Abstract
Cleavage of aromatic carbon–chlorine bonds is critical for the degradation of toxic industrial compounds. Here, we solved the X-ray crystal structure of chlorothalonil dehalogenase (Chd) from Pseudomonas sp. CTN-3, with 15 of its N-terminal residues truncated (ChdT), using single-wavelength anomalous dispersion refined to 1.96 Å resolution. Chd has low sequence identity (<15%) compared with all other proteins whose structures are currently available, and to the best of our knowledge, we present the first structure of a Zn(II)-dependent aromatic dehalogenase that does not require a coenzyme. ChdT forms a “head-to-tail” homodimer, formed between two α-helices from each monomer, with three Zn(II)-binding sites, two of which occupy the active sites, whereas the third anchors a structural site at the homodimer interface. The catalytic Zn(II) ions are solvent-accessible via a large hydrophobic (8.5 × 17.8 Å) opening to bulk solvent and two hydrophilic branched channels. Each active-site Zn(II) ion resides in a distorted trigonal bipyramid geometry with His117, His257, Asp116, Asn216, and a water/hydroxide as ligands. A conserved His residue, His114, is hydrogen-bonded to the Zn(II)-bound water/hydroxide and likely functions as the general acid-base. We examined substrate binding by docking chlorothalonil (2,4,5,6-tetrachloroisophtalonitrile, TPN) into the hydrophobic channel and observed that the most energetically favorable pose includes a TPN orientation that coordinates to the active-site Zn(II) ions via a CN and that maximizes a π–π interaction with Trp227. On the basis of these results, along with previously reported kinetics data, we propose a refined catalytic mechanism for Chd-mediated TPN dehalogenation.
Keywords: crystal structure, zinc, enzyme catalysis, enzyme mechanism, enzyme kinetics, bioremediation, Chd, chlorothalonil, dechlorination, dehalogenase
Introduction
Chlorinated aromatic hydrocarbons such as polychlorinated biphenyl, chlorobenzenes, and atrazine are important industrial starting materials for the manufacture of dyes, drugs, and pesticides/fungicides (1, 2). Aromatic carbon–chlorine bonds are typically very stable, and hence, these compounds persist in soil and contaminate groundwater. Their low water solubility, coupled with their toxicity, makes them particularly important targets for environmental remediation (3). Enzymatic dehalogenation of chlorinated natural organic matter by organohalide-respiring bacteria provides more soluble compounds that are less likely to bioaccumulate and more likely to be susceptible to further degradation or cellular recycling and, thus, less toxic (4–6). Therefore, cleavage of aromatic carbon–chlorine bonds is critical for the degradation of toxic industrial compounds. Although enzymatic dehalogenation offers a possible bioremediation solution for chlorinated aromatic compounds, a major impediment to understanding their biological role and bioremediation uses is the lack of a detailed understanding of their catalytic mechanisms.
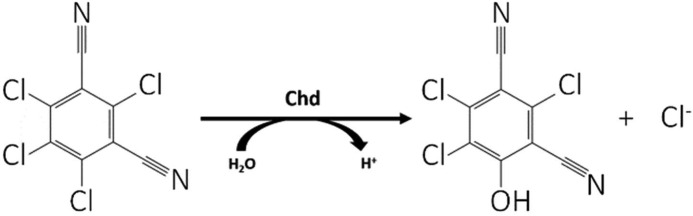
The prevailing dogma is that biological dechlorination reactions are catalyzed by either oxidative, reductive, or thiolytic dehalogenation processes (7–13). A relatively unknown biological dehalogenation process involves hydrolysis of a C-Cl bond (14). The chlorothalonil dehalogenase from Pseudomonas sp. CTN-3 (Chd) is a Zn(II)-dependent hydrolytic dehalogenase that selectively substitutes an aromatic chlorine-carbon moiety in chlorothalonil (2,4,5,6-tetrachloroisophtalonitrile, TPN) to an aromatic alcohol (4-hydroxytrichloro-isophthalonitrile, 4-OH-TPN) (Fig. 1) (15). TPN is one of the most commonly used fungicides in the United States with more than 5,000,000 kg sprayed on crops and fruits each year (16–19). TPN has low solubility in water (100 mg liter−1) but is strongly absorbed in soil, particularly soil with high organic matter such as those found in aquatic environments. TPN is highly toxic to fish and aquatic species, as well as birds and invertebrates (1, 15, 20). TPN is also a human skin and eye irritant that can cause severe gastrointestinal issues. Animal studies involving mice have shown that TPN can cause kidney cancer, so it has been classified by the U.S. Environmental Protection Agency as a probable human carcinogen (18). Given the widespread use of TPN and its toxicity, its biodegradation and environmental clean-up has become a topic of significant importance (22).
