Abstract
Differentiation of neuronal cells is crucial for the development and function of the nervous system. This process involves high rates of membrane expansion, during which the synthesis of membrane lipids must be tightly regulated. In this work, using a variety of molecular and biochemical assays and approaches, including immunofluorescence microscopy and FRET analyses, we demonstrate that the proto-oncogene c-Fos (c-Fos) activates cytoplasmic lipid synthesis in the central nervous system and thereby supports neuronal differentiation. Specifically, in hippocampal primary cultures, blocking c-Fos expression or its activity impairs neuronal differentiation. When examining its subcellular localization, we found that c-Fos co-localizes with endoplasmic reticulum markers and strongly interacts with lipid-synthesizing enzymes, whose activities were markedly increased in vitro in the presence of recombinant c-Fos. Of note, the expression of c-Fos dominant-negative variants capable of blocking its lipid synthesis–activating activity impaired neuronal differentiation. Moreover, using an in utero electroporation model, we observed that neurons with blocked c-Fos expression or lacking its AP-1–independent activity fail to initiate cortical development. These results highlight the importance of c-Fos–mediated activation of lipid synthesis for proper nervous system development.
Keywords: neurodifferentiation, neurodevelopment, c-Fos, lipid synthesis, phospholipid metabolism, hippocampus, AP-1, central nervous system, oncogene., neurodifferentiation, neurodevelopment, phospholipid metabolism, lipid synthesis, AP1 transcription factor (AP-1), central nervous system (CNS), oncogene
Neuronal differentiation, an intricate cellular process, comprises a series of complex and coordinated events, such as cell proliferation, migration, and differentiation (1–5). To reach its final structure and establish functional neuronal circuits, a neuron must promote neuritogenesis, neurite elongation, polarization, arborization, and synaptogenesis (6–8). The proper function of the nervous system heavily relies on the number of multiple and targeted synaptic contacts established by each neuron. Sprouting of axon branches along the axon shaft allows multiple innervation targets (9). The overall process of neuronal differentiation determines patterns of connectivity of neuronal circuits, whereas the disruption of these patterns may cause severe pathologies or developmental disorders (10, 11).
Outgrowth events involve cellular volume and surface area increase, thus demanding membrane material that must be supplied according to the temporal and spatial requirements of the cells in their different developmental stages (5, 6, 12–14). In fact, membrane expansion can reach an increment of up to 20% according to the different steps of neuronal differentiation (13).
Considering their architecture and size, it is reasonable to hypothesize that neurons will need temporal and domain-specific mechanisms to regulate the synthesis of the components required according to their local demands. In line with this hypothesis, Tsukita and Ishikawa (15) provided evidence of the three-dimensional structure of the endoplasmic reticulum (ER) in axonal processes. Several reports later convincingly demonstrated a synthetic machinery present in axons, with the capacity to regulate the levels of proteins, lipids, and other macromolecules (16, 17). Consequently, the mechanisms by which neurons supply components for membrane biogenesis, such as lipids in growing axons, have been the subject of several studies. Using compartmented culture systems, compelling evidence emerged demonstrating that axons carry out active lipid synthesis (18–20).
The expression of Fos, an oncogene of the immediate early gene family, is rapidly and transiently induced in different cell types by diverse stimuli (i.e. growth factors, neurotransmitters) (21–23). The proteins of the Fos family (c-Fos, Fos-B, ΔFos-B, Fra-1, Fra-2) heterodimerize mainly with those of the Jun family, comprising the well-known AP-1 transcription factors (24). Our laboratory described c-Fos as a moonlighting protein capable of achieving an additional, non-AP-1 function: it associates with the ER membranes and activates lipid synthesis through an interaction with specific lipid-synthesizing enzymes (25–31). Using PC12 cells, we distinguished between the two known functions of c-Fos (i.e. its AP-1 activity and its capacity to activate lipid synthesis). Initially, when cells are stimulated with nerve growth factor to differentiate into a sympathetic neuron phenotype, c-Fos is required in the nucleus to trigger the genomic program of differentiation. However, once the cells have been primed to differentiate, c-Fos is only required at the cytoplasm, associated to the ER, activating phospholipid synthesis, for cells to continue differentiating (32).
Herein, using hippocampal neurons, we have shown that c-Fos is capable of exerting its lipid synthesis activator capacity in the nervous system and that this AP-1–independent activity is crucial for normal neuronal differentiation. Furthermore, by in utero electroporation, we demonstrate that this role is involved in cortical development. In this direction, c-Fos could be an active player in different events required for neuronal plasticity, such as those involved in learning and memory formation.
Results
c-Fos expression during neuronal differentiation
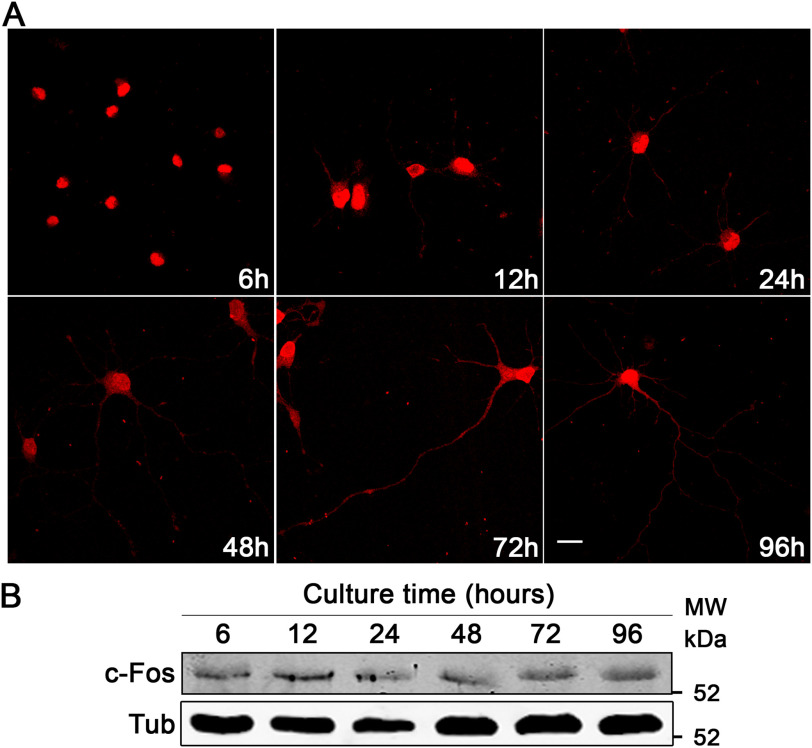
The presence of c-Fos in the brain, and more specifically in the hippocampus, was demonstrated almost 30 years ago by Dragunow et al. (33, 34). We initially evaluated c-Fos expression over time in primary cultures of rat hippocampal neurons. At all times examined, c-Fos was expressed at similar levels even after 96 h in culture, as determined by immunocytochemistry and Western blotting (Fig. 1). It is important to highlight that c-Fos expression is observed not only in the nucleus but also extending to the entire cytoplasm of the neurons.
Figure 1.
c-Fos is expressed in primary rat hippocampal neuronal cultures. A, rat primary hippocampal neurons were fixed at different times of culture and immunostained with an anti-c-Fos antibody (red). Scale bar, 20 μm. B, Western blotting of hippocampal cell lysates harvested at different times of culture. Membranes were immunostained with an anti c-Fos antibody (top) and an anti-tubulin (Tub) antibody (bottom) as a loading control and secondary antibodies suitable for near-IR fluorescence. The obtained images were converted to gray scale and inverted. The results of one of three independent experiments are shown.
We then evaluated whether c-Fos participates in the differentiation of rat hippocampal neurons. The method applied to determine the differentiation stage of the cultured cells was the one described by Dotti et al. (7). Briefly, cells in stage 1 of differentiation, shortly after they attach to the substrate, show motile lamellipodia developed around the periphery of the cell. At stage 2, these lamellipodia begin to transform into distinct minor processes that extend to a length of up to 10-15 µm. At stage 3, one of these minor processes begins to grow 5–10 times faster than the rest of them, indicating that the axon of the cell has been formed and the cell has polarized. At stage 4, a significant development of dendrites initiates, and finally, at differentiation stage 5, the axonal and dendritic arbors undergo a maturation process.
