Abstract
Children and adults with Philadelphia chromosome–like B cell acute lymphoblastic leukemia (Ph-like B-ALL) experience high relapse rates despite best-available conventional chemotherapy. Ph-like ALL is driven by genetic alterations that activate constitutive cytokine receptor and kinase signaling, and early-phase trials are investigating the potential of the addition of tyrosine kinase inhibitors (TKIs) to chemotherapy to improve clinical outcomes. However, preclinical studies have shown that JAK or PI3K pathway inhibition is insufficient to eradicate the most common cytokine receptor–like factor 2–rearranged (CRLF2-rearranged) Ph-like ALL subset. We thus sought to define additional essential signaling pathways required in Ph-like leukemogenesis for improved therapeutic targeting. Herein, we describe an adaptive signaling plasticity of CRLF2-rearranged Ph-like ALL following selective TKI pressure, which occurs in the absence of genetic mutations. Interestingly, we observed that Ph-like ALL cells have activated SRC, ERK, and PI3K signaling consistent with activated B cell receptor (BCR) signaling, although they do not express cell surface μ-heavy chain (μHC). Combinatorial targeting of JAK/STAT, PI3K, and “BCR-like” signaling with multiple TKIs and/or dexamethasone prevented this signaling plasticity and induced complete cell death, demonstrating a more optimal and clinically pragmatic therapeutic strategy for CRLF2-rearranged Ph-like ALL.
Keywords: Hematology, Oncology
Keywords: Leukemias, Protein kinases, Signal transduction
Introduction
Philadelphia chromosome–like B cell acute lymphoblastic leukemia (Ph-like B-ALL) occurs in 15% to 40% of older children, adolescents, and adults with B-ALL and is associated with high rates of relapse and poor long-term survival (1–6). Ph-like ALL is now known to be driven by more than 70 unique genetic rearrangements or fusions that activate oncogenic kinase signaling. Translocation or intrachromosomal deletion involving the cytokine receptor–like factor 2 gene (CRLF2; encoding the thymic stromal lymphopoietin receptor [TSLPR]) accounts for approximately 50% of Ph-like ALL cases and is frequently associated with concomitant JAK2 or JAK1 mutations. The TSLPR is a heterodimeric cytokine receptor composed of CRLF2 and IL-7R α chain (CD127) proteins that mimic IL-7 (the cytokine responsible for early B cell survival) signaling and induce phosphorylation of STAT5 upon binding to its ligand, thymic stromal lymphopoietin (TSLP) (7, 8). Identification of these cytokine receptor and kinase alterations suggests that patients with Ph-like ALL could benefit from tyrosine kinase inhibitor–based (TKI-based) therapies (9–18).
We previously identified that combinatorial JAK and PI3K pathway targeting with the JAK1/JAK2 inhibitor (JAK1/JAK2i) ruxolitinib and the dual PI3K/mTORi gedatolisib had superior antileukemia activity and partially circumvented compensatory reactivation of phosphorylated (p) STAT5 and AKT, but was insufficient to induce complete leukemic cell death (15). Our more recent studies have shown that CRLF2-rearranged (CRLF2-R) Ph-like ALL cells are not JAK oncogene addicted and rapidly overcome inhibitory effects of single-TKI treatment (19). These findings raise the question of how Ph-like ALL circumvents JAK pathway inhibition and highlight the need to elucidate other critical signaling dependencies. The present studies thus aimed to identify mechanisms of drug resistance to JAK inhibition, elucidate other essential signaling dependencies, identify potential ALL-associated cooperating genetic lesions, and develop new precision medicine therapeutic approaches for patients with CRLF2-R Ph-like ALL. We identified that CRLF2-R Ph-like ALL changes its genetic and phenotypic identity from cytokine receptor dependence to “B cell receptor–like” (“BCR-like”) dependence following targeted JAK inhibition. Genetic deletion or pharmacologic inhibition of activated signaling molecules also resulted in rapid adaptation and cell survival via compensatory mechanisms. As detailed below, we demonstrate that JAK, PI3K, and BCR-like signaling are required for Ph-like ALL leukemogenesis and that inhibition of all 3 pathways is necessary and sufficient to eradicate disease.
Results
Ph-like ALL is associated with poor clinical outcomes.
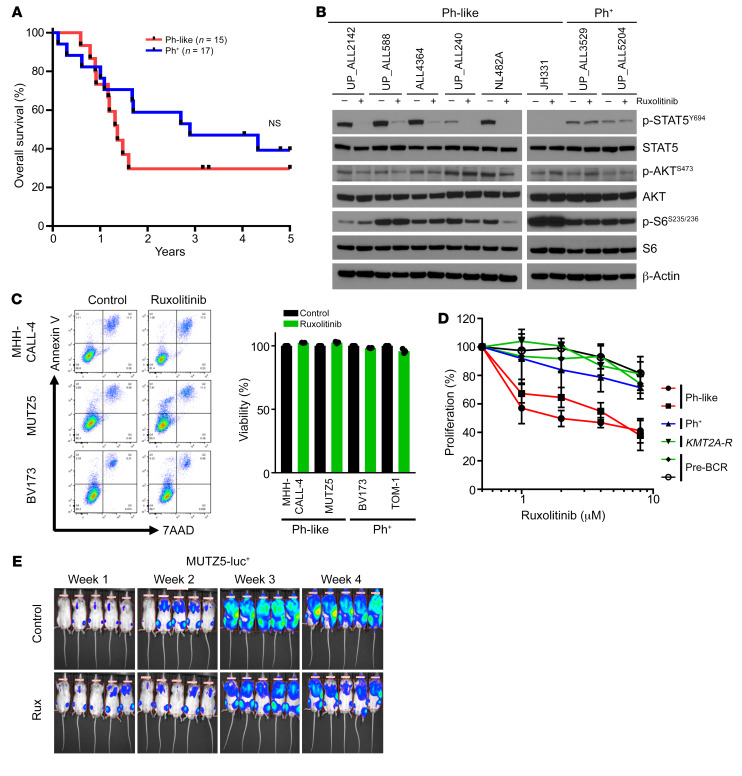
Our group and others previously reported that Ph-like ALL is common across the pediatric-to-adult age spectrum and is associated with high risk of relapse and poor long-term survival (1–4, 17, 20–22). To confirm these observations, we performed low-density microarray analysis, kinase fusion testing, and next-generation sequencing of diagnostic ALL bone marrow or peripheral blood specimens from all adults with available de novo B-ALL specimens banked at the University of Pennsylvania as described (4). Patient age, sex, race/ethnicity, diagnostic white blood cell (WBC) count, cytogenetic and molecular testing data, treatment, and clinical outcomes were tabulated where available (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI134424DS1). Clinical outcomes of patients with Ph-like ALL (treated with standard chemotherapy and without TKIs) were quite poor with rapid rates of relapse and 2-year overall survival of 29.6% (Figure 1A). Median survival of patients with Ph-like (n = 15) and Ph+ (n = 17) ALL was 1.4 years and 2.9 years, respectively. These data further validate the dismal clinical outcomes of patients with Ph-like ALL treated with conventional chemotherapy and emphasize need for more optimal treatment strategies.
Figure 1. Poor clinical outcomes and inadequate treatment effects of JAK inhibitor monotherapy in Ph-like ALL.
(A) Kaplan-Meier survival analysis of adult patients with Ph+ or Ph-like ALL treated at the University of Pennsylvania for whom outcome data were available (n = 49). (B) Western blot analysis of indicated proteins in 6 Ph-like ALL PDX cases and 2 Ph+ ALL PDX cases. (C) Ph-like ALL cell lines and Ph+ ALL cell lines were treated with 1 μM ruxolitinib for 72 hours (n = 3 independent experiments), and viability was assessed via flow cytometry. (D) B-ALL cell lines were treated with increasing concentrations of ruxolitinib for 72 hours (n = 3 independent experiments). Cell proliferation and viability were measured via XTT assay. (E) One million luciferase-labeled MUTZ5 cells were injected via tail vein into NSG mice and treated with control or ruxolitinib chow for 28 days. Data are represented as individual values with mean ± SEM bars. Significance for A was calculated by the log-rank (Mantel-Cox) test.
JAK inhibition is insufficient to kill Ph-like ALL.
Ph-like ALL is characterized by activated cytokine receptor signaling with high levels of p-STAT5 (4, 10, 15, 23), particularly in the most common subtype harboring CRLF2 rearrangements and JAK2 mutations. Protein analysis of multiple Ph-like and non–Ph-like ALL cell lines and human leukemia cells harvested from patient-derived xenograft (PDX) models confirmed high p-STAT5 expression levels in Ph-like ALL cells, although p-STAT5 levels were also expectedly high in Ph+ ALL PDX cases and cell lines (SUP-B15, TOM-1) and in wild-type FLT3-overexpressing KMT2A-R ALL cell lines (HB11;19 and SEM) (Figure 1B and Supplemental Figure 1A). Importantly, in vitro JAK inhibition of Ph-like ALL cells with ruxolitinib markedly decreased p-STAT5 at initial time points, as seen in earlier studies (10, 11, 24, 25). Surprisingly, we observed that p-STAT5 levels returned to baseline by 72 hours of continued drug treatment, suggesting rapid signaling adaptation. More moderate and transient reduction of AKT/mTOR signaling in ruxolitinib-treated cells was detected over time (Figure 1B, Supplemental Figure 1B, and Supplemental Figure 2A). To delineate more extensively the potential signaling and cytotoxic effects of JAK inhibition in Ph-like ALL, we incubated Ph-like and Ph+ ALL cell lines with ruxolitinib in vitro. While flow cytometry analysis did not detect increases in apoptosis or cell death with ruxolitinib exposure (Figure 1C), cell proliferation was moderately affected in Ph-like ALL cells (Figure 1D). We further confirmed partial but incomplete inhibition of leukemia proliferation in vivo in luciferase-expressing MUTZ5 (human Ph-like ALL cell line with IGH-CRLF2 translocation, JAK2R683G, and IKZF1 deletion) xenograft models treated with ruxolitinib for 28 days (Figure 1E), demonstrating that single-agent TKI therapy was insufficient for cure in CRLF2-R Ph-like ALL.
CRLF2 signaling is not required for survival of Ph-like ALL cells.
