Abstract
The current global pandemic COVID-19 caused by the SARS-CoV-2 virus has already inflicted insurmountable damage both to the human lives and global economy. There is an immediate need for identification of effective drugs to contain the disastrous virus outbreak. Global efforts are already underway at a war footing to identify the best drug combination to address the disease. In this review, an attempt has been made to understand the SARS-CoV-2 life cycle, and based on this information potential druggable targets against SARS-CoV-2 are summarized. Also, the strategies for ongoing and future drug discovery against the SARS-CoV-2 virus are outlined. Given the urgency to find a definitive cure, ongoing drug repurposing efforts being carried out by various organizations are also described. The unprecedented crisis requires extraordinary efforts from the scientific community to effectively address the issue and prevent further loss of human lives and health.
Keywords: Coronavirus, Covid-19, drug discovery, drug repurposing, drug target, SARS-CoV-2
Introduction
The advent of the devastating COVID-19 pandemic in 2019 has left more than 5.5 million people infected and more than 340,000 deaths all over the world (Zhou et al.2020a; https://www.who.int/emergencies/diseases/novel-coronavirus-2019). These numbers demonstrate the large-scale damage this virus has caused on a global scale. COVID-19, as the World Health Organization (WHO) has designated this disease, is caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) (Du Toit 2020) (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it). Since there is no absolute definitive drug or vaccine available that can contain the spread of this deadly virus, the management strategy for the disease is primarily aimed at treating the symptoms (Jin et al.2020). Lack of treatment options has only led to an increased number of fatalities due to the disease (Abbas et al.2020; Down et al.2020).
Academic labs and drug discovery organizations world over are working tirelessly to evaluate compounds that can inhibit the spread of SARS-CoV-2 in humans. To achieve this it is essential to first identify drug targets and subsequently identify and evaluate compounds and biologics that can effectively engage these targets and inhibit the spread (Alexander et al.2020a, b; Dong et al.2020). However, such efforts can be arduous and involve a painstakingly long process. Therefore in parallel, we should also evaluate known antivirals and repurpose them either as single agents or in combinations so that they can effectively contain the spread of the virus (Ahn et al.2020; Hijikata et al.2020; Jeon et al.2020). In order to do this, global concerted efforts are required, and rapid clinical trials need to be conducted to evaluate the role of potential candidate compounds in this particular disease and population setting.
Since the onset of this century, Coronaviruses have created a pandemic-like situation at least at two earlier events. First was the Severe Acute Respiratory Syndrome (SARS) pandemic at the beginning of the 21st century and then the Middle East Respiratory Syndrome (MERS) outbreak almost a decade before (Al-Omari et al.2019; Centers for Disease and Prevention 2013; Holmes 2003). Both these pandemics were caused by viruses belonging to the Coronavirus family of viruses (Benvenuto et al.2020). In fact, the symptoms of SARS and the current COVID-19 pandemic patients are quite similar, reaffirming the fact that both these viruses are quite similar in their genomic constitution and mode of transmission (Kandeel et al.2020; Kumar et al.2020; Xie and Chen 2020a). However, since the SARS epidemic, the SARS-CoV-2 virus has undergone mutations (Becerra-Flores and Cardozo 2020; Biswas et al.2020; Bzowka et al.2020; Poterico and Mestanza 2020; Yin 2020) and thus the drugs developed against the SARS virus might not very effective in containing the virus spread. In order to understand the drug targets and appreciate the ongoing efforts directed towards the identification of therapies against SARS-CoV-2, it is important to understand the virus biology, mode of transmission and replication cycle. This is especially important since any effective therapy against SARS-CoV-2 should preferably target the stages in the virus life cycle.
In this review, the potential drug targets for drug candidates against SARS-CoV-2 are discussed and an overview of the current status of drug development against SARS-CoV-2 infection is provided. The equally important efforts towards vaccine development are excluded from this review, as this topic is covered in a separate in-depth review (Mukherjee 2020).
Classification of coronaviruses
SARS-CoV-2 is a member of the Coronaviruses (CoV) class of viruses. CoV are essentially positive-stranded RNA viruses and display a crown-like appearance on the surface when observed under an electron microscope. It is due to the presence of this ‘crown-like structure’ that this class of viruses is called Coronaviruses (coronam being the Latin term for crown) (Alanagreh et al.2020; Rehman et al.2020). Structurally, these ‘crown-like structures’ are glycoprotein present on the viral envelope which facilitate virus entry in the host cells (Coutard et al.2020). There are four known subfamilies of coronaviruses, classified as Alphacoronavirus, Betacoronavirus, Gammacoronavorus and Deltacoronavirus (Li et al.2020a). Although the zoological evolution of coronaviruses is still an active research subject, it is widely accepted that Alpha- and Betacoronaviruses are predominant in bats and rodents whereas Delta- and Gammacoronavirus gene sources are the avian species (Brussow 2020a; Cascella et al.2020; Coronaviridae Study Group of the International Committee on Taxonomy of 2020).
The causative virus of COVID-19 pandemic, SARS-CoV-2 virus is a part Betacoronavirus subfamily and is believed to have crossed the species barrier to infect humans due to zoonotic transmission (Mackenzie and Smith 2020; Zimmermann and Curtis 2020). The hallmark of coronavirus transcription is the production of multiple sub-genomic RNAs containing sequences corresponding to both genomic ends (Song et al.2020). These viruses utilize RNA-dependent RNA synthesis to generate mRNAs transcribed by the host genome. Genetically, SARS-COV-2 is a positive-sense, single-stranded RNA virus with a genome size of 30 kb which encodes for two Open Reading Frames (ORFs) (Ceraolo and Giorgi 2020; Dabravolski and Kavalionak 2020; Yang et al.2005). These ORFs are designated as 1a and 1b, and code for protease 3ClPro and PLpro. These proteases then cleave the polypeptide into 16 non-structural proteins (Nsp) which are essentially viral enzymes involved in replication and packaging of the virus within the host cell and four structural proteins that contribute to the outer structure of the virus (Jean et al.2020; McKee et al.2020; van Boheemen et al.2012). Like other Coronaviruses, the outer surface of the SARS-CoV-2 virus is made of Spike (S) protein, envelope (E) protein, membrane (M) protein and the Nucleocapsid (N) protein. The M and E proteins are involved in virus morphogenesis and assembly. The Spike protein (S) is at the forefront of infection and interacts with the ACE-2 receptor on the host cell surface thereby promoting virus-cell membrane fusion during initiation of viral infection. The Envelope (E) and Membrane (M) protein constitute the cover outside the viral genetic material. Inside the shell of M and E proteins is the RNA which is guarded by the Nucleocapsid protein (Glebov 2020; Nieto-Torres et al.2011; Nieto-Torres et al.2015).
Molecular basis of disease transmission
It is a well-accepted fact that the SARS-CoV-2 transmission is facilitated by respiratory secretions in the form of droplet/aerosol when a person comes in close contact with the infected person (Guo et al.2020a, b). Recent reports suggest that the infection can also spread through stool, urine, and respiratory secretions (Casanova et al.2010; Ding et al.2020; Wang et al.2020b).
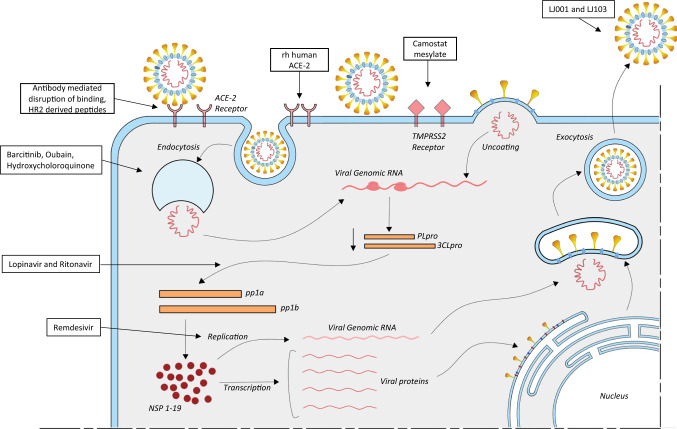
As soon as the SARS-CoV-2 virus enters the human body, it establishes a cycle of replication by binding to cell types such as enterocytes and pneumocytes. This virus can also infect tubular renal epithelial cells (Bao et al.2020), immune cells (Annweiler et al.2020) and cerebral neurons (Bilinska et al.2020; Xie and Chen 2020b). The Spike protein present on the surface of SARS-CoV-2 is responsible for the primary establishment of host-protein interaction. The Spike protein binds to the Angiotensin-converting enzyme-2 (ACE-2) receptor present on the host cell plasma membrane (Annweiler et al.2020; Hasan et al.2020b). Post receptor recognition, the viral genome including the Nucleocapsid is released into the cytoplasm of the host cell as shown in figure 1. As mentioned above, the SARS-CoV-2 viral genome has two ORFs: 1a and 1b. These ORFs translate to two polypeptides (PP) Pp1a and Pp1b which in turn hijack the host cellular ribosomes for their own translational process, thereby making a replication-transcription complex (Bojkova et al.2020; Dong et al.2020).
Figure 1.
Stages of SARS-CoV-2 life cycle. Along with the stages arrows point to drugs and candidate drugs active against the drug targets.
The polypeptide is processed by proteases and this processing results in 16 Non-Structural Proteins (NSPs), and each of these has its own specific function in replication and transcription (Hillen et al.2020; te Velthuis et al.2012). NSP1 and 2 are involved in the suppression of host gene expression. NSP5 is involved in replication whereas NSP4 and 6 are transmembrane proteins (Kandeel et al.2020; Stobart et al.2013; Zhang et al.2020b). NSP7 and 8 act as primases, while NSP9 is an RNA-binding protein. The dimeric form of NSP9 is critical for viral infection, and disruption of this dimerization could be a potential strategy to inhibit the infection. NSP10 is involved in replication and NSP12 is an RNA-dependent RNA polymerase. NSP12 has helicase activity, NSP14 demonstrates exonuclease activity, and NSP15 has endoribonuclease activity, while NSP16 possesses methyltransferase activity (Athmer et al.2017; Hillen et al.2020; Hu et al.2009; Jia et al.2019; Mirza and Froeyen 2020; Neogi et al.2020).
These NSPs with the help of host machinery translate the RNA coding for the viral Spike, Envelope, Nucleocapsid and Membrane proteins. These proteins then enter the endoplasmic reticulum (ER) – golgi apparatus and are involved in viral assembly and packaging. The viral genome binds to the Nucleocapsid (N) protein and results in the formation of the ribonucleoprotein complex (RNP) (Cong et al.2017; Gui et al.2017; Narayanan et al.2008; Narayanan et al.2015).
As depicted in figure 1, Coronaviruses exploit the host’s endosomal pathway to gain entry in the host cell. As such the virus entry into the host cell is an energetically unfavorable process (Brielle et al.2020), but viruses are able to overcome this barrier due to the low pH environment and pH-dependent endosomal cysteine protease (Du et al.2020; Simmons et al.2011; Zhang et al.2019). Other host proteases, such as transmembrane protease serine 2 (TMPRSS2) and TMPRSS11D (also known as airway trypsin-like protease), are involved in the processing of the Spike protein into its constituent subunits S1 and S2 and promote virus entry at the plasma membrane of the host (Iwata-Yoshikawa et al.2019; Maggio and Corsini 2020; Matsuyama et al.2010). Agents that modify the pH or inhibit these proteases can be potential drug targets for anti-coronavirus therapy (Baglivo et al.2020; Bein et al.2020). Finally, the virus particle is assembled, and it again exploits the host’s exosomal pathway and fuses with the plasma membrane resulting in the release of virus particles into the extracellular region. Upon infection, the viral load increases in the host body and this results in an increase in pro-inflammatory cytokines (Alosaimi et al.2020; Conti et al.2020; Magro 2020) and chemokines (Alosaimi et al.2020; Channappanavar and Perlman 2017; Gralinski et al.2018; Skinner et al.2019), which have the potential to damage the lung tissue(Tanaka et al.2013), leading to deterioration of lung function, and finally lung failure (Brussow 2020b).
Approaches for drug discovery targeting SARS-CoV-2
Antiviral drugs targeting the SARS-CoV-2 can be classified into two major classes, with the first group targeting virus–host interactions or inhibiting viral assembly (Zhou et al.2020b). The other approach would include drugs that modulate broad-spectrum host innate immune responses or interfere with signaling pathways involved in viral replication. These drugs may be capable of engaging host receptors or proteases utilized for viral entry or may impact the endocytosis pathway (2020; Channappanavar and Perlman 2017; Dong et al.2020; Hijikata et al.2020; Jeon et al.2020; Liu et al.2020b; McKee et al.2020; Sanders et al.2020).
