Abstract
Vaccination is a mainstay in preventive medicine, reducing morbidity and mortality from infection, largely by generating pathogen-specific neutralizing antibodies. However, standard immunization strategies are insufficient with increasing age due to immunological impediments, including defects in T follicular helper (Tfh) cells. Here, we found that Tfh generation is inversely linked to the expression of the ecto-NTPDase CD39 that modifies purinergic signaling. The lineage-determining transcription factor BCL6 inhibited CD39 expression, while increased Tfh frequencies were found in individuals with a germline polymorphism preventing transcription of ENTPD1, encoding CD39. In in vitro human and in vivo mouse studies, Tfh generation and germinal center responses were enhanced by reducing CD39 expression through the inhibition of the cAMP/PKA/p-CREB pathway, or by blocking adenosine signaling downstream of CD39 using the selective adenosine A2a receptor antagonist istradefylline. Thus, purinergic signaling in differentiating T cells can be targeted to improve vaccine responses, in particular in older individuals who have increased CD39 expression.
Keywords: Aging, Vaccines
Keywords: Adaptive immunity, Cellular senescence, T cells
Introduction
Vaccination is one of the most successful and safest interventions in modern medicine and has facilitated the extinction of the smallpox virus and the nearly complete eradication of other devastating viruses such as the poliomyelitis virus. Although vaccination programs have been extremely successful in children, they have been less beneficial in adults, particularly in older populations or individuals who have an underlying disease (1). The majority of vaccines provide protection through the induction of pathogen-specific antibodies (Abs) (2) that require the generation and expansion of follicular helper T (Tfh) cells. Tfh cells provide help to B cells and are essential for germinal center (GC) formation, affinity maturation, and the development of memory B cells. Harnessing the generation of Tfh cells is therefore pivotal for improving vaccination strategies.
Unlike effector CD4+ T cell lineages that develop from naive T cells with environmental cues from specific cytokines, there is no single event that defines Tfh cell differentiation. Rather, Tfh cell differentiation is a multistep, multisignal process (3). Signals driving Tfh differentiation are less restrictive than those for CD4+ effector T cells, permitting Ab production irrespective of the pathogen’s nature. Tfh cell generation occurs in the context of a wide variety of conditions, while effector T cell lineages develop in response to unique sets of cytokines and are tailored to the need to defend specific infections. As a result, Tfh differentiation necessarily accommodates a large degree of heterogeneity and signals can be redundant. Additionally, signals are species specific and Tfh lineage-determining cytokines differ between mouse and human, complicating the development of interventions that target Tfh generation.
Attempts to improve Tfh generation have so far focused on exploring novel adjuvants to modify the initial stimulation. These adjuvants may function by slowing the release of the antigen and/or stimulating innate cells to secrete mediators that drive differentiation toward effector cell lineages such as Th1 or Th17 versus Tfh cells. Slow-delivery immunization has been attractive because it mimics natural infection more accurately (4). A recent study in nonhuman primates showed that slow delivery using nonmechanical osmotic pumps or 2-week escalating-dose immunization induced higher frequencies of GC Tfh cells and generated protective Abs against difficult antigens such as the HIV envelope (5).
The alternative approach of directly targeting differentiating T cells has been less explored. Early determinants favoring Tfh cells include T cell receptor–major histocompatibility complex II dwell time, IL-6 (or IL-12 in humans), and inducible costimulator (ICOS), while IL-2 is a potent inhibitor of Tfh cell differentiation (6, 7). The lineage-determining transcription factor BCL6 functions by regulating genes involved in cell migration as well as repressing alternate states. In particular, BCL6 represses PRDM1 encoding BLIMP1 (8), which is strongly associated with CD4+ effector states. Strikingly, CD4+ T cells from older individuals have a propensity for differentiating into effector T cell lineages that preferentially express the transcription factors BLIMP1, RUNX3, and JUN/JUNB while lacking BCL6 and TCF1, and are inefficient in providing B cell help (9, 10). This bias in differentiation is at least in part determined by the sustained activation of signaling pathways due to the increased expression of miR-21 and is associated with the increased expression of the ecto-ATPase CD39 in older individuals (9, 11).
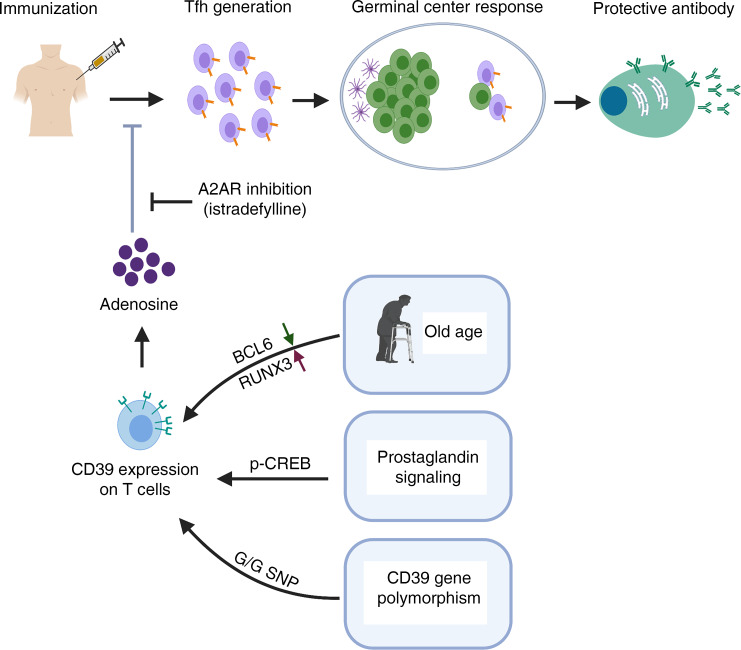
Here, we observed that individuals lacking CD39 expression due to a frequent germline polymorphism in close proximity to the ENTPD1 transcription start site have increased frequencies of circulating Tfh and greater induction of Tfh cell generation in vitro. We found that BCL6 and CD39 inversely cross-regulate each other, supporting the model that CD39 expression is an important checkpoint in Tfh differentiation. In 2 mouse models, infection with lymphocytic choriomeningitis virus (LCMV) or immunization with 4-hydroxy-3-nitrophenylacetyl hapten–conjugated ovalbumin (NP-OVA), CD39 exerted an inhibitory function on Tfh generation. We propose that this checkpoint can be targeted to improve vaccine responses, particularly in elderly individuals who have increased CD39 expression.
Results
Failure of Tfh cells to induce expression of CD39.
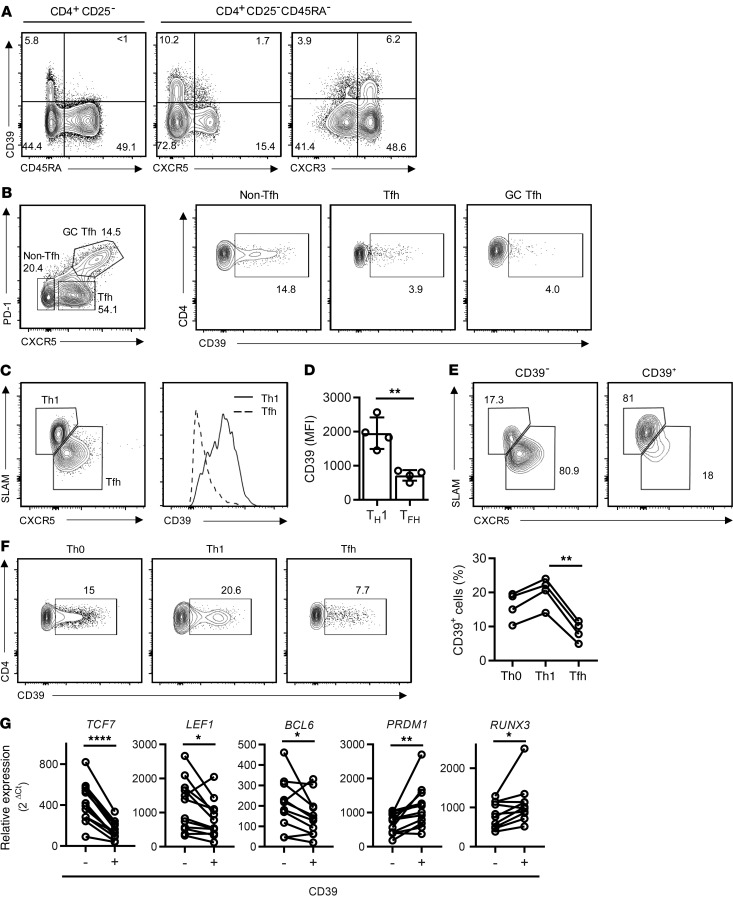
In addition to its constitutive expression on Tregs, CD39 is inducible in a subset of CD4+ and CD8+ T cells (9). To determine whether CD39 expression is lineage specific and may therefore be regulated by lineage-determining transcription factors, we examined the expression of CD39 on circulating human T cells. Excluding CD39+CD25+CD4+ Tregs (12), CD39 was preferentially expressed in the subset of CD45RA– Th1 memory cells that stained positive for CXCR3 (Figure 1A). In contrast, CD39 was absent on CXCR5+ circulating Tfh cells, consistent with our previous finding that activated CD39+ cells fail to help B cells to differentiate (9). A similar trend in lineage specificity was seen in T cells isolated from tonsil tissue (Figure 1B). CD39 was expressed on a subset of non-Tfh cells, while expression on Tfh and GC Tfh was infrequent. To provide direct evidence that Tfh differentiation and CD39 expression are inversely related, we used the Armstrong LCMV infection mouse model. B6 mice, adoptively transferred with LCMV-specific SMARTA CD4+ T cells, were infected and CD39 expression was assessed on SMARTA cells on day 8 after infection. Indeed, CD39 expression was higher on Th1 than on Tfh cells (Figure 1, C and D). The vast majority of CD39– cells displayed the phenotype of Tfh cells, while CD39+ cells were almost exclusively Th1 cells (Figure 1E). This pattern was reproduced with human naive CD4+ T cells cultured under Th1- or Tfh-polarizing conditions (Figure 1F). Transcription factor profiles of purified CD39+ and CD39–CD4+ T cells that were activated and cultured under nonpolarizing conditions were consistent with lineage-specific expression of CD39. CD39– T cells had higher expression of TCF7, LEF1, and BCL6 and lower expression of PRDM1 and RUNX3 than their CD39+ counterparts (Figure 1G).
