Abstract
Background
Sustained proliferation and active metastasis are hallmarks of cancer, and they pose major challenges to the development of treatments and a cure for hepatocellular carcinoma (HCC). Thus, the mechanisms of proliferation, migration, and invasion of cancer cells need to be investigated. Many studies indicate that dysregulation of microRNA plays important roles in the progression of HCC, but the role of placenta-specific microRNA (miR-512-3p) in HCC has not been systematically investigated.
Purpose
In the current study, the expression, biological function, and mechanisms of miR-512-3p involvement in HCC were investigated.
Methods
Real-time quantitative polymerase chain reaction assays were conducted to determine miR-512-3p levels in HCC tissues and cell lines. The StarBase V3.0 online platform was used to compare miR-512-3p levels in HCC tissues with TCGA data and to identify potential miR-512-3p target genes. Associations between miR-512-3p and clinicopathological characteristics were analyzed statistically. MTT, ethynyl deoxyuridine, and transwell assays were performed to assess cell viability, proliferation, migration, and invasion. The luciferase reporter gene assay was used to verify target genes. Recuse assays were performed to confirm whether large tumor suppressor kinase 2 (LATS2) participated in the regulatory effects of miR-512-3p on HCC cell proliferation and motility, and whether miR-512-3p mediated the tumor-promoting effects of hypoxia.
Results
miR-512-3p was upregulated in HCC and it was associated with worse survival and unfavorable clinicopathological characteristics. Functional assays indicated that miR-512-3p contributed to HCC cell proliferation, migration, and invasion. Mechanistically, LATS2—a downstream target of miR-512-3p—mediated the tumor-promoting effects of miR-512-3p in HCC. Hypoxia could elevate miR-512-3p levels in HCC cells, and miR-512-3p partially mediated the tumor-promoting effects of hypoxia.
Conclusion
Hypoxia-induced miR-512-3p contributes to HCC cell proliferation, migration, and invasion by targeting LATS2 and inhibiting the Hippo/yes-associated protein 1 pathways.
Keywords: hepatocellular carcinoma, invasion, migration, miR-512-3p, LATS2, proliferation
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer, and it is a major cause of cancer-related deaths worldwide.1–3 Approximately half of the total number of HCC cases and deaths worldwide occur in residents of China, and this is partly associated with the comparative prevalence of hepatitis B in that country.4,5 Due to a lack of effective early diagnostic indicators HCC is often at an advanced stage when it is diagnosed, after the optimal treatment time has passed.6 Recurrence and metastasis after surgical resection are associated with a poor prognosis.7 It is thus essential to identify diagnostic indicators and to investigate the molecular mechanisms involved in HCC progression, in an effort to improve the accuracy of diagnosis and the efficacy of treatment.
Numerous studies indicate that dysregulated microRNAs are involved in the progression of HCC because they regulate the functioning of genes involved in various cellular processes including proliferation, invasion,8–11 and migration,12–14 and metastasis15,16 by regulating key genes. In recent years the involvement of placenta-specific microRNA (miR-512-3p) has been identified in various human cancers. Zhu et al17 reported that inhibition of dedicator of cytokinesis 3 (DOCK3) by miR-512-3p contributed to suppression of metastasis in non-small cell lung cancer. In several other studies miR-512-3p has been upregulated in HCC.18,19 To date however, no study has systematically investigated the role of miR-512-3p in HCC.
Hippo signaling evidently has an inhibitory effect on HCC progression.20,21 The Hippo signaling pathway activates large tumor suppressor kinases, which phosphorylate yes-associated protein 1 (YAP), resulting in cytoplasmic YAP retention. Previous studies indicate that large tumor suppressor kinase 2 (LATS2), a key component of the Hippo signaling pathway, functions as a tumor suppressor gene in various cancers including lung cancer,22 glioma,23 endometrial cancer,24,25 colorectal cancer,26 breast cancer,27 esophageal squamous cell carcinoma,28 and HCC.29 LATS2 is regulated by several microRNAs. Xu et al30 reported that microRNA-302d promotes the proliferation of human pluripotent stem cell-derived cardiomyocytes by inhibiting LATS2 in the Hippo pathway. Cheng et al27 reported that miR-372 promotes breast cancer cell proliferation by directly targeting LATS2. Han et al31 reported that miR-103 promotes the metastasis and epithelial-mesenchymal transition of HCC by directly inhibiting LATS2. Notably however, relationships between LATS2 and miR-512-3p in HCC remain unknown.
Hypoxia is a main feature of HCC, and strong evidence suggests that it may promote HCC progression by regulating characteristics associated with malignancy, including cancer stem-like properties,32 proliferation,33,34 metastasis,35,36 and epithelial-mesenchymal transition37,38 in both hypoxia-inducible factor 1-alpha (HIF1-α)-dependent and HIF1-α-independent manners. Dou et al39 reported that hypoxia contributed to growth and metastasis by increasing the expression of tuftelin 1, which is an HCC oncoprotein. Zhou et al40 reported that hypoxia induced glycolysis in HCC cells by increasing the expression of long non-coding RNA retinoic acid early transcript 1K pseudogene, mediated by HIF1-α. Zheng et al41 recently reported that hypoxia drove tumorigenesis and metastasis in HCC by downregulating miR-196-5p, but relationships between hypoxia and miR-512-3p in HCC remain uncharacterized.
In the present study miR-512-3p levels in HCC tissues and cells were investigated. A series of functional experiments was then performed to explore the biological roles of miR-512-3p in HCC cell proliferation and motility. The downstream target gene mediating the effects of miR-512-3p on HCC cell proliferation and motility was then screened for and verified. Lastly, the effects of hypoxia on miR-512-3p expression were investigated.
Materials and Methods
Tissue Samples
Tissue samples were obtained from 45 patients who underwent liver resection at the Department of General Surgery at the First Affiliated Hospital of Nanchang University (Nanchang, China). None of the patients received any adjuvant therapy such as chemotherapy or radiotherapy before surgery. All HCC and non-tumor tissues were stored in liquid nitrogen after they were collected. All patients provided written informed consent, and the study was approved by the Ethics Committee of Nanchang University, China. The clinicopathological parameters of the patients are shown in Table 1.
Table 1.
