Abstract
AMPK is a key regulator at the molecular level for maintaining energy metabolism homeostasis. Mammalian AMPK is a heterotrimeric complex, and its catalytic α subunit exists in 2 isoforms: AMPKα1 and AMPKα2. Recent studies suggest a role of AMPKα overactivation in Alzheimer’s disease–associated (AD-associated) synaptic failure. However, whether AD-associated dementia can be improved by targeting AMPK remains unclear, and roles of AMPKα isoforms in AD pathophysiology are not understood. Here, we showed distinct disruption of hippocampal AMPKα isoform expression patterns in postmortem human AD patients and AD model mice. We further investigated the effects of brain- and isoform-specific AMPKα repression on AD pathophysiology. We found that repression of AMPKα1 alleviated cognitive deficits and synaptic failure displayed in 2 separate lines of AD model mice. In contrast, AMPKα2 suppression did not alter AD pathophysiology. Using unbiased mass spectrometry–based proteomics analysis, we identified distinct patterns of protein expression associated with specific AMPKα isoform suppression in AD model mice. Further, AD-associated hyperphosphorylation of eukaryotic elongation factor 2 (eEF2) was blunted with selective AMPKα1 inhibition. Our findings reveal isoform-specific roles of AMPKα in AD pathophysiology, thus providing insights into potential therapeutic strategies for AD and related dementia syndromes.
Keywords: Aging, Neuroscience
Keywords: Alzheimer’s disease, Protein kinases
Introduction
Alzheimer’s disease (AD) is the most common form of dementia and has become a global threat to public health. Currently, there is no effective intervention for curing AD or slowing disease progression, and completed clinical trials have not succeeded in identifying disease-modifying strategies (1, 2). There is an urgent need to understand the molecular mechanisms underlying AD pathophysiology and accordingly to identify novel therapeutic targets for this devastating neurodegenerative disease. Emerging evidence points to a role of AMPK in AD pathophysiology (3–6). AMPK is a Ser/Thr kinase that functions as a central energy sensor at the molecular level. Low energy states lead to activation of AMPK to maintain cellular energy homeostasis (7). Mammalian AMPK is a heterotrimeric complex consisting of a catalytic α subunit, a scaffolding β subunit, and a regulatory γ subunit (8). AMPK is activated by 2 general mechanisms: binding of AMP to the Bateman domain of the γ subunit, causing a conformational change at the α subunit, and phosphorylation of the α subunit at the Thr172 site, which involves either inhibition of protein phosphatases or activation of upstream kinases (9, 10). Dysregulation of energy metabolisms is implicated in the pathogenesis of multiple neurodegenerative diseases, including AD (11, 12). Recent studies indicate upregulation of brain AMPK activity in AD, as evaluated by AMPKα phosphorylation (3, 13). Moreover, AMPK overactivation is linked to the loss of hippocampal dendritic spines caused by amyloid β (Aβ) oligomers (4). Consistently, impairments of hippocampal synaptic plasticity associated with Aβ treatment or AD model mice are alleviated by repression of AMPK signaling (3). Nevertheless, whether AD-associated cognitive deficits can be improved by targeting AMPK is not known.
There are 2 isoforms of the AMPK catalytic α subunit: AMPKα1 and AMPKα2. These 2 isoforms are encoded by distinct genes located on different chromosomes: PRKAA1 on chromosome 5 and PRKAA2 on chromosome 1, respectively (14). Both α1 and α2 are expressed in most tissues for the regulation of energy homeostasis, and previous work indicates isoform-specific substrate specificity (15–18). However, the specific downstream targets and roles for AMPKα isoforms in the central nervous system remain unclear.
AMPK interacts with multiple signaling cascades and has many downstream effectors involved in biosynthesis (19). Among the downstream effects of AMPK, regulation of mRNA translation (de novo protein synthesis) is of particular interest. Substantial studies have demonstrated that long-term synaptic plasticity and memory formation are dependent on de novo protein synthesis (20–22). In agreement, defects of mRNA translation are implicated in cognitive syndromes associated with neurodegenerative diseases, including AD, prion disease, and frontotemporal dementia (FTD) (23–25). Briefly, AMPK activation results in the inhibition of mRNA translation through 2 potential mechanisms: (a) suppression of the mammalian target of rapamycin complex 1 (mTORC1) pathway, which controls synthesis of translational machinery and cap-dependent translation initiation, and (b) phosphorylation of eukaryotic elongation factor 2 (eEF2) kinase (eEF2K), leading to eEF2 phosphorylation and, consequently, disruption of the elongation step in mRNA translation (14). Moreover, recent studies indicate that interactions between AMPK and mTORC1 are dependent on eukaryotic initiation factor 2α subunit (eIF2α) activation levels (26). Phosphorylation of eIF2α by its kinase PKR-like ER kinase (PERK) results in suppression of general protein synthesis and has been linked to AD pathophysiology (23). Nevertheless, whether the role of AMPKα in the regulation of de novo protein synthesis is isoform specific is unknown.
In the present study, we reported isoform-specific dysregulation of AMPKα in postmortem human AD brain tissue (including both sporadic AD [sAD] and familial AD [FAD]). Using genetic approaches, we found that abnormal AMPKα1 upregulation plays a critical role in mediating AD-associated cognitive impairments and synaptic failure. Our findings reveal what we believe to be a previously unrecognized isoform-specific role of AMPK dysregulation in AD pathogenesis, which may provide insights into novel therapeutic avenues for AD and related dementia syndromes.
Results
Brain AMPKα isoform homeostasis is disrupted in AD.
To investigate regulation of AMPKα isoforms in AD, we first examined postmortem brain tissues from sAD patients (Table 1) and found that AMPKα1 levels were significantly increased, while AMPKα2 levels were markedly reduced in the hippocampus, as compared with those in age-matched controls (Figure 1A). Consistently, immunohistochemical staining of postmortem brain sections revealed AD-associated increased AMPKα1 and decreased AMPKα2 staining throughout the hippocampus, including both soma and neurites (Figure 1B). In contrast, no significant change in AMPKα isoforms was observed in the cerebellum (Figure1C). We further assayed postmortem hippocampal tissue from patients with neuropathologically confirmed Lewy body dementia (LBD) and FTD, 2 non-AD neuronal diseases with dementia syndromes (Tables 2 and 3). Notably, levels of AMPKα isoforms were not affected in either LBD or FTD, as compared with their respective age-matched controls (Figure 1, D and E). Interestingly, in postmortem hippocampal tissue from FAD patients (Table 4), we observed significantly elevated AMPKα1 expression, but no changes in AMPKα2 levels, as compared with tissue from age-matched controls (Figure 1F). Similarly, in hippocampal tissue from Tg19959 Tg mice that model FAD (27), there was a significant increase of AMPKα1 expression, but no changes in AMPKα2 levels (Figure 1G). Additionally, no changes were found in Tg mice in either AMPKβ or AMPKγ protein levels or phosphorylation of either AMPKα isoform (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI133982DS1). Finally, immunofluorescence combined with confocal imaging revealed elevated AMPKα1 levels in both soma and dendrites in the hippocampus of Tg mice (Figure1H). Taken together, these data demonstrate that AMPKα isoform homeostasis is disrupted in AD.
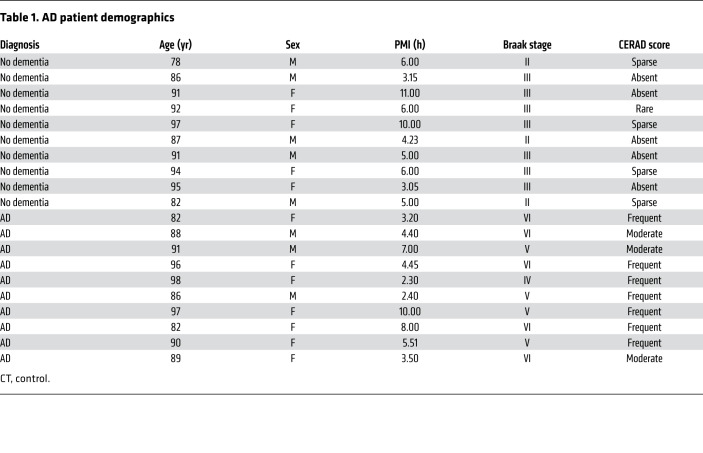
Table 1. AD patient demographics.
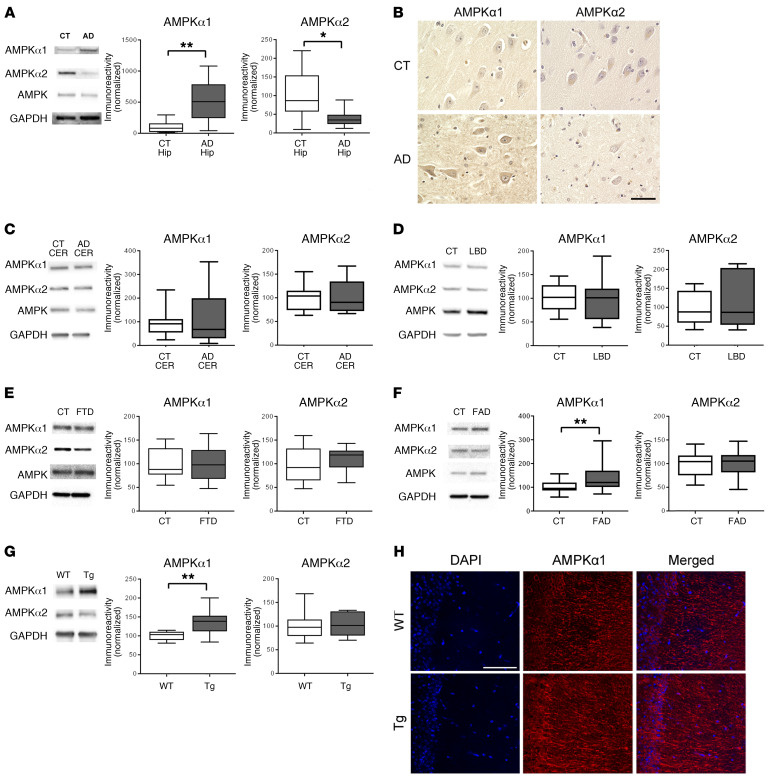
Figure 1. Expression of AMPKα isoforms is dysregulated in AD hippocampus.