Figure 1. Failure of Tfh cells to induce expression of CD39.
(A) CD39 expression in subsets of human peripheral blood CD4+CD25– T cells: representative contour plots. (B) CD39 expression on CD4+ T cell subsets in human tonsils; contour plots are representative of 3 samples. (C–E) SMARTA CD4+ T cells were transferred into B6 mice and analyzed 8 days after LCMV infection. CD39 expression was determined on Th1 (SLAMhiCXCR5lo) and Tfh (SLAMloCXCR5hi) SMARTA cells. Representative contour plot and histogram (C) and summary data from 1 experiment with 4 mice representative of 3 experiments; (D) data are shown as mean ± SEM. (E) Contour plots of Th1 or Tfh in gated CD39– and CD39+ SMARTA CD4+ cells. (F) Naive CD4 cells isolated from human PBMCs were activated with anti-CD3/anti-CD28 Dynabeads under nonpolarizing (Th0) or Th1- or Tfh-polarizing conditions; CD39 expression was assessed on day 5. (G) CD39– and CD39+ CD4+ T cells were isolated 5 days after stimulation with anti-CD3/anti-CD28 Dynabeads and profiled for the expression of selected transcription factors by qPCR. Data were compared by 2-tailed paired t test (D and G) or 1-way ANOVA with Tukey’s post hoc test (F). *P < 0.05; **P ≤ 0.01; ****P ≤ 0.0001.
Transcriptional regulation of ENTPD1 through RUNX3 and BCL6.
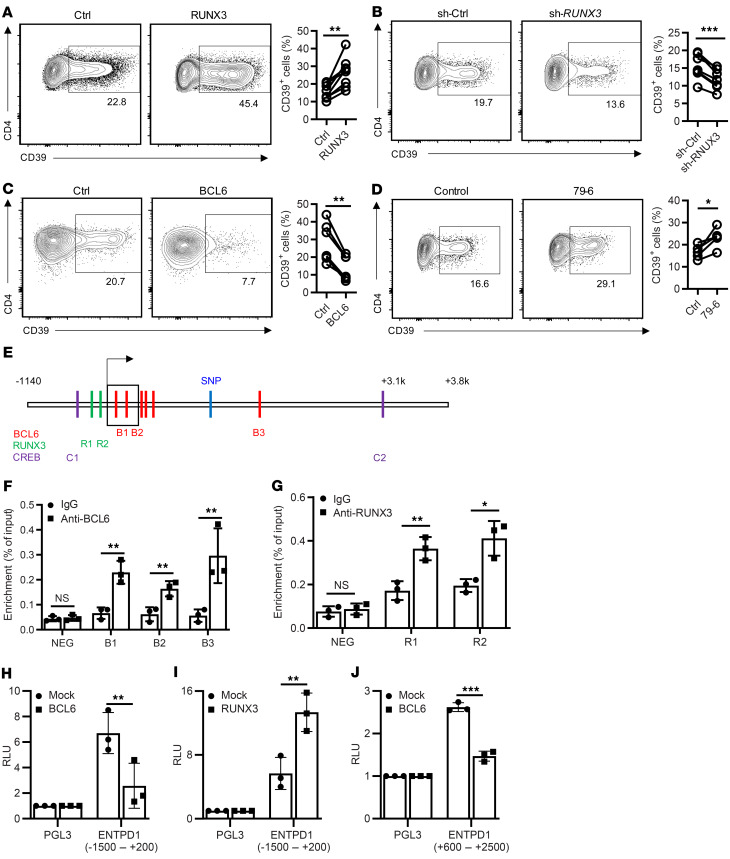
The correlation between CD39 expression and functional differentiation suggested that lineage-determining transcription factors are involved in transcriptional control of ENTPD1. Further indirect evidence for a causal relationship came from kinetic studies. CD39 expression progressively increased starting on day 3 after activation when RUNX3 increased and BCL6 decreased (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI132417DS1). RUNX3 expression was higher and BCL6 was lower in CD8+ than in CD4+ T cells, which may explain the observation that activation-induced expression of CD39 was more frequent in CD8+ than in CD4+ T cells (Supplemental Figure 1, D–F). Moreover, CD4+ T cells from older individuals that are more prone to induce CD39 expression upon activation had increased RUNX3 and decreased BCL6 expression on day 4 after activation (Supplemental Figure 1, G and H). We followed up on these correlative observations with mechanistic studies using overexpression and silencing of RUNX3 and BCL6. RUNX3 overexpression increased the frequencies of CD39+ cells for CD4+ cells (Figure 2A) and for CD8+ cells (Supplemental Figure 1I). Conversely, silencing of RUNX3 decreased the frequency of CD39+ T cells (Figure 2B). In contrast, ectopic expression of BCL6 repressed ENTPD1 transcription in both CD4+ and CD8+ cells (Figure 2C and Supplemental Figure 1J), while pharmacological inhibition of BCL6 activity increased the frequencies of CD39+CD4+ cells (Figure 2D).
Figure 2. Transcriptional regulation of ENTPD1.
(A–C) Human peripheral T cells were transduced with RUNX3-overexpressing lentivirus (A), RUNX3 shRNA lentivirus (B), or BCL6-overexpressing lentivirus (C). Cells were cultured for 5 days after anti-CD3/anti-CD28 Dynabead activation. CD39 expression in CD4+GFP+ cells was determined by flow cytometry. Results are shown as representative contour plots (left) and summary plots from 6 to 8 experiments (right). (D) Purified human naive CD4+ T cells were activated by anti-CD3/anti-CD28 Dynabeads in the presence or absence of the BCL6 inhibitor 79-6 for 5 days; CD39 expression was determined by flow cytometry. Representative contour plots and summary data from 5 experiments. (E) Predicted BCL6, RUNX3, and p-CREB binding sites adjacent to the ENTPD1 transcription start site. (F and G) Human memory CD4+ cells were activated by anti-CD3/anti-CD28 Dynabeads for 3 days. BCL6 (F) or RUNX3 (G) binding was determined by ChIP-PCR for indicated sites. Results are shown as mean ± SEM of 3 experiments. (H and I) HEK-293T cells were cotransfected with pGL3-basic or pGL3 reporter construct containing the ENTPD1 promoter together with mock or BCL6-overexpressing (H) or RUNX3-overexpressing (I) plasmids. Luciferase activity was determined after 48 hours. Results are shown as mean ± SEM of 3 experiments. (J) Reporter activity in HEK-293T cells cotransfected with pNL3.3-basic or the ENTPD1 enhancer construct together with BCL6 or mock plasmid. Data were compared by 2-tailed paired t test (A–D), 1-way ANOVA with post hoc Tukey’s test (F and G), or unpaired t test (H–J). *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001. NS, not significant.
To further examine the role of RUNX3 and BCL6 in ENTPD1 gene expression, we identified predicted binding motifs across the ENTPD1 locus using Matinspector software (Figure 2E) and performed ChIP-PCR. We found RUNX3 and BCL6 recruitment immediately upstream or downstream of the transcription start site (TSS); in addition, BCL6 bound to a site approximately 1.7 kb downstream of the TSS (Figure 2, F and G). To directly confirm BCL6 and RUNX3 involvement in regulating ENTPD1 transcription, we cloned the ENTPD1 promoter (–1500 to +200) and presumptive enhancer sequences (+600 to +2500), respectively, into pGL3 and pNL3.3 luciferase plasmids and performed reporter gene assays in HEK-293T cells with forced overexpression of BCL6 or RUNX3 (Figure 2, H–J). BCL6 repressed reporter activity of both the promoter and the enhancer construct, while RUNX3 upregulated promoter activity.
Inhibition of Tfh lineage differentiation by CD39 activity.
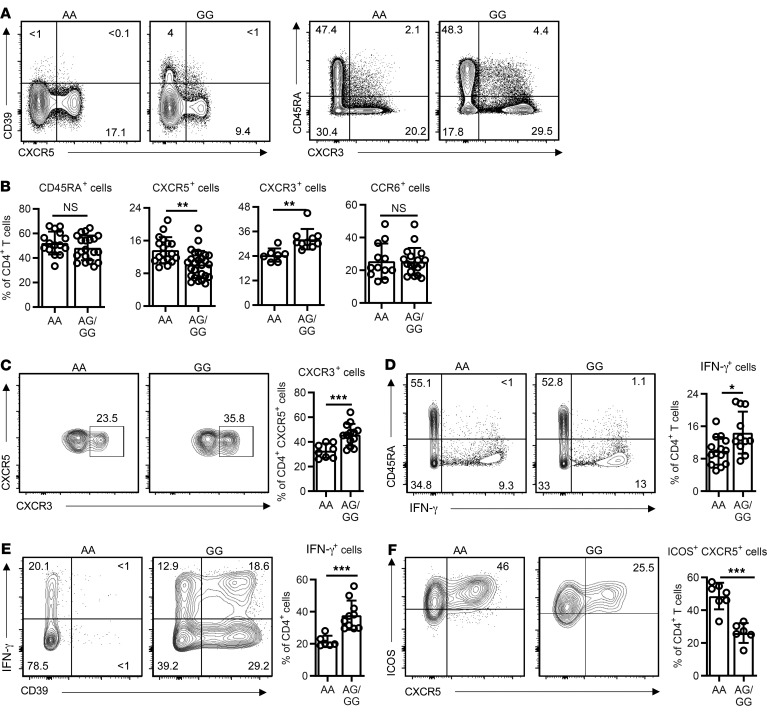
In our studies of CD39 expression, we noted that Tfh cells not only failed to express CD39, but that CD39 impaired Tfh differentiation as well. Transcription of ENTPD1 is regulated by an A/G SNP at position rs_10748643 downstream of the TSS, with individuals homozygous for A at this position largely lacking expression (13). The A and G polymorphisms are equally frequent, and the SNP is unrelated to BCL6 or RUNX3 binding sites. In initial studies, we found that individuals carrying the AA SNP had significantly higher frequencies of circulating CD4+CXCR5+ Tfh cells (cTfh) and fewer CD4+CXCR3+ Th1 memory cells, while the frequencies of total naive and CD4+CCR6+ cells were unchanged (Figure 3, A and B, and refs. 14, 15). Moreover, the CXCR3+ subset of cTfh cells that is poor in providing B cell help (15, 16) was smaller in AA individuals (Figure 3C). Consistent with the phenotypic data, frequencies of IFN-γ+CD4+ cells were lower in AA individuals (Figure 3D). Circulating Tfh cells had only minimal expression of the classical Tfh cell surface markers ICOS or PD-1, consistent with them being obtained from nonimmunized, noninfected individuals (Supplemental Figure 2A). To confirm that CD39 biases lineage differentiation, we cultured T cells from individuals characterized by their SNP under Th1 or Tfh conditions. As predicted, AA individuals only marginally upregulated CD39 expression (Supplemental Figure 2, B and C). Generation of IFN-γ–producing cells was decreased and generation of ICOS+CXCR5+ cells was increased in AA compared with AG/GG SNP–expressing CD4+ T cells (Figure 3, E and F). Similar differences in IFN-γ–producing cells were observed for CD8+ T cells (Supplemental Figure 2D).