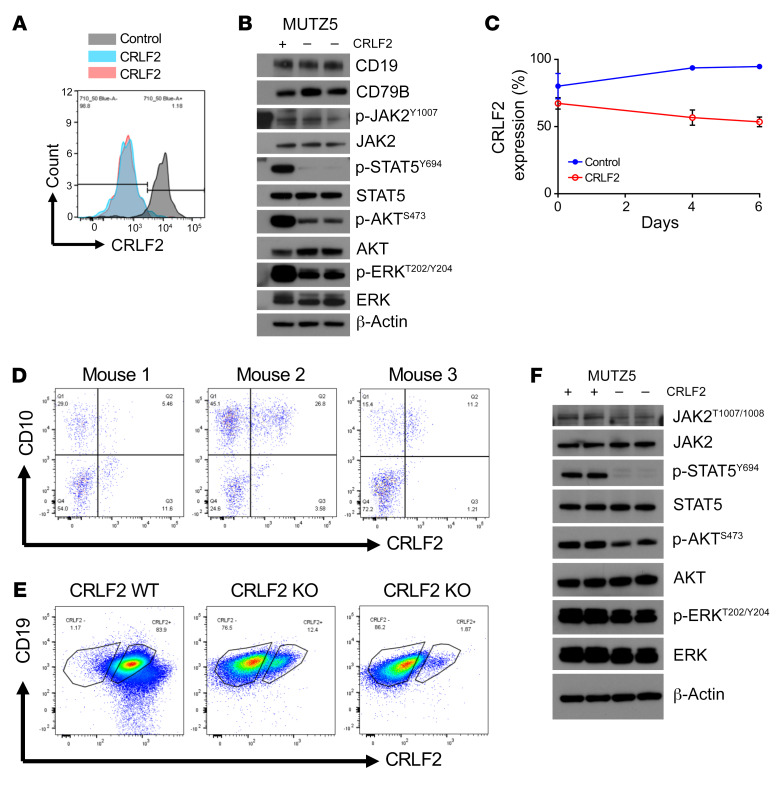
Given our observations of incomplete JAK addiction and the need to define other necessary signaling dependencies in CRLF2-R Ph-like ALL, we then studied the effects of in vitro JAK inhibition or stimulation of leukemia cells with TSLP (cytokine ligand for the TSLPR encoded by CRLF2) via broad-spectrum phospho-kinase arrays (Supplemental Figure 1C). TSLP stimulation of MHH-CALL-4 cells (human CRLF2-R/JAK2-mutant Ph-like ALL cell line) activated multiple signaling molecules, including p-STAT5, p-AKT, and p-ERK, and the transcription factor CREB, as reported in previous studies (26, 27). However, ruxolitinib treatment of MHH-CALL-4 cells in the absence of TSLP stimulation did not significantly decrease AKT, ERK, or CREB signaling. This observation suggests that multiple signaling pathways are regulated independently of JAK2 in CRLF2-R Ph-like ALL (Supplemental Figure 1C). To evaluate further whether signaling is regulated by CRLF2 in a JAK2-independent mechanism, we then tested MHH-CALL-4 cells coincubated with TSLP and ruxolitinib at 2 different time points. Interestingly, dual TSLP and ruxolitinib exposure did not alter AKT phosphorylation versus ruxolitinib-only-treated cells (Supplemental Figure 1D), demonstrating CRLF2-independent AKT signaling activation. To test further whether CRLF2 activates AKT or ERK signaling via a JAK2-independent mechanism, we generated CRLF2-null Ph-like cells. For this, we used Cas9 ribonucleoproteins and guide RNAs targeting CRLF2 for genetic deletion and validated loss of CRLF2 by flow cytometry (Figure 2A). Interestingly, CRLF2 deletion resulted in complete p-STAT5 dephosphorylation and moderate reduction in p-AKT and p-ERK levels, while no effects on p-JAK2 were detected (Figure 2B). To ascertain effects on cell proliferation, we mixed CRLF2-deleted MUTZ5 cells (human CRLF2-R/JAK2-mutant Ph-like ALL cell line) with nondeleted cells and assessed CRLF2 expression over time in vitro (Figure 2C). Nondeleted cell growth did not outcompete that of CRLF2-null cells over time, consistent with prior shRNA studies (28). Finally, we transplanted CRLF2-deleted (CRLF2/TSLPR protein–null) MUTZ5 cells into immunocompromised NSG mice and monitored the mice by flow cytometric analysis of venous blood samples for human cell engraftment and leukemia progression. Interestingly, CRLF2-null cells engrafted well in vivo with little or no outgrowth of native CRLF2+ cells observed (Figure 2D). CRLF2-null cells persisted and were detectable for several weeks in peripheral blood until mice became terminally ill and had to be sacrificed. As predicted, flow cytometry–sorted CRLF2-null MUTZ5 cells harvested from murine spleens (Figure 2E; nonmanipulated MUTZ5 cells added as gating controls) demonstrated decreased p-STAT5 compared with sorted CRLF2+ cells by Western blotting with minimal effects upon AKT and ERK signaling pathways (Figure 2F). These observations were validated via phospho–flow cytometry analysis of isolated MUTZ5 cells before cell sorting via separate gating of CRLF2-null and CRLF2+ cell populations (Supplemental Figure 1E). Taken together, CRLF2 genetic deletion recapitulates the effects of pharmacologic JAK inhibition with ruxolitinib (11), but does not appear necessary for Ph-like leukemogenesis. These data support a mechanism of signaling activation independent of CRLF2/TSLPR, which we explored further in the below experiments.
Figure 2. CRLF2 signaling is not required for survival of Ph-like ALL cells.
(A) MUTZ5 cells were electroporated with nontargeting control or ribonucleoproteins targeting CRLF2. CRLF2 deletion was validated via flow cytometry. (B) Western blot analysis of CRLF2+ and CRLF2-null MUTZ5 cells for the indicated proteins. (C) CRLF2-deleted (null or KO) MUTZ5 cells mixed with native CRLF2+ MUTZ5 control cells were monitored in vitro over time for outgrowth of CRLF2+ cells (n = 3 independent experiments). (D) Flow cytometry analysis of human CRLF2-deleted MUTZ5 cells in peripheral blood of engrafted NSG mice at 2 months after injection (n = 3). (E and F) End-study analysis of flow cytometry–sorted CRLF2-deleted (–) and CRLF2+ (+) MUTZ5 cells harvested from engrafted murine spleens by flow cytometry with CD19 and CRLF2/TSLPR surface staining (E) and Western blotting of the indicated proteins (F). Data are represented as individual values with mean ± SEM bars.
Murine CRLF2/JAK2-mutant bone marrow model mimics human Ph-like ALL signaling and sensitivity to JAK inhibition.
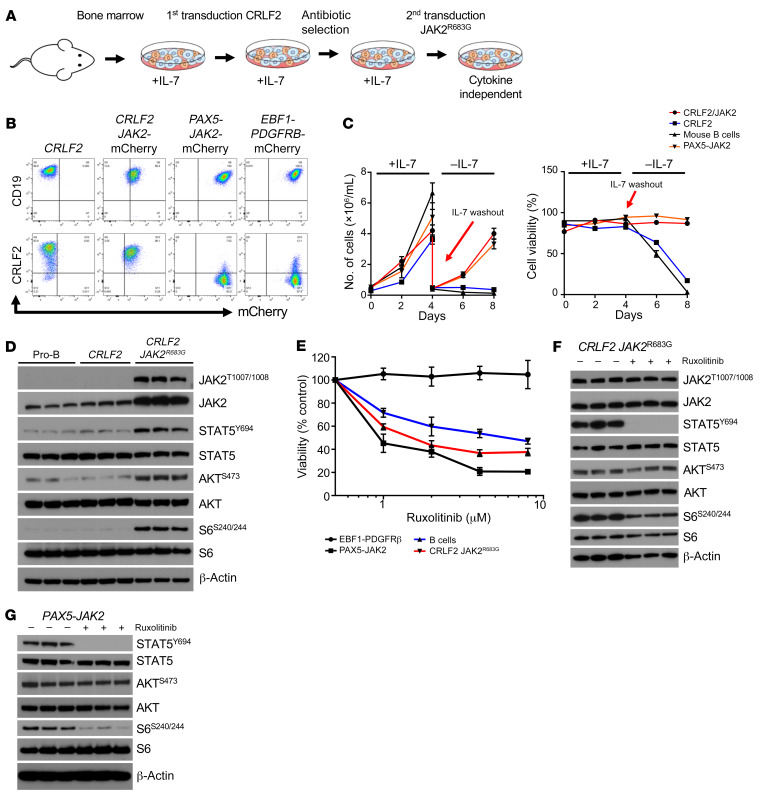
Long-term immortalized leukemia cell lines have potential for evolution of previously unidentified genetic alterations that can alter interpretation of experimental data. To validate our observed signaling phenomena mechanistically in a genetically “clean” Ph-like ALL model, we created murine bone marrow–derived CRLF2-overexpressing JAK2R683G-mutant (mCRLF2-JAK2) cells via 2-step lentiviral transduction for downstream experimental studies. We also transduced murine bone marrow cells with PAX5-JAK2 or EBF1-PDGFRB fusions for use as non–CRLF2-R Ph-like ALL control models (Figure 3, A and B). CRLF2 overexpression alone was insufficient to promote IL-7–independent cell growth of murine bone marrow cells, which, interestingly, instead required cotransduction with the JAK2R683G common comutation (Figure 3C). Western blot analysis of murine cell line lysates confirmed that both CRLF2 overexpression and JAK2 mutation were required for constitutive phosphorylation of STAT5, AKT, and S6 versus minimal signaling activation observed in CRLF2-only-overexpressing murine cells without JAK2 mutation (Figure 3D).
Figure 3. Genetic murine model recapitulates human Ph-like ALL signaling phenotype.
(A) Schematic illustrating the transduction procedure to generate murine Ph-like ALL models. (B) Flow cytometry analysis of CD19, CRLF2/TSLPR, and mCherry staining was performed on murine Ph-like ALL cells to confirm expression. (C) The left graph shows cell proliferation of the indicated murine cells in the presence of IL-7 and after IL-7 washout. The right graph shows the corresponding cell viability (n = 3 independent experiments). (D) Western blot analysis of the indicated target genes in IL-7–dependent pro-B cells, CRLF2+ cells, and CRLF2+ JAK2R683G-transduced (mCRLF2-JAK2) cells. (E) Viability analysis was performed on the indicated cell types with increasing concentrations of ruxolitinib for 72 hours (n = 3 independent experiments). (F and G) Western blot analysis of the indicated proteins in CRLF2/JAK2–transformed (F) and PAX5-JAK2–transformed (G) murine cells treated with 1 μM DMSO or ruxolitinib for 1 hour. Data are represented as individual values with mean ± SEM bars.
We next tested the sensitivity of the murine Ph-like ALL cell line models to ruxolitinib in vitro (Figure 3E). The mCRLF2-JAK2– and PAX5-JAK2–transduced mouse cells were moderately sensitive to JAK inhibition with ruxolitinib, similar to data from human Ph-like ALL cell lines (Figure 1D). Control IL-7–dependent murine pro–B cells also were sensitive to ruxolitinib, potentially via JAK1-mediated effects, while EBF1-PDGFRB–transduced cells were expectedly insensitive to ruxolitinib. In vitro ruxolitinib treatment of mCRLF2-JAK2 cells completely inhibited p-STAT5 and partially decreased p-S6 (Figure 3F), consistent with our observations in human Ph-like ALL cell lines and PDX cells (Figure 1B and Supplemental Figure 1B), but did not inhibit p-AKT even with prolonged exposure for 48 hours (Supplemental Figure 2A). Conversely, PAX5-JAK2–transduced murine bone marrow cells treated with ruxolitinib had marked decrement in p-STAT5 and p-S6 (Figure 3G), similar to our earlier observations in a BCR-JAK2 fusion ALL PDX model (NL482A in Figure 1B). These data are concordant with recent studies reporting greater oncogene addiction in Ph-like ALL with PAX-JAK2 rearrangement versus CRLF2 rearrangement (2, 29).
Ph-like ALL cells partially differentiate upon JAK2 inhibition.
