Abstract
In neurons, loading of neurotransmitters into synaptic vesicles uses energy from proton-pumping V-ATPases. These membrane protein complexes possess numerous subunit isoforms, complicating their analysis. We isolated homogenous rat brain V-ATPase through its interaction with SidK, a Legionella pneumophila effector protein. CryoEM allowed construction of an atomic model, defining the enzyme’s ATP:H+ ratio as 3:10 and revealing a homolog of yeast subunit f in the membrane region, which we tentatively identify as RNAseK. The c-ring encloses the transmembrane anchors for cleaved ATP6AP1/Ac45 and ATP6AP2/PRR, the latter being the (pro)renin receptor that, in other contexts, is involved in both Wnt signalling and the renin-angiotensin system that regulates blood pressure. The structure shows how ATP6AP1/Ac45 and ATP6AP2/PRR enable assembly of the enzyme’s catalytic and membrane regions.
Vesicular- or vacuolar-type ATPases (V-ATPases) are ATP-hydrolysis-driven proton pumps that are essential for acidification of endosomes, lysosomes, and the trans Golgi network, as well as for acid secretion by osteoclasts, kidney intercalated cells, and some tumour cells (1, 2). ATP hydrolysis in the V-ATPase catalytic V1 region drives rotation of a central rotor subcomplex and leads to proton translocation through the membrane-embedded VO region. V-ATPase activity is regulated by reversible separation of the V1 and VO regions, with ATP hydrolysis inhibited in the isolated V1 complex and the VO complex becoming impermeable to protons (3, 4). In neurons, V-ATPase activity energizes synaptic vesicle membranes, allowing transporters to load the vesicles with neurotransmitters (Fig. 1A) (1, 2). Fusion of synaptic vesicles with the presynaptic membrane requires separation of the V1 and VO regions but it is not known how these events are coordinated (5). The regulated release of neurotransmitters from synaptic vesicles into the synaptic cleft allows signal propagation from the axon terminal of a presynaptic neuron to the dendrite of a postsynaptic neuron. After release of neurotransmitter, subsequent endocytosis and regeneration of synaptic vesicles occurs via clathrin-independent and clathrin-mediated routes (6), with the formation of clathrin-coated vesicles temporarily blocking V-ATPase activity (7). In the Saccharomyces cerevisiae enzyme, which is the most thoroughly characterized to date, the V1 region contains subunits A3B3CDE3FG3H, and the VO region contains subunits ac8c′c″def and Voa1p (8, 9). The V1 regions of mammalian V-ATPases contain the same subunits as the yeast enzyme while mammalian VO regions are thought to be composed of acxc″de as well as ATP6AP1, also known as Ac45, and ATP6AP2, also known as the (pro)renin receptor (10, 11). ATP6AP2/PRR is involved in several signalling pathways (12) including the renin-angiotensin system for regulating blood pressure and electrolyte balance (10, 11, 13) and Wnt signalling in stem cells and embryo development (14). The precise arrangement of subunits in the mammalian VO region remains unclear. Further, mammals have multiple isoforms of some subunits in both V1 and VO that are expressed in a tissue-dependent and cellular-compartment-dependent way. These include two isoforms of subunit B, two of C, two of E, three of G, four of a, two of d, and two of e (15, 16). Mass spectrometry of purified rat-brain synaptic vesicles detected V-ATPase subunits A, B1, B2, C1, D, E1, F, G1, G2, a1, a4, c, d1, ATP6AP1/Ac45, and ATP6AP2/PRR (17).
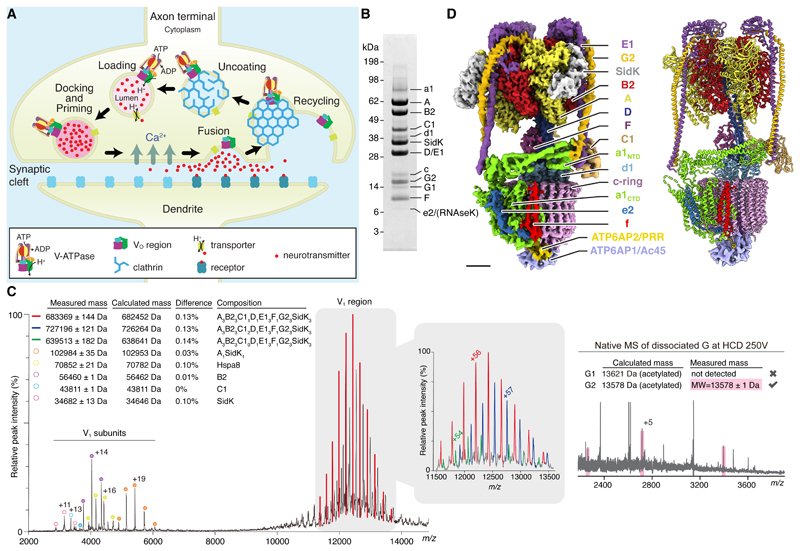
Fig. 1. Overall structure of brain V-ATPase.
(A) Cycle of synaptic vesicle loading, docking and priming, fusion, and recycling. (B) SDS-PAGE of rat brain V-ATPase isolated with 3×FLAG SidK1-278 and gel filtration chromatography. (C) Native mass spectrometry of V1 region (left) and native mass spectrometry of dissociated subunit G (right) at a higher-energy collisional (HCD) voltage of 250 V. The charge state for one peak per subunit is indicated. The table shows the measured mass for each peak (± SD of fit) and the calculated mass depending on subunit composition (Table S5). The difference between calculated and measured masses is indicated. (D) Composite cryoEM map (left) and atomic model (right) of brain V-ATPase in rotational state 1. Scale bar, 25 Å.