Figure 3. Influence of CD39 expression on T cell differentiation.
PBMCs were typed for the A/G single nucleotide polymorphism at position rs_10748643 associated with low or high expression of ENTPD1. (A) Representative contour plots showing expression of CD39 versus CXCR5 (Tfh) and CD45RA versus CXCR3 (Th1) in peripheral blood CD4+ T cells after gating for CD25– non-Treg cells from individuals with the AA low- and GG high-expresser ENTPD1 SNP. (B) Frequencies of CD45RA+, CXCR5+, CXCR3+, and CCR6+ cells within peripheral blood CD4+CD25– T cells in individuals stratified for the AA versus AG/GG ENTPD1 SNP. (C) CXCR3 expression by CXCR5+CD4+T cells from A. Representative contour plots and summary data from 8 AA and 13 AG/GG SNP individuals. (D) Total T cells characterized for the ENTPD1 SNP genotypes were stimulated with PMA/ionomycin for 3 hours; IFN-γ production by CD4+ cells was determined by flow cytometry. Representative contour plots and summary data from 14 AA and 11 AG/GG individuals. (E and F) T cells were cultured under Th1- (E) or Tfh-polarizing (F) conditions for 5 days; IFN-γ production (E) and ICOS+CXCR5+ expression (F) within CD4+ cells were determined by flow cytometry. Data are shown as mean ± SEM; comparisons were done by 2-tailed unpaired t test. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001. NS, not significant.
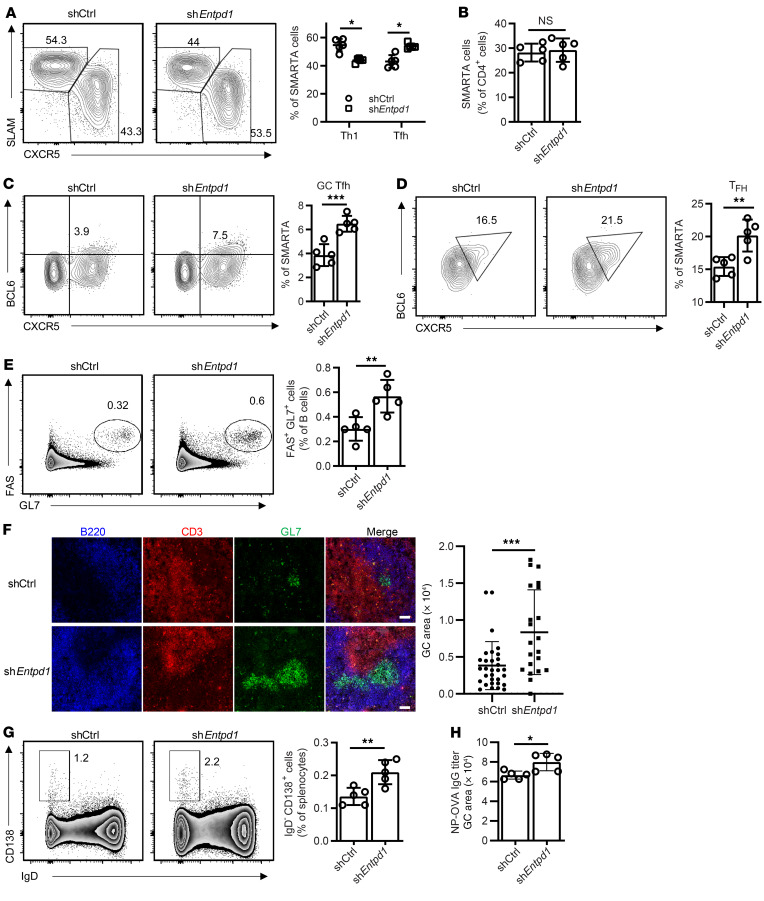
To examine whether CD39 influences Tfh differentiation in vivo, we retrovirally transduced SMARTA CD4+ T cells with an shRNAmir specific for Entpd1 or control shRNAmir (shCtrl) and transferred the cells into B6 mice followed by LCMV infection. Reduced CD39 expression in shEntpd1 SMARTA CD4+ T cells was confirmed by CD39 staining (Supplemental Figure 3A). Similar results were obtained with 2 different shRNAmirs, shEntpd1-1 (Figure 4A) and shEntpd1-2 (Supplemental Figure 3, B and C). Expansion of SMARTA cells was not affected by Entpd1 silencing (Figure 4B); however, CD39-deficient cells were biased to differentiate into Tfh rather than Th1 cells compared with control-silenced SMARTA cells (Figure 4A). This bias correlated with an increased fraction of GC Tfh within the shEntpd1+ SMARTA cells (Figure 4C and Supplemental Figure 3) and was functionally important, as percentages of BCL6+ B cells (Supplemental Figure 3) and FAS+GL7+ GC B cells were increased (Supplemental Figure 3F). Effects of CD39 silencing favored Tfh differentiation and was seen as early as on day 3 (Figure 4D).
Figure 4. Effect of Entpd1 silencing on murine T cell responses after LCMV infection.
SMARTA CD4+ T cells transduced with the indicated shRNAmir retrovirus were transferred into B6 mice. Spleen cells were analyzed on day 8 (A–C) and day 3 (D) after LCMV infection. (A) Frequencies of Tfh (CXCR5hiSLAMlo) and Th1 (SLAMhiCXCR5lo) SMARTA cells. (B) Frequencies of SMARTA CD4+ T cells as percentage of total CD4+ T cells. (C) Frequencies of GC Tfh (CXCR5+BCL6+) cells. All data are shown as representative contour plots and summary data from 1 of 2 experiments with 4 to 5 mice in each group. (D) CXCR5+BCL6+ Tfh cells were analyzed and quantified by flow cytometry. Data from 1 experiment with 5 mice in each group. (E–H) OT-II CD4+ T cells transduced with the indicated retrovirus were transferred into CD4-knockout mice and analyzed 12 days after immunization with NP-OVA in alum. (E) Frequencies of FAS+GL7+ cells as percentage of total B cells. (F) Histological analysis of spleens from immunized mice. Representative images of B220, CD3, and GL7 staining and comparison of GC areas in mice reconstituted with control and shEntpd1 OT-II T cells. Scale bars: 50 μm. (G) Frequencies of CD138+IgD– plasma cells. Representative contour plots and summary data from 1 experiment with 5 mice in each group. (H) NP-OVA–specific IgG titers. Data shown as mean ± SEM were compared by unpaired 2-tailed t test. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001. NS, not significant.
To determine whether CD39 expression on the responding T cell population has a similar effect in a classical immunization model as in the LCMV infectious disease model, shCtrl- or shEntpd1-transduced OT-II CD4 cells were transferred into CD4-knockout mice, followed by NP-OVA immunization. Similar to LCMV infection, shEntpd1+ OT-II cells differentiated preferentially into Tfh compared with shCtrl OT-II cells (Supplemental Figure 3, G and H). Consequently, frequencies of GC B cells, GC sizes (Figure 4, E and F), frequencies of plasma cells (Figure 4G), and NP-OVA–specific IgG Ab titers (Figure 4H) were increased in mice that received shEntpd1+ OT-II cells. Similar changes in T cell lineage differentiation were obtained by treating mice with the CD39 inhibitor ARL67156 for 8 days after LCMV infection (Supplemental Figure 4, A and B).
The effect of CD39 expression extended beyond the acute stage of the immune response. In the LCMV infectious disease model, total numbers of antigen-specific CD4+ and CD8+ T cells were increased on day 28 after infection (Supplemental Figure 4, C and D). These observations are consistent with our previous study showing that T cells gaining expression of CD39 in an immune response are short-lived effector T cells that undergo CD39-dependent cell death (9).
Mechanisms of CD39-mediated inhibition of Tfh differentiation.
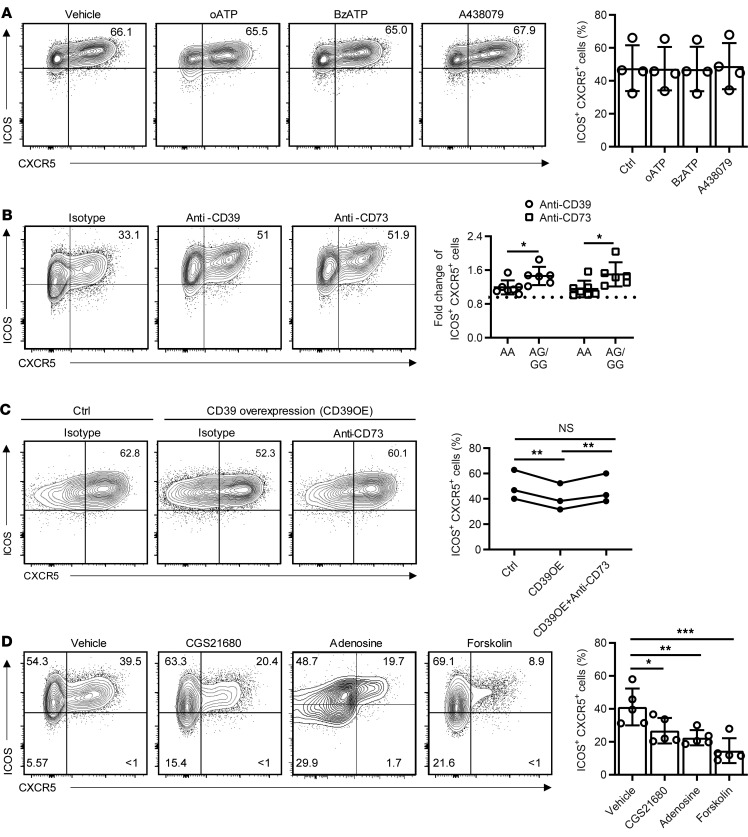
CD39 functions as an ectonucleotidase (NTPDase) and may therefore suppress Tfh differentiation by depleting extracellular ATP levels. ATP stimulates Ca2+ influx through receptors of the P2X family, of which P2X1, P2X4, P2X5, and P2X7 are expressed in human T cells (17). To examine the effect of P2X receptors on Tfh generation, naive CD4+ T cells were cultured under Tfh-polarizing conditions in the presence of the global P2X antagonist periodate-oxidized ATP (oATP) or the P2X7-specific antagonist A438079. Neither of the inhibitors reduced the generation of ICOS+CXCR5+ Tfh cells (Figure 5A). Moreover, culturing with the P2X7 agonist benzoylbenzoyl-ATP (BzATP) also did not have an impact on Tfh differentiation. Taken together, these observations suggest that CD39 does not influence lineage differentiation through depletion of ATP. However, CD39’s enzymatic function is clearly important because blocking its activity with an anti-CD39 Ab increased the generation of Tfh cells, more so in AG/GG individuals than in AA individuals (Figure 5B). Although the ATP breakdown products ADP and AMP do not have any direct signaling function in T cells, they can be further cleaved to adenosine by CD73, which is expressed on CD39– subsets of T cells and other cells present in PBMCs (18). Blocking CD73 increased Tfh cell generation to similar levels as blocking CD39; again, the effect was less pronounced with AA individuals (Figure 5B).