Essentially, three general approaches can be utilized for screening of antiviral compounds capable of inhibiting the COVID-19 infection:
Repurposing of antiviral compounds
The first approach is to check existing antiviral compounds and molecules and estimate their effect on viral replication and packaging. Molecules like interferon alpha, beta and gamma, ribavirin and chemical inhibitors of cyclophilin 8 (Ma-Lauer et al.2020; Zhang et al.2020a) can be evaluated for their antiviral activities. These known antivirals have a strategic advantage since they are in active clinical use and their pharmacokinetic and pharmacodynamic properties are well studied. On the flip side, such drugs might lack specificity against SARS-CoV-2, and thus may have severe adverse effects (Ahn et al.2020; Busquet et al.2020).
High-throughput screening of compounds
The second approach involves screening of chemical libraries that constitute compounds targeting transcriptional machinery of various cell lines. High-throughput screening technology has the potential to screen large libraries of ‘drug-likely’ chemical compounds for chemical entities having antiviral effects. Even libraries of existing drugs can be screened to support drug repurposing efforts, thereby leading to the identification of new functions of many known drug molecules (Berdigaliyev and Aljofan 2020; de Wilde et al.2014; Dyall et al.2014; Kindrachuk et al.2015; Lu et al.2014). Marketed drugs like Lopinavir/ritonavir which was earlier intended to be used in anti-HIV therapy and was subsequently used to treat SARS have emerged as a result of the successful execution of such screening programs (Chu et al.2004; Cvetkovic and Goa 2003). However, a serious disadvantage of this approach is that the ‘hits’ obtained from such screenings may have immunosuppressive or cytotoxic effects at higher concentrations. Another disadvantage is that the half-maximal effective concentration (EC50) of drugs required to be effective against the SARS-CoV-2 infection might exceed the highest serum concentration (Cmax) levels that can be achieved by pharmacological dosing (Mirza and Froeyen 2020).
Inhibition of SARS-CoV-2 replication mediated by siRNA
The third approach could involve the development of specific novel agents resulting from strong basic research around the genomic and biophysical understanding of the SARS-CoV-2 life cycle. siRNA molecules or inhibitors that have the capability to inhibit specific viral enzymes involved in viral replication cycle, or monoclonal antibodies targeting the host receptor ACE-2 could be the result of such an endeavor (Hasan et al.2020b). Such an approach has the potential to return a large number of virus-specific promising therapies against the SARS-CoV-2 virus. One of the major hurdles in such therapies is the specific drug delivery of these molecules and a lack of understanding of siRNA-based therapy (Lesch et al.2019; Sohrab et al.2018).
Approaches for drug repurposing
Due to the immense financial implications, resource implications and time implications involved in novel drug discovery process, pharmaceutical companies and researchers in the field are inclining towards and relying on ‘Drug Repurposing’ efforts (Ashburn and Thor 2004). As the name suggests, using this approach, a known drug or an investigational drug candidate drug is studied for new uses that are beyond their scope of original intended medical indication. Some researchers and institutions also term ‘Drug repurposing’ as Drug Repositioning, Drug re-profiling or Drug re-tasking depending on the final outcome of studies (Scannell et al.2012).
This strategy can considerably lower the risk of failure of investigational drugs since the toxicity profile of the drug is already well evaluated and in most cases its adverse effects are well documented (Pammolli et al.2011). More importantly, this strategy can help save time involved in Drug development since the preclinical testing, safety assessment and even formulation development has already been completed for repurposed drugs (Nosengo 2016). Also, since the drugs have undergone clinical trials earlier, repurposed drugs can potentially skip phases 1 and 2 trials, and based on therapeutic indication and adverse effect profile, repurposed drugs can be considered directly for large scale phase 3 trials (Breckenridge and Jacob 2019). Another important use of repurposed drugs is in drug combination therapy as the use of drugs can be modulated by effective novel drug combinations (Urquhart 2018). Also, initial drug repurposing experiments do not require elaborate laboratory settings and most often new indications and combinations can be postulated using in-silico approaches (Hurle 2013). At the same time, this exciting approach suffers from some pitfalls and cautious consideration is required before positioning a drug for a new therapeutic indication. Mostly drug repurposing studies are driven by drug targets and identified drug targets might demonstrate polypharmacology thereby leading to adverse side effects (Aguilera et al.2019; Cheng 2019; Karuppasamy et al.2019). Also in experimental screening studies, usually a higher dose is used and this might lead to misidentification of a compound as active while its corresponding pharmacological dose might be toxic. At the same time the effective plasma concentrations of drugs might be higher than the maximum tolerable pharmacological dose. M oreover, a substantial structural modification of a drug might change its toxicity profile thereby warranting fresh toxicity studies (Colson and Raoult 2016; Strittmatter 2014).
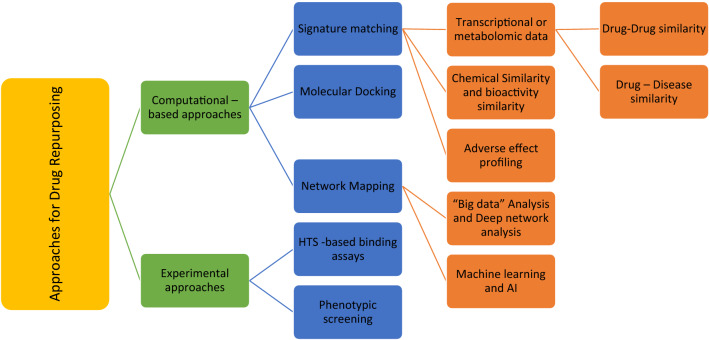
Many pharmaceutical companies are still shy of completely harnessing the potential of drug repurposing due to the attached intellectual property ‘burden’ and the associated costs (Farha and Brown 2019; Fetro and Scherman 2020; Talevi and Bellera 2020; Yildirim et al.2016). Nevertheless, the drug repurposing approaches can be broadly divided into two broad categories: (1) the computational approach, here bioinformatics tools are used to identify new indications for drugs already in use and the approach relies on ‘Big data’ analysis and Artificial Intelligence (AI) (Issa et al.2020; Ke et al.2020; Lee and Chen 2020). (2) The experimental approach, this is a more traditional approach and relies on in vitro experiments to postulate new applications of drugs (Cha et al.2018; Martorana et al.2016). Figure 2 shows a hierarchical view of approaches being used for drug repurposing
Figure 2.
Hierarchical view of approaches for drug repurposing.
Computational approach
Computational approaches for Drug repurposing are largely data-driven and involve a systematic analysis of gene expression, chemical structure, proteomic data or electronic healthcare records. The most commonly used computational approaches include Signature matching (Koudijs et al.2019), computational molecular docking (Pinzi and Rastelli 2019; Trosset and Cave 2019), Genomic association analysis (Cheng et al.2019; Nabirotchkin et al.2020), Pathway or network mapping (Infante et al.2020; Zhou et al.2020b) and retrospective analysis using electronic health records of approved drugs (Karaman and Sippl 2019; Pereira et al.2020; Shi et al.2020b).
‘Signature’ matching
Every drug or investigational drug candidate possesses some unique characteristics or ‘signature’ like its transcriptomic effect profile, structural or adverse effect profile and by matching these characteristics/signatures with another disease or drug, repurposing can be achieved (Karaman and Sippl 2019; Koudijs et al.2019). Using this approach for drug repurposing researchers rely on either drug–disease comparison or drug–drug comparison. In the first case, ‘signatures’ of a particular drug’s gene expression profile before and after treatment is compared with the differential gene expression profile obtained similarly by comparing profiles of healthy with the diseased (Jhamb et al.2019; Khosravi et al.2019). One such drug repurposing example that has taken this approach is the topiramate which is normally used as an antiepileptic drug and acts as an agonist for Gamma amino Butyric acid (GABA) activity (Dudley 2011; Dudley et al.2011). Based on studies by Dudley and co-workers using the drug-disease approach, it was established that topiramate can also be used for inflammatory bowel disease (IBD) as it had signatures comparable to prednisolone, the treatment of choice for IBD (Dudley 2011). The drug–drug similarity approach identifies a common mechanism of action for drugs that belong to different classes and are usually structurally dissimilar (Bakal et al.2019; Wu et al.2019; Yang et al.2019).
Computational molecular docking
Computational molecular docking is an indispensable tool for Drug repurposing activities (Chen et al.2012; Cicaloni et al.2019). Here, by using structure-based computational strategy, binding efficiency is predicted between the drug and the target molecule (Elfiky 2020). With this method large- and small-scale screens can be conducted with known drugs against a disease target (Brindha et al.2016; Kitchen et al.2004). However, this technique has its own limitations, such as, for many targets 3D structure is not elucidated or there is a lack of available screen-able macromolecular database that can provide structural information for a varied molecular class of drug (Xia 2017; Zhou et al.2019).
Network mapping
Molecular Pathway or Network mapping is one of the most commonly used methods for drug repurposing. Many identified drug targets are not directly druggable as their direct inhibition might lead to severe adverse effects and thus network mapping can inform about the upstream or downstream druggable targets thereby enabling drug repurposing (Oulas et al.2019; Zhou et al.2020b). Based on Gene expression pattern and disease pathology, drug and disease networks can be created using network mapping tools (Gns et al.2019; Janardhan et al.2018). Such maps and networks can open enormous possibilities for drug repurposing.
Artificial intelligence and drug repurposing
Advances in Information technology with Artificial Intelligence (AI) and ‘Big-data analysis’ are revolutionizing drug repurposing efforts and studies (Mucke 2018). With the help of machine learning tools, computational algorithms can be developed that can predict new drug target engagement with far greater accuracy than earlier used methods (Alvarez-Machancoses et al.2020; Kuang et al.2019; Luscher Dias et al.2020; Nabirotchkin et al.2020). Huge data generated by High-throughput Next Gen Sequencing (NGS) from numerous patients when combined with disease characteristics and treatment options can lead to the identification of new disease biomarkers and drug targets (Stupnikov et al.2018; Zai et al.2018). AI-driven supervised machine learning algorithms can implement multiomics and multitask learning to facilitate drug response elicited by engagement of multiple drug targets (Nascimento et al.2019; Nath et al.2018; Saberian et al.2019; Zhao and So 2019). The impact of this technology can be appreciated by evaluating a recent study where a computational methodology was developed that can utilize heterogeneous data from previously described Drug Target interactions to predict new interactions with even greater accuracy. The methodology known as ‘deepDTnet’ can integrate networks connecting multiple drugs with drug targets and disease database with the help of deep learning (a form of machine learning based on AI) (Brasil et al.2019; Monteiro et al.2020; Wen et al.2017; Zeng et al.2020).
Drug repurposing in antiviral drug discovery
The approaches for drug repurposing can also be utilized to scout for drugs that can be effective antivirals. By screening the database of small molecules against viral drug targets using computational methods, drugs or molecules can be identified may possess antiviral activity (Pizzorno et al.2019). Essentially three different scenarios can be discussed to facilitate antiviral drug repurposing: (1) Known target/new virus: In this scenario, an established antiviral drug targeting a specific protein/pathway is found to possess antiviral activity against other viruses. Known viral RNA polymerase Favipiravir and sofosbuvir were initially developed for the treatment of Influenza virus and Hepatitis C virus (HCV) infection and were repurposed for treatment of Ebola virus (Bai and Hsu 2019; Du et al.2020; Johansen et al.2015; Muthaiyan et al.2020; Salata et al.2019; Veljkovic et al.2015) and Zika virus infection (Abbasi 2016; Bernatchez et al.2020; Montes-Grajales et al.2020; Mumtaz et al.2017). (2) Known target/new indication: In this scenario, the pharmacological target is implicated to be affected in a new pathogenic infection. In such cases, drugs targeting these proteins can be repurposed as effective antiviral agents. One such example is about the repurposing of anti-cancer agent Imatinib. Cellular Abelson (ABL) kinase is the target of Imatinib and the same was shown to be active against coronaviruses (Ananthula et al.2018; Coleman et al.2016; Giuliani et al.2018). (3) New target/new indication: This scenario occurs when an approved drug with a specific target is found to target additional viral proteins or targets. Many antimicrobial agents like teicoplanin (Aziz et al.2018; Zhou et al.2016), ivermectin (Chaccour and Rabinovich 2019; Rabinovich 2018), itraconazole (Alhakamy and Md 2019; Pace et al.2016; Schloer et al.2019; Tsubamoto et al.2017) and nitazoxanide (Cao et al.2017; Rossignol 2016) were also found to be active against some viral infections.
