Abstract
Context
Loss-of-function mutations in the imprinted genes MKRN3 and DLK1 cause central precocious puberty (CPP) but whole gene deletions have not been reported. Larger deletions of the chromosome 15q11-13 imprinted locus, including MKRN3, cause Prader-Willi syndrome (PWS). CPP has been reported in PWS but is not common, and the role of MKRN3 in PWS has not been fully elucidated.
Objective
To identify copy number variants in puberty-related, imprinted genes to determine their role in CPP.
Methods
Probands with idiopathic CPP had chromosomal microarray (CMA) and targeted deletion/duplication testing for MKRN3 and DLK1.
Results
Sixteen female probands without MKRN3 or DLK1 variants identified by Sanger sequencing were studied. Whole gene deletions of MKRN3 were identified in 2 subjects (13%): a complete deletion of MKRN3 in Patient A (pubertal onset at 7 years) and a larger deletion involving MAGEL2, MKRN3, and NDN in Patient B (pubertal onset 5.5 years). Both were paternally inherited. Patient B had no typical features of PWS, other than obesity, which was also present in her unaffected family.
Conclusions
We identified 2 cases of whole gene deletions of MKRN3 causing isolated CPP without PWS. This is the first report of complete deletions of MKRN3 in patients with CPP, emphasizing the importance of including copy number variant analysis for MKRN3 mutation testing when a genetic diagnosis is suspected. We speculate that there is a critical region of the PWS locus beyond MKRN3, MAGEL2, and NDN that is responsible for the PWS phenotype.
Keywords: Precocious puberty, genetics, MKRN3, copy number variants, deletions, Prader-Willi syndrome
Central precocious puberty (CPP) is the premature activation of the hypothalamic-pituitary-gonadal (HPG) axis resulting in the development of secondary sexual characteristics before the age of 8 years in females and 9 years in males (1). Several genetic causes of idiopathic CPP (iCPP) have been described, including inactivating mutations in MKRN3 (2-4) and partial deletions and loss-of-function (LOF) mutations in DLK1 (5, 6). Both genes are imprinted and paternally expressed. While LOF variants in MKRN3 have been shown to cause CPP, copy number variants (CNVs) have yet to be reported.
Paternal deletions in the imprinted region on chromosome 15 where MKRN3 is located are also known to cause Prader-Willi syndrome (PWS), which is characterized by childhood behavior problems, intellectual disability, short stature, and obesity as a result of hyperphagia (reviewed in (7)). The chromosome 15q11-13 locus is referred to as the Prader-Willi syndrome imprinting region (PWS-IC). The reproductive phenotype in PWS is typically manifested by hypogonadotropic hypogonadism (HH) and delayed or absent puberty, but rarely, CPP has been reported in patients with PWS (8-10). Thus, the role of MKRN3 in PWS remains unknown.
In this study, we screened probands with precocious puberty for CNVs in the puberty-related imprinted genes. Deletions in the MKRN3 gene were compared with those reported in the literature to explore the role of this gene in the pubertal development of patients with PWS.
Methods
Study participants
Participants with a diagnosis of CPP were enrolled between August 2014 and January 2019 at the National Institutes of Health Clinical Center (NIH CC). All participants were female and were evaluated by a single pediatric endocrinologist at the time of enrollment. They were clinically confirmed to have breast development before 8 years in one of two ways. If they presented for enrollment around the time of pubertal onset, patients were clinically diagnosed with CPP at the NIH CC. If they were evaluated after progressing through pubertal stages while under the care of a pediatric endocrinologist in their local community, then historical data on pubertal milestones were collected from the medical records, including the clinician’s physical exam and biochemical evaluation, to confirm the diagnosis of CPP. Study criteria included clinical signs of progressive isosexual precocious puberty, pubertal level gonadotropins and absence of elevated TSH based on each institution’s published laboratory reference ranges, and rapid progression of puberty. Patients were excluded from participation if documentation of the above were unavailable, thus no participants were enrolled based on self/family recalled pubertal milestones alone. Bone ages were determined by the method of Greulich and Pyle and height predictions were calculated based on the method of Bayley and Pinneau (11). Records indicating an absence of pituitary adenoma or other intracranial lesions on magnetic resonance imaging (MRI) and lack of known exposure to sex steroids or cranial irradiation were required for participation. Patients with a known rare sequence variant in MKRN3 were also excluded.