(A) Hippocampus (Hip) lysate from sAD patients showed increased AMPKα1 and decreased AMPKα2 levels as compared with those of age-matched controls (CT). n = 10 with up to 4 technical replicates. *P = 0.0119; **P = 0.0014, unpaired t test. (B) Representative images of AMPKα isoform dysregulation in area CA1 of hippocampus in AD and age-matched control patients. Scale bar: 50 μm. Immunohistochemical experiments were replicated independently 3 times. (C) AMPKα isoform expression was unaffected in cerebellum (CER) samples from AD patients. n = 5 with up to 3 technical replicates. P = 0.8457 for AMPKα1; P = 0.9870 for AMPKα2, unpaired t test. (D) Hippocampal lysate from LBD patients had unaffected AMPKα isoform levels. n = 4 for control; n = 3 for LBD with 1 technical replicate. P = 0.9146 for AMPKα1; P = 0.5635 for AMPKα2, unpaired t test. (E) Levels of AMPK isoforms were unaltered in hippocampal tissue from FTD patients. n = 8 for control with 1 technical replicate; n = 5 for FTD with up to 3 technical replicates. P = 0.9283 for AMPKα1; P = 0.335 for AMPKα2, unpaired t test. (F) AMPKα1 levels were significantly increased in cortical lysates from FAD patients, while AMPKα2 levels were unaffected. n = 5 with 4 technical replicates. **P = 0.0060 for AMPKα1; P = 0.9412 for AMPKα2, unpaired t test. (G) AMPKα1 levels were significantly increased in hippocampal lysates from Tg19959 AD model mice compared with WT controls. AMPKα2 levels were unaffected. n = 7 with up to 2 technical replicates. **P = 0.0023 for AMPKα1; P = 0.9094 for AMPKα2, unpaired t test. Box-and-whisker plots represent the interquartile range, with the line across the box indicating the median. Whiskers show the highest and lowest values detected. (H) Immunofluorescent labeling of DAPI (blue) and AMPKα1 (red) distribution in area CA1 mouse hippocampal slices. n = 3. Scale bar: 200 μm.
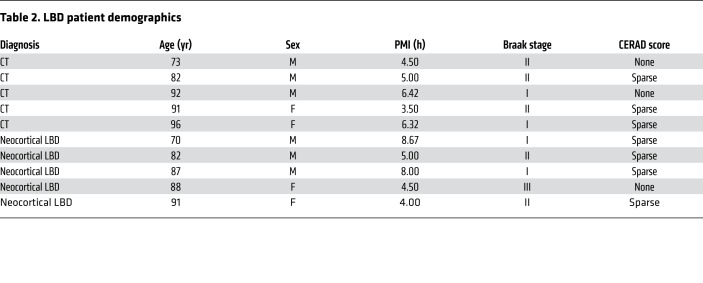
Table 2. LBD patient demographics.
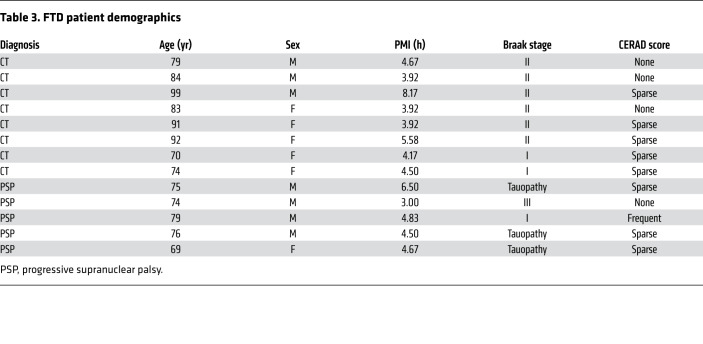
Table 3. FTD patient demographics.
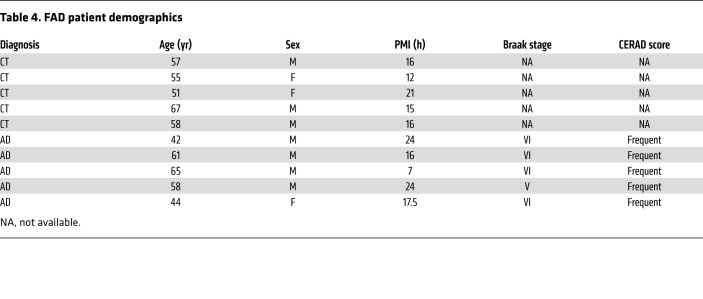
Table 4. FAD patient demographics.
Brain-specific suppression of AMPKα1 isoform alleviates learning and memory defects in Tg19959 AD model mice.
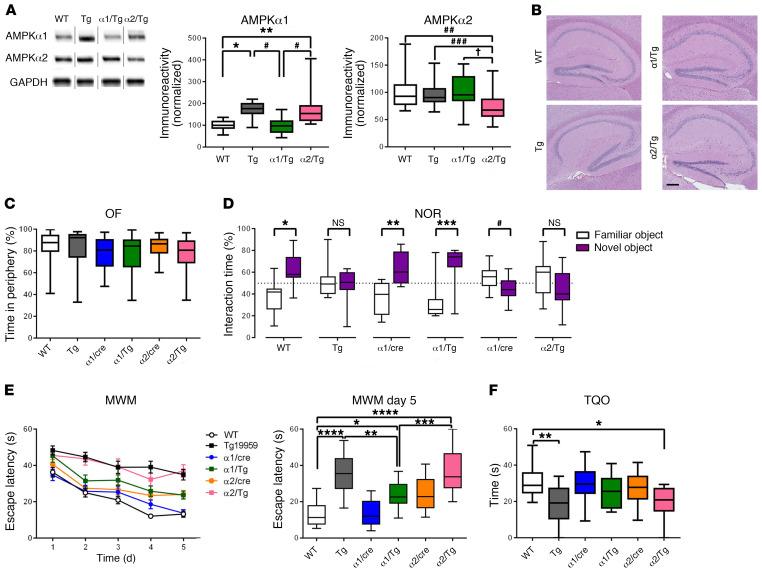
We further investigated whether there is an association between brain AMPK isoform dysregulation and AD pathophysiology. First, mice harboring loxP-flanked Prkaa1 or Prkaa2 were bred with mice expressing a brain-specific Cre recombinase (Camk2a-cre) (23) to generate heterozygous AMPKα1 or AMPKα2 conditional knockout mice (referred to as α1/cre and α2/cre respectively) (Supplemental Figure 1, D and E). Further, by crossing α1/cre and α2/cre mice with Tg19959 AD model mice, we generated Tg19959/AMPKα1+/– (α1/Tg) and Tg19959/AMPKα2+/– (α2/Tg) double-mutant mice along with 4 other experimental groups: WT, Tg19959 (Tg), α1/cre, and α2/cre mice. Western blot assay demonstrated that the increased levels of AMPKα1 in the hippocampi of Tg mice were restored to WT levels in α1/Tg mice, but remained elevated in α2/Tg mice (Figure 2A). Histological assays showed that the genetic reduction of AMPKα isoforms did not alter gross morphology of the hippocampus (Figure 2B and Supplemental Figure 1E).
Figure 2. Brain-specific suppression of AMPKα1 alleviates learning and memory defects in Tg19959 AD model mice.
(A) Brain-specific genetic reduction of AMPKα1 and AMPKα2 in Tg19959 AD model mice. Noncongruous WT, Tg19959 (Tg), AMPKα1+/–/Tg19959 (α1/Tg), AMPKα2+/–/Tg19959 (α2/Tg). n = 10, 10, 6, 7; up to 3 technical replicates. For AMPK α1: WT versus Tg, *P = 0.0212; WT versus α2/Tg, **P = 0.0029; Tg versus α1/Tg, ***P = 0.0130; α1/Tg versus α2/Tg, #P = 0.0016. One-way ANOVA with Tukey’s post hoc test, F = 8.218. For AMPKα2: WT versus α2/Τg, ##P = 0.005; Tg versus α2/Tg, ###P = 0.043; α1/Tg versus α2/Tg, †P = 0.007. One-way ANOVA with Tukey’s post hoc test. F = 7.585. (B) Representative H&E stain of hippocampal structure. n = 3. Scale bar: 50 μm. (C) Percentage of time spent in the periphery for the OF test. n = 25, 21, 17, 14, 19, and 13. (D) Percentage of time spent with familiar (white) and novel (purple) objects in the NOR task during the testing phase. Preference of less than 50% indicates cognitive impairment. n = 19, 13, 10, 9, 10, and 8. Statistical preference for novel or familiar object: WT, *P < 0.0001; Tg, P = 0.5523; α1/cre, **P = 0.0004; α1/Tg, ***P = 0.0008; α2/cre, #P = 0.0465; α2/Tg, P = 0.1497, unpaired t test. (E) Escape latency (s) over 5 days of training in the hidden platform MWM. Four trials/day, 5 days. n = 19, 17, 13, 17, 19, and 13. WT versus α1/Tg, *P = 0.0256; Tg versus α1/Tg, **P = 0.0094; α1/Tg versus α2/Tg, ***P = 0.0009; ****P < 0.0001, 1-way ANOVA with Tukey’s post hoc test. F = 18.16. (F) Percentage of time spent in the target quadrant during probe trial phase of MWM task. WT versus α2/Tg, *P = 0.0186; WT versus Tg, **P = 0.0027, 1-way ANOVA with Tukey’s post hoc test. F = 4.525. Box-and-whisker plots represent the interquartile range, with the line across the box indicating the median. Whiskers show the highest and lowest values detected.
To determine the consequence of selective AMPKα isoform reduction on AD-associated learning and memory impairments, we subjected the aforementioned mice to a series of behavioral tasks. First, we performed the open field (OF) test to assess general locomotor activity and baseline anxiety-like behavior and did not find any differences among all genotypes (Figure 2C and Supplemental Figure 1, F and G). We then tested the mice in the novel object recognition (NOR) task to evaluate their long-term hippocampus-dependent recognition memory (28). WT and α1/cre mice exhibited normal cognition, as indicated by their preference for the novel object over the familiar object on the test day (i.e., significantly more interaction with the novel object) (Figure 2D). In contrast, Tg mice spent similar amounts of time with either the familiar or novel objects, indicating deficits in learning and memory (Figure 2D and ref. 29). Notably, AD model mice with reduced AMPKα1 (α1/Tg) demonstrated normal cognition, as indicated by significantly more time spent with the novel objects (Figure 2D).
Next we assessed spatial learning and memory by testing mice with the hidden platform Morris water maze (MWM) behavioral task (28), a well-validated test of hippocampus-dependent spatial memory. Consistent with results from the NOR task, Tg mice displayed impaired learning and memory. During the acquisition (learning) phase, WT mice demonstrated marked day-to-day decrease in escape latency, whereas Tg mice exhibited longer escape latencies (Figure 2E). In the probe trial, Tg mice spent less time in the target quadrant compared with WT mice (Figure 2F). Notably, impairments of spatial learning and memory associated with Tg mice were alleviated by suppressing AMPKα1, as indicated by decreased escape latency and improved target quadrant occupancy (not significantly different from WT) by α1/Tg mice (Figure 2, E and F, and Supplemental Figure 1, H–J). In comparison, reduction of AMPKα2 did not alter cognitive deficits in Tg mice (Figure 2, E and F). In order to examine potential memory-independent effects of AMPKα isoform suppression, such as swimming ability or visual impairments, mice were also run through the visible platform (VP) task. No differences in latency to locate the visible platform were observed across all groups (Supplemental Figure 1K). Taken together, these data show that suppression of AMPKα1, but not AMPKα2, alleviates AD-associated learning and memory impairments.
Suppression of AMPKα isoforms does not alter AD-associated brain Aβ pathology or tau phosphorylation.
