Abstract
Emerging of the COVID-19 pandemic has raised interests in the field of biology and pathogenesis of coronaviruses; including interactions between host immune reactions specific, and viral factors. Deep knowledge about the interaction between coronaviruses and the host factors could be useful to provide a better support for the disease sufferers and be advantageous for managing and treatment of the lung infection caused by the virus. At this study, we reviewed the updated information on the pathogenesis of the COVID-19 and the immune responses toward it, with a special focus on structure, genetics, and viral accessory proteins, viral replication, viral receptors, the human immune reactions, cytopathic effects, and host-related factors.
Keywords: Coronavirus, COVID-19, Viral accessory proteins, Viral receptors, Immune reactions, Cytopathic effects
Introduction
Coronaviruses belong to the enveloped RNA viruses with a positive sense and non-segmented genome [1]. Until now, coronaviruses including Middle Eastern respiratory syndrome coronavirus (MERS-CoV), Severe acute respiratory syndrome coronavirus (SARS-CoV), and Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [Casituve agent of coronavirus 2019 (COVID-19)] are known to infect the human and cause respiratory disorders with high pathogenesis that led to regional and global outbreaks [2]. The coronaviruses are categorized into the Nidovirales orders, Coronaviridae family, and Coronavirinae subfamilies, which this subfamily include four genera: α-coronavirus, β-coronavirus, γ-coronavirus, and δ-coronavirus [3].
From November 2002 to June 2003, a viral respiratory disorder emerged in southern China and rapidly spread the other countries, resulting in more than 8000 confirmed cases [4]. SARS patients exhibit flu-like symptoms and pneumonia that in severe circumstances cause lethal respiratory failure and acute distress syndrome. In 2012, MERS-CoV was reported from Saudi Arabia as a causative agent of respiratory malady, similar to SARS, causing two major outbreaks in Saudi Arabia (2012) and South Korea (2015), resulting in over 2000 confirmed cases [5]. Previously, two coronaviruses (HCoV-229E and HCoV-OC43) have been identified to cause mild upper respiratory symptoms, although sometimes they cause severe low respiratory infection in the elderly, infants, as well as immunocompromised individuals. COVID-19 is a new infection that was first reported on 31 December 2019 (Wuhan, Hubei Province, China), that could cause a respiratory malady (influenza-like disorder) with symptoms such as dry cough, fever, tiredness, headache, diarrhea, sore throat and, in more severe cases it could induce pneumonia [6]. Until 31 May 2020, over 5,934,936 confirmed COVID-19 cases and 367,166 approved mortality were recorded worldwide by more than 216 countries, areas, or regions [7].
In spite of the not completely clear pathologies mechanism of COVID-19 infection, viral, and host-related factors are known to be crucial in the infection process. During COVID-19 infection, host factors stimulate the immune responses to the SARS-CoV-2 virus. Meanwhile, it has to be considered that immunopathogenesis is correlated with an out-of-control immune response, which may lead to pulmonary tissue injury. Therefore, there is an urgent need to better understanding of the interactions among host immune responses and viral factors of SARS-CoV-2 as well as host-related factors that will offer fundamental insights into the treatment and control of the disease. In this study, we first review the literature of the SARS-CoV-2 proliferation cycle, then summarize the current information of critical immune reactions activated during COVID-19 infection.
Structure, genetics and viral accessory proteins
The positive-sense RNA genome (30 kb) of SARS-CoV-2 contains a 5′ cap construct conjoint with a 3′ poly-A tail that allows performing as mRNA for translating replicate polyproteins. The 5′ end of the genome has a leader and untranslated region (UTR) sequence that comprises the compositions required for transcription and replication of RNA. The SARS-CoV-2 genome organization is 5′-leader-UTR-replicase-S-E-M–N-3′ UTR-poly A with intrinsic genes inside the structural genes at the 3′ end of the genome [3, 8, 9]. SARS-CoV-2 has a pleomorphic morphology with a diameter of 60–120 nm [3, 9]. The genome of SARS-CoV-2 is positive, sensitive, non-fragmented RNA, with an astonishingly large size in a range of 27 to 32 kb [8, 9].
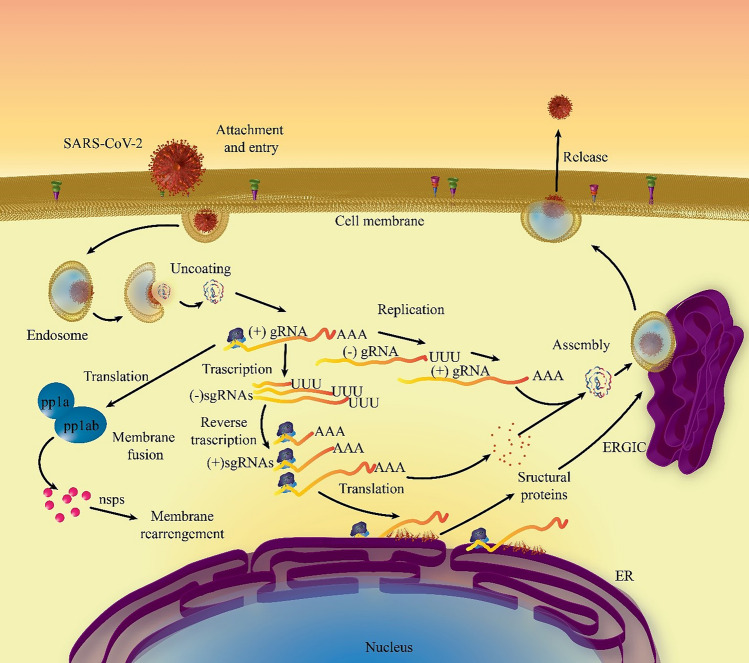
Genomic RNA of the SARS-CoV-2 virus contains five caped and 3-polyadenylated, including seven Open Reading Frames (ORF). ORF 1a and 1b comprise two-thirds of the virus genome, encoding unstructured poly-proteins. The rest of ORFs code structural proteins such as spike protein (S), envelopes protein (E), membrane protein (M) as well as nucleocapsid protein (N). Additionally, in some cases the hemagglutinin esterase (HE) gene is located between ORF 1b and ORF S. The structural proteins fold and enter into endoplasmic reticulum and transport to the transitional gap of endoplasmic Reticulum-Golgi. The SARS-CoV-2 replication cycle has steps including; attachment and penetrance, replication of virus replication, genome transcription and replication, structural protein translation, assemblage and discharging the virus [8–10]. Figure 1 presents a schematic overview of the SARS-CoV-2 replication cycle.
Fig. 1.