Data were reviewed for all participants to determine if there were any phenotypic differences between the participants with and without MKRN3 deletions. The clinical data were not normally distributed, except for age of pubarche, and therefore are reported as median (interquartile range). Body mass index (BMI) z-scores were calculated (12) for comparison across the age range of participants. To assess growth status, the z-score of each participant’s midparental target height (MPH) was also calculated (12) using a standardized age of 20 y/o for each parent’s height, and the difference between the MPH z-score and the height z-score of the participant at the time of the first visit with the pediatric endocrinologist was then calculated. Because some participants enrolled later in the course of pubertal development and were already being treated for precocious puberty with gonadotropin-releasing hormone (GnRH) antagonists, laboratory confirmation of CPP could not uniformly be performed at the NIH CC. Therefore, the hormone levels that were used to confirm diagnosis were measured at multiple laboratories with differing reference ranges. However, the relevant reference range for Tanner stage ≥2 for each assay was confirmed, and participants were only included if gonadotropin values were unequivocally in the pubertal range, as confirmed by a pediatric endocrinologist (A.D.). Since the clinical features of CPP are not the subject of this study, details of the multiple different hormone assays are not reported here. Given the retrospective data collection, the number of participants contributing to each data point is listed wherever there are missing data. Due to the small number of participants, no formal statistics were performed on the group, but the data are provided for descriptive purposes.
This study was approved by the Institutional Review Board of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Informed consent was obtained from parents, and when applicable, patients signed informed assent agreeing to participate in all aspects of the study.
Genetic testing
Genomic DNA from all probands was screened for rare sequence variants in MKRN3 for exclusion due to a known genetic etiology for their CPP prior to CNV testing. Sixteen probands were tested for CNVs and regions of homozygosity by Oligo-SNP Chromosomal Microarray Analysis (CMA; ClariSure, Quest Diagnostics; 2.67 million probes). The CMA consisted of 1.9 million copy number probes and 750,000 single nucleotide polymorphism (SNP) probes with an average inter-probe distance of 1,150 base pairs. The thresholds for genome-wide screening were set at greater than 200 kb for gains, greater than 50 kb for losses, and 2 or more segments greater than 5 Mb in size for regions of homozygosity.
Both MKRN3, an intronless gene, and individual exons of DLK1 are smaller than the limits of detection of the CMA, and therefore, CNVs involving these genes could be missed by CMA testing. To avoid this oversight, deletion and duplication testing was performed. DNAs from 15 (n = 15 of 16; 93.8%) probands were sent for a custom panel of MKRN3 and DLK1 next-generation sequencing (NGS) with deletion/duplication testing (Fulgent Genetics, Temple City, CA). Positive findings were confirmed by quantitative polymerase chain reaction (qPCR) with participants’ genomic DNA using 3 different primer sets (MKRN3-E1, MKRN3-E1B, and MKRN3-E1C), which were compared with 2 control loci (Autosomal [AC] and X chromosome [XC]; information about qPCR primers is available upon request from Fulgent Genetics). Results for both CMA and deletion/duplication testing were investigated for segregation in available family members.
Results
Clinical characteristics of iCPP participants
All participants were female (n = 16), the median age of thelarche (n = 15) was 6.8 (1.5-7.1) years, pubarche (n = 13) was 7.0 (6.5-7.5) years, and menarche (n = 6) was 3.3 (0.5-8.4) years (Table 1). Note that the number of participants who had experienced menarche at the time of enrollment was fewer than half of total participants. In addition, these participants presented at an earlier age (3.4 (0.5-8.5) years) with more rapid and advanced pubertal development than those who had not experienced menarche at the time of enrollment (median age 7.8 (7.3-7.9) years), most of whom had been treated prior to reaching menarche. This accounts for the lower age of menarche than thelarche in the overall group. Median skeletal maturation (n = 14) was advanced by 2.3 (1.5-2.8) years, consistent with the CPP phenotype. The median BMI standard deviation score (SDS) of patients who were at least 2 years of age at their first endocrine evaluation (n = 13) was 1.12 (0.62-1.72) with a range of −0.01 to 2.45. Of patients treated with GnRH agonists (n = 11), the median age of treatment initiation was 7.8 (4.7-8.3) years. None of the participants reported history of exposure to exogenous sex steroids or other estrogen-like compounds. In addition, none reported a history of hypotonia or feeding difficulties in infancy, nor evidence of hyperphagia, atypical eating behaviors, or other physical or cognitive features that are associated with PWS.