In order to isolate V-ATPase for structural analysis, we developed a purification strategy based on the high-affinity interaction of the enzyme with the Legionella pneumophila effector protein SidK (18, 19). While procedures capable of obtaining highly-purified synaptic vesicles have been described (17, 20) they enhance purity at the cost of yield, which would complicate or preclude structure determination. Instead, a two-step differential centrifugation procedure was used to collect rat synaptic vesicles and clathrin-coated vesicles along with other cell membranes. Membranes were solubilized with detergent and SidK (residues 1 to 278) fused to a C-terminal 3×FLAG tag was used with M2-agarose to purify V-ATPase. With this approach, a single ~2 g rat brain provided ~200 μg of highly purified V-ATPase (Fig. 1B). Mass spectrometry of tryptic fragments (Fig. S1A, Tables S1 and S2) identified the bands on the SDS-PAGE gel as V-ATPase subunits A, B2, C1, D, E1, F, G2, a1, c, and d1. A low-intensity band on the gel corresponding to subunit G1 was also detected, which from gel densitometry appears to comprise 17±3 % of subunit G in the preparation (±SD, three independent purifications). Subunit G1 may be a component of some V-ATPase complexes in synaptic vesicles (17). Alternatively, subunit G1 may be from copurifying lysosomal V-ATPase (21). The glycoprotein ATP6AP1/Ac45 and the small and hydrophobic proteins ATP6AP2/PRR and subunit e2, which do not stain clearly with Coomassie, were detected after cleavage with trypsin or chymotrypsin (Fig. 1B, Fig. S1A, Table S3). The subunit isoforms identified are all consistent with the V-ATPase from synaptic vesicle (17). This homogeneity could be due to the synaptic vesicle V-ATPase being the predominant form of the enzyme in the brain, or to V-ATPases from other cellular compartments in the brain having the same isoform composition as the synaptic vesicle enzyme. RNAseK, a hydrophobic protein recently found to associate with mammalian V-ATPase (22), was also detected after cleavage with trypsin or chymotrypsin (Fig. 1B, Fig. S1A, Table S3). Subunit H, which dissociates from bovine brain V-ATPase when treated with oxidizing agents (23), was not detected by mass spectrometry despite the absence of these reagents from the preparation. Subunit H is part of the mammalian V-ATPase (7, 17) and is needed for full enzyme activity (24). Loss of subunit H during purification of mammalian V-ATPase is markedly different from the subunit’s behaviour in S. cerevisiae (25). This difference is noteworthy because the protein’s physiological role of mechanically blocking ATP hydrolysis in the isolated V1 region likely requires a strong attachment to the enzyme (26, 27). While tryptic peptides from subunits B1, C2, a2, a3, and a4 were also detected (Table S4), integrated peak intensities for these peptides in extracted ion chromatograms were two to three orders of magnitude lower than for peptides from subunits B2, C1, and a1, indicating that their abundance is negligible for structural studies.
Native mass spectrometry further demonstrated the homogeneity of the enzyme preparation (Fig. 1C, red, and Fig. S1B and C). Spectra show the V1 region, presumably due to dissociation of the complex during analysis. The V1 region has a native mass of 683369 ± 144 Da, consistent with a subunit composition of A3B23C1DE13FG23SidK3 (Fig. 1C and S1B, Table S5). Fragmentation of the V1 region using a higher-energy collisional dissociation (HCD) voltage of 50 to 250 V confirmed the isoform composition of the V1 region as B2, C1, and E1 (Fig. S1B). Specifically, spectra for subunit G showed a native mass of 13578 ±1 Da (Fig. 1C, right), consistent with subunit G2 (13578 Da) but not G1 (13621 Da), both of which are N-terminally acetylated (Fig. S1C). Lower abundance peaks were also seen for the V1 region missing subunit C1 (Fig. 1C, green), and even with two copies of C1 (Fig. 1C, blue), the latter possibly being attributed to the loss of subunit H. The enzyme preparation had an ATP hydrolysis rate that decreased during hydrolysis, consistent with the complex lacking subunit H (24, 26) (Fig. S2). Further, the preparation did not demonstrate full coupling of ATPase activity in the V1 region to proton transport through the VO region: bafilomycin, which blocks the VO region, only partially inhibited ATPase activity. This lack of coupling could be due to free V1 complexes that remain active because subunit H is not present, or to the detergent-solubilized V-ATPase disassembling during ATP hydrolysis, as seen with the Manduca sexta enzyme (28). The extent of SidK inhibition on ATP hydrolysis activity could not be quantified because only the SidK-bound form of the enzyme was available, although previous work showed that SidK inhibits S. cerevisiae V-ATPase by ~40 % (19).
CryoEM of the preparation yielded three 3D maps, corresponding to ~120° rotations of the rotor subcomplex between states (25). These rotational states of the enzyme had overall resolutions of 3.9, 4.0, and 3.9 Å (Fig. S3 and S4). Focused refinement of rotational state 1 was able to improve the resolution to 3.8 Å resolution for the membrane region and 3.6 Å for the catalytic region of the complex. Together, these maps allowed construction of an atomic model for most of the complex, with a few components, including parts of subunits E1 and G2, the soluble N-terminal domain of subunit a1, subunit C1, and several luminal loops in the membrane region, modelled as backbone with truncated side chains (Fig. 1D, Table S6, Fig. S5). Similar to the yeast VO structure (8, 29), no density was apparent for the loop between residues 667 and 712 in subunit a1. Owing to the averaging that occurs during cryoEM image analysis, the minor population of complexes in the preparation possessing subunit G1 rather than G2 could produce a map that shows a weighted average of both isoforms. Alternatively, images of complexes containing subunit G1 may be excluded during 2D and 3D image classification if they do not average coherently with the major population of complexes to produce high-resolution map features. Where the map shows high-resolution features for subunit G, it accommodates subunit G2 better than G1 (Fig. S6), consistent with most of the V-ATPase complexes containing subunit G2. Interpolating between the three rotational states produced a movie (Movie S1) that shows the conformational changes in the enzyme that couple ATP hydrolysis in the V1 region to proton pumping through the VO region. These changes illustrate the flexibility of the enzyme, particularly in the peripheral stalks, subunit C1, and N-terminal domain of subunit a1.
