoral regions in patients with PDAC.
Supplementary Material
FIGURE S1 Respiratory binning scheme. (A) A narrow bar-shaped region of interest (ROI) in the head–foot direction was manually drawn on top of the liver–air interface. (B) The signal at each last saturation time within the ROI was averaged over left–right direction as a 1-dimensional (1D) projection. The 1D projection over the duration of the scan formed a 2D spatial-temporal profile showing the location changes of the liver dome. (C) The position of the liver dome was extracted and then divided into 6 respiratory bins
FIGURE S2 Simulated CA concentration curves and estimated CA concentration curves using the linear approach (LA) and T1-based Multitasking approach (MT) for all tissues. The CA concentration curves estimated by LA are different from the simulated curves, whereas the curves estimated by MT are the same with the simulated curves. The zoomed-in area in the image of the signal intensity of MT shows the SR curves for PDAC
FIGURE S3 CA concentration curves derived by LA and MT from a Multitasking DCE patient case for all tissues. The zoomed-in area in the image of the signal intensity of MT shows the SR curves for PDAC
FIGURE S4 Correlation between the size of PDAC and vp, Ktrans, ve, and Kep estimated by Multitasking DCE. The R and the p-value of the correlation are attached to the corresponding graph. No significant correlation (P = .35, .78, .41, and .54 for vp, Ktrans, ve, and Kep, respectively) was found for this data set
TABLE S1 Statistical result of vp, Ktrans, ve, and Kep for control group, tumor, and nontumoral regions in the patient group
TABLE S2 True value of vp, Ktrans, ve, and Kep in the simulation and the mean and SD measurement of each kinetic parameter estimated using LA and MT
TABLE S3 Values of vp, Ktrans, ve, and Kep estimated using LA and MT from a Multitasking DCE patient case
TABLE S4 Size and location of the tumor for all patients. The size of the tumor is defined as the largest diameter in axial CT images according to RECIST 1.1 criteria
VIDEO S1 Demonstration of the 6-dimensional images (3 spatial dimensions, 1 SR dimension, 1 respiratory dimension, and 1 DCE time dimension)
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the Supporting Information section.
REFERENCES
- 1.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Danet IM, Semelka RC, Nagase LL, Woosely JT, Leonardou P, Armao D. Liver metastases from pancreatic adenocarcinoma: MR imaging characteristics. J Magn Reson Imaging. 2003;18: 181–188. [DOI] [PubMed] [Google Scholar]
- 4.Wegner CS, Hauge A, Gaustad J-V, et al. Dynamic contrast-enhanced MRI of the microenvironment of pancreatic adenocarcinoma xenografts. Acta Oncol. 2017;56:1754–1762. [DOI] [PubMed] [Google Scholar]
- 5.Lee ES, Lee JM. Imaging diagnosis of pancreatic cancer: a state-of-the-art review. World J Gastroenterol. 2014;20:7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keleg S, Büchler P, Ludwig R, Büchler MW, Friess H. Invasion and metastasis in pancreatic cancer. Mol Cancer. 2003;2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oberstein PE, Olive KP. Pancreatic cancer: why is it so hard to treat? Therap Adv Gastroenterol. 2013;6:321–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheufele F, Hartmann D, Friess H. Treatment of pancreatic cancer—neoadjuvant treatment in borderline resectable/locally advanced pancreatic cancer. Transl Gastroenterol Hepatol. 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holzapfel K, Reiser-Erkan C, Fingerle AA, et al. Comparison of diffusion-weighted MR imaging and multidetector-row CT in the detection of liver metastases in patients operated for pancreatic cancer. Abdom Imaging. 2011;36:179–184. [DOI] [PubMed] [Google Scholar]
- 10.Koelblinger C, Ba-Ssalamah A, Goetzinger P, et al. Gadobenate dimeglumine–enhanced 3.0-T MR imaging versus multiphasic 64–detector row CT: prospective evaluation in patients suspected of having pancreatic cancer. Radiology. 2011;259:757–766. [DOI] [PubMed] [Google Scholar]
