Highlights
-
•
Major progress has been achieved with regard to the understanding of the phylogeny and genomic organization of SARS-CoV-2.
-
•
This review summarized crucial developments in the elucidation of the structure and function of key SARS-CoV-2 proteins.
-
•
The molecular details of SARS-CoV-2 infection and replication could improve the effective clinical treatment.
Keywords: COVID-19, SARS-CoV-2, Genome-encoded proteins, Structure-based screening, Drug-screening
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, the Middle Eastern respiratory syndrome coronavirus; NSP, non-structural protein; ORF, Open reading frame; RdRp, RNA-dependent RNA polymerase; Mpro, Main protease; 3CLpro, 3C-like protease; S protein, Spike glycoprotein; E protein, Envelope protein; M protein, Membrane protein; N protein, Nucleocapsid protein; gRNA, genomic RNA; sgRNA, subgenomic RNA; RMP, The remdesivir monophosphate; RBD, receptor-binding domain; RBM, receptor-binding motif; PD, peptidase domain; ACE2, angiotensin-converting enzyme 2; CatB/L, cysteine proteases-cathepsin B and L; HR1, heptad repeat 1; HR2, heptad repeat 2; 6-HB, six-helix bundle
Abstract
A severe form of pneumonia, named coronavirus disease 2019 (COVID-19) by the World Health Organization, broke out in China and rapidly developed into a global pandemic, with millions of cases and hundreds of thousands of deaths reported globally. The novel coronavirus, which was designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified as the etiological agent of COVID-19. On the basis of experience accumulated following previous SARS-CoV and MERS-CoV outbreaks and research, a series of studies have been conducted rapidly, and major progress has been achieved with regard to the understanding of the phylogeny and genomic organization of SARS-CoV-2 in addition its molecular mechanisms of infection and replication. In the present review, we summarized crucial developments in the elucidation of the structure and function of key SARS-CoV-2 proteins, especially the main protease, RNA-dependent RNA polymerase, spike glycoprotein, and nucleocapsid protein. Results of studies on their associated inhibitors and drugs have also been highlighted.
Graphical abstract
1. Introduction
In December 2019, a novel pneumonia named coronavirus disease 2019 (COVID-19), emerged in Wuhan, China (Zhu et al., 2020; Wu et al., 2020a). Its symptoms included dry cough, fever, headache, dyspnea, and pneumonia (Huang et al., 2020a; Liu et al., 2020; Wang et al., 2020a). Due to the high contagiousness and transmission rate of the virus during the presymptomatic phase (Hu et al., 2020; Huang et al., 2020b), COVID-19 became a pandemic and spread to more than 212 countries and territories, and community transmission took place in many countries including the United States, Germany, France, Spain, Japan, Singapore, South Korea, Iran, and Italy. Thus far, millions of cases and hundreds of thousands of deaths have been recorded, with rapidly increasing numbers globally.
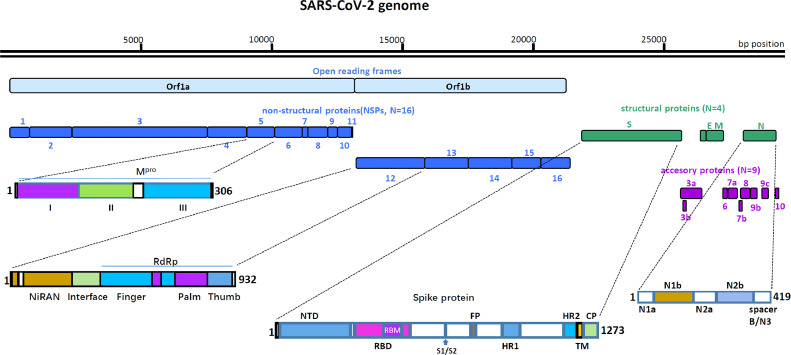
A previously unknown coronavirus, designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which belongs to β-coronaviruses and has a genomic sequence closely related to that of SARS-CoV, was determined as the etiological agent of this severe infection (Zhu et al., 2020; Coronaviridae Study Group of the International Committee on Taxonomy of V 2020; Zhou et al., 2020; Wu et al., 2020b). The Coronaviridae family includes two subfamilies: Letovirinae and Orthocoronavirinae, and the Orthocoronavirinae family consists of the α-coronavirus, β-coronavirus, γ-coronavirus, and δ-coronavirus genera (Cui et al., 2019). Many β-coronaviruses are human pathogens and cause severe respiratory diseases, including SARS-CoV, the Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV), and currently, SARS-CoV-2. Coronaviruses are enveloped, positive-sense, single-stranded RNA viruses with mammalian and avian hosts. The length of the SARS-CoV-2 genome is approximately 30 kb (Zhou et al., 2020), and it encodes at least 29 proteins, including 16 non-structural proteins (NSP), 4 structural proteins, and 9 accessory proteins (Fig. 1).
Fig. 1.