Table 1.
Clinical Characteristics of CPP Participants
| Characteristics | All participants* [n = 16] | Patient A | Patient B |
|---|---|---|---|
| Thelarche, years | 6.8 (1.5 to 7.1) [n = 15] | 7 | Unknown |
| Pubarche, years | 7.06 (6.5 to 7.5) [n = 13] | 7 | 6.7 |
| Menarche, years | 3.3 (0.5 to 8.4) [n = 6] | n/a | 6 |
| Family history of CPP, n (%) | 6 (37.5 %) | Yes | No |
| BMI SDS at diagnosis, >2 years of age | 1.12 (0.62 to 1.72) [n = 13] | 0.62 | 2.45 |
| Midparental target height, cm | 161.3 (159.4 to 165.1) [n = 15] | 161.3 | 158.8 |
| Midparental target height, z-score | −0.32 (−0.61 to 0.28) [n = 15] | -0.32 | -0.71 |
| [Height at initial endo evaluation z-score] – [Mid parental target height z-score] | 1.29 (1.10 to 1.78) [n = 13] | 1.19 | 2.09 |
| [Bone age] – [chronological age], years | 2.3 (1.5 to 2.8) [n = 14] | 2.8 | 2.7 |
| Age of first treatment, years | 7.8 (4.7 to 8.3) [n = 11] | 8.4 | 6.2 |
*median (25%-75%) unless otherwise noted; Bold, value outside interquartile range or above/below +/-2 standard deviations for all subjects.
Patient A
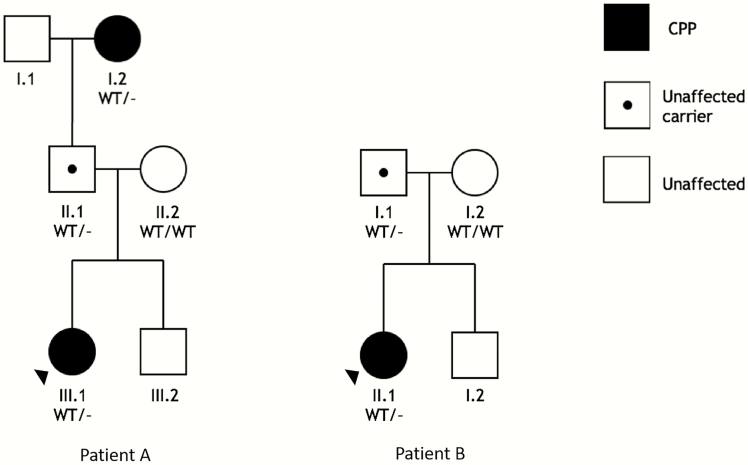
Patient A, a female of European descent, was 8 years 9 months of age at the time of enrollment. Her age of thelarche and pubarche were reportedly 7 years and she had also experienced some emotional lability (Table 1). She was initially evaluated at age 8 years 3 months due to rapidly progressing breast development, when the physical exam revealed Tanner stage 4 breasts and Tanner stage 3 pubic hair, and she was premenarchal. Her BMI was 17.3 kg/m2 (73rd percentile) at the time. Her basal luteinizing hormone (LH) concentration was 2.9 IU/L, follicle-stimulating hormone (FSH) was 6.8 UI/L, and estradiol was 19 pg/mL. A bone age was read as 11 years at age 8 years 3 months, with a predicted final adult height of 152.4 cm (midparental height 161.3 cm), and a brain MRI revealed no intracranial pathology. Patient A began treatment with leuprolide (11.25 mg injection/month) at 8 years 5 months. Patient A’s mother had menarche between 12 and 13 years old and her father reported normal pubertal timing, but the paternal grandmother had a history of early puberty with menarche between 8 and 9 years old. The paternal grandmother had a final adult height of 152 cm (Fig. 1).
Figure 1.
Families with MKRN3 deletions. Squares, male; circles, female. Filled symbols denote CPP phenotype; black center dots indicate unaffected carriers based on genotype; blank symbols denote unaffected individuals with normal or unknown genotype. Abbreviations: WT, wild type; -, deletion of locus: left, MKRN3; right, 15q11.2 locus containing MKRN3, MAGEL2, and NDN.