Figure 5. Mechanisms underlying CD39-mediated reduced generation of human Tfh cells.
(A) Human naive CD4+ T cells cultured under Tfh conditions in the presence or absence of P2X channel blockers (oATP, A438079) or the activator BzATP for 5 days. Frequencies of ICOS+CXCR5+ cells as percentage of CD4+ cells are shown as representative contour plots and summary data from 4 experiments. (B) Human total T cells of the different ENTPD1 SNP genotypes were cultured under Tfh conditions with indicated blocking antibody for 5 days. Frequencies of ICOS CXCR5+ cells as percentage of CD4+ cells are shown as contour plots for an individual typing AG for the ENTPD1 SNP (left); fold increases of ICOS+CXCR5+ T cell frequencies due to CD39 or CD73 blocking are compared for AA versus AG/GG donors (right). (C) Human naive CD4+ cells were activated with anti-CD3/anti-CD28 beads and in parallel transduced with Ctrl or ENTPD1-expressing lentivirus. On day 1.5, cells were enriched for successfully transduced GFP+ cells and then cultured on anti-CD3–coated plates in the presence of isotype control or anti-CD73 antibodies under Tfh condition for a total of 5 days. Results are shown as representative contour plots (left) and data from 3 experiments (right). (D) Human naive CD4+ cells were cultured under Tfh conditions with indicated small molecules for 5 days; cells were analyzed by flow cytometry. Representative contour plots and data from 5 experiments are shown. Data are shown as mean ± SEM and were compared by 2-tailed unpaired t test (A–C) or 1-way ANOVA with post hoc Tukey’s test (D). *P < 0.05, **P ≤ 0.01; ***P ≤ 0.001. NS, not significant.
The function of CD39 was further confirmed by overexpression studies. Naive CD4+ cells were infected with ENTPD1-expressing lentivirus. Successfully transduced GFP+ cells were isolated after 1.5 days and cultured under Tfh conditions with or without anti-CD73 for a total of 5 days. CD39 overexpression decreased Tfh frequencies that were rescued by CD73 inhibition (Figure 5C). To directly demonstrate the effect of adenosine on Tfh differentiation, we cultured naive CD4+ cells under Tfh-polarizing conditions in the presence of adenosine itself, the adenosine A2a receptor (A2aR) agonist CGS21680, the adenylate cyclase activator forskolin, or vehicle. Adenosine as well as CGS21680 significantly impaired Tfh differentiation (Figure 5D) without affecting cell survival and proliferation. Similarly, mimicking cAMP generation from A2aR signaling, forskolin inhibited Tfh cell generation (Figure 5D). Conversely, CGS21680, adenosine, and forskolin all increased IFN-γ+ cells as well as IL-2+ cells (Supplemental Figure 5).
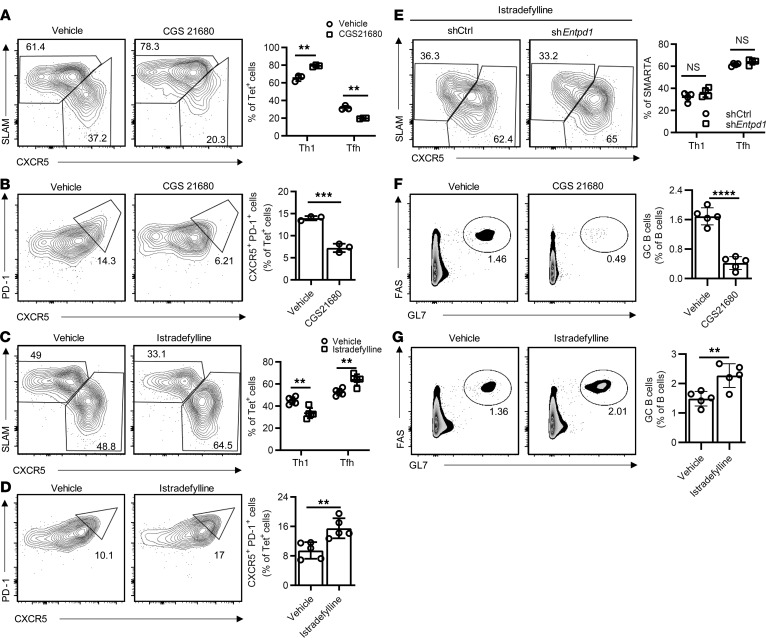
To examine whether adenosine A2aR signaling skews Th1 and Tfh differentiation in vivo, B6 mice were infected with LCMV followed by treatment with the A2aR agonist CGS21680 or vehicle. Activation of A2aR after LCMV infection significantly decreased Tfh and increased Th1 frequencies among GP66-77 IAb tetramer+ cells (Figure 6, A and B). Conversely, treatment with the selective A2aR antagonist istradefylline following infection with LCMV enhanced Tfh cells and attenuated Th1 cell differentiation (Figure 6, C and D). The effect of CD39 silencing on Tfh generation was no longer observed under istradefylline treatment (Figure 6E), confirming that CD39 influences Tfh differentiation through A2aR signaling. Similar effects on Tfh generation were seen in the OVA immunization model (Supplemental Figure 6). This effect was functionally important; GC activity decreased with CGS21680 as indicated by decreased frequencies of GC B cells (Figure 6F); conversely, GC B cells increased with istradefylline treatment (Figure 6G).
Figure 6. Effect of adenosine 2A receptor (A2aR) signaling on murine T cell responses after LCMV infection.
(A and B) B6 mice infected with LMCV were treated with vehicle or the A2aR agonist CGS21680 by twice daily intraperitoneal injections; splenocytes were examined after 8 days. Representative contour plots (left) of the expression of CXCR5, SLAM (A), and PD-1 (B) in GP66-77 IAb tetramer+ CD4+ T cells and frequency plots (right) of CXCR5hiSLAMlo (Tfh) and CXCR5loSLAMhi (Th1) CD4+ T cells (A) and PD-1+CXCR5+ GC Tfh (B). Data are from 3 mice in each group. (C and D) B6 mice infected with LMCV were treated by daily intraperitoneal injection of vehicle or istradefylline; splenocytes were analyzed on day 8 as described in A and B. Results on Tfh and Th1 (C) and GC Tfh (D) frequencies from 1 of 2 experiments with 4 to 5 mice in each group. (E) SMARTA CD4+ T cells transduced with the indicated shRNAmir retrovirus were transferred into B6 mice followed by LCMV infection. Starting on day 1 after infection, mice were treated with daily istradefylline. Contour and frequency plots of Tfh and Th1 cells on day 8 from 1 experiment with 4 mice in each group. (F and G) Wild-type mice immunized with NP-OVA were treated with CGS21680 (F) or istradefylline (G). GC B cell frequencies were determined on day 12 by flow cytometry: 5 mice in each group. Data are shown as mean ± SEM and were compared by 2-tailed unpaired t test. **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. NS, not significant.
A2aR is a G protein–coupled receptor signaling through the cAMP/PKA/p-CREB pathway. In a previous report, activation of this pathway in mice enhanced Th1 differentiation by inducing IL-12Rβ2 and IL-2Rβ expression (19). Indeed, we observed that CGS21680 and forskolin treatment significantly increased the expression of these receptors in human T cells, possibly explaining the bias toward generation of Th1 cells (Supplemental Figure 7A). Moreover, IL-2 production was increased by cAMP signaling even under Tfh-polarizing conditions (Supplemental Figure 7B). IL-2 signaling attenuates Tfh differentiation and enhances Th1 differentiation (7, 20). To determine whether increased IL-2 stimulation accounts for the inhibition of Tfh generation seen with adenosine, we performed Tfh polarization experiments with or without forskolin treatment and concomitant IL-2 neutralization. As expected, anti–IL-2 Abs increased Tfh generation; however, they were not able to counteract the forskolin-induced reduction of Tfh cells (Supplemental Figure 7C), suggesting that cAMP signaling impaired Tfh differentiation independently of increased IL-2 stimulation. Instead, we noted that several BCL6 target genes were upregulated by CGS21680 and forskolin, although BCL6 expression itself was not affected (Supplemental Figure 7D). We therefore quantified BCL6 recruitment to these loci by ChIP-PCR. As shown in Supplemental Figure 7E, increased cAMP signaling prevented BCL6 from binding to its target genes. Taken together, these data provide evidence for the model in which CD39/CD73-mediated generation of adenosine inhibits Tfh generation by stimulating the A2aR/cAMP pathway and preventing recruitment of the lineage-determining transcription factor BCL6.
Regulation of CD39 expression by the cAMP/PKA/p-CREB pathway.
