Highlights
-
•
Serological tests showed poor performances in early serum samples but improved in days 8–15 after onset of symptoms.
-
•
IgM band in LFI had low sensitivities and might be inadequate for acute diagnosis of COVID-19 infection.
-
•
Euroimmun commercial IgA ELISA was more sensitive but less specificic than IgG, which showed 100 % specificity.
-
•
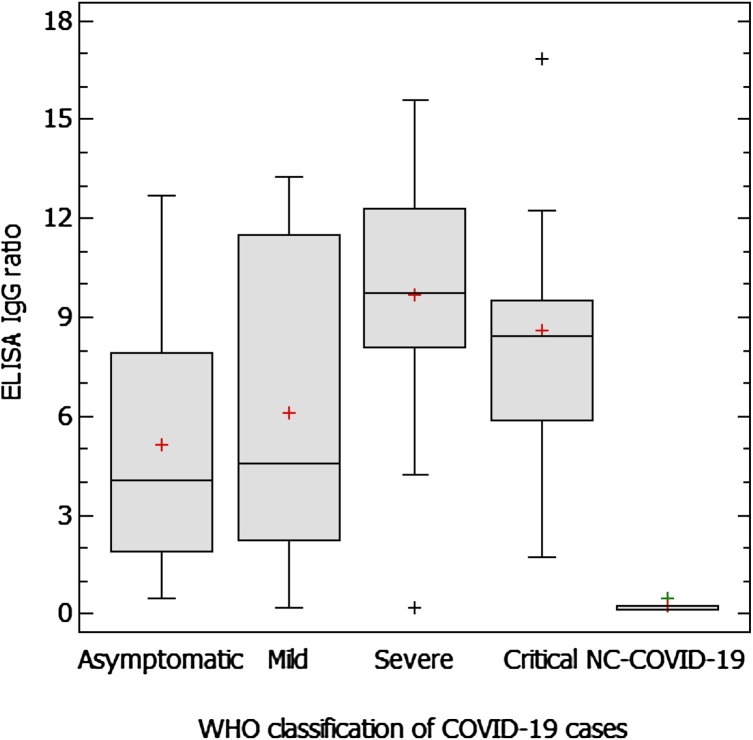
Ten days after symptoms’ onset, critical and severe COVID-19 patients had higher ELISA IgG ratios than asymptomatic or mild ones.
Keywords: SARS-CoV-2, COVID-19, Diagnosis, Antibody detection, Sensitivity, Specificity
Abstract
Background
COVID-19 pandemic has spread worldwide since December 2019. Serological tests for SARS-CoV-2 antibody testing are needed for detection of current or past infections. A wide range of commercial tests is available. However, most of them need to be validated.
Study design
The aim was to compare a commercial IgG and IgA ELISA (Euroimmun) with three lateral flow immunoassays (LFI): Hangzhou Alltest Biotech, Wuhan UNscience Biotechnology and Guangzhou Wondfo Biotech. Specificity was calculated with 62 available serum samples from 2018/19. The study included 152 sera from patients of which 109 were RT-PCR positive. Sensitivities for ELISA anti SARS-CoV-2 IgG and IgA were 81.5 % and 93.1 % and specificities 100 % and 80.6 %, respectively. LFI showed variable performances, overall results being better for Guangzhou Wondfo Biotech.
Conclusions
Commercial serological tests are useful for detection of antibodies in patients with COVID-19. ELISA presented better results than LFI. The results allowed to incorporate the most sensitive LFI to the daily workflow, combining with ELISA. Careful validation is encouraged before clinical laboratories start using these tests.
1. Introduction
On December 30th, 2019 the first few cases of a novel acute respiratory infectious disease were declared in Wuhan, China [1], which were promptly associated with a new beta-coronavirus, SARS-CoV-2, causing a disease that was later named COVID-19 [2]. Following the alarming increase of cases in and outside the country, the WHO declared the outbreak a pandemic on March 11th, 2020 [3]. Currently, COVID-19 has affected over 5 million people causing 340.000 deaths worldwide [4].
Reverse real-time PCR (RT-PCR) techniques have emerged as the (gold) standard diagnostic test for COVID-19 [5]. Nevertheless, in some situations, the sensitivity of RT-PCR tests has been worse than desired due to particular issues: variable viral loads depending on sample types and time of infection (i.e. nasopharyngeal vs. oropharyngeal, upper vs. lower respiratory tract); sample collection, conservation and transport; different gene targets [6]. In some of those “high-clinical-suspicion-RT-PCR-negative cases”, antibodies detection could be a helpful tool in COVID-19 diagnosis [[7], [8], [9], [10], [11]]. Serology plays a key role in contact tracing, epidemiological/seroprevalence studies, identification of convalescent plasma donors and evaluation of immune response to vaccines.