Patient B
At the time of enrollment, Patient B was a female aged 6 years 9 months. She is originally of Bolivian descent but now resides in the United States. She initially presented to medical attention in her local emergency department due to episodes of vaginal bleeding at 6 years old. Patient B had no known history of sex steroid exposures or vaginal/pelvic trauma. At the time of that evaluation, her basal LH concentration was 7.5 IU/L, FSH was 3.9 UI/L, and estradiol was 52 pg/mL. She was seen soon after, at age 6 years 2 months, for endocrine evaluation and was found to have Tanner stage 4 breasts, Tanner stage 1 pubic hair, and BMI of 23.4 kg/m2 (99th percentile). Gynecologic evaluation was not pursued because of Patient B’s Tanner stage and laboratory results at the time of menarche, which were consistent with CPP. A bone age at that time was read as 9 years 6 months, and a brain MRI was normal. Due to these findings, Patient B was diagnosed with idiopathic CPP and started on leuprolide treatment (11.25 mg injection/month) at 6 years 3 months of age. After initiating treatment, breast enlargement regressed slightly, and vaginal bleeding ceased. The mother of Patient B had menarche at age 13 and breast development at age 12. The father of Patient B reports normal timing of puberty and no known family history of early or late puberty in paternal grandparents.
MKRN3 deletions in CPP
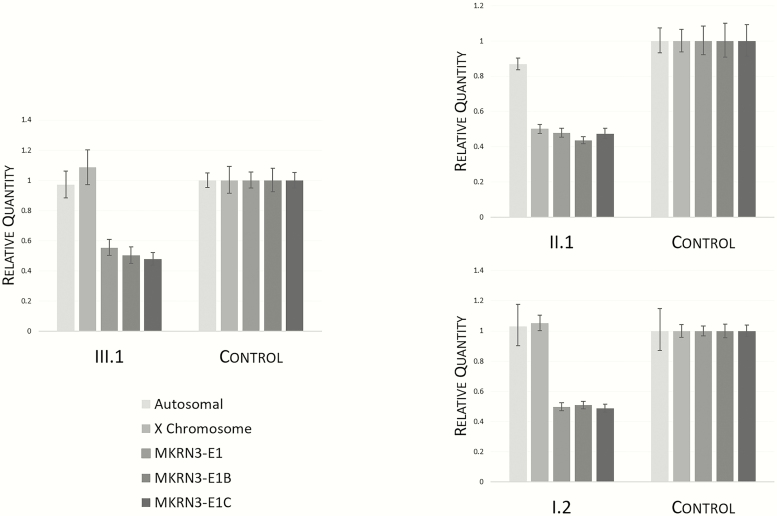
We identified whole gene deletions of MKRN3 in 2 unrelated patients with idiopathic CPP, while no CNVs in DLK1 were found by CMA or targeted deletion/duplication testing. A deletion of MKRN3 was detected in Patient A by the custom deletion/duplication panel. The genetic defect was inherited from her paternal grandmother via her unaffected father, consistent with the imprinted and paternally expressed inheritance pattern of the PWS-IC region (Fig. 1, left). The MKRN3 deletions were confirmed with qPCR, which revealed approximately half the relative expression of MKRN3 for all 3 carriers of the deletion, compared with control samples. (Fig. 2). Patient A (III.1) and her paternal grandmother (I.2) had similar relative expression of the X chromosome (XC) amplicon compared to the female control, while the relative expression of XC in the patient’s father (II.1) was approximately 0.5. These findings are consistent with a heterozygous deletion of MKRN3 in all 3 participants and the expected hemizygous X chromosome in Patient A’s father.
Figure 2.
Peripheral blood qPCR results for Family A: proband (III.1); father (II.1); paternal grandmother (I.2). Genomic DNA expression values are shown for 3 different MKRN3 primer sets (MKRN3-E1, MKRN3-E1B, and MKRN3-E1C) and control chromosomes (Autosomal [AC] and X chromosome [XC]), expressed as quantity relative to female control DNA (each graph, right).