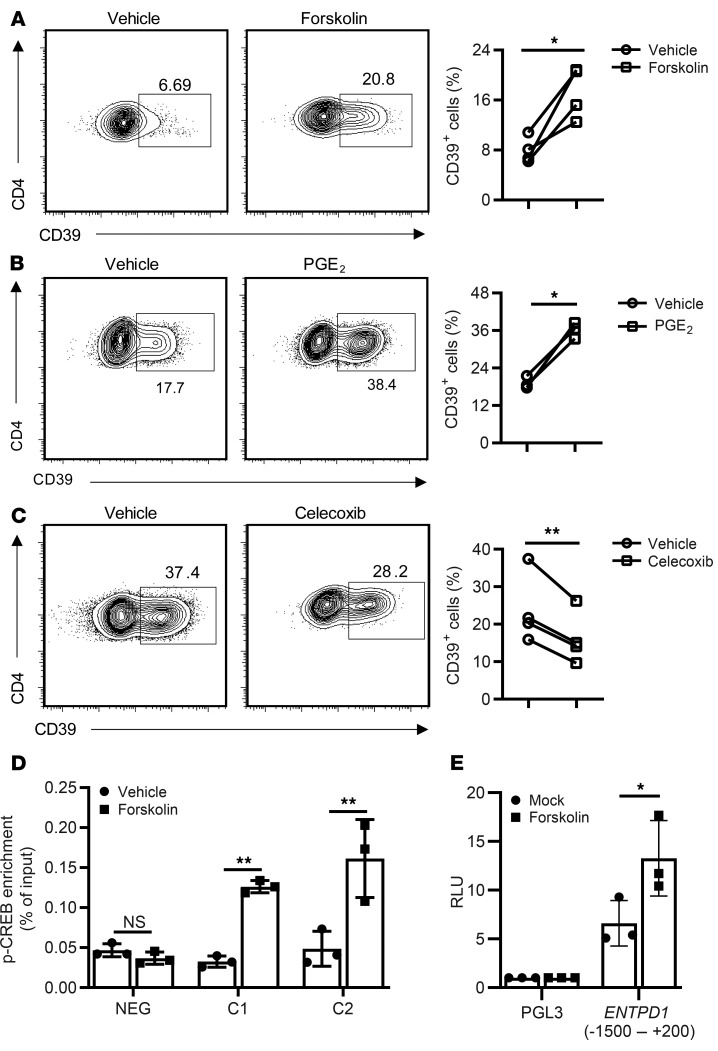
Because genomic variants reducing ENTPD1 transcription had an increased propensity to generate Tfh cells, we explored means to interfere with CD39 expression with the ultimate objective to improve vaccination-induced Tfh responses. We noted that ENTPD1 transcripts were approximately 10-fold increased upon treatment with the adenylyl cyclase activator forskolin, which may only in part derive from the impaired BCL6 binding to ENTPD1’s promoter and enhancer regions. As shown in Figure 7A, forskolin induced CD39 expression in CD4+ cells cultured with IL-7 in the absence of TCR stimulation. An endogenous activator of adenylate cyclase in T cells is PGE2. Addition of PGE2 to T cells activated with anti-CD3/anti-CD28 Dynabeads increased CD39 expression, while inhibition of PGE2 by the cyclooxygenase (COX) inhibitor celecoxib attenuated it (Figure 7, B and C). In previous studies, Liao and colleagues reported that p-CREB downstream of cAMP/PKA induces Entpd1 transcription by binding to its promoter in murine macrophage-like RAW cells (21). The human ENTPD1 gene includes potential CRE sites in the promoter region approximately 3.1 kb downstream of the TSS (Figure 2E). Recruitment of p-CREB to these sites was demonstrated by ChIP in forskolin-treated cells (Figure 7D). Furthermore, forskolin induced ENTPD1 promoter activity in reporter gene assays (Figure 7E). These observations suggest that stimulation of G protein–coupled receptors regulates ENTPD1 transcription by 2 mechanisms: transcriptional activation by p-CREB and prevention of BCL6 recruitment.
Figure 7. Regulation of ENTPD1 expression by the cAMP/PKA/p-CREB pathway.
(A–C) Contour plots of flow cytometry for CD39 expression and results from 3 to 4 experiments are shown. (A) CD39 expression on human CD4+ T cells cultured with IL-7 without TCR stimulation in the presence or absence of forskolin for 5 days. (B) CD4+ T cells activated by anti-CD3/anti-CD28 Dynabeads with or without PGE2 for 5 days. (C) Human CD4+ T cells activated by anti-CD3/anti-CD28 Dynabeads with or without celecoxib for 5 days. (D) CD4+ cells were cultured with IL-7 for 2 days followed by stimulation with forskolin or vehicle for 1 hour. ChIP was performed using anti–p-CREB (S133). Results are shown as mean ± SEM of 3 separate experiments. (E) HEK-293T cells were transfected with pGL3-basic or pGL3 containing the ENTPD1 promoter sequence. After 1 day, cells were treated with vehicle or forskolin. Luciferase activity was determined after an additional 24 hours. Results are shown as mean ± SEM of 3 independent experiments. Data were compared by 2-tailed paired t test (A–C) or 2-tailed unpaired t test (D and E). *P < 0.05, **P ≤ 0.01. NS, not significant.
COX-2 inhibition enhances Tfh differentiation through repressing CD39.
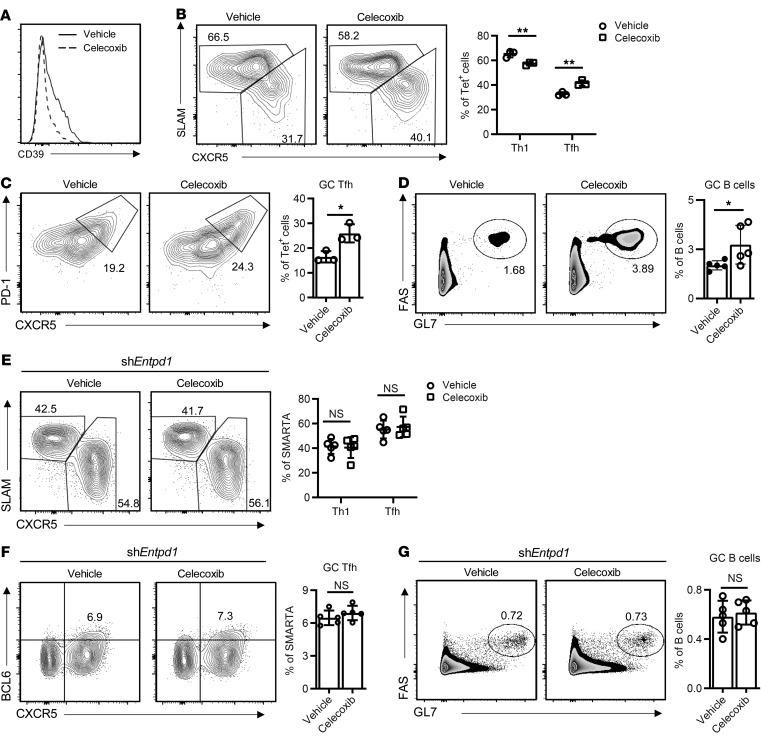
Our results indicated that cAMP/p-CREB activity represents a forward loop in increasing CD39 expression. PGE2 secreted upon T cell activation stimulates cAMP signaling that induces CD39 expression, which generates adenosine and further cAMP/p-CREB activation through A2aR. Inhibition of cAMP production should dampen this loop and therefore favor Tfh generation. To determine whether this can be accomplished by COX inhibition during the days following infection or vaccination, we treated B6 mice with celecoxib following LCMV infection and analyzed generation of virus-specific Th1 and Tfh cells by staining GP66-77 IAb tetramer–binding cells. Celecoxib downregulated CD39 expression in tetramer+ cells (Figure 8A) and enhanced Tfh and GC Tfh frequencies while attenuating Th1 differentiation (Figure 8, B and C). Again, similar results were obtained after OVA immunization; celecoxib increased Tfh frequencies (Supplemental Figure 8A), which contributed to increased GC B cell frequencies (Figure 8D). The effect of celecoxib on T cell differentiation was mediated by preventing CD39 expression. We silenced ENTPD1 in SMARTA cells that were adoptively transferred into B6 mice, followed by LCMV infection and celecoxib treatment. Silencing of ENTPD1 abrogated celecoxib-mediated changes in Th1 and Tfh cell generation (Figure 8E). Similarly, in the NP-OVA immunization model using OT-II cells transferred into CD4-knockout mice, the increase in frequencies of Tfh cells and GC B cells mediated by celecoxib was abrogated by CD39 silencing (Supplemental Figure 8B and Figure 8, F and G). Taken together, these data support the model that PGE2 production after T cell activation induces CD39 expression that then further inhibits Tfh generation through the production of adenosine.
Figure 8. COX-2 inhibition enhances Tfh differentiation through repressing CD39.
(A–C) B6 mice infected with LMCV were treated with vehicle or celecoxib i.p. every other day. Splenocytes were analyzed on day 8. (A) CD39 expression on gated GP66-77 IAb tetramer+ CD4+ T cells. Frequencies of CXCR5hiSLAMlo (Tfh) and CXCR5loSLAMhi (Th1) CD4+ T cells (B) and PD-1+CXCR5+ GC Tfh (C) from 1 representative of 2 experiments with 3 mice in each group. (D) B6 mice were immunized with NP-OVA. Celecoxib was given every other day starting on day 1 after immunization and splenocytes were analyzed by flow cytometry on day 12: results from 4 or 5 mice in each group. (E and F) SMARTA CD4+ T cells transduced with the Entpd1 shRNAmir retrovirus were transferred into B6 mice. Mice were infected with LCMV and treated with celecoxib every other day. Splenocytes were analyzed on day 8 after LCMV infection. Frequencies of Tfh (CXCR5hiSLAMlo) and Th1 (SLAMhiCXCR5lo) (E) and GC Tfh (CXCR5+BCL6+) (F) are shown as percentage of SMARTA CD4+ T cells. (G) OT-II CD4+ T cells transduced with indicated retrovirus were transferred into CD4-knockout mice. Mice were immunized with NP-OVA in alum and treated with celecoxib every other day. Splenocytes were analyzed on day 12 after immunization. Frequencies of GC B cells (FAS+GL7+) are shown as percentage of B cells. Data are representative of 2 experiments with 5 mice in each group. All data are shown as mean ± SEM and were compared by 2-tailed unpaired t test. *P < 0.05, **P ≤ 0.01. NS, not significant.
