Abstract
Alcohol has been classified as carcinogenic to humans by the International Agency for Research on Cancer (IARC). Studies have demonstrated that alcohol intake increases the risk of breast cancer, and alcohol also stimulates breast cancer cell growth. Deregulation of Pol III genes is tightly associated with tumour development. Transcription factor II-B (TFIIB)-related factor 1 (Brf1) is a transcription factor that specifically regulates Pol III gene transcription. Our in vivo and in vitro studies have indicated that alcohol enhances the transcription of Pol III genes to cause an alteration of cellular phenotypes, which is closely related with human breast cancer. Betaine is a vegetable alkaloid and has antitumour functions. Most reports about betaine show that the consumption level of betaine is inversely associated with a risk of breast cancer. Although different mechanisms of betaine against tumour have been investigated, nothing has been reported on the effect of betaine on the deregulation of Brf1 and Pol III genes. In this study, we determine the role of betaine in breast cancer cell growth and colony formation and explore its mechanism. Our results indicate that alcohol increases the rates of growth and colony formation of breast cancer cells, whereas betaine is able to significantly inhibit the effects of alcohol on these cell phenotypes. Betaine decreases the induction of Brf1 expression and Pol III gene transcription caused by ethanol to reduce the rates of cell growth and colony formation. Together, these studies provide novel insights into the role of betaine in alcohol-caused breast cancer cell growth and deregulation of Brf1 and Pol III genes. These results suggest that betaine consumption is able to prevent alcohol-associated human cancer development.
Keywords: Betaine, Alcohol, Cell growth, Colony formation, Brf1, Pol III genes
1. Introduction
Cancer seriously endangers human health and breast cancer causes deadly harm in women’s lives (1). Alcohol has been classified as carcinogenic to humans by the International Agency for Research on Cancer (IARC) (2–4). Alcohol consumption is associated with human cancers in different organs, such as breast, liver, stomach, pancreas, oral cavity, pharynx, oesophagus, larynx, colon, and ovary (4–9). Our previous study have showed that alcohol intake is associated with human breast cancer and alcohol stimulates breast cancer cell growth (4, 6). These studies are consistent with Baglia’s report (10).
Betaine is a vegetable alkaloid (11). Betaine plays many roles in preventing fatty liver, reducing blood pressure, improving ulcers, activating pancreatic β-cells, and repairing corneal and tumour tissues (12–17). Betaine intake can decrease chemical carcinogenesis (18). Meta-analysis and clinical research have showed that high betaine intake is associated with lower tumour incidence in breast cancer, colorectal adenoma, oesophageal adenocarcinoma, oesophageal squamous cell carcinoma, liver cancer, lung cancer, nasopharyngeal carcinoma, ovarian cancer, prostate cancer, colorectal cancer and so on (19–23). The role of betaine in breast cancer is interested in many scientists. The consumption level of betaine is inversely associated with breast cancer risk (24). High intake of betaine may be a promising strategy to prevent breast cancer development and reduce its mortality (25). Meanwhile, individual study has reported that high betaine intake does not reduce breast cancer risk among post-menopausal women (26). Therefore, we tried to verify the role of betaine in breast cancer and explore its mechanism.
Betaine plays the roles of anti-tumours through different ways. Betaine could form complex coacervates to reduce angiogenesis (27). It is also a methyl donor and could affect DNA methylation (24). Betaine has an anti-inflammatory effect (25, 28, 29). Betaine could reverse diethyl nitrosamine (DEN)-induced changes in oncogene c-myc and the tumour suppressor P-16 (30). In addition, betaine participates in fat metabolism, antioxidant events, and so on (30). However, no studies have explored its role in alcohol-induced deregulation of TFIIB-related factor 1 (Brf1) and RNA polymerase III-dependent gene (Pol III gene). While deregulation of Brf1 and Pol III genes is tightly linked to tumour development.
The products of Pol III genes are non-coding RNAs, such as 5S rRNAs and tRNAs. The roles of these non-coding RNAs are primarily involved in the biosynthesis of proteins. Deregulation of Pol III genes (5S rRNA and tRNAs) is closely related to cell transformation and tumourigenesis (15, 31–33). Brf1 is a specific transcription factor of Pol III genes. Our studies have demonstrated that alcohol increases Brf1 expression and Pol III gene transcription in vivo and in vitro (34–36). Upregulation of Pol III genes is tightly linked to cell proliferation, cell transformation and tumour development (34, 37).
Our previous studies in vivo and in vitro have demonstrated that alcohol treatment enhances the transcription of Pol III genes to promote an alteration of cellular phenotypes (35, 37, 38). This suggests that upregulation of Pol III genes caused by alcohol plays an important role in cancer development. Alcohol can be used as an agent to determine changes in these cellular phenotypes. Deregulation of Pol III genes is closely related to breast cancer (8, 35). Our studies demonstrate that estrogen receptor alpha (ERα) mediates Pol III gene transcription through Brf1, suggesting that ERα may play a critical role in alcohol-induced deregulation of Pol III genes in ER+ breast cancer development (35). Alcohol induces deregulation of Brf1 and Pol III genes in liver and breast cells, which share the common JNK1 signalling pathway to promote cell transformation. Through this common mechanism, alcohol-induced deregulation of Brf1 and Pol III genes brings about greater phenotypic changes (6). The interaction of Brf1 with ERα and Brf1 itself can be therapeutic targets for this disease (8). We have found that Tamoxifen inhibits tumour growth through its effect of repression on Brf1 expression and Pol III gene transcription (39). In addition, there are also reports that have claimed ErbB2/p38 and MCP-1 mediate alcohol-induced breast tumour growth (40–41).
In the present study, our results indicate that alcohol increases the rates of growth and colony formation of breast cancer cells. Betaine significantly inhibits the effect of alcohol on cell phenotypes. Betaine decreases the induction of Brf1 and Pol III genes caused by alcohol, resulting in the reduction of cell growth and colony formation. These studies provide a novel theoretical basis on the antitumor activity of betaine. This implies that betaine is able to serve as a prevention approach of alcohol-associated breast cancer.