Due to the presumed asymptomatic cases and the lack of large population studies, real seroprevalence remains unknown and is urgently needed to control the pandemic and to know the reliable infection rates. Multiple SARS-CoV-2 antibody detection tests have been commercialised in a short period of time with minimal validation requirements due to urgent need. Most of them detect IgM, IgA and/or IgG against the nucleocapsid protein (NP) or different domains of the spike glycoprotein (S1, S2 and RBD). Good performance has been shown to date with commercialised or in-house Enzyme-linked Immunosorbent Assay (ELISA) tests [7,8,10,12,13]. However, there is much concern about lateral flow immunoassay (LFI) tests, which are widespread due to their easy and fast performance but with no available proven sensitivity and specificity [13].
In this study, we aimed at comparing two commercial ELISA assays with three LFI tests to detect SARS-coV-2 antibodies.
2. Materials and methods
A total of 152 serum samples submitted to our laboratory for SARS-CoV-2 antibodies detection between 15th March and 23rd April 2020 from 130 patients were included in the study. We tested Euroimmun ELISA anti SARS-CoV-2 S1 domain IgA and IgG antibodies (Euroimmun Medizinische Labordiagnostika, Lübeck, Germany) and three LFI: Test 1 (Hangzhou Alltest Biotech Co., Ltd.), Test 2 (Wuhan UNscience Biotechnology Co., Ltd.), both with separated bands for IgM and IgG antibodies, and Test 3 (Guangzhou Wondfo Biotech Co., Ltd.), which detects total antibodies in a single band. Sixty-two sera from Jan–March 2018 and 2019, considered to be negative for SARS-CoV-2, were tested to calculate specificity. All tests were performed according to manufacturer’s instructions.
3. Results
One hundred and nine patients were microbiologically confirmed as COVID-19 cases (109/130, 84 %) since RT-PCR from nose/throat swab or other respiratory tract samples and/or IgG tested positive. Asymptomatic patients were detected by contact tracing. Twenty-one patients were not confirmed to be infected by SARS-CoV-2 (NC-COVID-19) after at least two RT-PCR and antibodies negative results.
Demographic data and severity of symptoms, according to the WHO criteria, are shown in Table 1 . Six cases (5.5 %) were diagnosed by serological assays. ELISA IgG ratios in different illness severity groups (>10 days after the onset of symptoms) and NC-COVID-19 are shown in Fig. 1 . Interestingly, the ANOVA test resulted in statistically significant differences between medians of asymptomatic/mild vs severe/critical pair of groups (5.1/6.1 vs. 9.7/8.6, respectively, p ≤ 0.05).
Table 1.
Demographic data according to WHO classification criteria of COVID-19.
| COVID-19 |
NC-COVID-19 | ||||
|---|---|---|---|---|---|
| Asymptomatic | Mild | Severe | Critical | ||
| No patients (%*) | 11 (10) | 54 (50) | 21 (19) | 23 (21) | 21 |
| Mean age, years (range) | 53 (9−93) | 65 (32−100) | 63 (31−87) | 70 (46−93) | 50 (18−91) |
| Gender, male/female (%) | 1/10 (9/91) | 26/28 (48/52) | 10/11 (4852) | 15/8 (65/35) | 9/12 (43/57) |
| SARS-CoV-2 RNA RT-PCR | |||||
| RNA detected (%**) | 3 (27) | 50 (93) | 20 (95) | 22 (96) | 0 |
| Med. days from symptom onset to 1st (range) | N/A | 7 (1−33) | 6 (2−26) | 7 (2−32) | N/A |
| Serology | |||||
| No of samples | 13 | 65 | 28 | 24 | 22 |
| Med. days from symptom onset (range) | N/A | 12 (2−30) | 21 (4−35) | 12 (2−22) | 7 (2−20) |
Percentages are calculated considering only the total number of SARS-CoV-2-confirmed positive patients (N = 109).
Percentage of RT-PCR positive result in each group of patients. N/A: not applied.
Fig. 1.
ELISA IgG ratios in different WHO classification COVID-19 groups of patients (over 10 days after onset of symptoms) and NC-COVID-19.
Fig. 1 Caption: Median values of the different groups were compared by the ANOVA test with 95 % of confidence interval. Statistically significant differences (p ≤ 0.05) were found among all the groups, except between “Asymptomatic” vs “Mild”, and “Severe” vs “Critical”.
Specificity results of the 62 sera from 2018/19 are shown in Table 2 . We also calculated the overall specificity, including the negative samples (N = 22) submitted to discard COVID-19. One sample was from a patient with a previous positive human Coronavirus OC43 RT-PCR result the week before.
Table 2.