Association Between miR-512-3p Expression and Clinicopathologic Features of Patients with Hepatocellular Carcinoma
| Characteristics | Number (n=45) | miR-512-3p Levels | P-value | |
|---|---|---|---|---|
| High (n=23) | Low (n=22) | |||
| Age (years) | 0.292 | |||
| <60 | 15 | 6 | 9 | |
| ≥60 | 30 | 17 | 13 | |
| Gender | 0.445 | |||
| Male | 33 | 18 | 15 | |
| Female | 12 | 5 | 7 | |
| HBV infection | 0.654 | |||
| Negative | 9 | 4 | 5 | |
| Positive | 36 | 19 | 17 | |
| Liver cirrhosis | 0.150 | |||
| Absent | 12 | 4 | 8 | |
| Present | 33 | 19 | 14 | |
| AFP (ng/mL) | 0.666 | |||
| <20 | 11 | 5 | 6 | |
| ≥20 | 34 | 18 | 16 | |
| Tumor size | 0.026* | |||
| <5cm | 21 | 7 | 14 | |
| ≥5cm | 24 | 16 | 8 | |
| Tumor multiplicity | 0.608 | |||
| Single | 29 | 14 | 15 | |
| Multiple | 16 | 9 | 7 | |
| Vascular invasion | 0.042* | |||
| No | 28 | 11 | 17 | |
| Yes | 17 | 12 | 5 | |
| Edmondson–Steiner grade | 0.399 | |||
| Ⅰ+Ⅱ | 30 | 14 | 16 | |
| III+Ⅳ | 15 | 9 | 6 | |
| TNM stage | 0.009* | |||
| Ⅰ+Ⅱ | 33 | 13 | 20 | |
| III+Ⅳ | 12 | 10 | 2 | |
Note: *P<0.05, statistically significant difference.
Abbreviations: HBV, hepatitis B virus; AFP, alpha-fetoprotein; TNM, tumor-node-metastasis.
Cell Culture
The HEK293T cell line, L02 cell line (human immortalized normal hepatic cell line), and human HCC cell lines (Hep3B, SMMC-7721, MHCC97-L, and HCCLM3) were purchased from the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Gibco, Grand Island, NY, USA) containing 1% penicillin-streptomycin (Sigma, St. Louis, MO, USA) and 10% fetal bovine serum (Gibco) in a 37°C incubator with 5% CO2. The hypoxic cell model was generated by culturing cells in a 37°C hypoxia incubator with 1% O2.
Cell Transfection
miR-512-3p mimics (miR-512-3p, miR10002823-1-5), control mimics (miR-control, miR1N0000001-1-5), miR-512-3p inhibitors (anti-miR-512-3p, miR20002823-1-5), and control inhibitors (anti-miR-NC, miR2N0000001-1-5) were purchased from RiboBio (Guangzhou, China). A LATS2 expression plasmid (LATS2, RC219394) and a negative control (EV, PS100001) were purchased from OriGene Technologies Inc. (Rockville, MD, USA). The Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was used in cell transfection assays, in accordance with the manufacturer’s instructions.
Real-Time Quantitative Polymerase Chain Reaction
RNA was extracted from tissues and cells with TRIzol purchased from Invitrogen and the miRVana microRNA Isolation Kit in accordance with the manufacturer’s instructions. Reverse transcription was then conducted using the TIANScript RT Kit (Tiangen Bio Inc., Beijing, China). Quantitative PCR was conducted with the SYBR Premix Ex TaqTM Kit (Takar Bio Inc., Kusatsu, Shiga, Japan) and TaqMan Human MiRNA Assay Kit (Genecopoeia Inc., Guangzhou, China). LATS2 and glyceraldehyde 3-phosphate dehydrogenase primers were purchased from Realgene (Nanjing, China). miR-512-3p and U6 primers were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Expression levels were quantified via the 2−ΔΔCt method. All primers used for quantitative real-time PCR (qRT-PCR) are shown in Table 2.
Table 2.
Primers Used in This Study
| Primers Name | Primer Sequence |
|---|---|
| LATS2 forward | 5ʹ- TGGCACCTACTCCCACAG-3’ |
| LATS2 reverse | 5ʹ- CCAAGGGCTTTCTTCATCT −3’ |
| TXNIP forward | 5ʹ- GAGCCAGCCAACTCAAGAGA-3’ |
| TXNIP reverse | 5ʹ- TAGCAGACACAGGTGCCATTA-3’ |
| GAPDH forward | 5ʹ- GAAGGTGAAGGTCGGAGTC −3’ |
| GAPDH reverse | 5ʹ- GAAGATGGTGATGGGATTTC −3’ |
| miR-512-3p forward | 5ʹ- CGGCGGCACTCAGCCTTGAGGG −3’ |
| miR-512-3p reverse | 5ʹ- GTGCAGGGTCCGAGGT −3’ |
| U6 forward | 5ʹ- CTCGCTTCGGCAGCACA-3’ |
| U6 reverse | 5ʹ- AACGCTTCACGAATTTGCGT −3’ |
MTT Assay
MTT was purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA), and MTT assays were conducted to assess cell viability. Absorbance was read using a microplate reader (Bio-Rad, Hercules, CA, USA).
Ethynyl Deoxyuridine Incorporation Assay
Ethynyl deoxyuridine (EdU) incorporation assays were conducted to assess cell proliferation ability, using Cell-LightTM EdU Apollo488 (RiboBio). A Zeiss fluorescence photomicroscope (Carl Zeiss, Oberkochen, Germany) was used to analyze the samples, and quantification was achieved by counting a minimum of five random fields per sample.
Transwell Assay
Transwell assays were conducted using transwell chambers (Millipore, Burlington, MA, USA), to assess the migration and invasion capacities of cells. In migration assays 3 × 104 HCC cells were cultured in the upper chamber with serum-free DMEM, and the lower chamber was filled with DMEM containing 20% serum. In invasion assays 3 × 104 HCC cells were seeded on Matrigel-coated membrane inserts, and the chamber was placed into a cell culture plate and incubated at 37°C for 24 h. Cells that had migrated or invaded across the transwell membrane were fixed in 4% paraformaldehyde for 30 min, then stained with 0.5% crystal violet for 30 min. A light microscope was used to analyze the samples, and quantification was achieved by counting a minimum of 10 random fields under 100x magnification.