Schematic overview of the SARS-CoV-2 replicative cycle. This replication cycle is divided into multiple steps: attachment and entry, replication of virus replication, genome transcription and replication, structural protein translation, and assemblage and release of the virus. SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, sgRNA single guide RNA, gRNA guide RNA, ER endoplasmic reticulum, ERGIC ER-Golgi intermediate compartment
The noticeable characteristic of coronaviruses is the spike proteins in a club-shaped form that give them the shape of a solar corona. The M protein is a common marker in the virion, which is a small (30 kDa) structure encompassing three transmembrane domains and a short N-terminal glycosylated ectodomain and a very larger C-terminal endodomain [8, 9]. Currently, it has been suggested that this protein could adopt two distinct conformations; enabling to improve membrane shape, along with attachment to the nucleocapsid [3].
Protein E of the SARS-CoV-2 virus (12 kDa) is a transmembrane structure with an N-terminal ectodomain and a C-terminal endodomain, having an ion channel action. Additionally, this construct promotes the assembly and discharge of the virus. The N protein is a unique construct within the nucleocapsid which includes one N-terminal and a C-terminal domain. Besides, this protein is phosphorylated which improves the affinity for RNA of viruses compared with the other non-viral RNAs. N protein attaches the viral genome with a beads-on-a-string type pattern. Another structural viral protein is HE which exists in β-coronaviruses and plays as a hemagglutinin that attaches to sialic acid on host glycoproteins and contains acetyl-esterase activity, which is suggested to enhance the cell entry mediated to S protein and viral spread via the host mucus [3, 8, 9, 11].
Viral replication
Following the receptor attachment, SARS-CoV-2 needs to reach the host cell cytosol. Binding is performed via S protein cleavage by a cathepsin, followed by cell membrane fusion. Cleavage is carried out at two sites in the S2 segment, and fusion occurs inside acidic endosomes, but some coronaviruses like murine coronavirus, so-called mouse hepatitis virus (MHV), could fuse into the host plasma membrane [3, 11–13]. The next step is the translation of the replicase gene (encodes two big ORFs) from genomic RNA in the virion [11].
These ORFs (rep1a and rep1b) form two polyproteins, pp1a and pp1ab, use a slippery sequence (5′-UUUAAAC-3′) and an RNA pseudoknot is performed which triggers the ribosomal frameshift of the rep1a reading frame to the rep1b. Polyproteins pp1a compromise the eleven non-structure proteins (NSPs) including nsps 1 to nsps 11, while pp1ab compromise nsps 1 to nsps 16. Coronaviruses encode proteases, for example, papain-like proteases, coded by nsp3, and a serine-type protease, the major protease, coded by nsp5 gene. At the next step, the nsps move inside the replicase-transcriptase complex to form an environment for synthesis and replication of RNA, as well as sub-genomic RNAs transcription. Synthesis of viral RNA forms sub-genomic and genomic RNAs. Sub-genomic RNAs provide mRNAs for transcription of the structural and accessory genes that are located downstream of the replicase polyproteins. Also, it has been shown that coronaviruses can recombine using non-homologous and homologous recombination [11–13].
Upon replication and subgenomic RNA synthesis, the structural proteins, including S, E, and M, are entered into the endoplasmic reticulum and migrate inside the secretory pathway within the Endoplasmic Reticulum-Golgi Intermediate Compartment (ERGIC), where genomes encapsidated by N protein fuse into ERGIC membranes which contains structural proteins, and form mature virions [3].
The M proteins, along with E proteins, prompt protein–protein cooperation that is necessary for coronaviruses assembly [11–13]. N proteins increase the production of virion like particles (VLPs), revealing that the integration of encapsidated genomes into the ERGIC promotes the development of the viral particle [3, 12, 13]. Moreover, the S proteins are incorporated into virions at this step. Besides, the M proteins bind to the nucleocapsid that improves the completion of the virions assembly. Finally, the virion is transferred outside of the cell and releases through exocytosis [3, 9].
Viral receptors
Penetrating to the target cell is a crucial point in the replication cycle of SARS-CoV-2. The primary marker of this stage is an adequate attachment of the S glycoprotein to a cell surface receptor [14, 15]. S1 ward is implicated in receptor attaching which includes N- and C-terminal domains that both may play as the receptor-binding domain (RBD), having significant determinants of cell tropism located in S1-CTD region. The elongated S2 domains of stalk form the spike trimer are essentially implicated in starting the integration of the viral envelope and membranes of the target cell. SARS-CoV-2 binds to special cellular receptors via the S protein, that makes deformation of the S proteins and then intercedes the fusion among the viral and cell membranes, leading to discharging the nucleocapsid into the host cell [16, 17].
Receptor attachment is the principal marker of the host spectrum and tissue tropism of coronavirus. Coronavirus has acquired cell surface factors as receptors, for example, aminopeptidase N (APN) for 229E, angiotensin-converting enzyme 2 (ACE2) for SARS as well as SARS-CoV-2, dipeptidyl peptidase 4 (DPP4) for MERS and O-acetylated sialic acid for OC43. Host proteases mediate the activation of S protein, for example, activation of S protein of SARS-COV-2 requires further cleavage by the endosomal cysteine protease cathepsin L and another trypsin-like serine protease [18–20].
Furthermore, S protein of SARS-CoV-2 encompasses two cleavage locations for furin which is a ubiquitously expressed protease [21]. Besides the above-mentioned proteases, extracellular, cell surface-related, and lysosomal ones have an essential impact on SARS-CoV-2 entry via triggering the integration of S protein [18–21]. In the case of SARS-CoV-2, the integration of viral and cellular membrane is initiated following the S protein cleavage via cell surface-related TMPRSS2) (transmembrane protease, serine 2) [22]. The same protease is required for cleavage and promotion of the 229E and MERS S protein [23].
Innate immunity
The innate immunity is a well-maintained defense mechanism for accelerated recognition and control of pathogens and subsequent promotion of the adaptive immune response. Sufficient trigger of the innate immunity depends on detecting the pathogen molecular patterns (PAMPs) via pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD‐like receptor (NLR), C‐type lectin‐like receptors, and free‐molecule receptors in the host cytoplasm [24].