- 11.Park HS, Lee JM, Choi HK, Hong SH, Han JK, Choi BI. Preoperative evaluation of pancreatic cancer: comparison of gad-olinium-enhanced dynamic MRI with MR cholangiopancreatography versus MDCT. J Magn Reson Imaging. 2009;30:586–595. [DOI] [PubMed] [Google Scholar]
- 12.Schima W, Függer R. Evaluation of focal pancreatic masses: comparison of mangafodipir-enhanced MR imaging and contrast-enhanced helical CT. Eur Radiol. 2002;12:2998–3008. [DOI] [PubMed] [Google Scholar]
- 13.Coolen BF, Poot DH, Liem MI, et al. Three-dimensional quantitative T1 and T2 mapping of the carotid artery: sequence design and in vivo feasibility. Magn Reson Med. 2016;75:1008–1017. [DOI] [PubMed] [Google Scholar]
- 14.Smith DS, Welch EB, Li X, et al. Quantitative effects of using compressed sensing in dynamic contrast enhanced MRI. Phys Med Biol. 2011;56:4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bali MA, Metens T, Denolin V, et al. Tumoral and nontumoral pancreas: correlation between quantitative dynamic contrast-enhanced MR imaging and histopathologic parameters. Radiology. 2011;261:456–466. [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, Lee JM, Park JH, et al. Solid pancreatic lesions: characterization by using timing bolus dynamic contrast-enhanced MR imaging assessment—a preliminary study. Radiology. 2013; 266:185–196. [DOI] [PubMed] [Google Scholar]
- 17.Akisik MF, Sandrasegaran K, Bu G, Lin C, Hutchins GD, Chiorean EG. Pancreatic cancer: utility of dynamic contrast-enhanced MR imaging in assessment of antiangiogenic therapy. Radiology. 2010;256:441–449. [DOI] [PubMed] [Google Scholar]
- 18.Naish JH, Hutchinson CE, Caunce A, et al. Multiple-bolus dynamic contrast-enhanced MRI in the pancreas during a glucose challenge. J Magn Reson Imaging. 2010;32:622–628. [DOI] [PubMed] [Google Scholar]
- 19.Ippoliti M, Lukas M, Winfried B, Schaeffter T, Makowski MR, Kolbitsch C. 3D nonrigid motion correction for quantitative assessment of hepatic lesions in DCE-MRI. Magn Reson Med. 2019;82:1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calcagno C, Mani V, Ramachandran S, Fayad ZA. Dynamic contrast enhanced (DCE) magnetic resonance imaging (MRI) of atherosclerotic plaque angiogenesis. Angiogenesis. 2010;13:87–99. [DOI] [PubMed] [Google Scholar]
- 21.Kim H. Variability in quantitative DCE-MRI: sources and solutions. J Nat Sci. 2018;4:e484. [PMC free article] [PubMed] [Google Scholar]
- 22.Wake N, Chandarana H, Rusinek H, et al. Accuracy and precision of quantitative DCE-MRI parameters: how should one estimate contrast concentration? Magn Reson Imaging. 2018;52:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christodoulou AG, Shaw JL, Nguyen C, et al. Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging. Nat Biomed Eng. 2018;2:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang N, Christodoulou AG, Xie Y, et al. Quantitative 3D dynamic contrast-enhanced (DCE) MR imaging of carotid vessel wall by fast T1 mapping using Multitasking. Magn Reson Med. 2019;81:2302–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, Liu Q, Christodoulou AG, Ma C, Lam F, Liang ZP. Accelerated high-dimensional MR imaging with sparse sampling using low-rank tensors. IEEE Trans Med Imaging. 2016;35:2119–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christodoulou AG, Redler G, Clifford B, Liang Z-P, Halpern HJ, Epel B. Fast dynamic electron paramagnetic resonance (EPR) oxygen imaging using low-rank tensors. J Magn Reson. 2016;270:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang Z-P. Spatiotemporal imaging with partially separable functions. In: Proceedings of the 4th IEEE International Symposium on Biomedical Imaging, Arlington, Virginia, 2007. pp 988–991. [Google Scholar]
- 28.Lingala SG, Hu Y, DiBella E, Jacob M. Accelerated dynamic MRI exploiting sparsity and low-rank structure: kt SLR. IEEE Trans Med Imaging. 2011;30:1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trzasko J, Manduca A, Borisch E. Local versus global low-rank promotion in dynamic MRI series reconstruction. In: Proceedings of the 19th Annual Meeting of ISMRM, Montréal, Canada, 2011. p 4371. [Google Scholar]
