Abstract
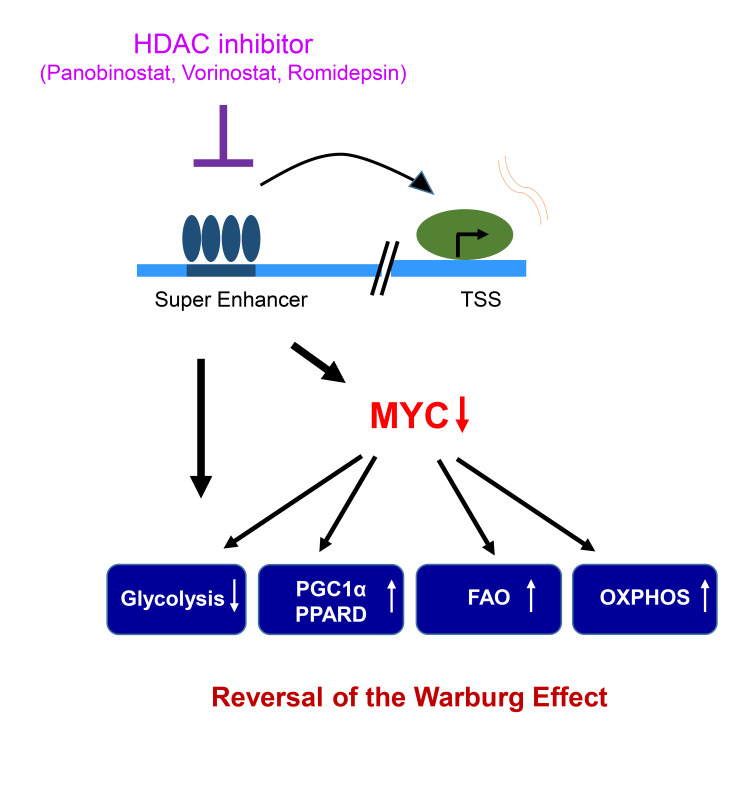
The Warburg effect is a tumor-related phenomenon that could potentially be targeted therapeutically. Here, we showed that glioblastoma (GBM) cultures and patients’ tumors harbored super-enhancers in several genes related to the Warburg effect. By conducting a transcriptome analysis followed by ChIP-Seq coupled with a comprehensive metabolite analysis in GBM models, we found that FDA-approved global (panobinostat, vorinostat) and selective (romidepsin) histone deacetylase (HDAC) inhibitors elicited metabolic reprogramming in concert with disruption of several Warburg effect–related super-enhancers. Extracellular flux and carbon-tracing analyses revealed that HDAC inhibitors blunted glycolysis in a c-Myc–dependent manner and lowered ATP levels. This resulted in the engagement of oxidative phosphorylation (OXPHOS) driven by elevated fatty acid oxidation (FAO), rendering GBM cells dependent on these pathways. Mechanistically, interference with HDAC1/-2 elicited a suppression of c-Myc protein levels and a concomitant increase in 2 transcriptional drivers of oxidative metabolism, PGC1α and PPARD, suggesting an inverse relationship. Rescue and ChIP experiments indicated that c-Myc bound to the promoter regions of PGC1α and PPARD to counteract their upregulation driven by HDAC1/-2 inhibition. Finally, we demonstrated that combination treatment with HDAC and FAO inhibitors extended animal survival in patient-derived xenograft model systems in vivo more potently than single treatments in the absence of toxicity.
Keywords: Oncology
Keywords: Intermediary metabolism
Introduction
Heterogeneous solid neoplasms, such as glioblastoma (GBM), remain highly challenging to treat due to multiple mechanisms including the deregulation of metabolism. The study of tumor cell metabolism has emerged over the past couple of years for a number of reasons (1, 2). The first central observation dates back to the 1920s, when Nobel laureate Otto Warburg discovered that tumor cells entertain a process called aerobic glycolysis that produces lactate from glucose in the presence of abundant oxygen (3). This basic observation in itself suggests that tumor cells have a higher dependency on glycolysis, potentially opening up tumor-specific therapeutic opportunities. In turn, glucose carbons are not oxidized and not lost in the form of CO2 but instead are kept within the body to be used for biosynthesis. In addition, what follows is that tumor cells use the TCA cycle not predominantly for energy generation but instead for shunting carbon into the cytosol via the ATP citrate lysate reaction (4), giving rise to acetyl-CoA in the cytosol, which in turn is used for biosynthesis of fatty acids and cholesterol to enable proliferation and cell division. Other more recent findings also position glycolysis at a central step in the production of amino acids including serine, which in turn fuels the synthesis of glycine to drive the folate cycle, supporting the production of nucleotides (5–10). At this point, it is not well understood how the Warburg effect is regulated at the epigenetic level.
Thus far, there is little information about the impact of histone deacetylases (HDACs) in the context of the regulation of tumor cell metabolism, and the concept that HDACs, such as HDAC1 and HDAC2 (HDAC1/-2), may control metabolism through regulation of an enhancer landscape is, we believe, unprecedented. HDAC inhibitors such as panobinostat (Pb) and vorinostat (Vr) or selective HDAC1/-2 inhibitors such as romidepsin (Ro) have been approved for the treatment of cutaneous T cell lymphoma and multiple myeloma, and clinical trials with Pb are ongoing for GBM.
Here, we report what we believe to be a novel strategy to reverse the Warburg effect. Our findings suggest that the Warburg effect is maintained by super-enhancers, which are larger aggregates of enhancer clusters that drive gene expression (11, 12). Our findings demonstrate that selective and broad HDAC inhibition disrupts super-enhancers globally and super-enhancers related to aerobic glycolysis, resulting in enhanced oxidative metabolism that is targetable with drug combination therapies in solid tumor cells and in patient-derived xenograft (PDX) models in vivo, with extension of overall survival. These findings are in part orchestrated by HDAC1/-2 inhibition–mediated suppression of c-Myc (disruption of Myc super-enhancer) followed by c-Myc–dependent upregulation of key transcription factors of oxidative metabolism, peroxisome proliferator–activated receptor γ coactivator 1 α (PGC1α), and peroxisome proliferator–activated receptor δ (PPARδ). These findings provide an efficient strategy for epigenetic targeting of the Warburg effect in solid tumors.
Results
Warburg effect–related genes are associated with super-enhancers in GBM cells and tissues.
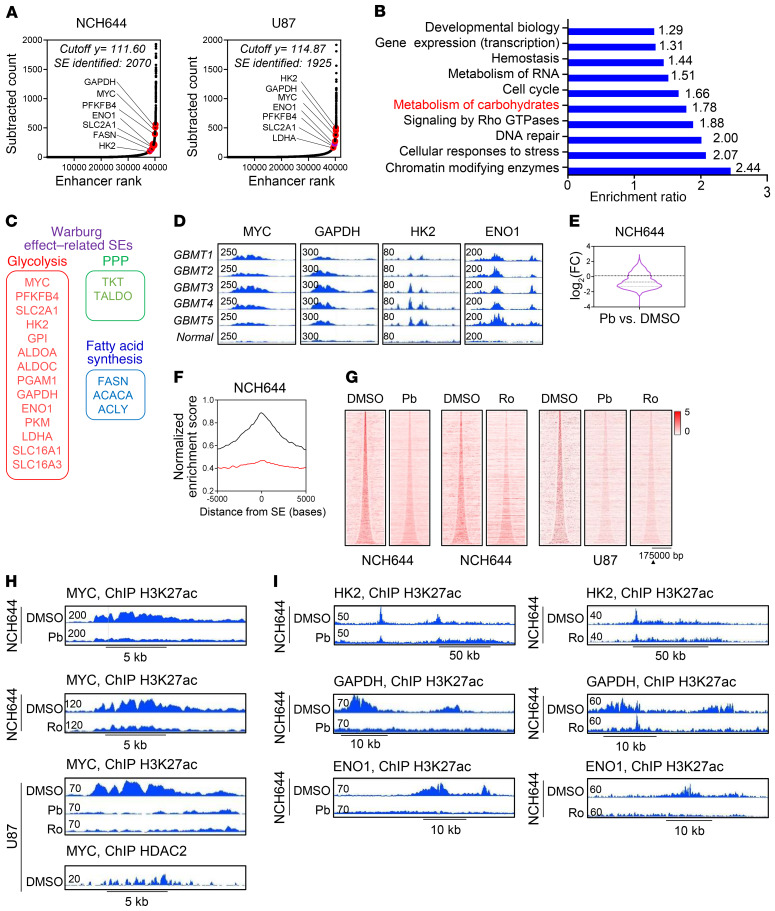
The Warburg effect consists of genes encoding for enzymes or transporters involved in glycolysis, the pentose phosphate pathway (PPP), and fatty acid synthesis (Figure 1, A–C). ChIP of acetylated H3K27 (H3K27ac) coupled with next-generation sequencing followed by computational analysis revealed super-enhancers across a number of these genes (Figure 1, A–C, and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI129049DS1). Similarly, when we analyzed published ChIP-Seq data, we noted that GBM tissues harbor super-enhancers related to genes linked to the Warburg effect as well (Figure 1D). In contrast, normal brain revealed significantly smaller peaks in the same regions, in keeping with the notion that the Warburg effect is a tumor-associated phenomenon that may be epigenetically targeted for therapy.
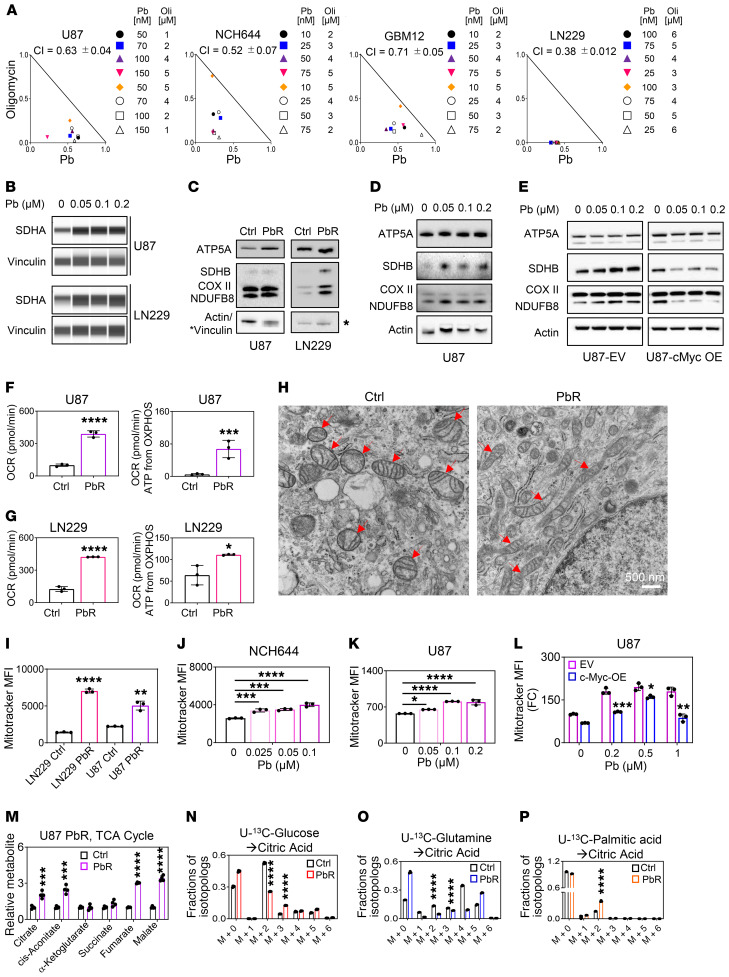
Figure 1. Identification of super-enhancers in the desert of Warburg effect–related genes that are disrupted by HDAC inhibitors.