Following the promotion via PAMPs, PRRs absorb adaptor proteins, that comprise complex signaling pathways involving several kinases [25]. This signaling pathway eventually results in the promotion of the essential transcription factors, such as the nuclear factor interferon regulatory factor 3 (IRF3), Nuclear factor-kappa B (NF-κB), and Activator Protein-1(AP-1) [25, 26]. Synergistically, these agents release type I interferons that are secreted and act nearby interferon α/β Receptor (IFNAR). The antiviral effect of interferon type I has interfered through different mechanisms [27]. Additionally, chemokines and cytokines are ordered to stimulate inflammatory reactions, which is responsible for extensive tissue damage [26]. PAMPs include lipids, lipoproteins, proteins, and nucleic acids of bacterial, viral, parasitic, and fungal origin detected by TLRs [28]. Also, PAMPs recognition via TLRs can happen in cell membranes, lysosomes, endosomes, as well as endocytolysosomes. Overall, various TLRs could trigger different biological reactions via subsequent stimulation of several adapter proteins, like myeloid differentiation factor 88 (MyD88), TIR-domain-containing adapter-inducing interferon-β (TRIP), Toll/interleukin-1 receptor (TIR) domain-containing adapter protein (TIRAP) as well as TRIF-related-adaptor-molecule (TRAM) [25, 29].
Currently, just a limited report is available on the host innate immune status of SARS-CoV-2 infected patients. In one report where 99 cases in Wuhan were examined, increased total neutrophils (38%), diminished total lymphocytes (35%), increased serum IL-6 (52%) and enhanced c-reactive protein (84%) were detected [30]. Increased neutrophils and diminished lymphocytes also associate with the disease severity and mortality in COVID-19 patients [6]. Furthermore, patients needing ICU care had raised plasma levels of several innate cytokines to include IP-10, MCP-1, MIP-1A, and TNFα [31]. These clinical characteristics intimated the likelihood of involvement of highly pro-inflammatory statuses in the disease progression and severity. This early sharp rise in the serum levels of pro-inflammatory cytokines has also been reported in SARS-CoV and MERS-CoV infection, implying a potential similar cytokine storm-mediated disease severity. The robust innate immune response toward viral infection relies profoundly on the interferon (IFN) type I responses and its downstream cascade that completes in managing viral replication and initiation of the effective adaptive immune response [32, 33].
Immune cells involvement
Macrophages and dendritic cells play a crucial role in the innate immune and adaptive immune responses. These cells can stimulate T lymphocytes and B lymphocytes, therefore induce coupling promoting innate and adaptive immunity. The immature dendritic cell has a high migration capacity, and mature dendritic cells could considerably trigger T cells in the central link of regulation and maintaining the immune responses. Dendritic precursor cells are differentiated into dendritic cells via adjusting inducers; such as granulocyte–macrophage colony-stimulating factor, interleukin-4 (IL‐4), and tumor necrosis factor-alpha (TNF‐α) [34, 35]. CD4 positive T cells and CD8 positive T cells have an essential function by adjusting immune reactions toward viruses and the risk of autoimmunity and inflammation [36]. CD4 positive T cells elevate the production of virus‐specific antibodies via stimulating T‐dependent B cell activation [37] and, CD8 positive T cells are cytotoxic and can kill the cells infected with the virus [38].
Additionally, T helper cells generate proinflammatory cytokines via the nuclear factor kappa B (NF‐kB) signaling pathway [39]. Interleukin 17 (IL‐17) cytokines call up monocytes and neutrophils to the infected area through inflammation and stimulate other downstream cytokine and chemokine cascades, including Interleukin 1 (IL‐1), Interleukin 6 (LL‐6), Interleukin 8 (IL‐8), Interleukin 21 (IL‐21), TNF‐beta, and monocyte chemoattractant protein-1 (MCP‐1) [40, 41]. Such information has been considered for related vaccine design and therapeutics developing studies [42]. Recently, it has been demonstrated that T cell response to SARS S protein and other structural proteins (e.g., the M and N proteins) is long‐lasting, suggesting that the design of vaccines based of viral structural proteins could stimulate dominant, effective, and long‐term memory responses toward virus [43]. A recent finding shows that humoral immunity is necessary for controlling the persistent phase of coronavirus malady [44, 45]. Current evidence well-indicate that Th1 type response is key to the reliable control of SARS-CoV and MERS-CoV and presumably valid for SARS-CoV-2 as well. Similar findings of distinct CD8 + T cells have been reported during the MERS-CoV clearance in a mouse model too. Consequently, this knowledge can be beneficial in the case of SARS-CoV-2, as well. However, in the case of SARS-CoV-2 recent reports propose that the PBMCs of SARS-CoV-2 infected individuals have shown adequate mitigation in the CD8 + and CD4 + T cell counts, which may result in compromised T memory cell production and persistence in SARS-CoV-2 survivors [46].
The complement system has a critical function in the immune responses to infection caused by the coronavirus since this system enables the detection and response to viral particles. Considering its potency to harm the host tissues, the complement system is regulated via inhibiting proteins of serum. C3a and C5a show a proinflammatory activity that could initiate the recruitment of the inflammatory cell, such as activation of neutrophil cells [47]. Blocking C3a and C5a could be a treatment for severe lung damage, and the anti‐C5a antibody has shown to save the mice from MERS infection [2]. Activation of complement and the contact system, through the formation of bradykinin, may perform a role in the SARS-CoV-2-induced pulmonary edema, and its recommended that further work is necessary to verify the information. Although complement is sufficient in defending against various viruses, it does not seem to be protective against the coronavirus. C3 knockout mice contaminated with SARS-CoV had fewer lung disease than wild-type mice, proposing, that complement may have a role in coronavirus pathogenesis. Some evidence suggests that revealed pulmonary edema may be bradykinin-induced and could be the reason that corticosteroids, antihistamines, and other traditional mediations for edema are not sufficient. ACE2 is a co-receptor for SARS-CoV-2, and studies so far have not achieved a benefit or risk correlated with the application of either ACE-inhibitors or angiotensin receptor antagonists [48].
Interferons
Macrophages are critical producers of type I interferons and other pro-inflammatory cytokines which trigger antiviral protection, even though they potentially contribute to immune pathology mediated to viral infections [49]. Interferons mediate direct antiviral influences that restrict viral replication via activating/regulating a group of well-defined antiviral effectors, such as protein kinase R (PKR) and Ribonuclease L (RNase L), while inflecting other facets of the innate and adaptive immune responses via stimulating a wide variety of interferon-inducible genes [50].