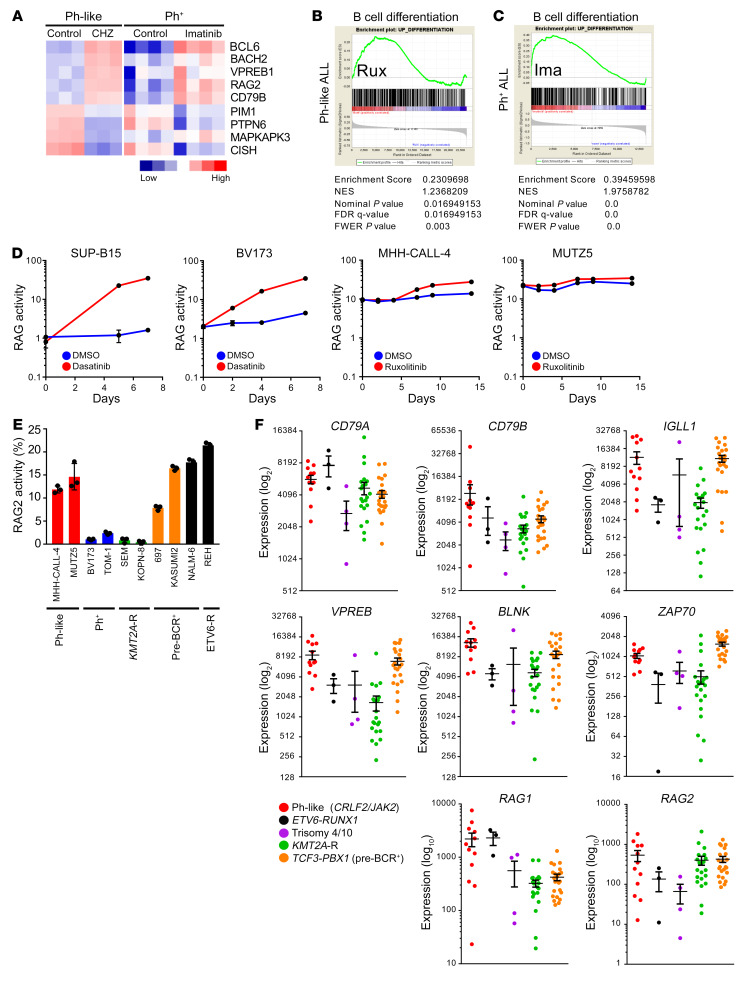
During normal development in the bone marrow, B cells differentiate from cytokine receptor dependence to pre-BCR dependence after successful rearrangement of their VHDJH gene segments (30, 31). Pre-BCR signaling is known to activate PI3K/AKT signaling and other pre-BCR–associated signaling molecules, such as SFKs, SYK, BTK, BLNK, and ERK (31–34), and pre-BCR activation drives cell survival in some subtypes of B-ALL (35). We next hypothesized that JAK2 inhibition could induce CRLF2-R Ph-like ALL differentiation based on prior reports of Ph+ ALL cell differentiation with imatinib exposure (36, 37). To determine whether Ph-like ALL cells differentiate and switch from a cytokine receptor–dependent to a pre-BCR–dependent phenotype, we first performed gene expression analyses and compared publicly available expression data of the Ph-like ALL cell line MHH-CALL-4 treated with the JAK2i CHZ868 (12) versus Ph+ ALL cell lines treated with imatinib (38) and identified that both ALL subtypes upregulate genes that are involved in cell differentiation and pre-BCR signaling upon short-term TKI exposure (Figure 4A and Supplemental Figure 3A, left). Similar effects were observed in CRLF2-R Ph-like ALL PDX samples treated with CHZ868 for 3 days in vivo (Supplemental Figure 3A, right, and ref. 12). The most prominently upregulated genes in Ph-like ALL cells included BCL6, a transcriptional repressor of TP53-induced apoptosis during V(D)J recombination; RAG1 and RAG2, encoding enzymes that initiate V(D)J recombination; and BLNK, an adaptor molecule required for pre-BCR signaling (36).
Figure 4. JAK inhibition induces partial differentiation of Ph-like ALL cells.
(A) Meta-analysis of gene expression data comparing MHH-CALL-4 cells treated with the type 2 JAK2 inhibitor CHZ868 with Ph+ ALL cells (BV173, NALM-1, SUP-B15, and TOM-1) treated with imatinib (12, 38). (B and C) Gene set enrichment analysis (GSEA) plots demonstrate enrichment of B cell differentiation gene sets in MHH-CALL-4 cells treated with ruxolitinib 1 μM for 12 hours (n = 3) (B) and Ph+ ALL treated with imatinib (38) (C); statistical analysis is shown. (D) Ph+ ALL cell lines and Ph-like ALL cell lines were lentivirally transduced with a RAG enzyme activity reporter construct. Ph+ and Ph-like ALL cells were treated with 100 nM dasatinib and 1 μM ruxolitinib, respectively. RAG enzyme activity was measured at the indicated times (n = 3 independent experiments). (E) Steady-state RAG enzyme activity was measured via flow cytometry in B-ALL cell lines (n = 3 independent experiments). (F) Supervised analysis of publicly available gene expression data from children with CRLF2-R/JAK2-mutant Ph-like ALL (n = 12), ETV6-RUNX1 ALL (n = 3), hyperdiploid ALL with trisomy 4 and 10 (n = 4), KMT2A-R ALL (n = 21), and TCF3-PBX1 ALL (n = 23) from the National Cancer Institute TARGET database. Data are represented as individual values with mean ± SEM bars.
To validate these data, we treated Ph-like ALL cell lines and PDX model cells in vitro with ruxolitinib and again observed overexpression of BCL6, BLNK, RAG1, and RAG2 mRNA (Supplemental Figure 3, B and C). To evaluate functionally whether Ph-like ALL cells shift their genetic identity from JAK/STAT dependence to pre-BCR dependence, we next performed gene set enrichment analysis (GSEA). First, we generated a B cell differentiation gene signature by taking the 500 most upregulated genes in mouse bone marrow cells after IL-7 withdrawal (Supplemental Table 8) and applied these to Ph-like ALL cells treated with ruxolitinib or Ph+ ALL cells treated with imatinib (Figure 4, B and C). Interestingly, ruxolitinib-exposed Ph-like ALL cells demonstrated a similar, albeit less profound, enrichment of B cell differentiation genes when compared with imatinib-exposed Ph+ ALL cell lines.
The defining feature of B cell maturation is induction of RAG activity, which initiates V(D)J recombination. To investigate whether ruxolitinib modulates RAG1/2 protein levels and enzymatic activity, we retrovirally transduced Ph-like ALL cells with a RAG enzyme reporter construct (39) and used dasatinib-treated Ph+ ALL cells as a positive control. As expected, RAG enzymatic activity significantly increased over time in Ph+ ALL cells following TKI treatment. However, incubation of Ph-like ALL cells with ruxolitinib only moderately increased RAG activity (Figure 4D). Further analysis of additional B-ALL subtypes showed that endogenous RAG enzyme activity appeared highest in Ph-like, ETV6-RUNX1, and TCF3-PBX1 ALL cell lines (Figure 4E). Elevated RAG enzyme activity has been previously described in ALL with ETV6-RUNX1 fusions, which may contribute to leukemogenesis (40, 41). To validate potential for partial ruxolitinib-induced cell differentiation, we assessed ruxolitinib-treated and nontreated MHH-CALL-4 cells for CD43 surface expression (a marker expressed during early B cell development), but did not detect decreased expression following JAKi exposure (Supplemental Figure 3E). Taken together, these data do not support a robust model of JAKi-induced differentiation in Ph-like ALL cells, although exposure to ruxolitinib or CHZ868 clearly shifts their gene expression signature toward a pre-BCR+ cellular identity.
Pre-BCR signaling molecules are highly expressed in Ph-like ALL.
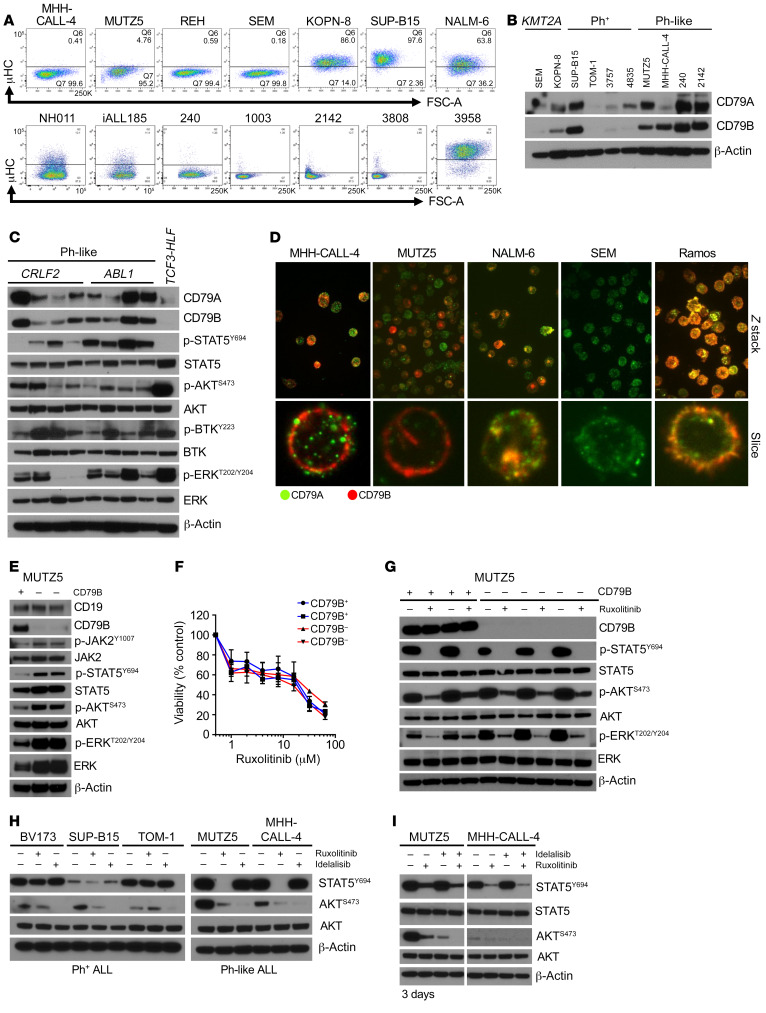
Given the unexpected finding of baseline-elevated RAG enzyme activity in Ph-like ALL cells, we then considered whether these cells at baseline may have activated pre-BCR signaling. First, we assessed whether Ph-like ALL cells have constitutive expression of pre-BCR signaling components via analysis of publicly available National Cancer Institute TARGET (Therapeutically Applicable Research to Generate Effective Treatments; http://ocg.cancer.gov/programs/target/projects/acute-lymphoblastic-leukemia) gene expression data of high-risk childhood B-ALL specimens (42). We focused on CRLF2-R Ph-like ALL cases with concomitant activating JAK2 point mutations and compared with data from other ALL subtypes with ETV6-RUNX1 fusion from t(12;21), hyperdiploidy with trisomies of chromosomes 4 and 10, KMT2A rearrangements at chromosome 11q23, or TCF3-PBX1 fusion from t(1;19) (35). Interestingly, Ph-like ALL samples showed elevated RNA expression of multiple pre-BCR signaling molecules comparable to levels in TCF3-PBX1 (pre-BCR+) ALL, including CD79A and CD79B, IGLL1, VPREB, BLNK, ZAP70, RAG1, and RAG2 (Figure 4F). High RAG2 RNA levels were validated in primary adult Ph-like ALL specimens and PDX model cells and compared with levels in Ph+ ALL cells (Supplemental Figure 3F). We further assessed the extent to which Ph-like ALL cell lines and patient samples express pre-BCR protein components. We surprisingly detected no expression of surface or intracellular μ-heavy chain (μHC) via flow cytometry analysis, suggesting that Ph-like ALL is arrested at a pro-B instead of pre-B stage (Figure 5A) that normally expresses very low levels of CD79A and CD79B (43). Ph-like and Ph+ ALL cells also had very high levels of CD79A and CD79B proteins, which were comparable to levels detected in pre-BCR+ KOPN-8 and SUP-B15 ALL cell lines (Figure 5B). Prior studies have reported CD79A- and CD79B-induced phosphorylation of downstream pre-BCR molecules, such as BTK and ERK, in the absence of BCR stimulation or the BCR itself (44–47). We thus assessed additional downstream pre-BCR signaling molecules and indeed identified that AKT, BTK, and ERK were highly phosphorylated in Ph-like ALL PDX cells (Figure 5C). To test whether CD79A and CD79B heterodimers are expressed on the cell surface of Ph-like ALL cells, we performed immunofluorescence microscopy experiments using the pre-BCR+ ALL NALM-6 and Burkitt lymphoma Ramos cell lines as positive controls (Figure 5D and Supplemental Figure 4). Interestingly, we visualized mixed populations of CD79A-only or CD79B-only expression in some Ph-like ALL cells, while other cells had coexpression of both molecules. CD79A appeared to be clustered together throughout the cell, while CD79B was more diffusely expressed near the cell surface. Conversely, the pre-BCR+ B-ALL cell line NALM-6 and the BCR+ lymphoma cell line Ramos showed clear colocalization of CD79A and CD79B as anticipated. Our data thus demonstrate that critical components of pre-BCR signaling are present in Ph-like ALL cells despite the absence of μHC and suggest potential sufficiency to activate downstream “BCR-like” signaling.
Figure 5. Expression of BCR signaling molecules in Ph-like ALL cells.