- 30.Zhao B, Haldar JP, Christodoulou AG, Liang ZP. Image reconstruction from highly undersampled (k, t)-space data with joint partial separability and sparsity constraints. IEEE Trans Med Imaging. 2012;31:1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolda TG, Bader BW. Tensor decompositions and applications. SIAM Rev. 2009;51:455–500. [Google Scholar]
- 32.Zhou R, Huang W, Yang Y, et al. Simple motion correction strategy reduces respiratory-induced motion artifacts for kt accelerated and compressed-sensing cardiovascular magnetic resonance perfusion imaging. J Cardiovasc Magn Reson. 2018;20:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higaki T, Tatsugami F, Fujioka C, et al. Visualization of simulated small vessels on computed tomography using a model-based iterative reconstruction technique. Data Brief. 2017;13:437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Lathauwer L, De Moor B, Vandewalle J. A multilinear singular value decomposition. SIAM J Matrix Anal Appl. 2000; 21:1253–1278. [Google Scholar]
- 35.Kanal E, Maravilla K, Rowley H. Gadolinium contrast agents for CNS imaging: current concepts and clinical evidence. Am J Neuroradiol. 2014;35:2215–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Y, Goerner FL, Snyder C, et al. T1 relaxivities of gadolinium-based magnetic resonance contrast agents in human whole blood at 1.5, 3, and 7 T. Invest Radiol. 2015;50:330–338. [DOI] [PubMed] [Google Scholar]
- 37.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T1-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. [DOI] [PubMed] [Google Scholar]
- 38.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. 2004;52:141–146. [DOI] [PubMed] [Google Scholar]
- 39.Russek S, Boss M, Jackson E, et al. Characterization of NIST/ISMRM MRI system phantom. In: Proceedings of the 20th Annual Meeting of ISMRM, Melbourne, Australia, 2012. p 2456. [Google Scholar]
- 40.Esserman L, Hylton N, George T, Weidner N. Contrast-enhanced magnetic resonance imaging to assess tumor histopathology and angiogenesis in breast carcinoma. Breast J. 1999;5:13–21. [DOI] [PubMed] [Google Scholar]
- 41.Su M-Y, Cheung Y-C, Fruehauf JP, et al. Correlation of dynamic contrast enhancement MRI parameters with microvessel density and VEGF for assessment of angiogenesis in breast cancer. J Magn Reson Imaging. 2003;18:467–477. [DOI] [PubMed] [Google Scholar]
- 42.de Lussanet QG, Backes WH, Griffioen AW, et al. Dynamic contrast-enhanced magnetic resonance imaging of radiation therapy-induced microcirculation changes in rectal cancer. Int J Radiat Oncol Biol Phys. 2005;63:1309–1315. [DOI] [PubMed] [Google Scholar]
- 43.Alonzi R, Padhani AR, Allen C. Dynamic contrast enhanced MRI in prostate cancer. Eur J Radiol. 2007;63:335–350. [DOI] [PubMed] [Google Scholar]
- 44.Feng L, Wen Q, Huang C, Tong A, Liu F, Chandarana H. GRASP-Pro: imProving GRASP DCE-MRI through self-calibrating subspace-modeling and contrast phase automation. Magn Reson Med. 2020;83:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yachida S, Iacobuzio-Donahue CA. The pathology and genetics of metastatic pancreatic cancer. Arch Pathol Lab Med. 2009;133:413–422. [DOI] [PubMed] [Google Scholar]
- 46.Embuscado EE, Laheru D, Ricci F, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Disibio G, French SW. Metastatic patterns of cancers: results from a large autopsy study. Arch Pathol Lab Med. 2008;132:931–939. [DOI] [PubMed] [Google Scholar]
- 48.Othman AE, Falkner F, Weiss J, et al. Effect of temporal resolution on diagnostic performance of dynamic contrast-enhanced magnetic resonance imaging of the prostate. Invest Radiol. 2016;51:290–296. [DOI] [PubMed] [Google Scholar]
- 49.Henderson E, Rutt BK, Lee T-Y. Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magn Reson Imaging. 1998;16:1057–1073. [DOI] [PubMed] [Google Scholar]
- 50.Li K-L, Buonaccorsi G, Thompson G, et al. An improved coverage and spatial resolution—using dual injection dynamic contrast-enhanced (ICE-DICE) MRI: a novel dynamic contrast-enhanced technique for cerebral tumors. Magn Reson Med. 2012;68:452–462. [DOI] [PubMed] [Google Scholar]
