Abstract
Herpesviruses infect virtually all humans and establish lifelong latency and reactivate to infect other humans. Latency requires multiple functions: maintaining the herpesvirus genome in the nuclei of cells; partitioning the viral genome to daughter cells in dividing cells; avoiding recognition by the immune system by limiting protein expression; producing noncoding viral RNAs (including microRNAs) to suppress lytic gene expression or regulate cellular protein expression that could otherwise eliminate virus-infected cells; modulating the epigenetic state of the viral genome to regulate viral gene expression; and reactivating to infect other hosts. Licensed antivirals inhibit virus replication, but do not affect latency. Understanding of the mechanisms of latency is leading to novel approaches to destroy latently infected cells or inhibit reactivation from latency.
Introduction
Eight herpesviruses infect humans (Table 1). With the exception of varicella-zoster virus (VZV), all human herpesviruses spread through saliva, mucosal or skin lesions, or genital secretions and infect oropharyngeal or genital tract mucosal epithelial cells or skin. VZV is spread by the respiratory route or by contact with vesicular lesions and infects epithelial cells in the oropharynx, upper respiratory tract, or conjunctiva. Three human alphaherpesviruses, herpes simplex viruses 1 and 2 (HSV-1 and HSV-2) and VZV, establish latency in sensory neurons and reactivate from neurons to infect epithelial cells in the mucosa or skin, resulting in a vesicular rash. Three human betaherpesviruses, human cytomegalovirus (HCMV), human herpesvirus-6 (HHV-6), and HHV-7, establish latency in mononuclear cells and can reactivate to cause visceral disease. Two human gammaherpesviruses, EBV and Kaposi’s sarcoma–associated herpesvirus (KSHV), establish latency in B cells. These two viruses are associated with several B cell and epithelial cell malignancies in which tumor cells are latently infected with EBV (Hodgkin lymphoma, Burkitt lymphoma, gastric carcinoma, nasopharyngeal carcinoma) or KSHV (primary effusion lymphoma, multicentric Castleman disease, Kaposi sarcoma). Many herpesviruses (e.g., HSV, EBV, HCMV, HHV-6, HHV-7) are shed in the saliva or genital secretions almost daily, usually in the absence of symptoms, indicating that latency is a dynamic process and that reactivation occurs very frequently. Unlike the other herpesviruses, VZV usually reactivates only once in a lifetime. Mathematical models for HSV (1) and EBV (2) reactivation from chronic carriers estimate that reactivation is a frequent event, but only a very small percentage of latently infected cells reactivate at any one time. The frequent shedding of most herpesviruses in salivary or genital secretions indicates that despite neutralizing antibody and potent cellular immunity, most of the herpesviruses have evolved mechanisms to evade these immune responses.
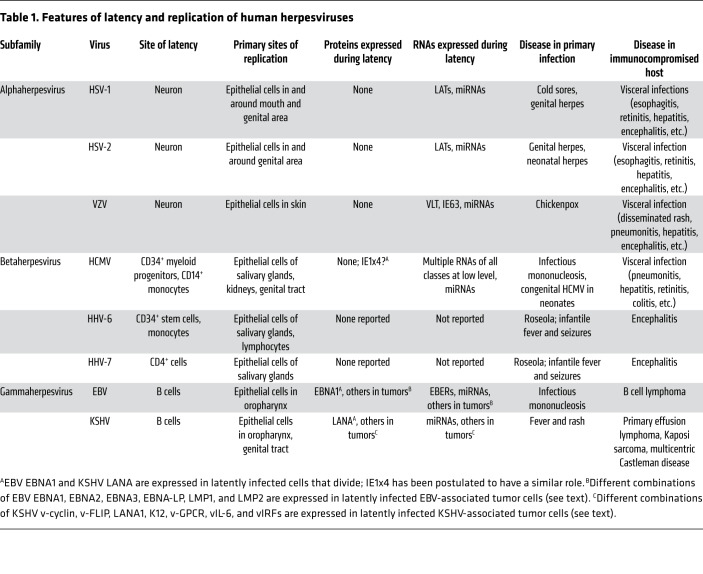
Table 1. Features of latency and replication of human herpesviruses.
Principles of herpesvirus latency
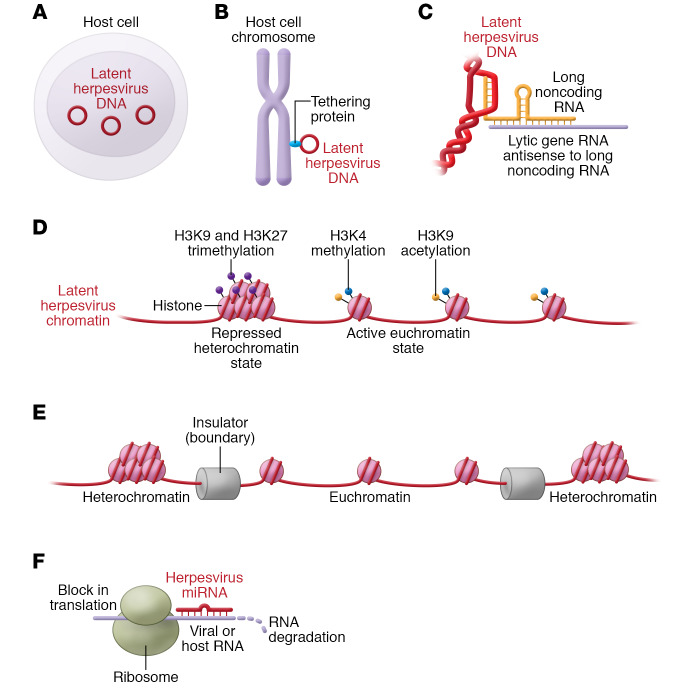
The success of the herpesviruses is largely due to their ability to establish latency and reactivate. Herpesvirus latency requires the virus to establish a persistent infection in cells without destruction by the immune system. Viral DNA is stably maintained in the nucleus of the cell as multiple copies of circular episomes (Figure 1A). Herpesviruses that maintain latency in dividing cells (e.g., HCMV, EBV, KSHV) express viral proteins when the cells divide that tether the viral genome to chromosomes so that episomes are partitioned to daughter cells (ref. 3 and Figure 1B). In latently infected cells, the virus must limit expression of viral proteins so as to avoid detection by the immune system. Many herpesviruses express long noncoding viral RNAs that contribute to establishment of latency (4). For example, alphaherpesviruses express long noncoding RNAs that are transcribed antisense to other viral RNAs expressed during lytic infection (Figure 1C). Herpesvirus genomes associate with histones in a complex termed chromatin both to regulate gene expression and to avoid damage by host proteins (5). Genes normally expressed during virus lytic replication are silenced by heterochromatic histone modifications (e.g., histone H3 lysine 9 [H3K9] and H3K27 trimethylation) during latency, while other genes are activated by euchromatic marks (e.g., H3K4 methylation and H3K9 acetylation) (Figure 1D). Polycomb repressive complex 2, which methylates histone H3K27, is important for latency of HSV, HCMV, EBV, and KSHV. KAP1 (also known as TRIM28) binds to viral chromatin and recruits a histone H3K9-methyltransferase to silence HCMV, EBV, and KSHV lytic genes to maintain latency. The epigenetic organization of the viral genome is also modulated by chromatin insulators, consisting of DNA sequences and the corresponding DNA-binding proteins and chromatin-modifying proteins, which function as boundaries to separate active (euchromatic) and inactive (heterochromatic) regions of the genome and thus help to regulate latency and reactivation (ref. 5 and Figure 1E). Herpesvirus microRNAs (miRNAs) that are expressed during latency inhibit expression of lytic genes (6–8) and regulate expression of host cell genes to evade recognition of latently infected cells by the host immune system (Figure 1F). Finally, viruses must be able to reactivate from latency to infect other hosts. This is usually initiated by host cell signaling in response to external signals (e.g., UV irradiation triggering the DNA damage response or allogeneic stimulation) or internal signals (e.g., apoptosis, hypoxia, or metabolic stress) that activate expression of immediate-early virus genes to initiate lytic replication.