Lessons from SARS: pharmacological interventions
Lessons from SARS and MERS epidemic can be used to develop some therapies for SARS-CoV-2 infection (Zumla et al.2016). Previously used antiviral drugs like oseltamivir, peramivir, zanamivir, ganciclovir, acyclovir and ribavirin are not recommended for COVI-19 treatment (Li et al.2020c; Wang et al.2020b). Also, systemic corticosteroid treatment such as methylprednisolone is not appreciated as a treatment option for SARS-CoV-2 infected patients (Veronese et al.2020). In such a scenario, given the similarity of SARS-CoV and MERS virus along with the SARS-CoV-2 virus, an insight into the treatment options available for SARS and MERS could provide valuable inspirations for Drug discovery and repurposing (Zumla et al.2016). However, for both SARS and MERS, no specific treatment was recommended; rather WHO recommended managing the disease based on patients’ symptoms (Graham et al.2013).
Ribavirin and corticosteroids
In the year 2003, the SARS pandemic, like COVID-19, caught the world by surprise. Early days in the SARS pandemic patients in Hong Kong and Canada were treated with a combination of ribavirin and corticosteroids (Rainer 2004; Tsang et al.2003). However, subsequent reports indicated towards ribavirin’s high rate of toxicity and lack of ability to control the infection spread (Booth and Stewart 2005; Sung et al.2004; Wong et al.2003). Use of corticosteroid, methylprednisolone is controversial as far as SARS was concerned. In multiple cases, dose-related toxicity was observed. The lower dose of methylprednisolone, 250–500mg/day demonstrated some improvement on a subset of critical SARS patients however a prolonged usage in the absence of any specific antimicrobial agent predisposed the patients to disseminated fungal infection (Chan et al.2003a, b; Tsang and Seto 2004; Tsang and Lam 2003) and avascular necrosis (Chan et al.2003a, b). Based on localized, single-center uncontrolled clinical studies, it was recommended that Corticosteroids should only be used as a ‘rescue therapy’ and not as a treatment as it might impair the host viral clearance (Sung et al.2004).
Interferons
Viral infections such as hepatitis B and C have been successfully treated by Interferon treatment (Christian et al.2004). Since Interferon is also broad-spectrum antiviral, it was used in some single-center, open-labelled, uncontrolled clinical settings against SARS. In these small trials, patients were stable post-treatment with Interferon in combination with corticosteroids. The results pointed out that perhaps with interferon treatment the lung deterioration can be delayed (although this could not be proven statistically) (Zhao et al.2003). Also, post this treatment, improved oxygen saturation levels and lower creatine phosphokinase levels were observed in critically ill patients. However, this warranted properly designed clinical trials globally (Loutfy et al.2003).
Ritonavir and lopinavir
A combination of viral protease inhibitor, ritonavir 400 mg and lopinavir 100 mg, when administered orally with 12 h interval for 10 to 14 days as standard therapy, yielded the most promising outcome in Hong Kong–based clinical studies (Chan et al.2003a, b). Here the subjects showed a reduction in steroid dependency and nosocomial infections. At the same time reduction in viral load and concomitant increase in peripheral lymphocyte count was observed. This combination appears to be promising and even for COVID-19 as well this combination is being tested in clinical trials. However, some serious adverse effects were also reported from patients undergoing treatment for SARS infection with this combination. Subjects were observed to suffer from pancreatitis, diarrhea, abdominal pain and liver dysfunction among other associated discomforts like abdominal pain, asthenia, headache, nausea, insomnia and skin rash (Chu et al.2004).
Key CoV targets for drug development and available therapies
As of date, no specific and definite antiviral drug is available for the treatment of CoV-associated pathologies (Barlow et al.2020; Martinez 2020a; McKee et al.2020; Wu et al.2020a). However, some therapeutic agents based on the biology of the virus and some potential drug targets have been identified.
Since the onset of previous global coronavirus pandemics like MERS and SARS, considerable research has gone into the search for suitable drug targets and subsequent drug candidates (Lou et al.2020). Based on this and life cycle stages of SARS-CoV virus, the therapies that have the potential to act on coronaviruses can be divided into five broad categories/approaches:
Inhibition of virus binding to the host receptor by either chemical compounds or monoclonal antibodies. These agents can block or effectively engage the host’s cell surface receptor thereby preventing virus binding and subsequent internalization (Li et al.2019; Salata et al.2019; Shang et al.2020; Tortorici and Veesler 2019).
Target the viral endocytosis. This process enables the virus to enter the host cell and release its genetic material for further replication and therefore blocking virus-mediated endocytosis is a logical target for antiviral therapy (Baglivo et al.2020; Delvecchio et al.2016; Glebov 2020; Omotade and Roy 2019; Praveen et al.2020).
Neutralize the virus particle. This can be accomplished by the compounds and antibodies acting on enzymes or functional proteins critical to virus replication and multiplication (Algaissi and Hashem 2020; Goo et al.2020; Pinto et al.2020; Wang et al.2020a; Wu et al.2020b).
Targeting the viral structural proteins like the membrane, envelope and Nucleocapsid protein thereby blocking virus repackaging (Hijikata et al.2020; Kato et al.2019; Mirza and Froeyen 2020; Saha et al.2020; Zhang et al.2020b).
Restoration of host’s innate immunity by the agents capable of producing virulent factors (Azkur et al.2020; Casanova et al.2020; Encinar and Menendez 2020; Gemmati et al.2020; Mantlo et al.2020; Tufan et al.2020).
Table 1 summarizes the drug targets and compounds active against SARS-CoV-2, their current development status and the pros and cons of the proposed therapy.
Table 1.
List of probable drug targets against SARS-CoV-2 and compounds/agents effective against these targets
| Targeted viral components | Examples | Mechanismofaction | Status | Pros | Cons |
|---|---|---|---|---|---|
| Inhibition of SARS -CoV-2 fusion/entry | |||||
| RBD of the S1 subunit of S | REGN3051 and REGN3048 mAbs | Antibodies target the RBD domain of the S1 subunit | Preclinical | Efficacy demonstrated in vitro | Narrow spectrum |
| S2 subunit of S | HR2P and P1 peptides | Antiviral peptides that inhibit fusion of S with host cell receptor | Preclinical | anti-HIV peptidel has been marketed | Narrow spectrum |
| TMPRSS2 | Camostat Mesylate | TMPRSS2 inhibitor that blocks the TMPRSS2-entry pathway | Marketed | Promising results in vitro. Effect on patients need to be tested | Broad spectrum. Developed for therapy against SARS |
| Inhibition of endocytosis | |||||
| Endosomal acidification | chloroquine | An antimalarial that sequesters protons in lysosomes to increase the intracellular pH | Marketed | Broad spectrum; many SARS-CoV-2 affected patients show good recovery | No concrete clinical data to suggest efficacy |
| Clathrin-mediated endocytosis | Oubain | ATP1A1- binding steroids; inhibits clathrin-mediated endocytosis | Marketed | Active against MERS-CoV | May have risk of cardiac toxicity |
| Inhibition of Viral Enzymes | |||||
| 3CLpro | Lopinavir | Inhibits 3CLpro activity | Marketed | Broad spectrum |
Toxicity Adverse impact on immune system |
| PLpro | GRL0617 | Inhibits PLpro activity | Preclinical | Narrow spectrum | No animal or clinical data available |
| RdRp | Remdesivir | Nucleotide analogue; Broad spectrum: many viral infections, inhibits viral RNA synthesis | Marketed | Active against SARS-CoV and MERS-CoV at high doses in vitro | Side effects are common and may be severe with high dose regimens |
| Inhibition of Viral envelope (E), membrane (M), Nucleocapsid (N) and accessory proteins | |||||
| E and M Protein | siRNA | Short chains of dsRNA that interfere with the expression of SARS-CoV proteins | Preclinical | Promising in vitro studies. | Optimal delivery method in humans uncertain |
| N Protein | Pj34 | Impairs viral replication | Preclinical |
Narrow spectrum Effective in vitro and in animal studies |
Optimal delivery method in humans is uncertain |
| Membrane and Accessory proteins | Lj001 and JL103 | Induces membrane damage | Preclinical | Broad spectrum |
Anti-CoV activity yet to be demonstrated Unstable physiologically and photo dependent |
Also, the present development stage as well as the pros and cons of therapies are listed
Inhibition of SARS -CoV-2 fusion/entry
SARS-CoV-2 utilizes the spike protein present on the viral surface to gain entry into the host cells. The protein–protein interaction that takes place between the subunits of the spike protein and the active site of the ACE-2 receptor can be targeted to identify an effective treatment strategy (Wrapp 2020). Like other viruses, the coronaviruses also outsmart drugs targeted against them by constantly mutating the active site of spike protein (Becerra-Flores and Cardozo 2020; Chang et al.2020; Goo et al.2020; Qing et al.2020). As a result, the SARS-CoV-2 spike protein recognizes the ACE-2 receptor more efficiently than the previously studied SARS virus (Albini et al.2020; Chen et al.2020; Dediego et al.2008; Hasan et al.2020a; Mathewson et al.2008). Specifically, the Receptor Binding Domain (RBD) of the spike protein is a critical target for antibody-mediated disruption of binding. Many antibodies demonstrating the ability to disrupt this binding are in preclinical stages of development (Chen et al.2020; Tai et al.2020; Tian et al.2020). Another strategy is to engage and overwhelm the ACE-2 receptor with recombinant human ACE-2 which is normally present on the cell surface. So delivering an excess of the soluble ACE2 helps to neutralize the virus, by competitively binding to SARS-CoV-2 (rhACE2; GSK2586881) (Ameratunga et al.2020; Basit et al.2020). An open-labeled, randomized, controlled pilot clinical trial is in progress to evaluate this approach (NCT04287686).
Apart from the ACE-2 receptor, cellular serine protease TMPRSS2 also plays an important role in facilitating the entry of the virus in host cells (Hoffmann 2020). A clinically proven chemical inhibitor of TMPRSS2, Camostat Mesylate is also able to significantly reduce infection in cell lines of human lung origin (McKee et al.2020; Rahman et al.2020; Shirato et al.2013; Sternberg et al.2020). In addition to this, the heptad repeat 1 (HR1) and heptad repeat 2 (HR2) present on the SARS-CoV-2 have also been implicated in the facilitation of cell membrane fusion. HR2–derived peptides exhibit effective fusion inhibitory activity (Bosch et al.2008; Huang et al.2019; Wang et al.2019; Xia et al.2019a; Xia et al.2019b).
Inhibition of endocytosis
It is known that post fusion of the spike protein with ACE-2 receptor, the virus is ingested in the cells in a pH and receptor-dependent endocytosis (Chu et al.2006; Glebov 2020). Targeting endocytosis can be another potential strategy towards developing potential drug candidates against SARS-CoV-2. Clathrin-mediated endocytosis is regulated by AP-2- associated protein kinase 1 (AAK1) (Uitdehaag et al.2019). Based on library screening, Janus kinase inhibitor Baricitinib was identified as a possible candidate drug for SARS-CoV-2 (Baglivo et al.2020; Cantini et al.2020; Lo Caputo et al.2020). Also, Oubain, another inhibitor of clathrin-mediated, is being tested for its efficacy in drug trials for SARS-CoV-2 positive patients (Sisk et al.2018). Recently, Chloroquine and its derivative hydroxychloroquine have garnered great interest as a therapy against SARS-CoV-2 infection (Alexander et al.2020a; Alia and Grant-Kels 2020; Arnold and Buckner 2020; Ballout et al.2020; Costanzo et al.2020). Several clinical trials are underway to assess the contribution of chloroquine therapy in inhibiting SARS-C0V-2 viral progression (Keshtkar-Jahromi and Bavari 2020). Also, it was shown in vitro, that a derivative of Chloroquine, hydroxychloroquine (EC50 = 0.72µM) is far more potent in inhibiting SARS-CoV-2 infection than Chloroquine (EC50 = 5.47 µM) (Savarino et al.2003; Yao et al.2020). Although the exact molecular mechanism of hydroxychloroquine in the treatment of COVID-19 remains elusive, it is believed that hydroxychloroquine may impair endosome-mediated viral entry or the late stages of viral replication (Devaux et al.2020).
Inhibition of viral enzymes
As a result of aggressive antiviral drug development and discovery programs undertaken in the past, multiple drugs have been developed against viral proteases, polymerases and helicases (Martinez 2020b). Drugs developed against other viral diseases such as Remdesivir (Wang 2020), Flavipiravir (Li et al.2020b), Lopinavir/Ritonavir (Beck et al.2020; Costanzo et al.2020; Gyebi et al.2020; Schoergenhofer et al.2020) are presently being evaluated in clinical trials for their efficacy in containing the COVID-19 pandemic. Remdesivir, an antiviral drug developed against Ebola, is an adenosine analog which inserts into viral RNA chains by RNA-dependent RNA polymerases (RdRps) and results in premature transcription termination (Cao et al.2020; Gordon et al.2020). Similarly, Favipiravir and Ribaviorin are guanine analogs and are approved for some viral diseases (Costanzo et al.2020; Fan et al.2020). EIDD-2801 is another oral antiviral drug that acts as a nucleotide analog, like Remdisivir, albeit with lower EC50, and is orally administrable (Agostini et al.2019). Other antiviral compounds Lopinavir and Ritonavir are protease inhibitors that target 3C–like protease (3CLpro) of SARS-CoV-2 (Bhatnagar et al.2020). The main coronavirus protease, 3CLpro is responsible for processing the polypeptide to NSPs. Using high-throughput screening for compounds against 3CLpro, four molecules, viz. Prulifloxacin, Tegobuvir, Bictegravirand and Nelfinavir, were identified (Barnard et al.2006; De Clercq 2006; Liu et al.2020b).
