Abstract
Novel coronavirus disease 2019 (COVID-19) is the biggest threat to human being globally. The first case was identified in a patient with flu symptoms along with severe acute respiratory syndrome in Wuhan, China in December 2019 and now it has spread in more than 200 countries. COVID-19 is more lethal in the elderly and people with an underlying condition such as asthma, cancer, diabetes. Here we performed bioinformatic analysis to investigate the interaction of S2 subunit protein of SARS-nCoV-2 of novel coronavirus with tumor suppressor proteins p53 and BRCA-1/2. In this short communication we report the interaction between S2 subunit proteins with tumor suppressor proteins for the first time. This preliminary result will open up a new direction to investigate the effect of a novel coronavirus in cancer patients.
In December 2019, an outbreak of pneumonia was reported in Wuhan, China which was caused by a new strain of coronavirus called novel coronavirus (nCoV). The novel coronavirus disease-2019 (COVID-19), which started spreading globally, was latter announced as a pandemic by WHO. Till April 30, 2020, there are 3,090,445 infections and 217,769 deaths worldwide [1]. Previously known SARS-CoV which caused severe acute respiratory syndrome (SARS) has 79.5% sequence similarity with SARS-nCoV-2 (nCoV). The pernicious nCoV has been causing severe flu-like symptoms along with pneumonia specially in the elderly and people with ailments like hypertension, asthma, cancer, diabetes, etc. [2,3] but detail understanding of infection is still lacking.
Coronaviruses (CoVs) belong to coronaviridae family and are the largest RNA viruses identified till date. SARS-nCoV-2 contains a spike (S) protein which consists of two subunits S1 and S2. S1 aids the virus to infect human cells by binding to human angiotensin-converting enzyme 2 (hACE2) and S2 mediates the membrane fusion process. S2 subunit is further divided (N-terminal to C-terminal) into fusion peptide (FP), hepted repeat 1 (HR-1), hepted repeat 2 (HR-2), transmembrane domain (TM), and cytoplasmic domain (CP). After infection to host receptor, the HR-1 and HR-2 domain of S2 subunit interact with each other to form six-helix bundle (6-HB) fusion core, bringing viral and cellular membrane into close proximity for fusion and infection [4]. That is why it is very important to study the interaction of S2 subunit with other proteins, to gain insight in to its function and interaction with other potential proteins which have a central role in human diseases. This would unravel the possible mechanism of COVID-19 infection and severity in humans who are already suffering from an ailment.
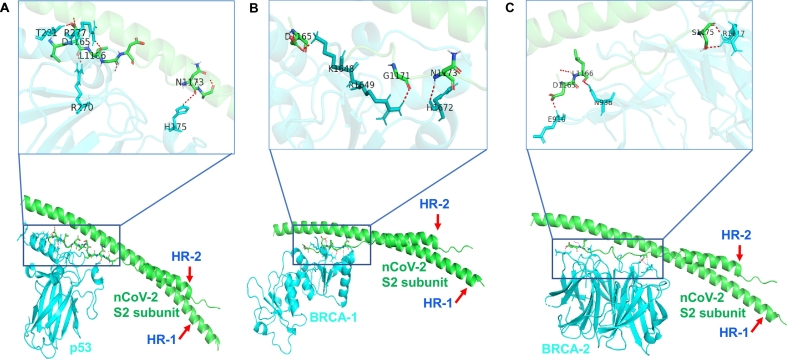
Here, we have investigated the interaction of S2 subunit to tumor suppressor and cell cycle-related proteins. HADDOCK 2.2 software [5,6] was used to analyze the interaction and found that S2 subunit of SARS-nCov-2 strongly interacts with p53 and BRCA-1/2 proteins (Figure 1). p53 and BRCA are the well-known tumor suppressor proteins, that regulate downstream genes in response to numerous cellular stress and are frequently mutated in human cancer [5,6]. Interestingly we found p53, BRCA-1 and BRCA-2 interact with heptic repeat-2 region of S2 subunit through C- terminal domain. PDB ID of these proteins was extracted from RCSB Protein Data Base (PDB) and details of crystal structure IDs and interacted amino acid residues are mentioned in the figure legend. This short bioinformatic analysis is a first time report and significant since COVID-19 is more fatal in people with underlying conditions specially lung diseases, diabetes and cancer. Therefore, further research is needed to understand COVID-19 effect in cancer patients and the detailed role of these interactions.
Figure 1.
Analysis of S2 subunit of SARS-nCoV-2 interaction with tumor suppressor proteins. (A) Interaction between S2 Subunit of nCoV (Green color PDB ID: 6LXT [4] asp 1165, leu 1166, asn 1173) with p53 protein (Cyan color PDB ID: 3EXJ [7] thr 281, arg 270, arg 277, his 175) (B) Interaction between S2 Subunit of nCoV (Green color PDB ID: 6LXT [4] asp 1165, gly 1171, asn 1173) with BRCA-1 protein (Cyan color PDB ID: 4Y18 [8] lys 1648, arg 1649, his 1672) (C) Interaction between S2 Subunit of nCoV (Green color PDB ID: 6LXT [4] asp 1165, leu 1166, ser 1175) with BRCA-2 protein (Cyan color PDB ID: 3EU7 [9] glu 916, leu 938, arg 1117). HR: hepted repeat. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Acknowledgments
Acknowledgement
We sincerely acknowledge HADDOCK2.2 software developed by Dr. Bonvin's lab Utrecht University, The Netherlands.
Author Statement
Nishant Singh: Conceptualization, Methodology, Software, correspondence; Anuradha Bharara Singh: Writing - Reviewing and Editing,
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Nishant Singh, Email: nishant900@gmail.com.
Anuradha Bharara Singh, Email: anuradha.bharara@gmail.com.
References
- 1.World Health Organization situation report-101. https://www.who.int/docs/default-source/coronavirus/situation-reports/20200430-sitrep-101-covid19.pdf?sfvrsn=2ba4e093_2
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Zundert G.C.P., Rodrigues J., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A. The HADDOCK2.2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Wassenaar T.A., van Dijk M., Loureiro-Ferreira N. WeNMR: Structural biology on the grid. J Grid Comput. 2012;10:743–767. doi: 10.1007/s10723-012-9246-z. [DOI] [Google Scholar]
- 7.Malecka K.A., Ho W.C., Marmorstein R. Crystal structure of a p53 core tetramer bound to DNA. Oncogene. 2009;28(3):325–333. doi: 10.1038/onc.2008.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Q., Paul A., Su D., Mehmood S., Foo T.K., Ochi T., Bunting E.L., Xia B., Robinson C.V., Wang B. Structure of BRCA1-BRCT/Abraxas complex reveals phosphorylation-dependent BRCT dimerization at DNA damage sites. Mol. Cell. 2016;61(3):434–448. doi: 10.1016/j.molcel.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver A.W., Swift S., Lord C.J., Ashworth A., Pearl L.H. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10(9):990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]