2. Materials and Methods
2.1. Cell Lines and Reagents
The human breast adenocarcinoma cell line MCF-7 (HTB-22) was from ATCC. The cell growth media and TRIzol reagents were from Life Technologies Inc. Absolute ethanol (200 proof) and betaine were from Sigma-Aldrich Co. The 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Cell Proliferation and Cytotoxicity Assay Kit was from Boster Biological Technology. Rabbit anti-Brf1 antibody (Catalog #: A301–228A) was from BETHYL Laboratory. Mouse monoclonal antibody of actin (Catalog #: sc-58673) was from Santa Cruz Biotechnology. Anti-rabbit IgG-HRP and anti-mouse IgG-HRP were from Cell Signalling. Chemiluminscence reagents were from Santa Cruz Biotechnology. PCR reagent kits were from Bio-Rad Biotech.
2.2. Cell Proliferation
MCF-7 cells were seeded in six-well plates in triplicate. The cells were treated with ethanol and betaine as indicated in figures. The confluences of the cells on the 6th day were approximately 85%−90%. The cell morphology was analysed by microscopy using a Nikon Eclipse TE300 and Metamorphic Program (Cell and Tissue Imaging Core of University of Southern California Research Center for Liver Diseases, P30 DK048522). Cells were assayed for viability and counted with a globulimeter and Coulter counter.
2.3. MTT assay
MCF-7 cells were cultured in 96-well plates. The ethanol and betaine were added as indicated in the figures, and OD values were detected using Omega 96 Well Plates Reader followed the protocol of MTT Cell Proliferation and Cytotoxicity Assay Kit: Remove the medium and place it with 100 μl of fresh culture medium. Add 10 μl of the MTT labelling reagent into each well, including a negative control of 100 μl medium alone. Incubate the microplate at 37°C for 4 hours in the humidified chamber (5% CO2). Add 100 μl of Formazan solubilization solution into each well and mix thoroughly and incubate the microplate at 37 °C for 18 hours in the humidified chamber (5% CO2). Measure the absorbance of the samples using Omega microplate reader at 570 nm. OD Value= (OD sample –OD negative)/6.
2.4. Cell colony formation
MCF-7 cells were suspended in 3 ml 0.25% (w/v) agar containing 10% FBS overlaid on and transferred to top of 3 ml 0.35% (w/v) agar in 6 well plates. The cells were treated with specified concentrations of ethanol and betaine at 1st and 4th day as indicated in the figures. Cells were fed with fresh complete media twice weekly. Colonies were stained with 0.005% (w/v) crystal violet, and then the colonies were counted under microscope and photographed at 8th day.
2.5. Western blot
MCF-7 cells were incubated with betaine for 2 hours and then treated with ethanol for 1 hour after starvation for 4 hours when the cell confluence was approximately 85–90%. Cells were collected with lysis buffer and sonicated. The suspensions were centrifuged to save the supernatants. Protein concentrations were determined by the Bradford method using Fluostar Omega spectrometer (Cell Biology Core Laboratory of University of Southern California Research Canter for Liver Diseases, P30DK DK48522). Lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Proteins were transferred from the sodium dodecyl sulfate–polyacrylamide electrophoresis gel to Hybond-P membrane, and immunoblot analysis was performed with specific antibodies of Brf1 or actin as indicated in figure 5. Bound primary antibody was visualized using horseradish peroxidase-conjugated secondary antibody and enhancing chemiluminescence reagents.
Fig 5.
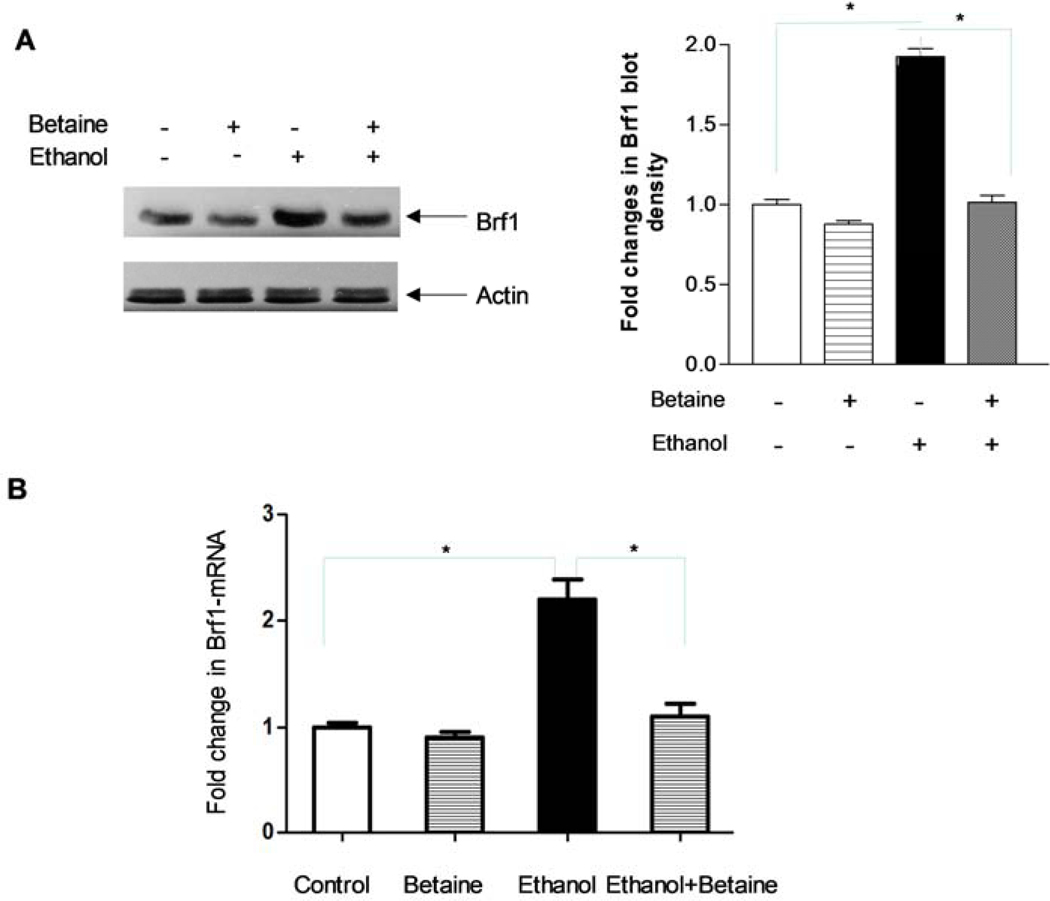
Betaine represses Brf1 expression. MCF-7 cells were pre-treated with betaine for 2 hours and then treated with ethanol for another 1 hour. (A) Immunoblot analysis was performed using protein lysates derived from these cells to detect the Brf1 protein level. Actin was used as a loading control. A represent immunoblot (A, Left panel). A densitometric analysis are revealed and the fold changes in Brf1 level were normalized by actin (A, Right panel). (B) Brf1 mRNA level. MCF-7 cells were treated with alcohol (50mM) and betaine as indicated above, to extract total RNA. mRNA levels of Brf1 were measured by RT-PCR. The fold changes are calculated by normalizing to the amount of GAPDH mRNA. The bars represent Mean ± SE of at least three independent determinations. These results indicate that betaine reduces the induction of Brf1 expression caused by alcohol.