There are two main pathways via them host cells sense invading viruses and trigger the interferon pathway; the cytoplasmic interferon induction pathways and TLR associated pathway [51]. TLRs including TLR3, TLR7, TLR8 as well as TLR9 could recognize viruses in endosomal compartments as they penetrate cells [52]. In contrast, cytoplasmic caspase activation and recruitment domain (CARD) RNA helicases, RIG-I, and melanoma differentiation-associated gene 5 (Mda5), identify viral RNAs in the cytoplasm [53]. Pathways are activated by PAMPs, including double-stranded RNA, that is a byproduct of virus replication or constructed single-stranded RNAs mediated to incoming viral genomes, being common objective [54]. The cytoplasmic interferon induction pathways and TLR use various adaptor proteins to mediate signaling, with the TLR dependent pathways applying MyD88 and TRIF. Downstream of the multiple adaptors, these pathways share some typical signaling molecules and transcription factors, for example; interferon regulatory factor 3 (IRF-3), activator protein-1 as well as NF-kB for initiation the type I interferon expression. Subsequently, type I interferons induce the Janus kinase/signal transducers and activators of transcription (JAK‐STAT) signal pathway, which enhances the transcription of genes induced by interferons [55]. Interferons restrict the spread of virus particles and have an immunomodulatory activity to improve the phagocytosis by macrophage cells, as well as restriction of natural killer’s function to infected target cells and T and B lymphocytes [56].
The viral nucleic acids subsequent infecting plasma‐like dendritic cells are detected via TLR7 and TLR9 to trigger the inflammatory cytokine formation and type I interferons associated with IRF as well as NF‐κB. Although coronaviruses are susceptible to interferon α and β, these viruses persist highly pathogenic in this situation [55]. However, the N proteins of SARS play as an antagonist for immune escape protein and interferon reactions of the host. Comparing with SARS-CoV, which is thought to induce inadequate interferon response, SARS-CoV-2 robustly triggers the expression of numerous IFN-inducible genes (ISGs). These ISGs display immunopathogenic potentials, defined by the overrepresentation of genes implicated in inflammation [57].
Chemokine and cytokine
The C-X-C motif of chemokine 10 (CXCL10) and C–C motif of Chemokine Ligand 3 (CCL3) are formed as the earliest chemokines following viral infection. CXCL10 overexpression in the central nervous system by a murine coronavirus demonstrated that exaggerated natural killer cells aggregation in immunodeficient recombination activating gene 1 (RAG1) negative/negative mice could be involved in the virus clearance. Despite the primary natural killer cell recruitment, there are few documents of a direct antiviral activity for wild type virus. Their potential to secrete interferon-γ can enhance the presentation of antigen through regulation of major histocompatibility complex class I (MHC 1) and class II (MHC 2) molecules [58, 59].
Augmented CCL3 has shown to be associated with the innate and adaptive immune reactions via inducing recruitment, promotion as well as the maturation of dendritic cells and T lymphocytes. Cytokines like IL-1α, IL-1β, IL-6, as well as IL-12 rapidly produce in the corona infected cells like. TNF-α, IL-12, as well as IL-1β are formed in resident host cells in reply to virus infection. Stimulation of adaptive immune reactions can change the composition and expression of infiltrating cells and chemokine in the viral infection site. Although C-X-C motif chemokine 9 (CXCL9), C–C Motif Chemokine Ligand 2 (CCL2), CCL3, C–C Motif Chemokine Ligand 4 (CCL4), C–C Motif Chemokine Ligand (CCL5), and C–C Motif Chemokine Ligand (CCL7) are formed in acute viral infection, CXCL9 and CXCL10 can gather the activated T cells and natural killers. Overall, CXCL10 is indicated to be the sustained and major chemokine in viral infection [59].
As shown by the recent studies, increased circulating levels of pro-inflammatory cytokines (e.g., Interferon γ, interleukin (IL-) 1B, IL-6, IL-12) and chemokines (CXCL10, and CCL2) are correlated with pulmonary inflammation and extensive lung involvement in SARS patients, similarly to what occurs in MERS-CoV infection [60]. Huang et al. [34] recently reported that infected patients with COVID 19 also show enhanced levels of pro-inflammatory cytokines and chemokines. The demonstration of increased levels of IL-1B, IFNγ, CXCL10, and CCL2 firmly pointed toward the activation of T-helper-1 (Th1) cell function. More importantly, the so-called “cytokine storm” emerged as the principal factor driving a more severe clinical course. This idea has been originated from the observation that COVID-19 patients requiring ICU admission revealed higher concentrations of CXCL10, CCL2, and TNFα as compared with those in which the infection was less severe and did not require ICU admission. To further complicate the issue, it should be highlighted that in patients with SARS-Cov-2 infection, as a difference from SARS-CoV infection, there is also an enhanced secretion Th2-immune-oriented cytokines such as IL-4 and IL-10, whose main effect is to repress inflammation [60].
Cytopathic effects
Although host airway epithelial cells provide a primary aim for infection, dendritic cells in the respiratory epithelium are sentinels of the host immune reactions that detect viral pathogens and present their antigens to T cells. It has been found that 229E triggers massive cytopathic effects (CPE) in dendritic cells, for example, the large syncytia and cell death. Monocyte is more resistant against viral infection and CPE of 229E because of the expression levels of CD13 (one membrane receptor for 229E) is similar, and cell death and CPE are acquired after 24 h of the 229E infection of monocytes. Cell death stimulated by 229E is independent from TNF-α, Fas ligand (FasL), TNF-related apoptosis-inducing ligand (TRAIL), as well as caspases, confirming that viral replication is the causative agent of the CPE. The result of dendritic cell death at the initial step of 229E infection might affect the central control of virus release and the establishment of persistent immune memory, therefore the individuals could be reinfected again by 229E [61].
Kidney cell lines from African green, cynomolgus, and rhesus monkey have demonstrated to be sensitive toward SARS infection. Additionally, in a study by Kaye et al. it was shown that kidney cells derived from a buffalo green monkey were actively infected with SARS, with titers up to 4.7 log10/mL higher than input SARS virus, like the levels in other monkey kidney cells. They showed that the most monkey kidney–derived cell lines like Vero E6, assisted SARS replication, having titers 3.9–4.7 log10/mL higher than input SARS virus titers [62, 63].
High titers of SARS available in these cells must be regarded while applying them for virus isolation, and proper safety guidelines must be pursued. The capacity of SARS to replicate effectively in kidney cell lines could be due to the presence of ACE-2 receptors in the kidney [64]. Also, ACE-2 is expressed in gastrointestinal, lung, as well as heart tissue [65]. The primate kidney cell lines are specifically sensitive toward SARS infection. This virus has been collected from the kidney of an infected human, so it has been proposed that human kidney cell lines could also provide replication of SARS [66]. In a report by Kaye et al., it was found that SARS could be collected in various cell lines, indicating that this virus could acquire high titers across multiple cell lines, even in the absence of CPE [62]. The relative contributions of virus replication and cytopathic effects or immunopathological host responses to the severe and fatal consequences of COVID-19 lung infections have as yet to be determined. In a pioneer study, Schifanella et al. showed that SARS-CoV-2 replication and cytopathic effects in type II alveolar pneumocytes make focal lung damage in a somebody with no history of pulmonary symptoms [67].