Discussion
In this study, we show that Tfh generation is linked to a lack of CD39 expression on the responding T cell population in 2 dimensions. CD39 prevents Tfh differentiation through a mechanism that involves the activation of the cAMP/PKA/p-CREB pathway, resulting in the inhibition of BCL6 binding and the further p-CREB–mediated upregulation of CD39. Conversely, BCL6, the lineage-determining transcription factor for Tfh cells, is a strong repressor of the ENTPD1 gene. As a consequence, Tfh polarization involves the successful repression of ENTPD1. Accordingly, generation of Tfh cells was shown to be superior in a subset of individuals who fail to transcribe ENTPD1 due to a frequent genomic variant (13). Preliminary studies in small populations have shown a trend for better vaccine responses in individuals carrying the ENTPD1 low-expresser SNP (9); appropriately powered studies are needed. The immediate application of these observations is that interference with CD39-dependent events can be used to increase the number of Tfh cells after vaccination and thus improve vaccine responses. Such an approach is particularly promising in older individuals, who have higher inducibility of CD39 in T cells, but defective vaccine responses (9). Because individuals carrying the ENTPD1 low-expresser SNP are healthy, short-term interventions targeting CD39 should be safe. We have identified 2 targets for such interference and have validated them in an infectious disease as well as an immunization mouse model. Treatment with a COX inhibitor improved Tfh generation through reducing CD39 expression. Likewise, treatment with the A2aR blocker istradefylline to block the activity of CD39/CD73-generated adenosine skewed the T cell response toward increasing Tfh cells.
Extracellular nucleotides are important regulators of cellular function. In particular, T cells express a multitude of nucleotide-sensing receptors (22). Owing to its ability to cleave ATP, CD39 has a checkpoint function in purinergic signaling (23). ATP is considered to derive from 2 major sources. It is released from cells undergoing cell death, leading to increased extracellular concentrations in tumor tissues and inflammatory infiltrates (24, 25). Alternatively, ATP is actively secreted through the PANNEXIN 1 channel by activated T cells. Extracellular ATP stimulates calcium influx through a variety of P2X receptors (17, 26). At high ATP concentrations, the calcium influx induces inflammasome activation and cell death. Such high concentrations may be reached in GCs when B cells undergo apoptosis. Indeed, P2X7 has been shown to be a negative regulator of Tfh cells, possibly by inducing pyroptosis of Tfh cells. In the intestinal environment, P2X7 stimulation reduces the number of Tfh cells in Peyer’s patches (27). Similar results were found in disease models of malaria infection and systemic lupus erythematosus (28, 29). In theory, CD39 could have a protective function in Tfh survival by reducing ATP and P2X signaling, if expressed in GCs. However, given the unequivocal repressor function of BCL6 in ENTPD1 transcription, CD39 is neither expressed on Tfh cells nor on GC B cells and therefore does not appear to be relevant in GCs. Indeed, individuals carrying the CD39 low-expresser SNP did not have increased cTfh cells. On the contrary, they had more cTfh cells due to increased Tfh generation.
Our studies in the mouse models with adoptively transferred T cells showed that CD39 expression on the differentiating antigen-specific T cell itself prevents Tfh generation. We considered the possibility that P2X receptors provide a positive signal in differentiation that is removed due to ATP cleavage by CD39. Indeed, ATP stimulation of P2X receptors has been shown to provide costimulatory signals in T cell activation (17, 30–32). Moreover, Stark et al. have shown that TCR stimulation modulates the effects of P2X signaling and that tissue-resident memory T cells are protected from the P2X7-induced cell death when recognizing antigen (33). Borges da Silva et al. were even able to show for mouse CD8+ T cells that P2X7-mediated signals were important in inducing metabolic changes that favor the induction of memory rather than effector T cells (34); in this study, increased mitochondrial activity led to increased ATP secretion and increased P2X7 activity. The resulting calcium influx facilitated activation of calcium-dependent kinases and phosphorylation of AMPK, which further increased mitochondrial activity. Previous studies have shown that the P2X antagonist oATP improves and the P2X7 agonist BzATP impairs Treg differentiation (35). In contrast, neither pharmacological inhibition nor activation of P2X channels at non-proapoptotic concentrations affected the generation of Tfh cells (Figure 5A).
Alternatively, CD39 regulates purinergic signaling by cleaving ATP to ADP and AMP, which is further cleaved to adenosine by CD73 (22). Although expression of CD39 and CD73 in human T cells is nearly mutually exclusive and, in contrast to mice, expression of CD73 in GC of human tonsils is low (data not shown), CD73 is expressed on a large variety of cells, suggesting that CD73 is not limiting in degrading AMP. Indeed, blocking CD73 abrogated the effect of CD39 on Tfh generation in the culture system with human total T cells. Moreover, we found that the effect of CD39 on reducing Tfh generation was due to the stimulation of A2aR by adenosine. Adenosine-mediated A2aR stimulation is a well-recognized pathway of Treg function; murine Treg cells frequently coexpress CD39 and CD73 and exert immunosuppressive function through the generation of adenosine (35–37). Moreover, adenosine signaling is important for Treg generation (38). However, adenosine’s effect in our studies was T cell subset specific and only affected Tfh, while generation of Th1 cells was even increased, unlike Treg-mediated broad immunosuppression. We also considered the possibility that CD39 favored the induction of regulatory Tfh (TFR) cells, which appears to be dependent on stimulation of A2aR (39); however, in our adoptive transfer experiments of wild-type and CD39-deficient SMARTA cells, we did not see a difference in TFR and the frequencies of FOXP3+ cells were equally low. Moreover, the inhibitory effect on Tfh differentiation was clearly linked to CD39 expression on the differentiating and expanding T cell population and not on any third-party cells, suggesting a direct effect on the differentiation program.
Although A2aR stimulation at high concentrations of adenosine is mostly immunosuppressive, adenosine can also have an immunomodulatory effect on T cell differentiation, as observed in recent mouse models and seen in the current study. Schmiel et al. examined the ability of an adenosine agonist to induce anergy in an immunization model and found Tfh generation and GC activity suppressed (40). Mechanistically, we now show that the G protein–mediated activation of the cAMP/PKA/p-CREB pathway prevented the binding of BCL6 to target regions and therefore directly affected Tfh generation and function (Supplemental Figure 7E). Moreover, we directly link Tfh generation to CD39 expression in humans, identifying strategies to improve immunization. Activation of the cAMP/PKA/p-CREB pathway, in response to activators of G protein–coupled receptors, directly induced the transcription of ENTPD1. As shown in Figure 8, inhibition of prostaglandin synthesis, and therefore PGE2 signaling, through EB2/EP4 receptors using a COX inhibitor improved Tfh generation in vivo in the LCMV mouse model. Importantly, prostaglandin-mediated inhibition of Tfh generation was dependent on the expression of CD39, implying an amplification loop in which PGE2 induced CD39 expression, resulting in more adenosine generation and further p-CREB signaling. In addition to A2aR inhibition, prevention of the activation-induced CD39 expression therefore appears to be a feasible intervention in vaccination strategies.
In addition to short-lived effector T cells in acute T cell responses, CD39 expression is also a hallmark of T cell exhaustion (41, 42); therefore, targeting CD39-dependent pathways may also be useful to treat exhaustion. Of particular interest, exhausted antigen-specific CD8+ T cells in the chronic LCMV infection model that are unresponsive to PD-1 checkpoint inhibition express CD39, while responsive T cells have transcriptional and epigenetic signatures of Tfh cells (43, 44). Future studies must explore the role of CD39 in inducing or maintaining exhaustion.
Our findings are of particular relevance for older individuals who have impaired vaccine responses. We have previously shown that CD4+ T cells from older individuals have increased expression of miR-21, reducing the expression of several negative signaling regulators such as PTEN, SPROUTY, and PDCD4 (11). As consequences, TCR-induced signaling is more sustained and T cell differentiation into short-lived effector T cells with increased expression of CD39 is favored. Previous murine studies have shown that mTORC inhibition could favorably shift the balance toward generation of Tfh and memory precursor cells (45). Rapalogs to inhibit disproportionate activation of the mTORC pathway are therefore being considered to improve immune health in the elderly (46), but are also of considerable risk given their broad effects on cell metabolism and associated antiproliferative and immunosuppressive functions. A less invasive and more targeted approach would be to inhibit CD39 expression, inhibit CD39 enzymatic activity, or block signaling of downstream-generated adenosine in older individuals who are able to express CD39. In the mouse model, we used istradefylline, a highly selective adenosine A2aR antagonist that has been used in clinical studies and is approved in Japan for the treatment of Parkinson’s disease (47). Istradefylline has been well tolerated in phase III treatment trials of more than a year duration without major toxicities. Such interventions, in particular, when given short-term in the context of vaccination, are likely to be safe, given that 25% of the general population are not able to express CD39 (9, 13).
Methods
Study population and cell purification.
Buffy coats and LRS chambers from 86 blood or platelet donors were purchased from the Stanford Blood Bank. In addition, PBMCs were obtained from 9 healthy volunteers. Total T or isolated CD4+ T cells were purified by human T or CD4 T cell enrichment cocktail (STEMCELL Technologies). Naive CD4+ T cells were further isolated from PBMCs by human naive CD4+ T cell isolation kit (STEMCELL Technologies). Genotyping for the ENTPD1 SNP at rs_10748643 SNP was done by PCR with 0.25 μL specific forward/reverse primers and the following probes — allele 1: VIC/MGB-NFQ; allele 2: FAM/MGB-NFQ (Applied Biosystems) — in quintuplicate in 384-well plates using the ABI 7900HT system. Palatine tonsils were obtained from the National Disease Research Interchange. Tonsil tissue was cut into small pieces with a scalpel. Five to 10 pieces of tonsil were gently grounded through a 70-μm filter to release mononuclear cells.
Cell culture.