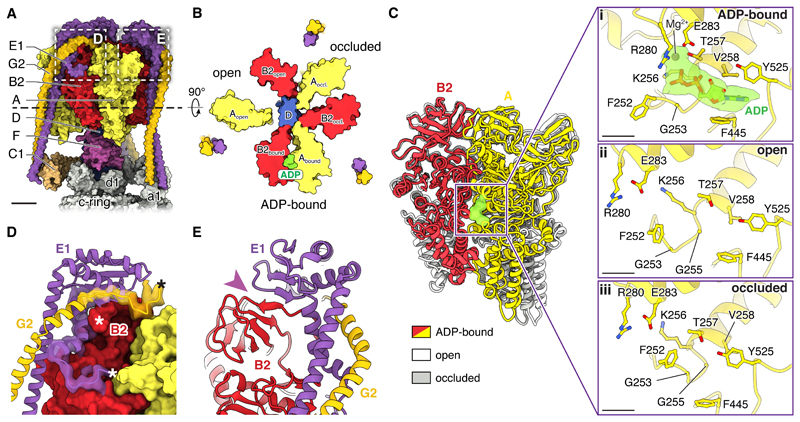
Previous high-resolution insight into the structure of the eukaryotic V1 region is limited to cryoEM of the intact yeast V-ATPase at ~7 Å resolution (19, 25, 29) and a 6.2 to 6.7 Å resolution crystal structure of the autoinhibited yeast V1(-C) complex (27). These maps only allowed visualization of α-helices in the structure. The current structure of the V1 region (Fig. 2A) enables comparison to atomic models of prokaryotic V/A-ATPase catalytic regions (30–32) as well as numerous atomic models of ATP synthase F1 regions (33). ATP synthases possess three catalytic and three non-catalytic nucleotide binding sites, found at the interfaces between subunits α (corresponding to V-ATPase subunit B) and β (corresponding to subunit A) (33). In contrast, and consistent with structures of the bacterial V/A-ATPase (31, 34), the mammalian V1 region lacks non-catalytic nucleotide binding sites and shows only a single bound nucleotide in one of the catalytic sites (Fig. 2B and 2C-i). This nucleotide could be modelled in the density as ADP, consistent with biochemical analysis of the M. sexta V-ATPase (28). The three pairs of catalytic subunits in F- and V-type ATPases interchange between three different conformations, originally described for the F1-ATPase as ATP-bound, ADP-bound, and empty (33). In the present structure, each catalytic AB pair similarly adopts one of three conformations (Fig. 2B and C). Comparison of the conformation of catalytic AB pairs to crystal structures of a prokaryotic V1/A1 region suggests that the ADP-bound site is in a post-hydrolysis state (Fig. 2B and C-i) (34) and that the site in an open conformation (clockwise from the ADP-bound site when viewed from V1 towards VO) is in a state with high affinity for ATP (31) (Fig. 2B and C-ii). The conformation of the third pair of AB subunits does not appear to correspond to previous structures but its nucleotide-binding pocket is occluded (Fig. 2B and C-iii), suggesting a low affinity for nucleotide (31). Therefore, the enzyme imaged here appears to be poised to bind ATP. Three copies of SidK1-278 are bound to the three A subunits (Fig. S7). The structural consequences of SidK binding to the mammalian V1 region are not known, but SidK perturbed the conformation of the yeast V1 region only subtly (19). Although lower-resolution structures of the yeast enzyme suggested that subunit G does not contact the rest of the V1 region (25, 27), the present structure shows that it does indeed participate in linking the EG heterodimers of the peripheral stalks to the catalytic A3B3 subcomplex in the mammalian enzyme (Fig. 2D). This connection relies on residues from the N terminus of subunit B2 and C-terminal residues of subunits E1 and G2 (Fig. 2D, asterisks) and includes a structure where β-strands from both subunit B2 and E1 form a single β-sheet (Fig 2E, purple arrowhead), also seen in a recent cryoEM map of a prokaryotic V/A-ATPase (32). Numerous mutations in subunits A, B1, B2, and E1 are linked to disease and can be mapped onto the structure (Fig. S8).
Fig. 2. Structure of the V1 region.
(A) Surface representation. Scale bar, 25 Å. (B) Cross-section through the V1 region viewed from V1 towards VO. ADP is shown in green. (C) Superposition of the catalytic AB pairs (left) and closeup of the conformations of the nucleotide binding sites (right). Scale bar, 5 Å. (D) Interaction of subunits B2, E1, and G2. N and C termini of models are indicated by ‘*’. (E) A continuous β-sheet is formed by E1 and B2 (purple arrowhead).