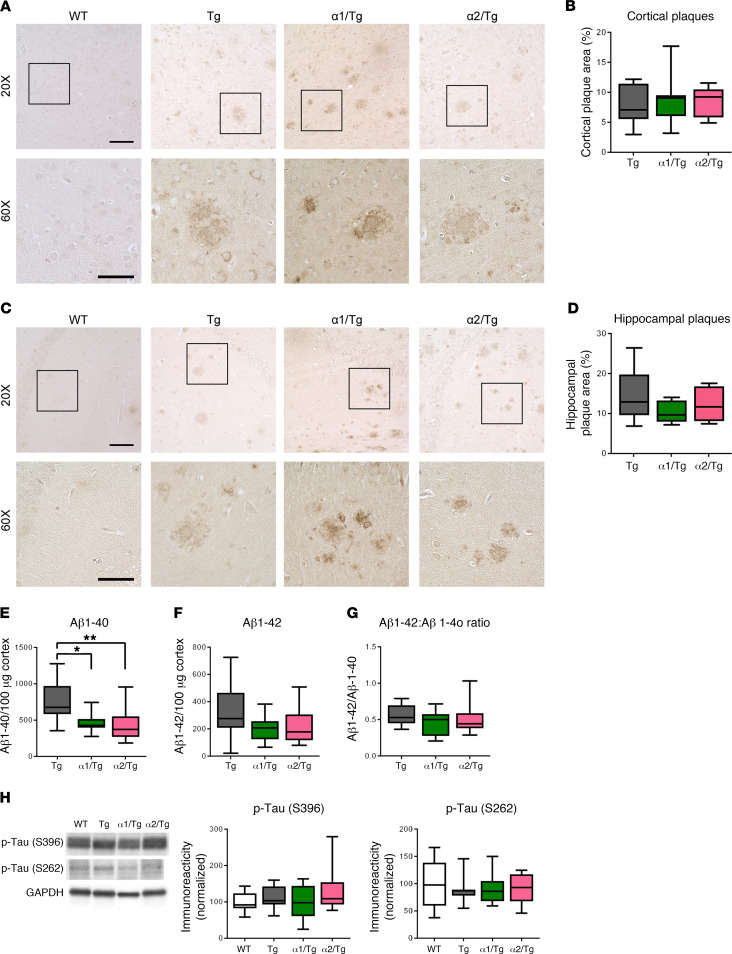
We went on to investigate potential mechanisms associated with the beneficial effects on cognition with AMPKα1 suppression in Tg mice. First, we analyzed brain Aβ pathology in the cortical and hippocampal brain areas and found similar Aβ plaque deposition in Tg and α1/Tg mice (Figure 3, A and B). Additionally, reduction of AMPKα2 did not alter brain Aβ plaque pathology either (Figure 3, C and D). We then used ELISA to quantify the brain levels of Aβ1-40 and Aβ1-42. Compared with Tg mice, both α1/Tg and α2/Tg mice showed decreased Aβ1-40 levels, whereas levels of Aβ1-42 were not altered, and neither was the ratio of Aβ42:40 (Figure 3, E–G). Furthermore, reduction of either AMPKα isoform did not alter levels of amyloid precursor protein (APP) and key APP-processing enzymes, including components of γ- and β-secretase (Supplemental Figure 2, A–C). Levels of hippocampal Aβ monomers assessed by Western blot were not changed with AMPKα isoform suppression (Supplemental Figure 2D). Interestingly, phosphorylation of tau at either S262 or S396 sites (known to be phosphorylated by AMPK; ref. 30), was unaffected by repression of either AMPKα isoform (Figure 3H). Levels of total tau were unaffected as well (Supplemental Figure 2E). These data indicate that alleviation of cognitive impairments in Tg mice by reducing AMPKα1 is unlikely to be associated with effects on brain Aβ pathology or tau phosphorylation.
Figure 3. Suppression of AMPKα does not alter AD-associated brain Aβ pathology or tau phosphorylation.
(A) Representative images of cortical Aβ plaque deposition in WT, Tg, α1/Tg, and α2/Tg mice. Scale bars: 100 μm (×20 images); 50 μm (×60 images). (B) Percentage of cortex covered by Aβ plaques in Tg, α1/Tg, and α2/Tg mice. n = 9 slices/3 mice. (C) Representative images of hippocampal Aβ plaque deposition in WT, Tg, α1/Tg, and α2/Tg mice. Scale bars: 100 μm (×20 images); 50 μm (×60 images). (D) Percentage of hippocampal area covered by amyloid plaques in Tg, α1/Tg, and α2/Tg mice. n = 9 slices/3 mice. (E) ELISA of prefrontal cortex lysate showed decreases of Aβ1-40 in both α1/Tg and α2/Tg mice, as compared with Tg mice. Tg versus α1/Tg, *P = 0.0356; Tg versus α2/Tg **P = 0.0226, 1-way ANOVA with Tukey’s post hoc test. F = 5.034. (F) ELISA showed no differences in levels of Aβ1-42 in Tg, α1/Tg, and α2/Tg mice. One-way ANOVA with Tukey’s post hoc test. F = 1.771. (G) The ratio of Aβ42:40 was unaltered in Tg, α1/Tg, and α2/Tg mice. One-way ANOVA with Tukey’s post hoc test. F = 1.635. Tg, n = 12; α1/Tg and α2/Tg, n = 9. (H) Western blot analysis of p-tau (S396 and S262) levels in hippocampus showed no differences among WT, Tg, α1/Tg, and α2/Tg mice. WT, n = 8; Tg, n = 6; α1/Tg, n = 6; α2/Tg, n =7. One-way ANOVA. Box-and-whisker plots represent the interquartile range, with the line across the box indicating the median. Whiskers show the highest and lowest values detected.
Reduction of brain AMPKα1 corrects AD-associated deficits in synaptic density and dendritic spine morphology.
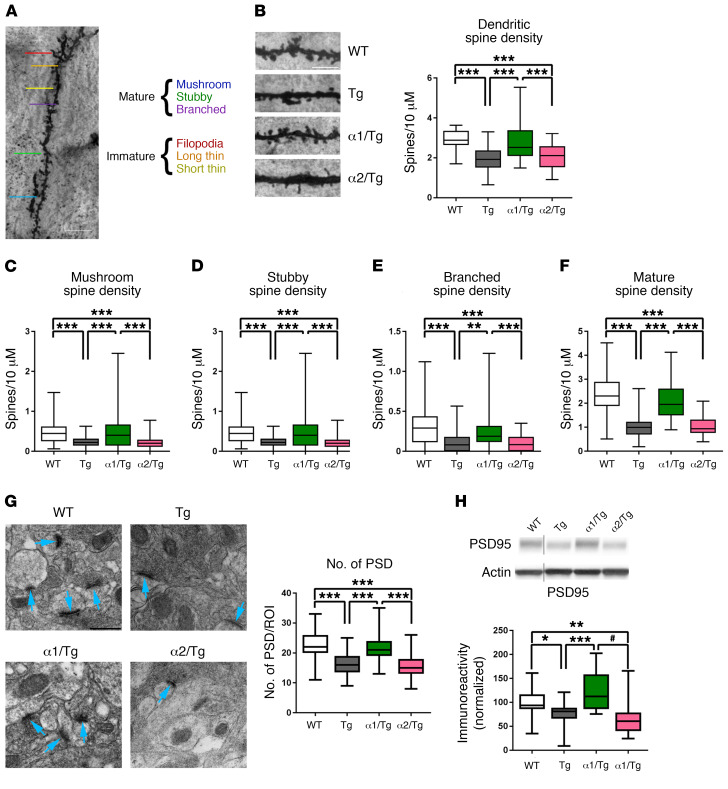
AD is considered a disease of synaptic failure (31). Loss of synapses or synaptic density correlates with memory impairments in both human AD patients and animal models of AD (12, 32, 33). Dendritic spine morphology is critical for synaptic integrity and closely associated with neural plasticity and memory formation (34, 35). We used the rapid Golgi-Cox staining technique to assess spine density and morphology changes of dendritic spines within area CA1 of the hippocampus. Consistent with a previous study (28), overall dendritic spine density in Tg mice was significantly decreased as compared with that in WT controls (Figure 4B). Importantly, AD-associated reduction of dendritic spine density was restored by suppressing AMPKα1, but not AMPKα2 (Figure 4B). We further analyzed spine morphology changes based on established guidelines on classification of “mature” and “immature” spine types (Figure 4A and ref. 36). We found that the density of mature spines (mushroom, stubby, and branched) in Tg mice was significantly decreased as compared with that of WT controls, and genetic reduction of AMPKα1 restored those defects in Tg mice (Figure 4, C–F). Interestingly, density of overall immature spines (thin and filopodia) was higher in Tg mice compared with that in WT controls, and that increase was blunted by suppressing AMPKα1 (Supplemental Figure 3, A–C).
Figure 4. Reduction of brain AMPKα1 corrects AD-associated deficits in synaptic density and dendritic spine morphology.
(A) Classification of mature (mushroom, stubby, and branched) and immature (filopodia, long thin, and short thin) spine types on dendrite. Scale bar: 20 μm. (B) Representative images from Golgi-Cox stain of area CA1 dendritic spines and quantification of total (mature and immature) spine density. Tg and α2/Tg spines have significantly decreased spine density as compared with those in WT and α1/Tg mice. Scale bar: 5 μm (×100). ***P < 0.0001, 1-way ANOVA with Tukey’s post hoc test. F = 107.2. (C–E) Quantification of subclassification of mature spines. Tg and α2/Tg mice have significantly fewer stubby (F = 18.84), branched (F = 20.87), and mushroom (F = 64.33) spines than WT and α1/Tg mice. **P < 0.005; ***P < 0.0001, 1-way ANOVA with Tukey’s post hoc test. (F) Quantification of mature spine density. WT, n = 4; Tg, α1/Tg, and α2/Tg, n = 3. Spine length (200 μm) analyzed from 5 regions of interest (ROIs) per slice, 3 to 7 slices per mouse. ***P < 0.0001, 1-way ANOVA with Tukey’s post hoc test. (G) Representative TEM from hippocampal CA1 dendrites and quantification of postsynaptic densities (PSDs). n = 3 animals per group. Scale bar: 500 μm. Blue arrows indicate PSDs. Number of PSDs is significantly decreased in Tg and α2/Tg mice as compared with WT and α1/Tg mice. ***P < 0.0001, 1-way ANOVA with Tukey’s post hoc test. F = 49.77. (H) Western blot analysis of hippocampal lysate showed significantly reduced levels of PSD95 in Tg and α2/Tg mice compared with WT and α1/Tg mice. WT, n = 10; Tg, n = 9; α1/Tg, n = 6; α2/Tg, n = 7. *P = 0.0305; **P = 0.0047; Tg versus α1/Tg, ***P = 0.0006; Tg versus α2/Tg, #P < 0.0001, 1-way ANOVA with Tukey’s post hoc test. F = 9.572, noncongruous. Box-and-whisker plots represent the interquartile range, with the line across the box indicating the median. Whiskers show the highest and lowest values detected.
Moreover, we investigated potential alterations of postsynaptic densities (PSDs) by using transmission electron microscopy (TEM). PSDs are located at the heads of spines and are vital for synaptic function (34, 37). Ultrastructural analysis revealed decreased PSD density in area CA1 of the hippocampus in Tg mice, which was restored in α1/Tg mice, but unaffected in α2/Tg mice (Figure 4G). Analysis of hippocampal PSD-95 levels by Western blot assay showed a similar deficit in Tg mice, which was also restored by genetic suppression of AMPKα1, but not AMPKα2 (Figure 4H). Taken together, these data suggest that genetic repression of AMPKα1 alleviated defects in hippocampal spine density/morphology and PSD formation associated with Tg19959 AD model mice. These findings are consistent with the results from the behavioral experiments described above (Figure 2).