Unless stated otherwise, isolated T cells were activated with anti-CD3/anti-CD28 Dynabeads (Thermo Fisher Scientific) at a bead-to-cell ratio of 1:5 in RPMI 1640 (MilliporeSigma) supplemented with 10% FBS, 100 U/mL penicillin, and 100 U/mL streptomycin (Thermo Fisher Scientific). Cells were cultured with 20 ng/mL human recombinant IL-12 (PeproTech) and 5 μg/mL anti–IL-4 (eBioscience) for Th1 polarization, and with 25 ng/mL IL-6, 10 ng/mL IL-1β, 25 ng/mL IL-23, and 5 ng/mL TGF-β for Tfh polarization (48). When indicated, CGS21680 (5 nM, Tocris Bioscience), adenosine (100 mM, MilliporeSigma), forskolin (10 μM, Tocris Bioscience), PGE2 (100 nM, Cayman Chemical), or 79-6 (BCL6 inhibitor, 100 μM, MilliporeSigma) was added on day 1 of culture; celecoxib (5 μM, Selleck Chemicals), oATP (50 μM, MilliporeSigma), BzATP (100 μM, MilliporeSigma), A438079 (P2X7 inhibitor, 25 μM, Tocris Bioscience), or anti–IL-2 Ab (5 μg/mL, AF-502-SP, R&D Systems) at the beginning of the culture. To block CD39 or CD73 enzymatic activity, anti-CD39 (3 μg/mL, A1, BioLegend), anti-CD73 (3 μg/mL, 7G2, Abcam), or isotype control Ab was added at the beginning of the culture.
Lentivirus production and transduction.
For overexpression studies, ENTPD1, BCL6, or RUNX3 was subcloned into pHAGE-CMV-IRES-ZsGreen-W (plasmid 26532, Addgene). Lentiviral vector containing RUNX3 shRNA was obtained from OriGene (TL309682). Lentivirus was packaged by transfection of a lentiviral vector, along with psPAX2 (plasmid 12260; Addgene) and pMD2.G (plasmid 12259; Addgene) expression vectors into HEK-293T cells by using FuGENE (Promega). Lentiviral particles were collected 72 hours after transfection, filtered through a 0.45-μm syringe filter (Millipore), concentrated using PEG-it solution (System Biosciences), and titered on HEK-293T cells. For lentiviral transduction, human total T cells were activated with anti-CD3/anti-CD28 beads at a bead-to-cell ratio of 1:2 and transduced with lentivirus in the presence of 8 μg/mL polybrene (MilliporeSigma) and 10 U/mL human IL-2 (PeproTech). In the polarizing experiments with overexpressed ENTPD1, human naive CD4+ cells were activated with beads in the presence of virus. After 1.5 days, GFP+ cells were sorted and cultured on plates coated with 5 μg/mL anti-CD3 and anti-CD28 Ab and Tfh-polarizing conditions for a total of 5 days.
Mice.
SMARTA TCR–transgenic (specific for LCMV gp66–77) mice were a gift from R. Ahmed (Emory University School of Medicine). Five- to 8-week-old CD4-knockout mice (B6.129S2-Cd4tm1Mak/J, 002663), OT-II mice (004194), and recipient C57BL/6J mice were purchased from The Jackson Laboratory. All mice were housed in the Stanford Research Animal Facility according to Stanford University guidelines.
Retroviral vectors, transductions, and cell transfers.
Entpd1 shRNAmirs (1: AGACATCTTATCCTTCAGTAA; 2: AGCTACCATTTGATCAGTTTC) were inserted into the pLMPd-Amt vector (49). Virion preparation was performed in the PLAT-E cell line; the medium was replaced after 10 hours and retrovirus-containing supernatant was collected after an additional 24 and 48 hours. For transduction, 2 × 106 CD4+ T cells were purified from SMARTA or OT-II splenocytes by negative selection (STEMCELL Technologies) and stimulated in 24-well plates precoated with 8 μg/mL anti-CD3 (145-2C11; eBioscience) and anti-CD28 (37.51; eBioscience) Ab in D-10 (DMEM + 10% FCS, with 2 ng/mL recombinant human IL-7 [PeproTech] and 50 μM β-mercaptoethanol [BME]). Twenty-four and 36 hours after stimulation, cells were transduced with retrovirus-containing supernatants supplemented with 50 μM BME and 8 μg/mL polybrene (MilliporeSigma) by centrifugation for 90 minutes at 1500 g at 32°C. Following transduction, cells were cultured with D-10 supplemented with 50 μM BME and 10 ng/mL human IL-2 (PeproTech). On day 3 after stimulation, CD4+ T cells were transferred into 6-well plates in D-10 plus 50 μM BME and 10 ng/mL IL-2. On day 5, the culture medium was replaced with D-10 supplemented with 50 μM BME and 2 ng/mL human IL-7 (PeproTech). On day 6, mAmetrine+ cells were sorted (FACSAria, BD Biosciences). Sorted SMARTA cells (1 × 104 for day 8 analysis or 4 × 105 for day 3 analysis) were transferred into recipient mice by intravenous tail vein injection. Host mice were rested for 3 days before being infected with LCMV-Armstrong virus. For NP-OVA immunization, 1 × 105 transduced OT-II CD4+ cells were transferred into CD4-knockout mice (B6.129S2-Cd4tm1Mak/J).
LCMV infection, NP-OVA immunization, and treatments.
Recipient mice were infected by i.p. injection of 5 × 105 plaque-forming units of LCMV-Armstrong for day 3 or 2.5 × 105 for day 8 analysis or immunized with 100 μg NP-OVA (Biosearch Technologies) precipitated in 100 μL 5% alum (aluminum potassium sulfate, MilliporeSigma). ARL 67156 (2 mg/kg in PBS, Tocris Bioscience) was given i.p. every other day; CGS-21680 (2.5 mg/kg in PBS, Tocris Bioscience) i.p. twice daily; celecoxib (15 mg/kg in DMSO, Selleck Chemicals) i.p. every other day; A2aR antagonist istradefylline (10 mg/kg in normal saline, MilliporeSigma) i.p. daily. PBS, saline, or DMSO (up to 20 μL) was given to control-treated mice.
Flow cytometry and Abs.
The following Abs and reagents were used for flow cytometric studies of murine cells: anti-CD4 (RM4-5, 1:400), anti-CD8 (53-6.7, 1:400), anti-CD39 (Duha59, 1:200), anti-B220 (RA3-6B2, 1:400), anti–PD-1 (29F.1A12, 1:400), anti-SLAM (TC15-12F12.2, 1:400), anti–GL-7 (GL7 1:200), anti-CD138 (281-2, 1:200), anti-IgD (11-26c.2a, 1:200) (all from BioLegend); anti-FAS (Jo2, 1:200), anti-CXCR5 (2G8; BD Biosciences), APC- or Pacific Blue–labeled streptavidin (eBioscience), and Fixable Viability Dye eFluor 506 (1:1000, BD Biosciences) were used. Cell surface staining was done for 30 minutes at 4°C in PBS supplemented with 1% FBS except for CXCR5. Cells were stained with biotinylated anti-CXCR5 for 1 hour followed by APC- or Pacific Blue–labeled streptavidin (eBioscience) at 4°C. For tetramer staining, cells were incubated with APC-labeled IAb/GP66–77 tetramers (provided by NIH tetramer core facility) at 37°C for 2 hours before cell surface staining. APC-labeled H-2Db/NP-396 tetramers were added together with Abs against surface markers. For intracellular staining, cells were fixed with BD Phosflow Lyse/Fix Buffer (BD Biosciences) and permeabilized with BD Phosflow Perm/Wash Buffer III (BD Biosciences), followed by intracellular staining with PerCP-cy5.5–conjugated Ab against BCL6 (clone K112-91; BD Biosciences, 1:25).
For cytometric studies of human cells, Abs against the following molecules were used: CD4 (RPA-T4, 1:400), CD8 (SK1, 1:400), CD39 (A1, 1:200), CD3 (HIT3a, 1:400), CXCR3 (G025H7, 1:200), CD25 (BC96, 1:400), CCR6 (G034E3, 1:200), ICOS (C398.4A, 1:200), CD122 (5H4, 1:200), IL-2 (MQ1-17H12, 1:200) (from BioLegend); CD45RA (HI100, 1:200), CXCR5 (RF8B2, 1:100), PD-1 (MIH4, 1:100), IFN-γ (B27, 1:100), IL-12Rβ2 (2B6/12beta2, 1:200) (all from BD Biosciences). IFN-γ and IL-2 intracellular staining was done using a Fixation/Permeabilization Solution Kit (BD Biosciences); for BCL6 staining, Cytofix buffer was used followed by Perm Buffer III buffer (BD Biosciences). Stained cells were analyzed using an LSRII or LSRFortessa (BD) and FlowJo software (Tree Star). Sorting was done on a FACSAria II or FACSAria III (BD Biosciences).
Immunohistology.
Spleens from immunized mice were frozen in OCT compound. Tissue was cut and sections were air-dried and fixed with paraformaldehyde. After washing, sections were permeabilized and blocked with 2% BSA in PBS, followed by incubation with FITC–anti-GL7 (GL7, 1:200), APC–anti-B220 (RA3-6B2, 1:500), and PE–anti-CD3 (17A2, 1:500) Abs (all from BioLegend).
ChIP-qPCR and quantitative RT-PCR.
ChIP assays were done using the ChIP-IT kit (Active Motif) according to the manufacturer’s instructions modified as follows. Cells were fixed with 1% formaldehyde and sonicated on ice using Active Motif sonicator to obtain DNA fragments sized between 100 and 1000 bp. Chromatin from 5 × 106 cells was precipitated with Abs (4 μg for each reaction) against BCL6 (N3, Santa Cruz Biotechnology), RUNX3 (9F4A17, BioLegend), or p-CREB Ser133 (87G3, Cell Signaling Technology). Normal rabbit (Santa Cruz Biotechnology) or normal mouse Ab (BioLegend) was used as control.
Total RNA was isolated using the RNeasy Plus Micro kit (QIAGEN) and reverse transcribed using Maxima First Strand cDNA Synthesis Kits (Thermo Fisher Scientific). Quantitative RT-PCR was done on the ABI 7900HT system (Applied Biosystems) using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific). The ChIP and qPCR primers are listed in Supplemental Table 1.
Luciferase reporter assay.
ENTPD1 promoter sequence (–1500 to +200) was inserted into the pGL3 basic plasmid; the putative ENTPD1 enhancer sequence (+600 to +2500) was cloned into the pNL3.3 plasmid. HEK-293T cells were cotransfected with luciferase reporter plasmid, thymidine kinase promoter–Renilla luciferase reporter plasmid, a BCL6- or RUNX3-expressing plasmid, or a control vector. After 48 hours, luciferase activities were determined by the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. To determine the effect of forskolin on ENTPD1 transcription, forskolin was added for the last 24 hours of culture before HEK-293T cells were assayed for reporter activity.