Western Blotting
RIPA Buffer (WB009A; Hat Biotechnology, Xi’an, China) was used to extract proteins in HCC cells or tissues. A BCA kit (WB003; Hat Biotechnology) was used to measure protein concentrations. All proteins were then electrophoresed in a 10% sodium dodecyl sulfate polyacrylamide gel and transferred to polyvinylidene difluoride membrane (Bio-Rad). The membranes were blocked with 10% non-fat milk, then incubated with specific primary antibodies at 4°C overnight. The antibodies used in the study were anti-LATS2 (1:500; bs-4081R; Beijing Bioss Biotechnology), anti-YAP (1:1000; #14,074; Cell Signaling Technology, Danvers, MA, USA), anti-phospho-YAP (ser127; 1:1000; #13,008; Cell Signaling Technology), anti-thioredoxin-interacting protein (1:1000; #14,715; Cell Signaling Technology), and anti-glyceraldehyde 3-phosphate dehydrogenase (1:1000, #5174; Cell Signaling Technology). The membranes were then incubated with secondary antibody (anti-rabbit #7074 or anti-mouse #7076; Cell Signaling Technology) for 2 h at room temperature. Lastly, enhanced chemiluminescence reagent (PierceTM ECL, Thermo ScientificTM, Waltham, MA, USA) was applied to detect the proteins.
Luciferase Reporter Assay
Luciferase reporter assays were performed to confirm direct binding between the LATS2 3ʹ-untranslated regions (UTRs) and miR-512-3p. Wild-type (WT) and mutant (MUT) 3ʹUTRs of LATS2 mRNA were synthesized and inserted downstream of the promoter in the pEZX-MT06 vector (Genecopoeia). Cells transfected with miR-512-3p mimics, inhibitors, or corresponding control vectors were also transfected with LATS2-3ʹUTR-WT and LATS2-3ʹUTR-MUT. The cells were then incubated for 48 h. Lastly, the Luc-PairTM Duo-Luciferase Assay Kit (Genecopoeia) was used to quantify luciferase activity.
Statistical Analysis
Data are presented as means ± the standard deviation, and at least three independent replicates were performed. One-way analysis of variance and two-tailed Student’s t-test were performed using SPSS software 24.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 7.0 (San Diego, CA, USA). Statistical significance was assessed via the Kaplan–Meier method, Pearson’s correlation analysis, and the Log rank test. Photoshop and Adobe Illustrator were used to generate images. p < 0.05 was deemed to indicate statistical significance.
Results
Clinical Outcomes and miR-512-3p in HCC
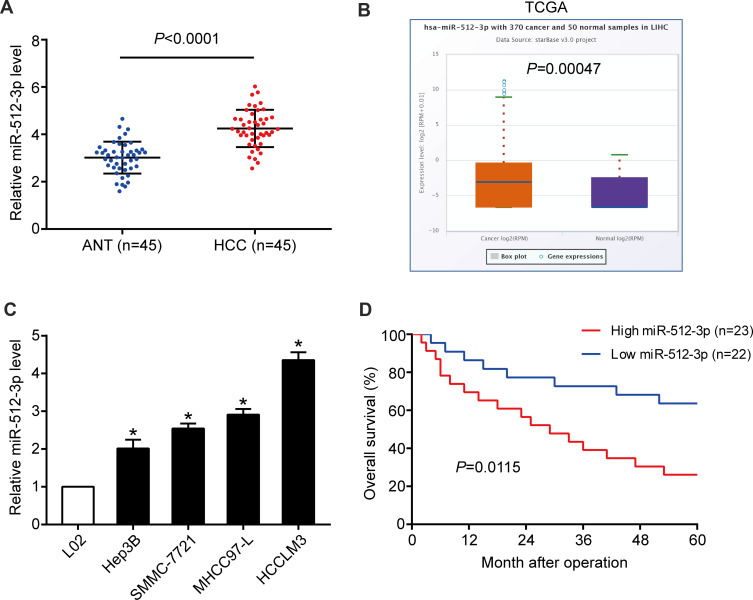
In HCC tissues miR-512-3p expression was higher than it was in non‑tumor tissues harvested in the study (p < 0.0001, Figure 1A), and it was higher than that reported in the TCGA data pertaining to normal liver tissues accessed via the StarBase V3.0 online platform (p = 0.00047, Figure 1B). Higher miR-512-3p levels were observed in HCC cell lines (Hep3B, SMMC-7721, MHCC97-L, and HCCLM3) than in the immortalized normal liver cell line L02 (Figure 1C). In miR-512-3p-high and miR-512-3p-low groups of HCC patients generated based on median miR-512-3p expression, high miR-512-3p was significantly correlated with tumor size (p = 0.026), vascular invasion (p = 0.042), and advanced tumor-node‑metastasis stage (p = 0.009) (Table 1). In Kaplan–Meier analysis HCC patients with high miR-512-3p expression exhibited worse overall survival (p = 0.0115, Figure 1D).
Figure 1.
Expression and prognostic value of miR-512-3p in HCC. (A) miR-512-3p levels in 45 human HCC samples and 45 adjacent normal tissue samples (p < 0.0001, Student’s t-test). (B) miR-512-3p levels were higher in the HCC tissues in the current study than the levels derived from normal liver tissues recorded in the TCGA database, accessed via the StarBase V3.0 platform (p = 0.0005, Student’s t-test). (C) qRT-PCR was performed to determine miR-512-3p levels in HCC cell lines (Hep3B, SMMC-7721, MHCC97-L, and HCCLM3) and L02 cells. *p < 0.05, Student’s t-test vs L02, n = 3. (D) In Kaplan–Meier survival analysis HCC patients with higher expression of miR-512-3p (n = 23) exhibited shorter overall survival than those with lower miR-512-3p expression (n = 22). p = 0.0115, Log rank test.