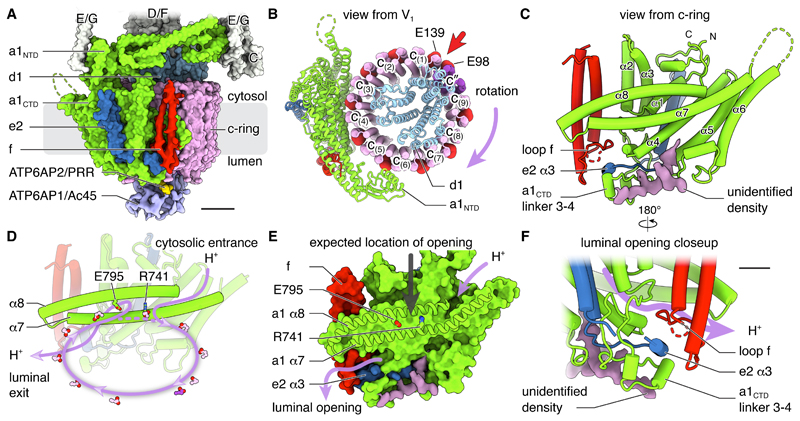
The VO region contains subunits a1, d1, e2, f, ATP6AP1/Ac45, ATP6AP2/PRR, and the c-ring (Fig. 3A). While previous high-resolution structures of S. cerevisiae VO regions were determined from auto-inhibited complexes separated from their V1 regions (8, 9, 29), the structure presented here is within the context of an assembled V-ATPase. Consequently, the soluble N-terminal domain of subunit a1 is found in its non-inhibitory conformation and does not make contact with subunit d1 (Fig. 3B, green). The membrane-embedded C-terminal domain of subunit a1 includes eight transmembrane α-helices (Fig. 3C) and creates the two offset half-channels that allow proton translocation (8). The subunit starts with a V-shaped insertion into the lipid bilayer formed by two short α-helices that do not entirely cross the membrane (α1 and α2), followed by four transmembrane α-helices (α3 to α6) and two highly tilted transmembrane α-helices (α7 and α8) that create the surface that contacts the c-ring. This fold closely follows yeast Vph1p and Stv1p (8, 29), with RSMDs between Cα atoms of 1 and 0.8 Å, respectively (Fig. S9A). The major difference between these structures is the insertion of a structured linker with α-helices connecting transmembrane α3 and α4 in subunit a1 (Fig. 3C and Fig. S9B). The c-ring contains nine copies of subunit c and one of subunit c″ (Fig. 3B, pink and purple). As expected from the lack of a homologous gene in mammals, the ring does not contain subunit c′ found in yeast. Conserved Glu residues from protomers of the c-ring are each capable of carrying a single proton during proton translocation (Fig. 3B, red spheres). As with the yeast VO region, subunit c″ breaks the pattern of conserved proton-carrying Glu residues occurring on alternating outer α-helices of the c-ring (Fig. 3B, red arrow) (8). The presence of ten protomers in the c-ring sets the ATP:H+ ratio at three ATP molecules hydrolyzed for every ten protons translocated. With a Gibbs free energy for ATP hydrolysis of approximately -60 kJ/mol (35), this ratio limits the transmembrane proton motive force established by the brain V-ATPase, , to ~18 kJ/mol, which is equivalent to ~3 pH units or ~180 mV (35). Similarity between the mammalian and yeast V-ATPase structures suggests they share the same mechanism for rotation-driven proton pumping (8) (Fig. 3D). In this mechanism, rotation of the c-ring drives abstraction of a proton from the cytoplasmic half-channel to neutralize the charge on a conserved c-ring Glu residue as it enters the lipid bilayer. Rotation brings a protonated Glu residue close to Arg741 of subunit a1, with formation of a salt bridge between the Arg and Glu causing release of the proton into the luminal half-channel that begins near Glu795 of subunit a1. While structures of ATP synthases (32, 36–38) have resolved the gap between the two tilted α-helices of subunit a where protons must pass, this opening cannot be seen here (Fig. 3E, grey arrow) or in other VO region structures (8, 9, 29). This difference is either because of limited resolution or because the opening only forms transiently in V-ATPases that, unlike ATP synthases, pump protons against a proton motive force. The surface of subunit a1 that contacts the c-ring has the pattern of a positive charge at the conserved Arg residue and negative charges at the mid-membrane termini of the two half channels (39) that has now been seen in all other rotary ATPases (Fig. S9C).
Fig. 3. Structure of the VO region.
(A) Surface representation with cryoEM density for subunits f, ATP6AP1/Ac45, and ATP6AP2/PRR. Scale bar, 25 Å. (B) Viewed from V1 with conserved proton-carrying Glu residues as red spheres. The direction of ATP-hydrolysis-driven rotation of the ring is indicated. (C) Cartoon representation viewed parallel to the plane of the lipid bilayer. (D) Proton path through the VO region. (E) Surface representation of a1CTD viewed parallel to the plane of the lipid bilayer. The grey arrow indicates the expected location of an opening between α7 and α8 leading towards the luminal half-channel. (F) Closeup view of the luminal terminus of the luminal half-channel, showing the interaction of subunits f, e2, a1, and the unidentified density. Scale bar, 10 Å.
Subunit e2 is adjacent to transmembrane α-helices 3 and 4 of subunit a1 and consists of an α-helical hairpin with a C-terminal tail, similar to the corresponding yeast protein Vma9p (8) (Fig. 3A and C, blue, and Fig. S10). However, unlike yeast Vma9p, subunit e2 has an extended C-terminal sequence that terminates in a third short α-helix (α3) encircled by the linker between α3 and α4 of subunit a1 and the luminal loop of subunit f (Fig. 3C and S10, blue arrow). The function of subunit e2 is unknown (40), but deletion of Vma9p causes the Vma-V-ATPase deficiency phenotype, where yeast can grow on medium buffered to pH 5.5 but not pH 7.5, are sensitive to extracellular calcium and a variety of heavy metals, and cannot grow on medium with typical concentrations of non-fermentable carbon sources (41). Recent analysis of both the vacuolar and Golgi forms of the S. cerevisiae V-ATPase identified the hypothetical protein YPR170W-B as a transmembrane α-helical hairpin subunit, named subunit f (8, 29). YPR170W-B is highly conserved in fungi but deletion of the gene did not cause the Vma-phenotype (8). The structure of the brain V-ATPase revealed density for a similar transmembrane α-helical hairpin in the VO region in a position corresponding to the yeast subunit f (Fig. 3, red). Bioinformatic analysis suggests RNAseK, a conserved metazoan protein (42), as a homolog of S. cerevisiae subunit f, sharing 32 % sequence identity and 52 % sequence similarity (Fig. S11A). Immunoprecipitation of RNAseK previously revealed that it is associated with V-ATPase (22) and mass spectrometry done here shows that RNAseK is present in this enzyme preparation (Tables S1 to S3). Consistent with a role in V-ATPase activity, RNAseK is necessary for viral endocytosis and replication (22, 43). The cryoEM density for subunit f, although lacking the necessary resolution to unambiguously identify the protein as RNAseK, is consistent with the expected size of RNAseK and accommodates most of its bulky side chains (Fig. S5B). Together, this evidence tentatively identifies RNAseK as subunit f in mammalian V-ATPases. Whether or not RNAseK functions as a ribonuclease within the V-ATPase is not known, but we note that the in vitro assays that established this protein and its homologs as ribonucleases (42, 44) were performed in the absence of detergents or other membrane mimetics despite clear transmembrane α-helices in hydropathy analysis of the protein’s amino acid sequence (Fig S11B). An additional unidentified protein-like density previously found in both the vacuolar and Golgi forms of the S. cerevisiae V-ATPase (9, 29) is also present in the mammalian complex (Fig. 3C to F, purple). All four of the membrane-embedded components from this part of the enzyme interact at the opening of the luminal proton half-channel, with the linker between α3 and α4 of subunit a1 acting as a scaffold for packing the luminal loop connecting the transmembrane α-helices of subunit f, the C-terminal sequence of subunit e2, and part of the unknown component (Fig. 3F). These four components are all elongated relative to their yeast counterparts in order to accommodate their interaction (Fig. S9 to S11).