Western blotting.
Cells were lysed in RIPA buffer containing PMSF and protease and phosphatase inhibitors (Santa Cruz Biotechnology) for 30 minutes on ice. Proteins were separated in denaturing 4%–15% SDS-PAGE gels (Bio-Rad Laboratories), transferred onto PVDF membranes (MilliporeSigma), and probed with Abs against RUNX3 (9F4A17; BioLegend) and β-actin (13E5; Cell Signaling Technology). Membranes were developed using HRP-conjugated secondary Abs and Pierce ECL Western blotting substrate (Thermo Fisher Scientific).
Statistics.
Statistical analysis was performed using Prism (GraphPad). Unless stated otherwise, data are presented as mean with error bars indicating the standard error of the mean (SEM). Paired or unpaired 2-tailed Student’s t tests were used for comparing 2 groups. One-way ANOVA with Tukey’s post hoc test was used for multigroup comparisons. P less than 0.05 was considered statistically significant.
Study approval.
Human studies were approved by the Stanford Institutional Review Board, and participants gave informed consent. All mouse studies were approved by the Stanford Administrative Panel on Laboratory Animal Care.
Author contributions
WC, FF, CK, CMW, and JJG designed the study. WC, FF, TG, CG, XL, and SL performed experiments. WC, FF, TG, CMW, and JJG analyzed and interpreted data. WC and JJG wrote the manuscript with all authors providing feedback. WC and FF contributed equally to this study. WC initiated this study and therefore is listed first.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01 AR042527, R01 HL117913, R01 AI108906, R01 HL142068, and P01 HL129941 to CMW, and R01 AI108891, R01 AG045779, U19 AI057266, and R01 AI129191 to JJG) and with resources and the use of facilities at the Palo Alto Veterans Administration Healthcare System. Tetramers were provided by the NIH Tetramer Core Facility supported by contract HHSN272201300006C from the NIAID. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Version 1. 05/26/2020
Electronic publication
Version 2. 07/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(7):3422–3436.https://doi.org/10.1172/JCI132417.
Contributor Information
Wenqiang Cao, Email: wenqcao@stanford.edu.
Fengqin Fang, Email: fqfang@stanford.edu.
Xuanying Li, Email: xul930@stanford.edu.
Chulwoo Kim, Email: cwkim@stanford.edu.
Simon Lambert, Email: smnlambert1@gmail.com.
Cornelia M. Weyand, Email: cweyand@stanford.edu.
Jörg J. Goronzy, Email: jgoronzy@stanford.edu.
References
- 1.Del Giudice G, et al. Fighting against a protean enemy: immunosenescence, vaccines, and healthy aging. NPJ Aging Mech Dis. 2018;4:1. doi: 10.1038/s41514-017-0020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17(7):1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirelli KM, Crotty S. Germinal center enhancement by extended antigen availability. Curr Opin Immunol. 2017;47:64–69. doi: 10.1016/j.coi.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirelli KM, et al. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. 2019;177(5):1153–1171.e28. doi: 10.1016/j.cell.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballesteros-Tato A, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36(5):847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209(2):243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang F, et al. Expression of CD39 on activated T cells impairs their survival in older individuals. Cell Rep. 2016;14(5):1218–1231. doi: 10.1016/j.celrep.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu M, et al. Signal inhibition by the dual-specific phosphatase 4 impairs T cell-dependent B-cell responses with age. Proc Natl Acad Sci U S A. 2012;109(15):E879–E888. doi: 10.1073/pnas.1109797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim C, et al. Activation of miR-21-regulated pathways in immune aging selects against signatures characteristic of memory T cells. Cell Rep. 2018;25(8):2148–2162.e5. doi: 10.1016/j.celrep.2018.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dwyer KM, Deaglio S, Gao W, Friedman D, Strom TB, Robson SC. CD39 and control of cellular immune responses. Purinergic Signal. 2007;3(1–2):171–180. doi: 10.1007/s11302-006-9050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman DJ, et al. From the cover: CD39 deletion exacerbates experimental murine colitis and human polymorphisms increase susceptibility to inflammatory bowel disease. Proc Natl Acad Sci U S A. 2009;106(39):16788–16793. doi: 10.1073/pnas.0902869106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson N, et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62(1):234–244. doi: 10.1002/art.25032. [DOI] [PubMed] [Google Scholar]
- 15.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34(1):108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt N, Bentebibel SE, Ueno H. Phenotype and functions of memory Tfh cells in human blood. Trends Immunol. 2014;35(9):436–442. doi: 10.1016/j.it.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woehrle T, et al. Pannexin-1 hemichannel-mediated ATP release together with P2X1 and P2X4 receptors regulate T-cell activation at the immune synapse. Blood. 2010;116(18):3475–3484. doi: 10.1182/blood-2010-04-277707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raczkowski F, et al. CD39 is upregulated during activation of mouse and human T cells and attenuates the immune response to Listeria monocytogenes. PLoS One. 2018;13(5):e0197151. doi: 10.1371/journal.pone.0197151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao C, Hirata T, Soontrapa K, Ma X, Takemori H, Narumiya S. Prostaglandin E2 promotes Th1 differentiation via synergistic amplification of IL-12 signalling by cAMP and PI3-kinase. Nat Commun. 2013;4:1685. doi: 10.1038/ncomms2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13(4):405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao H, Hyman MC, Baek AE, Fukase K, Pinsky DJ. cAMP/CREB-mediated transcriptional regulation of ectonucleoside triphosphate diphosphohydrolase 1 (CD39) expression. J Biol Chem. 2010;285(19):14791–14805. doi: 10.1074/jbc.M110.116905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cekic C, Linden J. Purinergic regulation of the immune system. Nat Rev Immunol. 2016;16(3):177–192. doi: 10.1038/nri.2016.4. [DOI] [PubMed] [Google Scholar]
- 23.Takenaka MC, Robson S, Quintana FJ. Regulation of the T cell response by CD39. Trends Immunol. 2016;37(7):427–439. doi: 10.1016/j.it.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii KJ, Akira S. Potential link between the immune system and metabolism of nucleic acids. Curr Opin Immunol. 2008;20(5):524–529. doi: 10.1016/j.coi.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Di Virgilio F, Sarti AC, Falzoni S, De Marchi E, Adinolfi E. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat Rev Cancer. 2018;18(10):601–618. doi: 10.1038/s41568-018-0037-0. [DOI] [PubMed] [Google Scholar]
- 26.Chekeni FB, et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature. 2010;467(7317):863–867. doi: 10.1038/nature09413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proietti M, et al. ATP-gated ionotropic P2X7 receptor controls follicular T helper cell numbers in Peyer’s patches to promote host-microbiota mutualism. Immunity. 2014;41(5):789–801. doi: 10.1016/j.immuni.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Faliti CE, et al. P2X7 receptor restrains pathogenic Tfh cell generation in systemic lupus erythematosus. J Exp Med. 2019;216(2):317–336. doi: 10.1084/jem.20171976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salles ÉM, et al. P2X7 receptor drives Th1 cell differentiation and controls the follicular helper T cell population to protect against Plasmodium chabaudi malaria. PLoS Pathog. 2017;13(8):e1006595. doi: 10.1371/journal.ppat.1006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filippini A, Taffs RE, Sitkovsky MV. Extracellular ATP in T-lymphocyte activation: possible role in effector functions. Proc Natl Acad Sci U S A. 1990;87(21):8267–8271. doi: 10.1073/pnas.87.21.8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenk U, et al. Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci Signal. 2008;1(39):ra6. doi: 10.1126/scisignal.1160583. [DOI] [PubMed] [Google Scholar]
- 32.Yip L, et al. Autocrine regulation of T-cell activation by ATP release and P2X7 receptors. FASEB J. 2009;23(6):1685–1693. doi: 10.1096/fj.08-126458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark R, et al. TRM maintenance is regulated by tissue damage via P2RX7. Sci Immunol. 2018;3(30):eaau1022. doi: 10.1126/sciimmunol.aau1022. [DOI] [PubMed] [Google Scholar]
- 34.Borges da Silva H, et al. The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8+ T cells. Nature. 2018;559(7713):264–268. doi: 10.1038/s41586-018-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schenk U, et al. ATP inhibits the generation and function of regulatory T cells through the activation of purinergic P2X receptors. Sci Signal. 2011;4(162):ra12. doi: 10.1126/scisignal.2001270. [DOI] [PubMed] [Google Scholar]
- 36.Antonioli L, Pacher P, Vizi ES, Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol Med. 2013;19(6):355–367. doi: 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mascanfroni ID, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat Med. 2015;21(6):638–646. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta A. A metabolic immune checkpoint: adenosine in tumor microenvironment. Front Immunol. 2016;7:109. doi: 10.3389/fimmu.2016.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbott RK, et al. The GS protein-coupled A2a adenosine receptor controls T cell help in the germinal center. J Biol Chem. 2017;292(4):1211–1217. doi: 10.1074/jbc.C116.764043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmiel SE, Yang JA, Jenkins MK, Mueller DL. Cutting edge: adenosine A2a receptor signals inhibit germinal center T follicular helper cell differentiation during the primary response to vaccination. J Immunol. 2017;198(2):623–628. doi: 10.4049/jimmunol.1601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta PK, et al. CD39 expression identifies terminally exhausted CD8+ T cells. PLoS Pathog. 2015;11(10):e1005177. doi: 10.1371/journal.ppat.1005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sade-Feldman M, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175(4):998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Im SJ, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537(7620):417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jadhav RR, et al. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci U S A. 2019;116(28):14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray JP, et al. The interleukin-2-mTORc1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity. 2015;43(4):690–702. doi: 10.1016/j.immuni.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mannick JB, et al. TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci Transl Med. 2018;10(449):eaaq1564. doi: 10.1126/scitranslmed.aaq1564. [DOI] [PubMed] [Google Scholar]
- 47.Pinna A. Adenosine A2A receptor antagonists in Parkinson’s disease: progress in clinical trials from the newly approved istradefylline to drugs in early development and those already discontinued. CNS Drugs. 2014;28(5):455–474. doi: 10.1007/s40263-014-0161-7. [DOI] [PubMed] [Google Scholar]
- 48.Schmitt N, et al. The cytokine TGF-β co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol. 2014;15(9):856–865. doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen R, et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity. 2014;41(2):325–338. doi: 10.1016/j.immuni.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.