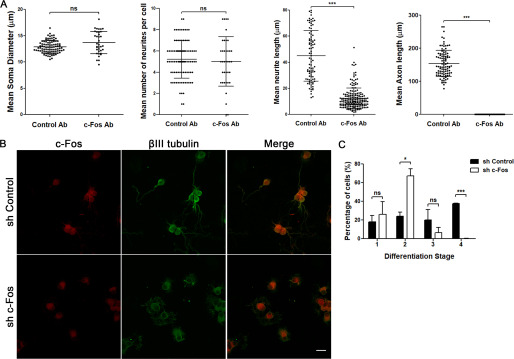
To evaluate the effect of blocking c-Fos activity during this complex process, primary cultures of rat hippocampal neurons were profected at 2 h of culture with anti-c-Fos antibodies or with a nonrelated antibody as a control using the BioPORTER system. Cells were examined 48 h later by immunocytochemistry but only using secondary antibodies in the case of the anti-c-Fos–profected cells. As can be seen in the graphs of Fig. 2A, blocking c-Fos activity impairs axon formation. We quantified different morphological features of the neurons, observing that there were no changes in soma size or in the mean number of neurites per cell when comparing cells lacking c-Fos activity and the control ones. It should be noted that we were not able to observe the development of axonal-like processes even after 48 h of culture when c-Fos activity was impaired, a phenomenon not observed in the control cells (Fig. 2A).
Figure 2.
Blocking the activity or the expression of c-Fos impairs neuronal differentiation. A, cells were profected with anti c-Fos (c-Fos Ab) or with a nonrelated anti-mouse IgG antibody as a control (Control Ab) 2 h after seeding using BioPORTER and then were fixed after 48 h of culture. The different morphological aspects were quantified from microscopy images, using ImageJ software, and are shown as the mean ± S.D. (error bars) in each case. Student's t test statistical analysis was performed using GraphPad Software. ***, p < 0.001; n.s., nonsignificant; in each experiment, n = 40 cells from each condition were examined. Results of one of three independent experiments performed are shown. B, cells were infected at the initiation of the culture with lentiviral particles designed to express an anti c-Fos shRNA or an shRNA with a scrambled sequence of c-Fos as a control. After 48 h of culture, cells were fixed and immunostained with an anti-c-Fos antibody (red, first column) and an anti βIII-tubulin antibody (green, second column). The third column shows the merge between both labels. Scale bar, 20 μm. C, morphological quantification of neuronal differentiation stages in both c-Fos and scrambled infected cells was performed using ImageJ software. The graph shows the mean number of cells ± S.D. in each case. Student's t test statistical analysis was performed using GraphPad software. *, p < 0.05; ***, p < 0.001; n.s., nonsignificant; n = 198 cells for scrambled shRNA–infected cells, n = 254 cells for c-Fos shRNA-infected cells. Results of one of three independent experiments performed are shown.
These results were confirmed by blocking c-Fos expression. When neurons were infected 2 h after seeding with lentiviral vectors designed to express a specific shRNA against c-Fos, impaired differentiation was clearly observed (Fig. 2B). When neurons were classified into the differentiation stages according to their morphology, more than 85% of the c-Fos-knocked-down neurons remained at stages 1 and 2 of differentiation as compared with the almost 50% of neurons in stages 3 and 4 of the control cultures (Fig. 2C).
c-Fos co-localizes with ER markers in neuronal processes
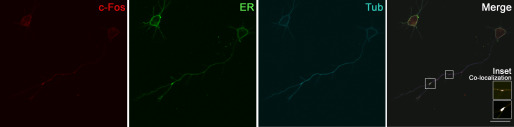
Previous studies from our laboratory have shown that upon a subcellular fractionation of PC12 cell homogenates, c-Fos and the ER membranes are predominantly collected in the same fraction (35). This led us to examine whether the c-Fos/ER co-localization observed previously can be evidenced in neurons of the central nervous system as well. To test this, we performed immunocytochemistry assays on primary rat hippocampal neurons after 48 h of culture, and, as expected, a strong co-localization of c-Fos with the ER marker calnexin was detected (Fig. 3). Surprisingly, c-Fos and the ER were found not only in the perinuclear region of the soma; defined co-localization dots at punctuate structures confined to axonal branching points were also very frequently observed.
Figure 3.
c-Fos co-localizes with ER markers. Immunocytochemistry of c-Fos (red), the ER marker calnexin (green), and βIII-tubulin (cyan) of rat hippocampal neurons at 48 h of culture. The fourth panel shows the merged images between the three labels. A Pearson's r value of 0.81 was calculated for co-localization of both labels (Coloc2, ImageJ). An inset is shown with a co-localization analysis between c-Fos and the ER, where the co-localizing pixels are colored in white (Colocalization Finder, ImageJ). Scale bar, 20 μm. The results of one of three independent experiments are shown.
c-Fos activation of lipid-synthesizing enzymes in neurons
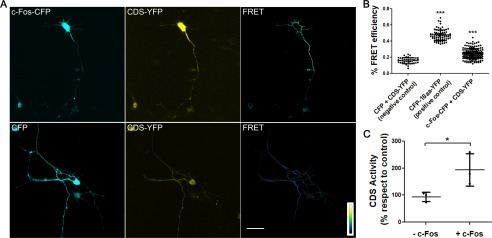
Previous results from our group have demonstrated that c-Fos physically interacts with and activates particular enzymes of the lipid synthesis pathways. Such is the case for CDP-diacylglycerol synthase (CDS), PI4KIIα (phosphatidylinositol 4 kinase II-α), and Lipin1 (29, 31). Taking this into consideration, it was hypothesized that c-Fos could be involved in neuronal differentiation by a lipid-activating mechanism that requires its physical interaction with specific enzymes. To this end, we examined the possible interaction of c-Fos with CDS, the enzyme that catalyzes the conversion of phosphatidic acid into CDP-diacylglycerol (CDP-DAG), the first step of phosphatidylinositol-phosphate synthesis in the ER. To infer protein-protein interactions, FRET experiments were performed (36). For this, rat primary hippocampal neurons were co-transfected to express c-Fos-mTurquoise2 and CDS-SYFP2 and examined by confocal microscopy. As shown in Fig. 4 (A and B), a positive FRET phenomenon is observed between both proteins, indicating that there is indeed a physical interaction between them.
Figure 4.
c-Fos activates and physically interacts with CDS. A, top row, neurons were co-transfected at 24 h of culture to express c-Fos-CFP (first panel) and CDS-YFP (second panel) and examined by confocal microscopy at 48 h. FRET images were obtained by the sensitized emission method and pseudocolored using ImageJ software (third panel). The bottom row shows a negative control with cells co-transfected with the CFP empty vector and CDS-YFP. The scale goes from no FRET (black) to maximum FRET (yellow). Scale bar, 30 μm. B, the graphic shows the quantification of the mean efficiencies ± S.D. (error bars) for the donor/acceptor pairs shown in the images, with one-way ANOVA and Tukey's post-test. ***, p < 0.001; n = 25 cells for each condition. The results of one of three independent experiments are shown. C, evaluation of CDS activity through measurement of the incorporation of [3H]CTP into CDP-DAG in neuron homogenates in the presence of c-Fos (+ c-Fos). Elution buffer was used as a control (– c-Fos). Results are the mean of three independent experiments performed in triplicate. The results are expressed as the mean ± S.D., with Student's t test analysis. *, p < 0.05.
To study whether this physical interaction leads to enzyme activation, CDS activity was assayed using cell homogenates obtained from rat hippocampal neurons as the enzyme source and [3H]CTP and dioleoyl-phosphatidic acid as substrates, with or without the addition of recombinant c-Fos to the incubates (29, 37, 38). At the times assayed, about 40% more CDP-DAG was synthetized in the +c-Fos incubates as compared with the control (Fig. 4C). This confirms that the lipid synthesis activation mediated by c-Fos observed previously also occurs in hippocampal neurons in culture. The association of c-Fos with CTP:phosphocholine cytidylyltransferase (CCT), the rate-limiting enzyme in phosphatidylcholine synthesis (Fig. S1), suggests that c-Fos could be also mediating the activation of other phospholipid pathways.