Figure 1.
Hydrolysis of TPN to 4-OH-TPN and chloride by Chd.
Several bacterial strains harbor Chd genes that exhibit remarkable sequence identity (>95%) (23–26). Chd was suggested to contain a conserved Zn(II)-binding domain similar to enzymes in the metallo-β-lactamase (MBL) superfamily and was proposed to be dimeric in solution (15). Although Zn(II) is required for Chd activity (26), its catalytic role remains unknown. A major limitation in understanding the catalytic mechanism of Chd and in developing small molecule catalysts that specifically target TPN, is a lack of knowledge regarding its structure and active site architecture. The only aromatic hydrolytic dehalogenase that has been structurally characterized to date is atrazine chlorohydrolase from Pseudomonas sp. (AtzA; PDB code 4V1X at 2.2 Å), which contains an iron ion coordinated by four histidine residues, an aspartate, and a water molecule (27). Although both AtzA and Chd hydrolytically dehalogenate aromatic compounds, they have relatively low sequence identity (>19%) and different metal ion requirements. Moreover, AtzA is hexameric, whereas Chd is dimeric (15, 27).
Herein, we report the 1.96 Å X-ray crystal structure of a 15 residue N-terminally truncated Chd enzyme that is fully active, using single-wavelength anomalous dispersion (SAD). A truncated form of Chd (ChdT), which lacked the first 15 N-terminal residues, was designed and utilized because of poor solubility and stability of the WT Chd enzyme at the concentrations required for crystallization. The X-ray structure reveals that Chd is a homodimer with a structural Zn(II) residing at the dimer interface and a mononuclear, trigonal bipyramidal (TBP) Zn(II) active site with His117, His257, Asp116, Asn216, and water/hydroxide as ligands. Based on this structure, substrate modeling, and previous kinetic studies (26), a refined catalytic mechanism is proposed.
Results
Overall structure of ChdT
ChdT crystals with dimensions 0.1 × 0.2 × 0.3 mm belonged to the primitive orthorhombic space group P212121 with unit cell parameters a = 90 Å, b = 80 Å, and c = 90 Å. The structure was solved via SAD using a wavelength of 1.28 Å, close to the absorption edge of zinc. The final model was refined to a resolution of 1.96 Å with Rfree and Rwork values of 19.80% and 16.22%, respectively. Final data processing and refinement statistics are listed in Table 1. A single homodimer was built in an asymmetric unit. The final refined model has been deposited to the PDB under code 6UXU.
Table 1.
Crystallographic parameters and statistics for ChdT (6UXU)
| Data processing | |
| Space group | P212121 |
| Cell dimension | |
| α, β, γ (°) | 90, 90, 90 |
| a, b, c (Å) | 54.8, 105.2, 122.4 |
| Processed resolution (Å) | 1.96 |
| I/σ(I) | 17.5 (1.1) |
| Resolution at I/σ(I) = 2a | 1.98 |
| Rmerge (%)b | 8.7 (117.6)c |
| Rpim (%)d | 2.7 (35.0) |
| CC½ (%)e | (63.1) |
| Completeness (%) | 98.0 (87.6) |
| Multiplicity | 7.2 |
| No. reflections | 362,906 |
| No. unique reflections | 50,386 |
| Refinement | |
| Rworkf/Rfreeg (%) | 15.79/19.75 |
| No. of atoms | |
| Protein | 4,693 |
| Ligand | 3 |
| Water | 379 |
| B factors (Å2) | |
| Protein | 23.7 |
| RMSDh | |
| Bond lengths (Å) | 0.010 |
| Bond angles (deg) | 1.014 |
| Ramachandran plot (%) | |
| Most favored | 96.14 |
| Allowed | 3.19 |
| Outliers | 0.67 |
a Provided resolution at I/σ = 2 for conventional assessment of data quality.
a Rmerge = Σ|Iobs − Iavg|/ΣIavg.
c The values for the highest-resolution bin are in parentheses.
d Precision-indicating merging R.
e Pearson correlation coefficient of two “half” data sets.
f Rwork = Σ|Fobs − Fcalc|/ΣFobs.
g 5% of the reflection data were selected at random as a test set, and only these data were used to calculate Rfree.
h RMSD, root-mean-square deviation.
The two monomers of the homodimer display a 2-fold noncrystallographic rotational axis that passes through a noncatalytic structural Zn(II) site (Fig. 2A). From a side view (Fig. 2B), the structural Zn(II) ion is close to one side of the dimer surface. For convenience purposes, we refer to this side of the dimer protein as the “top” side, i.e. where the structural Zn(II) ion resides, and the opposite side to as the “bottom.” The crystallographic packing was analyzed using PISA (28, 29) in the CCP4 suite, and the observed homodimer in the asymmetric unit was predicted by PISA to be the only stable quaternary structure in solution. The total predicted accessible surface area is 23,000 Å2, whereas the predicted buried surface area is 1,800 Å2.