(A) ChIP of H3K27ac coupled with next-generation sequencing of NCH644 and U87 GBM cells was performed followed by super-enhancer (SE) analysis. Shown are the super-enhancers of genes involved in glycolysis, the PPP, and fatty acid synthesis (Warburg effect–related genes). The peak located at the HK2 locus in the NCH644 cells is slightly below the cutoff and therefore a strong enhancer. (B) “Reactome analysis” of mutual super-enhancer genes in NCH644, U87, and LN229 GBM cells. FDR Q < 0.05. (C) The Warburg effect consists of genes encoding for enzymes or transporters involved in glycolysis, the PPP, or fatty acid synthesis. (D) Published ChIP-Seq (H3K27ac) data for GBMs and normal brain tissue (pileup values are indicated) (GSE101148 and GSE17312). (E and F) Representation of global disruption of the super-enhancer landscape of NCH644 cells treated with Pb. FC, fold change. (G) Heatmaps of super-enhancers in control- and HDAC inhibitor–exposed NCH644 and U87 GBM cells. Scale bar indicates the intensities. (H) ChIP-Seq (H3K27ac) was performed in NCH644 and U87 cells treated with vehicle (DMSO), Pb, or Ro. Shown are the respective tracks around the Myc locus (pileup values are indicated). (I) ChIP-Seq (H3K27ac) was performed in NCH644 cells treated with vehicle, Pb, or Ro. Shown are the respective tracks around HK2, GAPDH, and ENO1.
FDA-approved HDAC inhibitors disrupt super-enhancers related to the Warburg effect.
The recent literature suggests that HDAC inhibitors may potently disrupt super-enhancers (13, 14). Therefore, we performed tests to determine whether HDAC inhibitors are capable of interfering with the super-enhancer landscape in GBM model systems. We performed ChIP-Seq in the presence or absence of the pan-HDAC inhibitor Pb or the selective HDAC inhibitor Ro. We found that in both patient-derived stem-like NCH644 GBM cells and U87 GBM cells, Pb and Ro treatment led to a global disruption of the super-enhancer landscape with reduced binding of RNA polymerase II (Rpb1) (Figure 1, E–I, and Supplemental Figure 1, B–J), including Warburg effect–related genes such as MYC, hexokinase 2 (HK2), GAPDH, and enolase 1 (ENO1). HDAC2 ChIP-Seq on our cell lines revealed that this enzyme colocalized with the super-enhancer identified by H3K27ac ChIP-Seq (Figure 1H and Supplemental Figure 1E). Gene ontology (GO) analysis suggested an impaired RNA polymerase II–related transcription (Supplemental Figure 1J). Transcriptome analysis validated the suppression of many of these mRNAs (Supplemental Figure 1G).
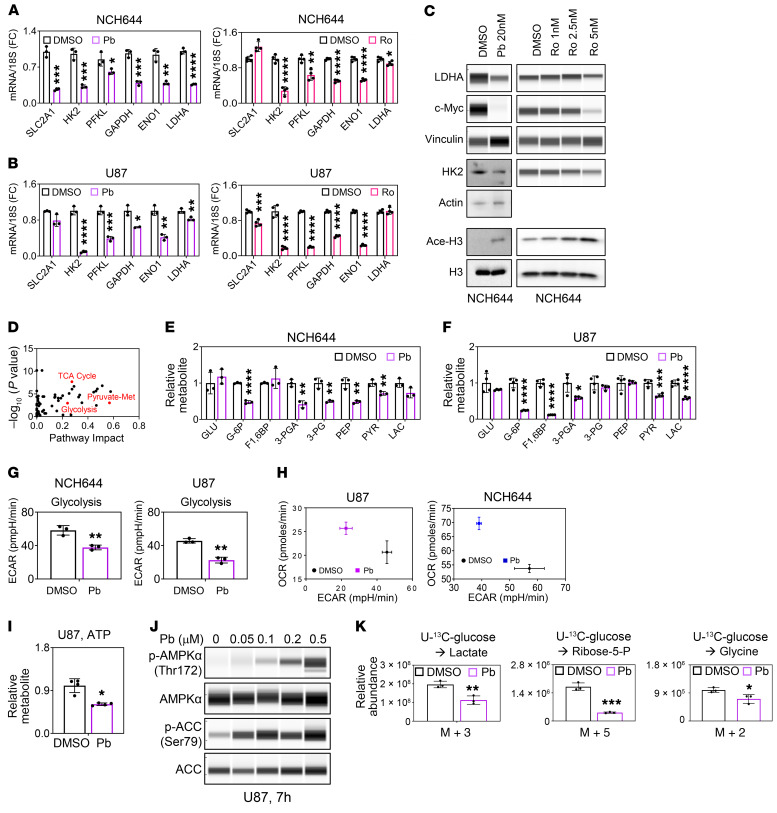
To confirm these changes, we evaluated glycolysis-related transcripts in the stem-like GBM cells NCH644 and NCH421k as well as in established U87 GBM cells following treatment with Pb or Ro (Figure 2, A and B, and Supplemental Figure 2, A and B). Our findings highlighted a suppression of key glycolytic transcripts at the mRNA level, including transporter genes encoding for glucose transporter 1 (SLC2A1) as well as key enzymes such as genes encoding for hexokinase 2 (HK2), ATP-dependent 6-phosphofructokinase, liver type (PFKL), GAPDH, ENO1, and lactate dehydrogenase A (LDHA), which was partially recapitulated by HDAC1/-2 silencing (Supplemental Figure 2C). We validated some of these key enzymes at the protein level and found that Pb and Ro reduced the protein expression of HK2, GLUT1, LDHA, and c-Myc accompanied by enhanced acetylation of H3K27 in NCH644 and U87 cells (Figure 2C and Supplemental Figure 2D).
Figure 2. HDAC inhibitors reverse the Warburg effect.
(A) Real-time PCR analysis of genes related to glycolysis from stem-like NCH644 GBM cells treated with 0.5 μM Pb or 2 nM Ro for 24 hours (n = 3–4). (B) Real-time PCR analysis of genes related to glycolysis from established U87 GBM cells treated with 0.5 μM Pb or 5 nM Ro for 24 hours (n = 3–4). (C) Analysis of protein lysate from NCH644 cells treated with the indicated concentration of Pb (LDHA, c-Myc, vinculin [loading control]: protein capillary electrophoresis [PCE]; HK2, actin [loading control]: standard Western blot gel; Ace-H3, H3 [loading control]: standard Western blot) or Ro for 24 hours (LDHA, c-Myc, HK2, vinculin [loading control]: PCE; Ace-H3, H3 [loading control]: standard Western blot). (D) U87 GBM cells were treated with 0.5 μM Pb for 24 hours and analyzed by LC/MS followed by metabolite (Met) pathway analysis. (E and F) Quantifications of glycolysis-related metabolites from NCH644 and U87 cells treated with 0.5 μM Pb for 24 hours (n = 3–4). GLU, glucose; G-6P, glucose-6-phosphate; F1,6BP, fructose-1,6-bisphosphate; 3-PGA, glyceraldehyde-3-phosphate; 3-PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; PYR, pyruvate; LAC, lactate. (G and H) NCH644 and U87 cells were exposed to 0.2 μM Pb, and the OCR and ECAR were recorded (n = 3). (I) U87 cells were treated and harvested as in E and F. Shown are the levels of ATP (determined by LC/MS). (J) PCE analysis of lysates from U87 cells treated with the indicated concentrations of Pb for 7 hours. (K) Quantifications of the relative abundances of the indicated 13C isotopologs from U-13C-glucose in U87 GBM cells treated with 0.5 μM Pb for 24 hours (n = 3). Data represent the mean ± SD. Statistical significance was determined by 2-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Given these genomic changes in metabolism, we continued with a polar metabolite analysis using liquid chromatography and mass spectrometry (LC/MS), and metabolic pathway analysis suggested impairment of glycolysis in both neurosphere NCH644 and established U87 GBM cells (Figure 2, D–F). Next, we determined whether these reduced expression levels of glycolytic enzymes indeed translated into reduced glycolysis rates. To this end, we performed extracellular flux analysis and confirmed that Pb as well as Vr reduced the extracellular acidification rate (ECAR) with a concurrent increase in the oxygen consumption rate (OCR), suggesting a potential compensatory mechanism for energy production (Figure 2, G and H, and Supplemental Figure 2, E–G). These changes in energy metabolism were associated with a reduction in ATP levels, suggesting that an HDAC inhibitor–mediated reduction in glycolysis leads to energy deprivation, which in turn leads to a compensatory enhancement of the OCR (as a surrogate for the activation of oxidative phosphorylation [OXPHOS]) (Figure 2I). Low levels of ATP were also accompanied by an activation of AMP-activated protein kinase A (AMPKA) (Figure 2J and Supplemental Figure 3A). A transcriptional signature of energy deprivation supported these findings as well (Supplemental Figure 3B). We also tested whether AMPKA activation is pivotal for survival by silencing AMPKA expression through a shRNA. Our results indicated that knockdown of AMPKA had no impact on cell death mediated by the HDAC inhibitor (Supplemental Figure 3, C and D).
To further validate these observations, we performed carbon tracing with U-13C-glucose and found a significant reduction of lactate (m+3), in keeping with our extracellular flux analysis (Figure 2K). Similarly, other glycolytic intermediates or associated metabolites (e.g., nucleotides or ratios of metabolites) displayed a reduction in labeling and total levels and revealed reduced glucose carbon labeling (Supplemental Figure 3, E–H, and Supplemental Figure 4, A–E).
Aside from glycolysis, the Warburg effect encompasses additional metabolic pathways, including the PPP and related ribose production for nucleotide biosynthesis, serine synthesis, the hexosamine biosynthetic pathway, and lipid synthesis via the TCA cycle. We noted decreased U-13C-glucose labeling of metabolites associated with these pathways (Supplemental Figure 4, A–F). Given the pivotal role of the PPP in tumor growth, we analyzed the levels of metabolites from the oxidative and nonoxidative PPP and related metabolites (e.g., NADPH2 and nucleotides) (Supplemental Figure 3, F–H). In like manner, we found a reduction of ribose-5-phosphate (m+5) and other related metabolites in U-13C-glucose carbon-tracing experiments (Figure 2K and Supplemental Figure 4B). Akin to glycolysis, we detected a pronounced reduction in the PPP, which was accompanied by a striking deregulation of the NADP/NADPH2 ratio (up to 24 times higher in Pb-treated cells), suggesting that these cells harbored a significant deficiency in biosynthesis as well as in detoxification of ROS (Supplemental Figure 3, E, G, and H). Consistently, we found a reduction of the GSH/GSSG (reduced/oxidized glutathione) ratio and a significant reduction in total GSH levels (Supplemental Figure 3, E and G).
In alignment with PPP suppression, we detected a pronounced reduction in ribulose-5-phosphate, which is the precursor of phosphoribosyl pyrophosphate (PRPP), the key molecule for nucleotide synthesis (Supplemental Figure 3F). It came as no surprise that we found a reduction in purine and pyrimidine nucleotides, which suggested an impairment in nucleic acid synthesis (Supplemental Figure 3, G and H). We consistently detected reduced U-13C-glucose carbon labeling in nucleotides (Supplemental Figure 4C). We also observed a reduction in glycine labeling by U-13C-glucose, suggesting an impairment of the serine/glycine synthesis pathway upon exposure to the pan-HDAC inhibitor (Figure 2K). Finally, glucose carbon labeling of lipid synthesis–related glycerol-3-phosphate as well as of the hexosamine biosynthesis–related acetyl-glucosamine-1-phosphate was reduced (Supplemental Figure 4, D and E). All in all, these characteristics support the notion that HDAC inhibition blocks the Warburg effect in GBM cells.
The c-Myc protein is involved in metabolic reprogramming elicited by HDAC inhibition.