Neuronal differentiation is impaired in cells lacking cytoplasmic c-Fos
The results shown so far support the need of c-Fos to normally complete the differentiation events. However, they do not allow us to discern whether the effect is due to the AP-1 or the lipid synthesis activator function of c-Fos. To activate lipid synthesis, c-Fos associates through its N-terminal domain (NA domain, amino acids 1-138 of full-length c-Fos) with the enzymes it activates (29, 31, 32). However, this portion of the protein is not sufficient to produce lipid synthesis activation because of the absence of the basic domain (BD, amino acids 139-159), a domain required for enzyme activation to occur. As the NA domain is not involved in c-Fos AP-1 functions, it seems reasonable to propose its use as a dominant negative of its lipid synthesis activator function. In other words, the NA should interfere only with the lipid-synthesizing function of c-Fos and not with its AP-1, transcriptional one. Furthermore, we have previously shown that recombinant NA is not able to activate total phospholipid synthesis and inhibits CDS activity (30, 38).
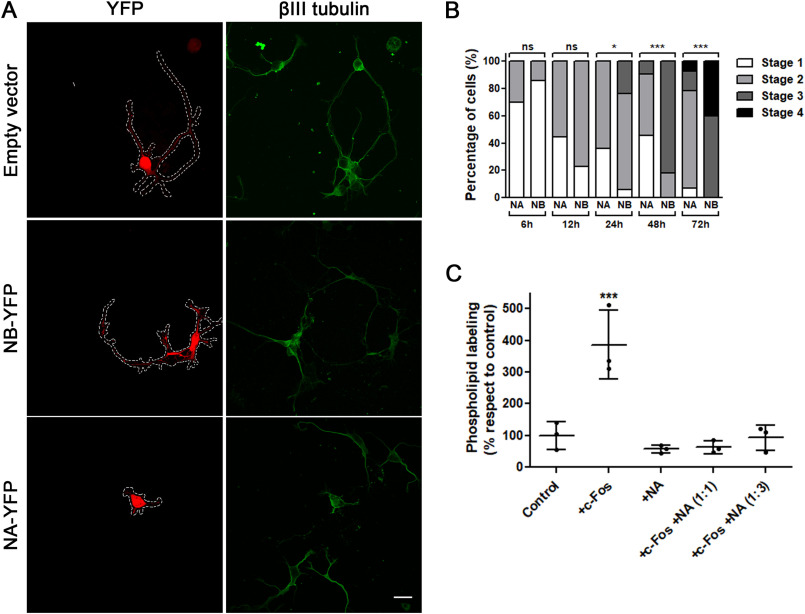
Taking this into consideration, we examined whether the impairment in neuronal differentiation could be linked to the lack of c-Fos–dependent lipid synthesis activity. Hippocampal neuronal cells were transfected with a construct that contains the NA domain, which does not activate lipid synthesis, or with the construct NB, which contains amino acids 1–159 (NB domain), which is capable of activating lipid synthesis at levels comparable with those of full-length c-Fos, both fused with YFP. The empty vector was used as a control. Then the differentiation stages of the transfected cells at different fixation times were analyzed. It can be seen in Fig. 5A and its quantification in Fig. 5B (cells fixed at 48 h) that the expression of the NA domain impairs neuronal differentiation as evidenced after a morphological analysis and stage quantification of the NA-transfected cells relative to the NB-transfected or nontransfected ones. Even more, it is clear that the impairment in differentiation promoted by the NA domain is comparable with that observed when c-Fos expression is blocked (Fig. 2, B and C).
Figure 5.
NA deletion mutant of c-Fos impairs differentiation in neuronal cultures and abrogates c-Fos-dependent lipid synthesis activation. A, neuronal cultures were transfected at seeding with NA-YFP (pseudocolored red, bottom row), NB-YFP (pseudocolored red, middle row), or the empty vector as a control (pseudocolored red, top row) and were fixed after 48 h of culture. Cells were subjected to immunofluorescence against βIII-tubulin (green, second column). Scale bar, 20 μm. B, morphological quantification of neuronal differentiation stages in both NA-YFP– and NB-YFP–transfected cells at different fixation times. The results of one of three independent experiments are shown. A normality Kolmogorov–Smirnov test was performed, where the deviation from the distribution with respect to the NB-transfected cells was evaluated. *, p < 0.05; ***, p < 0.001; n.s., nonsignificant; n = 15 from each condition were examined. C, evaluation of 32P-phospholipid labeling capacity of neuron homogenates in the presence of c-Fos (+c-Fos), NA (+NA), or both (+c-Fos +NA 1:1 and +c-Fos +NA 1:3). Elution buffer was used as a control (Control). Results are the mean of three independent experiments performed in triplicate. The results are expressed as the mean ± S.D. (error bars), with one-way ANOVA. ***, p < 0.001 with respect to control conditions.
To test whether this effect was due to the lipid synthesis activator capacity of c-Fos, we performed biochemical assays to assess total phospholipid labeling in homogenates obtained from hippocampal neurons primary cultures. Previous reports have shown that the γ-phosphate group of labeled [32P]ATP can be incorporated into different lipids of cell lysates assayed in vitro, mainly phosphatidylinositol and its derivatives and phosphatidic acid (39–44), and that recombinant c-Fos promotes an increase in their synthesis (45). When assaying this in neurons, it is clear that the addition of recombinant c-Fos to the assay promotes a significant increase in total phospholipid labeling, whereas recombinant NA does not (Fig. 5C). However, if both c-Fos and NA are added together, the activating effect of c-Fos is abolished. These results are compatible with a competition between c-Fos and NA for binding of c-Fos to the enzymes as shown previously in other systems (38). This is also compatible with the notion that differentiation is impaired as a consequence of the lack of the lipid synthesis activator capacity of c-Fos.
c-Fos is essential for cortical development
We next studied whether c-Fos participates in cortical development in vivo by using in utero electroporation. Briefly, the vertebrate cortex is organized into layers of neurons that share functions, morphology, and birthdates (46). During development, radial glia progenitors in the ventricular zone (VZ) divide asymmetrically to originate cortical pyramidal projecting neurons. Then they suffer a radial migration process toward the marginal zone and through the subventricular zone (SVZ) and lower intermediate zone (IZ). In the IZ, neurons acquire a transient multipolar morphology where they extend and retract multiple dynamic projections and move in apparently random directions (47–49). As cells approach the middle of the IZ, the genesis of the axon starts, and when they reach the upper region of the IZ, they change their morphology from multipolar to bipolar and carry on radial migration (46, 50, 51).
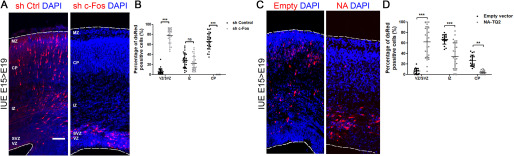
Using the electroporation model, it can be determined whether the manipulated neurons, which are destined to migrate to the upper layers of the brain cortex, suffer an impairment in this process or in the establishment of neuronal polarity. Cortical progenitors at embryonic day 15 (E15) were electroporated with a specific shRNA targeting c-Fos, and an analysis of the location and morphology of the progeny was performed at E19 after in vivo differentiation. Visualization of the electroporated cells was achieved by co-electroporating the shRNAs with a plasmid encoding the DsRed fluorescent protein.
We first analyzed differentiation of cells electroporated with a nonrelevant shRNA (shControl) plus DsRed at E19. About 10% of the neurons were located in the VZ/SVZ, 30% of cells were found migrating through the IZ, and the majority (60%) had reached the top of the cortical plate (Fig. 6A (left); quantification in Fig. 6B). Cells with knocked-down expression of c-Fos clearly showed an altered distribution and abnormal migration: around 80% of the cells remained arrested at the VZ/SVZ compared with the 10% in this localization in control experiments, and practically no electroporated cells were observed in the upper cortical plate (CP) (Fig. 6A (right); quantification in Fig. 6B).
Figure 6.
