SUMMARY
Understanding of NAD+ metabolism provides many critical insights into health and diseases; yet highly sensitive and specific detection of NAD+ metabolism in live cells and in vivo remains difficult. Here we present ratiometric, highly responsive genetically encoded fluorescent indicators, FiNad, for monitoring NAD+ dynamics in living cells and animals. FiNad sensors cover physiologically relevant NAD+ concentrations and sensitively respond to increases and decreases in NAD+. Utilizing FiNad, we performed a head-to-head comparison study of common NAD+ precursors in various organisms and mapped their biochemical roles in enhancing NAD+ levels. Moreover, we showed that increased NAD+ synthesis controls morphofunctional changes of activated macrophages, and directly imaged NAD+ declines during aging in situ. The broad utility of the FiNad sensors will expand our mechanistic understanding of numerous NAD+-associated physiological and pathological processes, and facilitate screening for drug or gene candidates that affect uptake, efflux, and metabolism of this important cofactor.
INTRODUCTION
Nicotinamide adenine dinucleotide (NAD+) and its reduced form NADH are key coenzymes for redox reactions and play central roles in energy metabolism(Verdin, 2015; Zhao et al., 2018). NAD+ is also a co-substrate of many regulatory enzymes, such as sirtuins (SIRTs), poly (ADP-ribose) polymerases (PARPs), and the cyclic ADP-ribose synthases (CD38 and CD157); and participates in a wide range of cellular processes(Canto et al., 2015; Rajman et al., 2018). Its levels are often altered in aging(Bonkowski and Sinclair, 2016; Imai and Guarente, 2014; Verdin, 2014), neurodegeneration(Verdin, 2015), kidney injury(Poyan Mehr et al., 2018), obesity(Canto et al., 2012), diabetes(Yoshino et al., 2011), adipogenesis(Ryu et al., 2018), cancer(Tateishi et al., 2015), and congenital malformations(Vander Heiden, 2017). In eukaryotic cells, the NAD+-synthesizing and NAD+-consuming enzymes are highly compartmentalized(Canto et al., 2015). For many years, the weak endogenous fluorescence of NADH has been used in studies of mitochondrial metabolic states; however, NAD+ lacks the intrinsic fluorescence, which hinders its direct imaging in living cells and in vivo. Traditional biochemical methods, such as enzymatic cycling assays, chromatography, and mass spectrometry, require cell lysis, making it challenging to obtain spatiotemporal information about NAD+ functions in single living cells and in vivo.
A genetically encoded fluorescent sensor for measuring NAD+ was previously reported by fusing a bacterial NAD+-binding DNA ligase (LigA) to cpVenus (abbreviated as LigA-cpVenus)(Cambronne et al., 2016). Although LigA-cpVenus represented a significant step forward for live cell NAD+ imaging, its dynamic response and specificity appeared to be limited. Very recently, a semisynthetic NAD+ indicator, denoted NAD-Snifit, was shown to have a large dynamic response in vitro(Sallin et al., 2018). NAD-Snifit, however, has no intrinsic fluorescence and requires exogenous dyes for labelling. Thus, the excess of dyes have to be removed by a multiple washing procedure and lengthy incubation, which is not only time intensive but also may render the analysis susceptible to artifacts. Considering the vital role that NAD+ metabolism plays in health and disease, a genetically encoded NAD+ sensor of superior performance remains to be developed.
Previously, we and others developed genetically encoded NADH sensors for monitoring the NADH/NAD+ redox state at subcellular resolution (Bilan et al., 2014; Hung et al., 2011; Zhao et al., 2015; Zhao et al., 2011). Among them, the SoNar sensor is unique as its fluorescence also markedly responds to the binding of NAD+; however, SoNar reports the NADH/NAD+ ratio rather than the absolute concentrations of the two nucleotides intracellularly. Herein, we report the development of a highly responsive, ratiometric, genetically encoded NAD+ sensor, denoted FiNad (fluorescent indicator of NAD+/AXP ratio), where AXP is the total pool of ATP and ADP. FiNad allows rapid, sensitive, specific, and real-time read-out of NAD+ dynamics in various organisms, thereby providing a powerful tool for NAD+ detection and bioimaging.
DESIGN
Circular permuted fluorescent proteins (cpFPs) have been utilized for the development of genetically encoded fluorescent sensors to monitor cellular metabolites. In these cpFPs, the original N- and C-termini are joined with a short peptide linker, and new N- and C-termini are created around the fluorophore of cpFPs, making its fluorescence highly sensitive to changes in metabolite levels. In our previous study (Zhao et al., 2015), cpYFP was inserted between amino acids residues located on surface loops of Rex protein from Thermus aquaticus (T-Rex). Among them, the chimera with cpYFP inserted after Phe189 of T-Rex showed significantly higher responses upon NADH binding or NAD+ binding than other chimeras (Zhao et al., 2015). To develop an NAD+ specific sensor, we then created a library of constructs with randomized linkers of 1–3 amino acid residues between T-Rex and cpYFP in the F189/L190 chimera, and selected for the variants not only with enhanced dynamic response to NAD+ and but also with reduced affinity toward NADH (Figures S1A and S1B). This library of variants was expressed in E. coli on solid media, and clones that exhibited bright green fluorescence were picked. Chimera variants expressed in these clones were then assayed for their fluorescence response to NAD+ and NADH, respectively (Figure S1A). Among more than 500 clones, we identified one variant exhibited the most dramatic increase in fluorescence excited at 485 nm upon NAD+ addition (Figure S1C). This variant, termed FiNad, was sequenced (Figure S1B; Table S1) and further characterized. As a genetically encoded sensor, FiNad can be easily introduced into cells, organelles, or organisms of interest by transfection, infection, or electroporation. In comparison, it would be very challenging to apply semisynthetic sensors such as NAD-Snifit(Sallin et al., 2018) for in vivo studies in animals, as it is difficult to remove unbound extraneous dyes, which lead to significant interference (the dye itself strong fluorescence). We, therefore, reasoned that FiNad might be a very useful reagent with which to monitor NAD+ fluctuations in live cells and in vivo.
RESULTS
A highly responsive genetically encoded NAD+/AXP ratio sensor
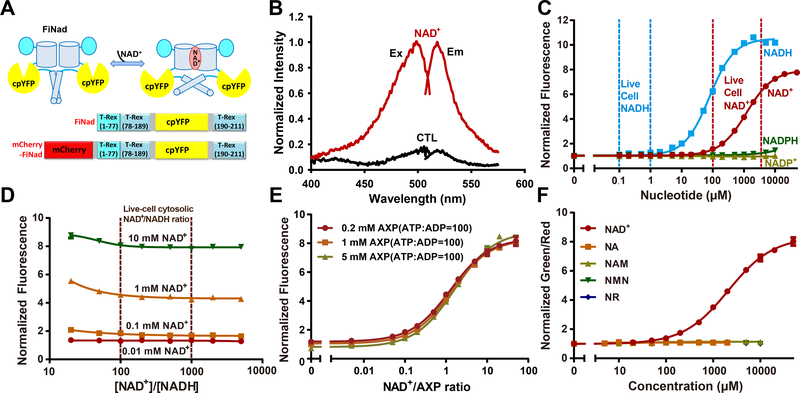
FiNad sensor was created by inserting cpYFP between residues 189 and 190 of the T-Rex monomer using short polypeptide linkers, Gly and Gly-Thr-Gly (Figures 1A and S1B), and manifested a dramatic dynamic range, i.e. 7-fold increase in fluorescence (ΔF/F) when excited at 485 nm in the presence of NAD+ (Figures 1B and S1C). FiNad’s fluorescence responded to NADH similarly, but not to other adenine nucleotides, such as NADP+, NADPH, ATP, or ADP (Figure S1D). Nevertheless and interestingly, we found that ATP or ADP, the adenine nucleotides most highly abundant in the cells, competes with the binding of NAD+ to FiNad, shifting the apparent Kd of FiNad for NAD+ from ~14 μM to ~1.3 mM (Figure S1E), which falls in the range of physiological NAD+ levels (Figure 1C). The competition between NAD+ and AXP is not surprising, as ADP is an intrinsic moiety of NAD+. In the presence of physiological adenine nucleotide concentrations, the Kd of FiNad for NADH shifted from ~1.5 μM to ~100 μM, which is far above cytosolic/nuclear free NADH concentration (0.1–1 μM)(Zhang et al., 2002; Zhao et al., 2011) (Figures 1C and S1D). Considering that cytosolic/nuclear NAD+/NADH ratio typically ranges from 100 to 1000(Zhao et al., 2016), thus, FiNad should respond to the intracellular NAD+ concentration, but not to the NAD+/NADH ratio in cytosol or nucleus of the cells (Figure 1D). According to this competitive scheme, FiNad’s steady-state fluorescence response should report the NAD+/AXP ratio rather than the absolute concentrations of these nucleotides (Figures 1E and S1F–S1J). Like ATP or ADP, AMP has similar competitive effect on NAD+ binding (Figure S1K); however, intracellular AMP concentration is two orders of magnitude less than the total pool of ATP and ADP(Park et al., 2016). Thus, its effect on the response of FiNad would be minimal within cells. Furthermore, our data show that FiNad’s response to NAD+ was not affected by NADP+, NADPH, and various ATP/ADP ratios, which determine the energy charge of the cell (Figures S1L and S1M). Considering that the total physiological pool of adenine nucleotides are usually maintained in homeostasis(Carling et al., 2011; Hardie et al., 2012), we surmised that the FiNad sensor actually reports intracellular NAD+ fluctuations.
Figure 1. A highly responsive genetically encoded NAD+/AXP ratio sensor.
(A) Schematic representation of the NAD+ sensor FiNad. Fluorescent protein cpYFP was inserted into a monomer of NAD+(NADH)-binding bacterial protein T-Rex. Binding of NAD+ induces changes in protein conformation and fluorescence. (B) Fluorescence spectra of purified FiNad in the control condition (black), and saturated NAD+ (dark red). The excitation spectrum recorded at an emission wavelength of 535 nm has a maximum at 500 nm; the emission spectrum recorded at an excitation wavelength of 490 nm has a maximum at 518 nm. (C) Fluorescence responses of FiNad with excitation at 485 nm in the presence of different concentrations of NAD+ and its analogs (n=3). Data are normalized to the initial value. (D) FiNad sensor responds to the indicated [NAD+], but not to the NAD+/NADH ratio (n=3). Data are normalized to the fluorescence in the absence of NAD+. (E) FiNad fluorescence plotted against NAD+/AXP ratio at the indicated total adenine nucleotide concentration (n=3). Data are normalized to the initial value (1mM AXP). (F) The green/red ratio of mCherry-FiNad towards NAD+ or its precursors (n=3). Data are normalized to the initial value. For C, D and F, the total adenine nucleotide concentration was 1 mM. Data are the mean ± s.d. See also Figure S1, Table S1 and Table S2.
For dual color ratiometric imaging, we fused FiNad with mCherry, a red fluorescent protein (Figure 1A). mCherry-FiNad responded to NAD+ very similarly to FiNad, suggesting the mCherry fusion did not interfere with FiNad’s NAD+ sensing function (Figure S1N). Therefore, the green/red fluorescence ratio of mCherry-FiNad can be used to report NAD+ dynamics specifically, unaffected by the sensor’s concentration. Further studies showed that the mCherry-FiNad sensor had high selectivity toward NAD+, without any apparent fluorescence changes toward or in the presence of NAD+ precursors such as nicotinic acid (NA), nicotinamide (NAM), nicotinamide mononucleotide (NMN), and nicotinamide riboside (NR) (Figures 1F, S1O, and S1P). In comparison, the LigA-cpVenus NAD+ sensor showed significant non-specificity, as it responded to two frequently used NAD+ precursors, NMN and NR, with affinity similar to that of NAD+ (Figure S1Q). As a result, the response of LigA-cpVenus to NAD+ was diminished in the presence of 0.5–5 mM NMN or NR (Figures S1R and S1S), the pharmacological concentrations widely used in studies of intracellular NAD+ metabolism (Ryu et al., 2018; Yoshino et al., 2018).