Repression of brain AMPKα1 restores abnormal eEF2 phosphorylation and de novo protein synthesis deficits in Tg19959 mice.
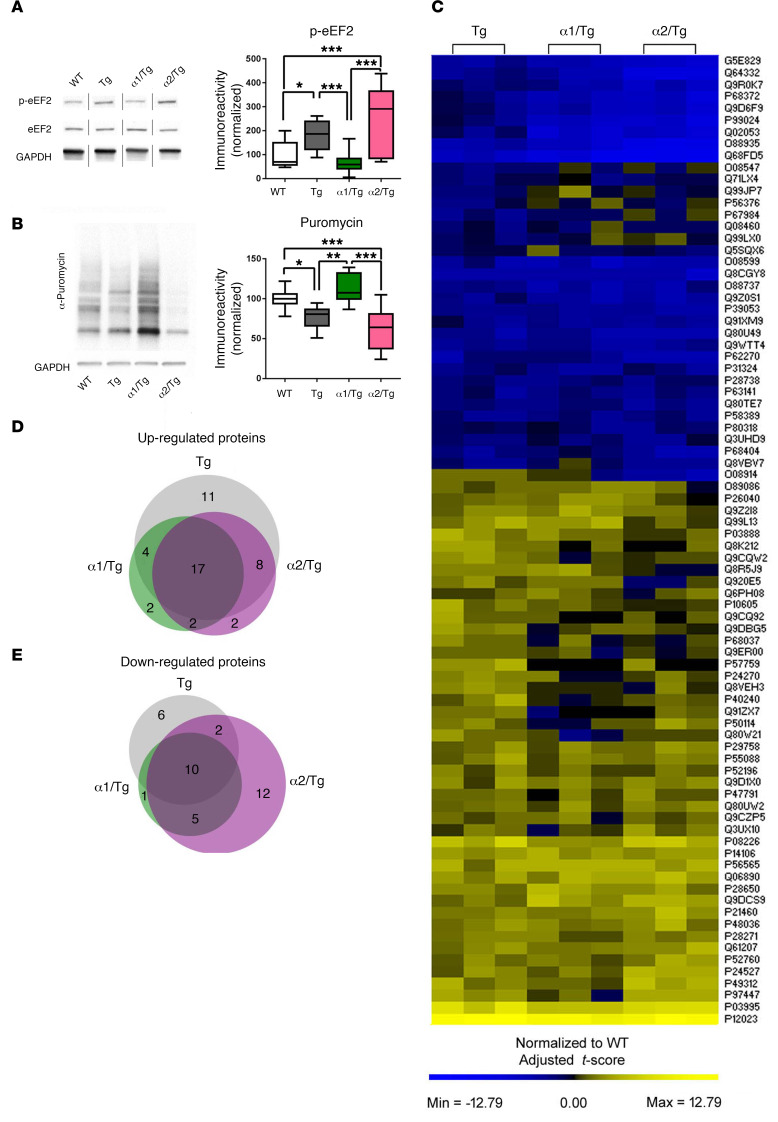
De novo protein synthesis is essential for memory formation and synaptic plasticity and is notably impaired in AD (23, 28, 38). As described in the Introduction, AMPK may regulate protein synthesis via 2 main downstream signaling pathways: mTORC1 and eEF2K/eEF2 (14). Biochemical assay showed that brain mTORC1 signaling was not affected by reducing either AMPKα1 or AMPKα2, as indicated by unaltered phosphorylation levels of mTOR and TSC2 in α1/Tg or α2/Tg mice, as compared with those in WT or Tg mice (Supplemental Figure 4, A and B). Additionally, we examined hippocampal protein levels of the structural components of protein phosphatase 2A (PP2A), a potential phosphatase and key regulator for AMPK activity (39, 40), and did not observe any alterations (Supplemental Figure 4, C–E). Levels of eEF2 phosphorylation were elevated in hippocampi of Tg mice, in agreement with recent studies (28, 41). Markedly, AD-associated eEF2 hyperphosphorylation was restored with selective AMPKα1, but not AMPKα2, suppression (Figure 5A).
Figure 5. Repression of brain AMPKα1 restores abnormal eEF2 phosphorylation and de novo protein synthesis deficits in Tg19959 mice.
(A) Genetic reduction of AMPKα1, but not AMPKα2, corrected eEF2 hyperphosphorylation in hippocampi of Tg19959 AD model mice. n = 5 with 3 technical replicates, noncongruous. WT versus Tg, *P = 0.0297; ***P < 0.001, 1-way ANOVA with Tukey’s post hoc test. F = 13.88. (B) Representative images and quantification for SUnSET puromycin incorporation assay. Assay was performed on area CA1 of hippocampus. WT, Tg, and α2/Tg, n = 5; α1/Tg, n = 3. *P = 0.0231; **P = 0.0027; ***P < 0.001, 1-way ANOVA with Tukey’s post hoc test, F = 12.85. Box-and-whisker plots represent the interquartile range, with the line across the box indicating the median. Whiskers show the highest and lowest values detected. (C) Patterns of protein expression in hippocampi of WT, Tg, α1/Τg, and α2/Tg mice, reported as normalized deviation from the mean. (D and E) Venn diagrams showing proteins that were significantly altered (upregulated or downregulated) in the Tg conditions that were restored to WT levels in α1/Tg, α2/Tg, or both α1/Tg and α2/Tg hippocampi.
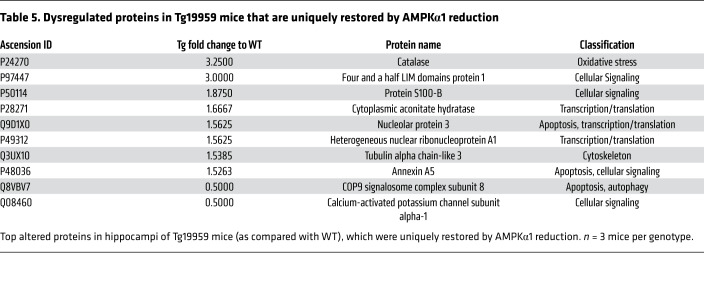
To directly measure de novo protein synthesis, we used the surface sensing of translation (SUnSET, a nonradioactive puromycin end-labeling assay) on living acute hippocampal slices (42, 43). In agreement with previous studies (44), de novo protein synthesis in hippocampal slices (assessed by puromycin labeling) was reduced in Tg mice as compared with WT controls (Figure 5B). Notably, AD-associated protein synthesis defects were alleviated by reduction of AMPKα1 (Figure 5B). AMPKα2 repression did not improve de novo protein synthesis deficiency in Tg mice (Figure 5B). We further examined potential isoform-specific effects of AMPKα repression on protein synthesis with mass spectrometry–based (MS-based) proteomic experiments. Altered proteins were summarized using a heatmap (Figure 5C). Briefly, proteins identified as significantly altered from the WT in other genotypes were pooled, and for each protein, a deviation from the mean was calculated and mapped using PermutMatrix software (45). Of all the proteins identified (2041 in total), 27 were commonly dysregulated (17 upregulated, 10 downregulated). From the pooled proteins, 10 proteins were identified as being uniquely restored with AMPKα1 repression, including proteins involved in apoptosis, Ca2+ signaling, cytoskeletal dynamics, and oxidative stress (Figure 5, C–E, and Table 5). Thus, AD-associated abnormal eEF2 phosphorylation and de novo protein synthesis deficits were lessened by genetic reduction of AMPKα1 isoform.
Table 5. Dysregulated proteins in Tg19959 mice that are uniquely restored by AMPKα1 reduction.
Reduction of brain AMPKα1 mitigates impairments of cognition and synaptic plasticity associated with APP/PS1 AD model mice.
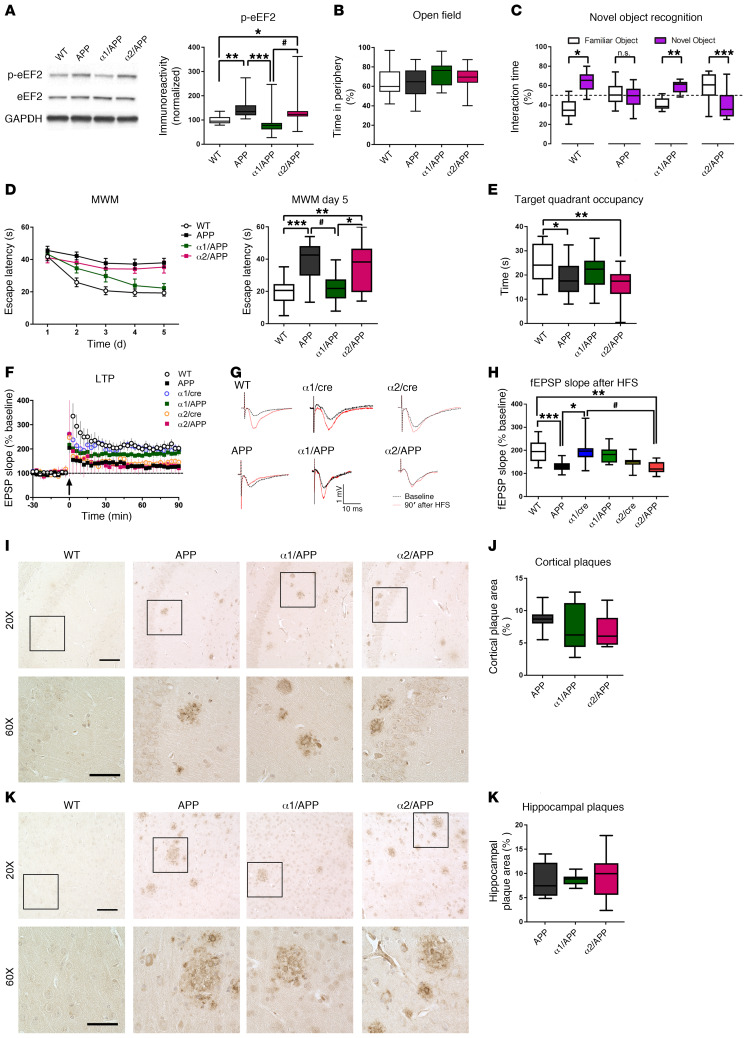
We further determined whether the above findings related to selective AMPKα isoform suppression can be replicated in another rodent model of AD. We crossed APP/PS1 AD model mice (46) with either α1/cre or α2/cre mice to generate APP/PS1/AMPKα1+/– (α1/APP) and APP/PS1/AMPKα2+/– (α2/APP) double-mutant mice. AMPKβ and AMPKγ levels were unaffected among different genotypes (Supplemental Figure 5, A and B). As was seen in the Tg mice, APP/PS1 mice also exhibited elevated levels of eEF2 phosphorylation, which was corrected by AMPKα1 (but not AMPKα2) repression (Figure 6A). To verify the effects of selective AMPKα isoform suppression on AD-associated learning and memory deficits, mice were subjected to the same behavioral battery as the Tg mice cohort. Briefly, baseline anxiety-like behavior, assessed by OF task, was indistinguishable among all groups (Figure 6B and Supplemental Figure 5, C and D). Compared with WT control mice, APP/PS1 mice displayed learning and memory impairments based on results from both NOR and hidden platform MWM behavioral tasks (Figure 6, C–E). Importantly, AD-associated cognitive deficits were alleviated by selective suppression of AMPKα1, as demonstrated by normal learning and memory performance in α1/APP mice shown in both NOR and MWM behavioral tests (Figure 6, C–E). In contrast, α2/APP mice showed similar impaired cognition in both tests, as compared with APP/PS1 mice (Figure 6, C–E). We also evaluated regulation of long-term synaptic plasticity in these mice by examining hippocampal long-term potentiation (LTP), one of the most intensively studied cellular models for learning and memory (47, 48). APP/PS1 mice showed LTP impairments that were improved by suppression of AMPKα1, but not AMPKα2 (Figure 6, F–H). Interestingly, suppression of AMPKα2 (but not AMPKα1) resulted in LTP impairment (Figure 6, F–H). Moreover, brain Aβ pathology in APP/PS1 mice was unaltered with repression of either AMPKα isoform (Figure 6, I–L, and Supplemental Figure 6, A–C). Additionally, there was no change in tau phosphorylation or total tau levels across groups (Supplemental Figure 6, D and E). These results are consistent with the findings in Tg19959 AD model mice described above, indicating that brain-specific genetic suppression of AMPKα1 is able to prevent AD-associated cognitive defects and synaptic plasticity impairments.