Inhibition of viral envelope, membrane, nucleocapsid and accessory proteins
SARS-CoV-2 envelope (E), membrane (M) and Nucleocapsid (N) protein are critical for virus survival and propagation, and therefore such structural proteins are the best drug targets. Since these viral proteins are structurally very different from the host proteins, drug targeting these proteins will have minimal adverse effects. Apart from protecting the viral genome, these structural proteins are also involved in suppressing the host immune system, thereby providing the virus a strategic advantage over the host (Bojkova et al.2020; Borgio et al.2020; Cherian et al.2020). The N protein acts to suppress RNA silencing and suppresses RNA interference-mediated by siRNA. Therefore many siRNA based therapeutics target viral E, M and N protein translation and inhibit viral replication, at least in vitro (Nur et al.2015; Sohrab et al.2018; Song et al.2019). However, siRNA-based therapies are still not available for human use due to inherent stability issues and the unavailability of reliable delivery methods (De Clercq 2006).
The E protein also serves as an ion channel and this action is inhibited by hexamethylene amiloride (Pervushin et al.2009). Another chemical inhibitor PJ34 targets the unique ribonucleotide-binding pocket at the N-terminal domain of N protein (Lin et al.2014). It is important to note that most of these inhibitors were designed against the SARS virus; due to the mutations in the SARS-CoV-2 virus, such inhibitors may not be as effective in fighting against the ongoing COVID-19 pandemic.
LJ001 and LJ003 are broad-spectrum antiviral compounds that not only inhibit viral entry in the host cells but also damage the viral membrane by producing singlet oxygen molecules. Unfortunately, LJ001 is physiologically unstable and is photo-dependent (Barlow et al.2020). Nevertheless, LJ001 defines a new class of antiviral compounds, and further research into this class of compounds will yield encouraging results.
Suppression of excessive inflammatory response
A well-orchestrated cytokine response is critical for the host immune response. It has been reported that some SARS-CoV-2 infected patients demonstrate a hyper-inflammatory response, possibly due to deregulated cytokine response. It was reported that COVID-19 patients in the ICU had high cytokines in plasma when compared with non-ICU patients, suggesting that cytokine dysregulation is involved in the severe form of COVID-19 disease (Channappanavar and Perlman 2020; Liu et al.2020c). Additionally, SARS-CoV-2-infected patients admitted in ICU display increased levels of GM-CSF and IL6+CD4+T cells when compared to ICU naïve patients (Boettcher et al.2007). The above facts point to the possibility that inhibition of excessive inflammatory response might reduce the severity of COVID-19 disease. Corticosteroids are known to have excellent pharmacological potential in suppressing systemic inflammation (Liu et al.2020a; Zha et al.2020). However, their use in COVID-19 patients is still debatable and requires a detailed study.
It has been demonstrated that after the onset of SARS-CoV-2 infection, CD4+T Cells are activated to produce GM-CSF and other inflammatory cytokines, thus resulting in further induction of CD14+CD16+ monocytes with high expression of interleukin 6 (IL-6) (Conti et al.2020; Liu et al.2020a). This observation leads to the possibility that by blocking the IL-6 receptor we could potentially reduce immune stress caused by SARS-CoV-2. In line with this observation, a multicenter, randomized, controlled clinical trial is currently underway using an IL-6 receptor-specific antibody Tocilizumab (Li et al.2020d) (NCT04315480).
Another recent advance in COVID -19 treatments is the Convalescent plasma treatment. With infection rates growing and no specific therapy available, therapy with convalescent plasma (CP) has been proposed as a principal treatment. In this therapy, the plasma obtained from a donor recovered from the disease is used to develop humoral immunity against SARS-CoV-2-infected patients. The plasma from the donor patient acts as a source of human antibodies against the infection (Jawhara 2020; Shi et al.2020a, b). However, large scale human trials need to be conducted to better understand and evaluate CP as a method of treatment for COVID-19.
Current efforts from top pharmaceutical companies
Many pharmaceutical giants have now jumped into the race to find a drug for SARS-CoV-2 infected patients. Given the high number of fatalities across the globe, even regulatory authorities are giving rapid approvals to conduct clinical trials for promising candidate drugs. Table 2 shows a list of companies that are conducting clinical trials or are seeking approval from regulatory authorities to conduct trials. Since the onset of the SARS epidemic, much knowledge was generated about drug targets and some candidate molecules were developed as well (Kumar et al.2017). However, these molecules could not be taken to clinical trials since there were not enough patients suffering from SARS virus by the time these drugs were developed. Nevertheless, this information is of tremendous use since SARS–CoV and SARS-CoV-2 share striking similarities in the genome, replication cycle and even symptoms experienced by patients (Su and Lai 2020). Systemic genomic comparisons have revealed a striking 79% similarity at the nucleotide level between SARS-CoV-2 and SARS-CoV. However, only 72% nucleotide similarity was observed in the spike (S) protein of both the viruses (Zhang and Holmes 2020). At the biochemical level both the virus display preferential binding to the ACE-2 receptor. Even at the clinical level, the chest X-rays of patients infected with either SARS-CoV or SARS-CoV-2 display multilobar ground glass like opacities. Similarly the CT scan of patients infected with either virus display lobar consolidations (Ceccarelli et al.2020).
Table 2.
List of companies actively involved in finding a drug to treat SARS-CoV-2 patients
| Company/organization | Candidate drug | Development phase | Current status and plans | Timelines |
|---|---|---|---|---|
| Gilead | Remdesivir | III | Remdesivir is now being tested in five Covid-19 clinical trials that have been set up at lightning speed | CT is anticipated to be completed by end of April 2020 |
| Sanofi | Hydroxychloroquine | Preclinical | Conduct additional CTs and supply millions of doses of an existing anti-malaria product | N.A. |
| Abbvie | Lopinavir/ritonavir combination | III | Collaboration ongoing with select health authorities and institutions globally to determine antiviral activity against SARS-CoV-2 | N.A. |
| Regeneron | Monoclonal antibody therapy | Preclinical | Aiming to select the top 2 antibodies for a cocktail therapy, which can either be administered to at-risk -vaccine naïve population or as treatment for those already infected | Potential to enter human CT by early summer 2020 |
| Ascletis | Combination of two antivirals | I | The Chinese company is testing a combination of antivirals, developed against HIV and the other approved for hepatitis C | N.A. |
| Takeda | Polyclonal antibody therapy | Preclinical | Collaboration with several health and regulatory agencies and health care partners across the globe on polyclonal antibody TAK-888 | Program initiated in March 2020 |
| Lilly | Antibody drug | Preclinical |
Eli Lilly developing antibody treatments for coronavirus infection. Using a blood sample from a coronavirus survivor Partner company AbCellera identified more than 500 antibodies that might protect against the virus |
CTs in humans to be started in the next four months of 2020 |
The information was collected from the company web sites or press release. CT Clinical Trial, N.A. status not known
However, since the SARS epidemic, the virus has mutated considerably, and as a result, it now has the Spike protein which is quite different from the previous version. This fact makes efforts to find drugs that inhibit virus entry to host cells quite difficult.
Companies are also relying on drug repurposing (Beck et al.2020; Wang et al.2020), like chloroquine from Sanofi is planned to be tested in Coronavirus patients world over. Meanwhile, other companies like Abbvie, Gilead and Regeneron are testing known antiviral in combinations in patients affected with the SARS-CoV-2 virus (https://www.abbvie.com/coronavirus.html). This approach provides a strategic advantage to the ongoing efforts, as the pharmacological effect of single drugs is well-studied on the human body. Also, we know the exact mechanism of action and dose regimen for these antivirals. Now using them in combination to inhibit more than 2 drug targets can be a winning combination (Borges do Nascimento et al.2020). Globally, scientists and health authorities are eagerly waiting to interpret the results obtained from these trials. In the meantime, companies like Takeda and Lilly are working on antibody therapy, and evaluating the efficacy of therapeutic molecules at breakneck speed. Under the aegis of WHO, ‘SOLIDARITY’ clinical trial is underway to evaluate the treatment opportunities available for COVID-19 patients. The trial is being conducted simultaneously in 30 countries and under this trial, four treatment options are being compared against the standard care options for SARS-CoV-2-infected patients. The trial will test the efficacies of Remdesivir and Chloroquine or Hydroxychloroquine as single agents, Lopinavir in combination with Ritonavir, and Lopinavir with Ritonavir plus Interferon beta-1a (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments).
Conclusion
Unfortunately, at the time of writing this article there are more than 22,00,000 confirmed patients suffering from SARS-CoV-2 with more than 152,000 reported deaths across the globe.
Novel infectious diseases resulting from the zoonotic transmission of ever-evolving and mutating coronaviruses will continue to pose a global threat to people’s lives and the global economy. Today, despite having suffered from two major coronavirus-related outbreaks like SARS and MERS, the world remains underprepared to formidably accept the challenge of a global pandemic like COVID-19. We are still clueless about the handling of the disease and far away from a definite line of treatment against the SARS-CoV-2 virus. It is extremely critical that a concerted effort across the globe is undertaken to find a robust cure for coronaviruses-related illness. Given the arduous and cost-sensitive road map for novel drug development, it is of utmost importance to develop broad-spectrum antivirals that act on common features of the coronavirus lifecycle. Drug repurposing should be broadened, and more combination drugs should be evaluated in patient trials to allow inhibition of the disease through more than one target. Although there is a lot to learn from the SARS and MERS epidemic, and a lot of research has been performed to develop a suitable cure, the same cannot be applied to COVID -19 due to viral evolution. Across the globe, serious efforts are ongoing to find compounds and drugs that can decrease COVID-19 progression and it is likely that all such prospects have not been covered in this review. WHO in collaboration with Microsoft maintains an active database of ongoing trials and compounds active against SARS-CoV-2 (https://www.who.int/ictrp/search/en/).
Extraordinary collaborations and technology exchange in the area of antiviral drug discovery and clinical trials will expedite patient access to more reliable drugs with improved therapeutic potential. This will also considerably reduce time-to-market for candidate drugs. The current ongoing research should lead to more collective drug discovery efforts, where partnership between research institutes and industries will be of paramount importance.
Acknowledgements
The author would like to acknowledge Dr Vasudharani Devanathan, Asst. Professor, IISER Tirupati, and Dr Harshini Chakravarthi, IISER Tirupati, for critical reading, and Mr Gowthaman Suresh for the figures.