Autoimmunity
Autoimmune diseases occur as a consequence of atypical immune reactions for the detection of non- and self-antigens [68]. Until now, up to 80 autoimmune diseases have been identified [69]. Many agents have been mediated to autoimmune diseases like age, genetics, and infection [70].
Interestingly, viral agents are being regarded as main infective agents which induce the autoimmune diseases in sensitive people, and some strategies have been offered to define the interrupt self-tolerance in the host by various viral infections. Multiple processes have been suggested for initiating and triggering the autoimmune disease induced virus. Molecular mimicry is the most probable process and happens when a virus antigen mimics a host antigen and stimulates cross-reactive T lymphocytes. Epitope spreading is another process, for example, the destruction of host tissue due to T lymphocyte activation unique to the virus has shown to induce promoting autoreactive T lymphocytes and liberation of self-antigens surrounded the inflammatory site. Bystander promotion is the stimulation of autoreactive T lymphocytes because of the cytokine formation in immune reactions targeted by the virus. Also, encrypted antigens from the host are formed from particular tissues during tissue damage targeted by the virus. Additionally, superantigens stimulate a wide variety of nonspecific T lymphocyte clones. It has been found that coronavirus, parainfluenza, and metapneumovirus are significantly mediated to rheumatoid arthritis. Rheumatoid arthritis is an autoimmune disorder contributing to interactions between environmental and genetic agents. It has been reported that rheumatoid arthritis due to the formation of autoantibodies in genetically susceptible individuals could be triggered by some driving factors like the virus [71]. However, an association between respiratory viral infections like coronavirus and developing rheumatoid arthritis is proposed [72].
It has been found that CD 4 positive and CD8 positive T lymphocytes can link the demyelination in vivo, for example, rodents Theiler’s encephalomyelitis viral infection and murine coronavirus [73]. Additionally, some reports have declared that CD8 T lymphocytes distinct to myelin could stimulate a progressive type of experimental autoimmune encephalomyelitis (EAE). Also, these cells account for major infiltrating immune cells in demyelinating lesions in multiple sclerosis individuals. Moreover, CD8 positive T lymphocytes stimulate demyelination in Theiler’s encephalomyelitis virus infection in vivo. Reduced level of demyelination has been identified in CD4 positive T lymphocytes deficient mice infected by murine coronavirus infection, which is consistent with an activity for CD8 positive T lymphocytes. In a study by Pewe et al. their findings support the notion that CD8 positive T lymphocytes can associate with demyelination independent to CD4 positive T lymphocytes. Also, they found that interferon-γ is significant for demyelination mediated to CD8 positive T lymphocyte. Demyelination was not diminished when depleted interferon-γ negative/negative splenocytes were translocated [74].
Interferon-γ has many impacts on inflammation in the central nervous system, for example, as a proinflammatory molecule and in adjusting the trafficking of leukocyte cells and the population dynamics of T lymphocyte [75]. Interferon-γ, in pro-inflammatory activity, is participated in roles including activation macrophage cells, adjusting MHC I and II transcription, and stimulating adhesion molecules necessary for their penetrating the central nervous system. Also, interferon-γ is contributed to the stimulation of chemokines expression, such as CCL2, CCL7, CCL4, CCL5, CXCL9, and CXC L10, that are chemoattractants for monocytes and lymphocytes and are up-regulated in infected mice central nervous system [74]. Overall, these findings show that interferon-γ is important for demyelination associated with CD8 positive T lymphocytes in mice infected by murine coronavirus because of macrophage activation, and migration inside the central nervous system.
Channappanavar et al. has reported a pathogenic cascade that drives to ARDS and death; this mechanism is possible operative in COVID-19 infections. That study suggest a scenario in which viral replication leads to a delayed interferon response, accumulation, and activation of monocyte/macrophages and neutrophils, which in aggregate result in an excessive immune response accountable for the alveolar and endothelial apoptosis [76]. There is certainly a complexity in the participation of type I IFNs concerning early and late responses. The former may limit virus replication through induction of interferon-stimulated genes. At the same time, the latter can worsen disease by heightening the recruitment and function of macrophages and other innate immune cells [77]. The dysregulation of inflammatory responses starts pulmonary injury that leads to ARDS. Patients admitted for severe COVID-19 infections should probably hold maintenance therapies; these judgments are properly left to the rheumatology/infectious disease teams using care of them. Rheumatologists and infectious disease experts should be managing concurrently in this epidemic as precise data are not accessible, thus making recommendations speculative.
Host factors
People at the highest risk for severe corona infection comprise individuals aged > 60 years and, those with underlying situations like diabetes, hypertension, cardiovascular disorder, cancer as well as a chronic respiratory disease. Surprisingly, COVID infection in infants noted to be relatively mild and rare, with up to 2.4% of the cases. Additionally, mortality increases with age, having the highest rate in individuals > 80 years. Based on findings and analysis, it has found that males are more at risk than females [78]. Although males and females have been infected at equal rates, the death number in males was 2.8%, compared with 1.7% among females. Additionally, the crude death ratio is higher among males (4.7%) in comparison with females (2.8%) [79].
When a woman is pregnant, her body and immune system naturally become weak, consequently, the mother could be infected more easily. Pregnant women experience immunologic and physiologic alterations, which might make them more susceptible to respiratory infections such as COVID19 [80]. Some individuals are at risk for corona infections, including chronic respiratory patients (like chronic obstructive pulmonary disease, asthma, emphysema, bronchitis), as well as chronic heart patients (like heart failure, chronic kidney disorder), people with chronic liver disorder (like hepatitis), and chronic neurological conditions patients (such as Parkinson, motor neuron disorder, multiple sclerosis) [81].
Obesity is a type of sustained low-grade inflammation. Along with serving as an energy-storage depot, adipose tissue has endocrine activities, forming adipokines with immunomodulatory influences [82]. Leptin is a proinflammatory adipokine expressed in the lung, predominantly produced by white adipose which contributes to physiological conditions including maturation [83]. It is also involved in respiratory control and pathological conditions, including Chronic Obstructive Pulmonary Disease (COPD), obstructive sleep apnoea, inside the respiratory system [84]. Obesity sufferers show high circulating rates of leptin that potentially mediated via the up-regulation of suppressor of cytokine signalling-3. Also, interferon expression happens by the same JAK-STAT signaling pathway which is negatively controlled through suppressor of cytokine signalling-3, suggesting a critical pathway by which interferon reactions are attenuated against respiratory viruses in obese individuals [85]. Table 1 presents some of the important host factors involved in the coronavirus pathogenesis.
Table 1.