Figure 6. Reduction of brain AMPKα1 mitigates impairments of cognition and synaptic plasticity associated with APP/PS1 AD model mice.
(A) Genetic reduction of AMPKα1, but not AMPKα2, corrected eEF2 hyperphosphorylation in hippocampi of APP/PS1 mice. n = 4. WT versus α2/APP, *P = 0.0464; WT versus APP, **P = 0.0029; α1/ΑPP versus α2/APP, #P = 0.006; α1/ΑPP versus APP, ***P = 0.0003; 1-way ANOVA with Tukey’s post hoc test. F = 8.455. (B) Unaltered periphery time in OF task. n = 28, 20, 13, and 20. (C) Preference for familiar (white) or novel (purple) objects in the NOR task. n = 12, 12, 10, and 10. WT, *P < 0.0001; APP, P = 0.7602; α1/APP, **P < 0.0001; α2/Tg, ***P = 0.0123, unpaired t test. (D) Escape latency or latencies in the hidden-platform MWM. Escape latency of day 5 was significantly higher in APP and α2/APP mice. n = 18, 18, 9, and 15. α1/ΑPP versus α2/APP, *P = 0.0259; WT versus α2/APP, **P = 0.0013; APP versus α1/APP, **P = 0.0064; WT versus APP, ***P < 0.0001. F = 10.53. (E) Target quadrant occupancy during probe trial of MWM task. WT versus APP, *P = 0.0298; WT versus α2/APP, **P = 0.003, 1-way ANOVA with Tukey’s post hoc test. F = 5.259. (F) Hippocampal LTP. HFS denoted by arrow. n = 18, 12, 12, 7, 8, and 9. (G) Representative fEPSP traces before and after HFS. (H) Quantification of the fEPSP slope 90 minutes after HFS. WT versus APP, ***P = 0.0009; WT versus α2/APP, **P = 0.0011; α1/cre versus APP, *P = 0.0017; α1/cre versus α2/APP, #P = 0.0017, 1-way ANOVA with Tukey’s post hoc test. F = 7.519. (I and J) Representative images and quantification of cortical Aβ plaque deposition. n = 9 slices/3 mice. (K and L) Representative images and quantification of hippocampal Aβ plaque deposition. n = 9 slices/3 mice. Scale bars: 100 μm (×20 images); 50 μm (×60 images). Box-and-whisker plots represent the interquartile range, with the line across the box indicating the median. Whiskers show the highest and lowest values detected.
Discussion
Among the top 10 causes of death, AD is the only disease without any effective therapeutic approach (1). The incidence of AD has been increasing dramatically with global aging, and it is imperative to identify novel therapeutic targets based on solid mechanistic studies. In the current study, we report that brain-specific suppression of AMPKα1 selectively alleviated cognitive deficits and synaptic failure displayed in 2 separate lines of AD model mice, revealing isoform-specific roles of AMPKα in AD pathophysiology. Energy metabolism dysregulation is linked to neurodegenerative diseases, including AD (11, 12, 49). AMPK is the central energy sensor at the molecular level and is considered to regulate all aspects of cellular function (19). As a master kinase functioning to maintain energy homeostasis, AMPK plays a key role in many biological processes. Considering all the downstream effectors of AMPK, dysregulation of AMPK signaling may be connected to many, if not all, cellular abnormalities and functional impairments in AD (5).
It is noteworthy that disruption of the AMPKα isoform expression levels in hippocampus was found in AD, but not in LBD or FTD, postmortem brain tissue (Figure 1). LBD cases are characterized by hippocampal α-synuclein without substantial levels of p-tau (50, 51). All FTD patients reported here were diagnosed with progressive supranuclear palsy, and such patients typically have increased levels of tau phosphorylation in hippocampus (52). Furthermore, neither LBD nor FTD cases exhibit significant hippocampal Aβ deposition (51, 52). Our data indicate that dysregulation of AMPKα isoform homeostasis might be specific to AD-related pathological processes, such as Aβ or phosphorylation of AD-type tau, but not related to α-synuclein, frontotemporal lobar degeneration–tau (FTLD-tau), or nonspecific conditions associated with neurodegenerative diseases in general. Impaired brain energy metabolisms have been implicated in many neurodegenerative diseases (11). Our current study suggests AD-specific dysregulation of brain AMPKα isoform expression, which may provide insights into development of novel biomarkers and therapeutic approaches for AD. Meanwhile, our findings do not exclude a role of (aberrant) AMPK signaling in pathogenesis of other neurodegenerative diseases. Furthermore, it is possible that upregulation of AMPKα1 expression in sAD is compensatory in response to downregulation of AMPKα2 expression to maintain homeostasis or vice versa. Interestingly, while increased levels of hippocampal AMPKα1 were observed in both sAD and FAD cases, reduction of hippocampal AMPKα2 levels was seen only in sAD, but not in FAD. Further, the pattern of hippocampal AMPKα isoform dysregulation in Tg19959 AD model mice, which are models of FAD, matches those of FAD patients. These differences could be attributed to factors such as aging (FAD cases are younger than sAD in our study) or certain comorbidities in sAD cases, and future studies are warranted to elucidate the nature and source of this deviation.
Multiple studies indicate a role of AMPK in AD-associated Aβ and tau pathology (5, 6, 53, 54). AMPK regulation may affect AD brain pathology, although controversy arises regarding whether Aβ accumulation and tau hyperphosphorylation can be alleviated or aggravated by the activation of AMPK (5, 6, 53, 54). On the other hand, increased AMPK activity (assessed by AMPKα phosphorylation) was observed in brain tissue from AD patients and Tg mouse models of AD and was induced by exogenous Aβ application (3, 13). Thus, there probably exists a reciprocal relationship between AMPK dysregulation and AD brain pathology. It is worth mentioning that these studies were conducted across multiple experimental models and species, which could contribute to the seemingly inconsistent findings (55). Although AMPK expression is conserved across many species, little is known about the exact roles of AMPK isoforms and their subcellular distribution in neuronal systems. To the best of our knowledge, none of the aforementioned studies actually clarifies potential distinct roles of AMPK isoforms in AD-associated impairments of both cognition and synaptic plasticity in their experimental paradigms. In the current study, reducing AMPKα1 or AMPKα2 did not alter brain levels of either Aβ42 or p-tau in AD model mice. Future studies to elucidate the complexity of AMPK isoforms in the central nervous system will provide insights into some of the controversy surrounding the effects of AMPK and AMPK-related drugs (e.g., metformin) on AD (56–58). The current study indicates that brain-specific repression of AMPKα2 does not improve cognitive deficits and synaptic plasticity impairments in AD model mice. It was previously reported that LTP failure induced by application of exogenous synthetic Aβ in acute hippocampal slices was prevented in slices derived from global AMPKα2 knockout mice (3). These results suggest different roles of AMPKα2 regulation in AD-related synaptic plasticity impairments under acute or chronic circumstances. Additionally, potential compensatory signaling mechanisms (e.g., upregulation of other AMPK isoforms) associated with global AMPKα2 knockout may contribute to the different performance for LTP.
The findings that AD-associated hippocampal eEF2 hyperphosphorylation was blunted by selectively reducing AMPKα1 is particularly interesting. As an mRNA translational factor, eEF2 mediates the translocation step during the elongation phase of protein synthesis, catalyzing movement of tRNA from the ribosomal A-site to the P-site (59, 60). Phosphorylation of eEF2 at Thr56 leads to disruption of new peptide growth and thus inhibits overall protein synthesis (59). AMPK is known to be activated in low energy status, leading to the inhibition of protein synthesis/mRNA translation (14). Repression of mRNA translation by eEF2 phosphorylation (under the condition of AMPK activation) may initially serve as a protective strategy to conserve energy resources (by turning off major energy-consuming processes, such as protein synthesis) for cells to cope with low energy states. However, a prolonged increase of eEF2 phosphorylation and inactivation (e.g., in the case of AD) would be detrimental for cognitive function, since de novo protein synthesis is indispensable for long-lasting forms of synaptic plasticity and memory consolidation. Indeed, cognitive impairments associated with multiple neurodegenerative diseases are linked to defects in capacity of de novo protein synthesis (23–25). Consistently, a recent study shows that repression of eEF2K and thus eEF2 phosphorylation is sufficient to improve multiple aspects of pathophysiology in AD model mice (28).
The results of the proteomic experiments may drive future research into AMPK isoforms in AD in several ways. Each of the proteins found to be uniquely restored with selective AMPKα1 repression indicates potential mechanisms through which AMPKα1 repression is beneficial in AD. A variety of pathways are also implicated, including those in apoptosis, cell signaling, mRNA translation, and oxidative stress (Table 5). Among the identified proteins restored in selective AMPKα1 repression is S100 calcium-binding protein B (S100B). S100B overexpression is seen in inflammation and astrocyte activation surrounding Aβ plaques and is thought to help drive disease progression (61, 62). Also identified in the proteomics screen were proteins involved in membrane and spine dynamics, including calcium-activated potassium channel subunit α-1 (KCa1.1). Levels of KCa1.1 were significantly reduced in Tg mice and were restored by suppression of AMPKα1, but not AMPKα2. KCa1.1 is activated by cytosolic Ca2+ increases or membrane depolarization and opens to export K+ and is implicated in intellectual and seizure disorders (63). Notably, aberrant intracellular calcium signaling has been linked to AD pathogenesis (64, 65).
In summary, our findings indicate that targeting brain dysregulation of AMPK isoforms, particularly AMPKα1, might be a feasible therapeutic strategy for AD-associated cognitive impairments. Future endeavors are critical for identifying small molecule modulators that are selective for AMPKα isoforms to be tested in AD models. Moreover, given the central role of AMPK in energy metabolism regulation and the complexity of the AMPK signaling network, future work is necessary to determine the detailed molecular mechanisms underlying the interactions between AMPK isoforms and AD pathogenesis at different stages (early vs. late) or forms (familial vs. sporadic).