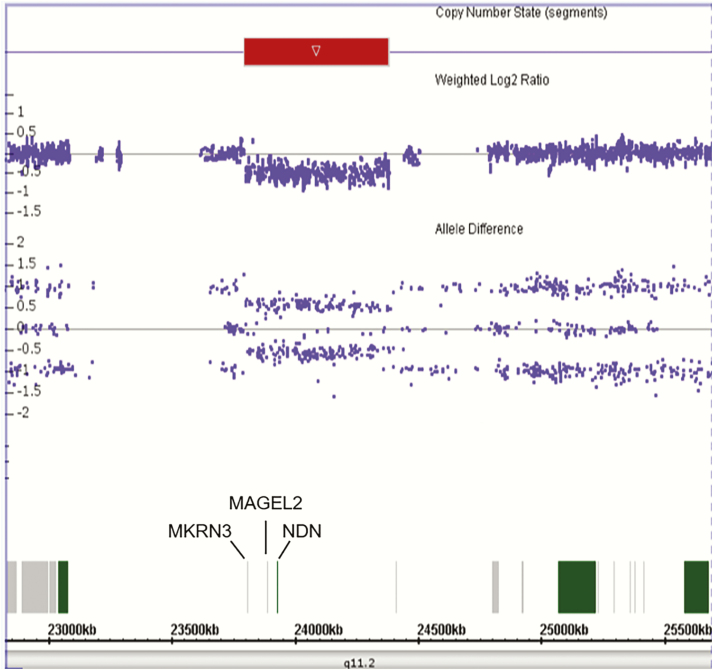
Patient B was found to have a 584 kb deletion of 15q11.2 (GRCh37/hg19 chr 15:23,798,088 – 24,382,443) involving MKRN3, MAGEL2, and NDN detected by CMA (Fig. 1, right; Fig. 3). The weighted log2 signal ratio and allele difference are approximately 0.5 (Fig. 3), consistent with a heterozygous deletion. The red arrow represents the region of deletion for Patient B. The deletion was confirmed to be paternally inherited by CMA in the father (data not shown), which is again consistent with the imprinted and paternally expressed inheritance pattern seen in this region. Patient B’s father had no personal or known family history of iCPP, but his parents were not available for evaluation. Although patient B had the highest BMI among this cohort of iCPP patients, she had no signs or symptoms suggestive of PWS, and she had a paternal family history of obesity in individuals who have no signs of PWS or abnormal puberty.
Figure 3.
Chromosomal microarray analysis (CMA) for Patient B identifies a deletion in chromosome 15q11.2. Red arrow indicates the location of the weighted Log2 Ratio and Allele Difference of approximately 0.5, consistent with a heterozygous deletion. Scale (bottom): chromosome 15 locus (exons, grey/green bars), chromosomal position and band (GRCh37/hg19).
Pubertal phenotype of PWS involving MKRN3 deletions is not always associated with CPP
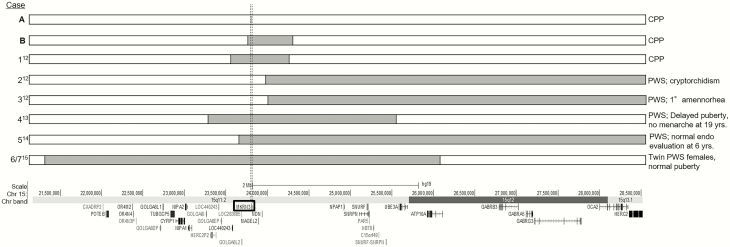
Because of the known association between deletions or imprinting defects in this region and PWS, we performed a literature search of all reported cases of PWS in which the coordinates of the defect were known (Fig. 4) (13-16). Kanber et al (13) reported a patient presenting with signs of PWS: obesity, high pain threshold, and developmental delay, with an unbalanced translocation [45, X, der(X) t(X; 15) (q28; q11.2)] resulting in the deletion of MKRN3, MAGEL2, and NDN. After further phenotypic evaluation, the patient was determined to have CPP with pubertal onset at 7 years 6 months of age and was not found to have PWS. The 2 other patients in this publication had phenotypes consistent with PWS, and their deletions did not include MKRN3, MAGEL2, or NDN but extended toward the telomeric end of the PWS-IC (13). The remaining 4 cases with PWS had deletions including MKRN3 but with no signs of precocious puberty (14-16). The deletions in patients 4 to 7 extended well beyond MKRN3, MAGEL2, and NDN, and they exhibited a range of pubertal phenotypes from normal (cases 6, 7) (16) to delayed (case 4 with delayed puberty and absence of menarche at 19 years old, consistent with HH) (14) to unknown (case 5, normal female for age, but too soon to tell definitively whether precocious or delayed puberty would develop) (15).