Expression of c-Fos is involved in cortical development. A, embryo brains were electroporated with a specifically designed shRNA to block c-Fos expression (sh c-Fos) (right) or a control scrambled shRNA (left) at E15 and analyzed at E19. IUE, in utero electroporation; MZ, marginal zone. Scale bar, 100 μm. B, quantification of the distribution of dsRed-positive cells in the CP, IZ, and VZ/SVZ. The mean ± S.D. of the quantification of at least 20 coronal cryosections from three independent experiments performed independently is shown, with two-way ANOVA with Bonferroni post-test. ***, p < 0.001; n.s., nonsignificant. C, embryo brains were electroporated with a vector designed to express the NA domain of c-Fos fused to CFP or the empty vector as a control at E15 and analyzed at E19. D, quantification of the distribution of dsRed-positive cells in the CP, IZ, and VZ/SVZ. The mean ± S.D. of the quantification of at least 20 coronal cryosections from three independent experiments performed independently is shown, with two-way ANOVA with Bonferroni post-test. ***, p < 0.001; **, p< 0.001.
Electroporation assays were then performed with the deletion mutants of c-Fos to evaluate whether its lipid synthesis activator function is responsible for the abnormal phenotype observed when c-Fos expression is blocked. Electroporated cells of control experiments using the empty vector show a localization similar to that shown in the controls of Fig. 6A (Fig. 6C (left); quantification in Fig. 6D). By contrast, cells transfected with c-Fos NA domain, which acts as a dominant negative of its lipid synthesis activator function, remained arrested at the VZ/SVZ and IZ (Fig. 6C (right); quantification in Fig. 6D). These results support the notion that the AP-1–independent function of c-Fos is in fact involved in cortical development.
Discussion
Although for a long time it was assumed that biosynthesis of most macromolecules in neurons was confined to the cell body (52, 53), an increasing body of molecular evidence has supported the presence of functional ER-resident components in axons (15–17, 54–62). Together with the understanding of the complex structural features of the axonal ER, different studies provided emerging evidence of active, axon-localized lipid synthesis (18, 19, 63–66). In fact, in the case of phosphatidylcholine synthesis, almost 50% has been found to be locally synthesized in distal axons, and it is required for axonal growth (20). Although this highlights the importance of the axonal ER in lipid homeostasis and in the maintenance of the complex structure of neurons, there have been no significant advances in recent years regarding the regulation of lipid metabolism in neurons.
Taking into consideration that the plasma membrane surface area of a developing neuron increases up to 20% per day (13) and that the compositional differences along the membrane due to the functional differences of axons and dendrites must necessarily be maintained, the molecular mechanisms regulating localized lipid synthesis must be carefully orchestrated. Because most of the local metabolic demands for axonal development, plasticity, or regeneration must be rapidly supplied, it seems reasonable to propose that, at least in part, the mechanisms involved in lipid synthesis regulation could be independent of the cells' nuclear activity. In the present work, we provide evidence that confirms the existence of the AP-1–independent function of c-Fos as a lipid synthesis activator in the nervous system, particularly in rat hippocampal neurons in culture and in the developing brain cortex during embryonic stages.
The fact that c-Fos is expressed during neuronal differentiation but is not in the adult brain in nonpathological conditions (67) implies that its expression is related to developmental events. c-Fos was described as a marker of neuronal activity, specially tied to learning and memory processes (68). In the experiments presented herein where c-Fos expression or its activity is blocked, neurons do not develop an axon and remain at early stages of differentiation without a distinguishable axon even after 48 h of culture. Until now, the effects of c-Fos on neuronal plasticity have always been linked to gene expression through its nuclear function, but given the evidence presented herein, it is possible that they are associated with processes that involve changes in lipid homeostasis. This can be particularly visualized in the experiments carried out with dominant negative of the lipid synthesis activator function of c-Fos, where the same results of impairment on differentiation in culture are observed although AP-1 activity should not be affected.
This hypothesis can be extended to cortical development: when performing in utero experiments, the cells with no c-Fos expression remain at the ventricular and subventricular zones instead of migrating to the superior layers of the cortex, an observation that implies a strong failure in normal cortical development. These results deepen our previous findings in the cerebral cortex of fos (−/−) mice, which show a strong reduction in the cortex thickness and a marked tendency of cells toward an undifferentiated phenotype (69). Because the in utero model does not have compensatory effects relying on other proteins that might fulfill the function of c-Fos, the phenotype observed in our experiments confirms the importance of this protein in cortex development. Even more, the same in utero experiments with dominant negatives of its AP-1–independent function show that this effect is directly related to its lipid synthesis activator role. However, the changes in the localization of the NA-transfected cells are not as extreme as those observed in the c-Fos-knocked-down cells. Two different interpretations arise from the intermediate phenotype observed in this case: both functions, AP-1 and lipid synthesis activation, are actively involved in cortical development, or the expression level of the NA domain is not enough to displace all of the endogenous c-Fos molecules. It remains to be determined whether the observed phenotype is a consequence of failures in polarization or migration of the cells.
c-Fos has been implicated in numerous physiological processes in the nervous system. It was the first transcription factor whose induction was proven to be dependent on the activity of the neuron (70, 71), a finding that rapidly transformed c-Fos in a marker of neuronal activity. In fact, it has been consistently demonstrated that its expression rises in the central nervous system after learning or memory trainings (68), although these changes are observed only during the first sessions of the protocols, indicating that this is probably an adaptive response. Given the results presented in this work, it seems reasonable to hypothesize that c-Fos lipid activator function could be involved in processes related to neuronal plasticity and learning processes by providing new membrane lipids for the establishment of new circuits that are no longer required once they are formed.
In light of all of the above-mentioned results, the role of AP-1–independent c-Fos is of vital importance for neuronal development. Its lipid synthesis activator capacity might contribute to the high membrane expansion rates necessary for the extension of the different neuronal processes that favor polarization and the correct establishment of synaptic connections for normal nervous system function.
Experimental procedures
Cell cultures, profection, and transfection
Dissociated hippocampal pyramidal neurons were prepared from fetal rat brain and cultured as described (72). Briefly, pregnant Rattus norvegicus Wistar rats at 18 days postfertilization were euthanized, and prenatal pups were excised from the uterus with sterile dissecting scissors. Pups were then decapitated with sterile scissors in a laminar flow hood, and the removed heads were placed in plates with sterile Hanks' balanced salt solution (Sigma–Aldrich) at 4 °C under a dissecting microscope. After hippocampus isolation and dissociation with 0.25% trypsin (Thermo Fisher Scientific), the tissue was dissociated with Pasteur pipettes. The cells obtained were plated onto acid-washed, polylysine-coated glass coverslips or polylysine-coated plates and maintained in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific) supplemented with 10% (v/v) horse serum (Thermo Fisher Scientific) for 2 h, after which the culture medium was replaced with serum-free Neurobasal medium supplemented with N2 and B27 supplements (Thermo Fisher Scientific). Cultures were maintained in a humidified 37 °C incubator with 5% CO2 for the indicated times.
Profections were performed using BioPORTER® Protein Delivery Reagent (Genlantis, San Diego, CA, USA) and transfections using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's protocol.
Production of shRNA containing lentiviral particles
Upon desired confluence, embryonic kidney epithelial HEK 293T cells (ATCC, Manassas, VA, USA) were co-transfected with MISSION® c-Fos custom shRNA plasmids (clones ID TRCN0000042680 and TRCN0000042678) cloned in pLKO.1-CMV-tGFP (Sigma–Aldrich) or with the control scrambled shRNA sequence and with compatible packaging plasmids using Lipofectamine 2000 (Thermo Fisher Scientific). Lentiviral titer was determined according to the manufacturer's protocol (MISSION®, Sigma–Aldrich).
Preparation of recombinant c-Fos and NA
His-tagged c-Fos and NA were expressed and recovered from pDS56-HisFos–transformed BL21 cells as described previously (30, 38, 73).