Schematic presentation of the SARS-CoV-2 genome organization, and the primary structures of Mpro, NSP12, S protein and N protein. The SARS-CoV-2 genome consists of ∼30 Kb RNA strand. There are 14 ORFs. The first two ORFs at 5′ untranslated regions code for polyprotein (pp1a/ab) that are segmented into 16 NSPs required for virus replication, followed by four structural proteins for spike glycoprotein(S), envelope protein(E), membrane protein(M), and nucleocapsid protein(N). At the 3′ terminus, there are nine accessory proteins (3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10). Mpro consists of three domains, Domains I (8–101 aa), II (102–184 aa) and III (201–303 aa). NSP12 has three domains, the RdRp domain (367–920 aa), NiRAN domain (4–28 aa and 69–249 aa) and interface domain (250–365 aa). The RdRp domain consists of three subdomains: the finger subdomain (66–581 and 621–679 aa), the palm subdomain (582–620 and 680–815 aa), and the thumb subdomain (816–920 aa). S glycoprotein is divided into two subunits by protease at the S1/S2 protease cleavage site, Its S1 subunit contains NTD (14–305 aa), RBD (319 –541 aa), and RBM (437–508 aa). Its S2 subunit contains FP (788–806 aa), HR1 (912–984 aa), HR2 (1163–1213 aa), TM (1214–1237 aa) and CP (1238–1273 aa). N protein encompasses two conserved domains, namely the N1b domain (49–175aa), and N2b domain (247–365aa), and three short regions [N1a (1–49aa), N2a (174–247aa), and spacer B/N3 (365–419aa)]. The white box represents loop or non-special structural domain connecting the two domains on either side.
Upon cell entry, the genomic RNA of SARS-CoV-2 is translated to produce two overlapping polyproteins, pp1a and pp1ab, from two open reading frames (ORFs), ORF1a and ORF1b, respectively. Subsequently, pp1ab is cleaved into 16 NSPs by viral proteases NSP3 and NSP5, which harbor a papain-like protease domain and a 3C-like protease domain, respectively (Fig. 1). The NSP12 (also known as the RNA-dependent RNA polymerase, RdRp) is the central component of the replication/transcription machinery in the synthesis of viral RNA, with the assistance of NSP7 and NSP8 as cofactors (Huang et al., 2020a; Subissi et al., 2014). SARS-CoV-2, as a positive-sense, single-stranded RNA virus, has the capacity to synthesize full-length negative-sense RNA chain, which serve as templates for further generation of positive-sense genomic RNA (gRNA) and subgenomic RNAs (sgRNAs). The gRNA is enveloped by structural proteins for progeny virion assembly, whereas the sgRNAs are relatively short and encode conserved structural proteins (spike [S] glycoprotein, envelope [E] protein, membrane [M] protein, and nucleocapsid [N] protein), and several accessory proteins. The nine accessory proteins (3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10) of SARS-CoV-2 participate in various processes, ranging from coronavirus replication to resistance against immune responses (Fig. 1) (Wu et al., 2020b; Kim et al., 2020).
Elucidating the structural and functional features of the major proteins encoded by the SARS-CoV-2 genome would facilitate the development of viral specific drug therapies and vaccines to combat the global pandemic. With the tireless efforts of researchers, the atomic resolution structure of some key SARS-CoV-2 proteins have been resolved using cryo-Electron Microscopy (cryo-EM) or X-ray methods (Table 1), and their corresponding biological functions have been reported, promoting the rapid discovery of antiviral agents with clinical potential. The present review summarizes current research findings and developments with regard to the structure and function of major proteins encoded by SARS-CoV-2, and the insights could promote drug and vaccine development for the treatment and management of COVID-19.
Table 1.
Overview of published structures of SARS-CoV-2 proteins.
| SARS-CoV-2 proteins | Protein Data Bank entry | Method | Resolution/ Å | Ligands |
|---|---|---|---|---|
| NSP12/NSP7/NSP8 | 6M71/7BTF (Gao et al., 2020a) | Cryo-EM | 2.9/2.95 | ZN |
| 7BV1/7BV2 (Yin et al., 2020) | Cryo-EM | 2.8/2.5 | F86, POP et al. | |
| 7BZF/7C2K (Wang et al., 2020c) | Cryo-EM | 3.26/2.93 | ZN | |
| 6YYT (Walls et al., 2019) | Cryo-EM | 2.90 | ZN | |
| NSP15 | 6VWW/6W01 (Callaway, 2020) | X-RAY | 2.2/1.9 | ACY et al. |
| Mpro | 6Y2G/6Y2F/6Y2E (Zhang et al., 2020) | X-RAY | 2.2/1.95/1.75 | GLY, O6K, DMS |
| 6LU7/6M03/7BQY (Jin et al., 2020b) | X-RAY | 2.16/2/1.7 | N3 | |
| 6LZE/6M0K (Dai et al., 2020) | X-RAY | 1.5/1.5 | 11a/11b | |
| S protein | 6VSB (Wrapp et al., 2020) | Cryo-EM | 3.46 | NAG |
| 6VYB/6VXX (Walls et al., 2020) | Cryo-EM | 3.2/2.8 | NAG | |
| S protein S2 subunit | 6LXT (Xia et al., 2020b) | X-RAY | 2.9 | PG4, ZN |
| S protein RBD-ACE2 | 6M17/6M18/6M1D (Yan et al., 2020) | Cryo-EM | 2.9/2.9/4.5 | NAG, ZN |
| 6VW1 (Shang et al., 2020) | X-RAY | 2.68 | BMA, NAG et al. | |
| 6M0J (Lan et al., 2020) | X-RAY | 2.45 | NAG, ZN, CL | |
| 6LZG (Gao et al., 2020b) | X-RAY | 2.5 | NAG, ZN | |
| S protein RBD-CR3022 | 6W41 (Yuan et al., 2020) | X-RAY | 3.08 | GOL, NAG, SO4 |
| Nucleocapsid protein N1b | 6M3M (Kang et al., 2020) | X-RAY | 2.70 | |
| Nucleocapsid protein N2b | 6WZO/6WZQ (Ye et al., 2020) | X-RAY | 1.42/1.45 |
2. Main protease (Mpro)
The functional polyproteins of SARS-CoV-2, pp1a and pp1ab, are cleaved into 16 NSPs, mainly by a 33.8-kDa main protease (Mpro), which is also referred to as 3C-like protease (3CLpro) (Anand et al., 2003; Hilgenfeld, 2014). There are at least 11 conserved Mpro restriction sites on the pp1ab protein, starting with its autolytic cleavage site (Hegyi and Ziebuhr, 2002). Because Mpro has no closely related homologues in humans, its specific inhibitor has been demonstrated to inhibit viral life cycle with no obvious biotoxicity (Pillaiyar et al., 2016; Hayden et al., 2003; Kim et al., 2016; Yang et al., 2005). Consequently, extensive studies on the structure of SARS-CoV-2 Mpro have been conducted rapidly following the COVID-19 outbreak (Anand et al., 2002; Yang et al., 2003). Jin et al., for the first time, reported the crystal structure of full-length SARS-CoV-2 Mpro in complex with N3 based on 2.1 Å X-ray diffraction data (Jin et al., 2020a). Almost at the same time, Zhang et al., determined the X-ray structures of the wild type SARS-CoV-2 Mpro protein at 1.75 Å, in addition to the structures of several inhibitor-bound Mpro proteins (Table 1) (Zhang et al., 2020).