As other cpFP-based sensors(Tao et al., 2017; Zhao et al., 2015; Zhao et al., 2011), mCherry-FiNad’s green fluorescence excited at 485 nm is sensitive to pH (Figure S1T), but its dynamic range, Kd, and red fluorescence excited at 590 nm are comparatively pH resistant (Figures S1T–S1V). When pH fluctuations do occur, the pH effects can be corrected by measuring mCherry-FiNad’s and mCherry-cpYFP’s fluorescence in parallel, owing to their very similarpH responses (Figure S1W). In addition, FiNad’s fluorescence is not significantly affected by temperature fluctuations between 20 °C and 40 °C (Figure S1X). Taken together, these data show that FiNad sensor displays excellent sensitivity, selectivity and a large dynamic range, which makes it a promising tool for live cell and in vivo NAD+ studies.
Imaging NAD+ metabolism in living bacteria
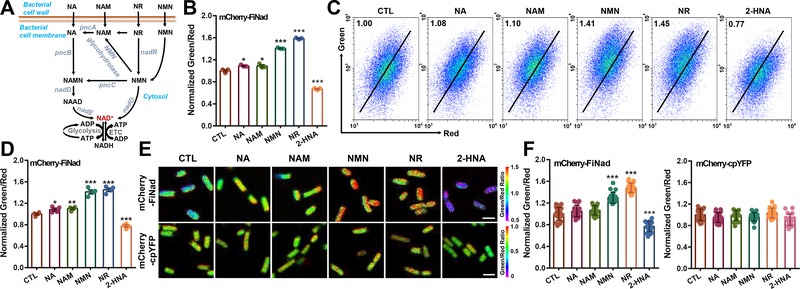
To assess the suitability of mCherry-FiNad in living bacteria, we expressed the sensor in the cytoplasm of E. coli BL21 (DE3) cells. FiNad manifested significant changes of its fluorescence when cellular NAD+ levels increased upon extraneous NAD+ precursor supplementation (e.g., NMN and NR), or when NAD+ levels decreased by nicotinic acid phosphoribosyltransferase (pncB) inhibitor, 2-hydroxynicotinic acid (2-HNA), treatment (Figures 2A and 2B). These data are consistent with the results of biochemical analysis of cellular NAD+ content (Figure S2A), and cellular AXP pool showed minimal changes (Figure S2B). In contrast, the LigA-cpVenus sensor showed minimal responses when cells were treated with NA, NAM, NMN, NR, or 2-HNA (Figures S2C and S2D). FiNad’s fluorescence can be monitored by flow cytometry analysis or confocal microscopy (Figures 2C–2F). As the control, mCherry-cpYFP’s fluorescence did not significantly change upon NAD+ precursors or 2-HNA treatment (Figures 2F, S2E, and S2F). These data excluded the possibility of interference by pH variations.
Figure 2. Imaging NAD+ metabolism in living bacteria.
(A) NAD+ biosynthesis from different precursors in bacteria. (B and C) Microplate assay (B, n=3) and flow cytometric analyses (C) of mCherry-FiNad fluorescence in E. coli BL21 (DE3) cells treated with NAD+ precursors or the pncB inhibitor 2-HNA. (D) Quantification of mCherry-FiNad fluorescence in panel C (n=4). (E and F) Fluorescence images (E) and quantification (F, n=20) of mCherry-FiNad or mCherry-cpYFP in E. coli BL21 (DE3) cells with NAD+ precursors or 2-HNA, scale bar, 2 μm. Data are the mean ± s.e.m (B, D) or mean ± s.d (F), normalized to the control condition (B, D, F). *P < 0.05, **P < 0.01, ***P < 0.001. See also Figure S2 and Table S3.
FiNad sensor reports NAD+ metabolism in living cells and in vivo
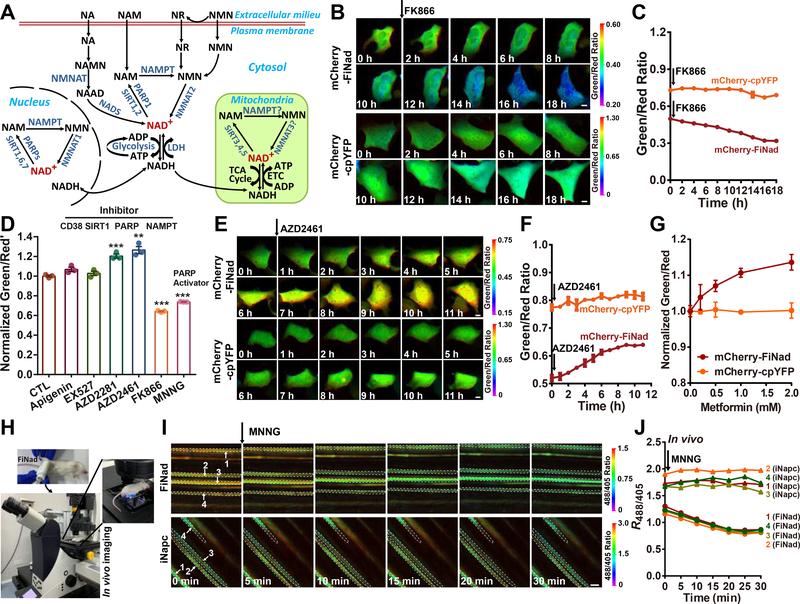
When stably expressed in human embryonic kidney 293 cells, the mCherry-FiNad sensor showed bright fluorescence and was mostly localized in the cytosol (Figure S3A). Agents that significantly affected SoNar fluorescence and cellular NADH/NAD+ redox states, e.g., the mitochondrial complex I inhibitor rotenone or pyruvate (Figure S3B), did not significantly alter FiNad’s fluorescence, consistent with our previous reports that these agents cause ratio fluctuation in cells, and that cellular NAD+ levels largely remain constant under these treatment conditions(Zhao et al., 2015; Zhao et al., 2016). When cells were treated with rotenone and oligomycin simultaneously, the fluorescence of PercevalHR, an ADP/ATP ratio biosensor, increased significantly (Figure S3C), suggesting an increase of cellular ADP/ATP ratio(Tantama et al., 2013); by contrast, mCherry-FiNad’s fluorescence remained constant. These data suggest that mCherry-FiNad is not sensitive to changes in intracellular ADP/ATP ratio or NADH/NAD+ ratio.
In mammalian cells, NAD+ is synthesized predominantly through the NAMPT-dependent salvage pathway(Canto et al., 2015). [For reference, we attach a diagram that explains the central nodes for cellular NAD+ metabolism in this study (Figure 3A).] We found that mCherry-FiNad correctly reported the decrease of cytosolic NAD+ level when NAMPT was inhibited by the pharmacological inhibitor, FK866, as indicated by the decrease of mCherry-FiNad green/red fluorescence ratio and cellular NAD+ pool (Figures 3B–3D, and S3D–S3F), whereas cellular AXP pool showed minimal changes (Figure S3F). Conversely, mCherry-FiNad’s fluorescence increased upon inhibition of three known NAD+-consuming enzymes concomitant with the expected increase in cellular NAD+ (Figures 3D–3F). Among them, inhibition of PARP1/2 (inhibitors, AZD2281 and AZD2461) was most effective at increasing cellular NAD+ level, followed by inhibition of CD38 (inhibitor, apigenin) and inhibition of SIRT1 (inhibitor, EX527) (Figures 3D–3F), suggesting PARPs are the major cellular NAD+ consumer in these cells. Interestingly, we also found that metformin, a drug used to treat type 2 diabetes mellitus (and possibly lifespan extension in some systems), induced a dose-dependent NAD+ increase (Figure 3G). In contrast, cells expressing mCherry-cpYFP instead of mCherry-FiNad showed no obvious fluorescence changes after metformin treatment, thereby excluding the pH effects (Figure 3G). FiNad was also able to report the rapid drop of cytosolic NAD+ levels during PARP1 activation by MNNG in living cells, ex vivo muscle tissues and live mice (Figures 3H–3J, and S3G–S3J). Consistent with this FiNad-based measurement, the measurement of the total NAD+ pool in cell lysates by a biochemical assay also showed that the cellular NAD+ level increased after PARP1/2, CD38, SIRT1 inhibition, or metformin treatment, and decreased with NAMPT inhibition or PARP activation, whereas cellular AXP pool showed minimal changes (Figures S3K–S3M). Only high concentrations of MNNG, the PARP activator, caused marked decrease of cellular AXP pool (Figure S3H), which was consistent with previous reports as massive ADP ribosylation reaction depleted AXP pool(Zong et al., 2004). Even under such extreme conditions, however, the decrease of NAD+ levels is still more significant than that of AXP levels, and FiNad correctly reported the decrease of the NAD+/AXP ratio. Collectively, these data suggest that cellular NAD+ is more sensitive to cellular activities and environmental changes, while adenine nucleotides have a strong tendency to maintain physiological homeostasis. We further expressed the FiNad sensor in the nucleus by tagging it with organelle-specific signal peptides (Figure S3A). The nuclear NAD+ level in resting cells or cells treated with PARP1/2 inhibitor was similar to that of cytosol (Figures S3A, S3N and S3O), as NAD+ diffuses freely between these two compartments. These data demonstrate the specific role of PARP1/2, CD38, SIRT1, and NAMPT as viable therapeutic targets for modulating NAD+ metabolism.
Figure 3. FiNad sensor reports NAD+ metabolism in living cells and in vivo.
(A) The central nodes for cellular NAD+ metabolism. (B and C) Fluorescence images (B) and quantification (C, n=3) of mCherry-FiNad or mCherry-cpYFP in HEK293 cells treated with 10 nM FK866, scale bar, 10 μm. (D) Effects of NAD+ metabolism pathway inhibitors and activators on cytosolic NAD+ measured by mCherry-FiNad fluorescence (n=3). (E and F) Fluorescence images (E) and quantification (F, n=3) of mCherry-FiNad or mCherry-cpYFP in HEK293 cells treated with 5 μM AZD2461, scale bar, 10 μm. (G) Fluorescence response of mCherry-FiNad in live cells to metformin (n=3). (H) General overview of the procedure for in vivo imaging of FiNad in muscle tissues of living mice. (I and J) In vivo fluorescence images (I) and quantification (J) of FiNad or iNapc in muscle tissues of living mice in response to MNNG indicating regions of interest (white dashed line). Images are pseudocolored by R488/405. Scale bar, 100 μm. Data are the mean ± s.e.m (D, G) or mean ± s.d. (C, F), normalized to the control condition (D, G). **P < 0.01, ***P < 0.001. See also Figure S3.