Figure 4.
Summary of published deletions in PWS-imprinting center region on chromosome 15. Dotted lines represent the borders of the MKRN3 locus. Shaded grey regions in bars represent the coordinates of published deletions. Left side: letter (present study, bold) or number (references cited) assigned to each case; right side: diagnoses published; bottom: scale indicating chromosome 15 position, band, with genes involved below (GRCh37/hg19). Abbreviations: CPP, central precocious puberty; PWS, Prader-Willi syndrome; Chr, chromosome.
Together, this analysis demonstrates a range of pubertal developmental trajectories among patients with deletions involving MKRN3. Patients with larger PWS-IC region deletions including MKRN3 and many other genes had PWS without evidence of CPP, while those with smaller deletions just including MKRN3 and in some cases a small number of additional genes did not have PWS but did have CPP. In addition, the deletions in some cases of PWS did not contain MKRN3, and none of these cases were reported to have CPP.
Discussion
This is the first report, to our knowledge, of nonsyndromic CPP due to heterozygous whole gene deletions of MKRN3. Our study of these patients adds to the growing data that LOF variants in MKRN3 cause CPP. The participants in the present study include both a girl with a single gene deletion of MKRN3 who inherited the defect from her obligate carrier father, precisely illustrating the expected imprinted, paternally expressed inheritance pattern, as well as a girl with a larger heterozygous deletion of MKRN3 including MAGEL2 and NDN, also inherited from the girl’s father. In the latter case (Patient B), DNA was not available to test the paternal grandparents, but the father did not have a history of CPP. We speculate that he likely inherited the deletion from his mother whose phenotype was not clear to her offspring, or that she was unaffected because her deletion was also on the maternal allele and therefore not expressed.
Mutations in the intronless MKRN3 gene have been associated with sporadic and familial CPP. The initial publication, by Abreu et al, identified 4 novel heterozygous variants in MKRN3 in 5 of 15 families with CPP (2). A later study of over 200 children with apparently sporadic CPP found 5 novel heterozygous variants in MKRN3 in 8 unrelated females (3). Segregation analysis performed in 5 of 8 families revealed that each of the variants were inherited from unaffected fathers. Since then, many studies have been published containing missense and LOF variants in MKRN3 causing CPP in girls and boys (reviewed in (17)), as well as a deletion in the promoter region (18). MKRN3 encodes a 55 kDa protein that belongs to the Makorin zinc-finger family and is proposed to have ubiquitin-protein ligase activity. The precise mechanism by which MKRN3 mutations cause CPP has yet to be elucidated, but its function is speculated to be an inhibitory effect on pubertal activation of the HPG axis with declining hypothalamic expression around the onset of puberty in mice (2). It has also been reported that circulating MKRN3 protein levels decline in male and female humans prior to the onset of puberty (19, 20), though whether this circulating protein plays a role in the initiation of puberty remains unconfirmed. Nonetheless, given the increasing evidence that MKRN3 is involved in the onset of mammalian pubertal development, it is not surprising that whole gene deletions of MKRN3 are associated with CPP.
It is notable that Patient B has a deletion that includes 2 additional genes in the PWS-IC locus. Taken together, our analysis of deletions in this region raises the question of which areas of the PWS-IC are critical for development of the typical features of PWS. The deletion in case 1 (Fig. 4 (13)) was very similar to that in Patient B, and both of these patients were obese. However, given the prevalence of obesity, we cannot draw any conclusions about the relevance of the deletion to this phenotype. To date, 16 additional cases have been reported to have PWS and CPP (8-10). These patients have been genetically confirmed to have PWS by fluorescence in situ hybridization (FISH) or methylation assay which show deletions of the PWS-IC region, but with insufficient details available to determine the exact genes included in each deletion. We speculate that there is a more specific critical region of the PWS locus beyond MKRN3, MAGEL2, and NDN that is responsible for the PWS phenotype. While PWS patients can have deletions involving these 3 genes that extend toward the telomeric end of the region, it is not required in order to see the PWS phenotype (Fig. 4, cases 2, 3 (13)). On the other hand, the inclusion of MKRN3 in the deletion causing PWS does not predict the pubertal phenotype (Fig. 4, cases 4 (14), 6, 7 (16)). Thus, MKRN3 and other microdeletions centromeric to the PWS-IC locus may not be sufficient to cause PWS alone, and PWS patients with deletions that include MKRN3 do not necessarily develop CPP. Given that the premature pubertal development in CPP requires early activation of the HPG axis, the most likely explanation for the lack of early puberty in PWS patients with MKRN3 deletions is the inability to secrete pubertal level gonadotropins due to concomitant HH in these patients. It is also possible that genes in this region located beyond NBN could play an undiscovered but critical role in the regulation of the HPG axis through interactions that are upstream or downstream of the GnRH neuron, or even on GnRH neuronal migration or function, thus contributing to the HH phenotype. More detailed evaluation of the genes involved and the long-term reproductive hormonal outcomes in individuals with PWS is necessary to further examine these possibilities.