Co-immunoprecipitation assays, electrophoresis, and Western blotting (WB)
For co-immunoprecipitation assays, 500 μg of total protein from cells treated as indicated were immunoprecipitated for 4 h at 4 °C with Protein G–Sepharose (GE Healthcare) with the desired antibody and washed, and immunodetection was performed as described below. Cell lysates (50 μg) or immunoprecipitates were fractionated through SDS-containing polyacrylamide gels (12%) and electrotransferred to a nitrocellulose membrane at 300 mA for 1 h. Immunodetection was carried out by blocking of membranes with 10 mm PBS containing 5% (w/v) nonfat dried milk for 1 h at RT, followed by incubation with the desired antibody overnight at 4 °C in PBS-0.1% (v/v) Tween 20 (Sigma–Aldrich). Membranes were washed three times (10 min) with PBS-0.1% (v/v) Tween 20 and incubated for 1 h at RT with secondary antibodies. Membranes were washed, and detection was performed using an ODYSSEY IR imaging system (LI-COR, Lincoln, NE, USA).
In vitro phospholipid labeling
In vitro phospholipid labeling capacity of neurons was assayed as described previously (32, 74). Briefly, reactions were incubated for 60 min at 37 °C in a final volume of 50 µl containing 50 μg of cell homogenate protein as the enzyme source, 2.8 mm NaCl, 100 mm KCl, 10 mm MgCl2, 112 mm glucose, HEPES buffer, pH 7.5, 1.5 μCi of [32P]ATP (specific activity 3000 Ci/mmol; PerkinElmer Life Sciences) and the indicated amounts of c-Fos or NA. Conditions of linearity with time and protein concentration were determined for the enzyme with 1 ng of recombinant proteins per μg of protein cell homogenate, suspended in 3 µl of elution buffer or an equal volume of elution buffer for control reactions. For the competition conditions, the ratios between c-Fos and NA were calculated according to the number of molecules of each protein. Reactions were stopped by the addition of TCA and phosphotungstic acid (PTA; 5:0.5% (w/v), respectively). Incubates were centrifuged, and the pellet was washed three more times with TCA-PTA, 5:0.5% (w/v). After a final washing step with water, the pellet was suspended in 1.5 ml of chloroform/methanol (2:1). In this phase partitioning, lipids remain in the organic phase. Phospholipid labeling was quantified in the organic phase by scintillation counting (30, 74, 75). A more detailed version of the protocol has been published (76).
Enzyme activity determinations
Total CDS activity was assayed as described by Lykidis et al. (37). All reactions were performed in an 80-µl final volume containing 100 μg of cell homogenate protein as the enzyme source, 0.69 µm [3H]CTP (PerkinElmer Life Sciences), and 2 mm phosphatidic acid (Avanti Polar Lipids, Alabaster, AL, USA). Conditions of linearity with time and protein concentration were determined for the enzyme with 0.5 ng of recombinant c-Fos per μg of protein cell homogenate, suspended in 3 µl of elution buffer or an equal volume of elution buffer for control reactions (29, 38). The reaction was started by the addition of 10 mm MgCl2, and assays were incubated at 37 °C for 1 h. Reactions were stopped by the addition of 180 µl of chloroform/methanol/HCl (1:2:0.02%, v/v/v). After the addition of 60 µl of chloroform and 60 µl of KCl (2 m), phases were separated by centrifugation. The amount of [3H]CDP-diacylglycerol synthesized was determined by liquid scintillation counting in the organic phase.
Immunofluorescence (IF) and microscopy
Cells grown on round, acid-washed coverslips were rinsed twice with ice-cold 10 mm PBS and fixed in para-formaldehyde 4% (w/v) (Sigma–Aldrich), sucrose 4% (w/v) (Sigma–Aldrich) in 10 mm PBS at 37 °C for 10 min. Cells were then permeabilized with Triton X-100 (0.1% (v/v)) (Sigma–Aldrich) in 10 mm PBS for 10 min and blocked with horse serum (2%, v/v) and bovine serum albumin (3%, w/v) in 10 mm PBS for 2 h at RT in a humid chamber. Samples were incubated overnight at 4 °C in blocking buffer containing the desired antibody, washed twice with 10 mm PBS, and incubated with secondary antibodies for 2 h at RT, washed, and mounted with FluorSave (Millipore, Burlington, MA, USA). When indicated, 4′,6-diamino-2-phenylindole, dihydrochloride (DAPI; Thermo Fisher Scientific) was used to visualize nuclear structures.
Antibodies
The following primary antibodies were used: rabbit polyclonal anti-c-Fos antibody (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) diluted 1:200 for IF; rabbit polyclonal anti-c-Fos antibody (Sigma–Aldrich) diluted 1:2000 for WB; mouse monoclonal anti-α-tubulin antibody (Sigma–Aldrich) diluted 1:3000 for WB; mouse monoclonal anti-βIII-tubulin antibody (Sigma–Aldrich) diluted 1:1500 for IF; goat polyclonal anti-calnexin antibody (ER marker, Abcam, Cambridge, UK) diluted 1:500 for IF; rabbit polyclonal anti-CCTβ2 antibody (Sigma–Aldrich) diluted 1:500 for IF. The following secondary antibodies were used: IRDye 800CW goat anti-mouse antibody and IRDye 800 goat anti-rabbit antibody (LI-COR) diluted 1:25,000 for WB; Alexa 488, Alexa 546, and Alexa 633 (Thermo Fisher Scientific) diluted 1:1000 for IF.
FRET analysis
Cells grown on round, acid-washed coverslips in 24 multiwell plates were transfected with c-Fos-mTurquoise2-N1, CDS-pSYFP2, CCTβ2-pSYFP2, pSYFP2-mTurquoise2, or the empty vectors using Lipofectamine 2000 according to the manufacturer's protocol. After 24 h of transfection, cells were rinsed twice with ice-cold 10 mm PBS and fixed in para-formaldehyde 4% (w/v) (Sigma–Aldrich), sucrose (4%, w/v) (Sigma–Aldrich) in 10 mm PBS at 37 °C for 10 min. Cells were then washed three times with PBS and rinsed with Milli-Q water. Coverslips were mounted with FluorSave (Millipore, Burlington, MA, USA), and cells were visualized using an Olympus FV1000 laser-scanning confocal microscope with Olympus Fluoview Software (Olympus, Shinjuku, Tokyo, Japan). For FRET determinations, the sensitized emission measurement approach was used (36). The mTurquoise (donor) and SYFP (acceptor) chimeric proteins were excited with an argon laser at 458 and 515 nm, respectively. The emission channel was 470–500 nm for the donor and 530–560 nm for the acceptor. Background values were determined independently for each channel from a coverslip with nontransfected cells and then subtracted using ImageJ software. Donor spectral bleed-through and acceptor cross-excitation were calculated and corrected from single transfected cells. Mean FRET efficiency values (%E) within a cell were obtained on a pixel-by-pixel basis (31). The resulting image was then pseudocolored to better illustrate the distribution of the calculated efficiencies in the cell.
In utero electroporation
In utero electroporation was performed as described previously (77) with minor modifications. Briefly, pregnant C57BL/6J mice at E15 days were anesthetized with ketamine/xylazine (Laboratorios Richmond, Ciudad Autónoma de Buenos Aires, Argentina). Needles for injections were pulled from P-97 Flaming/Brownglass capillaries (World Precision Instruments, Sarasota, FF, USA). shRNA solutions were mixed with trypan blue 1% (v/v) at a DNA concentration of 0.5–1.5 µg/µl for each construct and injected. Five pulses of 40 V (50 ms ON, 950 OFF) were applied using 5-mm electrodes and a specially manufactured electroporator (LIADE National University of Córdoba, Córdoba, Argentina). The embryos were placed back into the abdominal cavity, which was then sutured. 3 days after intervention, the mother was sacrificed by cortical dislocation, the embryos were removed, and the brains of the electroporated ones were extracted and fixed with 4% (w/v) para-formaldehyde (Sigma–Aldrich) for 24 h. Then they were cryoprotected by immersion in sucrose (30%, w/v) (Sigma–Aldrich) and finally embedded in medium for frozen tissue specimens to ensure optimal cutting temperature (O.C.T.) and frozen in nitrogen. After 3 days at −80 °C, coronal cryosections of 20 µm were obtained and processed for immunofluorescence.