Figure 1. Features of herpesvirus latency.
(A and B) Latent herpesvirus genomes are maintained in the nuclei of cells as circular episomes (A) and in dividing cells the viruses express proteins during cell division that partition the episomes to daughter cells (B). (C) Alphaherpesviruses encode long noncoding viral RNAs during latency that are transcribed antisense to viral genes expressed during lytic infection. (D) Latent herpesvirus DNA genomes are associated with histone proteins; genes normally expressed during virus lytic replication are silenced by methylation or other modifications of their histone tails during latency. (E) Chromatin insulators containing DNA sequences and the corresponding DNA-binding proteins and chromatin-modifying proteins act to separate regions of active euchromatin and repressed heterochromatin to regulate latency. (F) Herpesvirus microRNAs (miRNAs) produced during latency degrade or inhibit expression of virus lytic genes or host cell genes.
Features of latency of human herpesviruses
Alphaherpesviruses.
HSV-1, HSV-2, and VZV establish latency in nonreplicating cells (neurons), where the viral genome circularizes to form an episome. These viruses express only a few RNA transcripts, none of which are translated during latency. Since neurons do not divide, it is not necessary to express a viral protein to tether the viral genome to chromosomes during cell division of latently infected cells.
The primary HSV-1 latency-associated transcript (LAT) is an approximately 8.3-kb long noncoding RNA that is spliced to form stable 2.0 and 1.5 introns with lariat structures (9). The LAT has a neuron-specific enhancer that promotes its expression in the nucleus of latently infected cells. Multiple functions have been attributed to LAT in regulation of viral latency. The two LAT introns are expressed antisense to the HSV ICP0 immediate-early gene. Deleting the LAT from HSV reduces the frequency of reactivation (10, 11), and replacing the HSV-2 LAT with the HSV-1 LAT results in HSV-2 with a reactivation frequency similar to that of HSV-1 (12). Thus, HSV-2 with an HSV-1 LAT reactivates nearly as frequently as wild-type HSV-1 in the rabbit eye model and with a frequency similar to that of wild-type HSV-1 in the genital tract of guinea pigs. LAT has also been implicated in protecting latently infected neurons from apoptosis (13) and in contributing to modification of histones to regulate expression of latent and lytic genes (see below).
VZV expresses two viral transcripts in neurons during latency: the VZV latency-associated transcript (VLT) and ORF63 RNA (14). The VLT is a positional homolog of the HSV LAT introns in that it is antisense to VZV ORF61, the ortholog of HSV ICP0. Thus, the alphaherpesviruses have likely evolved a similar mechanism to regulate latency. While one isoform of VLT is expressed in latently infected ganglia, multiple different spliced forms of VLT (but not the isoform in latency) are expressed during lytic infection. Transfection of cells with plasmids expressing VLT and VZV ORF61 shows that VLT RNA represses transcription of ORF61. VLT proteins are detected during lytic infection, but not during latency. VZV ORF63 RNA is also expressed during latency, although at lower levels than VLT. While prior studies suggested that ORF63 protein may be expressed during latency, more recent studies indicate that the protein may only be expressed during lytic infection (15). Thus, like HSV, no proteins are thought to be expressed during VZV latent infection.
Changes in the epigenetic character of viral genomes are important for modulating latency and reactivation. During HSV latency, the LAT region has marks of transcriptionally active euchromatin, including acetylated histone H3 lysine 9 and 14 (H3K9ac, H3K14ac) (16). In contrast, immediate-early lytic genes (ICP0, ICP4, ICP27) have marks of transcriptionally repressed heterochromatin with H3K9 and H3K27 trimethylation, H3K9 dimethylation (17), and histone hypoacetylation (18). LAT enhances H3K27 trimethylation (17) and H3K9 dimethylation (18) to silence lytic gene expression and maintain lytic gene chromatin in a heterochromatic state. LAT also reduces the amount of H3K4 dimethylation to reduce expression of lytic genes (18).
The CoREST transcriptional corepressor complex, containing histone deacetylases (19), and Polycomb repressive complex 2, which methylates H3K27 (20), both inhibit expression of HSV lytic replication genes. When HSV reactivates, LAT expression is decreased, correlating with a reduction in histone acetylation at the LAT promoter, while the lytic genes are expressed with a corresponding increase in acetylated histones (21, 22). During reactivation, an activator complex containing the cellular coactivator HCF1 coupled with histone demethylases and methyltransferases is recruited to HSV immediate-early gene promoters and functions to remove repressive H3K9 methylation and install activating H3K4 trimethylation marks (23). HCF1, which resides in the nucleus of most cells and activates HSV immediate-early gene transcription during lytic infection, is sequestered in the cytoplasm of sensory neurons (the site of HSV latency) but is rapidly transported to the nucleus during the initiation of HSV reactivation (24). Expression of the viral immediate-early transcriptional activator VP16, which interacts with HCF1, may be involved in promoting complete reactivation (25).
During VZV latency, the viral immediate-early ORF63 promoter, which is expressed during latency, is maintained in a euchromatic state with acetylated H3K9, while the promoters for two lytic genes (ORF36 and ORF14) are in a heterochromatic state without acetylation at H3K9 (26). In contrast, during lytic infection, the ORF14, ORF36, ORF62, and ORF63 lytic gene promoters have euchromatic chromatin with acetylated H3K9 (26) and trimethylated H3K4 (27). Similarly, transfection experiments indicate that, in the presence of the immediate-early ORF62 activator protein, the immediate-early promoter has increased H3K4me3 and decreased H3K9me, while in the absence of ORF62 protein there is increased methylation of H3K9 (23). Thus, the same histone modifications of latent and lytic genes in HSV are used in VZV.