All the aforementioned atomic structures of SARS-CoV-2 Mpro proteins are almost identical and highly similar to other Mpro proteins resolved previously (Ren et al., 2013; Tan et al., 2005; Wang et al., 2016; Xue et al., 2008), which suggests that the conserved protein is essential for the coronavirus family. Its overall structure is a dimer with a crystallographic 2-fold symmetry axis. Each protomer consists of three domains, Domains I (residues 8–101), II (residues 102–184) and III (residues 201–303), and a long loop region (residues 185–200) connects domains II and III (Fig. 2). The chymotrypsin-like and picornavirus 3C protease-like Domains I and II both form an antiparallel β-barrel structure, and the substrate-binding site is located in a cleft between them. Domain III is composed of five α-helices arranged into a largely antiparallel globular cluster, which forms a salt-bridge interaction between Glu290 of one monomer and Arg4 of the other monomer to mediate dimerization of the Mpro (Shi and Song, 2006). In addition, the dimer interface predominantly involves interactions between domain II of one protomer and the N-terminal residues of the other, which could facilitate the shaping of the substrate-binding pocket of Mpro protein (Fig. 2).
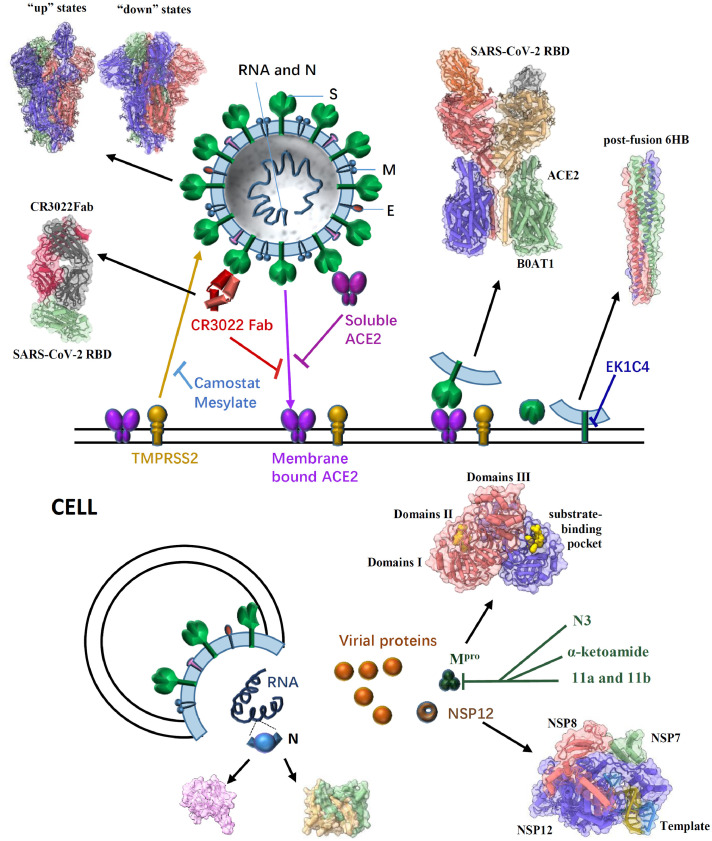
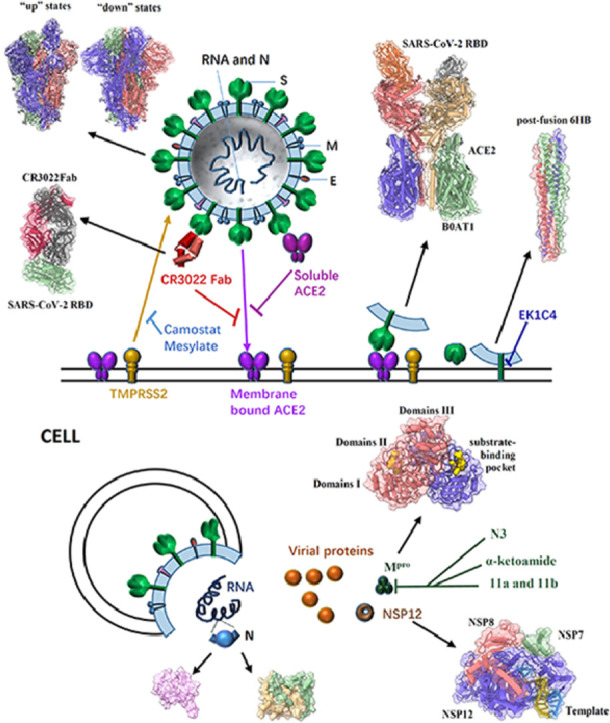
Fig. 2.