Mapping the different roles of NAD+ precursors in boosting NAD+ levels in various organisms
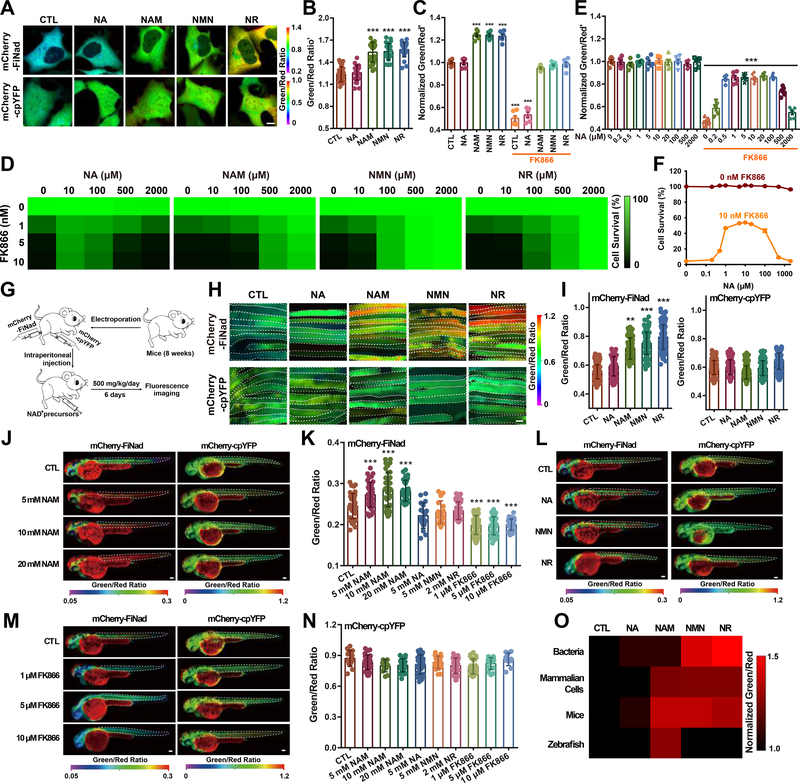
The administration of NAD+ precursors has long been known to promote a variety of beneficial effects in cells; however, how different NAD+ precursors are metabolized and regulated to protect cells remains unclear in mammalian cells (Figure 3A). As shown by FiNad’s fluorescence, NAD+ precursors such as NAM, NMN, and NR effectively increased cytosolic NAD+ levels (Figures 4A, 4B, S4A, and S4B), consistent with the results of biochemical analysis of cellular NAD+ content (Figure S4C), while cellular AXP levels remained constant (Figure S4D). Interestingly, NAD+ precursors such as NAM, NMN, and NR also reversed NAMPT inhibition-triggered NAD+ deficiency and cell death (Figures 4C, 4D, and S4E). The NAD+ precursor NA was much less effective at enhancing cytosolic NAD+ levels in resting HEK293 cells; however, NA was able to replenish cytosolic NAD+ levels in NAMPT-inhibited cells, and partially protected the cells from death (Figures 4E and 4F). These data suggest that the Preiss-Handler pathway for NAD+-synthesis from NA may be activated upon inhibition of the NAMPT-dependent salvage pathway. Interestingly, NA-mediated replenishment of the NAD+ level and improvement of cell survival in NAMPT-inhibited cells only occurred at low NA concentrations; higher concentrations of NA, e.g., 2 mM, failed to replenish the NAD+ level or protect the cells from death (Figures 4D–4F, and S4E). The explanation for this biphasic dose-dependent effect of NA in the presence of NAMPT inhibition on NAD+ (partial) restoration remains to be investigated.
Figure 4. Mapping the different roles of NAD+ precursors in boosting NAD+ levels in various organisms.
(A) Fluorescence images of mCherry-FiNad or mCherry-cpYFP in HEK293 cells treated with NAD+ precursors, scale bar, 10 μm. (B) Quantification of mCherry-FiNad fluorescence corrected by mCherry-cpYFP in panel A (n=25). (C) Effect of different NAD+ precursors (2 mM) on cytosolic NAD+ levels in the absence or presence of FK866 (n=6). (D) Effect of different NAD+ precursors on FK866-triggered cell death shown as a heatmap, with data from Figure S4E. (E and F) Effect of different concentrations of NA on cytosolic NAD+ levels (E) and cell viability (F) in the absence or presence of FK866 (n=6). (G) General overview of the procedure for NAD+ imaging in muscle tissues of mice. (H and I) Fluorescence images (H) and quantification (I, left: n=171, 83, 70, 155, 154; right: n=173, 130, 148, 115, 129) of mCherry-FiNad or mCherry-cpYFP in muscle tissues of mice in response to different NAD+ precursors indicating regions of interest (white dashed line). Images are pseudocolored by R488/561. Scale bar, 100 μm. (J-N) In vivo fluorescence imaging (J, L, M) and quantification (K, n=39, 36, 42, 28, 19, 16, 37, 42, 33, 24; N, n=15, 30, 12, 24, 28, 24, 34, 27, 15, 12) of zebrafish larvae expressing mCherry-FiNad or mCherry-cpYFP in response to different concentrations of NAM (J), NAD+ precursors (L), or FK866 (M) indicating regions of interest. (O) A head-to-head comparison study of four common NAD+ precursors in enhancing NAD+ levels in different species shown as a heatmap, with data from Table S3. Data are the mean ± s.e.m (C, E, F) or mean ± s.d. (B, I, K, N). **P < 0.01, ***P < 0.001. See also Figure S4 and Table S3.
To assess further the in vivo efficacy of four precursors in increasing NAD+ levels, we transferred a vector encoding mCherry-FiNad into mouse muscle tissues via electroporation. The following day, mice at 8 weeks of age were given intraperitoneal injections of vehicle control, NA, NAM, NMN, or NR (500 mg per kg daily) for 6 days. Notably, NAM, NMN, and NR treatment resulted in a significant NAD+ increase in muscle tissues compared with controls or NA treatment (Figures 4G–4I), consistent with the results of biochemical analysis of cellular NAD+ content (Figure S4F), while cellular AXP level remain constant (Figure S4G). As a control, minimal changes in fluorescence were observed in mCherry-cpYFP-expressing muscle tissues (Figures 4H and 4I). We further microinjected mCherry-FiNad sensor into the animal pole of zebrafish embryos and cultivated the embryos for ~30 h. Individual zebrafish were next exposed to NAD+ precursors for an additional ~24 h, and then narcotized for in vivo imaging. Upon NAM supplementation, we observed a dose-dependent increase in NAD+ level, as shown by mCherry-FiNad’s fluorescence ratio signal (Figures 4J and 4K); however, the other three NAD+ precursors, i.e., NA, NMN, and NR, were not able to boost NAD+ levels (Figures 4K and 4L). NAMPT inhibition with FK866 treatment induced a marked decrease in NAD+ level at both 28 h and 54 h post-fertilization in zebrafish (Figures 4K, 4M, S4H and S4I). These data suggest that the NAM-NAMPT axis accounts for NAD+ biosynthesis in zebrafish. As a control, minimal changes in fluorescence were observed in mCherry-cpYFP-expressing zebrafish with NAD+ precursors or FK866 treatment (Figures 4J, 4L–4N, and S4J). Consistent with this FiNad-based measurement, biochemical analysis also showed that total NAD+ level increased after NAM supplementation, and decreased with NAMPT inhibition (FK866), whereas cellular AXP pool showed minimal changes in zebrafish (Figures S4K and S4L). Taken together, we utilizing FiNad have performed a head-to-head comparison study of four common NAD+ precursors in various organisms and mapped their roles in enhancing NAD+ levels shown as a heatmap (Figure 4O).
Increased synthesis of NAD+ controls morphofunctional changes in activated macrophages
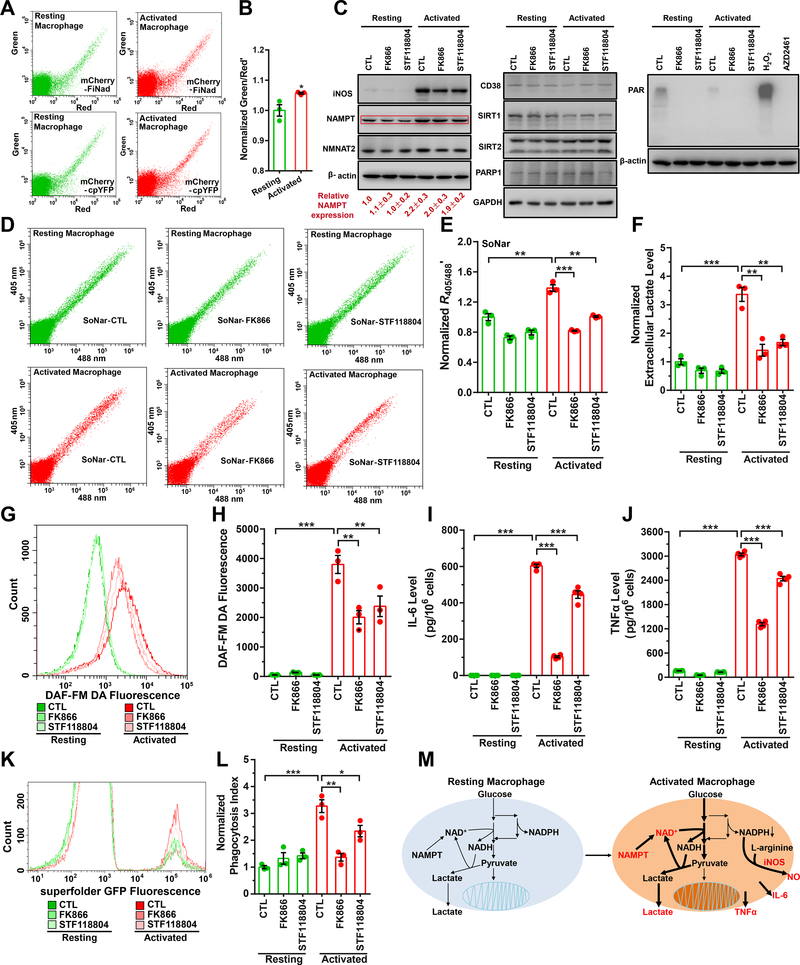
Macrophages function as ‘pathogen sensors,’ and are critical participants in inflammation and aging (Plowden et al., 2004; Solana et al., 2006). The bioenergetic demands of different macrophage populations have been studied extensively (Ghesquiere et al., 2014); however, much less is known about the relationship between macrophage activation and cellular NAD+ metabolism. Macrophage activation induced by lipopolysaccharide (LPS) and interferon-γ (IFN-γ) modestly increased cytosolic NAD+ level as shown by the fluorescence ratio of mCherry-FiNad (Figures 5A, 5B, S5A–S5C), consistent with the results of biochemical analysis (Figures S5D and S5E). To investigate the cause of the NAD+ increase, we studied enzymes associated with NAD+ synthesis (e.g., NAMPT, NMNAT2) and/or NAD+ consumption (e.g., PARP1, CD38, and SIRT1/2) by Western blotting. Among them, only NAMPT protein was elevated (~1-fold) in activated macrophages (Figure 5C). Treatment with specific NAMPT inhibitors, FK866 or STF118804, suppressed LPS/IFN-γ-stimulated glycolysis as shown by changes in the cytosolic NADH/NAD+ ratio (Figures 5D, 5E, and S5F–S5H) and lactate excretion (Figure 5F); nitric oxide (NO) production as shown by DAF-FM DA fluorescence (Figures 5G and 5H); production of the proinflammatory cytokines, interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNFα) (Figures 5I and 5J); and phagocytosis efficiency (Figures 5K and 5L). Collectively, these observations suggest that a global link exists between NAMPT-mediated NAD+ synthesis and macrophage activation. We then study effects of macrophage activation on NADPH using the NADPH sensor iNap1(Tao et al., 2017). LPS/IFN-γ treatment induced a significant decrease of NADPH levels in the cells (Figures S5I and S5J), particularly under glucose-deprived conditions (Figures S5K and S5L), which were consistent with our previous reports (Tao et al., 2017). To investigate the cause of the NADPH level changes, we studied the major enzymes associated with NADPH consumption (e.g., iNOS, FASN, GSR) and/or NADPH synthesis (e.g., G6PD, PGD, MTHFD1, ME1, and IDH1) by Western blotting. Among them, only iNOS protein was significantly induced in activated macrophages (Figure S5M). Treatment with a known iNOS inhibitor, L-NAME(Wang et al., 2015), suppressed LPS/IFN-γ-induced cytosolic NADPH decrease (Figures S5J and S5L), and nitric oxide production (Figures S5N and S5O); however, iNOS inhibition had minimal effects on the physiological function of activated macrophages, such as production of the proinflammatory cytokines (e.g., TNFα) (Figure S5P) and phagocytosis efficiency (Figure S5Q). These data are consistent with previous reports that macrophages synthesize nitric oxide via iNOS and consume NADPH upon activation (Noda and Amano, 1997). To clarify, a working model is provided in Figure 5M, showing that increased synthesis of NAD+ controls the morphofunctional changes of activated macrophages and upregulation of iNOS is associated with increased NO production and NADPH consumption.
Figure 5. Increased synthesis of NAD+ controls morphofunctional changes in activated macrophages.