Microscope image acquisition
Imaging was performed on an Olympus FV1000 laser-scanning confocal microscope using Olympus Fluoview software (Olympus, Shinjuku, Tokyo, Japan) and a ×60 (1.4 NA) oil objective or a ×10 (0.40 NA) air objective. Images were analyzed with ImageJ software.
Experimental design and statistical analysis
All of the statistical analyses were performed using GraphPad Prism software. Statistical significance was defined by a p value of <0.05. The statistical test performed and the number of replicates for each experiment are indicated in the corresponding figure legends.
Animal care
All of the procedures were performed according to the Guide for the Care and Use of Laboratory Animals (8th Edition) and the approved protocols of the Institutional Board for Animal Welfare (CICUAL, Facultad de Ciencias Químicas, Universidad Nacional de Córdoba, Argentina).
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
We thank Dr. Hugo Maccioni (CIQUIBIC, CONICET, Facultad de Ciencias Químicas, Universidad Nacional de Córdoba, Argentina) for helpful discussions and critical reading of the manuscript. We greatly acknowledge the technical and imaging assistance of Dr. María Cecilia Sampedro and Dr. Carlos Rubén Más from the Centro de Micro y Nanoscopía de Córdoba, CEMINCO (CONICET), Universidad Nacional de Córdoba, Córdoba, Argentina and the technical assistance with animal care and maintenance of Elvira Rosa Andrada (CIQUIBIC, CONICET).
This article contains supporting information.
Author contributions—L. R.-B., G. O. F., F. B. P., S. Q., and B. L. C. conceptualization; L. R.-B., G. O. F., F. B. P., A. M. C. G., and C. G. P. data curation; L. R.-B. software; L. R.-B., G. O. F., F. B. P., A. M. C. G., and C. G. P. formal analysis; L.R .-B. validation; L. R.-B., G. O. F., F. B. P., A. M. C. G., and C. G. P. investigation; L. R.-B., G. O. F., F. B. P., A. M. C. G., and C. G. P. visualization; L. R.-B., G. O. F., F. B. P., A. M. C. G., and C. G. P. methodology; L. R.-B. and B. L. C. writing-original draft; L. R.-B., G. O. F., F. B. P., A. M. C. G., C. G. P., S. Q., and B. L. C. writing-review and editing; S. Q. and B. L. C. resources; S. Q. and B. L. C. funding acquisition; B. L. C. supervision; B. L. C. project administration.
Funding and additional information—This work was supported by the Secretaría de Ciencia y Técnica, Universidad Nacional de Córdoba; the Fondo para la Investigación Científica y Tecnológica, Ministerio de Ciencia, Tecnología e Innovación, Argentina (Grants PICT 2017-2761, PICT 2012-2797, PICT 2013-2996, and PICT 2015-3702); and the Instituto Nacional del Cáncer, Ministerio de Salud, Argentina. Gabriel Orlando Ferrero, César Germán Prucca, Santiago Quiroga, and Beatriz Leonor Caputto are members of and Lucía Rodríguez Berdini, Florentyna Bustos Plonka, and Andrés Mauricio Cardozo Gizzi are fellows of CONICET (Consejo Nacional de Investigaciones Científicas y Tecnológicas), Ministerio de Ciencia, Tecnología e Innovación, Argentina.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ER
- endoplasmic reticulum
- CCT
- CTP:phosphocholine cytidylyltransferase
- CDP-DAG
- CDP-diacylglycerol
- CDS
- CDP-diacylglycerol synthase
- VZ
- ventricular zone
- SVZ
- subventricular zone
- IZ
- intermediate zone
- E
- embryonic day
- WB
- Western blotting
- RT
- room temperature
- PTA
- phosphotungstic acid
- IF
- immunofluorescence
- DAPI
- 4′,6-diamino-2-phenylindole, dihydrochloride
- NA
- numerical aperture
- ANOVA
- analysis of variance
- CP
- cortical plate.
References
- 1. Hatten M. E. (1999) Expansion of CNS precursor pools: a new role for Sonic Hedgehog. Neuron 22, 2–3 10.1016/S0896-6273(00)80668-6 [DOI] [PubMed] [Google Scholar]
- 2. Hatten M. E. (1999) Central nervous system neuronal migration. Annu. Rev. Neurosci. 22, 511–539 10.1146/annurev.neuro.22.1.511 [DOI] [PubMed] [Google Scholar]
- 3. Millet L. J., and Gillette M. U. (2012) Over a century of neuron culture: from the hanging drop to microfluidic devices. Yale J. Biol. Med. 85, 501–521 [PMC free article] [PubMed] [Google Scholar]
- 4. Spillane M., and Gallo G. (2014) Involvement of Rho-family GTPases in axon branching. Small GTPases 5, e27974 10.4161/sgtp.27974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Winkle C. C., and Gupton S. L. (2016) Membrane trafficking in neuronal development: ins and outs of neural connectivity. Int. Rev. Cell Mol. Biol. 322, 247–280 10.1016/bs.ircmb.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cáceres A., Ye B., and Dotti C. G. (2012) Neuronal polarity: demarcation, growth and commitment. Curr. Opin. Cell Biol. 24, 547–553 10.1016/j.ceb.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dotti C. G., Sullivan C. A., and Banker G. A. (1988) The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8, 1454–1468 10.1523/JNEUROSCI.08-04-01454.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schelski M., and Bradke F. (2017) Neuronal polarization: from spatiotemporal signaling to cytoskeletal dynamics. Mol. Cell. Neurosci. 84, 11–28 10.1016/j.mcn.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 9. Armijo-Weingart L., and Gallo G. (2017) It takes a village to raise a branch: cellular mechanisms of the initiation of axon collateral branches. Mol. Cell. Neurosci. 84, 36–47 10.1016/j.mcn.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bullmore E., and Sporns O. (2012) The economy of brain network organization. Nat. Rev. Neurosci. 13, 336–349 10.1038/nrn3214 [DOI] [PubMed] [Google Scholar]
- 11. Tracey T. J., Steyn F. J., Wolvetang E. J., and Ngo S. T. (2018) Neuronal lipid metabolism: multiple pathways driving functional outcomes in health and disease. Front. Mol. Neurosci. 11, 10 10.3389/fnmol.2018.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quiroga S., Bisbal M., and Cáceres A. (2018) Regulation of plasma membrane expansion during axon formation. Dev. Neurobiol. 78, 170–180 10.1002/dneu.22553 [DOI] [PubMed] [Google Scholar]
- 13. Pfenninger K. H. (2009) Plasma membrane expansion: a neuron's Herculean task. Nat. Rev. Neurosci. 10, 251–261 10.1038/nrn2593 [DOI] [PubMed] [Google Scholar]
- 14. Vance J. E., Campenot R. B., and Vance D. E. (2000) The synthesis and transport of lipids for axonal growth and nerve regeneration. Biochim. Biophys. Acta 1486, 84–96 10.1016/S1388-1981(00)00050-0 [DOI] [PubMed] [Google Scholar]
- 15. Tsukita S., and Ishikawa H. (1976) Three-dimensional distribution of smooth endoplasmic reticulum in myelinated axons. J. Electron Microsc. (Tokyo) 25, 141–149 10.1093/oxfordjournals.jmicro.a050013 [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez C., and Couve A. (2014) The axonal endoplasmic reticulum and protein trafficking: cellular bootlegging south of the soma. Semin. Cell Dev. Biol. 27, 23–31 10.1016/j.semcdb.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 17. Luarte A., Cornejo V. H., Bertin F., Gallardo J., and Couve A. (2018) The axonal endoplasmic reticulum: one organelle-many functions in development, maintenance, and plasticity. Dev. Neurobiol. 78, 181–208 10.1002/dneu.22560 [DOI] [PubMed] [Google Scholar]