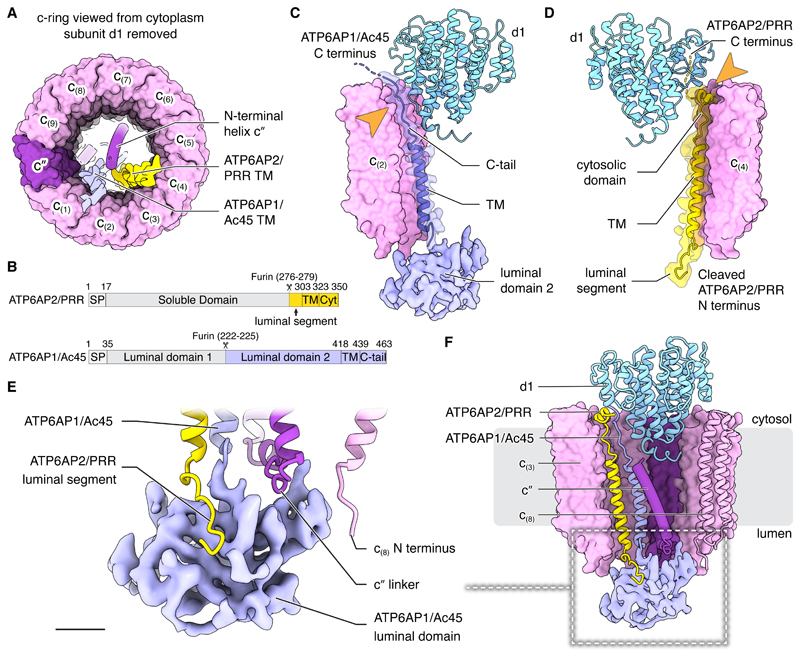
Unlike the fungal V-ATPase, three rather than two transmembrane α-helices are within the lumen of the c-ring (Fig. 4A). The centremost α-helix corresponds to the N-terminal α-helix of subunit c″, also seen in the yeast V-ATPase (8, 9, 29). The two other transmembrane α-helices are from ATP6AP1/Ac45 and ATP6AP2/PRR (Fig. 4A and S5C). ATP6AP1/Ac45 mutations in humans can lead to immunodeficiency, cognitive impairment, liver dysfunction, and abnormal protein glycosylation (45) while mutations in ATP6AP2/PRR can result in neurodegeneration as well as X-linked parkinsonism and epilepsy (12). Both proteins are frequently mutated in granular cell cancers (46). Immature ATP6AP1/Ac45 contains two soluble N-terminal domains on the luminal side of the membrane separated by a furin cleavage site (47) (Fig. 4B, bottom). In the map, the soluble region of APT6AP1/Ac45 appears as a single low-resolution domain ~30×20×30 Å in size (Fig. 4C, light blue), consistent with only the second luminal domain being in the complex. Further, although mass spectrometry detected peptides from uncleaved ATP6AP1/Ac45, it suggested that predominantly the cleaved form is present (Fig. S12A and C). The C-terminal transmembrane α-helix of ATP6AP1/Ac45 is homologous with that of the yeast V-ATPase subunit Voa1p (45) and is found in an equivalent position inside the c-ring (9). This transmembrane α-helix stretches approximately half-way across the thickness of the lipid bilayer terminating as a C-terminal tail that interacts with two of the subunits in the c-ring and subunit d1 before entering the cytoplasm (Fig. 4C, light blue). ATP6AP2/PRR is known to associate with mammalian V-ATPase (10, 11) and is essential for biogenesis of active V-ATPase (48). The strong density for its transmembrane α-helix in the map suggests that every complex includes the subunit (Fig. S5C). Consistent with an emerging role for V-ATPases in cell signalling (49), the presence of ATP6AP2/PRR in the complex has tied V-ATPases to the renin-angiotensin system for regulation of blood pressure and electrolyte balance (10, 11, 13), Wnt signalling (14), and other pathways (12). The gene for ATP6AP2/PRR encodes an N-terminal extracellular/luminal soluble domain and a transmembrane anchor (Fig. 4B, top). The soluble domain increases the angiotensin I generating activity of renin and can function both in a membrane-bound form and, when released by proteolysis, in a soluble form (50,51). In the structure, the renin-activating domain of ATP6AP2/PRR is missing (Fig. 4D, yellow). The transmembrane anchor consists of a long α-helix and a short α-helical turn connected by an extended linker with N- and C-terminal tails. The location of ATP6AP2/PRR’s transmembrane region, captured within the stable c-ring, dictates that after its incorporation into the c-ring the protein must remain associated with the V-ATPase. Mass spectrometry detected some intact ATP6AP2/PRR in the preparation but, consistent with the cryoEM density, the cleaved transmembrane region alone was much more abundant (Fig. S12B and D). In contrast to the renin-angiotensin system, Wnt signalling relies on ATP6AP2/PRR remaining membrane-anchored, with interaction of proteins with the extracellular/luminal part of ATP6AP2/PRR leading to signalling in the cytoplasm (14). The absence of the soluble domain of ATP6AP2/PRR from the structure suggests that ATP6AP2/PRR’s function in Wnt signalling involves either a subpopulation of intact ATP6AP2/PRR molecules that are not associated with V-ATPase, or a different population of V-ATPase or VO complexes containing intact ATP6AP2/PRR.
Fig. 4. Interaction of subunits within the membrane embedded rotor subcomplex.