(A) Flow cytometry analysis of immunoglobulin μ-heavy chain (μHC) expression in B-ALL cell lines (top) and Ph-like patient samples (bottom). The NALM-6 cell line and a primary pre-BCR+ B-ALL patient specimen (3958) were used as positive controls. (B) Western blot analysis of CD79A and CD79B expression in B-ALL cell lines and patient samples. (C) Western blot analysis of BCR signaling molecules in 8 Ph-like ALL PDX specimens and 1 TCF3-HLF ALL PDX case. (D) CD79A (green) and CD79B (red) expression was assessed via immunofluorescence microscopy in Ph-like ALL cell lines, pre-BCR+/BCR+ ALL positive controls, and a KMT2A-R B-ALL negative control. Original magnifications: ×63 for upper panel, ×158 for lower panel. (E) Western blot analysis of indicated proteins in control (+) and CD79B-deleted (–) MUTZ5 cells. (F) Sensitivity analysis of CD79B+ and CD79B– MUTZ5 cells to increasing concentrations of ruxolitinib (n = 3 independent experiments per sample). (G) Western blot analysis of CD79B+ and CD79B– MUTZ5 cells 3 weeks after electroporation treated with and without 1 μM ruxolitinib for 2 hours. (H) Ph+ ALL and Ph-like ALL cells were treated with 1 μM ruxolitinib for 1 hour or 1 μM idelalisib for 1 hour, and p-STAT5 and p-AKT levels were measured. (I) MUTZ5 and MHH-CALL-4 cells were treated with 1 μM ruxolitinib or 1 μM idelalisib for 72 hours alone or in combination, and protein expression of p-STAT5Y694 and p-AKTS473 was measured using the total proteins as controls. Data are represented as individual values with mean ± SEM bars.
We next focused on defining the extent to which CD79B-dependent signaling is involved in Ph-like ALL. Surprisingly, CD79B deletion in MUTZ5 cells via CRISPR guide RNAs resulted in hyperactivation of ERK signaling, but did not alter PI3K/AKT pathway phosphoproteins (Figure 5E). To test the effect of continuous CD79B deletion, we expanded CD79B– cells for 3 weeks in in vitro culture to study long-term effects of CD79B deletion. Interestingly, ruxolitinib treatment of CD79B-deleted versus nondeleted MUTZ5 cells did not induce differential signaling effects, and increased sensitivity of CD79B-deleted cells to ruxolitinib was not observed (Figure 5F). However, steady-state p-ERK levels continued to be elevated in CD79B-deleted MUTZ5 cells over time (Figure 5G). Taken together, these results show that CD79B is involved in ERK signaling regulation.
Prior studies have shown that murine Tp53–/– Lnk–/– B-ALL cells are sensitive to dual inhibition of JAK2 and PI3Kδ and may thus model similar kinase-dependent signaling as in human Ph-like ALL (48). As the PI3K pathway is another known critical mediator of BCR signaling (49, 50), we further tested the extent to which direct PI3K inhibition silences AKT signaling in Ph-like ALL. We treated Ph-like and Ph+ ALL cell lines with ruxolitinib or the isoform-selective PI3Kδi idelalisib and analyzed protein lysates by Western blotting. As anticipated, short-term ruxolitinib exposure inhibited p-STAT5 in Ph-like but not in Ph+ ALL cells and partially inhibited p-AKT levels as previously observed (11). Only idelalisib completely inhibited AKT signaling (Figure 5H). Interestingly, longer-term cotreatment of cells with JAKi and PI3Kδi for 72 hours was required to prevent signaling reactivation observed after single-agent ruxolitinib exposure (Figure 5I). These data further suggest that JAK2 and BCR-like activation of AKT occur redundantly and cooperatively in Ph-like ALL.
JAK and PI3K signaling pathways contribute to cell growth and survival in Ph-like ALL.
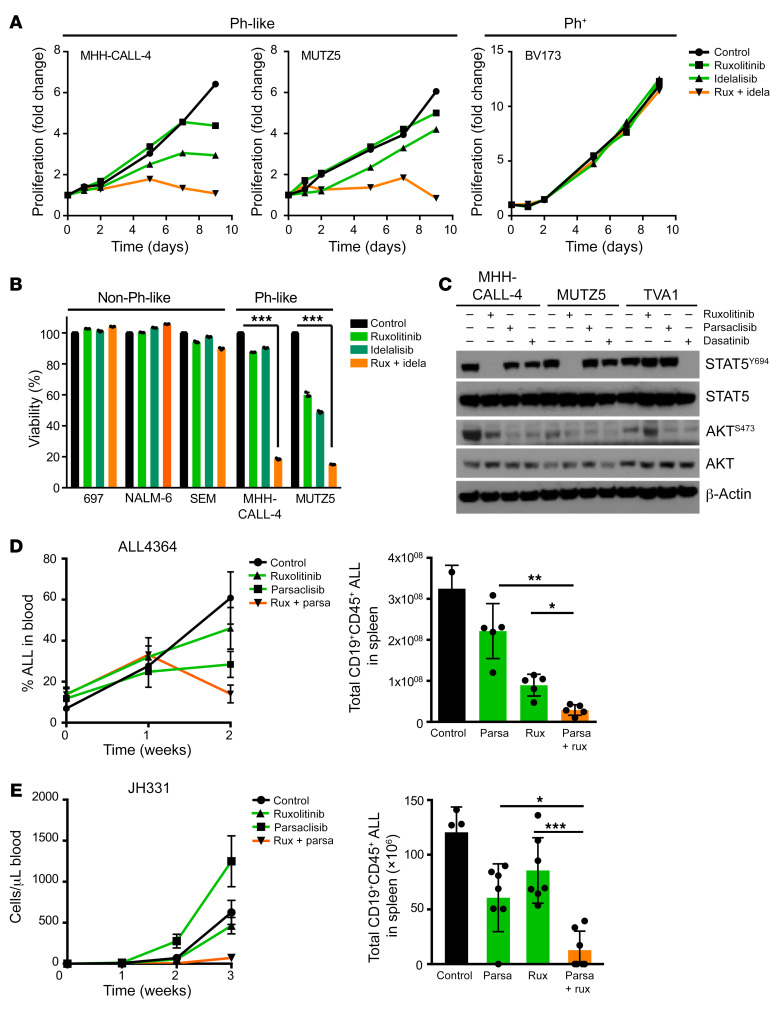
Earlier studies by our group and others have descriptively shown that dual inhibition of JAK2 and PI3K or mTOR inhibits leukemia proliferation in vivo in Ph-like ALL PDX models more effectively than single-agent TKIs, but is still inadequate to induce complete cure (15, 16). In the current studies, we first confirmed the effects of ruxolitinib and idelalisib in CRLF2-R/JAK2-mutant Ph-like (MHH-CALL-4, MUTZ5) and Ph+ (BV173) ALL cell lines. While single-TKI exposure for 9 days induced minimal effects on cell proliferation, combined JAK1/2 and PI3Kδ inhibition decreased cell proliferation to a greater extent in Ph-like ALL cells (Figure 6A). In additional viability assays, we observed reduced proliferation of Ph-like ALL cells treated with ruxolitinib and idelalisib for 3 or 7 days with minimal effects detected in Ph+ ALL cells (Figure 6B and Supplemental Figure 5A). Incidentally, we further measured B cell gene expression levels to determine whether dual signaling pathway inhibition could induce Ph-like cell differentiation as a possible mechanism of drug resistance (Supplemental Figure 5B). Interestingly, higher expression levels of RAG1, RAG2, BCL6, SYK, and BLNK were detected in cells from one dual-TKI-treated PDX model (ALL4364). However, RAG enzymatic activity was not increased in Ph-like cell lines after dual treatment (Supplemental Figure 5C), indicating a stronger transcriptional response potentially without functional consequences.
Figure 6. Combined JAK and PI3K inhibition induces cell death and halts proliferation.
(A) Indicated B-ALL cell lines were treated with DMSO, 1 μM JAKi ruxolitinib (rux), 1 μM PI3Kδi idelalisib (idela), or a combination of ruxolitinib and idelalisib (1 μM each) for 9 days (n = 1 for each cell line). Drugs and cell culture media were replaced every 3 days. Cell density was counted at indicated time points. (B) Indicated cell lines were subjected to cell viability analysis via flow cytometry after exposure to DMSO, 1 μM ruxolitinib, 1 μM idelalisib, or a combination of ruxolitinib and idelalisib (1 μM each) for 7 days (n = 3 independent experiments). (C) Western blot analysis of p-STAT5Y694 and p-AKTS473 levels in CRLF2-R (MUTZ5, MHH-CALL-4) or ABL1-R (TVA1) Ph-like ALL cells treated with 1 μM ruxolitinib, 1 μM INCB05465, and/or 100 nM dasatinib. (D and E) Two CRLF2-R Ph-like ALL PDX models (ALL4364 [n = 5 mice] and JH331 [n = 7 mice]) were randomized to treatment with vehicle (control), 1 mg/kg PI3Kδi parsaclisib (parsa, formerly INCB050465) orally twice daily, 2 g/kg ruxolitinib rodent chow continuously provided, or both inhibitors for the specified times with flow cytometric quantification of human ALL in murine peripheral blood (line graphs) and in end-study spleens (bar graphs). Data are represented as individual values with mean ± SEM bars. *P < 0.05; **P < 0.01; ***P < 0.001 by unpaired t test (B) or ANOVA with Dunnett’s post-test for multiple comparisons (D and E).
We then validated the necessity of PI3K pathway signaling in Ph-like ALL using our mCRLF2-JAK2 cells (shown in Figure 3) treated with increasing concentrations of ruxolitinib or idelalisib. We observed moderately diminished cell viability with TKI monotherapies, while dual JAKi and PI3Kδi exposure markedly decreased viability (Supplemental Figure 2B). We further confirmed that the selective PI3Kδi parsaclisib (formerly INCB050465 [ref. 51]) similarly inhibited AKT signaling in vitro in two CRLF2-R (MUTZ5, MHH-CALL-4) and one ETV6-ABL1 (TVA1 [ref. 52]) Ph-like ALL cell lines, using ruxolitinib and dasatinib (SRC/ABL/BTKi) as positive inhibitor controls. As expected, ruxolitinib decreased p-STAT5 in MHH-CALL-4 and MUTZ5 cells, but not in TVA1 cells, while dasatinib decreased p-STAT5 in TVA1 cells as expected. We further observed significant dephosphorylation of AKT in both CRLF2-R and ABL1-R Ph-like ALL cells treated with parsaclisib (Figure 6C).
Finally, we assessed the ability of dual JAK and PI3Kδ inhibition to inhibit leukemia proliferation in vivo in CRLF2-R/JAK2-mutant Ph-like ALL PDX models. As predicted (15), combined ruxolitinib and parsaclisib had more potent antileukemia activity than TKI monotherapies in PDX models ALL4364 and JH331 (P < 0.05 for all), but did not fully eradicate leukemia burden in treated animals (Figure 6, D and E). Western blotting analysis of splenic lysates (with >90% human ALL involvement) from control and TKI-treated mice was also performed to assess in vivo signaling inhibition. We observed consistently decreased p-STAT5 with ruxolitinib treatment and minimal p-S6 inhibition in parsaclisib-treated animals. Dual TKI treatment more strongly inhibited p-S6, but did not affect p-AKT (Supplemental Figure 5D). Overall, these data show enhanced but incomplete antileukemia effects of combined JAK and PI3Kδ inhibition and suggest that additional therapeutic targeting is necessary to eliminate Ph-like ALL fully.
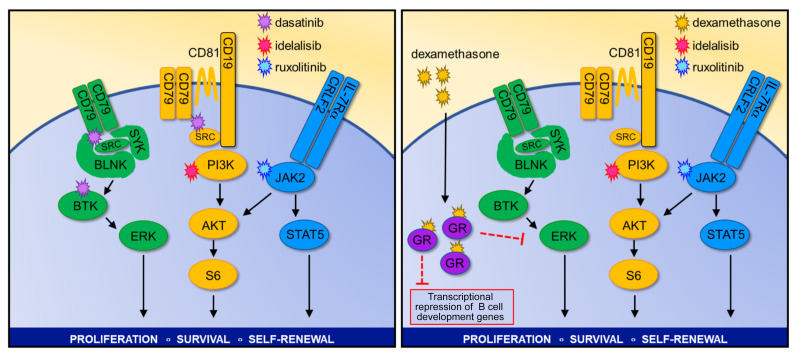
Complete inhibition of BCR-like signaling is additionally required to eradicate CRLF2-R Ph-like ALL.