Specificity of the 3 lateral flow immunoassays and the 2 ELISA assays for all and pre-COVID-19 (2018-2019) only serum samples.
| Lateral Flow Immunoassay |
ELISA |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Test 1 | Test 2 | Test 3 | |||||||
| Type of antibodies | IgM | IgG | IgM/IgG | IgM | IgG | IgM/IgG | IgM/IgG | IgA | IgG |
| No. tested samples (all) | 84 | 84 | 84 | 81 | 81 | 81 | 84 | 84 | 84 |
| Negative | 79 | 83 | 78 | 73 | 78 | 71 | 80 | 69 | 84 |
| Inconclusive/positive | 5 | 1 | 6 | 8 | 3 | 10 | 4 | 15 | 0 |
| Specificity (%) | 94 % | 98.8 % | 92.8 % | 90.1 % | 96.3 % | 87.7 % | 95.2 % | 81.2 % | 100 % |
| No. tested samples 2018/19 | 62 | 62 | 62 | 60 | 60 | 60 | 62 | 62 | 62 |
| Negative | 58 | 62 | 58 | 54 | 59 | 53 | 62 | 50 | 62 |
| Inconclusive/positive | 4 | 0 | 4 | 6 | 1 | 7 | 0 | 12 | 0 |
| Specificity (%) | 93.5 % | 100 % | 93.5 % | 90 % | 98.3 % | 88.3 % | 100 % | 80.6 % | 100 % |
Fifteen samples gave ratio values above the manufacturer’s cut-off (≥0.8) in the IgA ELISA. To improve specificity, we determined a new cut-off (OD ratio = 1.56) to be 3 times the mean value of the pre-COVID-19 serum samples [12]. We established an in-house grey-zone, 1.07–1.56, using the cut-off confidence interval and applied these new values to our samples: all NC-COVID-19 IgA ratio values were below the cut-off. Nine confirmed cases resulted IgA negative although six corresponded to early samples (≤7 days). IgG assay though showed 100 % specificity.
LFI specificity values varied: IgG bands in Tests 1 and 2 showed better results than their respective IgM bands. Combining IgG and IgM results, Test 3 showed the best specificity (100 %), followed by Test 2 and 1.
The ELISA and LFI sensitivities were overall calculated for different weeks after symptoms’ onset (Table 3 ). The IgA ELISA assay was the most sensitive test the first week after onset, although 1 out of 3 patients could have been misdiagnosed at this early stage of infection. Both IgA ELISA and Test 3 showed much better results between 8 and 14 days after onset. IgG could be detected by ELISA and Test 2 in most samples. From day 15 after onset, seroconversion was detected in almost all patients. ELISA IgG, LFI Test 3 and IgG band of Test 2 were positive for 96 %, 98 % and 100 % of COVID-19 patients respectively. Asymptomatic patients were poorly diagnosed by LFI IgM bands alone. ELISA IgA correctly detected all cases in this group. The results showed Test 1 to be less sensitive in all groups.
Table 3.
Sensitivity of the 3 LFI and 2 ELISA for different days after symptoms’ onset.
| Days after onset | ELISA Sensitivity (95 %CI) |
Lateral Flow Immunoassay Sensitivity (95 %CI) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Test 1 |
Test 2 |
Test 3 |
||||||||||||
| N | IgA | N | IgG | N | IgM | N | IgG | N | IgM | N | IgG | N | IgM/IgG | |
| 1−7 | 22/28 | 71.4 (63.4−93.8) | 15/28 | 53.5 (33.9−72.5) | 8/27 | 29.6 (13.8−50.2) | 13/27 | 48.1 (28.7−68.1) | 16/24 | 66.7 (44.7−84.4) | 14/24 | 58.3 (36.6−77.9) | 18/27 | 66.7 (46−83.5) |
| 8−14 | 38/39 | 97.4 (86.5−99.9) | 32/39 | 82.1 (66.5−92.5) | 17/39 | 43.6 (27.8−60.4) | 28/39 | 71.8 (55.1−85) | 24/32 | 75 (56.6−88.5) | 30/32 | 93.8 (79.2−99.2) | 38/39 | 97.4 (86.5−99.9) |
| 15−28 | 49/50 | 98 (89.4−99.9) | 48/50 | 96 (86.3−99.5) | 12/49 | 24.5 (13.3−38.9) | 41/49 | 83.7 (70.3−92.7) | 23/25 | 92 (74−99) | 25/25 | 100 (86.3−100) | 49/50 | 98 (89.4−99.9) |
| Asymptomatic | 13/13 | 100 (75.3−100) | 12/13 | 92.3 (64−99.8) | 4/11 | 36.3 (10.9−69.2) | 7/11 | 63.6 (30.8−89.1) | 7/11 | 63.6 (30.8−89.1) | 10/11 | 90.9 (58.7−99.8) | 11/13 | 84.6 (54.6−98.1) |
| Total | 121/130 | 93.1 (87.3−96.8) | 106/130 | 81.5 (73.8−87.8) | 41/126 | 32.5 (24.5−41.5) | 89/126 | 70.6 (61.9−78.4) | 70/92 | 76.1 (66.1−84.4) | 79/92 | 85.9 (77−92.3) | 116/129 | 89.9 (83.4−94.5) |
4. Discussion
Our results suggest that commercial ELISA assays and LFI tests can be used as complementary tools in COVID-19 diagnosis. Antibody testing allowed us to diagnose some COVID-19 cases with repeated negative RT-PCR results. As previously reported, we detected high sensitivity in serological assays from day eight after symptoms’ onset compared to the first week of illness [7,8,10]. We found significant differences in levels of IgG, between asymptomatic/mild and severe/critical patients, consistent with others reports [7,11], jeopardising long-term detection of asymptomatic/mild cases among the population, if the IgG level trend decreases over time.