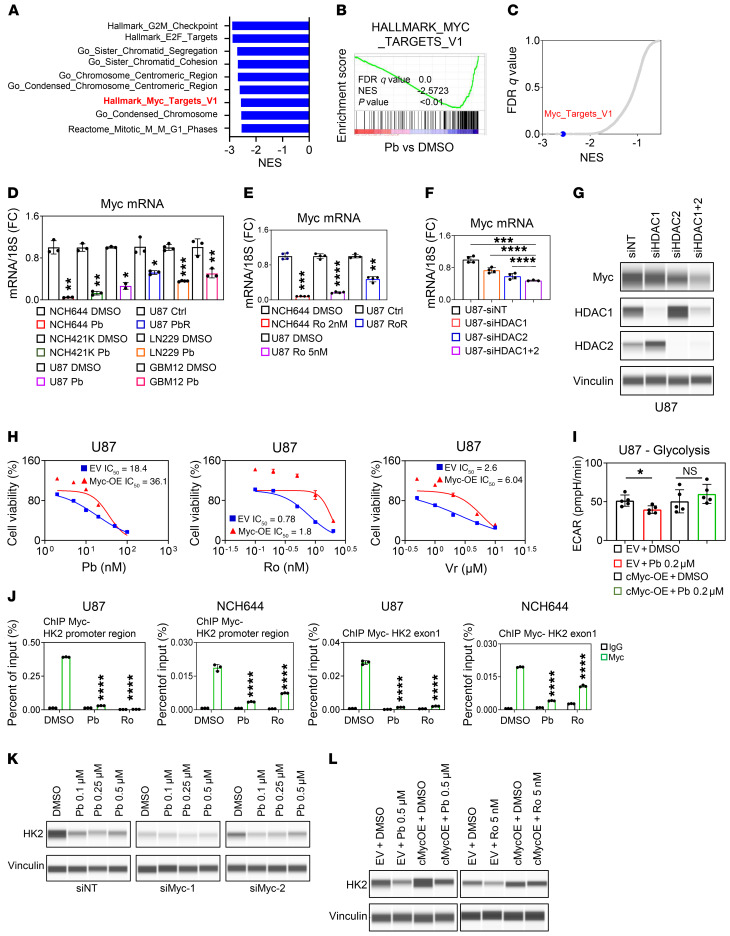
To elucidate the underlying mechanism by which the HDAC inhibitor orchestrates these changes in metabolism, we consulted our transcriptome data and found that both c-Myc and its targets were prominently suppressed in GBM cells treated with Pb (Figure 3, A–C). These observations were also confirmed by real-time PCR and protein expression analysis (Figure 3, D and E) and were highly linked to the earlier observation that HDAC inhibition resulted in disruption of super-enhancers. We assessed which HDACs were involved in the regulation of c-Myc transcriptional and protein levels and found involvement of both HDAC1 and HDAC2, respectively, in keeping with the observation that the HDAC1/-2 blocker Ro was sufficient to suppress c-Myc protein levels (Figure 3, F and G, and Supplemental Figure 5A). Next, we determined whether c-Myc was actually involved in the response to HDAC inhibitors and found that overexpression of c-Myc protected against a reduction in viability by Pb, Vr, and Ro (Figure 3H).
Figure 3. HDAC inhibitors suppress c-Myc protein levels and thereby reduce survival and glycolysis in GBM cells.
(A) The top 9 pathways identified by GSEA of NCH644 cells treated with 0.5 μM Pb for 24 hours (transcriptome analysis). (B) GSEA plot. (C) Graphical representation of the FDR Q values versus NES derived from the analysis in A and B. (D) GBM cells were treated with Pb or were chronically exposed to Pb (n = 3). (E) GBM cells were treated with Ro or were chronically exposed to Ro (n = 4). (F) U87 GBM cells were transfected with HDAC1 siRNA (siHDAC1), HDAC2 siRNA (siHDAC2), or a combination of both (siHDAC1+2) (n = 3–4). (G) PCE analysis of lysates from U87 cells transfected with HDAC1 siRNA, HDAC2 siRNA, or a combination of both. (H) U87 cells were treated with the indicated concentrations of Pb, Ro, or Vr for 72 hours, and cellular viability was determined. (I) U87 cells expressing a c-Myc construct were treated with 0.2 μM Pb for 24 hours, and a glycolysis stress test was performed (n = 5). (J) ChIP-qPCR of different locations around the HK2 gene (promoter and exon 1) from the indicated cell lysate with either a c-Myc or IgG antibody (n = 3). (K) PCE analysis of U87 cells that were transfected with an siRNA against Myc-1 or Myc-2, treated with the indicated concentration of Pb for 24 hours, and analyzed for the indicated protein. (L) PCE analysis of lysates from U87 cells that were transduced with a c-Myc construct, treated with 0.5 μM Pb or 5 nM Ro for 24 hours, and analyzed for HK2. Data represent the mean ± SD. Statistical significance was determined by 2-tailed Student’s t test (F, I, and J) or by 1-way ANOVA (G and H). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Ctrl, control; EV, empty vector; NES, normalized enrichment score; OE, overexpression; siNT, nontargeting siRNA .
Given that c-Myc is a transcription factor mastering aerobic glycolysis, we hypothesized that a HDAC inhibitor–mediated reduction of glycolysis occurs through c-Myc inhibition. We silenced c-Myc in 2 GBM cell lines using 2 c-Myc siRNAs and confirmed that silencing of c-Myc resulted in suppression of glycolytic key enzymes and transport proteins (Supplemental Figure 5B). Next, we analyzed the ectopic expression of c-Myc protein in 2 GBM cell lines. As anticipated, we found that c-Myc overexpression rescued HDAC inhibitor–mediated suppression of glycolysis and related parameters, supporting the idea of a primary role of c-Myc in regulating carbohydrate metabolism in the context of HDAC inhibitors, which we observed in 2 different cell lines of different genetic backgrounds (Figure 3I and Supplemental Figure 5, C and D). To establish a closer link between c-Myc and its function in regulating key glycolytic enzymes following treatment with HDAC inhibitors, we performed ChIP for c-Myc and assessed its binding to the promoter regions of HK2, Myc, and LDHA and found that c-Myc avidly bound to all these regions (Figure 3J and Supplemental Figure 5E). However, following HDAC inhibitor treatment, the interactions of c-Myc with these promoter regions were disrupted. Rescue experiments with c-Myc revealed that overexpression of c-Myc counteracted the HDAC inhibitor–mediated reduction in HK2, and when c-Myc levels were silenced by 2 siRNAs, no further reduction was noted following Pb treatment (Figure 3, K and L). As a side note and in keeping with the disruption of the Myc super-enhancer, we also noted a reduced presence of the H3K27ac mark within intron 1 of the Myc gene (Supplemental Figure 5F). All in all, these findings strongly suggest a key role of c-Myc in HDAC inhibitor–mediated regulation of glycolysis.
Acute and chronic HDAC inhibitor exposure leads to activation of oxidative metabolism.
We sought to determine how HDAC inhibitor–treated cells counteract glycolysis inhibition. Our extracellular flux analysis showed that acute HDAC inhibitor treatment resulted in an increase in the OCR (Figure 2H). This finding was confirmed in PDX GBM12 cells that, upon acute treatment with Pb, showed an increase in the OCR as well (Supplemental Figure 6, A and B). We evaluated the impact of OXPHOS on the survival of cells subjected to acute HDAC inhibitor treatment. To this purpose, we interfered with ATP synthesis through inhibition of respiratory complex V by oligomycin. We found that treatment with oligomycin along with Pb or Ro synergistically reduced the viability of several different model systems (Figure 4A and Supplemental Figure 6, C–E). These results suggest that broad and selective HDAC inhibitors render tumor cells more dependent on oxidative energy metabolism. We also evaluated HDAC inhibitor susceptibilities in cells that had been cultured in the presence of galactose, which over time renders cells more reliant on oxidative energy metabolism. As anticipated, galactose-treated cells were slightly more resistant to the HDAC inhibitors Pb and Vr (Supplemental Figure 6F). Similarly, chronically exposed GBM cells were more sensitive to oligomycin than were controls (Supplemental Figure 6G). Increased mitochondrial respiration is regulated at several levels. In turn, we determined protein expression levels of the 5 respiratory chain complexes and found that expression especially of complex I and complex II (succinate dehydrogenase complex iron sulfur subunit B [SDHB] and succinate dehydrogenase complex flavoprotein subunit A [SDHA]) was increased following HDAC inhibitor treatment (acute and chronic), enabling higher OXPHOS activity (Figure 4, B–D, and Supplemental Figure 6H). Given the earlier involvement of c-Myc in metabolic regulation following HDAC inhibition, we tested whether the increase in the components of the electron transport chain was dependent on c-Myc. Our results indicated that c-Myc overexpression partially counteracted the Pb-mediated increase in OXPHOS complexes (Figure 4E and Supplemental Figure 6I).
Figure 4. HDAC inhibitors drive oxidative energy metabolism.
(A) Isobolograms show the results for U87, NCH644, GBM12, and LN229 cells that were treated with Pb in the presence of oligomycin (Oli) for 72 hours. (B) PCE analyses of U87 and LN229 cells treated with Pb for 24 hours. (C) Western blots of the OXPHOS complex from parental U87 and LN229 GBM cells and U87 and LN229 GBM cells chronically exposed to Pb (PbR). (D) Western blots of the OXPHOs complex from U87 cells treated with Pb for 24 hours. (E) U87 cells were transduced with a c-Myc construct, treated with Pb for 24 hours, and analyzed for OXPHOS complexes. (F and G) OCR and OXPHOS-driven ATP production rates in U87 and LN229 cells chronically exposed to Pb (n = 3). (H) Electron microscopic images of parental U87 cells and U87 cells chronically Pb. Arrows highlight mitochondria. Scale bar: 500 nm. (I) Parental U87 and LN229 cells and U98 and LN229 cells chronically exposed to Pb were stained with MitoTracker and analyzed by flow cytometry (n = 3). (J and K) NCH644 and U87 cells were treated with Pb, stained with MitoTracker, and analyzed by flow cytometry (n = 3). (L) c-Myc construct–transduced U87 cells were treated with Pb for 24 hours, stained with MitoTracker, and analyzed by flow cytometry (n = 3). (M) TCA cycle metabolites in parental U87 cells or U87 cells chronically exposed to Pb (n = 3). (N) Parental U87 cells or U87 cells chronically exposed to Pb were cultured in DMEM media (25 mM U-13C-glucose, 4 mM glutamine) for 24 hours (n = 3). (O) U87 parental cells or U87 cells chronically exposed to Pb were cultured in DMEM media (25 mM glucose, 4 mM U-13C-glutamine) for 24 hours (n = 3). (P) Parental U87 cells or U87 cells chronically exposed to Pb were cultured in DMEM media (5 mM glucose, 1 mM glutamine, 100 μM U-13C-palmitic acid) for 24 hours (n = 3). Data represent the mean ± SD. Statistical significance was determined by 2-tailed Student’s t test (F–I and L–P) or 1-way ANOVA (J and K). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Aside from acute treatment, it is paramount to consider model systems in which cells have been chronically exposed to a drug compound. This will ultimately elucidate the mechanisms by which tumor cells manage to evade therapy and lead to the identification of targets to circumvent this phenomenon. The LN229 and U87 cells subjected to chronic Pb (PbR) treatment showed an increased number of OXPHOS complexes and larger, tubular-shaped mitochondria, accompanied by higher mtDNA levels, enhanced OCR, OXPHOS-related ATP production, and metabolites related to the TCA cycle (Figure 4, F–M, Supplemental Figure 6J, and Supplemental Figure 7, A–I). The increase in mitochondria size was already appreciable after 24 hours of treatment with HDAC inhibitors, albeit to a lesser extent (Figure 4, J and K). Akin to the electron transport subunits, c-Myc was involved in regulating the size of mitochondria following HDAC inhibitor treatment, since overexpression of c-Myc partially attenuated the HDAC inhibitor–mediated increase in mitochondria size (Figure 4L and Supplemental Figure 7J).