(A) Subunits ATP6AP1/Ac45, ATP6AP2/PRR, and c″possess transmembrane α-helices in the centre of the c-ring. (B) ATP6AP2/PRR (top) and ATP6AP1/Ac45 (bottom) possess luminal domains that are absent from the structure. SP, signal peptide; Cyt, cytosolic. (C and D) Subunit d1 interacts with the C termini of ATP6AP1/Ac45 (C) and ATP6AP2/PRR (D). (E) Subunits c″, c(8), and ATP6AP2/PRR interact with the second luminal domain of ATP6AP1/Ac45. Scale bar, 10 Å. (F) Subunits c″, ATP6AP1/Ac45, ATP6AP2/PRR, and d1 create a network of interactions that connect the vesicle lumen and the cytoplasm. Scale bar, 25 Å.
The structure shows numerous interactions between ATP6AP1/Ac45, ATP6AP2/PRR, subunits d1, c″, and multiple c-subunits (Fig. 4E and F). Because these proteins are all part of the rotor subcomplex, the interactions persist during rotation and are also found in rotational states 2 and 3 (Fig. S13). In the lumen, the soluble domain of ATP6AP1/Ac45 interacts with the N-terminal tail of cleaved ATP6AP2/PRR, the linker that connects subunit c″ to its N-terminal α-helix in the middle of the c-ring, and the N terminus of subunit c(8) (Fig. 4E). Near the cytoplasmic surface of the VO region, the C-terminal tail of ATP6AP1/Ac45 and the short C-terminal α-helix of ATP6AP2/PRR, including 12 of the 19 residues in its intracellular domain (14), are sandwiched between subunits of the c-ring and subunit d1 (Fig. 4C and D, arrow). VO complexes assemble in the endoplasmic reticulum (ER), with the incorporation of subunit d allowing ER release (52, 53) and subsequent binding of the V1 region (53). Free subunit d in the cytoplasm (54) does not bind the V1 complexes that are preassembled there (55), suggesting that the conformation of subunit d changes upon incorporation into VO, making it competent to interact with subunit D of V1 (Fig. 1D). Indeed, subunit d adopts a more open conformation when it engages subunit D of the central rotor in the intact V-ATPase and a more closed conformation when V1 is detached and subunits d and D are disconnected (Fig. S14). The C-terminal tails of ATP6AP1/Ac45 and ATP6AP2/PRR produce part of the surface onto which subunit d1 assembles (Fig. 4C, D, and F). This structure explains why mutations in ATP6AP1/Ac45 and ATP6AP2/PRR are associated with related disease phenotypes (12, 45, 46). It also explains why ATP6AP1/Ac45 and ATP6AP2/PRR appear to work together to allow subunit d binding, ER release, and subsequent V1 assembly onto VO in the mammalian V-ATPase (9, 56). It is tempting to speculate that ATP6AP1/Ac45 and ATP6AP2/PRR are also involved in the reverse process, with conformational changes within them altering the conformation of subunit d1, disrupting its interaction with subunit D, and triggering separation of the V1 and VO regions.
Supplementary Material
One Sentence Summary.
CryoEM and mass spectrometry reveal the subunit composition and how (pro)renin receptor works with Ac45 in V-ATPase assembly.
Acknowledgements
We thank Samir Benlekbir for assistance with cryoEM data collection, Thamiya Vasanthakumar for advice on ATPase assays and assistance with Fig. 1A, and Stephane Angers, Peter Brzezinsky, and members of the Rubinstein laboratory for discussions.
Funding
This work was supported by the Canadian Institutes of Health Research Grant PJT166152 (JLR), grant 69551-ENABLE from the European Research Council (CVR), and a Wellcome Trust Investigator Award 104633/Z/14/Z (CVR). YMA was supported by a Restracomp fellowship and a Canadian Institutes of Health Research Postdoctoral fellowship. JLR was supported by the Canada Research Chairs program. CryoEM data was collected at the Toronto High-Resolution High-Throughput cryoEM facility, supported by the Canada Foundation for Innovation and Ontario Research Fund.
Footnotes
Author Contributions: JLR conceived the project and experimental approach and supervised the research; SAB made the SidK expression vector and developed an initial protein purification strategy. YMA developed the final protein purification procedure, prepared cryoEM specimens, collected cryoEM data, calculated the cryoEM maps, built and refined the atomic models, and performed biochemical assays. DW performed proteomics and native mass spectrometry experiments and CVR supervised the mass spectrometry. JLR and YMA wrote the manuscript and prepared the figures with input from the other authors.
Competing interests: We declare no competing interests.
Data and materials availability: CryoEM maps are deposited in the Electron Microscopy Data Bank under accession numbers EMD-21317 to 21319 for maps of the intact complex and 21345 to 213453 for maps from focused refinement. Atomic models are deposited in the Protein Data Bank under accession numbers 6VQ6, 6VQ7, and 6VQ8 for composite models, and 6VQ9, 6VQA, 6VQB, 6VQC, 6VQG, 6VQH, 6VQI, 6VQJ, and 6VQK for models built into maps from focused refinement. The SidK-3×FLAG expression vector is available from the corresponding author.