Euroimmun IgA assay was less specific but more sensitive than IgG. Even if our IgA specificity value was better than was previously found [[12], [13], [14]], we decided to use a more restrictive cut-off value. This way, we consider it is a reliable early marker of COVID-19 infection, although it always needs to be confirmed by a posterior serum sample to detect seroconversion. The excellent specificity of IgG assay has led us to use it as a confirmation test for SARS-CoV-2 antibody testing.
We found variable performance of LFI tests as elsewhere [13]. For this reason, it might not be a sufficient strategy to use LFI tests alone for COVID-19 diagnosis. Instead, we decided to use Test 3, with acceptable specificity and sensitivity, combined with ELISA as a part of our daily workflow. For Test-3 positive samples we perform a second LFI test to obtain a preliminary and fast report, differentiating IgM and IgG, which is always confirmed by ELISA assay afterwards.
Nevertheless, our results are provided for diagnostic purposes in a specific pandemic situation and so predictive values of the assays may change depending on the COVID-19 prevalence. The estimated COVID-19 incidence rate in our province for this period was 242.2 cases/100,000 inhabitants [15]. To date only prevalence estimations have been performed [16] and some population studies are currently undergoing to accurately determine it. Little is known about serological response to SARS-CoV-2 or whether individuals who develop antibodies after infection remain protected against subsequent re-infection. Further antibodies studies are needed to better understand COVID-19 spread. We strongly recommend serological tests to be validated by specialists before being used in clinical laboratories.
Funding statement
None declared.
CRediT authorship contribution statement
Maria Martínez Serrano: Conceptualization, Methodology, Investigation, Validation, Data curation, Writing - original draft, Writing - review & editing. David Navalpotro Rodríguez: Conceptualization, Methodology, Investigation, Validation, Data curation, Writing - original draft, Writing - review & editing. Nuria Tormo Palop: Conceptualization, Methodology, Investigation, Validation, Data curation, Writing - original draft, Writing - review & editing. Roberto Olmos Arenas: Investigation, Methodology. Marta Moreno Córdoba: Investigation, Methodology. Mª Dolores Ocete Mochón: Methodology, Data curation. Concepción Gimeno Cardona: Supervision, Validation, Writing - review & editing.
Declaration of Competing Interest
None declared.
References
- 1.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Director-General’s opening remarks at the media briefing on COVID-19 -. 11 March 2020.
- 4.World Health Organization. Coronavirus disease (COVID-19) Situation Report-123, 2020.
- 5.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020 doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020:1–22. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q., Xu S., Zhu H., Xu Y., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020:1–8. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X., zhu Q., Liu L. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C.Y., Cai J.P., Chan J.M.C., Chik T.S.H., Lau D.P.L., Choi C.Y.C., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C.K., Poon R.W.S., Luo C.T., Cheng V.C.C., Chan J.F.W., Hung I.F.N., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020:1–4. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 12.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lassaunière R., Frische A., Harboe Z.B., Nielsen A.C., Fomsgaard A., Krogfelt K.A., Jørgensen C.S. Evaluation of nine commercial SARS-CoV-2 immunoassays. MedRxiv. 2020 doi: 10.1101/2020.04.09.20056325. 2020.04.09.20056325. [DOI] [Google Scholar]
- 14.Jääskeläinen A.J., Kekäläinen E., Kallio-kokko H., Mannonen L., Kortela E., Vapalahti O. Evaluation of commercial and automated SARS- CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Eurosurveillance. 2020:1–8. doi: 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Informe resumen COVID-19 . 2020. RedMIVA y AVE. Descriptivo RedMIVA Curva epidémica por Fecha primer resultado de laboratorio. [Google Scholar]
- 16.Ceylan Z. Estimation of COVID-19 prevalence in Italy, Spain, and France. Sci. Total Environ. 2020;729:138817. doi: 10.1016/j.scitotenv.2020.138817. [DOI] [PMC free article] [PubMed] [Google Scholar]