- 18. Vance J. E., Pan D., Vance D. E., and Campenot R. B. (1991) Biosynthesis of membrane lipids in rat axons. J. Cell Biol. 115, 1061–1068 10.1083/jcb.115.4.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vance J. E., Pan D., Campenot R. B., Bussière M., and Vance D. E. (1994) Evidence that the major membrane lipids, except cholesterol, are made in axons of cultured rat sympathetic neurons. J. Neurochem. 62, 329–337 10.1046/j.1471-4159.1994.62010329.x [DOI] [PubMed] [Google Scholar]
- 20. Posse de Chaves E., Vance D. E., Campenot R. B., and Vance J. E. (1995) Axonal synthesis of phosphatidylcholine is required for normal axonal growth in rat sympathetic neurons. J. Cell Biol. 128, 913–918 10.1083/jcb.128.5.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Curran T., and Morgan J. I. (1987) Memories of Fos. Bioessays 7, 255–258 10.1002/bies.950070606 [DOI] [PubMed] [Google Scholar]
- 22. Hughes P., and Dragunow M. (1995) Induction of immediate-early genes and the control of neurotransmitter-regulated gene expression within the nervous system. Pharmacol. Rev. 47, 133–178 [PubMed] [Google Scholar]
- 23. Caputto B. L., and Guido M. E. (2000) Immediate early gene expression within the visual system: light and circadian regulation in the retina and the suprachiasmatic nucleus. Neurochem. Res. 25, 153–162 10.1023/A:1007508020173 [DOI] [PubMed] [Google Scholar]
- 24. Angel P., and Karin M. (1991) The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072, 129–157 10.1016/0304-419X(91)90011-9 [DOI] [PubMed] [Google Scholar]
- 25. Guido M. E., de Arriba Zerpa G. A., Bussolino D. F., and Caputto B. L. (1996) Immediate early gene c-fos regulates the synthesis of phospholipids but not of gangliosides. J. Neurosci. Res. 43, 93–98 10.1002/jnr.490430112 [DOI] [PubMed] [Google Scholar]
- 26. Bussolino D. F., de Arriba Zerpa G. A., Grabois V. R., Conde C. B., Guido M. E., and Caputto B. L. (1998) Light affects c-fos expression and phospholipid synthesis in both retinal ganglion cells and photoreceptor cells in an opposite way for each cell type. Brain Res. Mol. Brain Res. 58, 10–15 10.1016/S0169-328X(98)00065-5 [DOI] [PubMed] [Google Scholar]
- 27. de Arriba Zerpa G. A., Guido M. E., Bussolino D. F., Pasquare S. J., Castagnet P. I., Giusto N. M., and Caputto B. L. (1999) Light exposure activates retina ganglion cell lysophosphatidic acid acyl transferase and phosphatidic acid phosphatase by a c-Fos-dependent mechanism. J. Neurochem. 73, 1228–1235 10.1046/j.1471-4159.1999.0731228.x [DOI] [PubMed] [Google Scholar]
- 28. Bussolino D. F., Guido M. E., Gil G. A., Borioli G. A., Renner M. L., Grabois V. R., Conde C. B., and Caputto B. L. (2001) c-Fos associates with the endoplasmic reticulum and activates phospholipid metabolism. FASEB J. 15, 556–558 10.1096/fj.00-0446fje [DOI] [PubMed] [Google Scholar]
- 29. Alfonso Pecchio A. R., Cardozo Gizzi A. M., Renner M. L., Molina-Calavita M., and Caputto B. L. (2011) c-Fos activates and physically interacts with specific enzymes of the pathway of synthesis of polyphosphoinositides. Mol. Biol. Cell 22, 4716–4725 10.1091/mbc.e11-03-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gil G. A., Silvestre D. C., Tomasini N., Bussolino D. F., and Caputto B. L. (2012) Controlling cytoplasmic c-Fos controls tumor growth in the peripheral and central nervous system. Neurochem. Res. 37, 1364–1371 10.1007/s11064-012-0763-8 [DOI] [PubMed] [Google Scholar]
- 31. Cardozo Gizzi A. M., Prucca C. G., Gaveglio V. L., Renner M. L., Pasquaré S. J., and Caputto B. L. (2015) The catalytic efficiency of lipin 1β increases by physically interacting with the proto-oncoprotein c-Fos. J. Biol. Chem. 290, 29578–29592 10.1074/jbc.M115.678821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gil G. A., Bussolino D. F., Portal M. M., Alfonso Pecchio A., Renner M. L., Borioli G. A., Guido M. E., and Caputto B. L. (2004) c-Fos activated phospholipid synthesis is required for neurite elongation in differentiating PC12 cells. Mol. Biol. Cell 15, 1881–1894 10.1091/mbc.e03-09-0705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dragunow M., Peterson M. R., and Robertson H. A. (1987) Presence of c-fos-like immunoreactivity in the adult rat brain. Eur. J. Pharmacol. 135, 113–114 10.1016/0014-2999(87)90767-9 [DOI] [PubMed] [Google Scholar]
- 34. Dragunow M., and Robertson H. A. (1987) Generalized seizures induce c-fos protein(s) in mammalian neurons. Neurosci. Lett. 82, 157–161 10.1016/0304-3940(87)90121-2 [DOI] [PubMed] [Google Scholar]
- 35. Silvestre D. C., Maccioni H. J., and Caputto B. L. (2009) Content of endoplasmic reticulum and Golgi complex membranes positively correlates with the proliferative status of brain cells. J. Neurosci. Res. 87, 857–865 10.1002/jnr.21915 [DOI] [PubMed] [Google Scholar]
- 36. Gordon G. W., Berry G., Liang X. H., Levine B., and Herman B. (1998) Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74, 2702–2713 10.1016/S0006-3495(98)77976-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lykidis A., Jackson P. D., Rock C. O., and Jackowski S. (1997) The role of CDP-diacylglycerol synthetase and phosphatidylinositol synthase activity levels in the regulation of cellular phosphatidylinositol content. J. Biol. Chem. 272, 33402–33409 10.1074/jbc.272.52.33402 [DOI] [PubMed] [Google Scholar]
- 38. Racca A. C., Prucca C. G., and Caputto B. L. (2019) Fra-1 and c-Fos N-terminal deletion mutants impair breast tumor cell proliferation by blocking lipid synthesis activation. Front. Oncol. 9, 544 10.3389/fonc.2019.00544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galliard T., Michell R. H., and Hawthorne J. N. (1965) Incorporation of phosphate into diphosphoinositide by subcellular fractions from liver. Biochim. Biophys. Acta 106, 551–563 10.1016/0005-2760(65)90071-8 [DOI] [PubMed] [Google Scholar]
- 40. Hayashi K., Yagihara Y., Nakamura I., Katagiri A., Arakawa Y., and Yamazoe S. (1967) Incorporation of 32P from [γ-32P]ATP into polyphosphoinositides and phosphatidic acid in subcellular particles of guinea pig brain. J. Biochem. 62, 15–20 10.1093/oxfordjournals.jbchem.a128630 [DOI] [PubMed] [Google Scholar]
- 41. Schibeci A., and Schacht J. (1977) Action of neomycin on the metabolism of polyphosphoinositides in the guinea pig kidney. Biochem. Pharmacol. 26, 1769–1774 10.1016/0006-2952(77)90344-6 [DOI] [PubMed] [Google Scholar]
- 42. Sun G. Y., and Lin T. N. (1989) Time course for labeling of brain membrane phosphoinositides and other phospholipids after intracerebral injection of [32P]ATP. Evaluation by an improved HPTLC procedure. Life Sci. 44, 689–696 10.1016/0024-3205(89)90475-X [DOI] [PubMed] [Google Scholar]
- 43. Wissing J. B., and Behrbohm H. (1993) Diacylglycerol pyrophosphate, a novel phospholipid compound. FEBS Lett. 315, 95–99 10.1016/0014-5793(93)81141-L [DOI] [PubMed] [Google Scholar]
- 44. Tong W., and Sun G. Y. (1994) Phosphorylation of lipids in rat primary glial cells and immortalized astrocytes (DITNC). Lipids 29, 385–390 10.1007/BF02537306 [DOI] [PubMed] [Google Scholar]
- 45. Ferrero G. O., Renner M. L., Gil G. A., Rodríguez-Berdini L., and Caputto B. L. (2014) c-Fos-activated synthesis of nuclear phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P(2)] promotes global transcriptional changes. Biochem. J. 461, 521–530 10.1042/BJ20131376 [DOI] [PubMed] [Google Scholar]