(A) NAD+ detection in resting or activated RAW264.7 mouse macrophages expressing mCherry-FiNad or mCherry-cpYFP by flow-cytometry. (B) Quantification of mCherry-FiNad fluorescence corrected by mCherry-cpYFP in panel A (n=3). (C) Immunoblots for enzymes associated with NAD+ synthesis or consumption in resting or activated RAW264.7 cells treated with NAMPT inhibitors. For NAMPT quantification, the means ± s.d. are shown (n=3). (D-L) Effect of NAMPT inhibitors, FK866 or STF118804, on cytosolic NADH/NAD+ ratio measured by SoNar (D, E, n=3), lactate excretion (F, n=3), nitric oxide production measured by DAF-FM DA fluorescence (G, H, n=3), production of proinflammatory cytokines, IL-6 (I, n=4) and TNFα (J, n=4), and phagocytosis efficiency (K, L, n=3) in resting or activated RAW264.7 mouse macrophages. (M) Working models for NAMPT and NAD+ level regulation in resting or activated RAW264.7 mouse macrophages. For B, E, F, H-J, and L, data were obtained from three or more independent assessments and expressed as the mean ± s.e.m, normalized to the control condition in the absence of compounds (B, E, F, L). *P < 0.05, **P < 0.01, ***P < 0.001. See also Figure S5.
Imaging NAD+ decline in cell senescence, in the aging mouse, and in human urine-derived stem cells at different ages
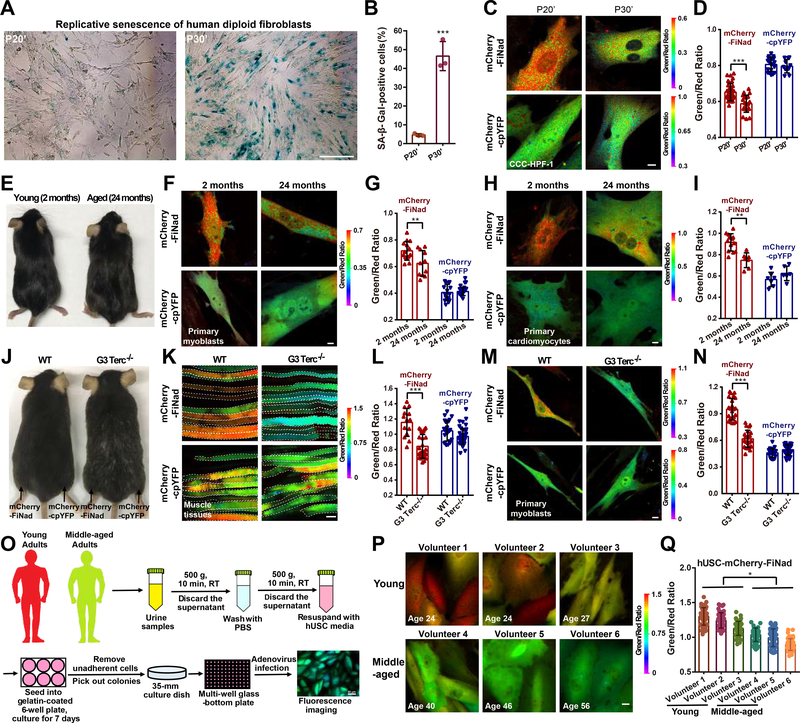
An increasing number of studies have shown that cellular NAD+ levels decline with age, and the deterioration of NAD+ metabolism promotes aging-associated diseases(Bonkowski and Sinclair, 2016; Verdin, 2015). To assess NAD+ dynamics during aging in situ, we applied the mCherry-FiNad sensor to four typical models of aging research, including replicative senescence of human fibroblasts, naturally aged mice, aged telomerase-deficient mice, and human urine-derived stem cells at different ages. Human diploid fibroblasts are widely used as a model system for cell senescence(Kraya et al., 2017). Indeed, human fibroblasts CCC-HPF-1 at passage 30 have a higher percentage of senescence-associated β-galactosidase (SA-β-Gal, a gold-standard marker of senescent cells) (Figures 6A and 6B) and lower NAD+ level, as shown by mCherry-FiNad’s fluorescence (Figures 6C and 6D) compared to passage 20. mCherry-FiNad fluorescence imaging also revealed the decrease of NAD+ levels in both myoblasts and cardiomyocytes freshly isolated from aged mice (~24 months), compared to that from young mice (~2 months) (Figures 6E–6I). To visualize NAD+ level in muscle tissues, we transferred mCherry-FiNad cDNA by electroporation into muscle of 10-month-old WT or the third generation of telomerase-deficient (G3Terc−/−, G3) mice (Figure 6J), which were characterized by PCR (Figure S6A) and have shown prominent premature aging symptoms as reported previously (Sahin et al., 2011). The mCherry-FiNad sensors expressed well in muscle tissue of these mice, and their fluorescence was visible 5 days after electroporation. There was a marked decrease of NAD+ level in muscle tissues of G3 mice compared to that of the WT mice, as shown by mCherry-FiNad’s fluorescence (Figures 6K and 6L). No obvious change in fluorescence was observed in mCherry-cpYFP-expressing muscle tissues (Figures 6K and 6L). Intriguingly, we also observed a remarkable decrease of NAD+ levels in myoblasts freshly isolated from G3 mice compared with that from WT mice (Figures 6M and 6N), suggesting that there is a direct connection between NAD+ decline and telomere attrition-driven aging. We isolated urine-derived stem cells (hUSC) (Zhou et al., 2012) from healthy young (24–27 years) and middle-aged (40–56 years) men to assess further age-related changes in NAD+ levels (Figure 6O). mCherry-FiNad fluorescence imaging showed a decrease of NAD+ levels in hUSC from middle-aged adults, compared to that from young adults (Figures 6P and 6Q). As a control, minimal changes in fluorescence were observed in mCherry-cpYFP-expressing hUSC at different ages (Figures S6B and S6C). We also measured AXP and NAD+ levels during aging by biochemical assays. The data showed that cellular AXP pool slightly decreased during aging, however, the decrease of NAD+ levels were much more significant (Figures S6D–S6K). Therefore, FiNad correctly reported the decrease of the NAD+/AXP ratio, based on four typical models of aging research.
Figure 6. Imaging NAD+ decline in cell senescence, in the aging mouse, and in human urine-derived stem cells at different ages.
(A and B) Senescence-associated β-galactosidase (SA-β-Gal) staining (A) and quantification (B, n=3) of human diploid fibroblasts (CCC-HPF-1) at late passages (P20, P30). Scale bar, 100 μm. (C and D) Fluorescence imaging (C) and quantification (D, n=34, 21, 20, 11) of CCC-HPF-1 cells expressing mCherry-FiNad or mCherry-cpYFP. Scale bar, 10 μm. (E) Young mice (~2 months) and aged mice (~24 months). (F-I) Fluorescence imaging (F, H) and quantification (G, n=14, 11, 15, 16; I, n=13, 7, 7, 6) of mCherry-FiNad or mCherry-cpYFP in myoblasts (F, G) and cardiomyocytes (H, I) freshly isolated from aged mice (~24 months) or young mice (~2 months). Scale bar, 10 μm. (J) 10-month-old WT and the third generation of telomerase-deficient (G3Terc−/−, G3) mice. (K-N) Fluorescence imaging (K, M) and quantification (L, n=14, 27, 23, 34; N, n=23, 24, 22, 30) of mCherry-FiNad or mCherry-cpYFP in muscle tissues of mice (K, L) and myoblasts (M, N) freshly isolated from WT mice or G3 mice indicating regions of interest (white dashed line). Scale bar, 100 μm (K) or 10 μm (M). (O) General overview of the procedure for isolation and NAD+ imaging of human urine-derived stem cells (hUSC) at different ages. (P and Q) Fluorescence images (P) and quantification (Q, n=50) of mCherry-FiNad in hUSC at different ages. Images are pseudocolored by R488/561. Scale bar, 10 μm. Data are presented as the mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. See also Figure S6.
DISCUSSION
Investigators have long explored the metabolic roles of NAD+ and AXP, which are important metabolic mediators. There also exists a close structural and functional link between NAD+ and adenine nucleotides. NAD+ itself is an adenine nucleotide, as ADP is an intrinsic moiety of NAD+ that is incorporated during its synthesis at the expense of ATP. Furthermore, of course, NAD+ and its reduced form (NADH) are necessary cofactors for many metabolic reactions, such as glycolysis, the tricarboxylic acid cycle, and fatty acid β-oxidation, and the conversion between them drives the production of ATP via anaerobic glycolysis and mitochondrial oxidative phosphorylation(Fang et al., 2017). Several studies have also shown that millimolar ATP could inhibit the activity of NAD+-substrate enzymes, such as SIRT1(Kang et al., 2017), PARP(Toledano et al., 2012), and CD38(Takasawa et al., 1993). In addition, AXP competes the binding of NAD+ to the NAD+/NADH sensing transcription factors such as Rex(Wang et al., 2008) or NadR(Grose et al., 2005). These reports suggested that the activities of these important enzymes and transcription factors might be determined actually by the NAD+/AXP ratio, but not the NAD+ level. Thus, FiNad sensor provides a unique yet powerful tool for investigating the role of the NAD+/AXP ratio, which may be more physiologically or pathophysiologically relevant as a key metabolic indicator or regulator of energy metabolism.
In practice, the total physiological pool of adenine nucleotides is usually maintained homeostatically for proper functioning of organisms(Carling et al., 2011; Hardie et al., 2012). Consistently, we found that cellular AXP pool showed minimal changes after PARP1/2, CD38, or SIRT1 inhibition, NAD+ synthesis inhibition or boosting. Thus, changes in FiNad sensor fluorescence actually reflect intracellular NAD+ fluctuations under these circumstances. As expected, the FiNad sensor responds well to increases or decreases of NAD+ in different contexts, such as extraneous NAD+ precursor supplementation, drug treatment, modulation of NAD+-synthesis or consumption, macrophage activation or aging, consistent with the biochemical analysis of cellular NAD+ content. The previously reported genetically encoded NAD+ sensor, LigA-cpVenus(Cambronne et al., 2016), seems to have significant non-specific responses to NMN and NR, the two NAD+ precursors that are frequently used to replenish NAD+ pools and to improve health or life span in many studies, and does not respond to NAD+ fluctuation in cells treated with pharmaceutical concentrations of these precursors, as shown in previous work(Cambronne et al., 2016; Ryu et al., 2018), as well as by our data. Such non-specificity may complicate live-cell or in vivo studies in NAD+ metabolism. By contrast, FiNad shows high specificity for NAD+ without response to NAD+ analogs or precursors. FiNad also displays a much improved dynamic range in vitro (7 folds over that of LigA-cpVenus), making it useful for tracking subtle differences in NAD+ metabolism (Table S2). Currently, FiNad is the only sensor that permits sensitive, specific, and real-time readout of NAD+ metabolism in vivo across different organisms. We therefore strongly believe FiNad sensor is a significant step toward resolving the biological function of NAD+.
Administration of NAD+ precursors has long been known to promote beneficial effects, including improving metabolic dysfunction, slowing the aging process, and protecting against certain metabolic diseases (e.g., obesity, diabetes mellitus, and neurodegeneration), prompting numerous clinical trials of NAD+ boosters in humans(Rajman et al., 2018). Our head-to-head comparison in vivo study to evaluate the efficacy of NAD+ precursors (Table S3) showed that NMN and NR appear rather effective in bacteria, mammalian cells, and mice, while NAM was more effective in vertebrate cells, zebrafish, and mice than in bacteria as means to increase NAD+. Especially, the NAM-NAMPT axis mainly accounts for NAD+ biosynthesis in zebrafish. It is well known that NAD+ boosters like NMN and NR can extend lifespan. NAM can be as effective as NMN or NR at enhancing NAD+ levels in mammalian cells as our data shown; however, it is intriguing that NAM supplementation has little effect on extending lifespan(Mitchell et al., 2018). We found that NMN and NR also efficiently enhance NAD+ level in bacteria, whereas NAM only slightly does so. Future studies might be needed to investigate whether NAD+ enhancement in gut microflora is also vital for extending lifespan, considering the importance of gut microflora in many aspects of human health. Surprisingly, NA is essentially ineffective at augmenting NAD+ in resting bacteria, mammalian cells, zebrafish, and mice; however, NA may help to replenish the NAD+ pool through the Preiss-Handler pathway when activated during inhibition of the NAMPT-dependent salvage pathway. Interestingly, NA works only at a low dosage in these conditions.