Methods
Postmortem tissue samples.
Postmortem human tissue was obtained from the University of Washington School of Medicine Brain Bank and the NIH NeuroBioBank affiliated with the University of Maryland (Baltimore, Maryland, USA). Diagnoses were based on cognitive testing, postmortem Braak (AD stages V–VI) stage, and Consortium to Establish a Registry for Alzheimer’s disease (CERAD) scores (66, 67). Studies were performed using hippocampal tissue from male and female patients clinically diagnosed with AD and age-matched controls. Mean age of death was 89.6 years. Postmortem interval (PMI) ranged between 2 and 10 hours, with a mean of 5.3 hours. For hippocampal tissue from FTD patients and age-matched controls, mean age of death was 80.4 years with a PMI between 3 and 9 hours and average of 4.8 hours. For hippocampal tissue from LBD patients and age-matched controls, mean age of death was 85.2 years with a PMI between 3.5 and 9 hours and an average of 5.6 hours. Patient information is presented in Table 1, Table 2, Table 3, and Table 4.
Mice.
All mice were housed at the Wake Forest School of Medicine barrier facility under the supervision of the Animal Research Program. Mice adhered to a 12-hour light/12-hour dark cycle with regular feeding, cage cleaning, and 24-hour food and water access. Both male and female mice were used for experimentation. Breeders for Tg19959 AD model mice were provided by George Carlson of McLaughlin Research Institute (Great Falls, Montana, USA) (27). APP/PS1 mice were purchased from the Jackson Laboratory and expressed human APP (KM670/671NL) and presenilin-1 (68). The following types of mice were purchased from the Jackson Laboratory: B6.Cg-Tg(Camk2a-cre)T29-1Stl/J (Camk2a-cre mice; stock 005359); Prkaa1tm1.1Sjm/J (loxP-flanked Prkaa1 mice; stock 014141); and Prkaa2tm1.1Sjm/J (loxP-flanked Prkaa2 mice; stock 014142). Mice harboring loxP-flanked Prkaa1 or Prkaa2 were bred with mice expressing a brain-specific Cre recombinase (Camk2a-cre) (23) to generate AMPKα1 or AMPKα2 conditional knockout mice (referred to as α1/cre and α2/cre). These mice were further crossed with AD model mice to generate the double-mutant mice. Genotypes were verified with PCR. Tg19959 mice age 6 to 9 months and APP/PS1 mice age 12 to 16 months were used for all experiments (28, 68).
Western blot assay.
Tissues were removed from appropriate structures and flash frozen on dry ice. Tissues were then homogenized in an appropriate lysis buffer and quantified as previously described (43). Samples were loaded on 4% to 15% TGX Precast Gels (Bio-Rad) and transferred to nitrocellulose membranes. Membranes were blocked and then probed overnight at 4°C using primary antibodies of interest. Blots were washed, and HRP-labeled secondary antibodies were added. Primary antibodies used were as follows: AMPKα1 (1:1000; Cell Signaling Technology, catalog 2795S), AMPKα2 (1:1000; Cell Signaling Technology, catalog 2757S), AMPKα1 (1:1000; Abcam, catalog ab3759), AMPKα2 (1:1000; Abcam, catalog 3760), p-tau (Ser396) (1:1000; Thermo Fisher, catalog 44-752G), p-tau Ser262 (1:1000; Thermo Fisher, catalog 44-750G), Aβ (6E10) (1:1000; MilliporeSigma, catalog Sig39320), APP (1:1000; Cell Signaling Technology, catalog CS2452), BACE1 (1:1000; Cell Signaling Technology, catalog CS5606), presenilin 2 (PS2) (1:1000; Cell Signaling Technology, catalog CS9979), PSD-95 (1:1000; Cell Signaling Technology, catalog Cs3450), puromycin (1:5000; Millipore, catalog AB3258), p-eEF2 (Thr 56) (1:1000; Cell Signaling Technology, catalog CS2331), eEF2 (1:1000; Cell Signaling Technology, catalog CS2332), PP2AA (1:1000; Cell Signaling Technology, catalog CS2041), PP2AB (1:1000; Cell Signaling Technology, catalog cs4953), PP2AC (1:1000; Cell Signaling Technology, catalog 2038), p-mTOR (Ser2448) (1:1000; Cell Signaling Technology, catalog CS2971), mTOR (1:1000; Cell Signaling Technology, catalog CS2983), p-TSC2 (Thr 1462) (1:1000; Cell Signaling Technology, catalog CS2983), β-actin (1:10000, MilliporeSigma, catalog A2228), GAPDH (1:10,000, Cell Signaling Technology, catalog CS5174), AMPKβ1 (1:1000; Abcam, catalog 71C10), AMPKγ (1:1000; Invitrogen, Thermo Fisher Scientific, catalog PA5-36314), tau (1:1000; MilliporeSigma, catalog T5530), catalase (1:1000; Cell Signaling Technology, catalog 14097S), and KCaα1 (1:1000; Abcam, catalog ab3586). All antibodies were diluted in either 5% w/v milk/TBST or 5% w/v BSA/TBST. The blots were visualized using chemiluminescence (Clarity ECL; Bio-Rad) and the Bio-Rad ChemiDoc MP Imaging System. Densitometric analysis was performed using ImageJ software (NIH). Data were normalized to β-actin or GAPDH (for total protein analysis) or relevant total proteins (for phospho-protein analysis) unless otherwise specified.
Immunohistochemistry for postmortem human tissue.
Postmortem tissue sections from patients were prepared at the University of Washington. Brains were fixed in 10% neutral buffered formalin. Hippocampal samples were embedded in paraffin and sectioned at 5-μm thickness. Sections were mounted on positively charged slides and baked for 30 minutes at 60°C. For staining, sections were deparaffinized in xylene and rehydrated through a graded alcohol series. Slides were boiled in citrate buffer (pH 6.0) for antigen retrieval. Endogenous peroxidase activity was blocked using 3% hydrogen peroxide for 25 minutes. Slides were incubated in a humid chamber in primary antibody for AMPKα1 (1:500; Cell Signaling Technology, catalog 2795S) or AMPKα2 (1:250, Cell Signaling Technology, catalog 2757S) overnight at 4°C. Sections were then incubated in biotinylated α-rabbit secondary antibody (1:200; Vector Labs, catalog PI-1000) for 30 minutes at room temperature followed by Vectastain Elite ABC reagent (Vector Labs) for another 30 minutes. Primary and secondary antibodies and ABC reagent were diluted in 1% BSA/PBS. Diaminobenzidine (DAB) was diluted in Tris buffer (pH 7.7) and 3% hydrogen peroxidase in a working DAB solution. Sections were developed in DAB for 10 minutes in a 42°C water bath. Slides were counterstained using Mayer’s hematoxylin and stained blue with 0.2% lithium carbonate. Negative controls were incubated in 1% BSA with rabbit IgG as the primary antibody. Sections were dehydrated in an alcohol series and cleared with xylene, coverslipped, and dried overnight. Slides were imaged at ×20 and ×60 on a Keyence BZ-X710 microscope.
Immunohistochemistry for mouse tissue.
Following euthanasia, mouse brains were hemisected and fixed overnight in ice-cold PFA and transferred to 70% EtOH. Paraffin embedding was performed by Wake Forest Pathology. Paraffin-embedded mouse sections were deparaffinized in xylene and rehydrated through a graded ethanol series. Antigen retrieval used citrate buffer (pH 6.0) in a standard 10-minute microwave procedure. Blocking was done for 2 hours with 10% NGS in 1%BSA/TBS. Slides were incubated in a humidified chamber in primary antibody for amyloid-β (6E10) (1:200) overnight at 4°C. Following a second 15 minutes of 3% hydrogen peroxide blocking, sections were incubated in biotinylated anti-mouse secondary antibody (1:200; Vector Labs) for 1 hour at room temperature, followed by Vectastain Elite ABC Reagent (Vector Labs) for another 30 minutes. Primary and secondary antibodies as well as ABC reagent were diluted in 1% BSA/TBS. Sections were developed in DAB (Vector Labs) for 30 seconds to 3 minutes with monitoring. Slides were counterstained using Mayer’s hematoxylin for 60 seconds and stained blue with 0.2% lithium carbonate for 20 seconds. In between each step of immunohistochemistry, sections were rinsed using distilled water or TBSTX (pH 7.4). Negative controls were incubated in 1% BSA with no primary antibody. Sections were dehydrated in an alcohol series and cleared with xylene, coverslipped, and dried overnight. Imaging was performed using BZ-X710 All-in-One Fluorescent Microscope (Keyence).
Immunofluorescence and confocal microscopy.
Hippocampal slices were fixed overnight in ice-cold 4% paraformaldehyde in PBS. Fixed free-floating sections were subsectioned to 40 μm using a Leica VT1200S vibratome and permeabilized with 0.3% Triton X-100. Sections were blocked with 10% normal goat serum and 0.1% sodium azide in 1% BSA in PBS for 5 hours and incubated overnight with primary antibody for AMPKα1 (12.5 μg; R&D Systems, Bio-Techne; catalog AF3197). Alexa Fluor 633 secondary antibodies (1:250 Thermo Fisher catalog A21082) were used. The sections were imaged using an Olympus FV1200 confocal microscope at original magnification ×20. All parameters (pinhole, contrast, gain, and offset) were held constant for all sections across the same experiment.
OF and NOR.
Mice were handled 7 days before behavioral testing and habituated to the behavior room for at least an hour before experimentation. Animals were placed in an opaque arena (40 cm3) for 15 minutes. The distance traveled, average velocity, and duration spent by the animal in the center were analyzed by Noldus software (Noldus Information Technology). Following OF, mice underwent 2 days’ familiarization with identical ceramic objects in the testing arena. Mice were given 5 minutes to explore the objects. Twenty-four hours following familiarization, animals were tested in the arena with 1 object randomly replaced with a novel object. Time spent with each object was manually calculated and reported as a percentage of total interaction time. An interaction of less than 50% with the novel object indicates memory impairment (29). Mice with fewer than 10 seconds total interaction time or more than 90% object preference were excluded. Data collection and analysis were performed blinded.
MWM.
MWM was performed as previously described (28). The training paradigm consisted of 4 trials (60 seconds maximum, 15-minute intertrial interval) with average escape latency or latencies assessed per trial over 5 consecutive days. Two hours following the final trial, mice underwent a probe trial. Probe trajectories, quadrant occupancy, velocity, and distance were monitored by Ethovision XT (Noldus Information Technology). The VP task consists of 4 trials per day (60 seconds maximum, 15-minute interval) for 2 days, with the escape platform marked by a visible cue and moved randomly throughout the quadrants. Data collection and analysis were performed blinded.
Hippocampal slice preparation and electrophysiology.