- 46. Kwan A. C., and Dan Y. (2012) Dissection of cortical microcircuits by single-neuron stimulation in vivo. Curr. Biol. 22, 1459–1467 10.1016/j.cub.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tabata H., and Nakajima K. (2003) Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J. Neurosci. 23, 9996–10001 10.1523/JNEUROSCI.23-31-09996.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bielas S., Higginbotham H., Koizumi H., Tanaka T., and Gleeson J. G. (2004) Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu. Rev. Cell Dev. Biol. 20, 593–618 10.1146/annurev.cellbio.20.082503.103047 [DOI] [PubMed] [Google Scholar]
- 49. Noctor S. C., Martínez-Cerdeno V., Ivic L., and Kriegstein A. R. (2004) Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- 50. Nadarajah B., and Parnavelas J. G. (2002) Modes of neuronal migration in the developing cerebral cortex. Nat. Rev. Neurosci. 3, 423–432 10.1038/nrn845 [DOI] [PubMed] [Google Scholar]
- 51. Stipursky J., Francis D., and Gomes F. C. (2012) Activation of MAPK/PI3K/SMAD pathways by TGF-β1 controls differentiation of radial glia into astrocytes in vitro. Dev. Neurosci. 34, 68–81 10.1159/000338108 [DOI] [PubMed] [Google Scholar]
- 52. Weiss P., and Hiscoe H. B. (1948) Experiments on the mechanism of nerve growth. J. Exp. Zool. 107, 315–395 10.1002/jez.1401070302 [DOI] [PubMed] [Google Scholar]
- 53. Ledeen R. W. (1985) Transport, Exchange, and Transfer of Phospholipids in the Nervous System, pp. 135–172, John Wiley & Sons, Inc., New York [Google Scholar]
- 54. Palay S. L. (1958) The morphology of synapses in the central nervous system. Exp. Cell Res. 14, 275–293 [PubMed] [Google Scholar]
- 55. Droz B., Rambourg A., and Koenig H. L. (1975) The smooth endoplasmic reticulum: structure and role in the renewal of axonal membrane and synaptic vesicles by fast axonal transport. Brain Res. 93, 1–13 10.1016/0006-8993(75)90282-6 [DOI] [PubMed] [Google Scholar]
- 56. Rambourg A., and Droz B. (1980) Smooth endoplasmic reticulum and axonal transport. J. Neurochem. 35, 16–25 10.1111/j.1471-4159.1980.tb12484.x [DOI] [PubMed] [Google Scholar]
- 57. Broadwell R. D., and Cataldo A. M. (1983) The neuronal endoplasmic reticulum: its cytochemistry and contribution to the endomembrane system. I. Cell bodies and dendrites. J. Histochem. Cytochem. 31, 1077–1088 10.1177/31.9.6309951 [DOI] [PubMed] [Google Scholar]
- 58. Lindsey J. D., and Ellisman M. H. (1985) The neuronal endomembrane system. III. The origins of the axoplasmic reticulum and discrete axonal cisternae at the axon hillock. J. Neurosci. 5, 3135–3144 10.1523/JNEUROSCI.05-12-03135.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Willis D., Li K. W., Zheng J. Q., Chang J. H., Smit A. B., Kelly T., Merianda T. T., Sylvester J., van Minnen J., and Twiss J. L. (2005) Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J. Neurosci. 25, 778–791 10.1523/JNEUROSCI.4235-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Valenzuela J. I., Jaureguiberry-Bravo M., and Couve A. (2011) Neuronal protein trafficking: emerging consequences of endoplasmic reticulum dynamics. Mol. Cell. Neurosci. 48, 269–277 10.1016/j.mcn.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 61. Merianda T., and Twiss J. (2013) Peripheral nerve axons contain machinery for co-translational secretion of axonally-generated proteins. Neurosci. Bull. 29, 493–500 10.1007/s12264-013-1360-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rao K., Stone M. C., Weiner A. T., Gheres K. W., Zhou C., Deitcher D. L., Levitan E. S., and Rolls M. M. (2016) Spastin, atlastin, and ER relocalization are involved in axon but not dendrite regeneration. Mol. Biol. Cell 27, 3245–3256 10.1091/mbc.E16-05-0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kumara-Siri M. H., and Gould R. M. (1980) Enzymes of phospholipid synthesis: axonal versus Schwann cell distribution. Brain Res. 186, 315–330 10.1016/0006-8993(80)90978-6 [DOI] [PubMed] [Google Scholar]
- 64. Gould R. M., Pant H., Gainer H., and Tytell M. (1983) Phospholipid synthesis in the squid giant axon: incorporation of lipid precursors. J. Neurochem. 40, 1293–1299 10.1111/j.1471-4159.1983.tb13569.x [DOI] [PubMed] [Google Scholar]
- 65. Gould R. M., Spivack W. D., Robertson D., and Poznansky M. J. (1983) Phospholipid synthesis in the squid giant axon: enzymes of phosphatidylinositol metabolism. J. Neurochem. 40, 1300–1306 10.1111/j.1471-4159.1983.tb13570.x [DOI] [PubMed] [Google Scholar]
- 66. Tanaka T., Yamaguchi H., Kishimoto Y., and Gould R. M. (1987) Lipid metabolism in various regions of squid giant nerve fiber. Biochim. Biophys. Acta 922, 85–94 10.1016/0005-2760(87)90248-7 [DOI] [PubMed] [Google Scholar]
- 67. Caubet J. F. (1989) c-fos proto-oncogene expression in the nervous system during mouse development. Mol. Cellular Biol. 9, 2269–2272 10.1128/MCB.9.5.2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gallo F. T., Katche C., Morici J. F., Medina J. H., and Weisstaub N. V. (2018) Immediate Early genes, memory and psychiatric disorders: focus on c-Fos, Egr1 and Arc. Front. Behav. Neurosci. 12, 79 10.3389/fnbeh.2018.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Velazquez F. N., Prucca C. G., Etienne O., D'Astolfo D. S., Silvestre D. C., Boussin F. D., and Caputto B. L. (2015) Brain development is impaired in c-fos -/- mice. Oncotarget 6, 16883–16901 10.18632/oncotarget.4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Morgan J. I., and Curran T. (1988) Calcium as a modulator of the immediate-early gene cascade in neurons. Cell Calcium 9, 303–311 10.1016/0143-4160(88)90011-5 [DOI] [PubMed] [Google Scholar]
- 71. Sagar S. M., Sharp F. R., and Curran T. (1988) Expression of c-fos protein in brain: metabolic mapping at the cellular level. Science 240, 1328–1331 10.1126/science.3131879 [DOI] [PubMed] [Google Scholar]
- 72. Banker G. A., and Cowan W. M. (1977) Rat hippocampal neurons in dispersed cell culture. Brain Res. 126, 397–425 10.1016/0006-8993(77)90594-7 [DOI] [PubMed] [Google Scholar]
- 73. Borioli G. A., Caputto B. L., and Maggio B. (2001) c-Fos is surface active and interacts differentially with phospholipid monolayers. Biochem. Biophys. Res. Commun. 280, 9–13 10.1006/bbrc.2000.4081 [DOI] [PubMed] [Google Scholar]
- 74. Ferrero G. O., Velazquez F. N., and Caputto B. L. (2012) The kinase c-Src and the phosphatase TC45 coordinately regulate c-Fos tyrosine phosphorylation and c-Fos phospholipid synthesis activation capacity. Oncogene 31, 3381–3391 10.1038/onc.2011.510 [DOI] [PubMed] [Google Scholar]
- 75. Portal M. M., Ferrero G. O., and Caputto B. L. (2007) N-terminal c-Fos tyrosine phosphorylation regulates c-Fos/ER association and c-Fos-dependent phospholipid synthesis activation. Oncogene 26, 3551–3558 10.1038/sj.onc.1210137 [DOI] [PubMed] [Google Scholar]
- 76. Rodriguez-Berdini L., and Ferrero G. O. (2016) A technique for the measurement of in vitro phospholipid synthesis via radioactive labeling. Bio-protocol 6 10.21769/BioProtoc.1705 [DOI] [Google Scholar]
- 77. Saito T., and Nakatsuji N. (2001) Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240, 237–246 10.1006/dbio.2001.0439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.