Larger studies are needed to determine if there are genotype-phenotype correlations between CPP patients without MKRN3 variants and those with deletions in MKRN3. While the sample size of the present study is insufficient to statistically assess this, pubertal milestones were similar among all participants, and the only clinically significant difference between individuals with an MKRN3 deletion and those without is obesity in Patient B. Upon careful questioning, she does not have any evidence of hyperphagia or any other signs or symptoms of PWS in her history or presentation. The age of first treatment was later in Patient A than the rest of the group, but this is not likely to be related to her deletion, as her pubertal milestones were similar to the rest of the participants, and the decision to treat is an individualized, clinician-dependent decision, especially in girls presenting after age 6 years.
MKRN3 is a small, single-exon gene which makes deletion and duplication testing difficult by chromosomal microarray, leading us to employ targeted deletion/duplication detection methods. This approach identified the MKRN3 whole gene deletion in Patient A, which was further confirmed using qPCR techniques, validating this finding. Our ability to make predictions about the prevalence of MKRN3 whole gene deletions in the population is limited by the small sample size in this study but even considering the small sample size, with this unbiased cohort (other than sex), the fact that we identified 2 patients with whole gene deletions of MKRN3 strongly suggests the importance of testing for whole gene deletion when a genetic etiology for CPP is being sought.
Therefore, our findings suggest that CPP patients undergoing genetic testing with negative MKRN3 sequencing results, and suspected nonmaternal inheritance (as paternal inheritance may be masked due to imprinting), should be tested for whole gene deletions for completeness of the genetic investigation. Due to the limits of detection for CMA arrays, smaller genes like MKRN3 should be tested by orthogonal methods like qPCR or multiplex ligation-dependent probe amplification (MLPA) to avoid false-negative results. While there were only 2 patients with MKRN3 deletions in this study, our results also suggest that there are no significant phenotypic differences between patients with deletions of MKRN3 and those without sequence variants in the gene. Further testing of patients with PWS using more detailed mapping of causative deletions is needed to define the role of other genes in the PWS-IC region contributing to the clinical features of the syndrome, including reproductive phenotypes.
Acknowledgments
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Development (NICHD), NIH and the American Lebanese Syrian Associated Charities. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to thank the patients and their families, the staff of the NIH Clinical Center, as well as Renius Owen, PhD, from Quest Diagnostics, for his support in providing data from commercial testing performed in the course of the study, and Youn Hee Jee, MD and Rebecca Persky, MD for their careful review of this manuscript.
Financial Support: This work was funded by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Development (NICHD), NIH and the American Lebanese Syrian Associated Charities.
Clinical Trial Information: ClinicalTrials.gov Registration no. NCT01500447.
Glossary
Abbreviations
- BMI
body mass index
- CMA
chromosomal microarray analysis
- CNV
copy number variant
- CPP
central precocious puberty
- FSH
follicle-stimulating hormone
- GnRH
gonadotropin-releasing hormone
- HH
hypogonadotropic hypogonadism
- HPG
hypothalamic-pituitary-gonadal
- iCPP
idiopathic CPP
- LH
luteinizing hormone
- LOF
loss-of-function
- MPH
midparental target height
- MRI
magnetic resonance imaging
- NIH
National Institutes of Health
- PWS
Prader-Willi syndrome
- PWS-IC
Prader-Willi syndrome imprinting center region
- qPCR
quantitative polymerase chain reaction
- SDS
standard deviation score
- SNP
single nucleotide polymorphism
Additional Information
Disclosure Summary: The authors have no conflicts of interest to disclose.