The TCA cycle serves as a siphon for carbons to be oxidized to CO2 or, alternatively, for their usage in biosynthesis (anaplerosis). In this vein, we determined the fate of U-13C-glucose, U-13C-glutamine, and U-13C-palmitic acid carbons in the context of chronic HDAC inhibitor treatment (Figures 4, M–P). Acute pan-HDAC inhibitor treatment resulted in an overall reduced labeling of TCA cycle metabolites by glucose carbons despite an increase in the OCR, suggesting that glucose oxidation was not the primary source that yielded the enhanced oxidative phenotype by acute pan-HDAC inhibitor treatment (Supplemental Figure 4F). This picture changes in the context of chronic pan-HDAC inhibitor treatment, which resulted in enhanced labeling of most TCA cycle metabolites by glucose carbons (Supplemental Figure 8, A and B). However, overall, labeling of citric acid was reduced, and a decrease in the m+2 citrate was noted, suggesting that glucose oxidation was decreased (Figure 4N). Instead, we detected a relative increase in the m+3 citrate isotopolog, pointing toward enhanced anaplerosis (Figure 4N). In turn, enhanced anaplerosis likely serves as a driver to enable oxidation of other potential substrates, such as fatty acids. Although the citrate showed decreased labeling from glutamine, it revealed enhanced labeling from carbons derived from palmitic acid, with increased labeling of the m+2 citric acid isotopolog, indicative of enhanced fatty acid oxidation (FAO) following HDAC inhibitor treatment (Figure 4, O and P, and Supplemental Figure 8, C and D). It is noteworthy that Ro appeared to elicit the most prominent increase in the m+2 citric acid isotopolog derived from palmitic acid, suggesting that HDAC1/-2 inhibition may be critical for the engagement in FAO (Supplemental Figure 8E).
FDA-approved HDAC inhibitors reprogram tumor cell oxidative metabolism through H3K27ac modification of the PGC1α promoter.
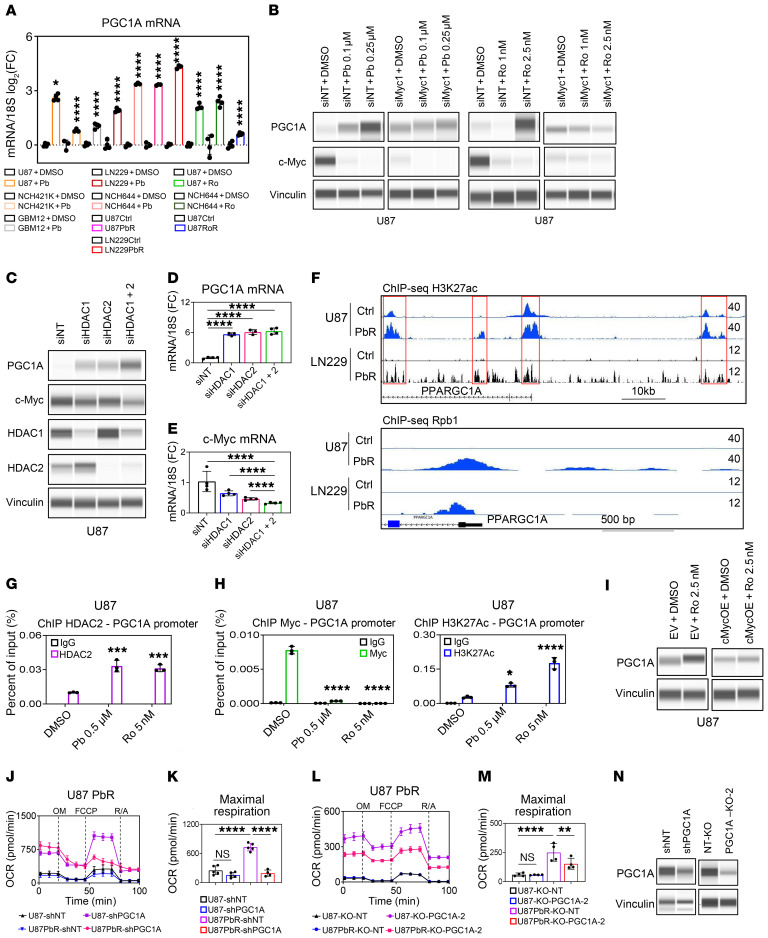
In seeking to identify regulators of the metabolic phenotype elicited by HDAC inhibition, we discovered that PGC1α, a master regulator of mitochondrial biogenesis (15), was increased on HDAC inhibitor–treated cells (Figure 5A and Supplemental Figure 9A). The increase in PGC1α mRNA and protein levels appeared to be inversely correlated with c-Myc levels (Figure 5, B–E). This finding is consistent with an earlier report (16) demonstrating that c-Myc suppresses PGC1α transcripts in model systems of pancreatic cancer stem cells.
Figure 5. Pan- and selective HDAC inhibitors upregulate PGC1α in a partially c-Myc–dependent manner to drive respiration.
(A) GBM cells were treated with Pb or Ro or chronically exposed to Pb or Ro (n = 3–4). (B) PCE analyses of U87 cells transfected with Myc siRNA and treated with Pb or Ro for 24 hours. (C) PCE analyses of U87 cells transfected with siRNA HDAC1, HDAC2, or a combination of both. (D and E) Real-time PCR analysis of U87 cells transfected with HDAC1 siRNA, HDAC2 siRNA, or a combination of both (n = 3–4). (F) ChIP-Seq profile of parental U87 and LN229 cells or U87 and LN229 cells chronically exposed to Pb with an antibody against H3K27ac or Rpb1. Shown are the respective tracks around the desert of the PPARGC1A (PGC1α) locus. (G) ChIP-qPCR (with anti-HDAC2 antibody) of the PGC1α promoter (c-Myc–binding region) from the indicated cell lysates (n = 3). (H) ChIP-qPCR of the PGC1α promoter (c-Myc–binding region) from the indicated cell lysates with either anti–c-Myc antibody or anti-H3K27ac antibody (n = 3). (I) PCE analysis of U87 cells transduced with a c-Myc construct and treated with 2.5 nM Ro for 24 hours. (J) Mitochondrial stress test of parental U87 cells or U87 cells chronically exposed to Pb and transduced with an shRNA against PGC1α (n = 4–5). O, oligomycin; F, FCCP; R/A, rotenone and antimycin A. (K) Maximal respiration data from the experiment in J. (L) Mitochondrial stress extracellular flux analysis of parental U87 cells or U87 cells chronically exposed to Pb and transduced with PGC1α sgRNAs (n = 4). (M) Maximal respiration data from the experiment in L. U87-KO-NT, nontargeting KO U87 cells; U87PbR-KO-NT, nontargeting KO U87 cells chronically exposed to Pb; U87-KO-PGC1A-2, PGC1A-2–KO U87 cells; U87PbR-KO-PGC1A-2, PGC1A-2–KO U87 cells chronically exposed to Pb. (N) PCE analysis of U87 cells transduced with an shRNA against PGC1α or PGC1α sgRNAs. Data represent the mean ± SD. Statistical significance was determined by 2-tailed Student’s t test (A) or 1-way ANOVA (D, E, G, H, K, and M). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We explored the mechanism by which HDAC inhibitors increase PGC1α and hypothesized that activation of histone marks might be involved in this process, since HDAC inhibitors mainly modulate histone proteins, as the name implies, by enhancing their acetylation. To this end, we performed ChIP and ChIP-Seq of H3K27ac and Rpb1. We noted H3K27ac and Rpb1 enrichments in the PGC1α promoter and several enhancer regions following pan-HDAC inhibitor treatment, in keeping with the transcriptional upregulation and increase in protein expression of PGC1α (Figure 5F). To further validate the hypothesis that HDAC inhibitor treatment increases the presence of H3K27ac at the promoter region of PGC1α, we conducted ChIP–quantitative PCR (ChIP-qPCR) assays with H3K27ac ChIP and amplified the PGC1α promoter region close to the transcription start site. We found that NCH644 cells acutely treated with the HDAC inhibitor Pb and LN229 and U87 cells chronically treated with Pb (PbR) had a marked increase in H3K27ac at the PGC1α promoter (Supplemental Figure 9B), coupled with an increase in PGC1α mRNA expression, in keeping with the hypothesis that HDAC inhibition increases PGC1α expression in part through enhanced H3K27 acetylation at the PGC1α promoter region. To account for global changes in the expression of activating histone marks, we performed Western blot analysis and noted an increase in H3K27ac levels in Pb-treated NCH644 cells and PbR-treated U87 cells, whereas in PbR-treated LN229 cells, there was a reduction in the H3K27ac mark (Supplemental Figure 9A). However, as mentioned, all cell cultures showed unequivocally enhanced binding of H3K27ac to the PGC1α promoter region, suggesting that global and local changes did not necessarily match. Since it has been reported that enhanced acetylation of H3K27 leads to diminished trimethylation of the bona fide repressive histone mark H3K27 (H3K27me3) (17), we tested whether a pan-HDAC inhibitor modulates H3K27me3 within the PGC1α promoter region. We noted a substantial suppression of H3K27me3 in PbR-treated LN229 cells, whereas PbR-treated U87 cells and Pb-treated NCH644 cells had less robust responses (Supplemental Figure 9B).
We wondered about the specificity of these effects and asked whether acute silencing of HDAC1, HDAC2, or their combination recapitulates the phenotype elicited by the inhibitors. Although silencing of HDAC1, HDAC2, or their combination suppressed c-Myc transcript and protein levels, we noted a sharp increase in PGC1α levels (Figure 5, C–E). An inverse relationship between c-Myc and PGC1α was also observed in xenografts following treatment with Pb (Supplemental Figure 9C). We continued to investigate the specific role of c-Myc and HDAC inhibitor–mediated increases in PGC1α by ChIP, silencing, and overexpression experiments (Figure 5, G–I, and Supplemental Figure 9, D–F). We identified several potential binding regions of c-Myc to the PGC1α promoter region and performed ChIP for c-Myc to demonstrate that c-Myc avidly binds to this region along with HDAC2, one of the targets of Pb and Ro (Figure 5, G and H, and Supplemental Figure 9, D and E). Notably, following HDAC inhibitor treatment, c-Myc binding to the PGC1α promoter region was drastically suppressed, coupled with enhanced acetylation of the same region (Figure 5H), suggesting that c-Myc may act as a suppressor of HDAC inhibitor–mediated increases in PGC1α. To further validate this hypothesis, we both silenced and overexpressed c-Myc in the context of HDAC inhibition. When c-Myc levels were down, neither Pb nor Ro upregulated PGC1α protein levels further (Figure 5B). Conversely, overexpression of c-Myc attenuated HDAC inhibitor–mediated increases in PGC1α (Figure 5I and Supplemental Figure 9F), in keeping with the notion that c-Myc is involved in regulating PGC1α and probably acts as a suppressor of PGC1α in the context of HDAC inhibitor treatment.
We assessed the impact of acute and chronic HDAC inhibitor treatment on the viability of cells transfected with PGC1α siRNA (Supplemental Figure 9, G–L). Silencing of PGC1α enhanced the cytotoxicity of Pb and Ro (Supplemental Figure 9, G and H). Similarly, silencing of PGC1α in GBM cells chronically exposed to Pb reduced their viability (Supplemental Figure 9I). Moreover, silencing of PGC1α counteracted the Pb-mediated increase in mitochondrial abundance and upregulation of isocitrate dehydrogenase subunit α (IDH3A) and SDHB (Supplemental Figure 9, J and K). Conversely, overexpression of PGC1α reduced the reduction of cellular viability elicited by Pb (Supplemental Figure 9, L and M). These findings establish PGC1α as a prosurvival factor in the context of HDAC inhibition.