Acute 400-μm transverse hippocampal slices were prepared using a Leica VT1200S vibratome as described (69). Briefly, slices were maintained at room temperature before experimentation (2 hours) in artificial cerebrospinal fluid (ACSF) containing the following: 118 mM NaCl, 3.5 mM KCl, 2.5 mM CaCl2, 1.3 mM MgSO4, 1.25 mM NaH2PO4, 5 mM NaHCO3, and 15 mM glucose, bubbled with 95% O2/5% CO2. For electrophysiology, slices were maintained at 32°C, and monophasic current stimuli of 100 μs were delivered with a bipolar silver electrode in the stratum radiatum of area CA3. Field excitatory postsynaptic potentials (fEPSPs) were recorded using a glass microelectrode from the stratum radiatum of area CA1. LTP was induced using high-frequency stimulation (HFS) comprising two 1-second 100 Hz trains, with a 60-second interval, delivered at 70% to 80% of evoked spike intensity.
Aβ ELISA.
Frozen mouse forebrain samples were sonicated as previously described (28). Samples were centrifuged at 16,000 g for 20 minutes at 4°C. The supernatant was collected for ELISA. Aβ 1-42 (Thermo Fisher Scientific, catalog KMB3441) and Aβ 1-40 (Thermo Fisher Scientific, catalog KMB3481) ELISAs were performed according to the manufacturer’s instructions. Ninety-six–well plates were read at 450 nm using an iMark microplate reader (Bio-Rad).
Golgi-Cox stain.
Brains were processed with the Rapid Golgi Kit (FD NeuroTechnologies, catalog PK401) in accordance with the manufacturer’s instructions. Transverse hippocampal sections (100 μm) were cut using a Leica VT1200S vibratome and mounted to gelatin-coated slides. Sections were developed and fixed in accordance with kit instructions. Imaging was performed using a BZ-X710 All-in-One Fluorescent Microscope (Keyence) with a ×100/NA 1.45 oil lens. Five different view fields containing approximately 200 μm of dendrites were imaged in the stratum radiatum of area CA1 per hippocampal slice. Dendritic spine density was calculated by the number of total spines per dendritic region of interest. Spines were morphologically classified based on published guidelines (34, 70). Spine analysis was done blinded.
TEM.
Samples for electron microscopy were prepared as previously described (44). Briefly, freshly dissected 1-mm transverse hippocampal sections had area CA1 dissected and fixed in 1% PFA + 2.5% glutaraldehyde in 0.1M Millonig’s phosphate buffer (pH 7.3) overnight. Samples were washed and postfixed in 1% osmium tetroxide in PBS for 1 hour. Samples were dehydrated through a graded ethanol series and incubated in propylene oxide for two 15-minute changes. Samples were subsequently infiltrated with Spurr’s resin and cured overnight at 70°C. Sections of 90 nm were made with a Reichert-Jung Ultracut E Ultramicrotome, stained with lead citrate and uranyl acetate, and viewed with a FEI Tecnai Spirit TEM operating at 80 kV (FEI Company). Images were obtained with a 2Vu CCD camera (Advanced Microscopy Techniques) at ×11,000. Three mice were used per genotype, and 20 images of the stratum radiatum of area CA1 were taken. ImageJ was used to analyze the PSD counts. Imaging and analysis were done blinded.
SUnSET assay.
Acute 400-μm transverse hippocampal slices were prepared using a Leica VT1200S vibratome as described (69). Slices were maintained at room temperature before experimentation (2 hours) in ACSF. Slices were incubated at 32°C for 1 hour in bubbling ACSF containing puromycin (1 μg/mL) and were flash frozen; area CA1 was microdissected for Western blot analysis (see above). Puromycin-labeled proteins were detected using an anti-puromycin antibody (1:5000; MilliporeSigma, catalog MABE343), and de novo protein synthesis was determined with the total lane density from 250 kDa to 15 kDa using ImageJ.
MS and proteomics analysis.
Whole hippocampi were flash frozen on dry ice, dissected in ice-cold PBS, and lysed immediately in 500 μL of PBS with protease/phosphatase inhibitor using a Bead Mill Homogenizer (Bead Ruptor, Omni International); 500 μL of ×2 RIPA buffer was added, and the mixture was incubated on ice for 30 minutes before centrifugation at 18,000 g for 10 minutes. Protein amount was measured in supernatant, and 50 μg of protein was subjected to tryptic digestion.
Reducing alkylation was performed in the presence of 10 mM dithiothreitol and 30 mM iodoacetamide. Four times the sample volume of cold acetone was added to the tube, which was incubated at –20°C overnight. Tubes were centrifuged at 14,000 g for 10 minutes to obtain protein pellet, which was resuspended in 50 mM ammonium bicarbonate; 1 μg of sequencing-grade modified trypsin was added (1:50 enzyme to substrate) and incubated at 37°C overnight. Tryptic digest was purified using a C18 desalting spin column and then prepared in 5% (v/v) ACN containing 1% (v/v) formic acid for liquid chromatography–tandem MS (LC-MS/MS) analysis.
The LC-MS/MS system consisted of a Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific) and a Dionex Ultimate-3000 Nano-UPLC System (Thermo Fisher Scientific) employing a Nanospray Flex Ion Source (Thermo Fisher Scientific). An Acclaim PepMap 100 (C18, 5 μm, 100 Å, 100 μm × 2 cm) trap column and an Acclaim PepMap RSLC (C18, 2 μm, 100 Å, 75 μm × 50 cm) analytical column were used for the stationary phase. Peptides were separated employing a linear gradient consisting of mobile phases A (water with 0.1% formic acid) and B (acetonitrile with 0.1% formic acid), where the gradient was from 5% B at 0 minutes to 40% B at 170 minutes. MS spectra were acquired by data-dependent scans consisting of MS/MS scans of the 20 most intense ions from the full MS scan with a dynamic exclusion option, which was 10 seconds.
Spectra were searched using the Sequest HT algorithm within the Proteome Discoverer, version 2.1 (Thermo Fisher Scientific), in combination with the mouse UniProt protein FASTA database (annotated 16,747 entries, December 2015). Search parameters were as follows; FT-trap instrument, parent mass error tolerance of 10 ppm, fragment mass error tolerance of 0.02 Da (monoisotopic), variable modifications of 16 Da (oxidation) on methionine, and fixed modification of 57 Da (carbamidomethylation) on cysteine.
Significantly altered proteins were calculated as previously described (29). Proteins with a fold change above 1.5 or below 0.667 and a P value below 0.08 were considered significant. Proteins significantly different from the WT condition were calculated, and the signal intensity was used to generate a heatmap using PermutMatrix (71) (Montpellier Bioinformatics Platform: http://www.atgc-montpellier.fr/permutmatrix). Venn diagrams were generated using BioVenn (http://www.biovenn.nl) (72).
Statistics.
Data are presented as box-and-whisker plots, which represent the interquartile range, with the line across the box indicating the median. Whiskers show the highest and lowest values detected. For comparisons between groups, a 2-tailed unpaired Student’s t test was used. For multiple groups, 1-way ANOVA with Tukey’s post hoc analysis (when applicable) was used. Error probabilities of P < 0.05 were considered statistically significant. Outliers were determined via Grubbs’ test. Statistics were performed using Prism 7 statistics software (GraphPad Software).
Study approval.
Mice were kept in compliance with the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). All animal experiments were performed in accordance with and with the approval of the Institutional Animal Care and Use Committee at Wake Forest University (protocol number A17-116). Postmortem human tissue samples were collected in accordance with approved Institutional Review Board protocols.
Author contributions
HRZ conceptualized experiments, collected and analyzed data, and wrote the manuscript. WY, BCB, NPK, XZ, and XW collected and analyzed data. JL and CMF collected and help analyze the proteomics data. CDK advised on pathology and provided human tissue samples. TM conceptualized experiments and wrote the manuscript. Order of co–first authors was determined by level of actual contribution to the paper.
Supplementary Material
Acknowledgments
We thank George Carlson of McLaughlin Research Institute for providing breeders for Tg19959 AD model mice. We thank NIH NeuroBioBank (University of Maryland) for providing human tissue. We thank Allison Beller of (University of Washington School of Medicine) for administrative support and technical help with human tissue samples. We thank Kenneth Grant for technical help on TEM imaging. We thank the Wake Forest School of Medicine Pathology Core for their help with tissue processing for immunohistochemistry. We would like to acknowledge the support provided by the Proteomics and Metabolomics Shared Resource of the Wake Forest Baptist Comprehensive Cancer Center (NIH/NCI P30 CA12197). We thank Ben Rowland for guidance on statistical analysis. This work was supported by NIH grants K99/R00 AG044469, R01 AG055581, R01 AG056622 (to TM), F31AG055264 (to HRZ), F31AG054113 (to BCB), and P50AG005136 (to CDK); Alzheimer’s Association grant NIRG-15-362799 (to TM); BrightFocus Foundation grant A2017457S; and the Nancy and Buster Alvord Endowment (to CDK).
Version 1. 03/26/2020
In-Press Preview
Version 2. 05/26/2020
Electronic publication
Version 3. 07/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(7):3511–3527.https://doi.org/10.1172/JCI133982.
See the related Commentary at Isoform-specific roles of AMPK catalytic α subunits in Alzheimer’s disease.
Contributor Information
Helena R. Zimmermann, Email: helena.zimmermann@northwestern.edu.
Wenzhong Yang, Email: wyang@wakehealth.edu.
Nicole P. Kasica, Email: nkasica@wakehealth.edu.
Xueyan Zhou, Email: xuzhou@wakehealth.edu.
Xin Wang, Email: xinwang@wakehealth.edu.
Brenna C. Beckelman, Email: brenna.beckelman@gmail.com.
Jingyun Lee, Email: jilee@wakehealth.edu.
Cristina M. Furdui, Email: cfurdui@wakehealth.edu.
C. Dirk Keene, Email: cdkeene@uw.edu.
Tao Ma, Email: tma@wakehealth.edu.