Chromatin insulators, which function as a boundary to separate regions of active and inactive chromatin, help to modulate HSV latency. A regulatory element (CTRL2) within HSV LAT that binds to CCCTC-binding factor (CTCF) as part of a chromatin insulator is important for establishment and maintenance of HSV latency (28), and/or reactivation of virus from latency (29). Furthermore, depletion of CTCF increases reactivation from latency (30). Additional CTCF binding sites are located at the boundary of the unique long and the internal long repeat regions of the HSV genome and are occupied by CTCF on latent viral genomes (31). CTCF binding sites have not been analyzed during VZV latency.
Viral and host cell miRNAs may also be important in modulating latency (32, 33). HSV expresses at least 29 miRNAs (34, 35). miRNAs expressed during latency are located within the LATs and in a region that crosses the junction of the HSV long and short repeat regions. HSV miR-H3 and miR-H4 target ICP34.5 (31, 32), which regulates HSV neurovirulence, and miR-H2 reduces expression of the ICP0 immediate-early transactivating gene (7, 36). miR-H6 inhibits ICP4, the major immediate-early transcriptional activator (7, 8). Cellular miRNAs also may play a role in maintaining latency. miR-138, a neuron-specific miRNA, represses expression of HSV ICP0 to promote latency (33). Expression of viral miRNAs decreases with HSV reactivation.
At least 24 miRNAs have been identified in VZV by next-generation sequencing; many of these miRNAs map to a region of the genome (between ORF56 and ORF63) that includes the VLT and ORF63 latency genes (37). Some miRNAs are antisense to ORF62, the major immediate-early gene, and one is antisense to the ORF63 latency gene. Seven have been confirmed by reverse transcriptase PCR in VZV-infected cells. One miRNA upstream of the VZV ORF63 latency gene regulates expression of ORF63 RNA.
Betaherpesviruses.
HCMV replicates in fibroblasts and epithelial cells and establishes latency primarily in CD34+ myeloid progenitor cells and CD14+ monocytes. The virus persists in and is shed from epithelial cells in the salivary glands. HHV-6 infects CD4+ T cells as well as B cells, T cells, NK cells, monocytes, and neural cells; the virus establishes a latent infection in CD34+ stem cells and monocytes, and a persistent infection in salivary glands. HHV-7 infects CD4+ T cells, where it establishes a latent infection; the virus also infects epithelial cells in the lungs and salivary glands.
While HCMV, HHV-6, and HHV-7 are maintained as episomes in the nucleus of latently infected cells, about 1%–2% of healthy people have HHV-6 integrated into their chromosomes near the subtelomeric/telomeric junction and the virus is transmitted through the germline (38, 39). At present, the clinical significance of integrated HHV-6 DNA is unknown.
HCMV, HHV-6, and HHV-7 establish latency in replicating cells; therefore, viral episomes require a protein for tethering to the host chromosome during cell division. An HCMV protein, IE1x4, is expressed in HCMV latently infected CD34+ hematopoietic progenitor cells, interacts with the terminal repeat element of the viral DNA, and is required for maintenance of the viral genome (40). This protein is postulated to be involved in tethering the HCMV genome to host cell chromosomes during viral replication, similar to the function of EBNA1 and LANA1 in EBV and KSHV, respectively. The crystal structure of the dimeric form of the carboxyl-terminal domain of the HHV-6A IE2 protein is similar to the structure of the EBV EBNA1 and KHSV LANA DNA-binding domains (41); however, it is unknown whether HHV-6A IE2 has a function similar to that of EBNA1 or LANA during replication of latent viral DNA.
Studies to determine latent gene expression in betaherpesviruses have been hampered by the very low frequency (1:10,000 to 1:100,000) of latently infected cells. The use of targeted probes to sequence RNA from human peripheral blood mononuclear cells or CD34+ hematopoietic stem cells showed low-level expression of a large number of HCMV transcripts of all kinetic classes (42). A separate study using RNA-Seq of naturally infected human tissues as well as single-cell RNA-Seq from in vitro latently infected CD14+ monocytes also found low levels of multiple RNA lytic transcripts with an increase in transcripts that are typically expressed late in infection (43). While earlier studies using experimental models for latent HCMV infection reported that only a few genes, predominantly US28, UL81-82 antisense transcript (LUNA), UL111A (viral IL-10), and UL138, were expressed during latency (44, 45), more recent studies have reported expression of many viral genes. Thus, with more sensitive assays and using naturally infected cells, HCMV latency appears to involve low-level expression of a large number of viral genes. Long noncoding RNAs, including RNAs 4.9 and 2.7, have also been detected in naturally and experimentally infected cells with latent HCMV (46). RNA4.9 interacts with the HCMV major immediate-early promoter and the Polycomb repressive complex, resulting in increased levels of H3K27me3, which represses lytic gene expression (46). RNA2.7 inhibits apoptosis (47).
Macrophages latently infected with HHV-6 in vitro were reported to express four HHV-6 latency-associated transcripts both within and in the same orientation as the coding region of the HHV-6 immediate-early 1 and 2 genes; however, the latency RNA transcriptional start sites and the 5′ noncoding regions differed from that of the immediate-early genes (48). HHV-6 U94 RNA was detected in peripheral blood mononuclear cells from healthy donors, suggesting that the gene might be expressed during latency (49). Analysis of a cell line containing integrated HHV-6A DNA showed no expression of viral genes (50).
As with other herpesviruses, the HCMV genome becomes associated with histones shortly after infection of cells. Using a cell line that becomes persistently infected with HCMV as a model for latency, lytic genes (e.g., UL32, UL54, UL83) were associated with repressive H3K9 methylation during latency (51). In contrast, during lytic replication, HCMV lytic genes are associated with H3K9ac, indicative of euchromatic chromatin. Similarly, the HCMV major immediate-early promoter is not associated with acetylated histones in naturally infected CD34+ cells, but becomes associated with acetylated histone H4 upon HCMV reactivation (52). The promoter for LUNA, which is expressed during latency in naturally infected cells, is associated with acetylated histones (53), while the HCMV major immediate-early promoter is associated with repressive H3K9me3, heterochromatin protein 1, and Daxx, which recruits histone deacetylases to repress the promoter (54). Polycomb repressive complex 2 methylates histone H3K27, resulting in H3K27me3, which suppresses expression of HCMV lytic replication genes (55). KAP1 (TRIM28) is also involved in repression of lytic gene expression by enhancing levels of repressive H3K9me3; phosphorylation of the KAP1 protein by mTOR or knockdown of KAP1 interrupts latency (56). In HHV-6, the chromosomal integrated genome is enriched in heterochromatin with H3K9me3 and H3K27me3, consistent with silencing of gene expression (50).
Other mechanisms contributing to HCMV latency include cytoplasmic sequestering of the HCMV pp71 immediate-early gene transcriptional activator (57) and the formation of a CTCF insulator in the first intron of the HCMV major immediate-early gene that functions to repress immediate-early gene expression (58). Reactivation of HCMV from latency involves expression of major immediate-early genes from an intronic promoter rather than the canonical immediate-early promoter (59). A sequence in the first intron of the HCMV major immediate-early gene binds to CTCF, and this binding inhibits expression of HCMV immediate-early gene expression and lytic replication (58).