2.6. RNA Isolation and RT-qPCR
MCF-7 cells were treated with ethanol and betaine as described above and Total RNA was isolated from the cells using single step extraction method with TRIzol reagent. RNA samples were quantified and reverse-transcribed in 20 μl reaction volume containing 1 × reverse transcription buffer. After first-strand cDNA synthesis, the cDNAs were diluted in DNase-free water, and real time qPCRs (RT-qPCRs) were performed with specific primers (Brf1: F: 5’-CCT CGG GCC TCT GCG GAG CAG-3’, R: 5’-TCA TCA ATG GTC AAC TGA CTG GTG G-3’, Pre-tRNA: F: 5’-GTC AGG ATG GCC GAG TGG TCT AAG-3’, R: 5’-CCA CGC CTC CAT ACG GAG AAC CAG AAG ACC C-3’, 5S rRNA: F: 5’-GGC CAT ACC ACC CTG AAC GC-3’, R:5’-CAG CAC CCG GTA TTC CCA GG-3’) and PCR reagent was from Bio-Rad Company. Brf1-mRNA, Pre-tRNA and 5S rRNA transcripts were measured by RT-qPCR in the ABI prism 7700 Sequence Detection System..
2.7. Statistical analysis
The data are expressed as the mean ± standard error of the mean for given samples and evaluated by Student’s t test or a one-way analysis of variance (ANOVA) using Statistical Package for Social Science (SPSS) 16 statistical software (SPSS Inc., Chicago, IL, USA). Each assay was repeated in triplicate. The level of significance was set at P<0.05.
3. Results
3.1. Betaine reduces the rate of MCF-7 cell growth
3.1.1. Betaine inhibition of cell growth is in a dose-dependent manner
Alcohol has been classified as carcinogenic to humans by IARC (2–4, 42); it induces the growth of tumour cells. Our previous studies have shown that the accumulation of the breast cancer cells of MCF-7 line was increased by ethanol at 25 mM and 50 mM (35). In this experiment, 50 mM ethanol is chosen to induce the growth of MCF-7 cells. The results indicate that ethanol obviously augments the growth of MCF-7 cells. The cells treated with ethanol alone or without ethanol were more confluent than the groups treated with betaine plus ethanol (Fig. 1). One of the main functions of betaine is antitumor activity. Different does of betaine were optimized for MCF-7 cell growth. The result indicates that betaine could reduce the induction of MCF-7 cell growth by ethanol. Compared with the ethanol group, the confluence and number of the MCF-7 cells were decreased with increasing doses of betaine (Fig. 1, Fig. 2B). The inhibiting role of betaine in MCF-7 cell growth was in a dose-dependent manner (Fig. 1).
Fig 1.
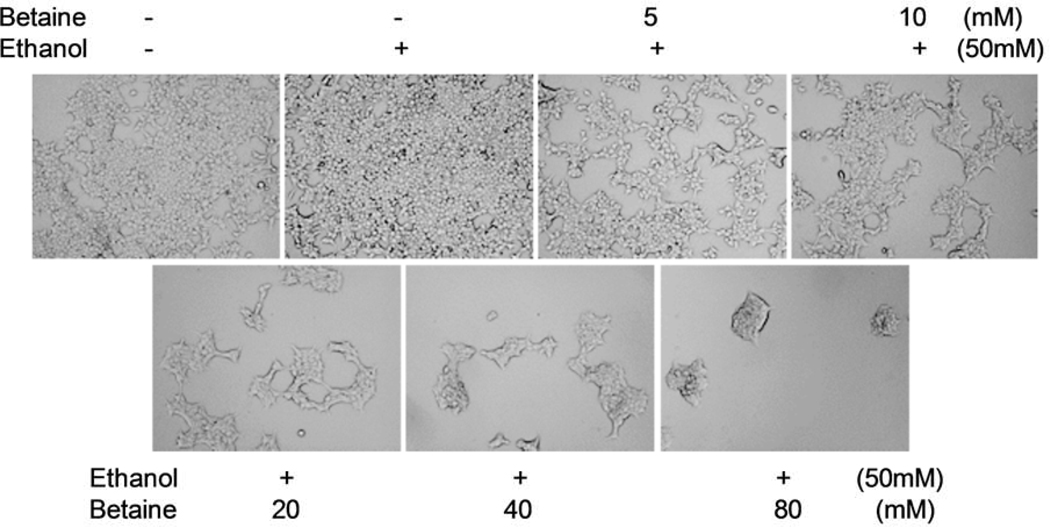
Role of betaine in inhibition of MCF-7 cell growth. 3000 MCF-7 cells were cultured with 3 ml medium per well in 6-well plates. Dose curve: ethanol (50 mM) and different amounts of betaine were added into the medium at the 0 hour, the 48th hour and the 96th hour, respectively. The pictures were taken under a microscope at the 6th day. Original magnification X 40.
Fig. 2.
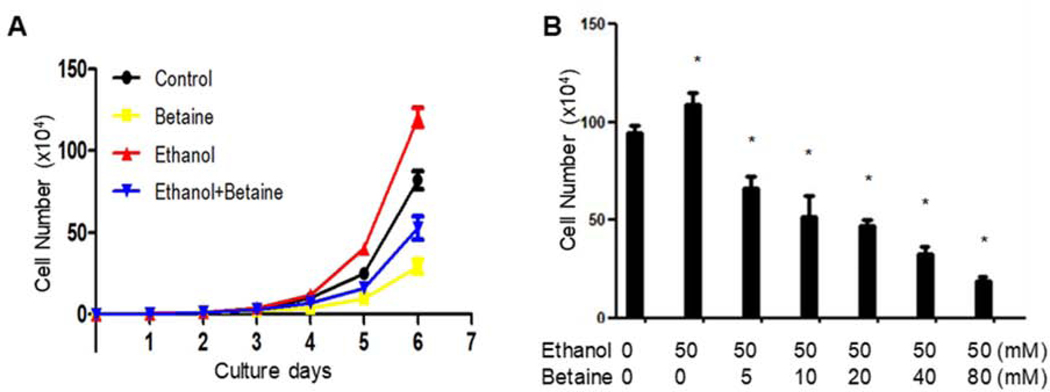
Betaine represses cell growth of human breast cancer. (A) Time curve, the ethanol (50 mM) and betaine (40 mM) were added in the medium, and the total cell numbers were counted from the first day to the sixth day. (B) The cells were treated with or without ethanol and different doses of betaine. The total cell numbers were counted after 6 days. * P<0.05 vs control (Ethanol=0, Betaine=0).