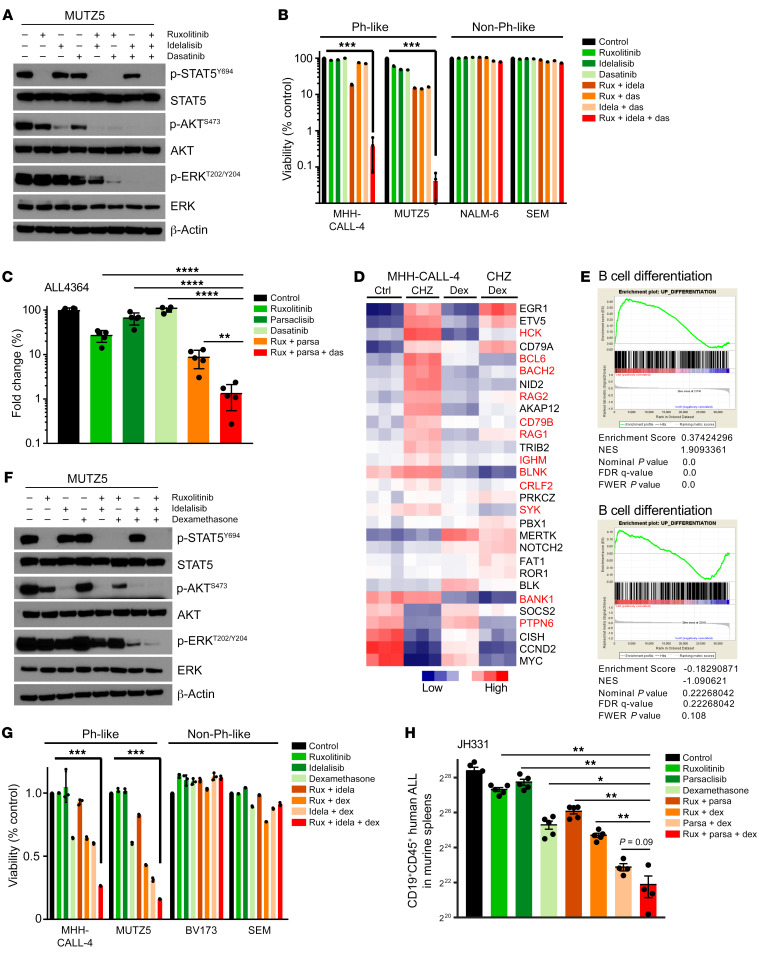
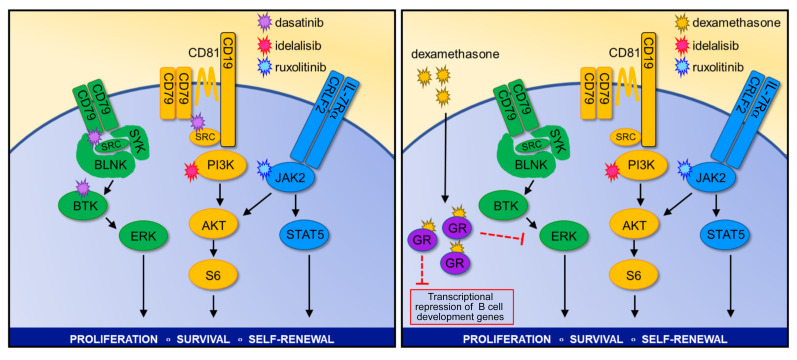
We next hypothesized that SFK-mediated signaling associated with BCR-like signaling could rescue Ph-like ALL cells treated with JAKi and PI3Ki based on 3 observations: (a) ruxolitinib treatment of CRLF2-R Ph-like ALL cells did not decrease p-SRC levels (Supplemental Figure 3D), (b) dasatinib treatment decreased p-AKT (Figure 6C), and (c) gene expression and protein analysis showed expression and activation of multiple pre-BCR signaling molecules (Figure 4F and Figure 5C). To test this hypothesis, we treated MUTZ5 cells in vitro with combinations of ruxolitinib, idelalisib, and/or dasatinib to ascertain whether triple kinase inhibition could more profoundly reduce hyperactive signaling and induce greater cell death. Strikingly, combined JAK, PI3K, and SRC/ABL inhibition silenced all 3 signaling pathways (Figure 7A) and markedly and preferentially inhibited viability of Ph-like ALL cell lines (Figure 7B). Strikingly, triple TKI therapy with ruxolitinib, idelalisib, and dasatinib effectively eliminated in vivo leukemia proliferation in a CRLF2-R Ph-like ALL PDX model (Figure 7C). Importantly, treated animals appeared to tolerate multi-kinase inhibition for several weeks of treatment without significant weight loss or other signs of toxicity. Finally, as proof of concept, we assessed the therapeutic potential of targeting the ERK signaling pathway directly with selumetinib in MHH-CALL-4 cells (in which we discovered a concomitant KRAS mutation; Supplemental Table 5) and MUTZ5 cells in combination with the JAKi ruxolitinib and the PI3Kδi idelalisib. This triple TKI combination also markedly decreased proliferation (Supplemental Figure 6A) and induced cell death (Supplemental Figure 6B), suggesting an effective alternative potential multi-TKI approach for CRLF2-R Ph-like ALL. Overall, these results support our hypothesis that signaling through an oncogenic CRLF2/JAK2 axis and signaling through a nononcogenic BCR-like axis work together to maintain Ph-like ALL cell viability and proliferation.
Figure 7. Triple targeting of kinase signaling is required to induce Ph-like ALL cell death.
(A) MUTZ5 cells were treated with 1 μM ruxolitinib (rux), 1 μM idelalisib (idela), and/or 1 μM SRC/ABLi dasatinib (das) in the indicated combinations for 1.5 hours before protein analysis. (B) Flow cytometric cell viability analysis of B-ALL cell lines treated with 1 μM ruxolitinib, 1 μM idelalisib, and/or 1 μM dasatinib for 7 days (n = 3 independent experiments). (C) A CRLF2-R Ph-like ALL PDX model (ALL4364) was treated with vehicle (control), 2 g/kg ruxolitinib chow continuously provided, 1 mg/kg parsaclisib orally twice daily, 10 mg/kg dasatinib orally twice daily, or multiple inhibitors for 14 days, with flow cytometric (FC) quantification of human ALL cells in end-study murine spleens (n = 5 mice per group). (D) Supervised meta-analysis of gene expression data of MHH-CALL-4 cells treated with 500 nM JAK2i CHZ868, 120 nM dexamethasone, or both drugs for 12 hours (12). (E) Top: GSEA plots for B cell differentiation gene sets in CHZ868- versus vehicle-treated MHH-CALL-4 cells. Bottom: Cotreatment of CHZ868 with dexamethasone. (F) MUTZ5 cells were treated with 1 μM ruxolitinib, 1 μM idelalisib, and/or 10 nM dexamethasone for 1.5 hours before Western blot analysis. (G) FC cell viability analysis of B-ALL cell lines treated with 1 μM ruxolitinib, 1 μM idelalisib, and/or 10 nM dexamethasone for 72 hours (n = 3 independent experiments) in the indicated combinations. (H) A CRLF2-R Ph-like ALL PDX model (JH331) was treated with vehicle, 2 g/kg ruxolitinib chow, 1 mg/kg parsaclisib (parsa) orally twice daily, 1 mg/kg dexamethasone (dex) i.p. once daily, or multiple drugs for 14 days, with FC quantification of human ALL cells in murine end-study spleens. Data are represented as individual values with mean ± SEM bars. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by unpaired t test (B and G) or ANOVA with Dunnett’s post-test for multiple comparisons (C and H).
As triple TKI therapy may not be clinically tolerable long-term in patients with Ph-like ALL, we also investigated the potential efficacy of alternative combinatorial treatment approaches. A recent study reported the ability of dexamethasone, a commonly used ALL chemotherapy drug, to repress critical molecules involved in BCR signaling and lymphoid development (4). We performed a supervised meta-analysis of MHH-CALL-4 cells treated with the JAK2i CHZ868, dexamethasone, or both drugs. Interestingly, CHZ868 increased expression of multiple B cell–associated genes versus vehicle-treated control cells, while dexamethasone monotherapy had minimal effects. Interestingly, exposure to both CHZ868 and dexamethasone appeared to prevent the genetic shift of Ph-like ALL cells toward a pre-BCR+ identity (Figure 7D, affected genes highlighted in red). To validate these findings, we performed gene set enrichment analysis (GSEA) and confirmed that indeed dexamethasone blocked the enrichment of genes involved in B cell differentiation in MHH-CALL-4 cells (Figure 7E) and in CRLF2-R ALL PDX cells (Supplemental Figure 6C). We thus treated MUTZ5 and MHH-CALL-4 cells in vitro with dexamethasone, ruxolitinib, or both to assess the extent to which steroid chemotherapy could reverse JAKi-mediated upregulation of B cell differentiation genes. We observed marked p-STAT5 inhibition with dexamethasone and no appreciable effects on p-AKT or BCL6 via Western blotting, while single-agent ruxolitinib exposure increased BCL6 expression. Similarly to the GSEA results, JAKi-induced BCL6 upregulation at a protein level was blocked with dexamethasone cotreatment (Supplemental Figure 6D). Expanded in vitro study of MUTZ5 cells treated with ruxolitinib, dexamethasone, and/or idelalisib demonstrated potent triple-pathway (JAK/STAT, PI3K, and BCR-like) signaling inhibition (Figure 7F) and induced greater cell death in Ph-like ALL versus non–Ph-like ALL cell lines (Figure 7G). Evaluation of the in vivo therapeutic potential of dual JAK and PI3K inhibition with dexamethasone in 2 CRLF2-R Ph-like ALL PDX models demonstrated near-curative effects similar to those of triple-TKI treatment (Figure 7H and Supplemental Figure 6E). Combination therapy with 2 TKIs and steroids again appeared tolerable in these preclinical models and further suggests the potential efficacy of this more clinically pragmatic approach.
In summary, data from our studies suggest that the most common CRLF2-R JAK-mutant subset of Ph-like ALL is driven by a highly integrated 3-pronged signaling network, which appears highly adaptable following single-node inhibition and suggests a lack of oncogene addiction that may ultimately lead to treatment failure of current single-TKI-based clinical strategies.
Discussion
Ph-like ALL is well established as a common high-risk subtype of B-ALL (53) associated with significant chemotherapy resistance, relapse, and poor overall survival across the age spectrum. Ph-like ALL is driven by a large variety of genetic mutations that lead to activated kinase signaling, but it is not yet known whether or not the addition of JAK inhibitors or ABL inhibitors to chemotherapy under current clinical evaluation (NCT02420717, NCT02723994, NCT03117751, NCT03571321, NCT01406756; ClinicalTrials.gov) will improve outcomes for patients with Ph-like ALL. In this preclinical study, we sought to define more precisely the signaling dysregulation that occurs in Ph-like ALL cells with a goal of potentially more optimal therapeutic exploitation.