HCMV encodes at least 26 miRNAs, and at least 12 are expressed in latently infected cells (60). These miRNAs regulate viral and cellular gene expression and likely promote maintenance of latency. HCMV miR-112-1 targets HCMV immediate-early mRNA to inhibit expression of the immediate-early protein (61). Host cell miRNA hsa-miR-200 regulates the HCMV immediate-early promoter and promotes latency (62).
While miRNAs that are antisense to the immediate-early genes of the virus have been detected in HHV-6B–infected cells undergoing lytic replication (63), and a miRNA has been identified in HHV-6A that targets the HHV-6A immediate-early U86 gene (64), the role of these miRNAs in latency has not been determined.
Gammaherpesviruses.
EBV establishes latent infection as an episome primarily in memory B cells in healthy people. The virus also establishes latent infections in B cells in Hodgkin or Burkitt lymphoma, in T cells or NK cells in patients with certain lymphomas or chronic active EBV, and in epithelial cells in nasopharyngeal and gastric carcinomas. Full-length EBV integrates into the genome in some lymphomas and nasopharyngeal carcinomas (38). KSHV establishes latency as an episome in B cells in healthy people and is latent in endothelial cells in Kaposi sarcoma and in B cells in primary effusion lymphoma and multicentric Castleman disease.
EBV EBNA1 tethers latent viral episomes to host chromosomes so that the viral genome is transmitted to daughter cells when latently infected B cells divide. KSHV LANA1 has a similar role in tethering the viral genome to host chromosomes during cell division.
Four primary patterns of latency have been reported for EBV, based on viral gene expression. Type 0 latency is observed in healthy carriers in whom only EBV-encoded RNAs (EBERs) and EBV miRNAs are expressed in circulating virus-infected B cells. EBERs are noncoding, nonpolyadenylated RNAs that interact with RNA-binding proteins, regulate PKR (which may be important for transformation) (65), and interact with PAX5 to bind to the terminal repeat of latent viral genomes (66). The other patterns of EBV latency are observed in virus-associated malignancies and include expression of the viral RNAs described above along with selected EBV proteins. Type 1 latency is seen in Burkitt lymphoma tissue with expression of EBNA1, while type 2 latency is observed in nasopharyngeal carcinoma and Hodgkin lymphoma with expression of EBNA1, LMP1, and LMP2. Type 3 latency is observed in EBV-transformed cells in culture and in patients with infectious mononucleosis and posttransplant lymphoproliferative disease with expression of all eight viral latency proteins (EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, EBNA-LP, LMP1, and LMP2) (67). In addition to its role in maintaining EBV episomes during cell division, EBNA1 upregulates its own expression and inhibits apoptosis (68). EBNA2 is a functional homolog of the Notch receptor and upregulates several viral (LMP1 and LMP2) and cellular proteins (CD21, CD23, c-myc) to increase B cell proliferation and inhibit apoptosis (69). The EBNA3s regulate EBNA2 and transactivate cellular genes (70). LMP1 is a functional homolog of CD40 and upregulates multiple proteins (NF-κB, JNK, STATs, BCL-2, A20, LFA-1, LFA-3) to promote B cell survival and inhibit apoptosis (71). LMP2 inhibits virus reactivation and activates β-catenin and Akt signaling pathways, contributing to epithelial cell transformation (72).
Studies of latency in KSHV have focused on tumor cells, including those from primary effusion lymphoma, Kaposi sarcoma, and multicentric Castleman disease. In KHSV latently infected primary effusion lymphoma cells, LANA1, kaposin A, viral interferon regulatory factor 3 (vIRF3), v-FLIP, and v-cyclin are expressed (67). In Kaposi sarcoma, LANA1, kaposin A, v-cyclin, v-FLIP, and viral G protein-coupled receptor (v-GPCR) are expressed. In multicentric Castleman disease, LANA1, v-FLIP, vIRF3, vIRF1, and vIL-6 are expressed. In addition to tethering the viral genome to host chromosomes during cell division, LANA1 inhibits Rb and p53, and upregulates survivin, c-myc, and β-catenin (73). Kaposin A is important for B cell transformation (74). The viral IRFs inhibit virus activation of IFN-α. vIRF1 inhibits MHC class I surface expression, while both vIRF1 and vIRF3 inhibit apoptosis (75). v-FLIP inhibits apoptosis and autophagy and activates NF-κB. v-Cyclin stimulates cell cycle progression from the G1 to the S phase and is not inhibited by cellular proteins, resulting in constitutive cell proliferation (74). v-GPCR activates Akt, NF-κB, and JNK. vIL-6 is a B cell growth factor, upregulates VEGF, and inhibits apoptosis (76). Thus, these proteins are important for cell proliferation, cell cycle transition, and preventing apoptosis.
As with other herpesviruses, the Polycomb repressive complex 2 plays a role in maintaining EBV latency by enhancing levels of repressive H3K27 trimethylation associated with lytic genes (77). In KSHV, LANA1 promotes silencing of lytic genes by recruiting Polycomb repressive complex 2 to viral genomes during latency (78). The complex dissociates from immediate-early and early genes during reactivation (79). KAP1/TRIM28 also contributes to both EBV and KSHV latency. In EBV, SUMOylated KAP1/TRIM28 binds to EBV OriLyt (the origin for lytic replication) and to immediate-early gene promoters, which suppresses immediate-early gene expression and promotes latency (80). EBV LMP1 promotes the binding of KAP1 to these sites, while KSHV LANA1–mediated SUMOylation of KAP1/TRIM28 enhances its binding to KSHV lytic promoters to facilitate latency (81, 82).
Binding of CTCF to EBV and KSHV genomes modulates virus latency. CTCF binds to EBV DNA in the region of the first intron of LMP2A and to EBV DNA in the 3′-untranslated region of LMP1 that is important for expression of latency genes. Deletion of the CTCF binding site results in reduction of LMP1 and LMP2A mRNAs, with a corresponding increase in LMP2B mRNA and loss of a stable DNA loop interaction between the EBV origin for virus replication (oriP) and LMP control regions (83). CTCF also binds to EBV DNA at the site of the EBNA1 Qp promoter, allowing it to function during latency (84). EBV DNA also has a CTCF binding site between oriP and the EBNA1 Cp promoter; binding of CTCF suppresses expression from the Cp promoter (85). In KSHV, CTCF binds to about 25 sites on the viral genome, and the protein activates KSHV lytic gene expression (86). The CTCF binding site at the 3′ end of the latency region of KSHV DNA is important for formation of a stable DNA loop between the latency region and the ORF50 immediate-early gene of KSHV (87).