Footnotes
This article is part of the Topical Collection: COVID-19: Disease Biology & Intervention
Corresponding editor: BJ Rao
References
- 2020 Coronavirus: Everyone wins when patents are pooled. Nature581 240 [DOI] [PubMed]
- Abbas AR, Abbas A, Ali Y, Memon SF. Important considerations regarding the future management of coronavirus (COVID-19) Int. J. Surg. 2020;79:6–7. doi: 10.1016/j.ijsu.2020.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi J. Repurposing Drugs to Treat Zika. JAMA. 2016;316:1636. doi: 10.1001/jama.2016.14757. [DOI] [PubMed] [Google Scholar]
- Agostini ML, Pruijssers AJ, Chappell JD, Gribble J, Lu X, et al. Small-Molecule Antiviral beta-d-N (4)-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019 doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera E, Alvarez G, Cerecetto H, Gonzalez M. Polypharmacology in the Treatment of Chagas Disease. Curr Med Chem. 2019;26:4476–4489. doi: 10.2174/0929867325666180410101728. [DOI] [PubMed] [Google Scholar]
- Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, et al. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19) J Microbiol Biotechnol. 2020;30:313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Omari A, Rabaan AA, Salih S, Al-Tawfiq JA, Memish ZA. MERS coronavirus outbreak: Implications for emerging viral infections. Diagn Microbiol Infect Dis. 2019;93:265–285. doi: 10.1016/j.diagmicrobio.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanagreh L, Alzoughool F, Atoum M. The Human Coronavirus Disease COVID-19: Its Origin, Characteristics, and Insights into Potential Drugs and Its Mechanisms. Pathogens. 2020 doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini A, Di Guardo G, Noonan DM, Lombardo M. The SARS-CoV-2 receptor, ACE-2, is expressed on many different cell types: implications for ACE-inhibitor- and angiotensin II receptor blocker-based cardiovascular therapies. Intern Emerg Med. 2020 doi: 10.1007/s11739-020-02364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander PE, Debono VB, Mammen MJ, Iorio A, Aryal K, et al. COVID-19 coronavirus research has overall low methodological quality thus far: case in point for chloroquine/hydroxychloroquine. J Clin Epidemiol. 2020 doi: 10.1016/j.jclinepi.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Armstrong J, Davenport AP, Davies J, Faccenda E et al. 2020 A rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development. IUPHAR Review 29. Br. J. Pharmacol.10.1111/bph.15094 [DOI] [PMC free article] [PubMed]
- Algaissi A, Hashem AM. Evaluation of MERS-CoV Neutralizing Antibodies in Sera Using Live Virus Microneutralization Assay. Methods Mol Biol. 2020;2099:107–116. doi: 10.1007/978-1-0716-0211-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhakamy NA, Md S. Repurposing Itraconazole Loaded PLGA Nanoparticles for Improved Antitumor Efficacy in Non-Small Cell Lung Cancers. Pharmaceutics. 2019 doi: 10.3390/pharmaceutics11120685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alia E and Grant-Kels JM 2020 Does hydroxychloroquine combat COVID-19? A timeline of evidence. J. Am. Acad. Dermatol.10.1016/j.jaad.2020.04.031 [DOI] [PMC free article] [PubMed]
- Alosaimi B, Hamed ME, Naeem A, Alsharef AA, AlQahtani SY, et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine. 2020;126:154895. doi: 10.1016/j.cyto.2019.154895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Machancoses O, DeAndres Galiana EJ, Cernea A, Fernandez de la Vina J, Fernandez-Martinez JL. On the Role of Artificial Intelligence in Genomics to Enhance Precision Medicine. Pharmgenomics Pers Med. 2020;13:105–119. doi: 10.2147/PGPM.S205082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameratunga R, Lehnert K, Leung E, Comoletti D, Snell R, et al. Inhaled modified angiotensin converting enzyme 2 (ACE2) as a decoy to mitigate SARS-CoV-2 infection. N Z Med J. 2020;133:112–118. [PubMed] [Google Scholar]
- Ananthula HK, Parker S, Touchette E, Buller RM, Patel G, et al. Preclinical pharmacokinetic evaluation to facilitate repurposing of tyrosine kinase inhibitors nilotinib and imatinib as antiviral agents. BMC Pharmacol Toxicol. 2018;19:80. doi: 10.1186/s40360-018-0270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annweiler C, Cao Z, Wu Y, Faucon E, Mouhat S, et al. Counter-regulatory ‘Renin-Angiotensin’ System-based Candidate Drugs to Treat COVID-19 Diseases in SARS-CoV-2-infected patients. Infect Disord Drug Targets. 2020 doi: 10.2174/1871526520666200518073329. [DOI] [PubMed] [Google Scholar]
- Arnold SLM and Buckner F 2020 Hydroxychloroquine for treatment of SARS-CoV-2 Infection? Improving our confidence in a model-based approach to dose selection. Clin. Transl. Sci.10.1111/cts.12797 [DOI] [PMC free article] [PubMed]
- Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Athmer J, Fehr AR, Grunewald M, Smith EC, Denison MR, et al. 2017 In situ tagged nsp15 reveals interactions with coronavirus replication/transcription complex-associated proteins. mBio810.1128/mbio.02320-16 [DOI] [PMC free article] [PubMed]
- Aziz DB, Teo JWP, Dartois V, Dick T. Teicoplanin - Tigecycline Combination Shows Synergy Against Mycobacterium abscessus. Front Microbiol. 2018;9:932. doi: 10.3389/fmicb.2018.00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020 doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglivo M, Baronio M, Natalini G, Beccari T, Chiurazzi P et al. 2020 Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: a possible strategy for reducing SARS-COV-2 infectivity? Acta Biomed.91 161–164 [DOI] [PMC free article] [PubMed]
- Bai JPF, Hsu CW. Drug Repurposing for Ebola Virus Disease: Principles of Consideration and the Animal Rule. J Pharm Sci. 2019;108:798–806. doi: 10.1016/j.xphs.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Bakal G, Kilicoglu H, Kavuluru R. Non-Negative Matrix Factorization for Drug Repositioning: Experiments with the repoDB Dataset. AMIA Annu Symp Proc. 2019;2019:238–247. [PMC free article] [PubMed] [Google Scholar]
- Ballout RA, Sviridov D, Bukrinsky MI and Remaley AT 2020 The lysosome: A potential juncture between SARS-CoV-2 infectivity and Niemann-Pick disease type C, with therapeutic implications. FASEB J. [DOI] [PMC free article] [PubMed]
- Bao L, Deng W, Huang B, Gao H, Liu J, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Barlow A, Landolf KM, Barlow B, Yeung SYA, Heavner JJ, et al. Review of Emerging Pharmacotherapy for the Treatment of Coronavirus Disease 2019. Pharmacotherapy. 2020 doi: 10.1002/phar.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DL, Day CW, Bailey K, Heiner M, Montgomery R, et al. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir Chem Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- Basit A, Ali T, Rehman SU. Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1768150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Flores M, Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract. 2020 doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bein B, Bachmann M, Huggett S, Wegermann P. SARS-CoV-2/COVID-19: Evidence-Based Recommendations on Diagnosis and Therapy. Geburtshilfe Frauenheilkd. 2020;80:491–498. doi: 10.1055/a-1156-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D, Giovanetti M, Ciccozzi A, Spoto S, Angeletti S, et al. The 2019-new coronavirus epidemic: Evidence for virus evolution. J Med Virol. 2020;92:455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdigaliyev N, Aljofan M. An overview of drug discovery and development. Future Med Chem. 2020;12:939–947. doi: 10.4155/fmc-2019-0307. [DOI] [PubMed] [Google Scholar]
- Bernatchez JA, Tran LT, Li J, Luan Y, Siqueira-Neto JL, et al. Drugs for the Treatment of Zika Virus Infection. J Med Chem. 2020;63:470–489. doi: 10.1021/acs.jmedchem.9b00775. [DOI] [PubMed] [Google Scholar]
- Bhatnagar T, Murhekar MV, Soneja M, Gupta N, Giri S, et al. Lopinavir/ritonavir combination therapy amongst symptomatic coronavirus disease 2019 patients in India: Protocol for restricted public health emergency use. Indian J Med Res. 2020 doi: 10.4103/ijmr.IJMR_502_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilinska K, Jakubowska P, Von Bartheld CS, Butowt R. Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age. ACS Chem Neurosci. 2020 doi: 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A, Bhattacharjee U, Chakrabarti AK, Tewari DN, Banu H, et al. Emergence of Novel Coronavirus and COVID-19: whether to stay or die out? Crit Rev Microbiol. 2020 doi: 10.1080/1040841X.2020.1739001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher IC, Steinberg T, Matiasek K, Greene CE, Hartmann K, et al. Use of anti-coronavirus antibody testing of cerebrospinal fluid for diagnosis of feline infectious peritonitis involving the central nervous system in cats. J Am Vet Med Assoc. 2007;230:199–205. doi: 10.2460/javma.230.2.199. [DOI] [PubMed] [Google Scholar]
- Bojkova D, Klann K, Koch B, Widera M, Krause D, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020 doi: 10.1038/s41586-020-2332-7. [DOI] [PubMed] [Google Scholar]
- Booth CM, Stewart TE. Severe acute respiratory syndrome and critical care medicine: the Toronto experience. Crit Care Med. 2005;33:S53–S60. doi: 10.1097/01.ccm.0000150954.88817.6. [DOI] [PubMed] [Google Scholar]
- Borges do Nascimento IJ, Cacic N, Abdulazeem HM, von Groote TC, Jayarajah U et al. 2020 Novel coronavirus infection (COVID-19) in humans: a scoping review and meta-analysis. J. Clin. Med.910.3390/jcm9040941 [DOI] [PMC free article] [PubMed]
- Borgio JF, Alsuwat HS, Al Otaibi WM, Ibrahim AM, Almandil NB, et al. State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2. Arch Med Sci. 2020;16:508–518. doi: 10.5114/aoms.2020.94567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch BJ, Rossen JW, Bartelink W, Zuurveen SJ, de Haan CA, et al. Coronavirus escape from heptad repeat 2 (HR2)-derived peptide entry inhibition as a result of mutations in the HR1 domain of the spike fusion protein. J Virol. 2008;82:2580–2585. doi: 10.1128/JVI.02287-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil S, Pascoal C, Francisco R, Dos Reis Ferreira V, Videira PA, et al. Artificial Intelligence (AI) in Rare Diseases: Is the Future Brighter? Genes (Basel) 2019 doi: 10.3390/genes10120978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge A, Jacob R. Overcoming the legal and regulatory barriers to drug repurposing. Nat Rev Drug Discov. 2019;18:1–2. doi: 10.1038/nrd.2018.92. [DOI] [PubMed] [Google Scholar]
- Brielle ES, Schneidman-Duhovny D, Linial M. The SARS-CoV-2 Exerts a Distinctive Strategy for Interacting with the ACE2 Human Receptor. Viruses. 2020 doi: 10.3390/v12050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindha S, Sundaramurthi JC, Velmurugan D, Vincent S, Gnanadoss JJ. Docking-based virtual screening of known drugs against murE of Mycobacterium tuberculosis towards repurposing for TB. Bioinformation. 2016;12:359–367. doi: 10.6026/97320630012368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H. The Novel Coronavirus - A Snapshot of Current Knowledge. Microb Biotechnol. 2020;13:607–612. doi: 10.1111/1751-7915.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H. The Novel Coronavirus - Latest Findings. Microb Biotechnol. 2020 doi: 10.1111/1751-7915.13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busquet F, Hartung T, Pallocca G, Rovida C, Leist M. Harnessing the power of novel animal-free test methods for the development of COVID-19 drugs and vaccines. Arch Toxicol. 2020 doi: 10.1007/s00204-020-02787-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzowka M, Mitusinska K, Raczynska A, Samol A, Tuszynski JA, et al. Structural and Evolutionary Analysis Indicate That the SARS-CoV-2 Mpro Is a Challenging Target for Small-Molecule Inhibitor Design. Int J Mol Sci. 2020 doi: 10.3390/ijms21093099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantini F, Niccoli L, Matarrese D, Nicastri E, Stobbione P, et al. Baricitinib therapy in COVID-19: A pilot study on safety and clinical impact. J Infect. 2020 doi: 10.1016/j.jinf.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao RY, Xu YF, Zhang TH, Yang JJ, Yuan Y et al. 2017 Pediatric drug nitazoxanide: a potential choice for control of Zika. Open Forum Infect. Dis.4 ofx009 10.1093/ofid/ofx009 [DOI] [PMC free article] [PubMed]
- Cao YC, Deng QX, Dai SX. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JL, Su HC, Effort CHG. A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020 doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova LM, Jeon S, Rutala WA, Weber DJ, Sobsey MD. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl Environ Microbiol. 2010;76:2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M, Rajnik M, Cuomo A, Dulebohn SC and Di Napoli R 2020 Features, evaluation and treatment coronavirus (COVID-19). In: StatPearls (Ed) Treasure Island (FL) [PubMed]
- Ceccarelli M, Berretta M, Venanzi Rullo E, Nunnari G and Cacopardo B 2020 Differences and similarities between Severe Acute Respiratory Syndrome (SARS)-CoronaVirus (CoV) and SARS-CoV-2. Would a rose by another name smell as sweet? Eur. Rev. Med. Pharmacol. Sci.24 2781–2783 [DOI] [PubMed]
- Centers for Disease C and Prevention Update: Severe respiratory illness associated with Middle East Respiratory Syndrome Coronavirus (MERS-CoV)–worldwide, 2012-2013. MMWR Morb Mortal Wkly Rep. 2013;62:480–483. [PMC free article] [PubMed] [Google Scholar]
- Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha Y, Erez T, Reynolds IJ, Kumar D, Ross J, et al. Drug repurposing from the perspective of pharmaceutical companies. Br J Pharmacol. 2018;175:168–180. doi: 10.1111/bph.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaccour C, Rabinovich NR. Advancing the repurposing of ivermectin for malaria. Lancet. 2019;393:1480–1481. doi: 10.1016/S0140-6736(18)32613-8. [DOI] [PubMed] [Google Scholar]