Innate and adaptive immunity undergoes severe deterioration with age, leading to increased susceptibility to infection and aging-related diseases (Solana et al., 2006). Macrophages play an important role in inflammatory responses, elimination of pathogens, and tissue repair. Utilizing FiNad, we show that the increase of NAD+ synthesis via the NAMPT-dependent salvage pathway is important for classical M1 activation of macrophages, including metabolic reprogramming, production of proinflammatory cytokines, and phagocytosis efficiency. Emerging evidence suggests that metabolic changes also play vital roles in the development and function of other types of immune cells, such as T lymphocytes(Tyrakis et al., 2016), B lymphocytes(Chan et al., 2017), and mast cells(Misto et al., 2019).
It has become clear that NAD+ levels steadily decline with age for all species, from yeast to humans; however, all of these reports are based on the enzymatic cycling assay, liquid chromatography, or mass spectroscopy assays, which require cell lysis. By using FiNad, we provide the evidence showing a direct demonstration of NAD+ decline during aging in situ, including replicative senescence, natural aging, telomerase deficient-derived premature aging, and age-related stem cell aging.
The ability to map NAD+ metabolism with the FiNad sensor is critical to the interrogation of NAD+’s roles in biologically and medically important processes, and in the ongoing search for NAD+-enhancing compounds, which may have important therapeutic or clinical implications. Given the significance and close link of NAD+/NADH and NADP+/NADPH and the current family of specific sensors for live cell/organism studies, investigators currently have the means available to develop a live-cell redox metabolism atlas using FiNad, Frex(Zhao et al., 2011), SoNar(Zhao et al., 2015), Apollo-NADP+(Cameron et al., 2016), and iNap sensors(Tao et al., 2017; Zou et al., 2018). Such an integrative approach would reveal the key determinants of redox currency in organisms.
Limitations
It should be noted that the FiNad, as an NAD+/AXP ratio sensor, does not directly provide quantitative information on total NAD+ concentration in the cells. When physiological pool of adenine nucleotides is maintained in homeostasis, FiNad can be used to measure free NAD+ dynamics. However, when there are global or localized changes of AXP, it may interfere with such free NAD+ measurement using FiNad sensor. Under such circumstances, free NAD+ levels may be calculated with the quantitative information of the total adenine nucleotide pool. We are currently developing genetically encoded sensors for total adenine nucleotide measurement. Once obtained, these sensors may be used together with the FiNad sensor, for the measuring of NAD+, AXP, and their ratios simultaneously in live cells.
As a genetically encoded fluorescent sensor, FiNad may be targeted to different subcellular compartments. However, since FiNad also binds with NADH, the sensor might not accurately measure NAD+ across all possible concentrations of NADH in subcellular compartments. For the cytosol/nucleus, free NADH concentration is 0.1–1 μM (Zhang et al., 2002; Zhao et al., 2011; Zhao et al., 2016), far below the apparent Kd of FiNad for NADH (~100 μM) in the presence of physiological adenine nucleotides concentrations. Therefore, it is unlikely that cytosolic/nuclear free NADH fluctuations would interfere with NAD+/AXP ratio measurement. Nevertheless, it would be challenging to use FiNad for tracing NAD+ dynamics in mitochondria, where FiNad’s response may be interfered by the high concentrations of free NADH (dozens of μM or even higher)(Joubert et al., 2004; Zhao et al., 2011). Therefore, a highly responsive, mitochondrial targeting NAD+ sensor is still desirable for such studies.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yuzheng Zhao (yuzhengzhao@ecust.edu.cn); and Yi Yang (yiyang@ecust.edu.cn). All these materials, including FiNad, mCherry-FiNad, mCherry-cpYFP, SoNar, iNap1, and iNapc, are available upon requests.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture, transfections, and infections
HEK293 cells (Cell Bank of Chinese Academy of Science) and Human fibroblasts CCC-HPF-1(Laboratory Animal Center of Hangzhou Normal University) were maintained in DMEM (high glucose) (Hyclone) with 10% FBS (Biological Industries) at 37 °C in a humidified atmosphere of 95% air and 5% CO2. Cells were plated in antibiotic-free DMEM (high glucose) supplemented with 10% FBS 16–24 h before transfection. We typically used 0.1 μg plasmids with 0.25 μL Hieff Trans™ liposomal transfection reagent for each well of a 96-well plate according to the manufacturer’s protocol.
RAW264.7 mouse macrophages (Cell Bank of Chinese Academy of Science) were grown in RPMI 1640 (Hyclone) supplemented with 10% FBS. RAW264.7 cells were transiently transfected with sensors using FuGENE HD transfection reagent (Promega). To activate macrophages, LPS (0.5 μg/mL) and IFN-γ (250 U/mL) were added 24 h after transfection for 14–16 h.
The entire coding sequence for mCherry-FiNad or mCherry-cpYFP was subcloned into a pLVX-Puro vector. Lentivirus was produced by co-transfecting two lentiviral packaging vectors (pMD2.G and psPAX2) in HEK293T cells. Lentiviral supernatants were collected 48 and 72 h after transfection. Primary cells in 96-well tissue culture plates were infected with lentivirus in media containing 8 μg/mL polybrene and centrifuged at 1,800 rpm for 1 h. Post-infection 6 h, virus was removed and cells were then cultured for 48–72 h. The recombinant adenoviruses of mCherry-FiNad or mCherry-cpYFP were generated from OBiO Technology (Shanghai). Human urine-derived stem cells were infected with adenoviruses for 36 h in 96-well glass bottom plates, and then imaged.
Isolation and culture of primary cells from mice
Primary cardiomyocytes were isolated as described by a Langendorff-free method (Ackers-Johnson et al., 2016). Briefly, the heart was sequentially perfused with 7 mL EDTA buffer, 5 mL perfusion buffer, 40 mL collagenase buffer and, finally, inhibition of enzymatic activity by adding 5 mL stop buffer. The heart was transferred to a 60-mm dish and teased apart into small pieces following with 15 times of gentle pipetting. The cells were passed through a 100 μm nylon mesh and underwent 4 sequential rounds of gravity settling, using 0, 0.3, 0.6, and 1.2 mM calcium recovery solution. The purified cardiomyocytes were immediately resuspended in plating medium (M199, 1 × penicillin/streptomycin, 5% FBS, 10 mM 2,3-butanedione monoxime) for 1 day and further cultured in culture media (M199, 1 × penicillin/streptomycin, 0.1% Bovine serum albumin, 10 mM 2,3-butanedione monoxime, 1 × Insulin/transferrin/selenium supplement (ITS) and 1 × chemically defined lipid (CD lipid) for further experiments.
Primary myoblasts were isolated from skeletal muscle as previous protocol(Hindi et al., 2017). Briefly, skeletal muscle tissues were minced and digested with collagenase II. The digested tissues were further pelleted by centrifugation, and cells were sequentially passed through a 70-μm and 40-μm filter. The cell suspension was spun down again and resuspended in myoblast growth media with fibroblast growth factor. After a 3-day culture on a matrigel-coated dish, cells were trypsinized and plating steps repeated until >95% myoblast purity was achieved.
Isolation of human urine-derived stem cells
Human urine-derived stem cells (hUSC) at different ages were obtained from anonymized healthy volunteers under protocols approved by the Institutional Review Board of Shanghai Jiaotong University Affiliated Sixth People’s Hospital, with the informed consent of all participants. The study enrolled 6 healthy male individuals at the age of 24, 24, 27, 40, 46 and 56 years. After mid and last stream urine was collected, and 5 ml antibiotic-antimycotic was added into 250 ml urine samples. Urine samples were centrifuged at 500 g for 10 min at room temperature, then the supernatant was discarded and the sediment was washed with phosphate-buffered saline (PBS). Cells were centrifuged again, and then the sediment was resuspended with USC culture medium (contained 44% Dulbecco’s modified Eagle medium, 0.25% Penicillin-Streptomycin, 5% fetal bovine serum, 10 ng/ml of human epidermal growth factor, 2 ng/ml of platelet-derived growth factor, 2 ng/ml of basic fibroblast growth factor, 0.5 % GlutaMax, 0.5 % non-essential amino acids solution and 52 % REGM BulletKit). The cell suspension was seeded into gelatin-coated 6-well plates and incubated at 37 °C in a humidified atmosphere with 5 % CO2. The medium was changed after 7 days of culture and the non-adherent cells were removed by washing with PBS. The colonies derived from single cells were marked, cultured, and further evaluated by the staining of stem cell surface markers according to the previously described methods(Zhang et al., 2008; Zhou et al., 2012).
Animal studies
C57BL/6J mice (refer as ‘wild type’ or ‘WT’) were provided from the Laboratory Animal Center of Hangzhou Normal University. Kunming mice, an outbred stock derived from Swiss albino mice, were purchased from Jiesijie Experimental Animal Co., Ltd (Shanghai, China). The third generation of telomerase-dysfunctional mice (refer as ‘G3Terc−/−’ or ‘G3’) were originally gifted from Prof. Lenhard K. Rudolph. Young (2 months) and old (24 months) WT mice, and G3 mice were all maintained in a specific-pathogen-free facility with a 12 h light / 12 h dark cycle and food and water available ad libitum. The Terc genotyping protocol is: 94 °C, 5 min; 34 cycles of (94 °C, 30 s; 56 °C, 30 s; 72 °C, 30 s); 72 °C, 5 min; and 4 °C hold, using the following primers: mTRWtF 5’-CTAAGCCGGCACTCCTTACAAG-3’, 5PPGK 5’-GGGGCTGCTAAAGCGCAT-3’, and mTRR 5’-TTCTGACCACCACCAACTTCAAT-3’. Size of fragments: wildtype ~ 200 bp and knockout ~ 180 bp. All procedures involving animals were approved by the Institutional Animal Care and Use Committee of Hangzhou Normal University, and East China University of Science and Technology.
Wild-type AB zebrafish embryos at one or two-cell stage were provided from zebrafish core facility, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. Embryos were cultured in a Petri dish with egg water (60 μg/mL Sea salts) at 28.5 °C, and then the fertilized egg developed into zebrafish larvae. The handling procedures were approved by the Institutional Animal Care and Use Committee of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
METHOD DETAILS
DNA cloning
For bacterial expression, FiNad was cloned into BamHI and XhoI sites of the pCDFDuet-1 vector (Novagen) (Table S1). For mammalian expression, the entire coding sequences of FiNad were subcloned into pcDNA3.1 Hygro(+) (Invitrogen) behind a Kozak sequence. For nuclear targeting (Table S1), the three-fold nuclear localization signal (3xNLS) DPKKKRKVDPKKKRKVDPKKKRKV was added to the C-terminus of sensors. For construction of lentivirus vectors, mCherry-FiNad and mCherry-cpYFP were ligated into the pLVX-IRES-Puro plasmid digested by XhoI and XbaI (Table S1). The PercevalHR sensors were obtained from Addgene. The gene encoding LigA-cpVenus was synthesized according to the amino acid sequence previously reported (Cambronne et al., 2016) and cloned into pCDFDuet-1 vector.
Protein expression and purification
All recombinant proteins with a 6× His-tag were expressed in Escherichia coli BL21 (DE3) by pCDFDuet-1 expression plasmid. Cultures were grown in Luria–Bertani (LB) media containing 50 μg/mL streptomycin at 37 °C until the cultures reached an OD600 of 0.3. Moreover, the growth temperature was shifted to 18 °C and protein expression was induced by the addition of 1 mM IPTG. After 24 h of incubation, bacteria were harvested and suspended in 50 mM potassium phosphate buffer, pH 7.4, containing 500 mM NaCl and 10 mM imidazole (buffer A), and lysed via ultrasonication. Cell lysate was centrifuged at 10,000 ×g for 30 min at 4 °C, and the supernatant was loaded onto a column of Ni2+ affinity chromatography. After washing with two column volumes of wash buffer (buffer A containing 30 mM imidazole), the proteins were eluted from the resin using 50 mM potassium phosphate buffer, pH 7.4, containing 500 mM sodium chloride and 300 mM imidazole. The protein preparations were then desalted and exchanged into 100 mM HEPES buffer containing 100 mM NaCl (pH 7.4) with a Sephadex column. Protein was diluted to the required concentration before assay.