General structure of a SARS-CoV-2 virion, the pre- and post-fusion states of the SARS-CoV-2 S glycoprotein, and some effective inhibitors targeting different proteins to impair SARS-CoV-2 infection. SARS-CoV-2 enters host cells mainly through the trimeric S glycoprotein (PDB ID: 6VSB, 6VYB/6VXX) containing RBD which binds to the human receptor ACE2 (PDB ID: 6M17). A previously characterized SARS-CoV monoclonal antibody CR3022 could bind to the SARS-CoV-2 RBD (PDB ID: 6W41); the epitope can potentially confer in vivo protection. The recombinant soluble human ACE2 can bind to RBD of the S protein to block the pre-fusion stage of SARS-CoV-2 infection. The TMPRSS2 for S protein priming can be inhibited by camostat mesylate, which in turn would block SARS-CoV-2 infection. After receptor binding of the S1 subunit to support membrane fusion and viral infection, the S2 subunit is exposed and undergoes a conformational change from a pre-fusion to a post-fusion state (PDB ID: 6LXT). The inhibitor EK1C4 targets the S2 to inhibit viral infection. In cells, SARS-CoV-2 RNA translates to different proteins, such as NSP12 (PDB ID: 7BV1), N protein (PDB ID: 6M3M & 6WZO) and Mpro (PDB ID: 6Y2G). Several compounds (N3, α-ketoamide,11a and 11b) have been identified to exhibit high inhibitory activity against SARS-CoV-2 Mpro activity and viral infection.
SARS-CoV-2 Mpro is an attractive and key target for antiviral reagent discovery. Several compounds that inhibit Mpro activity have been identified through a combination of structure-based virtual screening and high-throughput screening. For example, Wu et al., discovered a series of clinical drugs and natural products with antiviral, antibacterial, and anti-inflammatory activities that showed high affinity to SARS-CoV-2 Mpro by artificial intelligence-assisted computer virtual screening (Wu et al., 2020c). As reported previously, N3 could specifically inhibit multiple CoV Mpros, including those from SARS-CoV and MERS-CoV (Yang et al., 2005; Ren et al., 2013; Wang et al., 2016; Xue et al., 2008). Jin et al., resolved the crystal structure of SARS-CoV-2 Mpro in complex with N3 and demonstrated that N3 was a potent irreversible inhibitor of SARS-CoV-2 Mpro. Subsequently, using structure-based virtual and high-throughput screening, they identified six promising antiviral compounds out of over 10,000 compounds, including approved drugs, drug candidates in clinical trials, and other pharmacologically active compounds as Mpro inhibitors (Jin et al., 2020b). Zhang et al., presented the crystal structure of SARS-CoV-2 Mpro in complex with an α-ketoamide inhibitor and developed the lead compound into a potent inhibitor based on the structure, which displayed obvious lung tropism and suitability for inhalation (Zhang et al., 2020). In addition, Dai et al., designed and synthesized two peptide-like compounds (11a and 11b) that exhibited high inhibitory activity against SARS-CoV-2 Mpro and viral infection. More importantly, both compounds showed better PK properties and lower toxicity in vivo, suggesting that they are promising drug candidates. Furthermore 1.5 Å high-resolution structures of SARS-CoV-2 Mpro in complex with 11a or 11b, revealed the precise interaction and molecular mechanism between Mpro and compounds (Dai et al., 2020).
3. The RNA-dependent RNA polymerase (RdRp, or NSP12)
Coronavirus employs a multi-subunit replication/transcription machinery of viral NSPs to synthesize viral RNA (Ziebuhr, 2005). The RNA-dependent RNA polymerase (RdRp, or NSP12) is the core component and plays a central role in SARS-CoV-2 replication and transcription cycle (Ahn et al., 2012; Velthuis et al., 2010). NSP7 and NSP8 increase RdRp activity in template-prime binding and processivity as accessary factors (Subissi et al., 2014; Kirchdoerfer and Ward, 2019). Therefore, RdRp is considered as the primary target for nucleotide analog antiviral inhibitors such as remdesivir (Holshue et al., 2020; Wang et al., 2020b; Warren et al., 2016), whose triphosphate form (RTP) is the active drug within cells (Siegel et al., 2017), and shows the potential for use in COVID-19 therapy. Therefore, determining the three-dimensional structures of SARS-CoV-2 RdRp with RNA template or inhibitors can provide a basis for the design of novel antiviral therapeutics (Table 1).
Gao et al., for the first time, reported the high-resolution cryo-EM structure of SARS-CoV-2 full-length NSP12 (residues 1–932) in complex with NSP7 (residues 1–83) and NSP8 (residues 1–198) cofactors at 2.9 Å (Gao et al., 2020a). It comprises one NSP12 monomer, one NSP8 monomer and one NSP7-NSP8 pair, similar to SARS-CoV RdRp complex (Kirchdoerfer and Ward, 2019). The structure of the SARS-CoV-2 NSP12 has three domains: a “right hand” RdRp domain (residues 367–920), a nidovirus-unique N-terminal extension domain (residues 4–28 and 69–249, also named as NiRAN domain) and an interface domain (residues 250–365) (Fig. 1). The RdRp domain displays the canonical arrangement of the viral polymerase family (McDonald, 2013) and consists of three subdomains: the finger subdomain (residues 366–581 and 621–679), the palm subdomain (residues 582–620 and 680–815), and the thumb subdomain (residues 816–920). The NiRAN domain adopts a nidovirus RdRp-associated nucleotidyl-transferase (NiRAN) configuration (Lehmann et al., 2015) with eight helices and a five-stranded β-sheet. The interface domain connects the other two domains with three helices and five β-strands (Fig. 2). Different from SARS-CoV's, the RdRp structure of SARS-CoV-2 has an additional N-terminal β-hairpin (residues 29–50) which locates in the groove clamped by the NiRAN domain and the palm subdomain to stabilize the overall structure. The overall structure is further stabilized by the binding of NSP7 and NSP8, with one NSP8 monomer sitting on top of the finger subdomain and the NSP7-NSP8 pair interacting with the thumb-finger subdomains interface (Fig. 2).