References
- 1.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 2.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J Biol Chem. 1995;270:17025–17032. [PubMed] [Google Scholar]
- 4.Sumner JP, Dow JA, Earley FG, Klein U, Jager D, Wieczorek H. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J Biol Chem. 1995;270:5649–5653. doi: 10.1074/jbc.270.10.5649. [DOI] [PubMed] [Google Scholar]
- 5.Bodzeta A, Kahms M, Klingauf J. The Presynaptic v-ATPase Reversibly Disassembles and Thereby Modulates Exocytosis but Is Not Part of the Fusion Machinery. Cell Rep. 2017;20:1348–1359. doi: 10.1016/j.celrep.2017.07.040. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe S, Trimbuch T, Camacho-Pérez M, Rost BR, Brokowski B, Söhl-Kielczynski B, Felies A, Davis MW, Rosenmund C, Jorgensen EM. Clathrin regenerates synaptic vesicles from endosomes. Nature. 2014;515:228–233. doi: 10.1038/nature13846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farsi Z, Gowrisankaran S, Krunic M, Rammner B, Woehler A, Lafer EM, Mim C, Jahn R, Milosevic I. Clathrin coat controls synaptic vesicle acidification by blocking vacuolar ATPase activity. Elife. 2018;7:e32569. doi: 10.7554/eLife.32569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazhab-Jafari MT, Rohou A, Schmidt C, Bueler SA, Benlekbir S, Robinson CV, Rubinstein JL. Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase. Nature. 2016;539:118–122. doi: 10.1038/nature19828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roh S-H, Stam NJ, Hryc CF, Couoh-Cardel S, Pintilie G, Chiu W, Wilkens S. The 3.5-Å CryoEM Structure of Nanodisc-Reconstituted Yeast Vacuolar ATPase V o Proton Channel. Mol Cell. 2018;69:993–1004. doi: 10.1016/j.molcel.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huillier NL, Sharp MG, Dunbar DR, Mullins JJ. In: The Local Cardiac Renin Angiotensin- Aldosterone System. Frohlich ED, Richard NRE, editors. Springer International Publishing; New York: 2006. pp. 17–34. [Google Scholar]
- 11.Burcklé C, Bader M. Prorenin and Its Ancient Receptor. Hypertension. 2006;48:549–551. doi: 10.1161/01.HYP.0000241132.48495.df. [DOI] [PubMed] [Google Scholar]
- 12.Ichihara A, Yatabe MS. The (pro)renin receptor in health and disease. Nat Rev Nephrol. 2019 doi: 10.1038/s41581-019-0160-5. [DOI] [PubMed] [Google Scholar]
- 13.Ludwig J, Kerscher S, Brandt U, Pfeiffer K, Getlawi F, Apps DK, Schagger H. Identification and characterization of a novel 9.2-kDa membrane sector-associated protein of vacuolar proton-ATPase from chromaffin granules. J Biol Chem. 1998;273:10939–10947. doi: 10.1074/jbc.273.18.10939. [DOI] [PubMed] [Google Scholar]
- 14.Cruciat CM, Ohkawara B, Acebron SP, Karaulanov E, Reinhard C, Ingelfinger D, Boutros M, Niehrs C. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science (80-. ) 2010;327:459–463. doi: 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]
- 15.Toei M, Saum R, Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blake-Palmer KG, Su Y, Smith AN, Karet FE. Molecular cloning and characterization of a novel form of the human vacuolar H+-ATPase e-subunit: An essential proton pump component. Gene. 2007;393:94–100. doi: 10.1016/j.gene.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, Müller SA, et al. Molecular Anatomy of a Trafficking Organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 18.Xu L, Shen X, Bryan A, Banga S, Swanson MS, Luo ZQ. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, Beyrakhova K, Liu Y, Alvarez CP, Bueler SA, Xu L, Xu C, Boniecki MT, Kanelis V, Luo Z-Q, Cygler M, et al. Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein. PLoS Pathog. 2017;13:e1006394. doi: 10.1371/journal.ppat.1006394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed S, Holt M, Riedel D, Jahn R. Small-scale isolation of synaptic vesicles from mammalian brain. Nat Protoc. 2013;8:998–1009. doi: 10.1038/nprot.2013.053. [DOI] [PubMed] [Google Scholar]
- 21.Murata Y, Sun-Wada GH, Yoshimizu T, Yamamoto A, Wada Y, Futai M. Differential localization of the vacuolar H + pump with G subunit isoforms (G1 and G2) in mouse neurons. J Biol Chem. 2002;277:36296–36303. doi: 10.1074/jbc.M200586200. [DOI] [PubMed] [Google Scholar]
- 22.Perreira JM, Aker AM, Savidis G, Chin CR, McDougall WM, Portmann JM, Meraner P, Smith MC, Rahman M, Baker RE, Gauthier A, et al. RNASEK Is a V-ATPase-Associated Factor Required for Endocytosis and the Replication of Rhinovirus, and Dengue Virus. Cell Rep. 2015;12:850–863. doi: 10.1016/j.celrep.2015.06.076. [DOI] [PubMed] [Google Scholar]
- 23.Wilkens S, Inoue T, Forgac M. Three-dimensional structure of the vacuolar ATPase. Localization of subunit H by difference imaging and chemical cross-linking. J Biol Chem. 2004;279:41942–41949. doi: 10.1074/jbc.M407821200. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z, Bin Peng S, Crider BP, Andersen P, Xie XS, Stone DK. Recombinant SFD isoforms activate vacuolar proton pumps. J Biol Chem. 1999;274:15913–15919. doi: 10.1074/jbc.274.22.15913. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J, Benlekbir S, Rubinstein JL. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. doi: 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- 26.Parra KJ, Keenan KL, Kane PM. The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J Biol Chem. 2000;275:21761–21767. doi: 10.1074/jbc.M002305200. [DOI] [PubMed] [Google Scholar]
- 27.Oot RA, Kane PM, Berry EA, Wilkens S. Crystal structure of yeast V 1 -ATPase in the autoinhibited state. 2016;35:1–13. doi: 10.15252/embj.201593447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huss M, Wieczorek H. Influence of ATP and ADP on dissociation of the V-ATPase into its V1 and VO complexes. FEBS Lett. 2007;581:5566–5572. doi: 10.1016/j.febslet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Vasanthakumar T, Bueler SA, Wu D, Beilsten-Edmands V, Robinson CV. Structural comparison of the vacuolar and Golgi V- ATPases from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2019;116:7272–7277. doi: 10.1073/pnas.1814818116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagamatsu Y, Takeda K, Kuranaga T, Numoto N, Miki K. Origin of asymmetry at the intersubunit interfaces of V1-ATPase from Thermus thermophilus. J Mol Biol. 2013;425:2699–2708. doi: 10.1016/j.jmb.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 31.Arai S, Saijo S, Suzuki K, Mizutani K, Kakinuma Y, Ishizuka-Katsura Y, Ohsawa N, Terada T, Shirouzu M, Yokoyama S, Iwata S, et al. Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature. 2013;493:703–707. doi: 10.1038/nature11778. [DOI] [PubMed] [Google Scholar]