Characterization of FiNad in vitro
We stored the purified protein at −80 °C until experiments. Measurement of excitation and emission spectra of recombinant fluorescent sensor proteins was carried out as previously described (Zhao et al., 2015; Zhao et al., 2011). The purified sensor protein was placed into a cuvette containing 100 mM HEPES buffer with 100 mM sodium chloride (pH 7.4). Fluorescence was measured using a spectrofluorometer (EnSpire). Excitation spectra of FiNad and mCherry were recorded with an excitation range from 400 nm to 511 nm or 540 nm to 605 nm and an emission wavelength of 535 nm or 615 nm, respectively. For emission spectra, the emission range of FiNad and mCherry were 507–575 nm or 600–660 nm, while the excitation wavelength was 490 nm or 580 nm, respectively. Readings were taken every 1 nm with an integration time of 1 s, and the photomultiplier tube (PMT) voltage was set at 655 V (Green) or 600 V (Red).
For nucleotide titration, the protein was diluted in 100 mM HEPES buffer (pH 7.4) to a final concentration of 0.5 μM. The fluorescence value of FiNad or mCherry-FiNad, in the presence of nucleotides, was measured by a filter-based Synergy 2 Multi-Mode microplate reader using 485 BP 20 nm (or 420 BP 10 nm) or 590 BP 20 nm excitation, and 528 BP 20 nm or 645 BP 40 nm emission band-pass filters (BioTek). The stock solution of nucleotides was also prepared in 100 mM HEPES buffer (pH 7.4). Each assay was performed with 50 μL nucleotides and 50 μL protein in black 96-well flat bottom plate. Fluorescence intensity was measured immediately.
Fluorescence microscopy
For imaging of mammalian cells, cells were plated on 35 mm glass-bottom dish or 96-well glass-bottom plate. The mCherry-FiNad or mCherry-cpYFP sensors were expressed in subcellular compartments by tagging with organelle-specific signal peptides. Images were acquired using a Leica TCS SP8 SMD confocal laser scanning microscope system with super-sensitive HyD hybrid detectors. HC Plan Apo CS2 63×/1.40 NA oil objective was utilized. For dual-excitation ratio imaging, a 488-nm excitation laser and 500- to 550-nm emission range, and a 561-nm excitation laser and a 570- to 630-nm emission range were used. Raw data were exported to ImageJ software as 12-bit TIF for analysis. The pixel-by-pixel ratio of the 488 nm excitation green image by the 561 nm excitation red image of the same cell was used to pseudocolor the images in HSB color space as previously described(Zhao et al., 2015; Zhao et al., 2011).
For imaging of E. coli BL21 (DE3), cells were diluted to an OD600 of 0.4, and then seeded into 96-well glass-bottom plate pre-coated with 0.1 mg/mL poly-D-lysine. After ~10 min, cells were fixed by adding a drop of 1.5% low melting point agarose to each well. mCherry-FiNad fluorescence was visualized by a Leica TCS SP8 SMD confocal laser scanning microscope system with super-sensitive HyD hybrid detectors and an HC Plan Apo CS2 63×/1.40 NA oil objective.
Imaging NAD+ metabolism in muscle tissue and living mice
Mice were anesthetized intraperitoneally with sodium pentobarbital (60 mg/kg body weight). The mCherry-FiNad or mCherry-cpYFP sensor was transferred into muscle tissue via electroporation. Electrode needles were inserted into the muscle to a depth of 5 mm together with the syringe. 30–50 μg of purified DNA of mCherry-FiNad, mCherry-cpYFP, FiNad, or iNapc plasmid at 1 μg/μL in ddH2O were then injected into the leg skeleton muscles of mice with the syringe. Electric pulses were delivered immediately using an in vivo electroporator (TERESA-EPT I, Teresa) after removing the syringe. Six 50-msec-long pulses of 60 V voltage, 1 Hz were administered to each injection site at a rate of one pulse per sec. All procedures involving animals were approved by the Institutional Animal Care and Use Committee of East China University of Science and Technology.
For in vivo imaging of living mice, mice were anesthetized intraperitoneally with sodium pentobarbital at 5–7 days after electroporation. The electroporated muscle legs were depilated by shaver to reduce the fluorescence interference of hairs, and then placed on glass cover slips for imaging. For ex vivo imaging of muscle tissues, leg skeletal muscle was isolated from C57BL/6J mice or Kunming mice, and then sectioned with a Leica VT1200 S Fully automated vibrating blade microtome. The parameters were set to 3 mm amplitude, 1 mm/s speed, and 300 μm step size (thickness). Muscle sections were maintained in PBS and then placed evenly between two glass cover slips for imaging. Fluorescence was detected by a Leica TCS SP8 SMD confocal laser scanning microscope system with super-sensitive HyD hybrid detectors and an HC Plan APO CS2 20×/0.75 NA dry objective. For dual-excitation ratio imaging, a 488-nm excitation laser and 500- to 550-nm emission range, and a 561-nm excitation laser (or a 405-nm excitation laser) and a 570- to 630-nm (or 500- to 550-nm) emission range were used.
Imaging NAD+ metabolism in zebrafish larvae
One nL of mixture containing 4 mg/mL mCherry-FiNad or mCherry-cpYFP sensor protein in HEPES buffer was microinjected to the animal pole of wild-type AB zebrafish embryos at one or two-cell stage. Embryos were collected and placed in a Petri dish with egg water (60 μg/mL Sea salts) at 28.5 °C. The embryos were transferred into a 12-well culture plate (~20 embryos per well) at 6 h or 30 h post fertilization, and then incubated for ~24 h with NAD+ precursors or NAMPT inhibitor in egg water (60 μg/mL Sea salts). Zebrafish larvae were anesthetized with 0.6 mM tricaine in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) prior to imaging. If necessary, zebrafish larvae were dechorionated under the stereomicroscope using fine tweezers. The handling procedures were approved by the Institutional Animal Care and Use Committee of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
Live-cell fluorescence measurement using microplate reader
For E. coli BL21 (DE3), cells were seeded into glass tubes and cultured as the protein expression procedure. Bacteria cultures were diluted into an OD600 of 0.4, and then incubated with NAD+ precursors or NAPRT inhibitor 2-HNA for 2 h at 37 °C. After 2 h, bacteria were pelleted by centrifugation, washed twice with HEPES buffer, and then diluted into an OD600 of 0.2 for detection. Readings were recorded at 37 °C by a Synergy 2 Multi-Mode Microplate Reader (BioTek) with excitation filters 485 BP 20 nm and 590 BP 20 nm, and emission filter 528 BP 20 nm and 645 BP 40 nm, respectively.
For mammalian cells, cells were seeded in black 96-well flat bottom plate, transfected with the plasmid DNA of mCherry-FiNad or mCherry-cpYFP and expressed for 36 h. Cells were washed twice with PBS after carefully removing the medium. Data were recorded immediately after adding 100 μL PBS at 37 °C by a Synergy 2 Multi-Mode Microplate Reader (Bio Tek) with excitation filters 485 BP 20 nm and 590 BP 20 nm, and emission filter 528 BP 20 nm and 645 BP 40 nm, respectively. Fluorescence values were background corrected by subtracting the intensity of the cell samples not expressing sensors. Unless otherwise indicated, 25 mM glucose and 0.2 mM pyruvate was maintained in the buffer.
NAD+/NADH ratio and ATP/ADP ratio in live cells were measured using SoNar(Zhao et al., 2015) and PercevalHR (Tantama et al., 2013) as with FiNad described above, respectively.
Fluorescence-activated cell sorter analysis (FACS)
After treatment with NAMPT inhibitors, RAW264.7 mouse macrophages were scraped and monitored using FACS. FACS was performed with CytoFLEX-S flow cytometer (Beckman Coulter). Cells expressing mCherry-FiNad and mCherry-cpYFP sensors were excited using laser lines at 488 nm and 561 nm. Emission filters were 525 BP 40 nm and 610 BP 20 nm for the two excitation wavelengths separately. Cells expressing SoNar, iNap1 or iNapc sensors were excited using laser lines at 405 nm and 488 nm. Emission filters were 525 BP 40 nm for both excitation wavelengths.
E. coli BL21 (DE3) cells expressing mCherry-FiNad, mCherry-cpYFP, LigA-cpVenus, or cpVenus sensors were treated with NAD+ precursors, or NAPRT inhibitor 2-HNA for 2 h. Cells were pelleted by centrifugation, washed twice, and then resuspended into HEPES buffer. After 30 min, fluorescence was measured with FACS.
Western blot
For the Western blot analyses, cells were lysed in 1X SDS sample buffer supplemented with a protease/phosphatase inhibitor cocktail (Cell Signaling Technology). Equal amounts of total protein (30–60 μg) were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and electrotransferred onto PVDF membranes. Membranes were incubated with primary antibodies: NAMPT (sc-166946), NMNAT2 (sc-515206), CD38 (sc-374650), PAR (sc-56198), PARP1 (sc-74470), G6PD (sc-373886), PGD (sc-398977), IDH1 (sc-515396), ME1 (sc-365891), GSR (sc-133245), FASN (sc-48357), β-actin (sc-47778), and GAPDH (sc-365062) (Santa Cruz Biotechnologies); iNOS (#13120), SIRT1 (#9475), and SIRT2 (#12672) (Cell Signaling Technology); and MTHFD1 (10794–1-AP) (Proteintech), followed by secondary antibodies conjugated to horseradish peroxidase, and addition of chemiluminescence detection mixture (YEASEN, 36208ES60) and imaged.
ELISA
RAW264.7 mouse macrophages were cultured in 12-well plate and exposed to activators combined with NAMPT inhibitors for 12 h. The supernatant was removed and the fresh medium was added for 4 h incubation. The medium was then collected by centrifugation after 4 h. The content of IL-6 and TNF-α was determined immediately by using mouse IL-6 ELISA kit (R&D Systems, VAL604) and mouse TNF-α ELISA kit (Abbkine, KET7015) according to the manufacturer’s protocol, respectively.
Lactate assay
RAW264.7 mouse macrophages were exposed to activators combined with NAMPT inhibitors for 14 h. The supernatant was removed; and cells were washed once with fresh medium and was incubated for 1 h with NAMPT inhibitors. The medium was then collected to 1.5 mL EP tubes, and deproteined with equal volume of 0.5 M HClO4, neutralized with 3 M KOH, and centrifuged at 4 °C, 10, 000 g for 5 min. The supernatant was pipetted carefully to new EP tubes for assay. 50 μL samples were mixed with equal volume of reaction medium containing 20 μM resazurin, 0.4 U/mL diaphorase, 1 mM NAD+, and 1 U/mL lactate dehydrogenase. Changes in fluorescence excited at 540 nm and emitted at 590 nm were measured every 20 seconds for 10 min at 37 °C by a Synergy 2 Multi-Mode Microplate Reader.
NO detection
RAW264.7 mouse macrophages were cultured in 24-well plate and exposed to activators combined with NAMPT inhibitors for 14 h. After removing the medium, cells were washed once with PBS and then stained with 200 μL 5 μM DAF-FM DA probe (Beyotime, S0019) at 37 °C for 30 min. The cells were washed three times with PBS after 30 min, collected, and detected by flow cytometer (Beckman, CytoFLEX-S) with laser line at 488 nm and emission filter 525 BP 40 nm.
Phagocytosis assay
RAW264.7 mouse macrophages were seeded in 12 well-plate for ~24 h, and then washed three times with PBS and incubated with E. coli BL21(DE3) cells expressing superfolder GFP(Pedelacq et al., 2006) at 37 °C for 30 min in serum-free 1640 medium. The bacteria-to-cell ratio was approximately 50:1. The bacteria were removed after 30 min, and RAW264.7 cells were washed three times with PBS containing 1 mM EDTA. Cell samples were then resuspended with PBS-EDTA and analyzed by FACS. The phagocytic index of each sample was characterized as the mean superfolder GFP fluorescence multiplied by the percent of the positive population.