In a subsequent study, Yin et al. reported another two cryo-EM structures of the SARS-CoV-2 RdRp complex (Table 1). One is in apo form, at a resolution of 2.8 Å, and the other is in binding of a 50-base template-primer RNA and the monophosphate form of remdesivir (RMP) at a resolution of 2.5 Å (Yin et al., 2020). The overall structures of the two RdRp structures are highly similar to NSP12-NSP7-NSP8 complex solved by Gao et al. (2020a). In template-RMP RdRp complex, extensive protein-RNA interactions between the template-primer RNA and NSP12 were revealed, which explicates how the RdRp complex recognizes RNA rather than DNA, and suggests no sequence-specific RNA binding. RMP is covalently incorporated at the 3′ end of the primer strand and harbors the center of the catalytic active site, with extensive hydrogen bond networks. Two magnesium ions near the bound RMP form an active catalytic site. A pyrophosphate occupies the nucleotide entrance, which could make the active site inaccessible to nucleotide triphosphate (NTP) and in turn terminate replication process. In addition, Wang et al., reported near atomic-resolution structures of stalled pre-/post- translocated polymerase complexes, which revealed the molecular mechanism of SARS-CoV-2 RNA replication and its inhibition by remdesivir (Wang et al., 2020c).
4. Pre-fusion state of spike (S) protein
SARS-CoV-2 enters host cells mainly through the envelope-located trimeric spike (S) glycoprotein. S protein is translated as a large polypeptide, which is subsequently cleaved by host proteases to produce two functional subunits responsible for binding to the host cell receptor (N-terminal S1 subunit) and fusion of the viral and cellular membranes (C-terminal S2 subunit) (Bosch et al., 2003; Li, 2016; Walls et al., 2017). The receptor-binding domain (RBD) is located in the S1 subunit and can recognize human ACE2 receptor on target cell membranes (Figs. 1 and 2). In the pre-fusion state, the S1 subunit stabilizes the entire S protein trimer and protects the S2 protein from undergoing conformational change in advance. The S2 subunit contains a hydrophobic fusion peptide and two heptad repeat regions, which could form a six-helical bundle to mediate viral entry through fusion pores (Fig. 2).
The S protein is a key target for vaccines, inhibitors, neutralizing antibodies, and diagnostics, considering its indispensable function. In the infection process of CoVs, S is further cleaved by proteases (Madu et al., 2009; Millet and Whittaker, 2015); however, the cleaved S proteins always remain non-covalently associated in the metastable pre-fusion conformation (Belouzard et al., 2009; Burkard et al., 2014; Kirchdoerfer et al., 2016; Millet and Whittaker, 2014; Park et al., 2016; Tortorici and Veesler, 2019; Walls et al., 2016). Such cleavage has been proposed to activate the protein for membrane fusion via extensive irreversible conformational changes (Walls et al., 2017; Belouzard et al., 2009; Heald-Sargent and Gallagher, 2012). The entry into host cells of CoVs is very complicated that requires the concerted action of receptor-binding and proteolytic processing of the S protein to promote virus-cell fusion.
The single-particle cryo-EM structure of the SARS-CoV-2 spike trimer has recently been reported in two independent studies (Table 1) (Walls et al., 2020; Wrapp et al., 2020), both of which have demonstrated that the RBD can undergo a hinge-like conformational movement to shift between “up” or “down” states, as in other coronaviruses (Gui et al., 2017; Pallesen et al., 2017; Wrapp and McLellan, 2019; Yuan et al., 2017). The “down” conformation corresponds to the receptor-inaccessible state (hide), and the “up” conformation corresponds to a receptor accessible state (expose). Wrapp et al., for the first time, reported a 3.5 Å resolution cryo-EM structure of the SARS-CoV-2 S asymmetrical trimer, in which a single RBD is observed in the “up” conformation and two other RBDs are observed in the “down” conformation (Walls et al., 2020). The major difference between the two states is the distinct organization of the RBD within the S1 apex. Using the 3D variability feature in cryoSPARC v2 (Punjani et al., 2017), it is observed that RBD underwent a hinge-like conformational movement that transiently hide or expose the determinants of receptor binding, much like the breathing of S1 subunits.
Walls et al., also presented an asymmetric reconstruction of the trimer with a single S domain opened (the “up” conformation) at 3.2 Å resolution, and determined the closed (all “down” conformations) SARS-CoV-2 S ectodomain trimer at 2.8 Å resolution (applying 3-fold symmetry; Table 1) (Walls et al., 2020). Overall, in the “down” conformation, the SARS-CoV-2 S ectodomain is a 160 Å-long trimer with a triangular cross-section and the S1 subunit has a V-shaped architecture, resembling those of closely related β-coronaviruses, such as SARS-CoV S (Fig. 2).