3.1.2. Time curve
The time curve experiment was performed to observe the role of betaine in MCF-7 cell growth at different times (Fig. 2A). The above results indicate that betaine-repressed growth of MCF-7 cells by ethanol acts in a dose-dependent manner (Fig. 1 and Fig. 2B). At concentrations of 10 mM betaine or above, the numbers of the cells were significantly decreased (Fig. 1 and Fig. 2B). The morphology of MCF-7 cells was abnormal and cell viability is poor at 40mM of betaine, which was approaching to maximal inhibition (Fig. 1 and Fig. 2B). Therefore, to further optimize the time curve, we chose 40 mM betaine to pre-treat the cells. The results reveal that MCF-7 cells grew slowly in the first three days and then grew rapidly from the fourth to sixth day. The role of betaine in inhibiting MCF-7 cell growth induced by ethanol was not obvious in the first three days. However, the effect of betaine on repressing cancer cell growth became more and more clear from the fourth day (Fig 2A). This is consistent with MCF-7 cell growth conditions of the dose curve (Fig. 2B). Together, these results (Fig 1 and Fig 2) show that betaine is able to decrease the rate of breast cancer cell growth.
To further confirm the role of betaine in cell growth, we performed MTT assays to detect the growth curve of MCF-7 cells. As the basal space per well in 96-well plates is limited, the numbers of MCF-7 cells and the amounts of ethanol and betaine were reduced in proportion. The results show that the inhibition role of betaine in cell growth was elevated with increasing its dose (Fig 3A). The effect of betaine-inhibited cell growth becomes stronger and stronger with increasing time (Fig 3B). These results by MTT assay are consistent with the dose curve and time curve results shown above (Fig. 2A and 2B). This once again shows the inhibitory effect of betaine on the induction of MCF-7 cell growth by ethanol.
Fig 3.
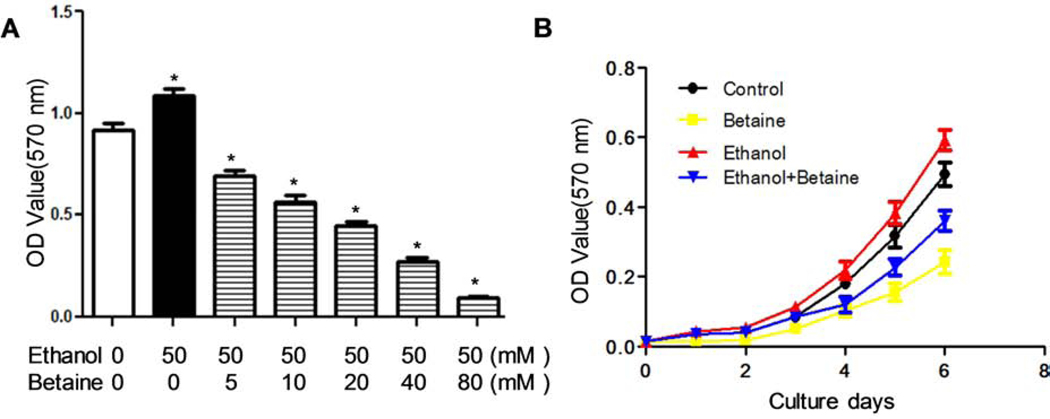
The role of betaine in MCF-7 cell growth by MTT assay. Approximately 300 MCF-7 cells were cultured with 300 μl medium per well in 96-well plates. (A) Dose curve. The ethanol (50 mM) and different concentrations of betaine were added into the medium at the 0 hour, the 48th hour and the 96th hour, respectively. OD value was detected after treated the cells for 6 days. (B) Time curve. The concentration of ethanol and betaine were 50 mM and 40 mM, respectively. OD value was detected every day after plating.
3.2. Betaine decreases the induction of MCF-7 cell colony formation by ethanol
Our previous data showed that ethanol treatment promotes the transformation of normal cells and increases colony formation of cancer cells (34, 43). Therefore, we further determined the effect of betaine on colony formation of the MCF-7 cells. We seeded MCF-7 cells in soft agar and treated the cells with the PBS, betaine, ethanol, and (ethanol + betaine), respectively. The results show that the numbers of colony formation was increased the ethanol group, but reduced in the (ethanol + betaine) group (Fig 4A). This indicates that ethanol can indeed elevate the rate of colony formation of MCF-7 cells, while betaine is able to reduce ethanol’s effect on the rate (Fig 4B).
Fig 4.
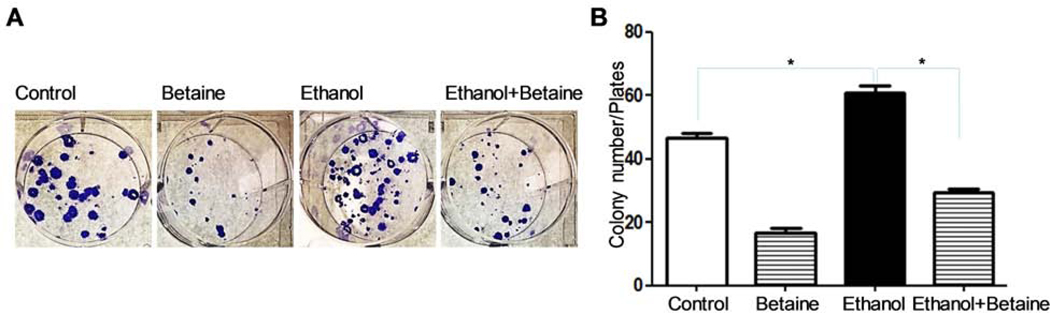
Colony formation assay. 1000 MCF-7 cells were planted in the upper agar in 6-well plates. Ethanol (50 mM) and betaine (40 mM) were added at the 1st and 4th, respectively. The result indicates that betaine is able to reduce the rate of MCF-7 cell colony formation caused by ethanol. *P < 0.01; **P < 0.01.