Next, we assessed the impact of PGC1α on HDAC inhibitor–mediated oxidative metabolic reprogramming (Figure 5, J–N). We performed silencing and CRISPR/Cas9-mediated knockout of PGC1α in PbR-exposed GBM cells, since those cells display the highest induction of PGC1α coupled with a substantially elevated OCR. Silencing of PGC1α was confirmed by protein capillary electrophoresis (Figure 5N). We subsequently used the control and PGC1α-targeted clones for mitochondrial stress extracellular flux analysis to determine the OCR (Figure 5, J–M). Our analysis revealed that genetic interference with PGC1α reduced the OCR, an effect that was most pronounced in the maximal respiration parameter in Pb-exposed cells. It is noteworthy that nontreated GBM cells revealed few OCR alterations following genetic modulation of PGC1α. Akin to the partial reversal of the prooxidative effect elicited by HDAC inhibitors, we noted a suppression of HDAC inhibitor–mediated increases in mitochondrial abundance (Supplemental Figure 9J). These results position PGC1α as a target of HDACs and as a master regulator of HDAC inhibitor–mediated metabolic reprogramming.
Acute and chronic HDAC inhibitors activate FAO in vitro and in PDX models in vivo.
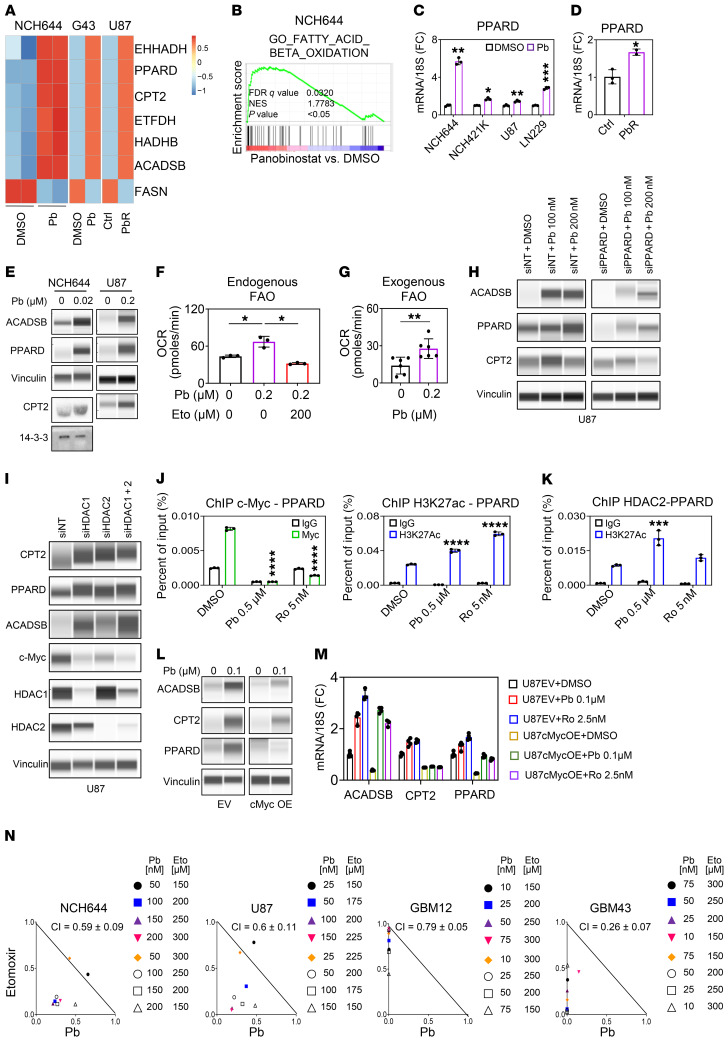
Our findings demonstrated that in NCH644 stem-like GBM cells, acute treatment with HDAC inhibitors led to a profound transcriptional reprogramming of lipid metabolism, with increases in gene sets related to catabolism of fatty acids, β-oxidation, and fatty acid transporters, as well as activation of FAO (Figure 6, A–D, and Supplemental Figure 10, A–D). These findings were independently confirmed by real-time PCR, which included upregulation of a master regulator of lipid metabolism, PPARD, mitochondrial fatty acid transporters, carnitine palmitoyltransferase 1A (CPT1A), and carnitine palmitoyltransferase II (CPT2), as well as enzymes related to the oxidation process of fatty acids, including very long-chain specific acyl-CoA dehydrogenase (ACADVL), short/branched chain–specific acyl-CoA dehydrogenase (ACADSB), electron transfer flavoprotein dehydrogenase (ETFDH), enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase (EHHADH), and hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit β (HADHB) (Supplemental Figure 10D). To exclude the possibility that these findings were not restricted to 1 condition per cell type, we extended our transcriptome analysis to a PDX model (GBM43) that was treated over several weeks with Pb in vivo as well as to PbR-treated U87 cell cultures (Figure 6A and Supplemental Figure 10C). With minor differences, the transcriptome data confirmed the findings from our earlier analysis in NCH644 GBM cells, positioning β-oxidation as a central pathway with potentially unique metabolic vulnerability following HDAC inhibitor exposure. We also validated the increase in FAO enzymes and transporters at the protein level (Figure 6E). Upstream of FAO, we noted an increase in the transcription factor PPARD at both mRNA and protein levels in several of our model systems (Figure 6E and Supplemental Figure 10D).
Figure 6. HDAC inhibitors drive lipid catabolism with activation of β-oxidation in a manner dependent on the transcription factor PPARD.
(A) Heatmap (mRNA) of NCH644 cells treated with 0.5 μM Pb for 24 hours, GBM43 (G43) cells treated with Pb (in vivo), and parent U87 cells and U87 cells chronically exposed to Pb. (B) GSEA plots of NCH644 cells treated with 0.5 μM Pb for 24 hours. (C) PPARD mRNA levels in GBM cells treated with 0.2 μM Pb for 24 hours (n = 3). (D) PPARD mRNA levels in parent U87 cells or U87 cells chronically exposed to Pb (n = 3). (E) PCE analysis of NCH644 cells and U87 cells treated with Pb for 24 hours (except for CPT2 and 14-3-3 [loading control] in NCH644 cells, which is standard Western blotting). (F) OCR in U87 GBM cells treated as indicated for 24 hours (n = 3). (G) OCR in U87 cells treated with 0.2 μM Pb in the presence of palmitate (n = 6). (H) U87 cells transfected with a siRNA against PPARD and treated with Pb for 24 hours. (I) U87 cells were transfected with siRNA against HDAC1, HDAC2, or a combination of both and analyzed by protein capillary electrophoresis. (J) ChIP-qPCR of the PPARD gene from the indicated cell lysates with either anti–c-Myc antibody or anti-H3K27ac antibody (n = 3). (K) ChIP-qPCR of the PPARD gene from the indicated cell lysates with anti-HDAC2 antibody (n = 3). (L) U87 cells were transduced with c-Myc and treated with 0.1 μM Pb for 24 hours. (M) mRNA levels in U87 cells transduced with c-Myc and treated with 0.1 μM Pb or 2.5 nM Ro for 24 hours (n = 3–4). (N) Isobolograms of NCH644, U87, GBM12, and GBM43 cells treated with Pb, with or without etomoxir, for 72 hours. Data represent the mean ± SD. Statistical significance was determined by 2-tailed Student’s t test (C, D, and G) or 1-way ANOVA (F, J, and K). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Eto, etomoxir.
We evaluated FAO in the context of HDAC inhibitor treatment. We performed extracellular flux analysis to determine whether the increase in the OCR related to HDAC inhibitors is attenuated through cotreatment with etomoxir, an inhibitor of FAO that interferes with CPT1A. Confirming this hypothesis, we found that both Vr- and Pb-mediated increases in the OCR were suppressed in the presence of etomoxir (Figure 6F and Supplemental Figure 10E). In a separate experiment, we evaluated the role of exogenous FAO in the presence or absence of an HDAC inhibitor through extracellular flux analysis. We found that HDAC inhibitor–treated cells had an increased OCR of exogenous FAO (Figure 6G). In alignment with the functional increase in FAO, we noted by LC/MS analysis that triglyceride levels were suppressed in cells exposed to Pb, implicating enhanced digestion of neutral lipids to fuel FAO (Supplemental Figure 10, F and G). Although FAO was increased, we also assessed whether fatty acid biosynthesis was affected by a pan-HDAC inhibitor. In agreement with our findings, chronic pan-HDAC inhibitor suppressed the 13C labeling of palmitic acid from glucose carbons (Supplemental Figure 10G).
The transcription factor PPARD is known to regulate FAO-related proteins. Therefore, we tested whether silencing of PPARD abrogates the increased expression of ACADSB and CPT2 and found that PPARD was involved in their upregulation (Figure 6H). To link a specific HDAC enzyme to the increase in PPARD, we silenced HDAC1, HDAC2, or their combination and found that silencing of either enzyme or a combination of both upregulated PPARD and related enzymes and suppressed c-Myc (Figure 6I). Next, we determined whether c-Myc acts upstream of PPARD. We performed ChIP for c-Myc and tested whether c-Myc binds to the promoter regions of PPARD, CPT2, and ACADSB. Indeed, we found that c-Myc protein interacted with these promoters and that this interaction was disrupted by HDAC inhibitors (Figure 6J and Supplemental Figure 11, A and B). Similarly, HDAC2 interacted with the PPARD promoter in the same location (Figure 6K). Following HDAC inhibitor treatment, we noticed an increased presence of H3K27ac at the Myc-binding region, consistent with a transcriptional increase in PPARD levels (Figure 6J and Supplemental Figure 11A). On the basis of these findings, we hypothesized that c-Myc acted as a suppressor of PPARD levels following HDAC inhibitor treatment. Indeed, forced expression of c-Myc attenuated the elevation of both PPARD and related targets at the protein and transcriptional levels following HDAC inhibitor treatment (Figure 6, L and M, and Supplemental Figure 11, C–F).
To demonstrate a prosurvival role of PPARD in the context of the HDAC inhibitor response, we silenced PPARD in GBM cells treated with HDAC inhibitors either acutely or chronically. Using a lentiviral shRNA targeting PPARD and through colony formation assays, we detected fewer colonies formed in cells transduced with PPARD shRNA compared with the cells transduced with nontargeting shRNA (Supplemental Figure 11, C and D). Similarly, silencing of PPARD enhanced the efficacy of the Pb-mediated reduction in cellular viability (Supplemental Figure 11E).
Given that we identified changes in both expression and function related to β-oxidation, we hypothesized that the increased reliance on FAO might affect the survival of HDAC inhibitor–treated cells and thus confer a unique vulnerability. To this end, we took advantage of the clinically validated drug etomoxir, which inhibits CPT1A and thereby FAO. The combination treatment of HDAC inhibitors plus etomoxir reduced the cellular viability of established, stem-like, and PDX GBM cells. (Figure 6N and Supplemental Figure 11, G–I). It is noteworthy that astrocytes were significantly less responsive to the combination treatment, suggesting that this combination treatment may exert a favorable toxicity profile (Supplemental Figure 11G).