We and others have previously described JAK/STAT and PI3K pathways as constitutively activated in CRLF2-R Ph-like ALL cells (4, 11, 20, 21). In our current studies, we surprisingly observed elevated RNA expression and protein levels of components of the pre-BCR signaling complex at similar levels to those of pre-BCR+ ALL despite absence of the μHC in Ph-like ALL. We posit that activation of this “BCR-like” signaling via CD79A and CD79B and SFKs is necessary and sufficient to constitute the third kinase pathway signaling requirement of Ph-like leukemogenesis and maintenance. Previous studies have shown that transforming oncogenes can directly activate multiple signaling molecules and mimic cytokine receptor and/or pre-BCR signaling to promote cell proliferation and survival (54). One of the most studied fusion oncogenes is BCR-ABL1, found in Ph+ ALL and chronic myeloid leukemia, which induces cytokine receptor signaling via STAT5 and PI3K and pre-BCR signaling via SYK, BTK, and ERK (54–59). Our studies show that these same signaling pathways are constitutively activated in CRLF2-R Ph-like ALL, albeit via different mechanisms. Although TKI treatment (imatinib, dasatinib, other ABL inhibitors) of Ph+ ALL cells results in dephosphorylation of all 3 signaling pathways and robust leukemic cell death, inhibition of JAK2 or genetic deletion of CRLF2 in Ph-like ALL primarily decreases STAT5 signaling (often temporarily) and only moderately inhibits leukemic cell proliferation. While Ph+ ALL is a classic example of oncogene addiction, our studies show that the similarly kinase-activated CRLF2-R Ph-like ALL is instead driven by a network of signaling molecules, many of which are activated independently of the major oncogenic CRLF2 rearrangement and appear highly adaptable to targeted kinase inhibition.
To investigate further potential mechanisms of AKT, BTK, and ERK signaling activation, we focused on the initial observation that CD79B RNA and protein levels in Ph-like ALL were highly expressed and at similar levels to those of pre-BCR+ ALL cells. However, analysis of the pre-BCR itself showed that Ph-like ALL cells are negative for μHC expression. Activation of downstream pre-BCR signaling by homodimerization of CD79B and coexpression of CD19 in the absence of μHC has been described (47), but is not typical of B cell signaling. In our studies, pharmacologic inhibition or genetic disruption of one of the 3 JAK/STAT, PI3K, or BCR-like components in Ph-like ALL cells led to rapid adaptation of signaling networks, establishing the dynamic ability of these cells to overcome targeted monotherapy and highlighting appreciable potential for rapid single-drug resistance. We thus hypothesized that BCR-like signaling in Ph-like ALL is activated by spontaneous association of cell surface proteins involved in normal BCR signaling. Previous studies in pre-BCR+ B-ALL suggest that CD79A and CD79B can cross-link and activate signaling in the absence of external factors (60), potentially via short-lived, homotypic pre-BCR interactions that provide survival signals (61). Demonstration of the exact complex and its mechanism of interaction with downstream signaling was beyond the scope of this study. Interestingly, however, we also detected decreased PTPN6 levels following prolonged JAK inhibition with ruxolitinib in some CRLF2-R Ph-like ALL cells (Supplemental Figure 3D), which suggests that a critical regulatory event in Ph-like ALL involves disruption of the phosphatase network that typically keeps JAK2 and BCR-like signaling in check. It is plausible that PTPN6 could be involved in induction of drug resistance in Ph-like ALL, although further studies are needed.
Our unexpected observation of ERK hyperactivation after CRISPR-mediated deletion of CD79B and insufficient inactivation of PI3K and ERK signaling after CRLF2 deletion may have broader therapeutic implications for patients with Ph-like ALL. Several CD19-targeting or CD22-targeting antibody-based or cellular immunotherapies have been approved by the US Food and Drug Administration or are under current clinical trial evaluation in children and adults with refractory B-ALL, and antigen-loss relapse has emerged as a major mechanism of immunotherapeutic resistance (62), including in patients with relapsed Ph-like ALL. A soon-to-open phase I clinical trial will assess the safety and preliminary efficacy of TSLPR-redirected chimeric antigen receptor T cells (63) in patients with relapsed/refractory CRLF2+ leukemias, and it is plausible that similar CRLF2– ALL relapses (analogous to data shown in Figure 2) will be observed over time in treated patients. Our preclinical genetic studies demonstrating intracellular kinase signaling activation in CRLF2-deleted Ph-like ALL cells recapitulate and validate effects of pharmacologic inhibition with ruxolitinib treatment of Ph-like ALL cells, demonstrating that inhibition of JAK2, PI3K, and SFK/BCR-like signaling is required to successfully kill CRLF2-R Ph-like ALL. As aforementioned, long-term triple kinase inhibition will likely not be clinically tolerable in patients, and we thus focused in subsequent experiments on identifying an alternative TKI-based strategy in combination with chemotherapy that could confer similar potent antileukemia activity. A previous study by Pufall and colleagues showed that dexamethasone represses genes involved in B cell development (e.g., ITGA4, IL7R, BCL6) and BCR signaling molecules (e.g., CD79B, BTK, FYN, PI3KCD) and that PI3K inhibition can resensitize Ph-like ALL cells to dexamethasone by interrupting a double-negative feedback loop (4). Our new data further extend this observation, as we demonstrate that dexamethasone strongly represses ruxolitinib-induced changes in B cell development and BCR signaling genes. Strikingly, dexamethasone together with ruxolitinib and idelalisib fully inhibited STAT5, PI3K, and ERK signaling and eliminated Ph-like ALL cells in vitro and in vivo in comparison with drug monotherapies or pairings, validating the clinical potential of dual TKIs in combination with routine ALL chemotherapy. It is thus plausible that such combinatorial approaches could effectively decrease relapse risk, increase cure rates, and perhaps even be used in combination with CRLF2-targeting immunotherapies to decrease potential immunotherapeutic resistance.
In summary, we describe a robust signaling network in CRLF2-R Ph-like ALL that combines oncogene-driven activation of cytokine receptor signaling via the TSLPR and JAK/STAT pathway with BCR-like signaling activation of PI3K and MAPK pathways. In contrast to Ph+ ALL, this Ph-like network appears to be highly redundant, as modulation of any single component minimally inhibits cell survival and is rapidly compensated by upregulation of other required “legs” of the system. We maintain that the apparent necessity of this highly robust signaling network and its swift “rewiring” observed with TKI monotherapy exposure could predict a priori potential failure of current clinical strategies that are assessing the efficacy of single-agent ruxolitinib or dasatinib with chemotherapy in patients with Ph-like ALL. Instead, our in vitro and in vivo results suggest that triple TKI therapy targeting JAK/STAT, PI3K, and SFK/BCR-like signaling will be required for complete eradication of Ph-like ALL, which may be alternatively clinically achievable via combination of 2 signaling inhibitors with the commonly used anti-ALL chemotherapy dexamethasone (Figure 8). These results demonstrate a novel mechanistic approach to kinase inhibition for Ph-like ALL that merits further evaluation and may inform development of the next generation of precision medicine trials for patients with Ph-like ALL with a goal of reducing relapse risk and improving long-term survival.
Figure 8. “Optimally lethal” combination therapeutic strategies for Ph-like ALL.
Methods
Primary human ALL specimens and cell lines.
Diagnostic bone marrow specimens from children and adults with Ph-like ALL were obtained via informed consent on Institutional Review Board–approved research protocols of the Children’s Hospital of Philadelphia (CHOP) and University of Pennsylvania (UPenn) in accordance with the Declaration of Helsinki (Supplemental Tables 1 and 2) as previously described (4, 15). Human Ph-like and non–Ph-like ALL cell lines (Supplemental Table 2) were purchased from the DSMZ biorepository (Braunschweig, Germany), validated by short tandem repeat analysis, and confirmed as Mycoplasma-free every 6 months. The Ph-like TVA1 cell line was initially created as a childhood Ph–like ALL patient-derived xenograft (PDX) model in immunocompromised mice and subsequently immortalized via in vitro culture (52). Archer fusionplex testing and next-generation sequencing of some cell lines and PDX models were performed at the CHOP Division of Genomic Diagnostics clinical core facility as previously described (64). The Ph-like kinase-activated gene expression signature was confirmed by a validated clinical low-density microarray analysis in relevant ALL cell lines and PDX models as previously described (6, 20, 65).
Human ALL cells harvested from murine spleens of some PDX models were cultured on OP9 stroma in Minimum Essential Medium (MEMα; Life Technologies) with GlutaMAX containing 20% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 1 mM sodium pyruvate. Cell lines were cultured in RPMI medium (Life Technologies) with GlutaMAX containing 10% or 20% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin (hereafter referred to as “B cell medium”) at 37°C in a humidified incubator with 5% CO2 and maintained in culture for fewer than 3 months to minimize infectious contamination and genetic drift.
Murine Ph-like ALL cell line models.
Bone marrow cells were harvested from 6-week-old C57BL/6J mice via flushing of femora and tibiae with a large-gauge needle and PBS. Bone marrow cells were filtered using a 70-μm cell strainer, and erythrocytes were depleted with ACK lysis buffer (Gibco). After additional PBS washing, bone marrow cells were cultured in IMDM (Gibco) with GlutaMAX supplemented with 20% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, 50 μmol/L β-mercaptoethanol (Gibco), and 10 ng/mL recombinant mouse IL-7 (PeproTech). To generate the murine Ph-like ALL models, we then retrovirally transduced murine bone marrow cells with CRLF2, ETV6-PDGFRB, or PAX5-JAK2 constructs 3 days later. CRLF2+ B-ALL cells were selected using blasticidin and additionally transduced with a JAK2R683G construct to generate a murine CRLF2-JAKR683G Ph-like ALL (mCRLF2-JAK2) cell line model. After transformation, mCRLF2-JAK2 cells were cultured in vitro without IL-7. Cell lines were cultured no longer than 3 months to avoid accumulation of additional mutations and infectious contamination and confirmed to be Mycoplasma-free.
TKIs and chemotherapy.
The following TKIs were used for in vitro and/or in vivo studies: JAK1/JAK2i ruxolitinib-infused rodent chow 2 g/kg (a gift from Matthew Stubbs and Bruce Ruggeri, Incyte Corp.), PI3Kδi parsaclisib (formerly INCB050465; Incyte), PI3Kδi idelalisib (SelleckChem), SRC/ABLi dasatinib (LC Labs), MEKi selumetinib (LC Labs), and ruxolitinib (SelleckChem) for in vitro use. Clinical-grade dexamethasone (Mylan) was purchased from the CHOP investigational pharmacy (Supplemental Table 3).
PDX modeling and in vivo preclinical drug trials.
All PDX model establishment and experimental animal studies were conducted on Institutional Animal Care and Use Committee–approved research protocols at CHOP and UPenn in accordance with NIH and American Veterinary Association Guidelines for the Euthanasia of Animals. Briefly, Ph-like and non–Ph-like ALL PDX models in 6- to 8-week-old male and female NOD/SCID/IL-2rγnull (NSG) mice were created via i.v. injection of primary human ALL cells as previously described with serial transplantation of harvested human leukemia cells from murine spleens into secondary or tertiary recipient NSG mice for experimental studies described herein (4, 14, 15, 66). After flow cytometric confirmation of more than 1% CD45+CD19+ (and CD10+ and TSLPR+ for CRLF2-R Ph-like) human ALL in peripheral blood of well-engrafted mice, animals were randomized to treatment with indicated combinations of (a) oral (p.o.) or intraperitoneal (i.p.) gavage vehicle and control chow, (b) ruxolitinib 2 g/kg rodent chow p.o. continuously provided, (c) parsaclisib 1 mg/kg p.o. twice daily, (d) dexamethasone 1 mg/kg p.o. once daily, and/or (e) dasatinib 10 mg/kg i.p. once daily for up to 14–21 days depending on rate of leukemia progression in control animals requiring study termination at a predetermined endpoint (4, 15, 19).
Flow cytometry analysis.
Human and mouse leukemia samples were stained with cell surface antibodies (Supplemental Table 4) according to the manufacturer’s instructions using Fc block and respective isotype controls. Annexin V, propidium iodide, 7-aminoactinomycin D, or DAPI (BD Biosciences) was used for apoptosis analyses. Flow cytometry was performed using FACSVerse or LSRII flow cytometers (BD Biosciences) with data analysis in Cytobank or FlowJo (Tree Star) and graphical display and statistical analysis in Prism (GraphPad) (see Statistics below).
Western blotting.