Host factors involved in the coronavirus pathogenesis
| Stage | Virus | Host factor | Function | References |
|---|---|---|---|---|
| Attachment and entry | SARS-CoV, SARS-CoV-2 | ACE2/ TMPRSS11D | Attachment/ S1/S2 cleavage to activate the S protein | [86] |
| Attachment and entry | HCoV-NL63 | ACE2 | Attachment and entry | [87] |
| Attachment and entry | MERS-CoV | dipeptidyl-peptidase 4 (DPP4), Furin | Attachment and entry | [88] |
| Attachment and entry | HCoV-OC43, HCoV-HKU1 | 9-O-acetylated sialic acid | Attachment and entry | [89] |
| Attachment and entry | IBV | Sialic acid/Furin | Attachment / S1/S2 cleavage to activate the S protein | [90] |
| Attachment and entry | HCoV-229E | APN /TMPRSS11D | Attachment/ S1/S2 cleavage to activate the S protein | [90] |
| Attachment and entry | HCoV-OC43 | acetyl-9-O-acetylneuraminic acid/ IFITM2 and IFITM3 | Attachment/ Promote virus entry | [91] |
| Translation of replicase and viral replication-transcription complexes (RTCs) Assembly | IBV | Annexin A2 | Modulate the frameshifting efficiency of viral RNA, Serve as the template for an anti-viral cellular protein | [92] |
| Translation of replicase and viral replication-transcription complexes (RTCs) Assembly | MHV | GBF1 and ARF1 | Required for efficient MHV RNA replication | [93] |
| Translation of structural proteins | SARS-CoV | N-linked glycosylation enzymes |
Post-translation modification of structural Proteins such as S, M |
[94] |
| Translation of structural proteins | MHV | O-linked glycosylation enzymes |
Post-translation modification of structural Proteins such as M protein |
[94] |
| Virion assembly and budding | HCoV-229E, HCoV-NL63 | β-tubulin |
β-tubulin interact with S proteins and reinforcement successful assembly and budding |
[94] |
| Virion assembly and budding | IBV | β-actin | β-actin interact with M proteins which involved in the virion assembly and budding | [95] |
SARS-CoV severe acute respiratory syndrome coronavirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, HCoV-NL63 human coronavirus NL63, MERS-CoV middle east respiratory syndrome coronavirus, HCoV-OC43 human coronavirus OC43, HCoV-HKU1 human coronavirus HKU1, IBV infectious bronchitis virus, HCoV-229E human coronavirus 229E, MHV mouse hepatitis virus, ACE2 angiotensin-converting enzyme 2, TMPRSS transmembrane protease, serine 2; APN aminopeptidase N, IFITM2 interferon induced transmembrane protein 2, IFITM3 interferon induced transmembrane protein 3, GBF1 golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1, ARF1 ADP ribosylation factor1
Genetics of hosts
Viruses usually demonstrate a difference in gaining sensitivity and clinical manifest, indicating that virus pathogenesis could have resulted from complicated intercommunication between viruses and hosts. This variability upon infection has been mediated to a range of agents like viral strain and sequence variation, age, immune status, and the genetic background of the host. Innate antiviral immunity prepares the primary line of protection toward pathogenic invasions by sensing PAMPs via PRRs. For example, activation of the cytoplasmic DNA sensor stimulates the formation of interferon to defend against viruses, which is also significant for the establishment of interferons in mousepox infection [96]. Interferons impede the replication of the virus via stimulating hundreds of Interferon-induced genes that control activities of antiviral influencers [97].
Variations in the genetic material of the multiple innate immune pathway genes, which cause antiviral effector activities influence the infection [98]. In the midst of interferon-stimulated genes, interferon-stimulated transmembrane proteins, expressed at the plasma membrane and membrane of endocytic vesicles, limit many other enveloped virus infections through inhibition of the viral envelope fusing with membranes of cells. Interferon-induced Transmembrane-3 variants that diminish gene expression or encode truncated forms are connected to a higher risk of influenza disease and a more serious health condition [99]. Host cells apply RNase L to destroy viral RNA and stimulate the formation of interferon through RIG-I to protect against RNA viruses [51].
Contrasted to another + RNA viruses, coronaviruses have a particularly large genome and employ a complex genome expression approach. Next to a role in fundamental virus replication or virus assembly, various of the coronavirus proteins expressed in the infected cell play a part into the coronavirus-host interplay. For instance, by interacting with the host cell to generate optimal conditions for coronavirus replication, via modifying host gene expression or by hindering the host’s antiviral defenses [100]. These SARS-CoV-2–host interplays are essential to viral pathogenesis and will eventually determine the consequence of infection. Due to the intricacy of the SARS-CoV-2 proteome and replication cycle, our understanding of host determinants implicated in coronavirus replication is yet in an initial step compared to what is understood for some other + RNA viruses.
Conclusion
The knowledge about coronavirus, including replication of the virus, cell biology, and pathogenesis has an accelerated progress rate because of the availability of reverse genetics systems. Currently, the raised interest in this class of viruses is due to the COVID-19 pandemic, and resources have been provided for studying these viruses. The impressive speed by which the COVID-19 was identified, and the genome was sequenced, along with the earlier accumulated data on the other members of the corona-virus family, has provided a lot of new insight about this universal health issue. There are still many significant and intriguing aspects remaining to be identified about COVID-19. Since coronaviruses appear with a periodical pattern, and unpredictably disperse quickly as well as stimulated severe disorders, they are regarded as a constant threat to human life. There are no approved vaccines or drugs for remedying coronaviruses disorders, and many animal reservoirs for coronaviruses and recombinant coronaviruses are existing. Future directions for COVID-19 studies include the further gaining knowledge of the replication mechanisms; clarifying the virulence molecular determinants and tropism, and the immune reactions. These studies should have special regard toward probable activities of group-specific proteins; developing vaccine techniques and antiviral drugs for human viruses. Although many new insights have been provided in regard to the COVID-19, it is still very critical to study about the isolation and definition of new coronaviruses with pathogenic potential for humans.
Acknowledgements
Thanks to guidance and advice from “Clinical Research Development Unit of Baqiyatallah Hospital”. There was a page limitation for all the other works, therefore we have to apologize from the authors whose works could not receive a citation in this manuscript.
Funding
No grant or financial support was devoted to this project by any organization.
Compliance with ethical standards
Conflict of interest
Nothing declared.
Research involving human participants and/or animals
No human or animal was involved in this study.
Informed consent
There was no human participant and consent was not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rasoul Mirzaei, Email: rasul.micro92@gmail.com.
Sajad Karampoor, Email: sajadkarampour1987@gmail.com.