To account for cell death, we treated NCH644, U87, and GBM12 cells with Pb in the presence or absence of etomoxir and stained the cells with annexin V and propidium iodide for apoptosis analysis. Our results showed that combination treatment with etomoxir and Pb, Vr, or Ro resulted in a significant enhancement of apoptosis in these cell cultures (Supplemental Figure 12, A–C). In order to ensure that these effects were not restricted to GBM cells and more broadly applicable, we conducted experiments in parallel using other solid tumors. We consistently obtained similar results in colon carcinoma cells (HCT116) and melanoma cells (A375), suggesting that the combination treatment of FAO inhibitors with HDAC inhibitors (Pb or Ro) is not restricted to GBM models and may be more broadly relevant to solid malignancies (Supplemental Figure 12A). We conducted Western blot analysis to assess the expression of markers related to apoptosis. The combination treatment resulted in substantial activation of effector and initiator caspases accompanied by enhanced cleavage of PARP, indicating cell death with apoptotic features (Supplemental Figure 12D). Since intrinsic apoptosis is primarily regulated by antiapoptotic B cell lymphoma 2 (Bcl-2) family members, we assessed expression levels of the most preeminent proteins in this group — Bcl-2, B cell lymphoma extra large (Bcl-xL), and myeloid cell leukemia 1 (Mcl-1) (18–23). Although expression of Bcl-2 and Bcl-xL was reduced by both Pb and the combination of etomoxir and Pb, we encountered a compensatory upregulation of Mcl-1 (Supplemental Figure 12E). To genetically verify that HDAC inhibitor–treated cells are more dependent on FAO, we silenced CPT1A, the target protein of etomoxir. Although silencing of CPT1A on its own elicited no apoptosis induction, the HDAC inhibitor caused more apoptotic cell death when CPT1A was silenced (Supplemental Figure 12, F–J). These findings provide additional specificity and suggest that etomoxir indeed enhanced the apoptotic effects of HDAC inhibitors by interfering with FAO.
Dual inhibition of HDAC and FAO leads to synergistic growth suppression of PDXs.
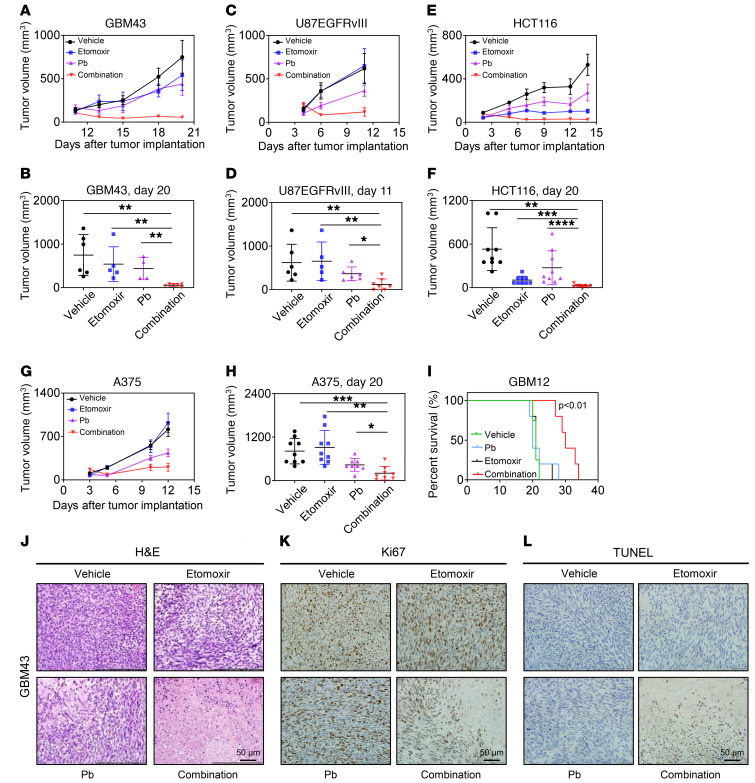
Finally, we evaluated our derived metabolic treatment strategy in PDX models. Given that Pb is an FDA-approved drug compound and etomoxir has reached the clinic as well, it was tempting to evaluate whether the 2 compounds in combination would elicit a synergistic growth reduction in vivo. To this purpose, we tested 1 PDX GBM model (GBM43), 1 conventional GBM model (U87 EGFRvIII), 1 colon carcinoma model (HCT116), and 1 BRAF V600E–mutated melanoma model (A375) (Figure 7, A–H, and Supplemental Figure 13, A–H). The variety of model systems related to solid organ malignancies ensured that the strategy of targeting aberrant metabolic pathways would be applicable to a broad range of solid cancers. In all 4 model systems tested, we found that the combination treatment of Pb and etomoxir synergistically reduced the growth of tumors compared with single-agent or vehicle-treated tumors, suggesting that the combination treatment was active in vivo. PDX orthotopic model systems are currently considered to be the most cutting-edge model systems. Thus, we tested our combination treatment in an orthotopic PDX GBM model system. We found that animals that received the combination treatment had a significantly longer overall survival than did vehicle-, Pb-, or etomoxir-treated mice, reinforcing the idea that this treatment may have potential clinical efficacy (Figure 7I). Histopathological examination demonstrated that the combination treatment led to a pronounced reduction in cellular density, a decrease in mitosis, and substantial induction of necrosis/apoptosis (Figure 7, J–L). Consistently, we found reduced Ki67 labeling with the combination treatment, whereas TUNEL staining was increased, reflecting the changes observed with conventional H&E staining. Collectively, our data provide a foundation for further evaluation of a drug combination therapy involving HDAC and FAO inhibitors.
Figure 7. Interference with oxidative energy metabolism along with HDAC inhibition synergistically reduces tumor growth in conventional and PDX models.
(A and B) GBM43 PDX tumor cells were implanted subcutaneously into immunocompromised mice. After tumor formation, 4 treatment groups were established: vehicle, etomoxir, Pb, or combined etomoxir and Pb treatment. Animals in the respective groups were treated 3 times a week (n = 3–6). (C and D) U87 EGFRvIII GBM cells were implanted subcutaneously into immunocompromised mice Mice were treated as indicated in A and B (n = 5–7). (E and F) HCT116 colon carcinoma cells were implanted subcutaneously into immunocompromised mice. Mice were treated in indicated in A and B (n = 9). (G and H) BRAF V600E–mutant A375 melanoma cells were implanted subcutaneously into immunocompromised mice. Mice were treated as indicated in A and B (n = 8–9). (I) GBM12 cells were implanted into the right striatum of nude mice that were then randomly divided into 4 treatment groups: vehicle, etomoxir, Pb, or combined treatment with etomoxir and Pb. Nine treatments were performed, and survival was analyzed using the Kaplan-Meier method. The log-rank test was used to assess statistical significance (n = 4–5). (J–L) At the end of the experiments in A and B, tumors from the individual groups were harvested for staining with H&E, TUNEL, or Ki67. Scale bars: 50 μm. Data represent the mean ± SD. Statistical significance was determined by 1-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Discussion
Targeting cancer cell metabolism as a strategy to identify novel treatments provides unique opportunities in the ever-growing landscape of biosynthetic and metabolic pathways and new drug discoveries (24, 25). ChIP-Seq and subsequent super-enhancer analysis in GBM cells informed us that the Warburg effect in tumors involves a landscape of super-enhancers (11, 26, 27) across genes related to glycolysis and associated biosynthetic pathways, such as fatty acid synthesis. We hypothesized that epigenetic targeting of the Warburg effect might be accomplished through HDAC inhibitors, since it was recently demonstrated that this class of compounds disrupts super-enhancers (28). In turn, we evaluated the metabolic vulnerabilities elicited by clinically approved selective and broad HDAC inhibitors. Indeed, our results showed that both Pb and Ro potently disrupted super-enhancers related to Warburg effect genes with associated transcriptional downregulation, providing evidence that HDAC inhibitors antagonize this anabolic metabolic pathway in tumor cells. Prior studies of HDAC inhibitors have shown some focused effects on tumor metabolism, such as modulation of the GLUT1 transporter and HIF-1α or fructose-bisphosphatase 1 (FBP1) transcription factors (29, 30), but have not provided an in-depth characterization of tumor cell metabolism.
Although HDAC inhibitors have shown efficacy in hematological malignancies, such as multiple myeloma and cutaneous T cell lymphoma (CTCL), the efficacy in solid tumors is less convincing (13, 31), necessitating strategies such as genetic and drug screens or metabolomics (as in this study). Through carbon tracing coupled with LC/MS analysis and extracellular flux experiments, our findings provide evidence that HDAC inhibitor treatment results in substantial metabolic reprogramming with a potent suppression of glycolysis and its associated biosynthetic pathways, thereby reversing the Warburg effect.
Mechanistically, we were able to show that broad and selective HDAC inhibition resulted in a profound suppression of mRNA and protein levels of c-Myc, a prosurvival factor and known master regulator of glycolysis (32, 33), and that, in turn, the reduction in c-Myc was involved in the HDAC inhibitor–mediated reduction of cellular viability and was a key regulator of HDAC inhibitor–mediated suppression of glycolysis. In addition, we found that the c-Myc locus contains a super-enhancer region (34) (as indicated by H3K27ac ChIP-Seq) in GBM tissue and cell cultures that is potently disrupted by Pb and Ro. Thus, our findings provide insights into the mechanisms by which broad and selective HDAC inhibitors control glycolysis. Notably, suppression of glycolysis was accompanied by an increase in the OCR, and HDAC inhibitor–treated cells became more dependent on OXPHOS for their survival. It has been consistently shown that increased activity of and reliance on OXPHOS are hallmarks of cancer stem and chemoresistant cells (15, 24, 35, 36).
Our ChIP-Seq and transcriptome analysis informed us about the potential mediator that primarily drives these oxidative metabolic changes. We noted an increase in H3K27ac in the promoter region of PPARGC1A, which encodes for a master regulator of oxidative metabolism, PGC1α (15, 16, 37). PGC1α has been implicated in oxidative metabolic reprogramming in the setting of the evolvement of therapeutic resistance (38). However, this has not been shown in the context of selective HDAC1/-2 inhibition in GBM models. Moreover, our findings suggest that an HDAC inhibitor–specific epigenetic mechanism facilitated the increase in PGC1α mRNA and protein levels. We were also able to link the HDAC inhibitor–mediated decline in c-Myc levels to the increase in PGC1α protein levels by demonstrating that c-Myc bound to the promoter region of PGC1α. Moreover, forced expression of c-Myc blocked the HDAC inhibitor–mediated increase in PGC1α, suggesting that c-Myc may act as a suppressor in this context. These findings are in line with an earlier study in pancreatic cancer stem cells that displayed a similar inverse relationship between these 2 transcription factors (25). Importantly, suppression of PGC1α rescued the oxidative metabolic phenotype elicited by HDAC inhibition and enhanced the reduction in cellular viability by HDAC inhibitors, suggesting that PGC1α is a key regulator in HDAC inhibitor–mediated metabolic reprogramming.
To fuel OXPHOS with reducing equivalents, HDAC inhibitor–treated cells or PDXs in vivo increased β-oxidation and reduced fatty acid biosynthesis. Consistently, HDAC inhibitor–treated cells and PDX models were more prone to cell death induction by etomoxir, an inhibitor of a major fatty acid transporter in the outer mitochondrial membrane. Mechanistically, we noted that the transcription factor PPARD was implicated in the metabolic effects elicited by selective HDAC inhibition and that, again, c-Myc acted as a suppressor of HDAC inhibitor–mediated upregulation of enzymes and transporters related to FAO.
These observations support a model system in which c-Myc drives aerobic glycolysis and suppresses OXPHOS and FAO, facilitating the transcription factors PGC1α and PPARD by directly binding to their promoter region. When c-Myc levels dropped following HDAC inhibitor treatment, the expression levels of PGC1α and PPARD rose. In turn, these 2 transcription factors appeared to mediate a prosurvival effect and oxidative phenotype in GBM cells. For future studies, it may be conceivable to target these 2 transcription factors to further enhance the efficacy of HDAC inhibitors and thereby abrogate metabolic reprogramming.