miR-512-3p and HCC Cell Proliferation, Migration, and Invasion
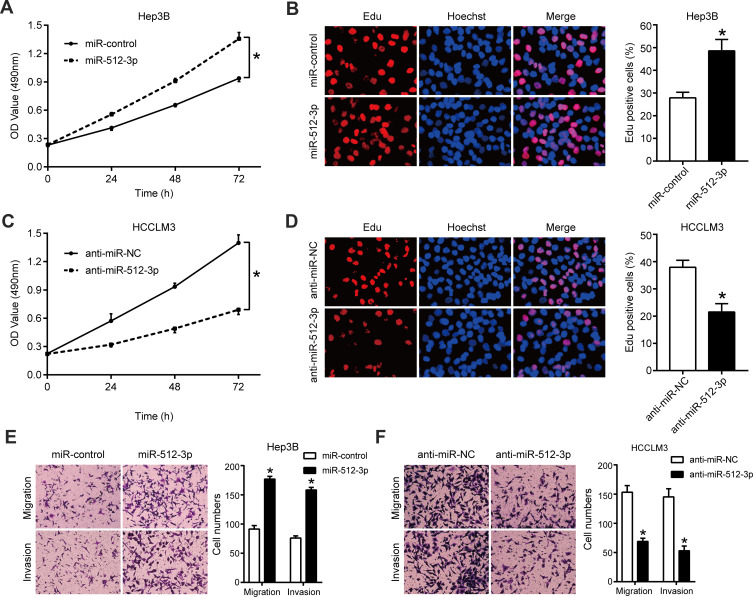
qRT-PCR results indicating the efficiency of transfection of Hep3B and HCCLM3 cells with miR-512-3p mimics and inhibitors are shown in Supplementary Figure 1. In MTT and EdU assays miR-512-3p mimics significantly enhanced the viability and proliferation of Hep3B cells, whereas miR-512-3p inhibitors reduced the viability and proliferation of HCCLM3 cells (p < 0.05, Figure 2A–D). In transwell migration and invasion assays miR-512-3p mimics markedly increased the number of Hep3B cells that passed through the membrane (p < 0.05, Figure 2E), and the number of MHCC97-H cells that passed through the membrane was significantly reduced by miR-512-3p inhibitors (p < 0.05, Figure 2F).
Figure 2.
miR-512-3p promotes HCC cell proliferation and motility. (A) In MTT assays miR-512-3p mimics increased Hep3B cell viability. *p < 0.05, analysis of variance, n = 3. (B) In EdU assays miR-512-3p mimics enhanced Hep3B cell proliferation. *p < 0.05, Student’s t-test; n = 3. (C) In MTT assays miR-512-3p inhibitors reduced HCCLM3 cell viability. *p < 0.05, analysis of variance, n = 3. (D) In EdU assays miR-1251-5p inhibitors suppressed MHCC97-H cell proliferation. *p < 0.05, Student’s t-test; n = 3. (E) In transwell migration and invasion assays miR-512-3p mimics enhanced the motility of Hep3B cells compared to control cells. *p < 0.05, Student’s t-test, n = 3. (F) In transwell migration and invasion assays miR-512-3p inhibitors impaired the motility of HCCLM3 cells compared to control cells. *p < 0.05, Student’s t-test; n = 3.
miR-512-3p and LATS2 Targeting in HCC
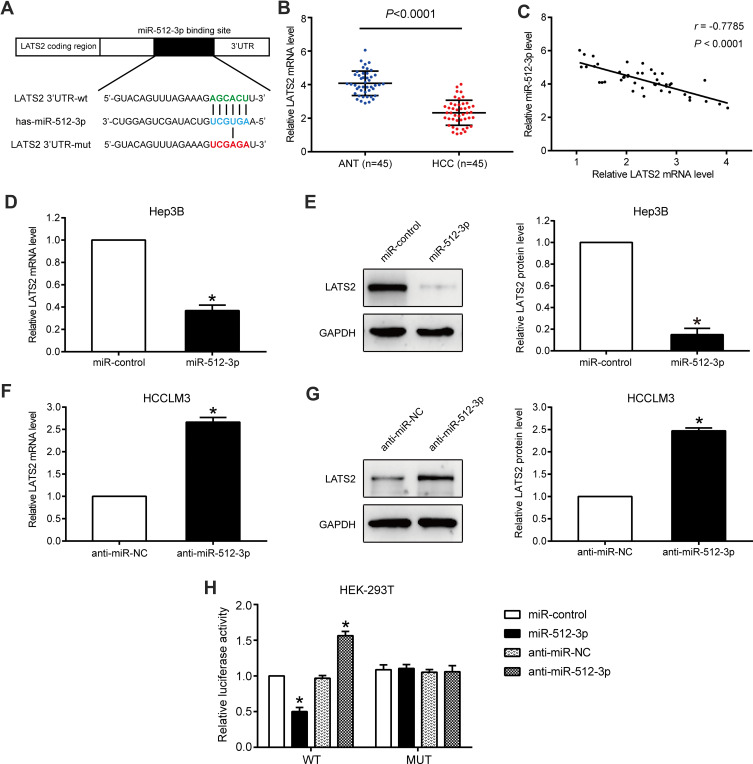
In qRT-PCR and Western blot assays conducted using HCC cells, only LATS2 was significantly downregulated by miR-512-3p (Figure 3D–G and Supplementary Figure 2A and B). Accordingly, LATS2 was selected as the target of miR-512-3p, and the complementary sequence between miR-512-3p and the 3ʹUTR of LATS2 is shown in Figure 3A. In qRT-PCR assays performed to detect LATS2 mRNA levels in 45 pairs of HCC tissues and adjacent non‑tumor tissues, LATS2 expression was lower in HCC tissues (p < 0.0001, Figure 3B). miR-512-3p expression was inversely correlated with LATS2 mRNA levels in HCC tissues (r = −0.7785, p < 0.0001, Figure 3C). qPCR and Western blot assays conducted to assess LATS2 levels in Hep3B cells treated with miR-512-3p mimics and HCCLM3 cells treated with miR-512-3p inhibitors indicated that LATS2 was significantly negatively regulated by miR-512-3p at the mRNA level and the protein level (p < 0.05, Figure 3D–G). In luciferase reporter gene assays miR-512-3p overexpression was suppressed but miR-512-3p knockdown enhanced the luciferase activity of the vector encoded with the WT-3ʹUTR of LATS2, but not the vector encoded with the MUT-3ʹUTR in HEK293T cells (p < 0.05, Figure 3H).