- 32.Zhou L, Sazanov LA. Structure and conformational plasticity of the intact Thermus thermophilus V/A-type ATPase. Science (80-. ) 2019;365:eaaw9144. doi: 10.1126/science.aaw9144. [DOI] [PubMed] [Google Scholar]
- 33.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki K, Mizutani K, Maruyama S, Shimono K, Imai FL, Muneyuki E, Kakinuma Y, Ishizuka-katsura Y, Shirouzu M, Yokoyama S, Yamato I, et al. Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor. Nat Commun. 2016;7 doi: 10.1038/ncomms13235. 13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholls DG, Ferguson SJ. Bioenergetics. ed 4th. Academic Press; London: 2013. [Google Scholar]
- 36.Hahn A, Vonck J, Mills DJ, Meier T, Kuhlbrandt W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science (80-. ) 2018;360:eaat4318. doi: 10.1126/science.aat4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo H, Suzuki T, Rubinstein JL. Structure of a bacterial ATP synthase. Elife. 2019;8:e43128. doi: 10.7554/eLife.43128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy BJ, Klusch N, Langer J, Mills DJ, Yildiz Ö, Kühlbrandt W. Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1-Fo. Science (80-.) 2019;364:eaaw9128. doi: 10.1126/science.aaw9128. [DOI] [PubMed] [Google Scholar]
- 39.Guo H, Bueler SA, Rubinstein JL. Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science (80-. ) 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bueler SA, Rubinstein JL. Vma9p Need Not Be Associated with the Yeast V-ATPase for Fully-Coupled Proton Pumping Activity in Vitro. Biochemistry. 2015;54:853–858. doi: 10.1021/bi5013172. [DOI] [PubMed] [Google Scholar]
- 41.Sambade M, Kane PM. The yeast vacuolar proton-translocating ATPase contains a subunit homologous to the Manduca sexta and bovine e subunits that is essential for function. J Biol Chem. 2004;279:17361–5. doi: 10.1074/jbc.M314104200. [DOI] [PubMed] [Google Scholar]
- 42.Rampias TN, Sideris DC, Fragoulis EG. Cc RNase: the Ceratitis capitata ortholog of a novel highly conserved protein family in metazoans. Nucleic Acids Res. 2003;31:3092–3100. doi: 10.1093/nar/gkg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hackett BA, Yasunaga A, Panda D, Tartell MA, Hopkins KC, Hensley SE. RNASEK is required for internalization of diverse acid-dependent viruses. Proc Natl Acad Sci U S A. 2015;112:7797–7802. doi: 10.1073/pnas.1424098112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Economopoulou MI, Fragoulis EG, Sideris DC. Molecular cloning and characterization of the human RNase kappa, an ortholog of Cc RNase. Nucleic Acids Res. 2007;35:6389–6398. doi: 10.1093/nar/gkm718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jansen EJR, Timal S, Ryan M, Ashikov A, Van Scherpenzeel M, Graham LA, Mandel H, Hoischen A, Iancu TC, Raymond K, Steenbergen G, et al. ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat Commun. 2016;7 doi: 10.1038/ncomms11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pareja F, Brandes AH, Basili T, Selenica P, Geyer FC, Fan D, Da A, Paula C, Kumar R, Brown DN, Gularte-mérida R, et al. Loss-of-function mutations in ATP6AP1 and ATP6AP2 in granular cell tumors. Nat Commun. 2018;9 doi: 10.1038/s41467-018-05886-y. 3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Louagie E, Taylor NA, Flamez D, Roebroek AJM, Bright NA, Meulemans S, Quintens R, Herrera PL, Schuit F, VanDe Ven WJM, Creemers WM. Role of furin in granular acidification in the endocrine pancreas: Identification of the V-ATPase subunit Ac45 as a candidate substrate. Proc Natl Acad Sci U. S. A. 2008;2 doi: 10.1073/pnas.0800340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinouchi K, Ichihara A, Sano M, Wada Y, Kurauchi-mito A, Bokuda K, Narita T, Oshima Y, Sakoda M, Tamai Y, Sato H, et al. The (Pro)renin Receptor/ATP6AP2 is Essential for Vacuolar H+-ATPase Assembly in Murine Cardiomyocytes. Circ Res. 2010;107:30–34. doi: 10.1161/CIRCRESAHA.110.224667. [DOI] [PubMed] [Google Scholar]
- 49.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H-ATPase. Science (80-. ) 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen G, Delarue F, Burcklé C, Bouzhir L, Giller T, Sraer J-D. Pivotal role of the renin / prorenin receptor in angiotensin II production and cellular responses to renin. J Clin Invest. 2002;109:1417–1427. doi: 10.1172/JCI14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cousin C, Bracquart D, Contrepas A, Corvol P, Muller L, Nguyen G. Soluble Form of the (Pro)Renin Receptor Generated by Intracellular Cleavage by Furin Is Secreted in Plasma. Hypertension. 2009;53:1077–1082. doi: 10.1161/HYPERTENSIONAHA.108.127258. [DOI] [PubMed] [Google Scholar]
- 52.Ryan M, Graham LA, Stevens TH. Voa1p Functions in V-ATPase Assembly in the Yeast Endoplasmic Reticulum. Mol Biol Cell. 2008;19:5131–5142. doi: 10.1091/mbc.E08-06-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graham LA, Hill KJ, Stevens TH. Assembly of the Yeast Vacuolar H+-ATPase Occurs in the Endoplasmic Reticulum and Requires a Vma12p/Vma22p Assembly Complex. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauerle C, Ho MN, Lindorfer MA, Stevens TH. The Saccharomyces cerevisiae VMA6 gene encodes the 36-kDa subunit of the vacuolar H+-ATPase membrane sector. J Biol Chem. 1993;268:12749–12757. [PubMed] [Google Scholar]
- 55.Doherty RD, Kane PM. Partial assembly of the yeast vacuolar H(+)-ATPase in mutants lacking one subunit of the enzyme. J Biol Chem. 1993;268:16845–16851. [PubMed] [Google Scholar]
- 56.Guida MC, Hermle T, Graham LA, Hauser V, Ryan M, Stevens TH, Simons M. ATP6AP2 functions as a V-ATPase assembly factor in the endoplasmic reticulum. Mol Biol Cell. 2018;29:2156–2164. doi: 10.1091/mbc.E18-04-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.