EBV encodes at least 44 miRNAs (88). The EBV BHRF1 miRNAs are expressed during infection and in type 3 latency when all the latent proteins are expressed. The EBV BART (Bam H1 A fragment rightward transcript) miRNAs are expressed in all forms of virus latency. miR-BART20-5p interacts with the 3′-untranslated region of the two immediate-early genes of EBV, BZLF1 and BRLF1, to inhibit their expression (89). BART miRNAs target multiple viral genes, including EBV LMP1 and LMP2, as well as several lytic genes. BART miRNAs also target at least 36 cellular genes, which are involved in histone demethylation, MAP kinase signaling, immune responses, miRNA production, apoptosis, tumor suppression, NF-κB signaling, and Wnt signaling. Recent studies indicate that the EBV BART miRNAs inhibit the activity of antiviral CD4+ and CD8+ cells (90, 91). Several cellular miRNAs are induced by EBV in latently infected B cells, including miR-155, which is critical for viral latency (92).
KSHV expresses 25 mature miRNAs that originate from the latency region of the genome within or between kaposin and v-FLIP. These miRNAs inhibit apoptosis, NK cell activity, p21 (which normally arrests the cell cycle), and interferon signaling; the miRNAs increase NF-κB signaling and regulate B cell differentiation (93). In addition, miRNA-K12-9-5p (94) and miRNA-K12-7-5p (95) inhibit translation of the immediate-early Rta protein and therefore block viral lytic replication.
EBV also encodes circular RNAs that originate from spliced transcripts in the BART region (96). These circular RNAs are expressed in Burkitt lymphoma, nasopharyngeal carcinoma, and other EBV lymphomas, indicating that they are expressed in different forms of EBV latency. Circular RNAs have also been detected from the LMP2 genes that are expressed in Burkitt and other EBV lymphoma cell lines. KSHV encodes a circular RNA expressed in latently infected primary effusion lymphoma and Kaposi sarcoma cells that originates from the vIRF4 gene, and encodes a circular RNA in multicentric Castleman disease and Kaposi sarcoma cells from the KSHV PAN/K7.3 locus (96). The function of these circular RNAs in latency is unknown at present.
Reactivation from latency
Reactivation allows herpesviruses to be transmitted to uninfected people, ensuring survival of the virus over time. Many herpesviruses are reactivated during periods of extreme stress such as during septic shock (97); clinically significant reactivation is common with immune suppression and loss of T cell immune surveillance. Reactivation of HSV is associated with UV radiation, stress, fever, and trauma to the nerves. HCMV reactivation can be induced by stimulation with allogeneic cells (98), which partially explains the strong association of HCMV reactivation with transplantation, especially with graft-versus-host disease. Differentiation of HCMV-infected monocytes into macrophages or dendritic cells, induced by proinflammatory cytokines, is also associated with HCMV reactivation. Similarly, differentiation of B cells to plasma cells, with expression of the transcriptional repressor BLIMP1 and transcription factor XBP-1, reactivates EBV from B cell latency (99).
Implications for therapy
Antiviral drugs inhibit virus replication, but they have no effect on latent viral DNA. Identification of viral proteins expressed during latency has been exploited to treat malignancies associated with latent herpesviruses. Virus-specific T cells that recognize EBV latency proteins have been used to kill lymphoma cells in patients (100). A small-molecule inhibitor of EBV EBNA1 inhibited growth of EBV-positive B cell and epithelial cell tumor xenografts in mice (101). A heat shock protein 90 inhibitor, which reduced the levels of EBNA1 and LMP1 in EBV-infected cells in vitro, lowered the percentage of virus-infected cells in the blood of a patient with chronic active EBV disease (102). Downregulation of a multidrug-resistant transporter protein during HCMV latency allowed a chemotherapeutic drug, vincristine, to effectively kill latently infected cells (103).
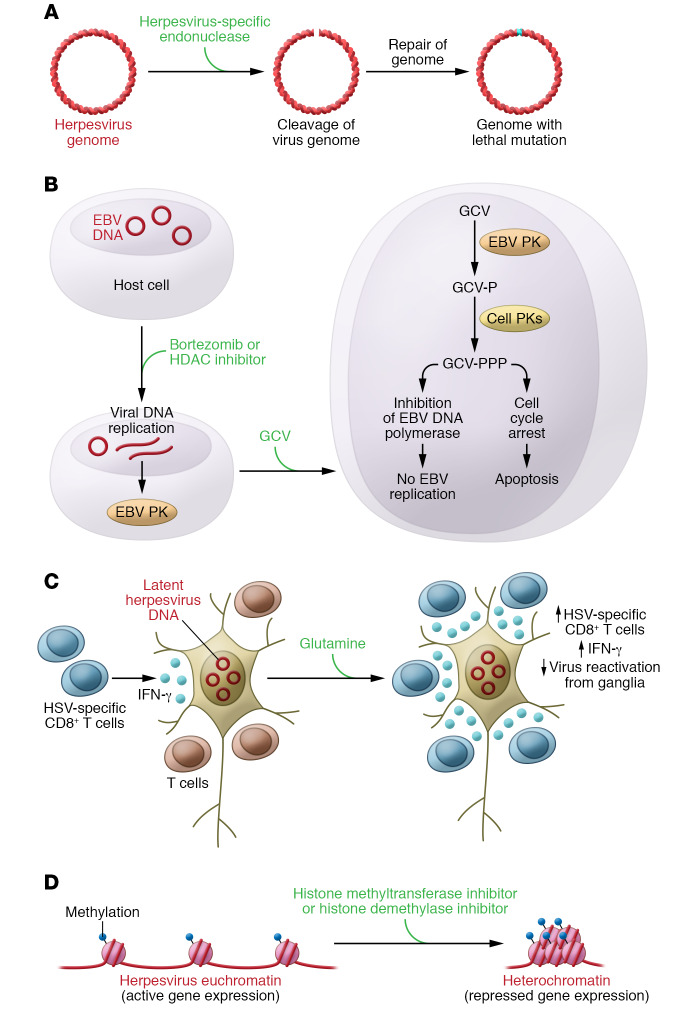
Engineered HSV-specific endonucleases (Figure 2A and ref. 104) or CRISPR/Cas (105) can cleave viral episomes in latently infected cells, and when DNA repair enzymes join the DNA ends, mutations in the viral genome may prevent latency or replication. One strategy to kill EBV lymphomas used induction of virus reactivation with a histone deacetylase inhibitor, arginine butyrate, which results in expression of the viral protein kinase; this was followed by treatment with ganciclovir, which the kinase phosphorylated to form a toxic molecule (ref. 106 and Figure 2B). Other drugs such as the proteasome inhibitor bortezomib can also induce virus reactivation; a study combining bortezomib with ganciclovir was performed in patients with EBV lymphomas (107). Compounds that inhibit reactivation from latency may also be useful. Mice given supplemental glutamine had higher numbers of HSV-specific IFN-γ–producing CD8+ T cells in their ganglia; glutamine was shown to reduce reactivation of HSV-1 and HSV-2 in mice and guinea pigs, respectively (Figure 2C and ref. 108). Inhibitors of histone methyltransferases (109) or histone demethylases (110, 111) resulted in enhanced levels of heterochromatin associated with viral genomes and blocked HSV and HCMV immediate-early gene expression (Figure 2D and ref. 111).