Figure 3.
LATS2 is a direct target of miR-512-3p in HCC. (A) miR-512-3p and its putative binding sequence in the 3ʹUTR of LATS2. The MUT LATS2 binding site was generated in the complementary site for the seed region of miR-512-3p. (B) LATS2 mRNA levels in 45 HCC samples and 45 samples from adjacent normal tissue. p < 0.0001, Student’s t-test. (C) In Pearson’s correlational analysis there was a significant inverse correlation between LATS2 mRNA and miR-512-3p in HCC tissues (r = −0.7785, p < 0.0001). (D) In qRT-PCR analyses LATS2 was significantly downregulated by miR-512-3p at the mRNA level in Hep3B cells and (F) HCCLM3 cells. *p < 0.05, Student’s t-test, n = 3. (E) In Western blot analysis LATS2 was significantly downregulated by miR-512-3p at the protein level in Hep3B cells and (G) HCCLM3 cells. *p < 0.05, Student’s t-test, n = 3. (H) miR-512-3p overexpression significantly suppressed the luciferase activity of WT but not MUT 3ʹUTRs of LATS2. miR-512-3p knockdown caused a dramatic increase in luciferase activity of WT but not MUT 3ʹUTRs of LATS2 in HEK293T cells. *p < 0.05, Student’s t-test, n = 3.
LATS2 Mediation of the Effects of miR-512-3p on Proliferation, Migration, and Invasion in HCC
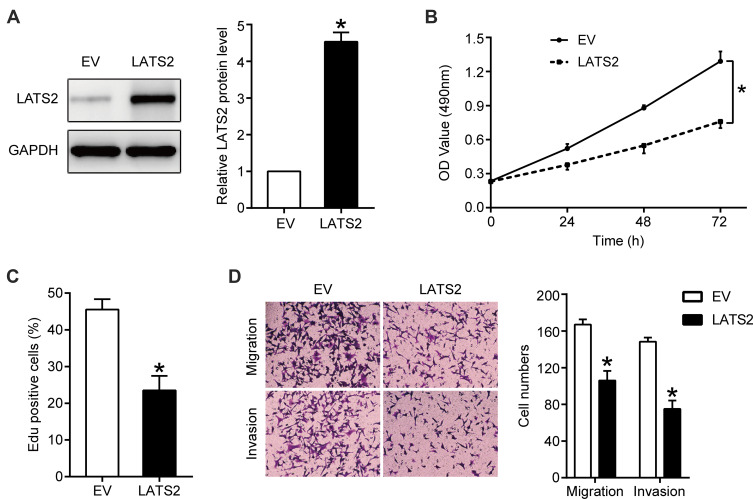
LATS2 was overexpressed in Hep3B cells overexpressing miR-512-3p via transfection with a LATS2 expression plasmid, and the transfection efficiency as confirmed by Western blotting is shown in Figure 4A. The results of MTT and EdU assays indicated that LATS2 overexpression partially abrogated the capacity of miR-512-3p to promote Hep3B cell viability and proliferation (p < 0.05, Figure 4B and C). In transwell assays the miR-512-3p mimic-induced enhanced motility of Hep3B cells was weakened after LATS2 overexpression (p < 0.05, Figure 4D).
Figure 4.
LATS2 overexpression partially abrogated the miR-512-3p overexpression-induced HCC cell proliferation, migration, and invasion. In Hep3B cells overexpressing miR-512-3p, LATS2 was overexpressed via transfection with a plasmid and the transfection efficiency was confirmed by Western blotting (A). *p < 0.05, Student’s t-test, n = 3. (B) MTT, (C) EdU, and (D) transwell assays were performed to assess the viability, proliferation, migration, and invasion capacities of HCCLM3 cells transfected with the corresponding vectors. *p < 0.05, analysis of variance or Student’s t-test, n = 3.
Effects of miR-512-3p on Hippo/YAP Signaling Pathways in HCC
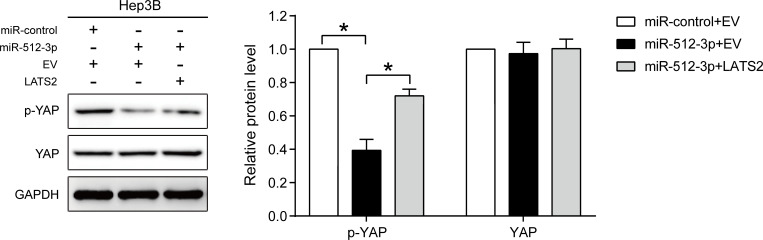
Western blot analysis indicated that miR-512-3p mimics reduced p-YAP expression, and that LATS2 overexpression partially abrogated the inhibitory effect of miR-512-3p mimics on p-YAP expression in Hep3B cells (p < 0.05, Figure 5).
Figure 5.
miR-512-3p inhibits the phosphorylation of YAP in HCC by targeting LATS2. Western blot indicating that miR-512-3p suppressed the phosphorylation of YAP in HCC, and that LATS2 overexpression reversed these effects. *p < 0.05, Student’s t-test, n = 3.
Hypoxia and miR-512-3p Expression in HCC
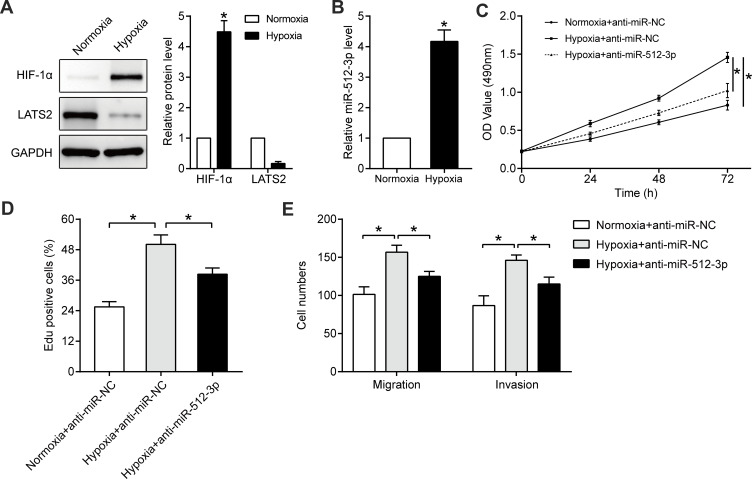
Hypoxia is an important feature of the microenvironment of solid tumors, and it promotes characteristics associated with malignancy such as growth39,42 and metastasis.43,44 In Hep3B cells cultured in hypoxic conditions (1% O2) for 24 h HIF1-α protein was significantly upregulated (p < 0.05, Figure 6A), confirming that the hypoxic cell model had been successfully generated. Hypoxia markedly increased miR-512-3p levels (p < 0.05, Figure 6B) and inhibited LATS2 expression (p < 0.05, Figure 6A). In functional assays hypoxia promoted the viability, proliferation, and mobility of Hep3B cells, and miR-512-3p inhibitors partially reversed the tumor-promoting effects of hypoxia (p < 0.05, Figure 6C–E).