Mohammad Sholeh, Email: mohammad.sholeh.mail@gmail.com.
Pouya Moradi, Email: Pooya.viro92@gmail.com.
Reza Ranjbar, Email: ranjbar@bmsu.ac.ir.
Faezeh Ghasemi, Email: ghasemi_808@yahoo.com.
References
- 1.Denison MR, et al. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol (Clifton, NJ) 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To KKW, et al. From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis. 2013;5(Suppl 2):S103–S108. doi: 10.3978/j.issn.2072-1439.2013.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skariyachan S, et al. Recent aspects on the pathogenesis mechanism, animal models and novel therapeutic interventions for Middle East respiratory syndrome coronavirus infections. Front Microbiol. 2019;10:569. doi: 10.3389/fmicb.2019.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Organization, T.W.H. (2019) Coronavirus disease (COVID-19) outbreak situation. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 8.Weiss SR, Leibowitz JL. Coronavirus pathogenesis. Adv Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Y, et al. Highlight of immune pathogenic response and hematopathologic effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 infection. Front Immunol. 2020;11:1022. doi: 10.3389/fimmu.2020.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skariyachan S, et al. Recent aspects on the pathogenesis mechanism, animal models and novel therapeutic interventions for middle east respiratory syndrome coronavirus infections. Front Microbiol. 2019;10:569–569. doi: 10.3389/fmicb.2019.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mousavizadeh L, Ghasemi S (2020) Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect S1684-1182(20)30082–7. [DOI] [PMC free article] [PubMed]
- 12.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belouzard S, et al. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol (Orlando, Fla) 2020;215:108427–108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen WH, Hotez PJ, Bottazzi ME (2020) Potential for developing a SARS-CoV receptor-binding domain (RBD) recombinant protein as a heterologous human vaccine against coronavirus infectious disease (COVID)-19. Hum Vaccine Immunother p. 1–4. [DOI] [PMC free article] [PubMed]
- 18.Bosch BJ, Smits SL, Haagmans BL. Membrane ectopeptidases targeted by human coronaviruses. Curr Opin Virol. 2014;6:55–60. doi: 10.1016/j.coviro.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan Y, et al. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, D.X., J.Q. Liang, and T.S. Fung, Human Coronavirus-229E, -OC43, -NL63, and -HKU1. Reference Module in Life Sciences, 2020: p. B978-0-12-809633-8.21501-X.
- 21.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell [DOI] [PMC free article] [PubMed]
- 22.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell [DOI] [PMC free article] [PubMed]
- 23.de Wilde AH, et al. Host factors in coronavirus replication. In: Trip RA, Tompkins SM, et al., editors. Roles of host gene and non-coding RNA expression in virus infection. New York: Springer; 2017. pp. 1–42. [Google Scholar]
- 24.Frieman M, Heise M, Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133(1):101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22(2):240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwanaszko M, Kimmel M. NF-κB and IRF pathways: cross-regulation on target genes promoter level. BMC Genomics. 2015;16(1):307–307. doi: 10.1186/s12864-015-1511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perry AK, et al. The host type I interferon response to viral and bacterial infections. Cell Res. 2005;15(6):407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- 28.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto M, Takeda K, Akira S. TIR domain-containing adaptors define the specificity of TLR signaling. Mol Immunol. 2004;40(12):861–868. doi: 10.1016/j.molimm.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahallawi WH, et al. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 34.Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125(2 Suppl 2):S3–S23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiasa M, et al. GM-CSF and IL-4 induce dendritic cell differentiation and disrupt osteoclastogenesis through M-CSF receptor shedding by up-regulation of TNF-α converting enzyme (TACE) Blood J Am Soc Hematol. 2009;114(20):4517–4526. doi: 10.1182/blood-2009-04-215020. [DOI] [PubMed] [Google Scholar]
- 36.Mason D. T-cell-mediated control of autoimmunity. Arthritis Res. 2001;3(3):133–135. doi: 10.1186/ar289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flynn S, et al. CD4 T cell cytokine differentiation: the B cell activation molecule, OX40 ligand, instructs CD4 T cells to express interleukin 4 and upregulates expression of the chemokine receptor, Blr-1. J Exp Med. 1998;188(2):297–304. doi: 10.1084/jem.188.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35(2):161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu T, et al. NF-κB signaling in inflammation. Signal Transduct and Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci. 2019;20(23):6008. doi: 10.3390/ijms20236008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner MD, et al. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta. 2014;1843(11):2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Sheikhshahrokh A, et al. Frontier therapeutics and vaccine strategies for sars-cov-2 (COVID-19): a review. Iran J Public Health. 2020;49:18–29. doi: 10.18502/ijph.v49iS1.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu WJ, et al. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dittel BN. CD4 T cells: balancing the coming and going of autoimmune-mediated inflammation in the CNS. Brain Behav Immun. 2008;22(4):421–430. doi: 10.1016/j.bbi.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuwabara T, Ishikawa F, Kondo M, Kakiuchi T (2017) The role of IL-17 and related cytokines in inflammatory autoimmune diseases. Mediat Inflamm 2017 [DOI] [PMC free article] [PubMed]
- 46.Kumar S, et al. Host immune response and immunobiology of human SARS-CoV-2 infection. Coronavirus Disease 2019 (COVID-19) 2019;2020:43–53. [Google Scholar]
- 47.Abou-El-Hassan H, Zaraket H. Viral-derived complement inhibitors: current status and potential role in immunomodulation. Exp Biol Med (Maywood, NJ) 2017;242(4):397–410. doi: 10.1177/1535370216675772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maglakelidze N, Manto KM, Craig TJ (2020) A review: does complement or the contact system have a role in protection or pathogenesis of COVID-19? Pulm Ther 1 [DOI] [PMC free article] [PubMed]
- 49.Vadiveloo PK, et al. Role of type I interferons during macrophage activation by lipopolysaccharide. Cytokine. 2000;12(11):1639–1646. doi: 10.1006/cyto.2000.0766. [DOI] [PubMed] [Google Scholar]
- 50.Sang Y, Miller LC, Blecha F. Macrophage polarization in virus-host interactions. J Clin Cell Immunol. 2015;6(2):311. doi: 10.4172/2155-9899.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee BL, Barton GM. Trafficking of endosomal toll-like receptors. Trends Cell Biol. 2014;24(6):360–369. doi: 10.1016/j.tcb.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barral PM, et al. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol Ther. 2009;124(2):219–234. doi: 10.1016/j.pharmthera.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol. 2014;426(6):1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nan Y, Wu C, Zhang Y-J. Interplay between Janus Kinase/signal transducer and activator of transcription signaling activated by type I interferons and viral antagonism. Front Immunol. 2017;8:1758–1758. doi: 10.3389/fimmu.2017.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, Guo L, Yang J, Wang C, Jiang S (2020) Overly exuberant innate immune response to SARS-CoV-2 infection
- 58.Phares TW, et al. Astrocyte-derived CXCL10 drives accumulation of antibody-secreting cells in the central nervous system during viral encephalomyelitis. J Virol. 2013;87(6):3382–3392. doi: 10.1128/JVI.03307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergmann CC, et al. Coronavirus immunity: from T cells to B cells. In: Perlman S, Gallagher T, Snijder EJ, et al., editors. The nidoviruses. New York: Springer; 2006. pp. 341–349. [Google Scholar]
- 60.Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M (2020) The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev [DOI] [PMC free article] [PubMed]
- 61.Mesel-Lemoine M, et al. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J Virol. 2012;86(14):7577–7587. doi: 10.1128/JVI.00269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaye M, et al. SARS–associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12(1):128. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Müller MA, et al. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio. 2012;3(6):e00515-12. doi: 10.1128/mBio.00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eckerle I, et al. In-vitro renal epithelial cell infection reveals a viral kidney tropism as a potential mechanism for acute renal failure during Middle East Respiratory Syndrome (MERS) Coronavirus infection. Virol J. 2013;10:359–359. doi: 10.1186/1743-422X-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamming I, et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus: a first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaye M. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12(1):128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schifanella L et al (2020) Massive viral replication and cytopathic effects in early COVID-19 pneumonia. arXiv:2005.00004.