5. Receptor recognition of S protein
During SARS-CoV infection, the RBD domain in its S protein has a core and a receptor-binding motif (RBM), which could directly bind to the peptidase domain (PD) of the angiotensin-converting enzyme 2 (ACE2), a type I membrane protein expressed in the lungs, heart, kidney, and intestine (Fig. 2) (Kirchdoerfer et al., 2018; Li et al., 2005; Song et al., 2018; Towler et al., 2004). Afterward, the S2 subunit is exposed and undergoes a conformational change from a pre-fusion to a post-fusion state. Therefore, binding to the ACE2 receptor is essential for viral infection (Belouzard et al., 2009; Simmons et al., 2005). Recent studies have demonstrated that SARS-CoV-2 S and SARS-CoV S share the same functional host cell receptor ACE2 (Li et al., 2003; Wan et al., 2020; Hoffmann et al., 2020a); however, the SARS-CoV-2 RBD has a significantly higher ACE2-binding affinity than SARS-CoV RBD, based on biochemical data (Wrapp et al., 2020; Wang et al., 2020d).
Yan et al., for the first time, reported cryo-EM structures of full-length human ACE2 in complex with a neutral amino acid transporter B0AT1 at 2.9 Å as well as the complex structure of SARS-CoV-2 RBD bound ACE2 at an overall resolution of 2.9 Å (local resolution of the ACE2-RBD interface was 3.5 Å) (Table 1) (Yan et al., 2020). The ACE2-B0AT1 complex is assembled as a dimer of heterodimers, in which the collectrin-like domain of ACE2 is the key mediator of homo-dimerization, and the extracellular PD contributes a minor interface. According to the rotation state of the PD, the ACE2-B0AT1 complex displays open or closed conformations, whereas the rest of the complex is largely unaltered. However, in the RBD-ACE2-B0AT1 ternary complex, only the closed state of ACE2 is observed.
In a separate study, Lan et al., determined the crystal structure of the wild-type SARS-CoV-2 RBD in complex with ACE2 at 2.45 Å resolution (Lan et al., 2020). Similarly, Shang et al., resolved the crystal structure of the SARS-CoV-2 RBD (a chimera protein contains the SARS-CoV RBD core and the SARS-CoV-2 RBM) bound to the ACE2 at 2.68 Å resolution (Table 1) (Shang et al., 2020). The overall structures of the SARS-CoV-2 RBD/ACE2 complexes are all similar to that of SARS-CoV, in which the RBM motif forms a gently concave surface with a ridge on one side and contacts the exposed outer surface of the claw-like (or arch shaped) structure of ACE2. Such atomic-level structural information provides important insights into the molecular basis for coronavirus recognition and infection, and could facilitate ongoing vaccine design and inhibitor screening efforts.
The S proteins of SARS-CoV-2 and SARS-CoV share a high sequence similarity of approximately 77% (Zhou et al., 2020); therefore, cross-reactive epitopes and neutralizing antibodies may exist. A previously characterized SARS-CoV monoclonal antibody CR3022 could bind to the SARS-CoV-2 RBD with a KD of 6.2 nM. Yuan et al., determined the crystal structure of CR3022 in complex with the SARS-CoV-2 RBD at 3.1 Å, and they observed that CR3022 targets a highly conserved epitope away from the ACE2-binding site (Fig. 2). Structural modeling further demonstrates that the epitope of CR3022 is only accessible when at least two RBD on the trimeric S protein are in the “up” conformation and slightly rotated so that all clashes can be avoided (Yuan et al., 2020).
Proteins that interact with SARS-CoV-2 S are also important targets for the design of novel antiviral therapies. Endosomal cysteine proteases-cathepsin B and L (CatB/L) (Simmons et al., 2005) and serine protease TMPRSS2 (Glowacka et al., 2011; Matsuyama et al., 2010; Shulla et al., 2011) can be used for SARS-CoV S priming in cell lines. However, only TMPRSS2 activity is essential for viral entry into primary target cells and for viral spread in infected hosts, whereas CatB/L activity is dispensable (Iwata-Yoshikawa et al., 2019; Shirato et al., 2017; Shirato et al., 2018; Zhou et al., 2015). Hoffmann et al., demonstrate that the SARS-CoV-2 S protein is primed by TMPRSS2, and camostat mesylate, a TMPRSS2 inhibitor, could block the infection of lung cells with SARS-CoV-2 and is a potential treatment option (Fig. 2) (Hoffmann et al., 2020b). Recently, it has been demonstrated that recombinant soluble human ACE2 (hrsACE2) can block the early stages of SARS-CoV-2 infection significantly. Clinical-grade hrsACE2 can reduce SARS-CoV-2 recovery from Vero cells by a factor of 1000–5000, and block SARS-CoV-2 infections in engineered human blood vessel organoids and human kidney organoids (Monteil et al., 2020).
6. Post-fusion state of spike (S) protein
After the binding of the S1 subunit to the ACE2 receptor, the S2 subunit undergoes conformational changes to insert the hydrophobic fusion peptide into the host target cell membrane. The heptad repeat 1 (HR1) domain in the S2 subunit forms a homotrimer, which exposes three highly conserved hydrophobic grooves on the surface that binds the heptad repeat 2 (HR2) domain in the S2 subunit; therefore, HR1 and HR2 domains interact to form a stable six-helix bundle (6-HB) fusion core. The 6-HB fusion core structure helps bring the viral and cellular membranes into close proximity for membrane fusion and viral infection (Xia et al., 2020a). SARS-CoV-2 and SARS-CoV S2 subunits share high sequence similarities, with 92.6% and 100% overall identities in HR1 and HR2 domains, respectively. Xia et al., found that HR1and HR2 domains in SARS-CoV-2 can interact to form 6-HB (Xia et al., 2020a), and they resolved the X-ray crystal structure of SARS-CoV-2′s 6-HB (Xia et al., 2020b). The overall 6-HB structure of SARS-CoV-2 is similar to that of SARS-CoV (Xia et al., 2019). Although the amino acid sequences of the HR2 domain from SARS-CoV-2 and SARS-CoV are identical, the HR1 domain has eight different residues, which may enhance HR1 and HR2 interactions and stabilize 6-HB conformation.