Figure 6.
miR-512-3p mediates the tumor-promoting effects of hypoxia. Western blotting was performed to quantify HIF1-α and LATS2 levels in Hep3B cells cultured under normoxic or hypoxic conditions (A). *p < 0.05, Student’s t-test, n = 3. (B) qRT-PCR was conducted to quantify miR-512-3p levels in Hep3B cells cultured under normoxic or hypoxic conditions. *p < 0.05, Student’s t-test, n = 3. (C) MTT, (D) EdU, and (E) transwell assays were performed to assess the viability, proliferation, migration, and invasion capacities of Hep3B cells with or without miR-512-3p knockdown under hypoxic conditions. *p < 0.05, analysis of variance or Student’s t-test, n = 3.
Discussion
MicroRNAs are involved in the regulation of HCC progression.45,46 The roles of miR-512-3p have recently been investigated in various tumors. Duan et al47 reported that miR-512-3p regulated malignant tumor behavior and multi-drug resistance in breast cancer cells by targeting Livin. Zhu et al17 reported that inhibition of RAC1-GEF DOCK3 by miR-512-3p contributed to the suppression of metastasis in non-small cell lung cancer. Notably however, the biological function of miR-512-3p in HCC remains unclear. In the current study miR-512-3p was significantly elevated in HCC, and elevated miR-512-3p was associated with worse survival and unfavorable clinicopathological characteristics including tumor size, vascular invasion, and advanced tumor-node‑metastasis stages. In functional experiments miR-512-3p promoted HCC cell proliferation and mobility. Collectively these results suggest that miR-512-3p functions as an oncogene in HCC.
In HCC Hippo signaling acts as a cancer suppression pathway by inhibiting tumor-related processes, including proliferation,48 migration,49 and invasion.50 In the Hippo pathway LATS kinases phosphorylate YAP and cause the cytoplasmic retention and degradation of YAP. In previous studies various microRNAs have been linked to LATS2 regulation in human cancers, including miR-372 in breast cancer,27 miR‑492 in retinoblastoma,51 miR-135b in cutaneous melanoma,52 miR-363 in ovarian cancer,53 and miR-103 in HCC.31 Relationships between LATS2 and miR-512-3p have not been investigated in HCC. The present study generated substantial evidence that LATS2 is a direct functional target of miR-512-3p in HCC. miR-512-3p level was inversely correlated with LATS2 expression in HCC tissues. miR-512-3p downregulated LATS2 expression in HCC cells at the mRNA level and the protein level, and miR-512-3p affected the luciferase activity of the WT-3ʹUTR of LATS2 but not the MUT-3ʹUTR of LATS2. Lastly, in recuse assays restoration of LATS2 reversed the effects of miR-512-3p on HCC cell proliferation and mobility by activating the Hippo/YAP pathway. These results confirm that LATS2 is a direct functional target of miR-512-3p.
Hypoxia is an important feature of the microenvironment of solid tumors, and promotes malignancy.32–36,44 In several previous studies microRNAs have reportedly mediated the cancer-promoting effects of hypoxia in various tumor types.54 Zheng et al41 suggested that hypoxia could drive tumorigenesis and metastasis in HCC by downregulating miR-196-5p. In the present study relationships between hypoxia and miR-512-3p expression were investigated for the first time. Hypoxia could upregulate miR-512-3p expression in HCC, and hypoxia-induced miR-512-3p partially mediated the tumor-promoting effects of hypoxia. The mechanism involved in the elevated miR-512-3p expression induced by hypoxia in HCC will be investigated in future studies.
In summary, the current study suggests that miR-512-3p functions as an oncogene in HCC, and promotes the proliferation and mobility of HCC cells. Upregulated miR-512-3p expression was associated with reduced survival and unfavorable clinicopathological characteristics. Mechanistically miR-512-3p promotes tumorigenic characteristics by targeting LATS2, which results in reduced Hippo signaling. Hypoxia elevated miR-512-3p levels in HCC cells, and miR-512-3p partially mediated the tumor-promoting effects of hypoxia. Results of the present study suggest that miR-512-3p may be a novel therapeutic target for HCC treatment.
Acknowledgments
This work was supported by a grant from the Science and Technology Research Project of Jiangxi Provincial Education Department (No. 170031).
Abbreviations
HCC, hepatocellular carcinoma; HIF1-α, hypoxia-inducible factor 1-alpha; LATS2, large tumor suppressor kinase 2; miR-512-3p, placenta-specific microRNA; MUT, mutant; qRT-PCR, quantitative real-time PCR; UTR, untranslated region; WT, wild-type; YAP, yes-associated protein 1.