β-galactosidase staining
Human fibroblasts CCC-HPF-1 were fixed in cold 4 % paraformaldehyde for 3 h, then washed with PBS and stained at 37 °C for 18 h in the presence of 1 mM X-gal. RGB images were taken by high-performance fluorescent microscopy system equipped with Nikon Eclipse Ti-E automatic microscope and Nikon DS-Fi2 camera.
Measurement of total NAD+ levels
NAD+ levels were measured by the enzymatic cycling assay according to a previously described method(Szabo et al., 1996; Ying et al., 2001). NAD+ levels were assessed based on the reduction of MTT to formazan through phenazine methosulfate facilitation. Cells were treated with different reagents for 24 h (MNNG treatment for 30 min) and then extracted and assayed. Relative NAD+ level was calculated by normalizing cell number or protein level.
Detection of cellular ATP and ADP levels
The ATP assay is based on CellTiter-Glo luminescent cell viability assay kit (Promega). Briefly, luciferase catalyzes the conversion of ATP and D-luciferin to light, which is measured by a Synergy 2 Multi-Mode Microplate Reader. The ADP level is measured by its conversion to ATP through Pyruvate kinase (Sigma, P1506) that is subsequently detected using the same reaction. Relative AXP level ([ATP]+[ADP]) was calculated by normalizing cell number or protein level.
Cell viability assay
Cells were plated at a density of 8×103 cells in triplicate in 96-well plates. After ~20 h, cells were treated with different compounds. The cells were then cultured for 48 h, and cell viability was evaluated by Cell Counting Kit-8 (CCK-8).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented either as a representative example of a single experiment repeated at least in triplicate or as three or more experiments. Data are presented as mean ± s.d. or mean ± s.e.m. All P values were obtained using unpaired two-tailed Student’s t-test. Values of P < 0.05 were considered statistically significant (*P < 0.05, **P < 0.01 and ***P < 0.001).
DATA AND SOFTWARE AVAILABILITY
All data supporting the findings of this study are available within the article and its Supplemental Information files. All constructs, including FiNad, mCherry-FiNad, mCherry-cpYFP, SoNar, iNap1 and iNapc, are available on request.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-iNOS (D6B6S) Rabbit mAb | Cell Signaling Technology | Cat# 13120 |
| Anti-PBEF (H-11) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-166946 |
| Anti-NMNAT-2 (B-10) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-515206 |
| Anti-CD38 (H-11) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-374650 |
| Anti-SirT1 (D1D7) Rabbit mAb | Cell Signaling Technology | Cat# 9475 |
| Anti-SirT2 (D4S6J) Rabbit mAb | Cell Signaling Technology | Cat# 12672 |
| Anti-pADPr (10H) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-56198 |
| Anti-PARP-1 (B-10) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-74470 |
| Anti-β-Actin (C4) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-47778 |
| Anti-GAPDH (G-9) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-365062 |
| Anti-G6PD (G-12) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-373886 |
| Anti-PGD (G-2) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-398977 |
| Anti-IDH1 (F-3) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-515396 |
| Anti-ME1 (C-6) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-365891 |
| Anti-GSR (C-10) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-133245 |
| Anti-FASN (G-11) Mouse mAb | SANTA CRUZ BIOTECHNOLOGY | Cat# sc-48357 |
| Anti-MTHFD1 Rabbit pAb | Proteintech | Cat# 10794-1-AP |
| Bacterial and Virus Strains | ||
| BL21(DE3) Chemically Competent Cell | TRANSGEN BIOTECH | CD601 |
| Trans5α Chemically Competent Cell | TRANSGEN BIOTECH | CD201 |
| Biological Samples | ||
| Mouse Primary Cardiomyocytes | This paper | N/A |
| Mouse Primary Myoblasts | This paper | N/A |
| Human Urine-derived Stem Cells | Shanghai Jiaotong University affiliated Sixth People’s Hospital | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| NAD+ | Shanghai YEASEN | Cat# 60323ES08 |
| NADH | Shanghai YEASEN | Cat# 60301ES03 |
| NADP+ | Shanghai YEASEN | Cat# 60324ES03 |
| NADPH | Shanghai YEASEN | Cat# 60302ES01 |
| ADP | Shanghai YEASEN | Cat# 60604ES03 |
| ATP | Sigma-Aldrich | Cat# A26209 |
| AMP | Shanghai YEASEN | Cat# 60603ES08 |
| Sodium Oxamate | Sigma-Aldrich | Cat# O2751 |
| Rotenone | Sigma-Aldrich | Cat# R8875 |
| Oligomycin | Aladdin | Cat# O111756 |
| Apigenin | Sigma-Aldrich | Cat# A3145 |
| FK866 Hydrochloride Hydrate | Sigma-Aldrich | Cat# F8557 |
| Nicotinic Acid (NA) | Sigma-Aldrich | Cat# N4126 |
| Nicotinamide (NAM) | Sigma-Aldrich | Cat# N3376 |
| β-Nicotinamide Mononucleotide (β-NMN) | Shanghai YEASEN | Cat# 60303ES80 |
| Nicotinamide Riboside (NR) | Shanghai Yuanye Bio-technology | Cat# S24896 |
| Ethyl 3-Aminobenzoate Methanesulfonate (MS-222) | Sigma-Aldrich | Cat# E10521 |
| Selisistat (EX 527) | Selleckchem | Cat# S1541 |
| Olaparib (AZD2281) | Selleckchem | Cat# S1060 |
| AZD2461 | Selleckchem | Cat# S7029 |
| 1-Methyl-3-Nitro-1-Nitrosoguanidine (MNNG) | TCI | Cat# M0527 |
| 2-Hydroxynicotinic Acid (2-HNA) | Energy Chemical | Cat# A010247 |
| Sodium Pyruvate | Thermo Fisher Scientific | Cat# 11360070 |
| Lipopolysaccharides (LPS) | Shanghai YEASEN | Cat# 60322ES10 |
| Recombinant Mouse IFN-γ | Absin Bioscience | Cat# abs01021 |
| Nω-Nitro-L-Arginine Methyl Ester Hydrochloride (L-NAME) | J&K Scientific | Cat# 257155 |
| Fugene HD Transfection Reagent | Promega | Cat# E2311 |
| Hieff Trans™ Liposomal Transfection Reagent | Shanghai YEASEN | Cat# 40802ES03 |
| DMEM Media (High Glucose) | HyClone | Cat# SH30243.01 |
| RPMI Medium Modified (1640) | HyClone | Cat# SH30809.01B |
| PBS | HyClone | Cat# SH30013.04 |
| Opti-MEM | Invitrogen | Cat# 31985-070 |
| Hexadimethrine Bromide | Sigma-Aldrich | Cat# H9268 |
| Poly-D-Lysine | Huaweiruike Chemical Co., Ltd | Cat# HWG24863 |
| Sodium Pentobarbital | Sigma-Aldrich | Cat# P3761 |
| Protease/phosphatase Inhibitor Cocktail | Cell Signaling Technology | Cat# 5872 |
| Super ECL Detection Reagent | Shanghai YEASEN | Cat# 36208ES60 |
| Resazurin | Sigma-Aldrich | Cat# R7017 |
| Diaphorase | Aladdin | Cat# D128545 |
| Phosphoenolpyruvic Acid Monopotassium Salt | Alfa Aesar | Cat# B20358.06 |
| Phenazine Methosulfate | Macklin | Cat# P815754 |
| Thiazolyl Blue Tetrazolium Bromide | Aladdin | Cat# T100896 |
| DAF-FM DA Probe | Beyotime Biotechnology | Cat# S0019 |
| Fetal Bovine Serum (FBS) | Biological Industries | Cat# 04-001-1ACS |
| Trypsin-EDTA | Gibco | Cat# 25200-072 |
| Human Fibronectin | Shanghai YEASEN | Cat# 40105ES08 |
| Lactate Dehydrogenase | Aladdin | Cat# L128329 |
| Pyruvate Kinase | Sigma-Aldrich | Cat# P1506 |
| Alcohol Dehydrogenase | Aladdin | Cat# A124756 |
| Critical Commercial Assays | ||
| Cell Counting Kit-8 (CCK-8) | Shanghai YEASEN | Cat# 40203ES60 |
| Mouse IL-6 ELISA Kit | R&D Systems | Cat# VAL604 |
| Mouse TNF-α ELISA Kit | Abbkine | Cat# KET7015 |
| CellTiter-Glo® Luminescent Cell Viability Assay Kit | Promega | Cat# G7573 |
| Senescence β-Galactosidase Staining Kit | Beyotime Biotechnology | Cat# C0602 |
| Experimental Models: Cell Lines | ||
| HEK293T | Cell Bank of Chinese Academy of Science | GNHu17 |
| HEK293 | Cell Bank of Chinese Academy of Science | GNHu43 |
| RAW264.7 Mouse Macrophages | Cell Bank of Chinese Academy of Science | TCM13 |
| Human Fibroblasts CCC-HPF-1 | Laboratory Animal Center of Hangzhou Normal University | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J Mice | Laboratory Animal Center of Hangzhou Normal University | N/A |
| Kunming Mice | Jiesijie Experimental Animal Co., Ltd | N/A |
| The Third Generation of Telomerase-dysfunctional Mice (refer as ‘G3Terc−/−’ or ‘G3’) | from Prof. Lenhard K. Rudolph | N/A |
| Zebrafish Larvae | Zebrafish core facility, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences | N/A |
| Oligonucleotides | ||
| Primer sequences for cloning or sequencing, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pcDNA3.1-FiNad | This paper | N/A |
| pcDNA3.1-mCherry-FiNad | This paper | N/A |
| pcDNA3.1-mCherry-cpYFP | This paper | N/A |
| pLVX-mCherry-FiNad | This paper | N/A |
| pLVX-mCherry-cpYFP | This paper | N/A |
| pcDNA3.1-SoNar | Zhao et al., 2015 Cell Metabolism | N/A |
| pcDNA3.1-cpYFP | Zhao et al., 2015 Cell Metabolism | N/A |
| pcDNA3.1-iNap1 | Tao et al., 2017 Nature Methods | N/A |
| pcDNA3.1-iNapc | Tao et al., 2017 Nature Methods | N/A |
| pCDFDuet-mCherry-FiNad | This paper | N/A |
| pCDFDuet-mCherry-cpYFP | This paper | N/A |
| psPAX2 | Addgene | Cat# 12260 |
| pMD2.G | Addgene | Cat# 12259 |
| pCDFDuet-LigA-cpVenus | Cambronne et al., 2016 Science | N/A |
| pCDFDuet-cpVenus | Cambronne et al., 2016 Science | N/A |
| GW1-PercevalHR | Addgene | #49082 |
| Software and Algorithms | ||
| Image J 1.50d | Fiji | http://fiji.sc/ |
| GraphPad Prism V6.01 | GraphPad Software | https://download.cnet.com/s/graphpad-prism-6-01/ |
| SigmaPlot 12.0 | SigmaPlot Software | https://sigmaplot.software.informer.com/12.0/ |
| Gen5 1.05 | Biotek | https://www.biotek.com/products/software-robotics/ |
| CytExpert 2.0 | Beckman Coulter | https://www.beckman.com/flow-cytometry/instruments/cytoflex/software |
| NIS-Elements AR Analysis Version 5.11.01 | Nikon | https://www.nikon.com/products/microscope-solutions/support/download/software/imgsfw/nis-ar_v5110164.htm |
| Other | ||
| 35 mm Glass Bottom Dish | Cellvis | D35-20-1-N |
| 96-well Glass Bottom Plate | Cellvis | P96-1-N |
ACKNOWLEDGMENTS
We thank Gary Yellen for the PercevalHR vector; Fei Wang, Ping Hu, Yimin Lao, Jing Yi, Chiqi Chen, Junke Zheng, Peili Ni, Yunbin Zhang, Yihong Wang, Xie Li for technical assistance; and Stephanie C. Tribuna for secretarial assistance. This research is supported by National Key Research and Development Program of China (2017YFA050400, 2019YFA0904800), NSFC (91649123, 91857202, 31722033, 31671484, 21937004, 91749203, 81525010, 81420108017), the Shanghai Science and Technology Commission (18JC1411900, 16430723100, 19YF1411400, 19YF1411300), Research Unit of New Techniques for Live-cell Metabolic Imaging(Chinese Academy of Medical Sciences, 2019RU01), Major Program of Development Fund for Shanghai Zhangjiang National Innovation Demonstration Zone(Stem Cell Strategic Biobank and Stem Cell Clinical Technology Transformation Platform, ZJ2018-ZD-004), Innovative research team of high-level local universities in Shanghai, Young Elite Scientists Sponsorship Program by Cast, Shanghai Young Top-notch Talent, the State Key Laboratory of Bioreactor Engineering, the Fundamental Research Funds for the Central Universities, the China Postdoctoral Science Foundation (2019M651413), and US National Institutes of Health (HL061795, HG007690, and GM107618), and the American Heart Association (D700382).