Figure 2. New approaches to killing latently infected cells or inhibiting reactivation from latency.
(A) Herpesvirus-specific endonucleases cleave viral DNA, and DNA repair enzymes rejoin the DNA that can result in inability to maintain latency or reactivate from latency. (B) Treatment of EBV latently infected cells with bortezomib or histone deacetylase (HDAC) inhibitors activates virus replication and production of the viral protein kinase, which phosphorylates ganciclovir, resulting in cell death (panel adapted with permission from ref. 115. © the American Society of Hematology). (C) Supplemental glutamine increases IFN-γ–producing HSV-specific T cells that reduce virus reactivation from ganglia. (D) Inhibition of histone methyltransferase or histone demethylase converts euchromatin to repressed heterochromatin and reduces immediate-early gene expression to inhibit reactivation. Glanciclovir, GCV; glanciclovir monophosphate, GCV-P; glanciclovir triphosphate, GCV-PPP; protein kinase, PK.
Future directions
Our knowledge of the mechanisms of herpesvirus latency is still evolving. Over 30 years after the discovery of the first RNA associated with herpesvirus latency (112), the mechanisms by which HSV LAT contributes to latency are still unclear. Further identification and characterization of viral RNAs and proteins expressed during herpesvirus latency and, in the case of KSHV and EBV, RNAs and proteins important for cell transformation, should lead to novel therapeutics to target latency and virus-associated malignancies. miRNAs expressed during latency in herpesviruses and circular RNAs identified in EBV and KHSV latency are especially promising targets for therapeutics. Functional screening of miRNAs, similar to the approach used to promote a specific phenotype in cells with a library of miRNA mimics (113), may help to identify viral and cellular miRNAs critical for herpesvirus latency. With the recent development of miRNA inhibitors, including miRNA mimics and anti-miRNAs, which are in clinical trials for human diseases (114), modulation of latent miRNAs or circular RNAs may be effective to inhibit herpesvirus latency.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases. I thank Thomas Kristie for reviewing the manuscript.
Version 1. 05/04/2020
Electronic publication
Version 2. 07/01/2020
Print issue publication
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(7):3361–3369.https://doi.org/10.1172/JCI136225.
References
- 1.Schiffer JT, et al. Frequent release of low amounts of herpes simplex virus from neurons: results of a mathematical model. Sci Transl Med. 2009;1(7):7ra16. doi: 10.1126/scitranslmed.3000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hadinoto V, Shapiro M, Sun CC, Thorley-Lawson DA. The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 2009;5(7):e1000496. doi: 10.1371/journal.ppat.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Leo A, Calderon A, Lieberman PM. Control of viral latency by episome maintenance proteins. Trends Microbiol. 2020;28(2):150–162. doi: 10.1016/j.tim.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed W, Liu ZF. Long Non-coding RNAs: novel players in regulation of immune response upon herpesvirus infection. Front Immunol. 2018;9:761. doi: 10.3389/fimmu.2018.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman PM. Epigenetics and genetics of viral latency. Cell Host Microbe. 2016;19(5):619–628. doi: 10.1016/j.chom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piedade D, Azevedo-Pereira JM. The role of microRNAs in the pathogenesis of herpesvirus infection. Viruses. 2016;8(6):E156. doi: 10.3390/v8060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83(3):1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu TT, Su YH, Block TM, Taylor JM. Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J Virol. 1996;70(9):5962–5967. doi: 10.1128/JVI.70.9.5962-5967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leib DA, et al. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989;63(7):2893–2900. doi: 10.1128/JVI.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause PR, et al. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J Exp Med. 1995;181(1):297–306. doi: 10.1084/jem.181.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshikawa T, Hill JM, Stanberry LR, Bourne N, Kurawadwala JF, Krause PR. The characteristic site-specific reactivation phenotypes of HSV-1 and HSV-2 depend upon the latency-associated transcript region. J Exp Med. 1996;184(2):659–664. doi: 10.1084/jem.184.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perng GC, et al. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287(5457):1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 14.Depledge DP, et al. A spliced latency-associated VZV transcript maps antisense to the viral transactivator gene 61. Nat Commun. 2018;9(1):1167. doi: 10.1038/s41467-018-03569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerboni L, et al. Apparent expression of varicella-zoster virus proteins in latency resulting from reactivity of murine and rabbit antibodies with human blood group a determinants in sensory neurons. J Virol. 2012;86(1):578–583. doi: 10.1128/JVI.05950-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubat NJ, Tran RK, McAnany P, Bloom DC. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J Virol. 2004;78(3):1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cliffe AR, Garber DA, Knipe DM. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol. 2009;83(16):8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A. 2005;102(44):16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du T, Zhou G, Khan S, Gu H, Roizman B. Disruption of HDAC/CoREST/REST repressor by dnREST reduces genome silencing and increases virulence of herpes simplex virus. Proc Natl Acad Sci U S A. 2010;107(36):15904–15909. doi: 10.1073/pnas.1010741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cliffe AR, Coen DM, Knipe DM. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio. 2013;4(1):e00590-12. doi: 10.1128/mBio.00590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amelio AL, Giordani NV, Kubat NJ, O’neil JE, Bloom DC. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J Virol. 2006;80(4):2063–2068. doi: 10.1128/JVI.80.4.2063-2068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol. 2007;81(23):13248–13253. doi: 10.1128/JVI.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15(11):1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristie TM, Vogel JL, Sears AE. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci U S A. 1999;96(4):1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawtell NM, Thompson RL. De novo herpes simplex virus VP16 expression gates a dynamic programmatic transition and sets the latent/lytic balance during acute infection in trigeminal ganglia. PLoS Pathog. 2016;12(9):e1005877. doi: 10.1371/journal.ppat.1005877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gary L, Gilden DH, Cohrs RJ. Epigenetic regulation of varicella-zoster virus open reading frames 62 and 63 in latently infected human trigeminal ganglia. J Virol. 2006;80(10):4921–4926. doi: 10.1128/JVI.80.10.4921-4926.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A. 2007;104(26):10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Washington SD, et al. The CCCTC binding factor, CTRL2, modulates heterochromatin deposition and the establishment of herpes simplex virus 1 latency in vivo. J Virol. 2019;93(13):e00415-19. doi: 10.1128/JVI.00415-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JS, Raja P, Pan D, Pesola JM, Coen DM, Knipe DM. CCCTC-binding factor acts as a heterochromatin barrier on herpes simplex viral latent chromatin and contributes to poised latent infection. mBio. 2018;9(1):e02372-17. doi: 10.1128/mBio.02372-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Washington SD, et al. Depletion of the insulator protein CTCF results in herpes simplex virus 1 reactivation in vivo. J Virol. 