3.3. Betaine inhibits Brf1 expression and Pol III gene transcription
3.3.1. Betaine inhibits the Brf1 expression of MCF-7 cells
Deregulation of Pol III genes is closely related to cell transformation and tumorigenesis (35–37). Brf1 is a transcription factor which specifically regulates Pol III gene transcription. Enhancement of Brf1 can upregulate the transcription of Pol III genes to promote the occurrence and development of cancer. In contrast, reducing Brf1 expression inhibits the transcription of Pol III genes, repressing cell transformation and tumour growth (35, 43–45). Alcohol is a carcinogen which upregulates Pol III gene transcription(35, 38). Thus, we speculate that betaine may reduce the ethanol-induced Brf1 expression of MCF-7 cells to downregulate the activity of Pol III genes and the cell growth. The result indicates that the levels of Brf1 protein (Fig 5A) and Brf1 mRNA (Fig 5B) of MCF-7 cells are enhanced by alcohol, while betaine is able to reduce the induction of Brf1 expression in MCF-7 cells treated by ethanol.
3.3.2. Betaine represses Pol III gene transcription
We have demonstrated that enhancement of Pol III gene transcription promotes cell proliferation and cell transformation (34, 37). Diluted alcohol at 25 – 50 mM increased Pol III gene transcription in cell culture models (35, 38). Thus, we hypothesize that betaine may reduce the rates of MCF-7 cell growth and colony formation through inhibiting Pol III gene transcription. The results show that alcohol increases the transcription levels of pre-tRNALeu (Fig 6A) and 5S rRNA (Fig 6B), but betaine decreases the levels of Pol III genes caused by alcohol (Fig. 6).
Fig 6.
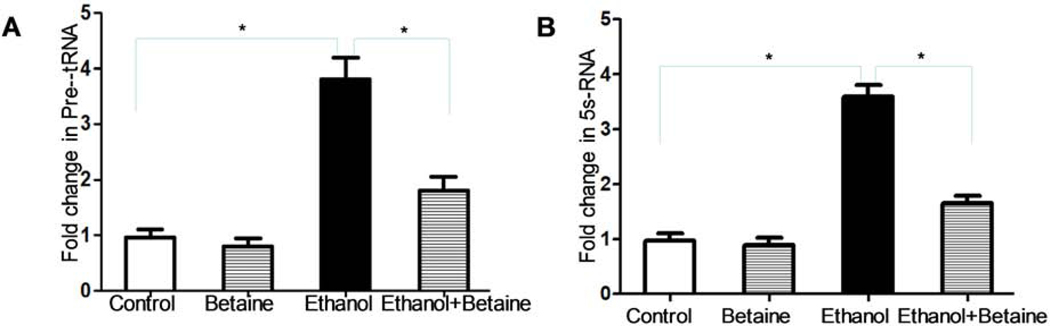
Betaine mediates Pol III gene transcription. MCF-7 cells were treated as indicated above. The amounts of pre-tRNALeu (A) and 5S rRNA (B) were measured by RT-qPCR. The fold changes are calculated by normalizing to the amount of GAPDH mRNA. The bars represent Mean ± SE of at least three independent determinations.
4. Discussion
Alcohol is a very common beverage in life. However, it is known as a carcinogen (2–4). Alcohol intake is associated with many human cancers, including breast cancer. Betaine is widely consumed in many kinds of food. Betaine appears as an anti-carcinogen that can prevent the occurrence of the tumour or slow down the progress of the tumour (18–23). There is still some controversy about betaine’s effect on tumours, especially in breast cancer (26, 46–48). In general, betaine consumption is negatively corelated with the occurrence and progression of breast cancer (48)46), but some scholars believe that betaine is not significantly correlated with the occurrence and progression of tumours, including in prostate cancer and breast cancer (26, 46–48). In the present study, we detected the effect of betaine on alterations of cellular phenotypes and Pol III genes. Our results confirm that alcohol can increase the rates of growth and colony formation of breast cancer cells, while betaine is able to inhibit these effects of alcohol. There is no obvious change in osmolarity at this condition of 40mM betaine. The breast cancer MCF-7 cells grow to almost full confluence under the induction of ethanol, whereas betaine represses cell growth and colony formation of the MCF-7 line caused by ethanol. The inhibiting roles of betaine in MCF-7 cell growth are in a dose-dependent and time-dependent manners (Fig. 1). This is because betaine represses Brf1 expression and Pol III gene transcription, which is strong evidence supporting the opinion that betaine is negatively correlated with breast cancer (Fig 7).
Fig 7.
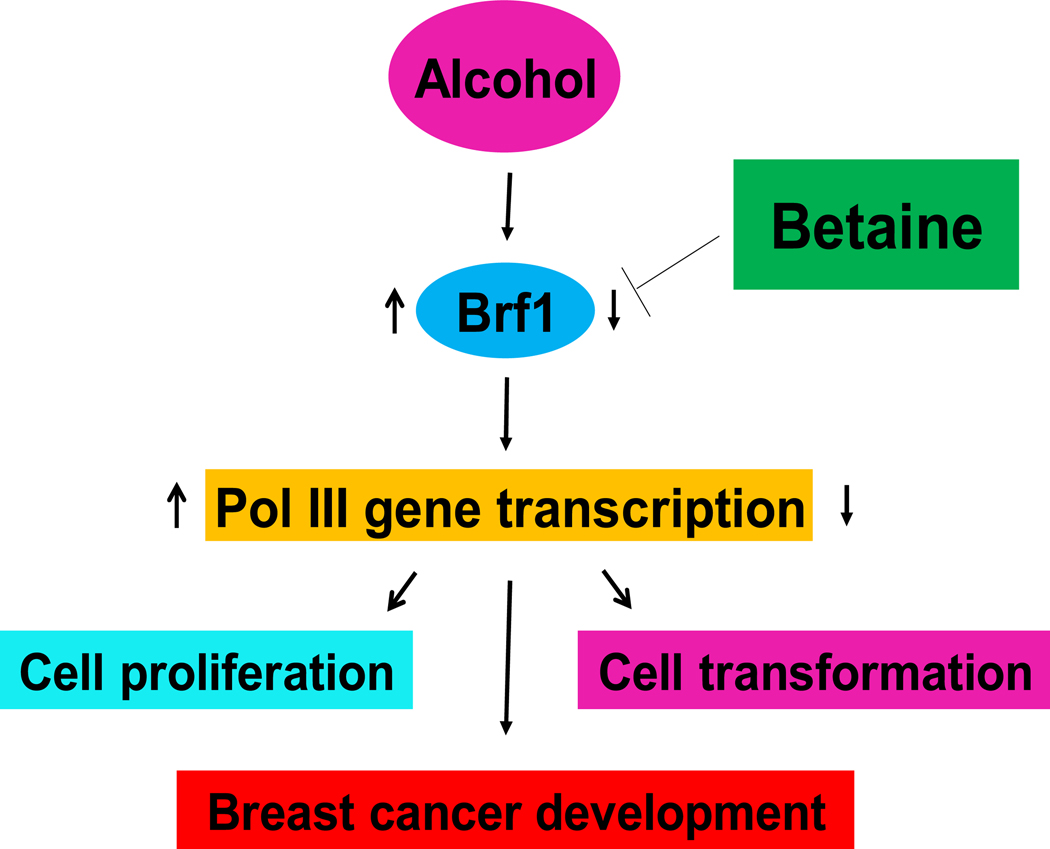
Schematic illustration of the role of betaine in Brf1 expression and Pol III gene transcription. Alcohol increases Brf1 expression to elevate Pol III gene transcription and to promote cell proliferation and transformation, eventually resulting in breast cancer development. In contrast, betaine inhibits the induction of these genes caused by alcohol to decrease Brf1 expression and Pol III gene transcription, leading to repression of breast cancer cell growth and tumor development.