- 51.Teixeira T, Hafyane T, Stikov N, Akdeniz C, Greiser A, Friedrich MG. Comparison of different cardiovascular magnetic resonance sequences for native myocardial T1 mapping at 3T. J Cardiovasc Magn Reson. 2016;18:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD-GRASP: golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med. 2016;75:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436. [DOI] [PubMed] [Google Scholar]
- 54.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coenegrachts K, Van Steenbergen W, De Keyzer F, et al. Dynamic contrast-enhanced MRI of the pancreas: initial results in healthy volunteers and patients with chronic pancreatitis. J Magn Reson Imaging. 2004;20:990–997. [DOI] [PubMed] [Google Scholar]
- 56.Johnson PT, Outwater EK. Pancreatic carcinoma versus chronic pancreatitis: dynamic MR imaging. Radiology. 1999;212:213–218. [DOI] [PubMed] [Google Scholar]
- 57.Tajima Y, Kuroki T, Tsutsumi R, Isomoto I, Uetani M, Kanematsu T. Pancreatic carcinoma coexisting with chronic pancreatitis versus tumor-forming pancreatitis: diagnostic utility of the time-signal intensity curve from dynamic contrast-enhanced MR imaging. World J Gastroenterol. 2007;13:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang T-T, Wang LI, Liu H-H, et al. Differentiation of pancreatic carcinoma and mass-forming focal pancreatitis: qualitative and quantitative assessment by dynamic contrast-enhanced MRI combined with diffusion-weighted imaging. Oncotarget. 2017;8:1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adelson JW, Miller PE. Heterogeneity of the exocrine pancreas. Am J Physiol Gastrointest Liver Physiol. 1989;256:G817–G825. [DOI] [PubMed] [Google Scholar]
- 60.Schoennagel BP, Habermann CR, Roesch M, et al. Diffusion-weighted imaging of the healthy pancreas: apparent diffusion coefficient values of the normal head, body, and tail calculated from different sets of b-values. J Magn Reson Imaging. 2011;34:861–865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Respiratory binning scheme. (A) A narrow bar-shaped region of interest (ROI) in the head–foot direction was manually drawn on top of the liver–air interface. (B) The signal at each last saturation time within the ROI was averaged over left–right direction as a 1-dimensional (1D) projection. The 1D projection over the duration of the scan formed a 2D spatial-temporal profile showing the location changes of the liver dome. (C) The position of the liver dome was extracted and then divided into 6 respiratory bins
FIGURE S2 Simulated CA concentration curves and estimated CA concentration curves using the linear approach (LA) and T1-based Multitasking approach (MT) for all tissues. The CA concentration curves estimated by LA are different from the simulated curves, whereas the curves estimated by MT are the same with the simulated curves. The zoomed-in area in the image of the signal intensity of MT shows the SR curves for PDAC
FIGURE S3 CA concentration curves derived by LA and MT from a Multitasking DCE patient case for all tissues. The zoomed-in area in the image of the signal intensity of MT shows the SR curves for PDAC
FIGURE S4 Correlation between the size of PDAC and vp, Ktrans, ve, and Kep estimated by Multitasking DCE. The R and the p-value of the correlation are attached to the corresponding graph. No significant correlation (P = .35, .78, .41, and .54 for vp, Ktrans, ve, and Kep, respectively) was found for this data set
TABLE S1 Statistical result of vp, Ktrans, ve, and Kep for control group, tumor, and nontumoral regions in the patient group
TABLE S2 True value of vp, Ktrans, ve, and Kep in the simulation and the mean and SD measurement of each kinetic parameter estimated using LA and MT
TABLE S3 Values of vp, Ktrans, ve, and Kep estimated using LA and MT from a Multitasking DCE patient case
TABLE S4 Size and location of the tumor for all patients. The size of the tumor is defined as the largest diameter in axial CT images according to RECIST 1.1 criteria
VIDEO S1 Demonstration of the 6-dimensional images (3 spatial dimensions, 1 SR dimension, 1 respiratory dimension, and 1 DCE time dimension)