We acknowledge several limitations of our studies. Although we demonstrated the key findings in several model systems, including patient-derived cells, several mechanistic experiments were performed in established GBM cell cultures for technical reasons. Another limitation lies in the fact that we did not perform rescue experiments in in vivo settings. In addition, certain rescue experiments showed only a partial rescue, which indicates that our identified key players were not the sole mediators of resistance and response following HDAC inhibitor treatment. An implication for future study is to further develop the precise mechanism of enhancer formation around the PGC1α locus. Although our studies have shown that c-Myc regulates PGC1α expression at the promoter level, we have not determined whether enhancer regions of PGC1α are regulated by Myc or additional factors. Moreover, given that HDACs have multiple targets, we cannot exclude the possibility that pathways other than metabolism are involved in HDAC inhibitor response and resistance.
Collectively, through dissection of the alterations in tumor cell metabolism elicited by HDAC inhibitors, we were able to identify specific metabolic changes that in turn can be targeted for tumor therapy. Thus, this work further strengthens the appeal of analyzing metabolomics for the design of more sophisticated anticancer therapies.
Methods
Cell cultures and growth conditions.
All cell lines were incubated at 37°C and were maintained in an atmosphere containing 5% CO2. U87, LN229, HCT116, and A375 cell lines were obtained from the American Type Culture Collection (ATCC). The U87 EGFRvIII cell line was provided by Frank Furnari (UCSD, La Jolla, California, USA). GBM12, GBM14, and GBM43 cells were obtained from Jann Sarkaria (Mayo Clinic, Rochester, Minnesota, USA). Cells were cultured in DMEM (Thermo Fisher Scientific, MT10013CV) supplemented with 10% FBS (Gemini) and 100 μg/mL Primocin (InvivoGen, ant-pm-1). NCH644 and NCH421K stem-like glioma cells (Cell Line Services, 820403) were cultured in GBM-MG with 100 μg/mL Primocin. The PbR-treated U87 and LN229 cells were exposed to 100 nM Pb for 1 week. The respective cell line depository authenticated the cells. All cell lines were obtained between 2014 and 2019.
Cell viability assays.
Viability assays were performed as previously described (39, 40). Cells were seeded at 3000 cells/well in 96-well plates and treated with different doses of the indicated drugs for 72 hours. Tests were performed in triplicate according to the manufacturer’s instruction (CellTiter-Glo Luminescent Cell Viability Assay, Promega). The combination index (CI) was calculated using the drug-synergism concept based on the median-effect equation (Chou-Talalay), which serves as the foundation for calculating normalized isobolograms (41, 42). Based on the computational analysis, CI values of less than 1 indicate synergy, a CI value of 1 represents additivity, and CI values of more than 1 indicate antagonism.
Measurement of apoptosis and mitochondrial membrane potential.
Fifty thousand cells were seeded in each well of 12-well plates 1 day before treatment. Cells were treated either DMSO or the indicated drugs for 24 to 48 hours and harvested for staining following the manufacturer’s instructions. For apoptosis, the Annexin V Apoptosis Detection Kit (BD Pharmingen, BD 556419) was used. Mitochondrial membrane potential was stained with tetramethylrhodamine, ethyl ester (TMRE) (Mitochondrial Membrane Potential Kit, Cell Signaling Technology [CST], 13296S). The data were analyzed with FlowJo software, version 8.7.1 (Tree Star). In addition, mitochondria were stained with MitoTracker Red CMXRos (Thermo Fisher Scientific, M7512) and analyzed by flow cytometry or by fluorescence microscopy.
Transfection of siRNAs and transduction of lentiviral particles using shRNA and CRISPR/Cas9.
Fifty thousand cells were seeded 1 day before transfection in each well of 12-well plates. Transfections were performed with Lipofectamine RNAiMAX (Invitrogen, Thermo Fisher Scientific, 13778075) according to the manufacturer’s instructions. The following siRNAs were purchased from Dharmacon: PGC1α siRNA (L-005111-00-0005), PPARD siRNA (L-003435-00-0005), HDAC1 siRNA (L-003493-00-0005), HDAC2 siRNA (L-003495-02-0005), CPT1A siRNA (L-009749-00-0005), and CPT1A-4 siRNA (J-009749-09-0005). Myc-1 siRNA (CST, 6341) and Myc-2 siRNA (CST, 6552) were purchased from CST. The following shRNAs were purchased from Santa Cruz Biotechnology: PGC1α (sc-38884-V), AMPKA (sc-29673-V), and PPARD (sc-36305-V). The CRISPR/Cas9 PGC1α-knockout lentivirus (NM_013261) was purchased from Applied Biological Materials. Cells were infected in the presence of 8 μg/mL polybrene and were selected with puromycin.
Extracellular flux analysis and FAO assay.
The Seahorse XFe24 analyzer was used for extracellular flux analysis, including the mitochondrial stress assay, the glycolysis stress assay, and the FAO assay following the manufacturer’s instructions (Agilent Technologies). GBM cells were seeded in XFe24 cell culture microplates (Agilent Technologies) at 30,000 cells/well in 250 μL DMEM containing 5 mM glucose, 1 mM glutamine, and 10% FBS and were allowed to attach overnight. Treatments were performed using the indicated compounds or the corresponding solvents in DMEM 5 mM glucose, 1 mM glutamine, and 1.5% FBS for 24 hours. Mitochondrial stress assays were run under the following media conditions: 10 mM glucose, 2 mM glutamine, and 1 mM pyruvate in assay medium, and 2 μM oligomycin, 2 μM trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP), and 0.5 μM rotenone/antimycin A were injected during the measurements. During the assay, 10 mM glucose was injected followed by 1 μM oligomycin and finally by 50 mM 2-deoxy-d-glucose (2-DG). ECAR reads were taken and used to calculate the glycolytic reserve and glycolytic capacity according to the manufacturer’s algorithms. The FAO assay was run under the following media conditions: 0.5 mM glucose, 1 mM glutamine, 0.5 mM l-carnitine, and palmitic acid conjugated to BSA according to the manufacturer’s instructions.
Western blot analysis and protein capillary electrophoresis.
Specific protein expression in cell lines was determined by Western blot analysis or protein capillary electrophoresis on a Wes instrument (ProteinSimple) as described previously (40). Briefly, 20–30 μg protein was loaded onto gradient precast gels supplied by Invitrogen (Thermo Fisher Scientific, NP0321BOX). Primary antibody incubations were performed overnight at 4°C. For standard Western blotting, the following antibodies were applied: acetyl–histone H3 (Lys27) (D5E4) (CST, 8173, 1:500), histone H3 (CST, 14269; 1:500), H3K27Me3 (CST, 9733; 1:500), SDHA (Abcam, ab123545; 1:500), OXPHOS (Abcam, ab110411;1:500), 14-3-3 (Santa Cruz Biotechnology, sc-59419; 1:500), PARP (CST, 9532; 1:500), cCP9 (CST, 7237; 1:500), cCP3 (CST, 9665; 1:500), Bcl-xL (CST, 2764; 1:500), Bcl-2 (CST, 4223, 1:500), Mcl-1 (CST, 5453; 1:500), CPT1A (CST, 12252; 1:500), and β-actin (MilliporeSigma, A1978, clone AC15; 1:2000). For protein capillary electrophoresis, the following antibodies were used: HK2 (CST, 2106S, 1:25); PGC1α (Novus Biologicals, NBP1-04676, 1:25); GLUT1 (CST, 12939; 1:25), LDHA (CST, 3582, 1:25); GAPDH (CST, 5174; 1:100); phosphorylated AMPKA (p-AMPKA) (CST, 2531; 1:25); AMPKA (CST, 5831; 1:25); p–acetyl-CoA carboxylase (p-ACC) (CST, 3661; 1:25); ACC (CST, 3662S; 1:25); HDAC1 (CST, 34589; 1:25); HDAC2 (CST, 57156;1:25), ACADSB (Thermo Fisher Scientific, PA5-29958; 1:25); CPT2 (Thermo Fisher Scientific, PA5-12217; 1:25); PPARD (Abcam, ab23673; 1:25); c-Myc (CST, 13987, 1:25); and vinculin (Abcam, ab129002, 1:500). The HRP-linked secondary antibodies were from Santa Cruz Biotechnology. Western blots were visualized on the Azure (C300) imaging system.
Subcutaneous xenograft model.
A total of 1 × 106 U87-EGFRvIII GBM cells, HCT116 colon carcinoma cells, A375 melanoma cells, or GBM12 and GBM43 PDX tumor cells were implanted subcutaneously into the flanks of 6- to 8-week-old SCID SHO mice as described before (40). Intraperitoneal treatments and tumor measurements were performed 3 times a week on the days indicated in each figure. Drugs were dissolved in a mixture of drug, cremophor EL (MilliporeSigma, 61791-12-6), ethyl alcohol (Pharmco-Aaper, 200 Proof, Gamma Irradiated Sterile), and PBS at a ratio of 10:32:8:50 (v/v/v/v). The drug doses used: 5 mg/kg Pb and 20 mg/kg etomoxir. Tumor size was measured with a caliper and calculated as follows: (length × width2)/2. Mouse body weights were monitored at each time point. At every final time point, the mice were sacrificed for harvesting of tumors, and representative tumors were photographed.
Orthotopic GBM PDX model.
For the GBM12 orthotopic model, 300,000 cells were intracranially injected as described earlier (3 mm lateral, 1 mm anterior of the bregma, and 3 mm down) (40). The drugs were dissolved in a mixture of drug, cremophor EL, ethyl alcohol, and PBS at a ratio of 10:32:8:50 (v/v/v/v). The following drug doses were used: 5 mg/kg Pb and 20 mg/kg etomoxir. Nine treatments were performed starting from day 5 until day 19 after cell implantations.
TUNEL and Ki67 staining.
The paraffin-embedded sections were dewaxed, rehydrated, and incubated in proteinase K (Agilent Technologies, Dako) for 5 minutes at 37°C. For TUNEL staining, the slides were exposed to TUNEL reaction mixture for 1 hour at 37°C, and the reaction was terminated in the converter peroxidase (POD) solution for 30 minutes at 37°C. TUNEL staining was highlighted with diaminobenzidine, and hematoxylin was used for nonspecific nuclear staining. For Ki67 staining, antigen retrieval was performed using citrate buffer followed by heating and incubation with Ki67 (Dako GA626) for 90 minutes at room temperature (RT). The slides were incubated with horse anti–mouse IgG (1:200) for 30 minutes and then incubated in ABC-peroxidase solution (1:50) for 30 minutes at RT.
LC/MS analysis, ATP levels, and isotope tracing.
LC/MS was performed in accordance with the isolation procedures established by the White Head Institute Core facilities as described previously (43, 44). Briefly, cells were isolated by adding a mixture of methanol/water/chloroform (600 μL/300 μL/400 μL in 6-well plates). The methanol contained internal standards (Metabolomics Amino Acid Mix Standard, MSK-A2-1.2, Cambridge Isolotope Laboratories). The polar and nonpolar layers were separated and dried under nitrogen. The polar samples were dissolved in water, whereas the lipid fractions were solubilized in a mixture of acetonitrile, 2-propanol, and water (65:30:5, v/v/v). The samples were analyzed by liquid chromatography–high-resolution mass spectrometry (LC/HRMS). Nucleotides, including ATP, were measured by LC/MS.
For isotope tracing experiments, the cells were starved in nutrient-free DMEM for 1 hour. Thereafter, the cells were exposed to media containing either 25 mM (U-13C6) d-glucose (Cambridge Isotope Laboratories), 4 mM (U-13C5) l-glutamine (Cambridge Isotope Laboratories), or 100 μM (U-13C16) palmitic acid (Cambridge Isotope Laboratories) for 24 hours in the presence of 1.5% dialyzed FBS (Thermo Fisher Scientific). Polar metabolites were extracted and read on an Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) linked to a Vanquish Ultra-High-Performance Liquid Chromatography (UHPLC) System (Thermo Fisher Scientific). A script developed by the Metabolomics Core Facility at Weill Cornell Medicine (New York, New York, USA) was used for data analysis (45). The energy status of cells was tested by measuring ATP levels. The LC/MS data were normalized to protein levels with a Pierce BCA Protein Assay Kit (23225, Thermo Fisher Scientific).