Based on the results above, betaine displays the effects of repression on cell proliferation and colony formation on MCF-7 cells. There are many explanations about betaine’s biological function: reducing angiogenesis, altering DNA methylation, anti-inflammatory effects, reversing oncogenic c-myc transcription, antioxidant effects and so on (24, 25, 27–30). However, nothing is reported about the roles of betaine in the transcription of Brf1 and Pol III genes. The products of the RNA Pol III gene are non-coding RNAs, such as 5S rRNA and tRNA. The functions of these non-coding RNAs are primarily involved in the biosynthesis of proteins. The deregulation of Pol III genes (5S rRNA and tRNA) is closely related to cell transformation and tumorigenesis (15, 31–33). Alcohol can induce the deregulation of Pol III gene transcription. Upregulation of Pol III genes is tightly linked to cell proliferation, cell transformation and tumour development (34, 37). In this study, we found that betaine inhibits the upregulation of Pol III gene transcription caused by alcohol. It indicates that betaine inhibit the breast cancer cell growth and colony formation through the downregulation of Brf1 and Pol III genes.
Brf1 is a specific transcription factor of Pol III genes (tRNAs and 5S rRNA); it positively modulates Pol III gene transcription to promote the occurrence of cancer. Studies have demonstrated that reducing Brf1 expression decreases Pol III gene transcription and limits cell transformation and tumour growth (35, 43–45). Therefore, exploring the role of these genes in the occurrence of breast cancer and the mechanism of their imbalance will provide a basis for the development of new therapeutic strategies. In the present study, we found that betaine can reverse the induction of Pol III gene transcription in alcohol-treated MCF-7 cells. We further detected the influence of betaine on Brf1 expression. The result showed that betaine could reduce the increase in Brf1 expression, at both the protein and mRNA levels, caused by alcohol. These studies demonstrate that betaine decreases the rates of growth and colony formation induced by alcohol in MCF-7 cells through the repression of Brf1 expression and Pol III gene transcription.
5. Conclusions
In summary, alcohol augments the rates of growth and colony formation of MCF-7 cells, while betaine can reduce the effects of alcohol on the cellular phenotypes. Repression of cell growth and colony formation of the MCF-7 line by betaine is attributed to its function in reducing the activities of Brf1 and Pol III genes (Fig 7). Together, these studies provide novel insights into betaine in alcohol-caused breast cancer cell growth and deregulation of Brf1 and Pol III genes. The study suggest that betaine consumption is able to prevent alcohol-associated breast cancer development.
Highlight:
Betaine decreases the rates of breast cancer cell proliferation and colony formation caused by alcohol.
The inhibiting role of betaine in the alcohol-increased the rates of breast cancer cell growth was in a dose-dependent manner.
Betaine represses Brf1 expression and Pol III gene transcription, resulting in the inhibition of the cell growth and colony formation.
Acknowledgements
We want to thank Drs. M. R. Stallcup and Danial Levy (University of Southern California) for scientific discussions. We thank Dr. Kusum Kharbanda (University of Nebraska Medical Center) for technical help. This work was supported by NIAAA grants AA021114 and AA02324 to S.Z. and China Postdoctoral Science Foundation (Grant No.2018M643051) to Z.H.
We would like to thank the reviewers for their thoughtful comments that guided us in our revision, resulting in a significantly improved manuscript, whose title is “Role of betaine in inhibiting the induction of RNA Pol III gene transcription and cell growth caused by alcohol”. The tracking number is CHEMBIOINT_2019_921. We have provided new data and answered the reviewer’s concerns point by point as detailed below.
The reviewers have no conflict of interest with any of the authors of this paper being resubmitted. This paper (or closely related research data) has not been published or accepted for publication. It is not under consideration at another journal.