- Chan KS, Lai ST, Chu CM, Tsui E, Tam CY, et al. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med J. 2003;9:399–406. [PubMed] [Google Scholar]
- Chan PK, Ip M, Ng KC, Rickjason CW, Wu A, et al. Severe acute respiratory syndrome-associated coronavirus infection. Emerg Infect Dis. 2003;9:1453–1454. doi: 10.3201/eid0911.030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TJ, Yang DM, Wang ML, Liang KH, Tsai PH, et al. Genomic Analysis and Comparative Multiple Sequence of SARS-CoV2. J Chin Med Assoc. 2020 doi: 10.1097/JCMA.0000000000000335. [DOI] [PubMed] [Google Scholar]
- Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R, Perlman S. Evaluation of Activation and Inflammatory Activity of Myeloid Cells During Pathogenic Human Coronavirus Infection. Methods Mol Biol. 2020;2099:195–204. doi: 10.1007/978-1-0716-0211-9_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Morrow JK, Tran HT, Phatak SS, Du-Cuny L, et al. From laptop to benchtop to bedside: structure-based drug design on protein targets. Curr Pharm Des. 2012;18:1217–1239. doi: 10.2174/138161212799436386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Hotez PJ, Bottazzi ME. Potential for developing a SARS-CoV receptor-binding domain (RBD) recombinant protein as a heterologous human vaccine against coronavirus infectious disease (COVID)-19. Hum Vaccin Immunother. 2020 doi: 10.1080/21645515.2020.1740560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. In Silico Oncology Drug Repositioning and Polypharmacology. Methods Mol Biol. 2019;1878:243–261. doi: 10.1007/978-1-4939-8868-6_15. [DOI] [PubMed] [Google Scholar]
- Cheng F, Lu W, Liu C, Fang J, Hou Y, et al. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat Commun. 2019;10:3476. doi: 10.1038/s41467-019-10744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian SS, Agrawal M, Basu A, Abraham P, Gangakhedkar RR, et al. Perspectives for repurposing drugs for the coronavirus disease 2019. Indian J Med Res. 2020;151:160–171. doi: 10.4103/ijmr.IJMR_585_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian MD, Poutanen SM, Loutfy MR, Muller MP, Low DE. Severe acute respiratory syndrome. Clin Infect Dis. 2004;38:1420–1427. doi: 10.1086/420743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CM, Cheng VC, Hung IF, Wong MM, Chan KH, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004 doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VC, McElroy LJ, Chu V, Bauman BE, Whittaker GR. The avian coronavirus infectious bronchitis virus undergoes direct low-pH-dependent fusion activation during entry into host cells. J Virol. 2006;80:3180–3188. doi: 10.1128/JVI.80.7.3180-3188.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicaloni V, Trezza A, Pettini F, Spiga O. Applications of in Silico Methods for Design and Development of Drugs Targeting Protein-Protein Interactions. Curr Top Med Chem. 2019;19:534–554. doi: 10.2174/1568026619666190304153901. [DOI] [PubMed] [Google Scholar]
- Coleman CM, Sisk JM, Mingo RM, Nelson EA, White JM, et al. Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion. J Virol. 2016;90:8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, Raoult D. Fighting viruses with antibiotics: an overlooked path. Int J Antimicrob Agents. 2016;48:349–352. doi: 10.1016/j.ijantimicag.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Kriegenburg F, de Haan CAM, Reggiori F. Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci Rep. 2017;7:5740. doi: 10.1038/s41598-017-06062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti P, Ronconi G, Caraffa A, Gallenga CE, Ross R et al. 2020 Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents3410.23812/conti-e [DOI] [PubMed]
- Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, De Giglio MAR and Roviello GN 2020 SARS CoV-2: Recent reports on antiviral therapies based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other drugs for the treatment of the new coronavirus. Curr. Med. Chem.10.2174/0929867327666200416131117 [DOI] [PubMed]
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic RS, Goa KL. Lopinavir/ritonavir: a review of its use in the management of HIV infection. Drugs. 2003 doi: 10.2165/00003495-200363080-00004. [DOI] [PubMed] [Google Scholar]
- Dabravolski SA, Kavalionak YK. SARS-CoV-2: Structural diversity, phylogeny, and potential animal host identification of spike glycoprotein. J Med Virol. 2020 doi: 10.1002/jmv.25976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev Anti Infect Ther. 2006;4:291–302. doi: 10.1586/14787210.4.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde AH, Jochmans D, Posthuma CC, Zevenhoven-Dobbe JC, van Nieuwkoop S, et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dediego ML, Pewe L, Alvarez E, Rejas MT, Perlman S, et al. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio R, Higa LM, Pezzuto P, Valadao AL, Garcez PP, et al. Chloroquine, an Endocytosis Blocking Agent, Inhibits Zika Virus Infection in Different Cell Models. Viruses. 2016 doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Lu P, Fan Y, Xia Y and Liu M 2020 The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J. Med. Virol.10.1002/jmv.25781 [DOI] [PMC free article] [PubMed]
- Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- Down B, Kulkarni S, Ahmed Khan AH, Barker B, Tang I. Novel coronavirus (COVID-19) infection: What a doctor on the frontline needs to know. Ann Med Surg (Lond) 2020 doi: 10.1016/j.amsu.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Toit A. Outbreak of a novel coronavirus. Nat Rev Microbiol. 2020;18:123. doi: 10.1038/s41579-020-0332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Zuo X, Meng F, Wu F, Zhao X, et al. Combinatorial screening of a panel of FDA-approved drugs identifies several candidates with anti-Ebola activities. Biochem Biophys Res Commun. 2020;522:862–868. doi: 10.1016/j.bbrc.2019.11.065. [DOI] [PubMed] [Google Scholar]
- Dudley JT 2011 Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 3 [DOI] [PMC free article] [PubMed]
- Dudley JT, Deshpande T and Butte AJ 2011 Exploiting drug-disease relationships for computational drug repositioning. Brief Bioinform.12 [DOI] [PMC free article] [PubMed]
- Dyall J, Coleman CM, Hart BJ, Venkataraman T, Holbrook MR, et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky AA. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinar JA, Menendez JA. Potential Drugs Targeting Early Innate Immune Evasion of SARS-Coronavirus 2 via 2’-O-Methylation of Viral RNA. Viruses. 2020 doi: 10.3390/v12050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HH, Wang LQ, Liu WL, An XP, Liu ZD, et al. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus (2019-nCoV) related coronavirus model. Chin Med J (Engl) 2020 doi: 10.1097/CM9.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farha MA, Brown ED. Drug repurposing for antimicrobial discovery. Nat Microbiol. 2019;4:565–577. doi: 10.1038/s41564-019-0357-1. [DOI] [PubMed] [Google Scholar]
- Fetro C, Scherman D. Drug repurposing in rare diseases: Myths and reality. Therapie. 2020;75:157–160. doi: 10.1016/j.therap.2020.02.006. [DOI] [PubMed] [Google Scholar]
- Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G et al. 2020 COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the Double X-chromosome in Females Be Protective against SARS-CoV-2 compared to the single X-chromosome in males? Int. J. Mol. Sci.2110.3390/ijms21103474 [DOI] [PMC free article] [PubMed]
- Giuliani S, Silva AC, Borba J, Ramos PIP, Paveley RA, et al. Computationally-guided drug repurposing enables the discovery of kinase targets and inhibitors as new schistosomicidal agents. PLoS Comput Biol. 2018;14:e1006515. doi: 10.1371/journal.pcbi.1006515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebov OO. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 2020 doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gns HS, Gr S, Murahari M, Krishnamurthy M. An update on Drug Repurposing: Re-written saga of the drug’s fate. Biomed Pharmacother. 2019;110:700–716. doi: 10.1016/j.biopha.2018.11.127. [DOI] [PubMed] [Google Scholar]
- Goo J, Jeong Y, Park YS, Yang E, Jung DI, et al. Characterization of novel monoclonal antibodies against MERS-coronavirus spike protein. Virus Res. 2020;278:197863. doi: 10.1016/j.virusres.2020.197863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski LE, Sheahan TP, Morrison TE, Menachery VD, Jensen K et al. 2018 Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio910.1128/mbio.01753-18 [DOI] [PMC free article] [PubMed]
- Gui M, Liu X, Guo D, Zhang Z, Yin CC, et al. Electron microscopy studies of the coronavirus ribonucleoprotein complex. Protein Cell. 2017;8:219–224. doi: 10.1007/s13238-016-0352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Huang Z, Lin L, Lv J. Coronavirus Disease 2019 (COVID-19) and Cardiovascular Disease: A Viewpoint on the Potential Influence of Angiotensin-Converting Enzyme Inhibitors/Angiotensin Receptor Blockers on Onset and Severity of Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J Am Heart Assoc. 2020;9:e016219. doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi GA, Ogunro OB, Adegunloye AP, Ogunyemi OM, Afolabi SO. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CL(pro)): an in silico screening of alkaloids and terpenoids from African medicinal plants. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A, Paray BA, Hussain A, Qadir FA, Attar F, et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A, Paray BA, Hussain A, Qadir FA, Attar F, et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata A, Shionyu-Mitsuyama C, Nakae S, Shionyu M, Ota M, et al. Knowledge-based structural models of SARS-CoV-2 proteins and their complexes with potential drugs. FEBS Lett. 2020 doi: 10.1002/1873-3468.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020 doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Hoffmann M. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KV. SARS coronavirus: a new challenge for prevention and therapy. J Clin Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Zhang Y, Li L, Wang K, Chen S, et al. Two adjacent mutations on the dimer interface of SARS coronavirus 3C-like protease cause different conformational changes in crystal structure. Virology. 2009;388:324–334. doi: 10.1016/j.virol.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Li M, Xu Y, Zhang J, Meng X, et al. Novel Gold Nanorod-Based HR1 Peptide Inhibitor for Middle East Respiratory Syndrome Coronavirus. ACS Appl Mater Interfaces. 2019;11:19799–19807. doi: 10.1021/acsami.9b04240. [DOI] [PubMed] [Google Scholar]
- Hurle MR 2013 Computational drug repositioning: from data to therapeutics. Clin. Pharmacol. Ther.93 [DOI] [PubMed]
- Infante T, Del Viscovo L, De Rimini ML, Padula S, Caso P, et al. Network Medicine: A Clinical Approach for Precision Medicine and Personalized Therapy in Coronary Heart Disease. J Atheroscler Thromb. 2020;27:279–302. doi: 10.5551/jat.52407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa NT, Stathias V, Schurer S, Dakshanamurthy S. Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol. 2020 doi: 10.1016/j.semcancer.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, et al. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J Virol. 2019 doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janardhan S, Dubovskaja V, Lagunin A, Rao BV, Sastry GN, et al. Recent Advances in the Development of Pharmaceutical Agents for Metabolic Disorders: A Computational Perspective. Curr Med Chem. 2018;25:5432–5463. doi: 10.2174/0929867324666171002120647. [DOI] [PubMed] [Google Scholar]
- Jawhara S. Could Intravenous Immunoglobulin Collected from Recovered Coronavirus Patients Protect against COVID-19 and Strengthen the Immune System of New Patients? Int J Mol Sci. 2020 doi: 10.3390/ijms21072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean SS, Lee PI, Hsueh PR. Treatment options for COVID-19: The reality and challenges. J Microbiol Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S, Ko M, Lee J, Choi I, Byun SY, et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother. 2020 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhamb D, Magid-Slav M, Hurle MR, Agarwal P. Pathway analysis of GWAS loci identifies novel drug targets and repurposing opportunities. Drug Discov Today. 2019;24:1232–1236. doi: 10.1016/j.drudis.2019.03.024. [DOI] [PubMed] [Google Scholar]
- Jia Z, Yan L, Ren Z, Wu L, Wang J, et al. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Cai L, Cheng ZS, Cheng H, Deng T et al. 2020 A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Mil. Med. Res.7 [DOI] [PMC free article] [PubMed]
- Johansen LM, DeWald LE, Shoemaker CJ, Hoffstrom BG, Lear-Rooney CM et al. 2015 A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci. Transl. Med.7 290ra289 [DOI] [PubMed]
- Kandeel M, Ibrahim A, Fayez M, Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol. 2020 doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman B, Sippl W. Computational Drug Repurposing: Current Trends. Curr Med Chem. 2019;26:5389–5409. doi: 10.2174/0929867325666180530100332. [DOI] [PubMed] [Google Scholar]