Microarray and gene set enrichment analysis.
Transcriptome and gene set enrichment analysis (GSEA) was performed as previously described (39). Data from the experiment were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE108958, GSE103961, and GSE108957).
ChIP-qPCR and ChIP-Seq.
Enzymatic ChIP assays were performed in accordance with the manufacturer’s instructions manual (SimpleChIP Enzymatic Chromatin IP Kit, CST, 9003). For each immunoprecipitation, approximately 4 × 106 GBM cells were cross-linked using 1% formaldehyde in complete cell medium for 10 minutes at RT and subsequently quenched with glycine for 5 minutes. Nuclei preparation and chromatin digestion were performed according to standard protocols. ChIP experiments were performed using anti-H3K27ac antibody (CST, 4535, 10 μL/sample), anti–rabbit IgG antibody (CST, 2729, 2 μL/sample), or anti-Rpb1 antibody (CST, 14958, 10 μL/sample). ChIP DNA was eluted, purified, and tested by real-time PCR. The primer sequences are shown in Supplemental Table 1. The signal over input was calculated by a standard formula as described in the manufacturer’s instruction manual (CST). Input and ChIP H3K27ac or Rbp1 data were submitted for next-generation sequencing (Illumina HiSeq 4000, single-read, 50 bp [SR50]) followed by library preparation. Computational analysis of ChIP-Seq data is provided in the Supplemental Methods.
Electron microscopy.
GBM cells were fixed in buffered 2.5% glutaraldehyde in cacodylate buffer for 1 hour at RT. Osmium tetroxide (1%) was used for extended fixation in the same buffer. After dehydration, Lx-112 (Ladd Research Industries) and Embed 812 (Electron Microscopy Sciences [EMS]) were used for the embedment process of the relevant GBM cells. Sections (60-nm) were cut on the MT-Power-Trome XL ultramicrotome. Uranyl acetate and lead citrate were used for staining. Analysis of the slides was performed with a JEOL JEM-1200 EXII electron microscope. ORCA-HR digital camera (Hamamatsu) was used to record the images.
Statistics.
Statistical significance was assessed by a 2-tailed Student’s t test or ANOVA (for multiple comparisons) using GraphPad Prism, version 8.0 (GraphPad Software). Three replicates were performed unless otherwise described. A P value of 0.05 or less was set as the level of statistical significance. For drug synergism analysis, CompuSyn software (ComboSyn) was used to compute the CI (CI <1 synergistic, CI = 1 additive and CI >1 as antagonistic). IC50 values were calculated by nonlinear regression.
Study approval.
All procedures were in accordance with animal welfare regulations and approved by the IACUC of the Columbia University Medical Center.
Author contributions
TTTN, YZ, and MDS designed research studies. TTTN, YZ, ES, CS, CT, EB, A. Mela, NH, and A. Mahajan conducted the experiments. TTTN, YZ, JZ, AOH, ZL, MMC, and MDS analyzed data. TTTN, YZ, MAW, GKM, JNB, PC, and MDS helped with the writing, review, and/or revision of the article. CMQ provided technical and material support (Seahorse Analyzer). TTTN and YZ share the first authorship. TTTN appears first in the author list because she was primarily involved with the preparation and revision of the manuscript until its publication.
Supplementary Material
Acknowledgments
MDS is supported by the National Institute of Neurological Disorders and Stroke (NINDS), NIH (R01NS095848, R01NS102366, and K08NS083732); the Louis V. Gerstner Jr. Scholars Program (2017–2020); and an American Brain Tumor Association Discovery Grant 2017 (DG1700013). TTTN is supported by an American Brain Tumor Association Basic Research Fellowship in Memory of Katie Monson (BRF1900018). Transcriptome analysis was supported by a NIH Clinical and Translational Science Award (CTSA) (UL1-TR001430, to the Boston University Microarray and Sequencing Resource Core Facility). These studies used the resources of the Cancer Center Flow Core Facility funded in part through center grants P30CA013696 from the National Cancer Institute (NCI), NIH, and S10RR027050 from the Office of the Director, NIH. The metabolomics analysis shown in Figure 2, E, F, and I, Figure 4M, and Supplemental Figure 3, E–H, was performed by the Whitehead Institute’s Metabolite Profiling Facility (Cambridge, Massachusetts, USA).
Version 1. 04/21/2020
In-Press Preview
Version 2. 06/08/2020
Electronic publication
Version 3. 07/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(7):3699–3716.https://doi.org/10.1172/JCI129049.
Contributor Information
Yiru Zhang, Email: yiruzh@gmail.com.
Enyuan Shang, Email: enyuan.shang@bcc.cuny.edu.
Chang Shu, Email: cs485@columbia.edu.
Consuelo Torrini, Email: ct2920@cumc.columbia.edu.
Junfei Zhao, Email: jz2835@cumc.columbia.edu.
Elena Bianchetti, Email: eb2985@cumc.columbia.edu.
Angeliki Mela, Email: am2904@cumc.columbia.edu.
Nelson Humala, Email: nh2560@cumc.columbia.edu.
Aayushi Mahajan, Email: am4518@cumc.columbia.edu.
Arif O. Harmanci, Email: Arif.O.Harmanci@uth.tmc.edu.
Zhengdeng Lei, Email: leizhengdeng@hotmail.com.
Mark Maienschein-Cline, Email: mmaiensc@gmail.com.
Mike-Andrew Westhoff, Email: Andrew.westhoff@uni-ulm.de.
Georg Karpel-Massler, Email: Georg.Karpel@uniklinik-ulm.de.
Jeffrey N. Bruce, Email: jnb2@columbia.edu.
Peter Canoll, Email: pc561@cumc.columbia.edu.
Markus D. Siegelin, Email: ms4169@cumc.columbia.edu.
References
- 1.Masui K, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18(5):726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuang R, et al. GLUT3 upregulation promotes metabolic reprogramming associated with antiangiogenic therapy resistance. JCI Insight. 2017;2(2):e88815. doi: 10.1172/jci.insight.88815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21(6):805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Sivanand S, et al. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol Cell. 2017;67(2):252–265.e6. doi: 10.1016/j.molcel.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, et al. PHGDH Defines a metabolic subtype in lung adenocarcinomas with poor prognosis. Cell Rep. 2017;19(11):2289–2303. doi: 10.1016/j.celrep.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 6.Pacold ME, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12(6):452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeNicola GM, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47(12):1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532(7598):255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagaraja S, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017;31(5):635–652.e6. doi: 10.1016/j.ccell.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez GJ, et al. Genome-wide dose-dependent inhibition of histone deacetylases studies reveal their roles in enhancer remodeling and suppression of oncogenic super-enhancers. Nucleic Acids Res. 2018;46(4):1756–1776. doi: 10.1093/nar/gkx1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vazquez F, et al. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23(3):287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancho P, et al. MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22(4):590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Pasini D, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of polycomb group target genes. Nucleic Acids Res. 2010;38(15):4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong J, Tan S, Zou F, Yu J, Zhang L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene. 2017;36(6):787–796. doi: 10.1038/onc.2016.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan R, et al. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell. 2017;32(6):748–760.e6. doi: 10.1016/j.ccell.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karpel-Massler G, Ishida CT, Zhang Y, Halatsch ME, Westhoff MA, Siegelin MD. Targeting intrinsic apoptosis and other forms of cell death by BH3-mimetics in glioblastoma. Expert Opin Drug Discov. 2017;12(10):1031–1040. doi: 10.1080/17460441.2017.1356286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elgendy M, et al. Dual modulation of MCL-1 and mTOR determines the response to sunitinib. J Clin Invest. 2017;127(1):153–168. doi: 10.1172/JCI84386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faber AC, et al. Assessment of ABT-263 activity across a cancer cell line collection leads to a potent combination therapy for small-cell lung cancer. Proc Natl Acad Sci USA. 2015;112(11):E1288–E1296. doi: 10.1073/pnas.1411848112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preuss E, Hugle M, Reimann R, Schlecht M, Fulda S. Pan-mammalian target of rapamycin (mTOR) inhibitor AZD8055 primes rhabdomyosarcoma cells for ABT-737-induced apoptosis by down-regulating Mcl-1 protein. J Biol Chem. 2013;288(49):35287–35296. doi: 10.1074/jbc.M113.495986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viale A, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gimple RC, et al. Glioma stem cell-specific superenhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov. 2019;9(9):1248–1267. doi: 10.1158/2159-8290.CD-19-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack SC, et al. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature. 2018;553(7686):101–105. doi: 10.1038/nature25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gryder BE, et al. Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat Commun. 2019;10(1):3004. doi: 10.1038/s41467-019-11046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemoine M, et al. The pan-deacetylase inhibitor panobinostat induces cell death and synergizes with everolimus in Hodgkin lymphoma cell lines. Blood. 2012;119(17):4017–4025. doi: 10.1182/blood-2011-01-331421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, et al. Inhibiting histone deacetylases suppresses glucose metabolism and hepatocellular carcinoma growth by restoring FBP1 expression. Sci Rep. 2017;7:43864. doi: 10.1038/srep43864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DH, Ryu HW, Won HR, Kwon SH. Advances in epigenetic glioblastoma therapy. Oncotarget. 2017;8(11):18577–18589. doi: 10.18632/oncotarget.14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolpaw AJ, Dang CV. MYC-induced metabolic stress and tumorigenesis. Biochim Biophys Acta Rev Cancer. 2018;1870(1):43–50. doi: 10.1016/j.bbcan.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Mair R, et al. Metabolic imaging detects low levels of glycolytic activity that vary with levels of c-Myc expression in patient-ferived xenograft models of glioblastoma. Cancer Res. 2018;78(18):5408–5418. doi: 10.1158/0008-5472.CAN-18-0759. [DOI] [PubMed] [Google Scholar]
- 34.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24(6):777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang G, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest. 2016;126(5):1834–1856. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farge T, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7(7):716–735. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo C, et al. A PGC1α-mediated transcriptional axis suppresses melanoma metastasis. Nature. 2016;537(7620):422–426. doi: 10.1038/nature19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23(3):302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karpel-Massler G, et al. Induction of synthetic lethality in IDH1-mutated gliomas through inhibition of Bcl-xL. Nat Commun. 2017;8(1):1067. doi: 10.1038/s41467-017-00984-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karpel-Massler G, et al. Inhibition of mitochondrial matrix chaperones and antiapoptotic Bcl-2 family proteins empower antitumor therapeutic responses. Cancer Res. 2017;77(13):3513–3526. doi: 10.1158/0008-5472.CAN-16-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen TTT, et al. Activation of LXR receptors and inhibition of TRAP1 causes synthetic lethality in solid tumors. Cancers (Basel) 2019;11(6):E788. doi: 10.3390/cancers11060788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 43.Franco J, Balaji U, Freinkman E, Witkiewicz AK, Knudsen ES. Metabolic reprogramming of pancreatic cancer mediated by CDK4/6 inhibition elicits unique vulnerabilities. Cell Rep. 2016;14(5):979–990. doi: 10.1016/j.celrep.2015.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gui DY, et al. Environment dictates dependence on mitochondrial complex I for NAD+ and aspartate production and determines cancer cell sensitivity to Metformin. Cell Metab. 2016;24(5):716–727. doi: 10.1016/j.cmet.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goncalves MD, et al. Fenofibrate prevents skeletal muscle loss in mice with lung cancer. Proc Natl Acad Sci USA. 2018;115(4):E743–E752. doi: 10.1073/pnas.1714703115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.