References
- 1.Alzheimer’s Association. 2019 Alzheimer’s disease facts and figures. Alzheimers Dement. 2019;15(3):321–387. doi: 10.1016/j.jalz.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Sci Transl Med. 2011;3(77):77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma T, et al. Inhibition of AMP-activated protein kinase signaling alleviates impairments in hippocampal synaptic plasticity induced by amyloid β. J Neurosci. 2014;34(36):12230–12238. doi: 10.1523/JNEUROSCI.1694-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mairet-Coello G, Courchet J, Pieraut S, Courchet V, Maximov A, Polleux F. The CAMKK2-AMPK kinase pathway mediates the synaptotoxic effects of Aβ oligomers through Tau phosphorylation. Neuron. 2013;78(1):94–108. doi: 10.1016/j.neuron.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Zimmermann HR, Ma T. Therapeutic potential of AMP-activated protein kinase in Alzheimer’s disease. J Alzheimers Dis. 2019;68(1):33–38. doi: 10.3233/JAD-181043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marinangeli C, Didier S, Vingtdeux V. AMPK in neurodegenerative diseases: implications and therapeutic perspectives. Curr Drug Targets. 2016;17(8):890–907. doi: 10.2174/1389450117666160201105645. [DOI] [PubMed] [Google Scholar]
- 7.Hardie DG. AMPK--sensing energy while talking to other signaling pathways. Cell Metab. 2014;20(6):939–952. doi: 10.1016/j.cmet.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao B, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449(7161):496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 9.Burkewitz K, Zhang Y, Mair WB. AMPK at the nexus of energetics and aging. Cell Metab. 2014;20(1):10–25. doi: 10.1016/j.cmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao B, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472(7342):230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 12.Ma T, Klann E. Amyloid β: linking synaptic plasticity failure to memory disruption in Alzheimer’s disease. J Neurochem. 2012;120(suppl 1):140–148. doi: 10.1111/j.1471-4159.2011.07506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vingtdeux V, Davies P, Dickson DW, Marambaud P. AMPK is abnormally activated in tangle- and pre-tangle-bearing neurons in Alzheimer’s disease and other tauopathies. Acta Neuropathol. 2011;121(3):337–349. doi: 10.1007/s00401-010-0759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg GR, Kemp BE. AMPK in health and disease. Physiol Rev. 2009;89(3):1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 15.Michell BJ, et al. Isoform-specific purification and substrate specificity of the 5’-AMP-activated protein kinase. J Biol Chem. 1996;271(45):28445–28450. doi: 10.1074/jbc.271.45.28445. [DOI] [PubMed] [Google Scholar]
- 16.Woods A, Salt I, Scott J, Hardie DG, Carling D. The alpha1 and alpha2 isoforms of the AMP-activated protein kinase have similar activities in rat liver but exhibit differences in substrate specificity in vitro. FEBS Lett. 1996;397(2-3):347–351. doi: 10.1016/S0014-5793(96)01209-4. [DOI] [PubMed] [Google Scholar]
- 17.Viollet B, et al. The AMP-activated protein kinase alpha2 catalytic subunit controls whole-body insulin sensitivity. J Clin Invest. 2003;111(1):91–98. doi: 10.1172/JCI16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jørgensen SB, et al. Knockout of the alpha2 but not alpha1 5’-AMP-activated protein kinase isoform abolishes 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranosidebut not contraction-induced glucose uptake in skeletal muscle. J Biol Chem. 2004;279(2):1070–1079. doi: 10.1074/jbc.M306205200. [DOI] [PubMed] [Google Scholar]
- 19.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25(18):1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5(12):931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 21.Alberini CM. The role of protein synthesis during the labile phases of memory: revisiting the skepticism. Neurobiol Learn Mem. 2008;89(3):234–246. doi: 10.1016/j.nlm.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61(1):10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma T, et al. Suppression of eIF2α kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat Neurosci. 2013;16(9):1299–1305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno JA, et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 2012;485(7399):507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radford H, Moreno JA, Verity N, Halliday M, Mallucci GR. PERK inhibition prevents tau-mediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathol. 2015;130(5):633–642. doi: 10.1007/s00401-015-1487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sujobert P, et al. Co-activation of AMPK and mTORC1 induces cytotoxicity in acute myeloid leukemia. Cell Rep. 2015;11(9):1446–1457. doi: 10.1016/j.celrep.2015.04.063. [DOI] [PubMed] [Google Scholar]
- 27.Chishti MA, et al. Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J Biol Chem. 2001;276(24):21562–21570. doi: 10.1074/jbc.M100710200. [DOI] [PubMed] [Google Scholar]
- 28.Beckelman BC, et al. Genetic reduction of eEF2 kinase alleviates pathophysiology in Alzheimer’s disease model mice. J Clin Invest. 2019;129(2):820–833. doi: 10.1172/JCI122954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antunes M, Biala G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn Process. 2012;13(2):93–110. doi: 10.1007/s10339-011-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thornton C, Bright NJ, Sastre M, Muckett PJ, Carling D. AMP-activated protein kinase (AMPK) is a tau kinase, activated in response to amyloid β-peptide exposure. Biochem J. 2011;434(3):503–512. doi: 10.1042/BJ20101485. [DOI] [PubMed] [Google Scholar]
- 31.Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298(5594):789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 32.Walsh DM, Selkoe DJ. Deciphering the molecular basis of memory failure in Alzheimer’s disease. Neuron. 2004;44(1):181–193. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Terry RD, et al. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30(4):572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 34.Hering H, Sheng M. Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci. 2001;2(12):880–888. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 35.Sala C, Segal M. Dendritic spines: the locus of structural and functional plasticity. Physiol Rev. 2014;94(1):141–188. doi: 10.1152/physrev.00012.2013. [DOI] [PubMed] [Google Scholar]
- 36.Risher WC, Ustunkaya T, Singh Alvarado J, Eroglu C. Rapid Golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS One. 2014;9(9):e107591. doi: 10.1371/journal.pone.0107591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okabe S. Molecular anatomy of the postsynaptic density. Mol Cell Neurosci. 2007;34(4):503–518. doi: 10.1016/j.mcn.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Ma T, Klann E. PERK: a novel therapeutic target for neurodegenerative diseases? Alzheimers Res Ther. 2014;6(3):30. doi: 10.1186/alzrt260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viollet B, et al. AMPK inhibition in health and disease. Crit Rev Biochem Mol Biol. 2010;45(4):276–295. doi: 10.3109/10409238.2010.488215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S, Scheffler TL, Rossie SS, Gerrard DE. AMPK activity is regulated by calcium-mediated protein phosphatase 2A activity. Cell Calcium. 2013;53(3):217–223. doi: 10.1016/j.ceca.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Jan A, et al. eEF2K inhibition blocks Aβ42 neurotoxicity by promoting an NRF2 antioxidant response. Acta Neuropathol. 2017;133(1):101–119. doi: 10.1007/s00401-016-1634-1. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6(4):275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 43.Zimmermann HR, et al. Genetic removal of eIF2α kinase PERK in mice enables hippocampal L-LTP independent of mTORC1 activity. J Neurochem. 2018;146(2):133–144. doi: 10.1111/jnc.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckelman BC, et al. Genetic reduction of eEF2 kinase alleviates pathophysiology in Alzheimer’s disease model mice. J Clin Invest. 2019;129(2):820–833. doi: 10.1172/JCI122954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin S, et al. Hippocampal metabolomics using ultrahigh-resolution mass spectrometry reveals neuroinflammation from Alzheimer’s disease in CRND8 mice. Anal Bioanal Chem. 2013;405(15):5105–5117. doi: 10.1007/s00216-013-6825-1. [DOI] [PubMed] [Google Scholar]
- 46.Jankowsky JL, Slunt HH, Ratovitski T, Jenkins NA, Copeland NG, Borchelt DR. Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol Eng. 2001;17(6):157–165. doi: 10.1016/S1389-0344(01)00067-3. [DOI] [PubMed] [Google Scholar]
- 47.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361(6407):31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 48.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44(1):5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 49.Arnold SE, et al. Brain insulin resistance in type 2 diabetes and Alzheimer disease: concepts and conundrums. Nat Rev Neurol. 2018;14(3):168–181. doi: 10.1038/nrneurol.2017.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Outeiro TF, et al. Dementia with Lewy bodies: an update and outlook. Mol Neurodegener. 2019;14(1):5. doi: 10.1186/s13024-019-0306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Esparcia P, et al. Dementia with Lewy Bodies: molecular pathology in the frontal cortex in typical and rapidly progressive forms. Front Neurol. 2017;8:89. doi: 10.3389/fneur.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackenzie IR, Neumann M. Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J Neurochem. 2016;138(suppl 1):54–70. doi: 10.1111/jnc.13588. [DOI] [PubMed] [Google Scholar]
- 53.Salminen A, Kaarniranta K, Haapasalo A, Soininen H, Hiltunen M. AMP-activated protein kinase: a potential player in Alzheimer’s disease. J Neurochem. 2011;118(4):460–474. doi: 10.1111/j.1471-4159.2011.07331.x. [DOI] [PubMed] [Google Scholar]
- 54.Cai Z, Yan LJ, Li K, Quazi SH, Zhao B. Roles of AMP-activated protein kinase in Alzheimer’s disease. Neuromolecular Med. 2012;14(1):1–14. doi: 10.1007/s12017-012-8173-2. [DOI] [PubMed] [Google Scholar]
- 55.Dasgupta B, Chhipa RR. Evolving lessons on the complex role of AMPK in normal physiology and cancer. Trends Pharmacol Sci. 2016;37(3):192–206. doi: 10.1016/j.tips.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y, et al. Antidiabetic drug metformin (GlucophageR) increases biogenesis of Alzheimer’s amyloid peptides via up-regulating BACE1 transcription. Proc Natl Acad Sci USA. 2009;106(10):3907–3912. doi: 10.1073/pnas.0807991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta A, Bisht B, Dey CS. Peripheral insulin-sensitizer drug metformin ameliorates neuronal insulin resistance and Alzheimer’s-like changes. Neuropharmacology. 2011;60(6):910–920. doi: 10.1016/j.neuropharm.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 58.Moore EM, et al. Increased risk of cognitive impairment in patients with diabetes is associated with metformin. Diabetes Care. 2013;36(10):2981–2987. doi: 10.2337/dc13-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269(22):5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 60.Proud CG. Regulation and roles of elongation factor 2 kinase. Biochem Soc Trans. 2015;43(3):328–332. doi: 10.1042/BST20140323. [DOI] [PubMed] [Google Scholar]
- 61.Mrak RE, Griffinbc WS. The role of activated astrocytes and of the neurotrophic cytokine S100B in the pathogenesis of Alzheimer’s disease. Neurobiol Aging. 2001;22(6):915–922. doi: 10.1016/S0197-4580(01)00293-7. [DOI] [PubMed] [Google Scholar]
- 62.Yardan T, Erenler AK, Baydin A, Aydin K, Cokluk C. Usefulness of S100B protein in neurological disorders. J Pak Med Assoc. 2011;61(3):276–281. [PubMed] [Google Scholar]
- 63.Yeşil G, Aralaşmak A, Akyüz E, İçağasıoğlu D, Uygur Şahin T, Bayram Y. Expanding the phenotype of homozygous KCNMA1 mutations; dyskinesia, epilepsy, intellectual disability, cerebellar and corticospinal tract atrophy. Balkan Med J. 2018;35(4):336–339. doi: 10.4274/balkanmedj.2017.0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bezprozvanny I, Mattson MP. Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci. 2008;31(9):454–463. doi: 10.1016/j.tins.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mustaly-Kalimi S, Littlefield AM, Stutzmann GE. Calcium signaling deficits in glia and autophagic pathways contributing to neurodegenerative disease. Antioxid Redox Signal. 2018;29(12):1158–1175. doi: 10.1089/ars.2017.7266. [DOI] [PubMed] [Google Scholar]
- 66.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 67.Mirra S, Heyman A, McKeel D, Sumi S, Crain B, Brownlee L, et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology. 1991;41(4):479–486. doi: 10.1212/WNL.41.4.479. [DOI] [PubMed] [Google Scholar]
- 68.Radde R, et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 2006;7(9):940–946. doi: 10.1038/sj.embor.7400784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang W, Zhou X, Zimmermann HR, Cavener DR, Klann E, Ma T. Repression of the eIF2α kinase PERK alleviates mGluR-LTD impairments in a mouse model of Alzheimer’s disease. Neurobiol Aging. 2016;41:19–24. doi: 10.1016/j.neurobiolaging.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Risher WC, Ustunkaya T, Singh Alvarado J, Eroglu C. Rapid Golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS ONE. 2014;9(9):e107591. doi: 10.1371/journal.pone.0107591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21(7):1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- 72.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.