Disclosure
The authors declare that there are no conflicts of interest pertaining to this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 4.Islami F, Chen W, Yu XQ, et al. Cancer deaths and cases attributable to lifestyle factors and infections in China, 2013. Ann Oncol. 2017;28:2567–2574. doi: 10.1093/annonc/mdx342 [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 6.Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut. 2015;64:842–848. doi: 10.1136/gutjnl-2014-307990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Wang L, Yao B, Liu Q, Guo C. miR-1307-3p promotes tumor growth and metastasis of hepatocellular carcinoma by repressing DAB2 interacting protein. Biomed Pharmacother. 2019;117:109055. doi: 10.1016/j.biopha.2019.109055 [DOI] [PubMed] [Google Scholar]
- 9.Niu Y, Tang G. miR-185-5p targets ROCK2 and inhibits cell migration and invasion of hepatocellular carcinoma. Oncol Lett. 2019;17:5087–5093. doi: 10.3892/ol.2019.10144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han B, Zheng Y, Wang L, et al. A novel microRNA signature predicts vascular invasion in hepatocellular carcinoma. J Cell Physiol. 2019;234:20859–20868. doi: 10.1002/jcp.28690 [DOI] [PubMed] [Google Scholar]
- 11.Ye Y, Song Y, Zhuang J, et al. MicroRNA-302a-3p suppresses hepatocellular carcinoma progression by inhibiting proliferation and invasion. Onco Targets Ther. 2018;11:8175–8184. doi: 10.2147/OTT.S167162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Yin X, Yu X, Dai C, Zhou F. Long noncoding LINC01551 promotes hepatocellular carcinoma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of microRNA-122-5p to regulate ADAM10 expression. J Cell Biochem. 2019;120:16393–16407. doi: 10.1002/jcb.28549 [DOI] [PubMed] [Google Scholar]
- 13.Tian XP, Wang CY, Jin XH, et al. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics. 2019;9:1965–1979. doi: 10.7150/thno.30958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Lu J, Cao J, Ma B, Gao C, Qi F. MicroRNA-18a promotes hepatocellular carcinoma proliferation, migration, and invasion by targeting Bcl2L10. Onco Targets Ther. 2018;11:7919–7934. doi: 10.2147/OTT.S180971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Huang WJ, Xi HL, et al. Tumor-suppressive miR-3650 inhibits tumor metastasis by directly targeting NFASC in hepatocellular carcinoma. Aging. 2019;11:3432–3444. doi: 10.18632/aging.101981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Guo Y, Li Y, et al. miR300 regulates tumor proliferation and metastasis by targeting lymphoid enhancer-binding factor 1 in hepatocellular carcinoma. Int J Oncol. 2019;54:1282–1294. doi: 10.3892/ijo.2019.4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X, Gao G, Chu K, et al. Inhibition of RAC1-GEF DOCK3 by miR-512-3p contributes to suppression of metastasis in non-small cell lung cancer. Int J Biochem Cell Biol. 2015;61:103–114. doi: 10.1016/j.biocel.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2012;18:5442–5453. doi: 10.3748/wjg.v18.i38.5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis. 2012;33:1113–1120. doi: 10.1093/carcin/bgs113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manmadhan S, Ehmer U. Hippo signaling in the liver - A long and ever-expanding story. Front Cell Dev Biol. 2019;7:33. doi: 10.3389/fcell.2019.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, Yang C, Yang S, Cheng F, Rao J, Wang X. miR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis. 2018;9:954. doi: 10.1038/s41419-018-0978-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Xie Y, Lv Y, Zhang Y, Liang Z, Han L, Xie Y. LATS2 promotes apoptosis in non-small cell lung cancer A549 cells via triggering Mff-dependent mitochondrial fission and activating the JNK signaling pathway. Biomed Pharmacother. 2019;109:679–689. doi: 10.1016/j.biopha.2018.10.097 [DOI] [PubMed] [Google Scholar]
- 23.Guo C, Liang C, Yang J, Hu H, Fan B, Liu X. LATS2 inhibits cell proliferation and metastasis through the Hippo signaling pathway in glioma. Oncol Rep. 2019;41:2753–2761. doi: 10.3892/or.2019.7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Sun D, Gao J, et al. MicroRNA-373 promotes the development of endometrial cancer by targeting LATS2 and activating the Wnt/beta-Catenin pathway. J Cell Biochem. 2018;120(5):8611–8618. [DOI] [PubMed] [Google Scholar]
- 25.Mitamura T, Watari H, Wang L, et al. microRNA 31 functions as an endometrial cancer oncogene by suppressing Hippo tumor suppressor pathway. Mol Cancer. 2014;13:97. doi: 10.1186/1476-4598-13-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao W, Zhu S, Li P, Zhang S. Large tumor suppressor kinase 2 overexpression attenuates 5-FU-resistance in colorectal cancer via activating the JNK-MIEF1-mitochondrial division pathway. Cancer Cell Int. 2019;19:97. doi: 10.1186/s12935-019-0812-3 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Cheng X, Chen J, Huang Z. miR-372 promotes breast cancer cell proliferation by directly targeting LATS2. Exp Ther Med. 2018;15:2812–2817. doi: 10.3892/etm.2018.5761 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Wang L, Wang L, Chang W, Li Y, Wang L. MicroRNA-373 promotes the development of esophageal squamous cell carcinoma by targeting LATS2 and OXR1. Int J Biol Markers. 2019;34:148–155. doi: 10.1177/1724600819827964 [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Li S, Wang R, Chen C, Ma W, Cai H. Anti-tumor effect of LATS2 on liver cancer death: role of DRP1-mediated mitochondrial division and the Wnt/β-catenin pathway. Biomed Pharmacother. 2019;114:108825. doi: 10.1016/j.biopha.2019.108825 [DOI] [PubMed] [Google Scholar]
- 30.Xu F, Yang J, Shang J, et al. MicroRNA-302d promotes the proliferation of human pluripotent stem cell-derived cardiomyocytes by inhibiting LATS2 in the Hippo pathway. Clin Sci. 2019;133:1387–1399. doi: 10.1042/CS20190099 [DOI] [PubMed] [Google Scholar]