2018;92(11):e00173-18. doi: 10.1128/JVI.00173-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amelio AL, McAnany PK, Bloom DC. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol. 2006;80(5):2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011;25(18):1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan D, et al. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe. 2014;15(4):446–456. doi: 10.1016/j.chom.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L, Li Q. The miRNAs of herpes simplex virus (HSV) Virol Sin. 2012;27(6):333–338. doi: 10.1007/s12250-012-3266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han Z, et al. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc Natl Acad Sci U S A. 2016;113(7):E894–E901. doi: 10.1073/pnas.1525674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A. 2008;105(31):10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markus A, Golani L, Ojha NK, Borodiansky-Shteinberg T, Kinchington PR, Goldstein RS. Varicella-zoster virus expresses multiple small noncoding RNAs. J Virol. 2017;91(24):e01710-17. doi: 10.1128/JVI.01710-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morissette G, Flamand L. Herpesviruses and chromosomal integration. J Virol. 2010;84(23):12100–12109. doi: 10.1128/JVI.01169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arbuckle JH, et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A. 2010;107(12):5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarrant-Elorza M, Rossetto CC, Pari GS. Maintenance and replication of the human cytomegalovirus genome during latency. Cell Host Microbe. 2014;16(1):43–54. doi: 10.1016/j.chom.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Nishimura M, Wang J, Wakata A, Sakamoto K, Mori Y. Crystal structure of the DNA-binding domain of human herpesvirus 6A immediate early protein 2. J Virol. 2017;91(21):e01121-17. doi: 10.1128/JVI.01121-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng S, Caviness K, Buehler J, Smithey M, Nikolich-Žugich J, Goodrum F. Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc Natl Acad Sci U S A. 2017;114(49):E10586–E10595. doi: 10.1073/pnas.1710522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shnayder M, et al. Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. mBio. 2018;9(2):e00013-18. doi: 10.1128/mBio.00013-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mason GM, et al. Human cytomegalovirus latency-associated proteins elicit immune-suppressive IL-10 producing CD4+ T cells. PLoS Pathog. 2013;9(10):e1003635. doi: 10.1371/journal.ppat.1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeves M, Sinclair J. Regulation of human cytomegalovirus transcription in latency: beyond the major immediate-early promoter. Viruses. 2013;5(6):1395–1413. doi: 10.3390/v5061395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossetto CC, Tarrant-Elorza M, Pari GS. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013;9(5):e1003366. doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316(5829):1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 48.Kondo K, Shimada K, Sashihara J, Tanaka-Taya K, Yamanishi K. Identification of human herpesvirus 6 latency-associated transcripts. J Virol. 2002;76(8):4145–4151. doi: 10.1128/JVI.76.8.4145-4151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotola A, Ravaioli T, Gonelli A, Dewhurst S, Cassai E, Di Luca D. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc Natl Acad Sci U S A. 1998;95(23):13911–13916. doi: 10.1073/pnas.95.23.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saviola AJ, et al. Chromatin profiles of chromosomally integrated human herpesvirus-6A. Front Microbiol. 2019;10:1408. doi: 10.3389/fmicb.2019.01408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ioudinkova E, et al. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene. 2006;384:120–128. doi: 10.1016/j.gene.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 52.Reeves MB, MacAry PA, Lehner PJ, Sissons JG, Sinclair JH. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc Natl Acad Sci U S A. 2005;102(11):4140–4145. doi: 10.1073/pnas.0408994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reeves MB, Sinclair JH. Analysis of latent viral gene expression in natural and experimental latency models of human cytomegalovirus and its correlation with histone modifications at a latent promoter. J Gen Virol. 2010;91(pt 3):599–604. doi: 10.1099/vir.0.015602-0. [DOI] [PubMed] [Google Scholar]
- 54.Saffert RT, Kalejta RF. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J Virol. 2007;81(17):9109–9120. doi: 10.1128/JVI.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abraham CG, Kulesza CA. Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models. J Virol. 2013;87(24):13193–13205. doi: 10.1128/JVI.02420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rauwel B, Jang SM, Cassano M, Kapopoulou A, Barde I, Trono D. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. Elife. 2015;4:e06068. doi: 10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalejta RF, Bechtel JT, Shenk T. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol Cell Biol. 2003;23(6):1885–1895. doi: 10.1128/MCB.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martínez FP, et al. CTCF binding to the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication. J Virol. 2014;88(13):7389–7401. doi: 10.1128/JVI.00845-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins-McMillen D, et al. Alternative promoters drive human cytomegalovirus reactivation from latency. Proc Natl Acad Sci U S A. 2019;116(35):17492–17497. doi: 10.1073/pnas.1900783116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lau B, et al. The expression of human cytomegalovirus microRNA MiR-UL148D during latent infection in primary myeloid cells inhibits activin A-triggered secretion of IL-6. Sci Rep. 2016;6:31205. doi: 10.1038/srep31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy E, Vanícek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105(14):5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Connor CM, Vanicek J, Murphy EA. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J Virol. 2014;88(10):5524–5532. doi: 10.1128/JVI.00481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuddenham L, Jung JS, Chane-Woon-Ming B, Dölken L, Pfeffer S. Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B. J Virol. 2012;86(3):1638–1649. doi: 10.1128/JVI.05911-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nukui M, Mori Y, Murphy EA. A human herpesvirus 6A-encoded microRNA: role in viral lytic replication. J Virol. 2015;89(5):2615–2627. doi: 10.1128/JVI.02007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharp TV, et al. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21(19):4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160(4):607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cohen JI. Herpesviruses. In: Bast RC, et al., eds. Cancer Medicine. 9th ed. Hoboken, New Jersey, USA: Wiley; 2017:353–359. [Google Scholar]
- 68.Frappier L. EBNA1. Curr Top Microbiol Immunol. 2015;391:3–34. doi: 10.1007/978-3-319-22834-1_1. [DOI] [PubMed] [Google Scholar]
- 69.Kempkes B, Ling PD. EBNA2 and its coactivator EBNA-LP. Curr Top Microbiol Immunol. 2015;391:35–59. doi: 10.1007/978-3-319-22834-1_2. [DOI] [PubMed] [Google Scholar]
- 70.Styles CT, Paschos K, White RE, Farrell PJ. The cooperative functions of the EBNA3 proteins are central to EBV persistence and latency. Pathogens. 2018;7(1):E31. doi: 10.3390/pathogens7010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang LW, Jiang S, Gewurz BE. Epstein-Barr virus LMP1-mediated oncogenicity. J Virol. 2017;91(21):e01718-16. doi: 10.1128/JVI.01718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cen O, Longnecker R. Latent membrane protein 2 (LMP2) Curr Top Microbiol Immunol. 2015;391:151–180. doi: 10.1007/978-3-319-22834-1_5. [DOI] [PubMed] [Google Scholar]