Abbreviation
- IARC
International Agency for Research on Cancer
- TFIIB
Transcription factor II B
- Brf1
TFIIB-related factor 1
- Pol III gene
RNA polymerase III-dependent gene
- MTT
3-(4,5dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- DEN
Diethyl-nitrosamine
- ERα
Estrogen receptor alpha
- RT-qPCR
Real time qPCR
- ErbB2
Receptor tyrosine-protein kinase
- P38
p38 mitogen-activated protein kinases
- MCP-1
Monocyte chemoattractant protein-1
Footnotes
Declaration:
The authors declare no competing financial interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Conflict of Interest
All authors read and approved the final manuscript. The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J Clin, (2018). 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- [2].I.W.G.o.t.E.o.C.R.t. Humans, IARC monographs on the evaluation of carcinogenic risks to humans. Ingested nitrate and nitrite, and cyanobacterial peptide toxins, IARC Monogr Eval Carcinog Risks Hum, 94 (2010) v-vii, 1–412 [PMC free article] [PubMed] [Google Scholar]
- [3].Cogliano VJ, Baan R, Straif K, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Wild CP, Preventable exposures associated with human cancers, J Natl Cancer Inst, 103 (2011) 1827–1839. 10.1093/jnci/djr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shi G, Zhong S, Alcohol-associated cancer and deregulation of Pol III genes, Gene, 612 (2017) 25–28. 10.1016/j.gene.2016.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Connor J, Alcohol consumption as a cause of cancer, Addiction, 112 (2017) 222–228. 10.1111/add.13477 [DOI] [PubMed] [Google Scholar]
- [6].Yi Y, Huang C, Zhang Y, Tian S, Lei J, Chen S, Shi G, Wu Z, Xia N, Zhong S, Exploring a common mechanism of alcohol-induced deregulation of RNA Pol III genes in liver and breast cells, Gene, 626 (2017) 309–318. 10.1016/j.gene.2017.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lei J, Chen S, Zhong S, Abnormal expression of TFIIIB subunits and RNA Pol III genes is associated with hepatocellular carcinoma, Liver Res, 1 (2017) 112–120. 10.1016/j.livres.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fang Z, Yi Y, Shi G, Li S, Chen S, Lin Y, Li Z, He Z, Li W, Zhong S, Role of Brf1 interaction with ERalpha, and significance of its overexpression, in human breast cancer, Mol Oncol, 11 (2017) 1752–1767. 10.1002/1878-0261.12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang Y, Wu H, Yang F, Ning J, Li M, Zhao C, Zhong S, Gu K, Wang H, Prognostic Value of the Expression of DNA Repair-Related Biomarkers Mediated by Alcohol in Gastric Cancer Patients, Am J Pathol, 188 (2018) 367–377. 10.1016/j.ajpath.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baglia ML, Malone KE, Tang MC, Li CI, Alcohol Intake and Risk of Breast Cancer by Histologic Subtype and Estrogen Receptor Status Among Women Aged 55 to 74 Years, Horm Cancer, 8 (2017) 211–218. 10.1007/s12672-017-0297-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Craig SA, Betaine in human nutrition, Am J Clin Nutr, 80 (2004) 539–549. 10.1093/ajcn/80.3.539 [DOI] [PubMed] [Google Scholar]
- [12].Perugini V, Best M, Kumar S, Guildford AL, Bone AJ, Macfarlane WM, Santin M, Phillips GJ, Carboxybetaine-modified succinylated chitosan-based beads encourage pancreatic beta-cells (Min-6) to form islet-like spheroids under in vitro conditions, J Mater Sci Mater Med, 29 (2017) 15. 10.1007/s10856-017-6018-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Deng R, Su Z, Hua X, Zhang Z, Li DQ, Pflugfelder SC, Osmoprotectants suppress the production and activity of matrix metalloproteinases induced by hyperosmolarity in primary human corneal epithelial cells, Mol Vis, 20 (2014) 1243–1252 [PMC free article] [PubMed] [Google Scholar]
- [14].Day CR, Kempson SA, Betaine chemistry, roles, and potential use in liver disease, Biochim Biophys Acta, 1860 (2016) 1098–1106. 10.1016/j.bbagen.2016.02.001 [DOI] [PubMed] [Google Scholar]
- [15].Zhong Q, Shi G, Zhang Y, Lu L, Levy D, Zhong S, Alteration of BRCA1 expression affects alcohol-induced transcription of RNA Pol III-dependent genes, Gene, 556 (2015) 74–79. 10.1016/j.gene.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dou X, Xia Y, Chen J, Qian Y, Li S, Zhang X, Song Z, Rectification of impaired adipose tissue methylation status and lipolytic response contributes to hepatoprotective effect of betaine in a mouse model of alcoholic liver disease, Br J Pharmacol, 171 (2014) 4073–4086. 10.1111/bph.12765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang W, Huang L, Gao J, Wen S, Tai Y, Chen M, Huang Z, Liu R, Tang C, Li J, Betaine attenuates chronic alcoholinduced fatty liver by broadly regulating hepatic lipid metabolism, Mol Med Rep, 16 (2017) 5225–5234. 10.3892/mmr.2017.7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zeng FF, Xu CH, Liu YT, Fan YY, Lin XL, Lu YK, Zhang CX, Chen YM, Choline and betaine intakes are associated with reduced risk of nasopharyngeal carcinoma in adults: a case-control study, Br J Cancer, 110 (2014) 808–816. 10.1038/bjc.2013.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Youn J, Cho E, Lee JE, Association of choline and betaine levels with cancer incidence and survival: A meta-analysis, Clin Nutr, (2018). 10.1016/j.clnu.2018.01.042 [DOI] [PubMed] [Google Scholar]
- [20].Sun S, Li X, Ren A, Du M, Du H, Shu Y, Zhu L, Wang W, Choline and betaine consumption lowers cancer risk: a meta-analysis of epidemiologic studies, Scientific reports, 6 (2016) 35547. 10.1038/srep35547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ying J, Rahbar MH, Hallman DM, Hernandez LM, Spitz MR, Forman MR, Gorlova OY, Associations between dietary intake of choline and betaine and lung cancer risk, PLoS One, 8 (2013) e54561. 10.1371/journal.pone.0054561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bingula R, Dupuis C, Pichon C, Berthon JY, Filaire M, Pigeon L, Filaire E, Study of the Effects of Betaine and/or C-Phycocyanin on the Growth of Lung Cancer A549 Cells In Vitro and In Vivo, J Oncol, 2016 (2016) 8162952. 10.1155/2016/8162952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lu MS, Fang YJ, Pan ZZ, Zhong X, Zheng MC, Chen YM, Zhang CX, Choline and betaine intake and colorectal cancer risk in Chinese population: a case-control study, PLoS One, 10 (2015) e0118661. 10.1371/journal.pone.0118661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang CX, Pan MX, Li B, Wang L, Mo XF, Chen YM, Lin FY, Ho SC, Choline and betaine intake is inversely associated with breast cancer risk: a two-stage case-control study in China, Cancer Sci, 104 (2013) 250–258. 