Over the past few decades, the viral HR1 domain in the S2 subunit has been proved to be another important target for the development of viral fusion and entry inhibitors. The pan-coronavirus entry inhibitor EK1 peptide has been demonstrated to target the HR1s of SARS-CoV and MERS-CoV and to inhibit viral infection (Xia et al., 2020a; Liu et al., 2004; Lu et al., 2014). Xia et al., generated several lipopeptides by conjugating the cholesterol molecule to the EK1 peptide. One of the lipopeptides, EK1C4, possesses the most potent inhibitory activity and is approximately 241 and 149 times more potent than the original EK1 peptide against SARS-CoV-2 S protein-mediated membrane fusion and pseudovirus infection with IC50s of 1.3 and 15.8 nM, respectively (Fig. 2). EK1C4 has also showed broad-spectrum inhibitory activity against infection of SARS-CoV, MERS-CoV and other HCoVs, suggesting EK1C4 can be used for the prevention and treatment of SARSr-CoVs infection (Xia et al., 2020b).
7. Nucleocapsid (N) protein
The nucleocapsid (N) protein of coronavirus is a multifunctional RNA-binding protein and forms a helical filament structure that is required for the assembly of viral genomic RNA into a ribonucleoprotein complex. The packaging of the viral genomic RNA regulates viral replication/transcription and modulates infectious cell metabolism (Cong et al., 2020; Masters and Sturman, 1990; McBride et al., 2014; Nelson et al., 2000; Stohlman et al., 1988). The N protein of SARS-CoV-2 is highly expressed and induces early antibody responses in infected patients, similar to in the case of SARS-CoV N protein (Ahmed et al., 2020; Liu et al., 2006; Shang et al., 2005; Hachim et al., 2020). Consequently, an enhanced understanding of the structural and functional features of SARS-CoV-2 N protein could facilitate the development of sensitive and specific immunological tests.
The common domain architecture of beta-coronavirus N protein encompasses several conserved parts: an ordered RNA-binding (N1b) domain, a dimerization (N2b) domain, and several short regions with high predicted disorder (N1a, N2a, and spacer B/N3; Fig. 1). The structures of CoVs N1b have been extensively reported, and some key residues involved in RNA binding and influencing virus infectivity have been identified (Grossoehme et al., 2009; Keane et al., 2012; Saikatendu et al., 2007; Tan et al., 2006). Kang et al., determined the crystal structure of the SARS-CoV-2 N1b domain at 2.7 Å resolution and revealed distinct surface electrostatic potential characteristics between them (Kang et al., 2020) (Table 1). The overall structure is a ‘right-hand shape’ composed of a β-hairpin, a β-sheet core and a long-loop region, which resembles a protruded basic finger, a basic palm, and an acidic wrist. The β-sheet core contains five antiparallel β-strands with a short α-helix and the long β-hairpin is a large protruding loop between β2 and β5 (Fig. 2).
The N2b domain mediates self-association into a dimer, which subsequently assembles into a higher-order helical filament, potentially involving cooperative interactions. Ye et al., reported two crystal structures of the SARS-CoV-2 N2b domain, exhibiting a tightly intertwined dimer that is similar to that of SARS-CoV (Table 1) (Chen et al., 2007; Takeda et al., 2008; Yu et al., 2006; Ye et al., 2020). The dimer interface consists of two β-strands and a short α-helix from each protomer that extend toward the opposite protomer and pack against its hydrophobic core (Fig. 2). In addition, a combination of multi-angle light scattering and hydrogen-deuterium exchange mass spectrometry techniques has been used to explore the self-assembly mechanism of the SARS-CoV-2 nucleocapsid. Such studies enable researchers to identify small molecules that could inhibit nucleocapsid self-assembly processes, and in turn minimize the infection severity and pathogenic infectivity.
8. Conclusion
Following the COVID-19 outbreak, SARS-CoV-2 was rapidly identified as the etiological agent. Although our understanding of coronaviruses has improved substantially following previous SARS-CoV and MERS-CoV outbreaks, in 2003 and 2012, respectively, effective antiviral drugs for SARS-CoV-2 treatment and vaccines for its prevention remain unavailable. Extensive studies on the structures and functions of the major proteins involved in SARS-CoV-2 has been conducted, which have enhanced our general understanding of the mechanisms of its infection and replication. Furthermore, combined with structure-based predictions, experiments at cellular and animal levels in addition to high-throughput screening activities have revealed numerous compounds, including clinical drugs, drug candidates, and other pharmacologically active compounds, to be inhibitors of viral key proteins; these are potential antiviral drugs for the treatment of COVID-19 disease.
The atomic structures of SARS-CoV-2 proteins not only help to research and develop antiviral drugs, but also provide important basis for vaccine design. Among four major structural proteins, the S glycoprotein plays crucial roles in viral entry and pathogenesis, which is a key target for vaccines, but it is highly glycosylated and antigenically variable. 20 N-linked glycosylation sequons per protomer are conserved among SARS-CoV-2 S and SARS-CoV S (Walls et al., 2020), and these oligosaccharides might modulate antibody recognition in SARS-CoV (Pallesen et al., 2017; Walls et al., 2019). Consequently, the precise structure of the S glycoprotein and the organization of these N-Linked Glycans will provide atomic-level information to help the design and development of vaccines. At present, multiple SARS-CoV-2 vaccine types are under development, including inactivated or weakened vaccines, viral vectored vaccines, DNA- or RNA-based vaccines, adenovirus-based vaccines and protein-based vaccines, which are based on the S antigen (Callaway, 2020; Gao et al., 2020b; Wu, 2020).