- 68.Mackay IR, Leskovsek NV, Rose NR. Cell damage and autoimmunity: a critical appraisal. J Autoimmun. 2008;30(1–2):5–11. doi: 10.1016/j.jaut.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davidson A, Diamond B. Autoimmune diseases. N Engl J Med. 2001;345(5):340–350. doi: 10.1056/NEJM200108023450506. [DOI] [PubMed] [Google Scholar]
- 70.Ercolini AM, Miller SD. The role of infections in autoimmune disease. Clin Exp Immunol. 2009;155(1):1–15. doi: 10.1111/j.1365-2249.2008.03834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smatti MK, et al. Viruses and autoimmunity: a review on the potential interaction and molecular mechanisms. Viruses. 2019;11(8):762. doi: 10.3390/v11080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joo YB, et al. Respiratory viral infections and the risk of rheumatoid arthritis. Arthritis Res Ther. 2019;21(1):199. doi: 10.1186/s13075-019-1977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oleszak EL, et al. Theiler's virus infection: a model for multiple sclerosis. Clin Microbiol Rev. 2004;17(1):174–207. doi: 10.1128/CMR.17.1.174-207.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pewe L, Perlman S. Cutting edge: CD8 T cell-mediated demyelination is IFN-γ dependent in mice infected with a neurotropic coronavirus. J Immunol. 2002;168(4):1547–1551. doi: 10.4049/jimmunol.168.4.1547. [DOI] [PubMed] [Google Scholar]
- 75.Rauch I, Müller M, Decker T. The regulation of inflammation by interferons and their STATs. JAK-STAT. 2013;2(1):e23820. doi: 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Channappanavar R, Perlman S (2017) Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. In: Seminars in immunopathology. Springer, New York [DOI] [PMC free article] [PubMed]
- 77.Askanase AD, Khalili L, Buyon JP. Thoughts on COVID-19 and autoimmune diseases. Lupus Sci Med. 2020;7(1):e000396–e000396. doi: 10.1136/lupus-2020-000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Times, T.N.Y. Coronavirus threatens Americans with underlying conditions. https://www.nytimes.com/2020/03/12/health/coronavirus-midlife-conditions.html.
- 79.The World Health Organization (2020) Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19)
- 80.Zhao X et al (2020) Analysis of the susceptibility to COVID-19 in pregnancy and recommendations on potential drug screening. Eur J Clin Microbiol Infect Dis 1–12. [DOI] [PMC free article] [PubMed]
- 81.Inform, N (2020) Coronavirus (COVID-19): social distancing. https://www.nhsinform.scot/illnesses-and-conditions/infections-and-poisoning/coronavirus-covid-19/coronavirus-covid-19-social-distancing.
- 82.Mraz M, Haluzik M. The role of adipose tissue immune cells in obesity and low-grade inflammation. J Endocrinol. 2014;222(3):R113–R127. doi: 10.1530/JOE-14-0283. [DOI] [PubMed] [Google Scholar]
- 83.Lago F, et al. The emerging role of adipokines as mediators of inflammation and immune responses. Cytokine Growth Factor Rev. 2007;18(3–4):313–325. doi: 10.1016/j.cytogfr.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 84.Tkacova R (2010) Systemic inflammation in chronic obstructive pulmonary disease: may adipose tissue play a role? Review of the literature and future perspectives. Mediat Inflamm 2010 [DOI] [PMC free article] [PubMed]
- 85.Almond MH, et al. Obesity and susceptibility to severe outcomes following respiratory viral infection. Thorax. 2013;68(7):684–686. doi: 10.1136/thoraxjnl-2012-203009. [DOI] [PubMed] [Google Scholar]
- 86.Meng T et al (2020) The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS. bioRxiv
- 87.Abdul-Rasool S, Fielding BC. Understanding human coronavirus HCoV-NL63. Open Virol J. 2010;4:76. doi: 10.2174/1874357901004010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hulswit RJ, et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci. 2019;116(7):2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lim YX, et al. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4(3):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao X, et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci. 2014;111(18):6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwak H, Park MW, Jeong S. Annexin A2 binds RNA and reduces the frameshifting efficiency of infectious bronchitis virus. PLoS ONE. 2011;6(8):e24067. doi: 10.1371/journal.pone.0024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Verheije MH, et al. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4(6):e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fung TS, Liu DX. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 95.Wang J, et al. Interaction of the coronavirus infectious bronchitis virus membrane protein with β-actin and its implication in virion assembly and budding. PLoS ONE. 2009;4(3):e4908. doi: 10.1371/journal.pone.0004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.An P, Guo J-T, Winkler CA. Editorial: Host Genetics in Viral Pathogenesis and Control. Frontiers in genetics. 2019;10:1038–1038. doi: 10.3389/fgene.2019.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoffmann H-H, Schneider WM, Rice CM. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36(3):124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343(6175):1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kenney AD, et al. Human genetic determinants of viral diseases. Annu Rev Genet. 2017;51:241–263. doi: 10.1146/annurev-genet-120116-023425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.de Wilde AH, et al. Host factors in coronavirus replication. Curr Top Microbiol Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]