Data Availability: The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Carel JC, Léger J. Clinical practice. Precocious puberty. N Engl J Med. 2008;358(22):2366-2377. [DOI] [PubMed] [Google Scholar]
- 2. Abreu AP, Dauber A, Macedo DB, et al. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013;368(26):2467-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Macedo DB, Abreu AP, Reis AC, et al. Central precocious puberty that appears to be sporadic caused by paternally inherited mutations in the imprinted gene makorin ring finger 3. J Clin Endocrinol Metab. 2014;99(6):E1097-E1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bessa DS, Macedo DB, Brito VN, et al. High frequency of MKRN3 mutations in male central precocious puberty previously classified as idiopathic. Neuroendocrinology. 2017;105(1):17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dauber A, Cunha-Silva M, Macedo DB, et al. Paternally inherited DLK1 deletion associated with familial central precocious puberty. J Clin Endocrinol Metab. 2017;102(5):1557-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomes LG, Cunha-Silva M, Crespo RP, et al. DLK1 is a novel link between reproduction and metabolism. J Clin Endocrinol Metab. 2019;104(6):2112-2120. [DOI] [PubMed] [Google Scholar]
- 7. Angulo MA, Butler MG, Cataletto ME. Prader-willi syndrome: a review of clinical, genetic, and endocrine findings. J Endocrinol Invest. 2015;38(12):1249-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee HS, Hwang JS. Central precocious puberty in a girl with prader-willi syndrome. J Pediatr Endocrinol Metab. 2013;26(11-12):1201-1204. [DOI] [PubMed] [Google Scholar]
- 9. Ludwig NG, Radaeli RF, Silva MM, et al. A boy with prader-willi syndrome: unmasking precocious puberty during growth hormone replacement therapy. Arch Endocrinol Metab. 2016;60(6):596-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu ML, Li J, Ding Y, et al. [Endocrine and metabolic features of female children with Prader-Willi syndrome: an analysis of 4 cases]. Zhongguo Dang Dai Er Ke Za Zhi. 2017;19(5):514-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greulich WW, Pyle SI.. Radiographic atlas of skeletal development of the hand and wrist. et al. ed. Stanford, CA: Stanford University Press; 1959. [Google Scholar]
- 12. Pediatric Z-score Calculator https://zscore.research.chop.edu/index.php. Published 2019. Accessed September 13, 2019.
- 13. Kanber D, Giltay J, Wieczorek D, et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet. 2009;17(5):582-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shao XY, Zhang R, Hu C, et al. Precise microdeletion detection of Prader-Willi Syndrome with array comparative genome hybridization. Biomed Environ Sci. 2010;23(3):194-198. [DOI] [PubMed] [Google Scholar]
- 15. Han JY, Park J, Jang W, Chae H, Kim M, Kim Y. A twin sibling with Prader-Willi syndrome caused by type 2 microdeletion following assisted reproductive technology: A case report. Biomed Rep. 2016;5(1):18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jehee FS, de Oliveira VT, Gurgel-Giannetti J, et al. ; Baylor-Hopkins Center for Mendelian Genomics Dual molecular diagnosis contributes to atypical Prader-Willi phenotype in monozygotic twins. Am J Med Genet A. 2017;173(9):2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Valadares LP, Meireles CG, De Toledo IP, et al. MKRN3 mutations in central precocious puberty: a systematic review and meta-analysis. J Endocr Soc. 2019;3(5):979-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Macedo DB, França MM, Montenegro LR, et al. Central precocious puberty caused by a heterozygous deletion in the MKRN3 promoter region. Neuroendocrinology. 2018;107(2):127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Busch AS, Hagen CP, Almstrup K, Juul A. Circulating MKRN3 levels decline during puberty in healthy boys. J Clin Endocrinol Metab. 2016;101(6):2588-2593. [DOI] [PubMed] [Google Scholar]
- 20. Hagen CP, Sørensen K, Mieritz MG, Johannsen TH, Almstrup K, Juul A. Circulating MKRN3 levels decline prior to pubertal onset and through puberty: a longitudinal study of healthy girls. J Clin Endocrinol Metab. 2015;100(5):1920-1926. [DOI] [PubMed] [Google Scholar]