All cells were lysed in CelLytic buffer (MilliporeSigma) supplemented with 1% HALT (protease inhibitor and phosphatase inhibitor cocktail; Thermo Fisher Scientific). Protein samples were loaded on NuPAGE 4%–12% Bis-Tris gradient gels (Invitrogen) and transferred on PVDF membranes (Millipore). For detection of mouse and human proteins, primary antibodies were used in combination with anti-rabbit or anti-mouse HRP-linked secondary antibodies (Cell Signaling Technology) and Amersham ECL Western blotting detection reagent (GE Life Sciences) (Supplemental Table 4).
Phosphoarray.
MHH-CALL-4 cells (human Ph-like ALL cell line with IGH-CRLF2 translocation, JAK2I682F, and KRASL23R; Supplemental Table 5) were treated in vitro with human thymic stromal lymphopoietin (TSLP) 25 nM for 30 minutes; ruxolitinib 1 μM for 1, 4, 25, or 48 hours; or DMSO (control), and subjected to protein isolation as described above. Phosphoprotein expression was detected using a human phospho-kinase array kit according to the manufacturer’s protocol (R&D Systems).
Immunoprecipitation studies.
ALL cells were lysed in cell lysis buffer (Cell Signaling Technology) and PMSF and precleared by incubation with protein A-agarose (Invitrogen) for 10 minutes. Cells were then incubated with human CD19 antibody (Cell Signaling Technology) (Supplemental Table 4) and protein A-agarose at 4°C overnight. Immunoprecipitates were washed 5 times the next day in cell lysis buffer before loading onto NuPAGE 4%–12% Bis-Tris gradient gels and transferring to PVDF membranes. Development and detection following immunoprecipitation were performed by Western blot as above.
Cell proliferation and viability assays.
One hundred thousand human ALL cells were seeded in a volume of 100 μL B cell medium per well (as described above) on Optilux 96-well plates (BD Biosciences). The JAKi ruxolitinib, the PI3Kδi idelalisib, or the PI3Kδi parsaclisib was diluted in medium and added at the indicated concentration in a total culture volume of 150 μL. After culturing for 72 hours, cell proliferation and viability were measured by XTT (Cell Signaling Technology) according to the manufacturer’s instructions with fluorescence read at 450 nm. Fold changes were calculated using baseline values of untreated cells as a reference (set to 100%) and displayed graphically in Prism.
Retroviral and lentiviral transduction.
Transfections of 293FT cells with retroviral and lentiviral constructs were performed using Lipofectamine 2000 with Opti-MEM media (both from Invitrogen). Viral supernatants were produced by cotransfection of 293FT cells with the viral gag-pol and packaging vectors together with the Cas9 or sgRNA constructs (Supplemental Table 6). Cultivation was performed in high-glucose DMEM (Invitrogen) with GlutaMAX containing 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, 25 mM HEPES, 1 mM sodium pyruvate, and 0.1 mM nonessential amino acids. Regular media were replaced after 16 hours by growth media containing 3 mM caffeine. After 24 hours, the viral supernatant was harvested and filtered through a 0.45-μm filter. Retroviral transductions were performed by loading of the viral supernatant on 50 μg/mL retronectin-coated (Takara) nontissue 6-well plates before centrifugation (2000 g, 90 minutes at 32°C) 2 times. Subsequently, 1 × 106 to 2 × 106 cells were loaded and transduced per well by centrifugation at 600 g for 30 minutes and maintained for 72 hours at 37°C with 5% CO2 before transfer into culture flasks. Lentiviral transductions were performed by loading of viral supernatant on 50 μg/mL retronectin-coated nontissue 6-well plates with 1 × 106 to 2 × 106 cells. Cells were centrifuged for 30 minutes at 600 g and maintained for 16 hours at 37°C with 5% CO2 before replacement of media with fresh RPMI and GlutaMAX containing 20% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin. Cells were then cultured at 37°C with 5% CO2 before analysis.
CRISPR-mediated gene deletion.
Stably transduced Cas9-expressing ALL cell lines or PDX specimens were generated via lentiviral Cas9 transduction and targeting guide RNAs following the protocol above and using chemically synthesized single guide RNAs purchased predesigned from Integrated DNA Technologies. Recombinant Cas9 (TrueCut Cas9 Protein v2, Thermo Fisher Scientific) was mixed with guide RNA (2.5 μg/5 μg guide RNA to Cas9 protein) to produce RNA-ribonucleoprotein complexes. Electroporation was performed using pulse code EO-117 on a Lonza 4D electroporation system. Nontargeting (NT) guide RNAs were used as controls.
Quantitative real-time PCR.
Total RNA from leukemia cells was extracted using RNeasy isolation kit (Qiagen). cDNA was generated using the qScript cDNA SuperMix (Quanta). Quantitative real-time PCR was performed with Power SYBR Green Master Mix (Applied Biosystems) and 7500 Real-time PCR System (Applied Biosystems) according to standard PCR conditions. Primers for quantitative real-time PCR are listed in Supplemental Table 7.
Microarray target preparation and hybridization.
Microarray services were provided by the UPenn Molecular Profiling Facility, including quality control tests of the total RNA samples by Agilent Bioanalyzer and NanoDrop spectrophotometry. All protocols were conducted as described in the Affymetrix WT Plus reagent kit manual and the Affymetrix GeneChip Expression Analysis technical manual. Briefly, 250 ng of total RNA was converted to first-strand cDNA using reverse transcriptase primed by poly(T) and random oligomers incorporating the T7 promoter sequence. Second-strand cDNA synthesis was followed by in vitro transcription with T7 RNA polymerase for linear amplification of each transcript. The resulting cRNA was converted to cDNA, fragmented, assessed by the Bioanalyzer, and biotinylated by terminal transferase end labeling. Five-and-a-half micrograms of labeled cDNA was added to Affymetrix hybridization cocktails, heated at 99°C for 5 minutes, and hybridized for 16 hours at 45°C to Clariom D Human Arrays using the GeneChip Hybridization Oven 645. The microarrays were then washed at low (6× SSPE) and high (100 mM MES, 0.1 M NaCl) stringency and stained with streptavidin-phycoerythrin. Fluorescence was amplified by addition of biotinylated anti-streptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A GeneChip 3000 7G scanner was used to collect fluorescence signal. Affymetrix Command Console and Expression Console were used to quantitate expression levels for targeted genes; default values provided by Affymetrix were applied to all analysis parameters. Microarray data are available from the NCBI’s Gene Expression Omnibus database (GEO GSE132486).
Immunofluorescence studies.
Coverslips were coated overnight with 100 μg/mL poly-l-lysine solution (MilliporeSigma) at 4°C and washed twice with PBS before addition of cells. For each condition, 300,000 cells were plated on a single coverslip and allowed to spread in the incubator for 40 minutes at 37°C in 5% CO2. After visual confirmation of attachment, cells were washed in PBS and fixed in 3% paraformaldehyde/PBS. After quenching with 50 mM NH4Cl, cells were treated with Fc block (clone 2.4G2), permeabilized with 0.3% Triton X-100/PBS, and blocked with PBS/0.01% saponin/0.25% fish skin gelatin. Cells were labeled with primary mouse anti-CD79A and rabbit anti-CD79B antibodies (Supplemental Table 4) and secondary donkey anti-mouse–Alexa Fluor 488 or goat anti-rabbit–Alexa Fluor 568 antibodies for 1 hour at room temperature. Following antibody labeling, coverslips were mounted onto holding plates and kept immersed in PBS for immediate imaging. Imaging was performed on a PerkinElmer Ultraview spinning disk confocal system using a ×63 Plan Apo, 1.4NA objective mounted on an Axiovert 200 M microscope platform (Carl Zeiss) equipped with an Orca ER CCD camera (Hamamatsu). Images were acquired using Volocity 6.3 imaging software (PerkinElmer) and analyzed using ImageJ software (NIH).
Statistics.
Data display and statistical analyses were performed using Prism 7 (GraphPad). Significance values for in vitro and in vivo studies were calculated using unpaired 2-tailed Student’s t test (2 groups) or ANOVA with Dunnett’s or Tukey’s post-test for multiple comparisons (3 or more groups) to determine statistical significance. Survival analysis distribution between 2 groups for clinical patient samples (Figure 1A) was determined by the log-rank test. A P value of less than 0.05 was considered significant. All values are expressed as mean ± SEM.
Study approval.
Viably cryopreserved leukemia cells from children, adolescents, and young adults with de novo Ph-like ALL were obtained from the Children’s Oncology Group (Monrovia, California, USA) for xenotransplantation studies as previously described (12, 14, 32). Additional specimens from children and adults with Ph-like ALL and other ALL subtypes were obtained from the Children’s Hospital of Philadelphia (CHOP, protocol 7767) and University of Pennsylvania (protocol 703158) leukemia biorepositories under approved CHOP and University of Pennsylvania institutional research protocols after obtainment of written informed consent in accordance with the Declaration of Helsinki. Animal studies were conducted with approval from the Institutional Animal Care and Use Committee at the Children’s Hospital of Philadelphia (protocol 232).
Author contributions
CH designed and directed the study, performed experiments, analyzed and interpreted data, and wrote the manuscript. GBW, JPL, DB, AL, YL, BM, AB, and KDC performed experiments and analyzed data. JKB provided key experimental reagents and technical expertise. AEP provided key experimental reagents. MC and SKT oversaw and directed the study, designed experiments, analyzed and interpreted data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Matthew Stubbs and Bruce Ruggeri at Incyte Corp. for provision of ruxolitinib and INCB05465 drugs for these studies. We also thank Tasleema Patel at the Children’s Hospital of Philadelphia (CHOP) Center for Childhood Cancer Research for assistance with human leukemia biorepository specimens and clinical annotations. We acknowledge David Fruman and Thang-Tran Vo at the University of California, Irvine, for sharing the TVA1 cell line and our patient at CHOP for sharing his cells to create the ALL1807 PDX model and subsequent immortalized STL-CHPB1719 cell line. This study was supported by Alex’s Lemonade Stand Foundation (Young Investigator award to CH and Center of Excellence Scholar in Developmental Therapeutics award to SKT), the American Cancer Society (Scholar award to AEP), the American Society of Hematology (Scholar award to CH), the Children’s Oncology Group Research Foundation (to SKT), the University of Pennsylvania Institute for Translational Medicine and Therapeutics Transdisciplinary Program (to CH, MC, and SKT), the Rally Foundation for Childhood Cancer Research (to SKT), the Simutis Family Childhood Leukemia Research Foundation in memory of Andrew David Simutis (to SKT), the V Foundation for Cancer Research (to SKT), and NIH awards NIH/NCCR UL1RR024134, NIH/NCI R21-CA198621 (to AEP and MC), R01-CA198089 (to MC), and K08-CA184418 and NIH/NCI 1U01CA232486 (to SKT), and Department of Defense Translational Team Science Award CA180683P1 (to SKT).
Version 1. 03/19/2020
In-Press Preview
Version 2. 06/02/2020
Electronic publication
Version 3. 07/01/2020
Print issue publication
Funding Statement
University of Pennsylvania Institute for Translational Medicine and Therapeutics [ITMAT] Transdisciplinary Program award to CH, MC, SKT
award to SKT
award to SKT
research support to SKT
R21CA198621 (award to AEP and MC), R01CA198089 (award to MC), and K08CA184418 (award to SKT)
Footnotes
Conflict of interest: AEP receives research funding from Astellas Pharmaceuticals, Daiichi Sankyo, Novartis, and FujiFilm and is a consultant/advisory board member for Astellas, Daiichi Sankyo, Novartis, Arog, Pfizer, Takeda, Agios, and AbbVie. MC receives research funding from Astellas Pharmaceuticals and Incyte Corp. and is a consultant/advisory board member for Janssen Pharmaceuticals. SKT receives research funding from Incyte Corp. and Gilead Sciences and is a scientific advisory board member for Aleta Biotherapeutics.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(7):3637–3653.https://doi.org/10.1172/JCI134424.
Contributor Information
Christian Hurtz, Email: churtz@pennmedicine.upenn.edu.
Gerald B. Wertheim, Email: gwerthei@gmail.com.