- Karuppasamy R, Veerappapillai S, Maiti S, Shin WH, Kihara D. Current progress and future perspectives of polypharmacology : From the view of non-small cell lung cancer. Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Takami Y, Kumar Deo V, Park EY. Preparation of virus-like particle mimetic nanovesicles displaying the S protein of Middle East respiratory syndrome coronavirus using insect cells. J Biotechnol. 2019;306:177–184. doi: 10.1016/j.jbiotec.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke YY, Peng TT, Yeh TK, Huang WZ, Chang SE, et al. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed J. 2020 doi: 10.1016/j.bj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshtkar-Jahromi M, Bavari S. A Call for Randomized Controlled Trials to Test the Efficacy of Chloroquine and Hydroxychloroquine as Therapeutics against Novel Coronavirus Disease (COVID-19) Am J Trop Med Hyg. 2020 doi: 10.4269/ajtmh.20-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A, Jayaram B, Goliaei B, Masoudi-Nejad A. Active repurposing of drug candidates for melanoma based on GWAS, PheWAS and a wide range of omics data. Mol Med. 2019;25:30. doi: 10.1186/s10020-019-0098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindrachuk J, Ork B, Hart BJ, Mazur S, Holbrook MR, et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother. 2015;59:1088–1099. doi: 10.1128/AAC.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen DB, Decornez H, Furr JR and Bajorath J 2004 Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov.3 [DOI] [PubMed]
- Koudijs KKM, Terwisscha van Scheltinga AGT, Bohringer S, Schimmel KJM, Guchelaar HJ. Transcriptome Signature Reversion as a Method to Reposition Drugs Against Cancer for Precision Oncology. Cancer J. 2019;25:116–120. doi: 10.1097/PPO.0000000000000370. [DOI] [PubMed] [Google Scholar]
- Kuang Z, Bao Y, Thomson J, Caldwell M, Peissig P, et al. A Machine-Learning-Based Drug Repurposing Approach Using Baseline Regularization. Methods Mol Biol. 2019;1903:255–267. doi: 10.1007/978-1-4939-8955-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virusdisease. 2020;31:13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Shin JS, Shie JJ, Ku KB, Kim C, et al. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CL(Pro) inhibitors. Antiviral Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Chen YP. Prediction of drug adverse events using deep learning in pharmaceutical discovery. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa040. [DOI] [PubMed] [Google Scholar]
- Lesch M, Luckner M, Meyer M, Weege F, Gravenstein I, et al. RNAi-based small molecule repositioning reveals clinically approved urea-based kinase inhibitors as broadly active antivirals. PLoS Pathog. 2019;15:e1007601. doi: 10.1371/journal.ppat.1007601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yang Y, Ren L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect Genet Evol. 2020;82:104285. doi: 10.1016/j.meegid.2020.104285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC, Wang XJ, Wang HR. Repurposing host-based therapeutics to control coronavirus and influenza virus. Drug Discov Today. 2019;24:726–736. doi: 10.1016/j.drudis.2019.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang YM, Xu JY and Cao B 2020b Potential antiviral therapeutics for 2019 Novel Coronavirus. Chin. J. Tuberc. Respir. Dis.43 [DOI] [PubMed]
- Li H, Wang YM, Xu JY, Cao B. Potential antiviral therapeutics for 2019 Novel Coronavirus. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:170–172. doi: 10.3760/cma.j.issn.1001-0939.2020.03.004. [DOI] [PubMed] [Google Scholar]
- Li H, Zhou Y, Zhang M, Wang H, Zhao Q, et al. Updated approaches against SARS-CoV-2. Antimicrobial Agents and Chemotherapy. 2020 doi: 10.1128/AAC.00483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Liu CL, Chang YM, Zhao J, Perlman S, et al. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li M, Zhou Z, Guan X, Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zheng Q, Wang Z. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang C, Huang F, Yang Y, Wang F et al. 2020c 2019-novel coronavirus (2019-nCoV) infections trigger an exaggerated cytokine response aggravating lung injury
- Lo Caputo S, Corso G, Clerici M, Santantonio TA. Baricitinib: a chance to treat COVID-19? J Med Virol. 2020 doi: 10.1002/jmv.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J, Tian SJ, Niu SM, Kang XQ, Lian HX et al. 2020 Coronavirus disease 2019: a bibliometric analysis and review. Eur. Rev. Med. Pharmacol. Sci.24 3411–3421 [DOI] [PubMed]
- Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, et al. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA. 2003;290:3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- Lu L, Liu Q, Zhu Y, Chan KH, Qin L, et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher Dias T, Schuch V, Beltrao-Braga PCB, Martins-de-Souza D, Brentani HP, et al. Drug repositioning for psychiatric and neurological disorders through a network medicine approach. Transl Psychiatry. 2020;10:141. doi: 10.1038/s41398-020-0827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma-Lauer Y, Zheng Y, Malesevic M, von Brunn B, Fischer G, et al. Influences of cyclosporin A and non-immunosuppressive derivatives on cellular cyclophilins and viral nucleocapsid protein during human coronavirus 229E replication. Antiviral Res. 2020;173:104620. doi: 10.1016/j.antiviral.2019.104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, Smith DW. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don’t. Microbiol Aust. 2020 doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio R, Corsini GU. Repurposing the mucolytic cough suppressant and TMPRSS2 protease inhibitor bromhexine for the prevention and management of SARS-CoV-2 infection. Pharmacol Res. 2020;157:104837. doi: 10.1016/j.phrs.2020.104837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro G. SARS-CoV-2 and COVID-19: is interleukin-6 (IL-6) the ‘culprit lesion’ of ARDS onset? What is there besides Tocilizumab? SGP130Fc. Cytokine X. 2020 doi: 10.1016/j.cytox.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MA. Compounds with Therapeutic Potential against Novel Respiratory 2019 Coronavirus. Antimicrob Agents Chemother. 2020 doi: 10.1128/AAC.00399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MA. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob Agents Chemother. 2020 doi: 10.1128/AAC.00399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorana A, Perricone U, Lauria A. The Repurposing of Old Drugs or Unsuccessful Lead Compounds by in Silico Approaches: New Advances and Perspectives. Curr Top Med Chem. 2016;16:2088–2106. doi: 10.2174/1568026616666160216153457. [DOI] [PubMed] [Google Scholar]
- Mathewson AC, Bishop A, Yao Y, Kemp F, Ren J, et al. Interaction of severe acute respiratory syndrome-coronavirus and NL63 coronavirus spike proteins with angiotensin converting enzyme-2. J Gen Virol. 2008;89:2741–2745. doi: 10.1099/vir.0.2008/003962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, et al. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza MU, Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro NRC, Ribeiro B, Arrais J. Drug-Target Interaction Prediction: End-to-End Deep Learning Approach. IEEE/ACM Trans Comput Biol Bioinform. 2020 doi: 10.1109/TCBB.2020.2977335. [DOI] [PubMed] [Google Scholar]
- Montes-Grajales D, Puerta-Guardo H, Espinosa DA, Harris E, Caicedo-Torres W, et al. In silico drug repurposing for the identification of potential candidate molecules against arboviruses infection. Antiviral Res. 2020;173:104668. doi: 10.1016/j.antiviral.2019.104668. [DOI] [PubMed] [Google Scholar]
- Mucke HAM. Drug Repurposing and Artificial Intelligence: From Liaison to Marriage. Assay Drug Dev Technol. 2018;16:231. doi: 10.1089/adt.2018.29078.edi. [DOI] [PubMed] [Google Scholar]
- Mukherjee R 2020 Global efforts on vaccines for COVID-19: since, sooner or later, we all will catch the coronavirus. J. Biosci.45 [DOI] [PMC free article] [PubMed]
- Mumtaz N, Jimmerson LC, Bushman LR, Kiser JJ, Aron G, et al. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antiviral Res. 2017;146:161–163. doi: 10.1016/j.antiviral.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Muthaiyan M, Pushan SS, Naorem LD, Venkatesan A. Understanding of Zaire ebolavirus-human protein interaction for drug repurposing. Virusdisease. 2020;31:28–37. doi: 10.1007/s13337-020-00570-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabirotchkin S, Peluffo AE, Rinaudo P, Yu J, Hajj R, et al. Next-generation drug repurposing using human genetics and network biology. Curr Opin Pharmacol. 2020 doi: 10.1016/j.coph.2019.12.004. [DOI] [PubMed] [Google Scholar]
- Narayanan K, Huang C, Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K, Ramirez SI, Lokugamage KG, Makino S. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento ACA, Prudencio RBC, Costa IG. A Drug-Target Network-Based Supervised Machine Learning Repurposing Method Allowing the Use of Multiple Heterogeneous Information Sources. Methods Mol Biol. 2019;1903:281–289. doi: 10.1007/978-1-4939-8955-3_17. [DOI] [PubMed] [Google Scholar]
- Nath A, Kumari P, Chaube R. Prediction of Human Drug Targets and Their Interactions Using Machine Learning Methods: Current and Future Perspectives. Methods Mol Biol. 2018;1762:21–30. doi: 10.1007/978-1-4939-7756-7_2. [DOI] [PubMed] [Google Scholar]
- Neogi U, Hill KJ, Ambikan AT, Heng X, Quinn TP, et al. Feasibility of Known RNA Polymerase Inhibitors as Anti-SARS-CoV-2 Drugs. Pathogens. 2020 doi: 10.3390/pathogens9050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres JL, Dediego ML, Alvarez E, Jimenez-Guardeno JM, Regla-Nava JA, et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres JL, Verdia-Baguena C, Jimenez-Guardeno JM, Regla-Nava JA, Castano-Rodriguez C, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534:314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- Nur SM, Hasan MA, Amin MA, Hossain M, Sharmin T. Design of Potential RNAi (miRNA and siRNA) Molecules for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Gene Silencing by Computational Method. Interdiscip Sci. 2015;7:257–265. doi: 10.1007/s12539-015-0266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omotade TO, Roy CR. Manipulation of Host Cell Organelles by Intracellular Pathogens. Microbiol Spectr. 2019 doi: 10.1128/microbiolspec.BAI-0022-2019. [DOI] [PubMed] [Google Scholar]
- Oulas A, Minadakis G, Zachariou M, Sokratous K, Bourdakou MM, et al. Systems Bioinformatics: increasing precision of computational diagnostics and therapeutics through network-based approaches. Brief Bioinform. 2019;20:806–824. doi: 10.1093/bib/bbx151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JR, DeBerardinis AM, Sail V, Tacheva-Grigorova SK, Chan KA, et al. Repurposing the Clinically Efficacious Antifungal Agent Itraconazole as an Anticancer Chemotherapeutic. J Med Chem. 2016;59:3635–3649. doi: 10.1021/acs.jmedchem.5b01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammolli F, Magazzini L, Riccaboni M. The productivity crisis in pharmaceutical R&D. Drug Discov: Nat. Rev; 2011. p. 10. [DOI] [PubMed] [Google Scholar]
- Pereira CA, Saye M, Reigada C, Silber AM, Labadie GR, et al. Computational approaches for drug discovery against trypanosomatid-caused diseases. Parasitology. 2020;147:611–633. doi: 10.1017/S0031182020000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Tan E, Parthasarathy K, Lin X, Jiang FL, et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:e1000511. doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020 doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci. 2019 doi: 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A, Padey B, Terrier O, Rosa-Calatrava M. Drug Repurposing Approaches for the Treatment of Influenza Viral Infection: Reviving Old Drugs to Fight Against a Long-Lived Enemy. Front Immunol. 2019;10:531. doi: 10.3389/fimmu.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poterico JA, Mestanza O. Genetic variants and source of introduction of SARS-CoV-2 in South America. J Med Virol. 2020 doi: 10.1002/jmv.26001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen D, Puvvada RC, Va M. Janus kinase inhibitor baricitinib is not an ideal option for management of COVID-19. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing E, Hantak M, Perlman S and Gallagher T 2020 Distinct roles for sialoside and protein receptors in coronavirus infection. mBio1110.1128/mbio.02764-19 [DOI] [PMC free article] [PubMed]
- Rabinovich NR. Ivermectin: repurposing an old drug to complement malaria vector control. Lancet Infect Dis. 2018;18:584–585. doi: 10.1016/S1473-3099(18)30176-2. [DOI] [PubMed] [Google Scholar]
- Rahman N, Basharat Z, Yousuf M, Castaldo G, Rastrelli L, et al. Virtual Screening of Natural Products against Type II Transmembrane Serine Protease (TMPRSS2), the Priming Agent of Coronavirus 2 (SARS-CoV-2) Molecules. 2020 doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainer TH. Severe acute respiratory syndrome: clinical features, diagnosis, and management. Curr Opin Pulm Med. 2004;10:159–165. doi: 10.1097/00063198-200405000-00003. [DOI] [PubMed] [Google Scholar]
- Rehman SU, Shafique L, Ihsan A, Liu Q. Evolutionary Trajectory for the Emergence of Novel Coronavirus SARS-CoV-2. Pathogens. 2020 doi: 10.3390/pathogens9030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol JF. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J Infect Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberian N, Peyvandipour A, Donato M, Ansari S, Draghici S. A new computational drug repurposing method using established disease-drug pair knowledge. Bioinformatics. 2019;35:3672–3678. doi: 10.1093/bioinformatics/btz156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P, Banerjee AK, Tripathi PP, Srivastava AK, Ray U. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of Coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci Rep. 2020 doi: 10.1042/BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salata C, Calistri A, Alvisi G, Celestino M, Parolin C, et al. Ebola Virus Entry: From Molecular Characterization to Drug Discovery. Viruses. 2019 doi: 10.3390/v11030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020 doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases? Lancet Infect Dis. 2003 doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell JW, Blanckley A, Boldon H and Warrington B 2012 Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. [DOI] [PubMed]