This review is a brief summary of recent developments toward the understanding of the structural and functional features of major proteins involved in SARS-CoV-2 infection and replication and corresponding drug development activities. Such molecular insights on SARS-CoV-2 infection and replication mechanisms could facilitate the effective clinical treatment and epidemiological control of the COVID-19 disease.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Ping Shan and Ruigang Su (F.S. lab) for their assistance in lab management. This work was supported by grants from National Natural Science Foundation of China (31830020) and National Key Research and Development Program of China (2017YFA0504700 and 2018YFA0901102).
Contributor Information
Yun Zhu, Email: zhuyun@ibp.ac.cn.
Fei Sun, Email: feisun@ibp.ac.cn.
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12 doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.G., Choi J.K., Taylor D.R., Oh J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J., Bosch B.J., de Haan C.A. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580:576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Ulasli M., Schepers H., Mauthe M., V'Kovski P., Kriegenburg F., Thiel V., de Haan C.A.M., Reggiori F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Rapid development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., 3rd Leibowitz JL, Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachim A., Kavian N., Cohen C.A., Chin A.W., Chu D.K., Mok C.K.P., Tsang O.T., Yeung Y.C., Perera R.A., Poon L.L., et al. Beyond the Spike: identification of viral targets of the antibody response to SARS-CoV-2 in COVID-19 patients. medRxiv. 2020 [Google Scholar]
- Hayden F.G., Turner R.B., Gwaltney J.M., Chi-Burris K., Gersten M., Hsyu P., Patick A.K., Smith G.J., 3rd, Zalman L.S. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003;47:3907–3916. doi: 10.1128/aac.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020 [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Song C., Xu C., Jin G., Chen Y., Xu X., Ma H., Chen W., Lin Y., Zheng Y., et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci. China Life Sci. 2020;63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Xia J., Chen Y., Shan C., Wu C. A family cluster of SARS-CoV-2 infection involving 11 patients in Nanjing, China. Lancet Infect. Dis. 2020;20:534–535. doi: 10.1016/S1473-3099(20)30147-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019 doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S.C., Liu P., Leibowitz J.L., Giedroc D.P. Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. J. Biol. Chem. 2012;287:7063–7073. doi: 10.1074/jbc.M111.287763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Liu H., Kankanamalage A.C., Weerasekara S., Hua D.H., Groutas W.C., Chang K.O., Pedersen N.C. Correction: reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., et al. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J., Leng C.H., Lien S.P., Chi H.Y., Huang C.Y., Lin C.L., Lian W.C., Chen C.J., Hsieh S.L., Chong P. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Fang Y.Y., Deng Y., Liu W., Wang M.F., Ma J.P., Xiao W., Wang Y.N., Zhong M.H., Li C.H., et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin. Med. J. (Engl.) 2020;133:1025–1031. doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y., Wang Q., Chan J.F., Du L., Yu F., et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Sturman L.S. Background paper. Functions of the coronavirus nucleocapsid protein. Adv. Exp. Med. Biol. 1990;276:235–238. doi: 10.1007/978-1-4684-5823-7_32. [DOI] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald S.M. Vol. 4. 2013. RNA synthetic mechanisms employed by diverse families of RNA viruses; pp. 351–367. (Wiley Interdiscip. Rev. RNA). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G.W., Stohlman S.A., Tahara S.M. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J. Gen. Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Jr., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. USA. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Ren Z., Yan L., Zhang N., Guo Y., Yang C., Lou Z., Rao Z. The newly emerged SARS-like coronavirus HCoV-EMC also has an "Achilles' heel": current effective inhibitor targeting a 3C-like protease. Protein Cell. 2013;4:248–250. doi: 10.1007/s13238-013-2841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B., Wang X.Y., Yuan J.W., Vabret A., Wu X.D., Yang R.F., Tian L., Ji Y.Y., Deubel V., Sun B. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem. Biophys. Res. Commun. 2005;336:110–117. doi: 10.1016/j.bbrc.2005.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273:1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J. Virol. 2017 doi: 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-Nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohlman S.A., Baric R.S., Nelson G.N., Soe L.H., Welter L.M., Deans R.J. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J. Virol. 1988;62:4288–4295. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M., Chang C.K., Ikeya T., Guntert P., Chang Y.H., Hsu Y.L., Huang T.H., Kainosho M. Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J. Mol. Biol. 2008;380:608–622. doi: 10.1016/j.jmb.2007.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Verschueren K.H., Anand K., Shen J., Yang M., Xu Y., Rao Z., Bigalke J., Heisen B., Mesters J.R., et al. pH-dependent conformational flexibility of the SARS-CoV main proteinase (M(pro)) dimer: molecular dynamics simulations and multiple X-ray structure analyses. J. Mol. Biol. 2005;354:25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Chen C., Tan W., Yang K., Yang H. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 2016;6:22677. doi: 10.1038/srep22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020 doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., McLellan J.S. The 3.1-angstrom cryo-electron microscopy structure of the porcine epidemic diarrhea virus spike protein in the prefusion conformation. J. Virol. 2019;93 doi: 10.1128/JVI.00923-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Ho W., Huang Y., Jin D.Y., Li S., Liu S.L., Liu X., Qiu J., Sang Y., Wang Q., et al. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet. 2020;395:949–950. doi: 10.1016/S0140-6736(20)30557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.C. Progress and concept for COVID-19 vaccine development. Biotechnol. J. 2020;15 doi: 10.1002/biot.202000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.K., Wang Q., Du L., Tan W., Wilson I.A., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav4580. eaav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol. Immunol. 2020 doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. bioRxiv. 2020 doi: 10.1101/2020.05.17.100685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Jr., Nunneley J.W., Barnard D., Pohlmann S., McKerrow J.H., Renslo A.R., et al. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]