Joseph P. Loftus, Email: loftusjp@email.chop.edu.
Daniel Blumenthal, Email: daniel.blumenthal.chop@gmail.com.
Anne Lehman, Email: talehman@pennmedicine.upenn.edu.
Yong Li, Email: weixi1989@gmail.com.
Asen Bagashev, Email: bagasheva@email.chop.edu.
Bryan Manning, Email: bmanning@sas.upenn.edu.
Katherine D. Cummins, Email: cumminsk@pennmedicine.upenn.edu.
Janis K. Burkhardt, Email: jburkhar@pennmedicine.upenn.edu.
Alexander E. Perl, Email: Alexander.Perl@uphs.upenn.edu.
Martin Carroll, Email: carroll2@mail.med.upenn.edu.
Sarah K. Tasian, Email: TasianS@email.chop.edu.
References
- 1.Herold T, et al. Adults with Philadelphia chromosome-like acute lymphoblastic leukemia frequently have IGH-CRLF2 and JAK2 mutations, persistence of minimal residual disease and poor prognosis. Haematologica. 2017;102(1):130–138. doi: 10.3324/haematol.2015.136366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain N, et al. Ph-like acute lymphoblastic leukemia: a high-risk subtype in adults. Blood. 2017;129(5):572–581. doi: 10.1182/blood-2016-07-726588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts KG, et al. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol. 2017;35(4):394–401. doi: 10.1200/JCO.2016.69.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruth KA, et al. Suppression of B-cell development genes is key to glucocorticoid efficacy in treatment of acute lymphoblastic leukemia. Blood. 2017;129(22):3000–3008. doi: 10.1182/blood-2017-02-766204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts KG, et al. Outcomes of children with BCR-ABL1–like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol. 2014;32(27):3012–3020. doi: 10.1200/JCO.2014.55.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts KG, et al. Genomic and outcome analyses of Ph-like ALL in NCI standard-risk patients: a report from the Children’s Oncology Group. Blood. 2018;132(8):815–824. doi: 10.1182/blood-2018-04-841676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin SD, et al. Thymic stromal lymphopoietin: a cytokine that promotes the development of IgM+ B cells in vitro and signals via a novel mechanism. J Immunol. 1999;162(2):677–683. [PubMed] [Google Scholar]
- 8.Isaksen DE, et al. Uncoupling of proliferation and Stat5 activation in thymic stromal lymphopoietin-mediated signal transduction. J Immunol. 2002;168(7):3288–3294. doi: 10.4049/jimmunol.168.7.3288. [DOI] [PubMed] [Google Scholar]
- 9.Mullighan CG, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009;106(23):9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoda A, et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2010;107(1):252–257. doi: 10.1073/pnas.0911726107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tasian SK, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120(4):833–842. doi: 10.1182/blood-2011-12-389932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu SC, et al. Activity of the type II JAK2 inhibitor CHZ868 in B cell acute lymphoblastic leukemia. Cancer Cell. 2015;28(1):29–41. doi: 10.1016/j.ccell.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weigert O, et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. J Exp Med. 2012;209(2):259–273. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maude SL, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120(17):3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tasian SK, et al. Potent efficacy of combined PI3K/mTOR and JAK or ABL inhibition in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2017;129(2):177–187. doi: 10.1182/blood-2016-05-707653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, et al. Inhibition of mTORC1/C2 signaling improves anti-leukemia efficacy of JAK/STAT blockade in CRLF2 rearranged and/or JAK driven Philadelphia chromosome-like acute B-cell lymphoblastic leukemia. Oncotarget. 2018;9(8):8027–8041. doi: 10.18632/oncotarget.24261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Den Boer ML, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10(2):125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang H, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115(7):1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurtz C, et al. Redundant JAK, SRC and PI3 kinase signaling pathways regulate cell survival in human Ph-like ALL cell lines and primary cells. Blood. 2017;130(suppl 1):717. doi: 10.1182/blood.V130.Suppl_1.717.717. [DOI] [Google Scholar]
- 20.Tasian SK, Loh ML, Hunger SP. Philadelphia chromosome-like acute lymphoblastic leukemia. Blood. 2017;130(19):2064–2072. doi: 10.1182/blood-2017-06-743252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts KG, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371(11):1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey RC, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115(26):5312–5321. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Bodegom D, et al. Differences in signaling through the B-cell leukemia oncoprotein CRLF2 in response to TSLP and through mutant JAK2. Blood. 2012;120(14):2853–2863. doi: 10.1182/blood-2012-02-413252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullighan CG, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41(11):1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertzberg L, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115(5):1006–1017. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- 26.Zhong J, et al. TSLP signaling network revealed by SILAC-based phosphoproteomics. Mol Cell Proteomics. 2012;11(6):M112.017764. doi: 10.1074/mcp.M112.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong J, et al. TSLP signaling pathway map: a platform for analysis of TSLP-mediated signaling. Database (Oxford) 2014;2014:bau007. doi: 10.1093/database/bau007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell LJ, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114(13):2688–2698. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- 29.Schinnerl D, et al. The role of the Janus-faced transcription factor PAX5-JAK2 in acute lymphoblastic leukemia. Blood. 2015;125(8):1282–1291. doi: 10.1182/blood-2014-04-570960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goetz CA, Harmon IR, O’Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol. 2004;172(8):4770–4778. doi: 10.4049/jimmunol.172.8.4770. [DOI] [PubMed] [Google Scholar]
- 31.Yasuda T, et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity. 2008;28(4):499–508. doi: 10.1016/j.immuni.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Fu C, Turck CW, Kurosaki T, Chan AC. BLNK: a central linker protein in B cell activation. Immunity. 1998;9(1):93–103. doi: 10.1016/S1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]
- 33.Kurosaki T, et al. Syk activation by the Src-family tyrosine kinase in the B cell receptor signaling. J Exp Med. 1994;179(5):1725–1729. doi: 10.1084/jem.179.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takata M, Kurosaki T. A role for Bruton’s tyrosine kinase in B cell antigen receptor-mediated activation of phospholipase C-γ2. J Exp Med. 1996;184(1):31–40. doi: 10.1084/jem.184.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geng H, et al. Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell. 2015;27(3):409–425. doi: 10.1016/j.ccell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duy C, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207(6):1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4(1):31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 38.Duy C, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473(7347):384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trancoso I, et al. A novel quantitative fluorescent reporter assay for RAG targets and RAG activity. Front Immunol. 2013;4:110. doi: 10.3389/fimmu.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46(2):116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swaminathan S, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16(7):766–774. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harvey RC, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116(23):4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koyama M, Ishihara K, Karasuyama H, Cordell JL, Iwamoto A, Nakamura T. CD79α/CD79β heterodimers are expressed on pro-B cell surfaces without associated mu heavy chain. Int Immunol. 1997;9(11):1767–1772. doi: 10.1093/intimm/9.11.1767. [DOI] [PubMed] [Google Scholar]
- 44.Kouro T, et al. Bruton’s tyrosine kinase is required for signaling the CD79b-mediated pro-B to pre-B cell transition. Int Immunol. 2001;13(4):485–493. doi: 10.1093/intimm/13.4.485. [DOI] [PubMed] [Google Scholar]
- 45.Fuentes-Pananá EM, Bannish G, Karnell FG, Treml JF, Monroe JG. Analysis of the individual contributions of Igα (CD79a)- and Igβ (CD79b)-mediated tonic signaling for bone marrow B cell development and peripheral B cell maturation. J Immunol. 2006;177(11):7913–7922. doi: 10.4049/jimmunol.177.11.7913. [DOI] [PubMed] [Google Scholar]
- 46.Luger D, et al. Expression of the B-cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS One. 2013;8(10):e76115. doi: 10.1371/journal.pone.0076115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X, et al. Continuous signaling of CD79b and CD19 is required for the fitness of Burkitt lymphoma B cells. EMBO J. 2018;37(11):e97980. doi: 10.15252/embj.201797980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng Y, et al. LNK/SH2B3 regulates IL-7 receptor signaling in normal and malignant B-progenitors. J Clin Invest. 2016;126(4):1267–1281. doi: 10.1172/JCI81468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srinivasan L, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139(3):573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aiba Y, Kameyama M, Yamazaki T, Tedder TF, Kurosaki T. Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood. 2008;111(3):1497–1503. doi: 10.1182/blood-2007-08-109769. [DOI] [PubMed] [Google Scholar]
- 51.Ramchandren R, et al. Ongoing phase 1/2 study of INCB050465 for relapsed/refractory (R/R) B-cell malignancies (CITADEL-101) J Clin Oncol. 2017;35(15 suppl):7350. doi: 10.1200/JCO.2017.35.15_suppl.7530. [DOI] [Google Scholar]
- 52.Gotesman M, et al. mTOR inhibition enhances efficacy of dasatinib in ABL-rearranged Ph-like B-ALL. Oncotarget. 2018;9(5):6562–6571. doi: 10.18632/oncotarget.24020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 54.Feldhahn N, et al. Mimicry of a constitutively active pre-B cell receptor in acute lymphoblastic leukemia cells. J Exp Med. 2005;201(11):1837–1852. doi: 10.1084/jem.20042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klejman A, et al. The Src family kinase Hck couples BCR/ABL to STAT5 activation in myeloid leukemia cells. EMBO J. 2002;21(21):5766–5774. doi: 10.1093/emboj/cdf562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Modi H, Li L, Chu S, Rossi J, Yee JK, Bhatia R. Inhibition of Grb2 expression demonstrates an important role in BCR-ABL-mediated MAPK activation and transformation of primary human hematopoietic cells. Leukemia. 2011;25(2):305–312. doi: 10.1038/leu.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pendergast AM, et al. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell. 1993;75(1):175–185. doi: 10.1016/S0092-8674(05)80094-7. [DOI] [PubMed] [Google Scholar]
- 58.Puil L, et al. Bcr-Abl oncoproteins bind directly to activators of the Ras signalling pathway. EMBO J. 1994;13(4):764–773. doi: 10.1002/j.1460-2075.1994.tb06319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skorski T, et al. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood. 1995;86(2):726–736. doi: 10.1182/blood.V86.2.726.bloodjournal862726. [DOI] [PubMed] [Google Scholar]
- 60.Fuentes-Pananá EM, Bannish G, Shah N, Monroe JG. Basal Igalpha/Igbeta signals trigger the coordinated initiation of pre-B cell antigen receptor-dependent processes. J Immunol. 2004;173(2):1000–1011. doi: 10.4049/jimmunol.173.2.1000. [DOI] [PubMed] [Google Scholar]
- 61.Erasmus MF, et al. Dynamic pre-BCR homodimers fine-tune autonomous survival signals in B cell precursor acute lymphoblastic leukemia. Sci Signal. 2016;9(456):ra116. doi: 10.1126/scisignal.aaf3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shah NN, Fry TJ. Mechanisms of resistance to CAR T cell therapy. Nat Rev Clin Oncol. 2019;16(6):372–385. doi: 10.1038/s41571-019-0184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qin H, et al. Eradication of B-ALL using chimeric antigen receptor-expressing T cells targeting the TSLPR oncoprotein. Blood. 2015;126(5):629–639. doi: 10.1182/blood-2014-11-612903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Surrey LF, et al. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Genome Med. 2019;11(1):32. doi: 10.1186/s13073-019-0644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harvey RC, et al. Development and validation of a highly sensitive and specific gene expression classifier to prospectively screen and identify B-precursor acute lymphoblastic leukemia (ALL) patients with a Philadelphia chromosome-like (“Ph-like” or “BCR-ABL1-like”) signature for therapeutic targeting and clinical intervention. Blood. 2013;122(21):826. doi: 10.1182/blood.V122.21.826.826. [DOI] [Google Scholar]
- 66.Loftus JP, et al. Combinatorial efficacy of entospletinib and chemotherapy in patient-derived xenograft models of infant acute lymphoblastic leukemia [online May 15, 2020]. Haematologica. doi: 10.3324/haematol.2019.241729. doi: 10.3324/haematol.2019.241729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.