10.1111/cas.12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xu X, Gammon MD, Zeisel SH, Bradshaw PT, Wetmur JG, Teitelbaum SL, Neugut AI, Santella RM, Chen J, High intakes of choline and betaine reduce breast cancer mortality in a population-based study, FASEB J, 23 (2009) 4022–4028. 10.1096/fj.09-136507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cho E, Holmes MD, Hankinson SE, Willett WC, Choline and betaine intake and risk of breast cancer among post-menopausal women, Br J Cancer, 102 (2010) 489–494. 10.1038/sj.bjc.6605510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hwang MP, Ding X, Gao J, Acharya AP, Little SR, Wang Y, A biocompatible betaine-functionalized polycation for coacervation, Soft Matter, 14 (2018) 387–395. 10.1039/c7sm01763d [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim DH, Sung B, Kang YJ, Jang JY, Hwang SY, Lee Y, Kim M, Im E, Yoon JH, Kim CM, Chung HY, Kim ND, Anti-inflammatory effects of betaine on AOM/DSSinduced colon tumorigenesis in ICR male mice, Int J Oncol, 45 (2014) 1250–1256. 10.3892/ijo.2014.2515 [DOI] [PubMed] [Google Scholar]
- [29].Du YP, Peng JS, Sun A, Tang ZH, Ling WH, Zhu HL, Assessment of the effect of betaine on p16 and c-myc DNA methylation and mRNA expression in a chemical induced rat liver cancer model, BMC Cancer, 9 (2009) 261. 10.1186/1471-2407-9-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee SY, Ko KS, Protective Effects of S-Adenosylmethionine and Its Combinations With Taurine and/or Betaine Against Lipopolysaccharide or Polyinosinic-polycytidylic Acid-induced Acute Hepatotoxicity, J Cancer Prev, 21 (2016) 152–163. 10.15430/JCP.2016.21.3.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Goodfellow SJ, Innes F, Derblay LE, MacLellan WR, Scott PH, White RJ, Regulation of RNA polymerase III transcription during hypertrophic growth, The EMBO journal, 25 (2006) 1522–1533. 10.1038/sj.emboj.7601040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].White RJ, RNA polymerase III transcription and cancer, Oncogene, 23 (2004) 3208–3216. 10.1038/sj.onc.1207547 [DOI] [PubMed] [Google Scholar]
- [33].Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, Johnson DL, PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex, Mol Cell Biol, 28 (2008) 4204–4214. 10.1128/MCB.01912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhong S, Johnson DL, The JNKs differentially regulate RNA polymerase III transcription by coordinately modulating the expression of all TFIIIB subunits, Proc Natl Acad Sci U S A, 106 (2009) 12682–12687. 10.1073/pnas.0904843106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Q, Jin J, Zhong Q, Yu X, Levy D, Zhong S, ERalpha mediates alcohol-induced deregulation of Pol III genes in breast cancer cells, Carcinogenesis, 34 (2013) 28–37. 10.1093/carcin/bgs316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhong Q, Xi S, Liang J, Shi G, Huang Y, Zhang Y, Levy D, Zhong S, The significance of Brf1 overexpression in human hepatocellular carcinoma, Oncotarget, 7 (2016) 6243–6254. 10.18632/oncotarget.6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhong S, Fromm J, Johnson DL, TBP is differentially regulated by c-Jun N-terminal kinase 1 (JNK1) and JNK2 through Elk-1, controlling c-Jun expression and cell proliferation, Mol Cell Biol, 27 (2007) 54–64. 10.1128/MCB.01365-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhong S, Machida K, Tsukamoto H, Johnson DL, Alcohol induces RNA polymerase III-dependent transcription through c-Jun by co-regulating TATA-binding protein (TBP) and Brf1 expression, J Biol Chem, 286 (2011) 2393–2401. 10.1074/jbc.M110.192955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhong Q, Shi G, Zhang Q, Lu L, Levy D, Zhong S, Tamoxifen represses alcohol-induced transcription of RNA polymerase III-dependent genes in breast cancer cells, Oncotarget, 5 (2014) 12410–12417. 10.18632/oncotarget.2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xu M, Ren Z, Wang X, Comer A, Frank JA, Ke ZJ, Huang Y, Zhang Z, Shi X, Wang S, Luo J, ErbB2 and p38gamma MAPK mediate alcohol-induced increase in breast cancer stem cells and metastasis, Mol Cancer, 15 (2016) 52. 10.1186/s12943-016-0532-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang S, Xu M, Li F, Wang X, Bower KA, Frank JA, Lu Y, Chen G, Zhang Z, Ke Z, Shi X, Luo J, Ethanol promotes mammary tumor growth and angiogenesis: the involvement of chemoattractant factor MCP-1, Breast Cancer Res Treat, 133 (2012) 1037–1048. 10.1007/s10549-011-1902-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].I.W.G.o.t.E.o.C.R.t. Humans, IARC monographs on the evaluation of carcinogenic risks to humans. Volume 97. 1,3-butadiene, ethylene oxide and vinyl halides (vinyl fluoride, vinyl chloride and vinyl bromide), IARC Monogr Eval Carcinog Risks Hum, 97 (2008) 3–471 [PMC free article] [PubMed] [Google Scholar]
- [43].Johnson SA, Dubeau L, Johnson DL, Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation, J Biol Chem, 283 (2008) 19184–19191. 10.1074/jbc.M802872200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang Q, Zhong Q, Evans AG, Levy D, Zhong S, Phosphorylation of histone H3 serine 28 modulates RNA polymerase III-dependent transcription, Oncogene, 30 (2011) 3943–3952. 10.1038/onc.2011.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhong Q, Shi G, Zhang Q, Zhang Y, Levy D, Zhong S, Role of phosphorylated histone H3 serine 10 in DEN-induced deregulation of Pol III genes and cell proliferation and transformation, Carcinogenesis, 34 (2013) 2460–2469. 10.1093/carcin/bgt219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rajaie S, Esmaillzadeh A, Dietary choline and betaine intakes and risk of cardiovascular diseases: review of epidemiological evidence, ARYA Atheroscler, 7 (2011) 78–86 [PMC free article] [PubMed] [Google Scholar]
- [47].Kotsopoulos J, Hankinson SE, Tworoger SS, Dietary betaine and choline intake are not associated with risk of epithelial ovarian cancer, Eur J Clin Nutr, 64 (2010) 111–114. 10.1038/ejcn.2009.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lee JE, Giovannucci E, Fuchs CS, Willett WC, Zeisel SH, Cho E, Choline and betaine intake and the risk of colorectal cancer in men, Cancer Epidemiol Biomarkers Prev, 19 (2010) 884–887. 10.1158/1055-9965.EPI-09-1295 [DOI] [PMC free article] [PubMed] [Google Scholar]