- 73.Uppal T, Banerjee S, Sun Z, Verma SC, Robertson ES. KSHV LANA — the master regulator of KSHV latency. Viruses. 2014;6(12):4961–4998. doi: 10.3390/v6124961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ueda K. KSHV genome replication and maintenance in latency. Adv Exp Med Biol. 2018;1045:299–320. doi: 10.1007/978-981-10-7230-7_14. [DOI] [PubMed] [Google Scholar]
- 75.Brulois K, Jung JU. Interplay between Kaposi’s sarcoma-associated herpesvirus and the innate immune system. Cytokine Growth Factor Rev. 2014;25(5):597–609. doi: 10.1016/j.cytogfr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cannon M. The KSHV and other human herpesviral G protein-coupled receptors. Curr Top Microbiol Immunol. 2007;312:137–156. doi: 10.1007/978-3-540-34344-8_5. [DOI] [PubMed] [Google Scholar]
- 77.Woellmer A, Arteaga-Salas JM, Hammerschmidt W. BZLF1 governs CpG-methylated chromatin of Epstein-Barr virus reversing epigenetic repression. PLoS Pathog. 2012;8(9):e1002902. doi: 10.1371/journal.ppat.1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toth Z, Papp B, Brulois K, Choi YJ, Gao SJ, Jung JU. LANA-mediated recruitment of host polycomb repressive complexes onto the KSHV genome during de novo infection. PLoS Pathog. 2016;12(9):e1005878. doi: 10.1371/journal.ppat.1005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toth Z, et al. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 2010;6(7):e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bentz GL, Moss CR, Whitehurst CB, Moody CA, Pagano JS. LMP1-induced sumoylation influences the maintenance of Epstein-Barr virus latency through KAP1. J Virol. 2015;89(15):7465–7477. doi: 10.1128/JVI.00711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cai Q, Cai S, Zhu C, Verma SC, Choi JY, Robertson ES. A unique SUMO-2-interacting motif within LANA is essential for KSHV latency. PLoS Pathog. 2013;9(11):e1003750. doi: 10.1371/journal.ppat.1003750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun R, Liang D, Gao Y, Lan K. Kaposi’s sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency. J Virol. 2014;88(13):7331–7344. doi: 10.1128/JVI.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen HS, et al. Epigenetic deregulation of the LMP1/LMP2 locus of Epstein-Barr virus by mutation of a single CTCF-cohesin binding site. J Virol. 2014;88(3):1703–1713. doi: 10.1128/JVI.02209-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog. 2010;6(8):e1001048. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tempera I, Klichinsky M, Lieberman PM. EBV latency types adopt alternative chromatin conformations. PLoS Pathog. 2011;7(7):e1002180. doi: 10.1371/journal.ppat.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li D, Mosbruger T, Verma D, Swaminathan S. Complex Interactions between cohesin and CTCF in regulation of Kaposi’s sarcoma-associated herpesvirus lytic transcription. J Virol. 2020;94(2):e01279-19. doi: 10.1128/JVI.01279-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kang H, Wiedmer A, Yuan Y, Robertson E, Lieberman PM. Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS Pathog. 2011;7(8):e1002140. doi: 10.1371/journal.ppat.1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skalsky RL, Cullen BR. EBV noncoding RNAs. Curr Top Microbiol Immunol. 2015;391:181–217. doi: 10.1007/978-3-319-22834-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jung YJ, Choi H, Kim H, Lee SK. MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J Virol. 2014;88(16):9027–9037. doi: 10.1128/JVI.00721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Albanese M, et al. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc Natl Acad Sci U S A. 2016;113(42):E6467–E6475. doi: 10.1073/pnas.1605884113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tagawa T, et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J Exp Med. 2016;213(10):2065–2080. doi: 10.1084/jem.20160248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol. 2010;84(22):11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qin J, Li W, Gao SJ, Lu C. KSHV microRNAs: tricks of the devil. Trends Microbiol. 2017;25(8):648–661. doi: 10.1016/j.tim.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6(6):570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin X, Liang D, He Z, Deng Q, Robertson ES, Lan K. miR-K12-7-5p encoded by Kaposi’s sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS One. 2011;6(1):e16224. doi: 10.1371/journal.pone.0016224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Toptan T, et al. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A. 2018;115(37):E8737–E8745. doi: 10.1073/pnas.1811728115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ong DSY, et al. Epidemiology of multiple herpes viremia in previously immunocompetent patients with septic shock. Clin Infect Dis. 2017;64(9):1204–1210. doi: 10.1093/cid/cix120. [DOI] [PubMed] [Google Scholar]
- 98.Söderberg-Nauclér C, Fish KN, Nelson JA. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91(1):119–126. doi: 10.1016/S0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 99.Reusch JA, Nawandar DM, Wright KL, Kenney SC, Mertz JE. Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters. J Virol. 2015;89(3):1731–1743. doi: 10.1128/JVI.02781-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bollard CM, et al. Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein-Barr virus latent membrane proteins. J Clin Oncol. 2014;32(8):798–808. doi: 10.1200/JCO.2013.51.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Messick TE, et al. Structure-based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth. Sci Transl Med. 2019;11(482):eaau5612. doi: 10.1126/scitranslmed.aau5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shatzer A, et al. Ganetespib, an HSP90 inhibitor, kills Epstein-Barr virus (EBV)-infected B and T cells and reduces the percentage of EBV-infected cells in the blood. Leuk Lymphoma. 2017;58(4):923–931. doi: 10.1080/10428194.2016.1213823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weekes MP, et al. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science. 2013;340(6129):199–202. doi: 10.1126/science.1235047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aubert M, et al. In vivo disruption of latent HSV by designer endonuclease therapy. JCI Insight. 2016;1(14):e88468. doi: 10.1172/jci.insight.88468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci U S A. 2014;111(36):13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perrine SP, et al. A phase 1/2 trial of arginine butyrate and ganciclovir in patients with Epstein-Barr virus-associated lymphoid malignancies. Blood. 2007;109(6):2571–2578. doi: 10.1182/blood-2006-01-024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bortezomib and Ganciclovir in Treating Patients With Relapsed or Refractory Epstein Barr Virus-Positive Lymphoma. NCT00093704. http://clinicaltrials.gov Updated October 2, 2015. Accessed March 10, 2020.
- 108.Wang K, et al. Glutamine supplementation suppresses herpes simplex virus reactivation. J Clin Invest. 2017;127(7):2626–2630. doi: 10.1172/JCI88990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arbuckle JH, et al. Inhibitors of the histone methyltransferases EZH2/1 induce a potent antiviral state and suppress infection by diverse viral pathogens. mBio. 2017;8(4):e01141-17. doi: 10.1128/mBio.01141-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hill JM, et al. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med. 2014;6(265):265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio. 2013;4(1):e00558-12. doi: 10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235(4792):1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 113.Eulalio A, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 114.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 115.Bollard CM, Cohen JI. How I treat T-cell chronic active Epstein-Barr virus disease. Blood. 2018;131(26):2899–2905. doi: 10.1182/blood-2018-03-785931. [DOI] [PMC free article] [PubMed] [Google Scholar]