Footnotes
DECLARATION OF INTERESTS
Y.Y., Y.Zhao, and Q.H. have filed a related patent by East China University of Science and Technology. The remaining authors declare no competing financial interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and three tables and can be found with this article.
REFERENCES
- Ackers-Johnson M, Li PY, Holmes AP, O’Brien SM, Pavlovic D, and Foo RS (2016). A Simplified, Langendorff-Free Method for Concomitant Isolation of Viable Cardiac Myocytes and Nonmyocytes From the Adult Mouse Heart. Circ Res 119, 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilan DS, Matlashov ME, Gorokhovatsky AY, Schultz C, Enikolopov G, and Belousov VV (2014). Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments. Biochim Biophys Acta 1840, 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowski MS, and Sinclair DA (2016). Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat Rev Mol Cell Biol 17, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne XA, Stewart ML, Kim D, Jones-Brunette AM, Morgan RK, Farrens DL, Cohen MS, and Goodman RH (2016). Biosensor reveals multiple sources for mitochondrial NAD(+). Science 352, 1474–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron WD, Bui CV, Hutchinson A, Loppnau P, Graslund S, and Rocheleau JV (2016). Apollo-NADP(+): a spectrally tunable family of genetically encoded sensors for NADP(+). Nat Methods 13, 352–358. [DOI] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. (2012). The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab 15, 838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Menzies KJ, and Auwerx J (2015). NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab 22, 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling D, Mayer FV, Sanders MJ, and Gamblin SJ (2011). AMP-activated protein kinase: nature’s energy sensor. Nat Chem Biol 7, 512–518. [DOI] [PubMed] [Google Scholar]
- Chan LN, Chen Z, Braas D, Lee JW, Xiao G, Geng H, Cosgun KN, Hurtz C, Shojaee S, Cazzaniga V, et al. (2017). Metabolic gatekeeper function of B-lymphoid transcription factors. Nature 542, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Lautrup S, Hou Y, Demarest TG, Croteau DL, Mattson MP, and Bohr VA (2017). NAD(+) in Aging: Molecular Mechanisms and Translational Implications. Trends Mol Med 23, 899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghesquiere B, Wong BW, Kuchnio A, and Carmeliet P (2014). Metabolism of stromal and immune cells in health and disease. Nature 511, 167–176. [DOI] [PubMed] [Google Scholar]
- Grose JH, Bergthorsson U, and Roth JR (2005). Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J Bacteriol 187, 2774–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, and Hawley SA (2012). AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindi L, McMillan JD, Afroze D, Hindi SM, and Kumar A (2017). Isolation, Culturing, and Differentiation of Primary Myoblasts from Skeletal Muscle of Adult Mice. Bio-protocol 7, e2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, and Yellen G (2011). Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab 14, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai SI, and Guarente L (2014). NAD and sirtuins in aging and disease. Trends Cell Biol 24, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert F, Fales HM, Wen H, Combs CA, and Balaban RS (2004). NADH enzyme-dependent fluorescence recovery after photobleaching (ED-FRAP): applications to enzyme and mitochondrial reaction kinetics, in vitro. Biophys J 86, 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Oka S, Lee DY, Park J, Aponte AM, Jung YS, Bitterman J, Zhai P, He Y, Kooshapur H, et al. (2017). Sirt1 carboxyl-domain is an ATP-repressible domain that is transferrable to other proteins. Nat Commun 8, 15560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraya T, Neumann L, Paelecke-Habermann Y, Deschauer M, Stoevesandt D, Zierz S, and Watzke S (2017). Cognitive impairment, clinical severity and MRI changes in MELAS syndrome. Mitochondrion 44, 53–57. [DOI] [PubMed] [Google Scholar]
- Misto A, Provensi G, Vozella V, Passani MB, and Piomelli D (2019). Mast Cell-Derived Histamine Regulates Liver Ketogenesis via Oleoylethanolamide Signaling. Cell Metab 29, 91–102. [DOI] [PubMed] [Google Scholar]
- Mitchell SJ, Bernier M, Aon MA, Cortassa S, Kim EY, Fang EF, Palacios HH, Ali A, Navas-Enamorado I, Di Francesco A, et al. (2018). Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab 27, 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, and Amano F (1997). Differences in nitric oxide synthase activity in a macrophage-like cell line, RAW264.7 cells, treated with lipopolysaccharide (LPS) in the presence or absence of interferon-gamma (IFN-gamma): possible heterogeneity of iNOS activity. J Biochem 121, 38–46. [DOI] [PubMed] [Google Scholar]
- Park JO, Rubin SA, Xu YF, Amador-Noguez D, Fan J, Shlomi T, and Rabinowitz JD (2016). Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat Chem Biol 12, 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, and Waldo GS (2006). Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24, 79–88. [DOI] [PubMed] [Google Scholar]
- Plowden J, Renshaw-Hoelscher M, Engleman C, Katz J, and Sambhara S (2004). Innate immunity in aging: impact on macrophage function. Aging Cell 3, 161–167. [DOI] [PubMed] [Google Scholar]
- Poyan Mehr A, Tran MT, Ralto KM, Leaf DE, Washco V, Messmer J, Lerner A, Kher A, Kim SH, Khoury CC, et al. (2018). De novo NAD(+) biosynthetic impairment in acute kidney injury in humans. Nat Med 24, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajman L, Chwalek K, and Sinclair DA (2018). Therapeutic potential of NAD-boosting molecules: the in vivo evidence. Cell Metab 27, 529–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, and Kraus WL (2018). Metabolic regulation of transcription through compartmentalized NAD(+) biosynthesis. Science 360, eaan5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. (2011). Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallin O, Reymond L, Gondrand C, Raith F, Koch B, and Johnsson K (2018). Semisynthetic biosensors for mapping cellular concentrations of nicotinamide adenine dinucleotides. eLife 7, e32638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana R, Pawelec G, and Tarazona R (2006). Aging and innate immunity. Immunity 24, 491–494. [DOI] [PubMed] [Google Scholar]
- Szabo C, Zingarelli B, O’Connor M, and Salzman AL (1996). DNA strand breakage, activation of poly (ADP-ribose) synthetase, and cellular energy depletion are involved in the cytotoxicity of macrophages and smooth muscle cells exposed to peroxynitrite. Proc Natl Acad Sci U S A 93, 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasawa S, Tohgo A, Noguchi N, Koguma T, Nata K, Sugimoto T, Yonekura H, and Okamoto H (1993). Synthesis and hydrolysis of cyclic ADP-ribose by human leukocyte antigen CD38 and inhibition of the hydrolysis by ATP. J Biol Chem 268, 26052–26054. [PubMed] [Google Scholar]
- Tantama M, Martinez-Francois JR, Mongeon R, and Yellen G (2013). Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nature communications 4, 2550. [Google Scholar]
- Tao R, Zhao Y, Chu H, Wang A, Zhu J, Chen X, Zou Y, Shi M, Liu R, Su N, et al. (2017). Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat Methods 14, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Wakimoto H, Iafrate AJ, Tanaka S, Loebel F, Lelic N, Wiederschain D, Bedel O, Deng G, Zhang B, et al. (2015). Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 28, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano E, Ogryzko V, Danchin A, Ladant D, and Mechold U (2012). 3’−5’ phosphoadenosine phosphate is an inhibitor of PARP-1 and a potential mediator of the lithium-dependent inhibition of PARP-1 in vivo. Biochem J 443, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrakis PA, Palazon A, Macias D, Lee KL, Phan AT, Velica P, You J, Chia GS, Sim J, Doedens A, et al. (2016). S-2-hydroxyglutarate regulates CD8(+) T-lymphocyte fate. Nature 540, 236–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG (2017). Metabolism and congenital malformations-NAD’s effects on development. N Engl J Med 377, 509–511. [DOI] [PubMed] [Google Scholar]
- Verdin E (2014). The many faces of sirtuins: Coupling of NAD metabolism, sirtuins and lifespan. Nat Med 20, 25–27. [DOI] [PubMed] [Google Scholar]
- Verdin E (2015). NAD(+) in aging, metabolism, and neurodegeneration. Science 350, 1208–1213. [DOI] [PubMed] [Google Scholar]
- Wang E, Bauer MC, Rogstam A, Linse S, Logan DT, and von Wachenfeldt C (2008). Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol Microbiol 69, 466–478. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhao Y, Wang C, Zhu Q, Du Z, Hu A, and Yang Y (2015). Organelle-Specific Nitric Oxide Detection in Living Cells via HaloTag Protein Labeling. PLoS One 10, e0123986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Sevigny MB, Chen Y, and Swanson RA (2001). Poly(ADP-ribose) glycohydrolase mediates oxidative and excitotoxic neuronal death. Proc Natl Acad Sci U S A 98, 12227–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Baur JA, and Imai SI (2018). NAD(+) intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab 27, 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, and Imai S (2011). Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab 14, 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, and Goodman RH (2002). Regulation of corepressor function by nuclear NADH. Science 295, 1895–1897. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McNeill E, Tian H, Soker S, Andersson KE, Yoo JJ, and Atala A (2008). Urine derived cells are a potential source for urological tissue reconstruction. J Urol 180, 2226–2233. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hu Q, Cheng F, Su N, Wang A, Zou Y, Hu H, Chen X, Zhou HM, Huang X, et al. (2015). SoNar, a Highly Responsive NAD+/NADH Sensor, Allows High-Throughput Metabolic Screening of Anti-tumor Agents. Cell Metab 21, 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Jin J, Hu Q, Zhou HM, Yi J, Yu Z, Xu L, Wang X, Yang Y, and Loscalzo J (2011). Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab 14, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang A, Zou Y, Su N, Loscalzo J, and Yang Y (2016). In vivo monitoring of cellular energy metabolism using SoNar, a highly responsive sensor for NAD(+)/NADH redox state. Nat Protoc 11, 1345–1359. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang Z, Zou Y, and Yang Y (2018). Visualization of nicotine adenine dinucleotide redox homeostasis with genetically encoded fluorescent sensors. Antioxid Redox Signal 28, 213–229. [DOI] [PubMed] [Google Scholar]
- Zhou T, Benda C, Dunzinger S, Huang Y, Ho JC, Yang J, Wang Y, Zhang Y, Zhuang Q, Li Y, et al. (2012). Generation of human induced pluripotent stem cells from urine samples. Nat Protoc 7, 2080–2089. [DOI] [PubMed] [Google Scholar]
- Zong WX, Ditsworth D, Bauer DE, Wang ZQ, and Thompson CB (2004). Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev 18, 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Wang A, Shi M, Chen X, Liu R, Li T, Zhang C, Zhang Z, Zhu L, Ju Z, et al. (2018). Analysis of redox landscapes and dynamics in living cells and in vivo using genetically encoded fluorescent sensors. Nat Protoc 13, 2362–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its Supplemental Information files. All constructs, including FiNad, mCherry-FiNad, mCherry-cpYFP, SoNar, iNap1 and iNapc, are available on request.