- Schloer S, Goretzko J, Kuhnl A, Brunotte L, Ludwig S et al. 2019 The clinically licensed antifungal drug itraconazole inhibits influenza virus in vitro and in vivo. Emerg. Microbes Infect.8 80–93 [DOI] [PMC free article] [PubMed]
- Schoergenhofer C, Jilma B, Stimpfl T, Karolyi M, Zoufaly A. Pharmacokinetics of Lopinavir and Ritonavir in Patients Hospitalized With Coronavirus Disease 2019 (COVID-19) Ann Intern Med. 2020 doi: 10.7326/M20-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Wan Y, Liu C, Yount B, Gully K, et al. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16:e1008392. doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhou C, He P, Huang S, Duan Y, et al. Successful treatment of plasma exchange followed by intravenous immunogloblin in a critically ill patient with 2019 novel coronavirus infection. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Chen X, Deng L. A Review of Recent Developments and Progress in Computational Drug Repositioning. Curr Pharm Des. 2020 doi: 10.2174/1381612826666200116145559. [DOI] [PubMed] [Google Scholar]
- Shirato K, Kawase M, Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013;87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Bertram S, Glowacka I, Steffen I, Chaipan C, et al. Different host cell proteases activate the SARS-coronavirus spike-protein for cell-cell and virus-cell fusion. Virology. 2011;413:265–274. doi: 10.1016/j.virol.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisk JM, Frieman MB, Machamer CE. Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J Gen Virol. 2018;99:619–630. doi: 10.1099/jgv.0.001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner D, Marro BS, Lane TE. Chemokine CXCL10 and Coronavirus-Induced Neurologic Disease. Viral Immunol. 2019;32:25–37. doi: 10.1089/vim.2018.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrab SS, El-Kafrawy SA, Mirza Z, Kamal MA, Azhar EI. Design and Delivery of Therapeutic siRNAs: Application to MERS-Coronavirus. Curr Pharm Des. 2018;24:62–77. doi: 10.2174/1381612823666171109112307. [DOI] [PubMed] [Google Scholar]
- Song F, Shi N, Shan F, Zhang Z, Shen J, et al. Emerging 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology. 2020;295:210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Yang Y, Wang L, Wang K, Ran L, et al. EIF4A2 interacts with the membrane protein of transmissible gastroenteritis coronavirus and plays a role in virus replication. Res Vet Sci. 2019;123:39–46. doi: 10.1016/j.rvsc.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg A, McKee DL, Naujokat C. Novel Drugs Targeting the SARS-CoV-2/COVID-19 Machinery. Curr Top Med Chem. 2020 doi: 10.2174/1568026620999200517043137. [DOI] [PubMed] [Google Scholar]
- Stobart CC, Sexton NR, Munjal H, Lu X, Molland KL, et al. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J Virol. 2013;87:12611–12618. doi: 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter SM. Overcoming Drug Development Bottlenecks With Repurposing: Old drugs learn new tricks. Nat Med. 2014;20:590–591. doi: 10.1038/nm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupnikov A, O’Reilly PG, McInerney CE, Roddy AC, Dunne PD, et al. Impact of Variable RNA-Sequencing Depth on Gene Expression Signatures and Target Compound Robustness: Case Study Examining Brain Tumor (Glioma) Disease Progression. JCO Precis Oncol. 2018 doi: 10.1200/PO.18.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YJ, Lai YC. Comparison of clinical characteristics of coronavirus disease (COVID-19) and severe acute respiratory syndrome (SARS) as experienced in Taiwan. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JJ, Wu A, Joynt GM, Yuen KY, Lee N, et al. Severe acute respiratory syndrome: report of treatment and outcome after a major outbreak. Thorax. 2004;59:414–420. doi: 10.1136/thx.2003.014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W, He L, Zhang X, Pu J, Voronin D, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020 doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevi A and Bellera CL 2020 Challenges and opportunities with drug repurposing: finding strategies to find alternative uses of therapeutics. Expert Opin. Drug Discov.15 397–401 [DOI] [PubMed]
- Tanaka Y, Sato Y, Sasaki T. Suppression of coronavirus replication by cyclophilin inhibitors. Viruses. 2013;5:1250–1260. doi: 10.3390/v5051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis AJ, van den Worm SH, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Li C, Huang A, Xia S, Lu S et al. 2020 Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect.9 382–385 [DOI] [PMC free article] [PubMed]
- Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trosset JY, Cave C. In Silico Drug-Target Profiling. Methods Mol Biol. 2019;1953:89–103. doi: 10.1007/978-1-4939-9145-7_6. [DOI] [PubMed] [Google Scholar]
- Tsang K, Seto WH. Severe acute respiratory syndrome: scientific and anecdotal evidence for drug treatment. Curr Opin Investig Drugs. 2004;5:179–185. [PubMed] [Google Scholar]
- Tsang KW, Ho PL, Ooi GC, Yee WK, Wang T, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- Tsang KW, Lam WK. Management of severe acute respiratory syndrome: the Hong Kong University experience. Am J Respir Crit Care Med. 2003;168:417–424. doi: 10.1164/rccm.2305012. [DOI] [PubMed] [Google Scholar]
- Tsubamoto H, Ueda T, Inoue K, Sakata K, Shibahara H, et al. Repurposing itraconazole as an anticancer agent. Oncol Lett. 2017;14:1240–1246. doi: 10.3892/ol.2017.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufan A, Avanoglu Guler A, Matucci-Cerinic M. COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turk J Med Sci. 2020;50:620–632. doi: 10.3906/sag-2004-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitdehaag JCM, Kooijman JJ, de Roos J, Prinsen MBW, Dylus J, et al. Combined Cellular and Biochemical Profiling to Identify Predictive Drug Response Biomarkers for Kinase Inhibitors Approved for Clinical Use between 2013 and 2017. Mol Cancer Ther. 2019;18:470–481. doi: 10.1158/1535-7163.MCT-18-0877. [DOI] [PubMed] [Google Scholar]
- Urquhart L. Market watch: top drugs and companies by sales in 2017. Drug Discov: Nat. Rev; 2018. p. 17. [DOI] [PubMed] [Google Scholar]
- van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS et al. 2012 Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio310.1128/mbio.00473-12 [DOI] [PMC free article] [PubMed]
- Veljkovic V, Loiseau PM, Figadere B, Glisic S, Veljkovic N et al. 2015 Virtual screen for repurposing approved and experimental drugs for candidate inhibitors of EBOLA virus infection. F1000Res4 34 [DOI] [PMC free article] [PubMed]
- Veronese N, Demurtas J, Yang L, Tonelli R, Barbagallo M, et al. Use of Corticosteroids in Coronavirus Disease 2019 Pneumonia: A Systematic Review of the Literature. Front Med (Lausanne) 2020;7:170. doi: 10.3389/fmed.2020.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Hua C, Xia S, Li W, Lu L, et al. Combining a Fusion Inhibitory Peptide Targeting the MERS-CoV S2 Protein HR1 Domain and a Neutralizing Antibody Specific for the S1 Protein Receptor-Binding Domain (RBD) Showed Potent Synergism against Pseudotyped MERS-CoV with or without Mutations in RBD. Viruses. 2019 doi: 10.3390/v11010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li W, Drabek D, Okba NMA, van Haperen R, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hu B, Hu C, Zhu F, Liu X et al. 2020b Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed]
- Wang M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Manli, Ruiyuan Cao, Leike Zhang, Xinglou Yang, Jia Liu, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research. 2020 doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen M, Zhang Z, Niu S, Sha H, Yang R, et al. Deep-Learning-Based Drug-Target Interaction Prediction. J Proteome Res. 2017;16:1401–1409. doi: 10.1021/acs.jproteome.6b00618. [DOI] [PubMed] [Google Scholar]
- Wong KT, Antonio GE, Hui DS, Lee N, Yuen EH, et al. Severe acute respiratory syndrome: radiographic appearances and pattern of progression in 138 patients. Radiology. 2003;228:401–406. doi: 10.1148/radiol.2282030593. [DOI] [PubMed] [Google Scholar]
- Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Liu J, Yue X. Prediction of drug-disease associations based on ensemble meta paths and singular value decomposition. BMC Bioinformatics. 2019;20:134. doi: 10.1186/s12859-019-2644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wang L, Kuo HD, Shannar A, Peter R, et al. An Update on Current Therapeutic Drugs Treating COVID-19. Curr Pharmacol Rep. 2020 doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li C, Xia S, Tian X, Kong Y, et al. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Lan Q, Pu J, Wang C, Liu Z, et al. Potent MERS-CoV Fusion Inhibitory Peptides Identified from HR2 Domain in Spike Protein of Bat Coronavirus HKU4. Viruses. 2019 doi: 10.3390/v11010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Yan L, Xu W, Agrawal AS, Algaissi A et al. 2019b A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv.5 eaav4580 [DOI] [PMC free article] [PubMed]
- Xia X. Bioinformatics and Drug Discovery. Curr Top Med Chem. 2017;17:1709–1726. doi: 10.2174/1568026617666161116143440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Chen Q. Insight into 2019 novel coronavirus - An updated interim review and lessons from SARS-CoV and MERS-CoV. Int J Infect Dis. 2020;94:119–124. doi: 10.1016/j.ijid.2020.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Chen Q. Insight into 2019 novel coronavirus - an updated intrim review and lessons from SARS-CoV and MERS-CoV. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Xie W, Xue X, Yang K, Ma J, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Luo H, Li Y, Wu FX, Wang J. Overlap matrix completion for predicting drug-associated indications. PLoS Comput Biol. 2019;15:e1007541. doi: 10.1371/journal.pcbi.1007541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Ye F, Zhang M, Cui C, Huang B, et al. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Gottwald M, Schuler P, Michel MC. Opportunities and Challenges for Drug Development: Public-Private Partnerships, Adaptive Designs and Big Data. Front Pharmacol. 2016;7:461. doi: 10.3389/fphar.2016.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020 doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zai CC, Tiwari AK, Zai GC, de Luca V, Shaikh SA, et al. Sequence Analysis of Drug Target Genes with Suicidal Behavior in Bipolar Disorder Patients. Mol Neuropsychiatry. 2018;4:1–6. doi: 10.1159/000488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Zhu S, Hou Y, Zhang P, Li L, et al. Network-based prediction of drug-target interactions using an arbitrary-order proximity embedded deep forest. Bioinformatics. 2020;36:2805–2812. doi: 10.1093/bioinformatics/btaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha L, Li S, Pan L, Tefsen B, Li Y, et al. Corticosteroid treatment of patients with coronavirus disease 2019 (COVID-19) Med J Aust. 2020 doi: 10.5694/mja2.50577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Han Y, Shi H, Chen J, Zhang X et al. 2020a Swine acute diarrhea syndrome coronavirus-induced apoptosis is caspase- and cyclophilin D- dependent. Emerg. Microbes Infect.9 439–456 [DOI] [PMC free article] [PubMed]
- Zhang S, Yi C, Li C, Zhang F, Peng J, et al. Chloroquine inhibits endosomal viral RNA release and autophagy-dependent viral replication and effectively prevents maternal to fetal transmission of Zika virus. Antiviral Res. 2019;169:104547. doi: 10.1016/j.antiviral.2019.104547. [DOI] [PubMed] [Google Scholar]
- Zhang WF, Stephen P, Theriault JF, Wang R, Lin SX. Novel Coronavirus Polymerase and Nucleotidyl-Transferase Structures: Potential to Target New Outbreaks. J Phys Chem Lett. 2020 doi: 10.1021/acs.jpclett.0c00571. [DOI] [PubMed] [Google Scholar]
- Zhang YZ, Holmes EC. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, So HC. Drug Repositioning for Schizophrenia and Depression/Anxiety Disorders: A Machine Learning Approach Leveraging Expression Data. IEEE J Biomed Health Inform. 2019;23:1304–1315. doi: 10.1109/JBHI.2018.2856535. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang F, Xu M, Huang K, Zhong W, et al. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol. 2003;52:715–720. doi: 10.1099/jmm.0.05320-0. [DOI] [PubMed] [Google Scholar]
- Zhou L, Li Z, Yang J, Tian G, Liu F, et al. Revealing Drug-Target Interactions with Computational Models and Algorithms. Molecules. 2019 doi: 10.3390/molecules24091714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Pan T, Zhang J, Li Q, Zhang X, et al. Glycopeptide Antibiotics Potently Inhibit Cathepsin L in the Late Endosome/Lysosome and Block the Entry of Ebola Virus, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) J Biol Chem. 2016;291:9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang X, Wang X, Hu B, Zhang L et al. 2020a A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature57910.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed]
- Zhou Y, Hou Y, Shen J, Huang Y, Martin W, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Curtis N. Coronavirus Infections in Children Including COVID-19: An Overview of the Epidemiology, Clinical Features, Diagnosis, Treatment and Prevention Options in Children. Pediatr Infect Dis J. 2020;39:355–368. doi: 10.1097/INF.0000000000002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]