- 31.Han LL, Yin XR, Zhang SQ. miR-103 promotes the metastasis and EMT of hepatocellular carcinoma by directly inhibiting LATS2. Int J Oncol. 2018;53:2433–2444. doi: 10.3892/ijo.2018.4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L, Wang L, Chen T, et al. LncRNA RUNX1-IT1 which is downregulated by hypoxia-driven histone deacetylase 3 represses proliferation and cancer stem-like properties in hepatocellular carcinoma cells. Cell Death Dis. 2020;11:95. doi: 10.1038/s41419-020-2274-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y, Min Z, Zhou Z, et al. Hypoxia-induced exosomes promote hepatocellular carcinoma proliferation and metastasis via miR-1273f transfer. Exp Cell Res. 2019;385:111649. doi: 10.1016/j.yexcr.2019.111649 [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70:1298–1316. doi: 10.1002/hep.30671 [DOI] [PubMed] [Google Scholar]
- 35.Hu W, Zheng S, Guo H, et al. PLAGL2-EGFR-HIF-1/2alpha signaling loop promotes HCC progression and Erlotinib insensitivity. Hepatology. 2020. doi: 10.1002/hep.31293 [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Ren H, Zhou Y, et al. The hypoxia conditioned mesenchymal stem cells promote hepatocellular carcinoma progression through YAP mediated lipogenesis reprogramming. J Exp Clin Cancer Res. 2019;38:228. doi: 10.1186/s13046-019-1219-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma P, Tang WG, Hu JW, et al. HSP4 triggers epithelial-mesenchymal transition and promotes motility capacities of hepatocellular carcinoma cells via activating AKT. Liver Int. 2020;40:1211–1223. doi: 10.1111/liv.14410 [DOI] [PubMed] [Google Scholar]
- 38.Cao MQ, You AB, Cui W, et al. Cross talk between oxidative stress and hypoxia via thioredoxin and HIF-2α drives metastasis of hepatocellular carcinoma. FASEB J. 2020;34:5892–5905. doi: 10.1096/fj.202000082R [DOI] [PubMed] [Google Scholar]
- 39.Dou C, Zhou Z, Xu Q, et al. Hypoxia-induced TUFT1 promotes the growth and metastasis of hepatocellular carcinoma by activating the Ca(2+)/PI3K/AKT pathway. Oncogene. 2019;38:1239–1255. doi: 10.1038/s41388-018-0505-8 [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, Huang Y, Hu K, Zhang Z, Yang J, Wang Z. HIF1A activates the transcription of lncRNA RAET1K to modulate hypoxia-induced glycolysis in hepatocellular carcinoma cells via miR-100-5p. Cell Death Dis. 2020;11:176. doi: 10.1038/s41419-020-2366-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H, Bi FR, Yang Y, et al. Downregulation of miR-196-5p induced by hypoxia drives tumorigenesis and metastasis in hepatocellular carcinoma. Horm Cancer. 2019;10:177–189. doi: 10.1007/s12672-019-00370-5 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Liu Y, Yan W, Tohme S, et al. Hypoxia induced HMGB1 and mitochondrial DNA interactions mediate tumor growth in hepatocellular carcinoma through Toll-like receptor 9. J Hepatol. 2015;63:114–121. doi: 10.1016/j.jhep.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang J, Wang GZ, Wang Y, Huang HZ, Li WT, Qu XD. Hypoxia-induced HMGB1 expression of HCC promotes tumor invasiveness and metastasis via regulating macrophage-derived IL-6. Exp Cell Res. 2018;367:81–88. doi: 10.1016/j.yexcr.2018.03.025 [DOI] [PubMed] [Google Scholar]
- 44.Liu Z, Wang Y, Dou C, et al. Hypoxia-induced up-regulation of VASP promotes invasiveness and metastasis of hepatocellular carcinoma. Theranostics. 2018;8:4649–4663. doi: 10.7150/thno.26789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang YJ, Pan Q, Yu Y, Zhong XP. microRNA-519d induces autophagy and apoptosis of human hepatocellular carcinoma cells through activation of the AMPK signaling pathway via Rab10. Cancer Manag Res. 2020;12:2589–2602. doi: 10.2147/CMAR.S207548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi DM, Shi XL, Xing KL, Zhou HX, Lu LL, Wu WZ. miR-296-5p suppresses stem cell potency of hepatocellular carcinoma cells via regulating Brg1/Sall4 axis. Cell Signal. 2020;72:109650. doi: 10.1016/j.cellsig.2020.109650 [DOI] [PubMed] [Google Scholar]
- 47.Duan WJ, Bi PD, Ma Y, Liu NQ, Zhen X. MiR-512-3p regulates malignant tumor behavior and multi-drug resistance in breast cancer cells via targeting Livin. Neoplasma. 2020;67:102–110. doi: 10.4149/neo_2019_190106N18 [DOI] [PubMed] [Google Scholar]
- 48.Mao J, Tian Y, Wang C, et al. CBX2 Regulates Proliferation and Apoptosis via the Phosphorylation of YAP in Hepatocellular Carcinoma. J Cancer. 2019;10:2706–2719. doi: 10.7150/jca.31845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ou H, Chen Z, Xiang L, et al. Frizzled 2-induced epithelial-mesenchymal transition correlates with vasculogenic mimicry, stemness, and Hippo signaling in hepatocellular carcinoma. Cancer Sci. 2019;110:1169–1182. doi: 10.1111/cas.13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng Y, Hou T, Ping J, Chen T, Yin B. LMO3 promotes hepatocellular carcinoma invasion, metastasis and anoikis inhibition by directly interacting with LATS1 and suppressing Hippo signaling. J Exp Clin Cancer Res. 2018;37:228. doi: 10.1186/s13046-018-0903-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Z, Zhang A, Zhang L. Inhibition of microRNA492 attenuates cell proliferation and invasion in retinoblastoma via directly targeting LATS2. Mol Med Rep. 2019;19:1965–1971. doi: 10.3892/mmr.2018.9784 [DOI] [PubMed] [Google Scholar]
- 52.Hu Y, Wang Q, Zhu XH. MiR-135b is a novel oncogenic factor in cutaneous melanoma by targeting LATS2. Melanoma Res. 2019;29:119–125. doi: 10.1097/CMR.0000000000000524 [DOI] [PubMed] [Google Scholar]
- 53.Mohamed Z, Hassan MK, Okasha S, et al. miR-363 confers taxane resistance in ovarian cancer by targeting the Hippo pathway member, LATS2. Oncotarget. 2018;9:30053–30065. doi: 10.18632/oncotarget.25698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karshovska E, Wei Y, Subramanian P, et al. HIF-1α (Hypoxia-Inducible Factor-1α) promotes macrophage necroptosis by regulating miR-210 and miR-383. Arterioscler Thromb Vasc Biol. 2020;40:583–596. doi: 10.1161/ATVBAHA.119.313290 [DOI] [PubMed] [Google Scholar]