Figure 2.
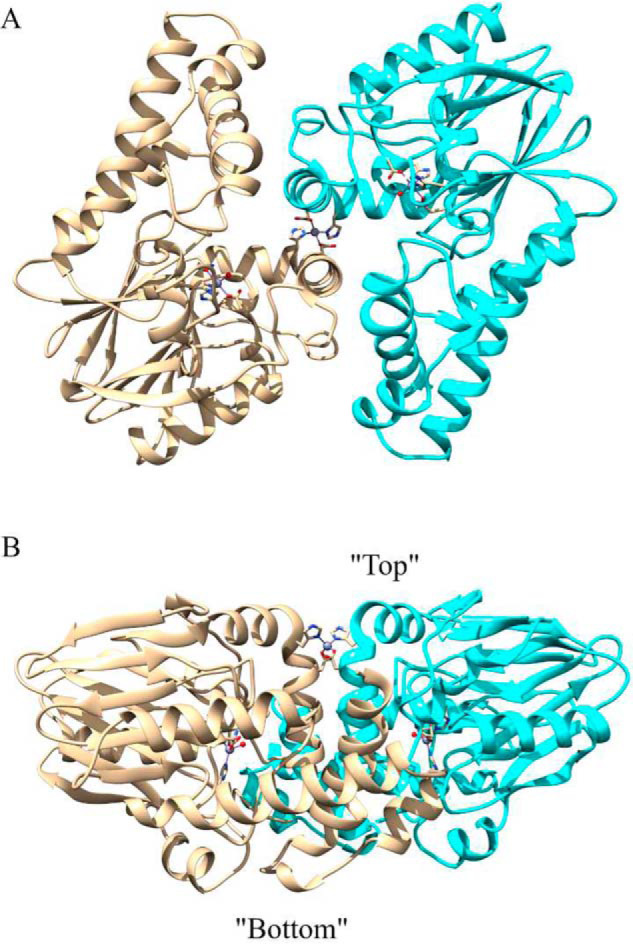
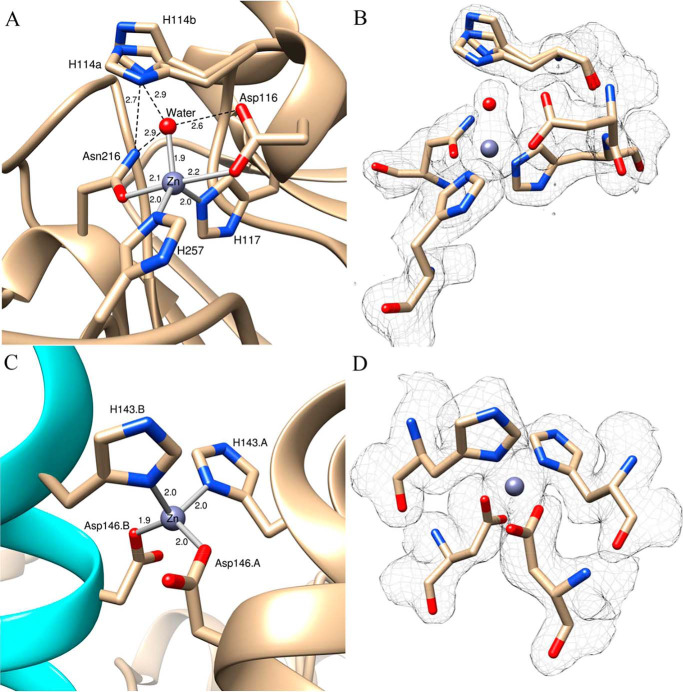
A, homodimer of ChdT. Three Zn(II) ions are shown as balls, and the coordination residues are shown as sticks. B, a side view of the ChdT homodimer, with defined top and bottom according to the relative positions to the structural Zn(II) ion.
Each monomer contains a catalytic Zn(II) ion that is solvent-accessible from both the top and the bottom of the protein (Fig. 3). From the bottom, a Y-shaped channel exists with two branched channels that lead to the catalytic Zn(II) ions and share a single large (8.5 × 17.8 Å) opening to bulk solvent. On the other hand, the structural Zn(II) ion is not accessible to the bulk solvent despite being located close to the top surface. From the top, the structural features surrounding the structural Zn(II) ion help to form two bent channels that provide access the catalytic Zn(II) ions.
Figure 3.
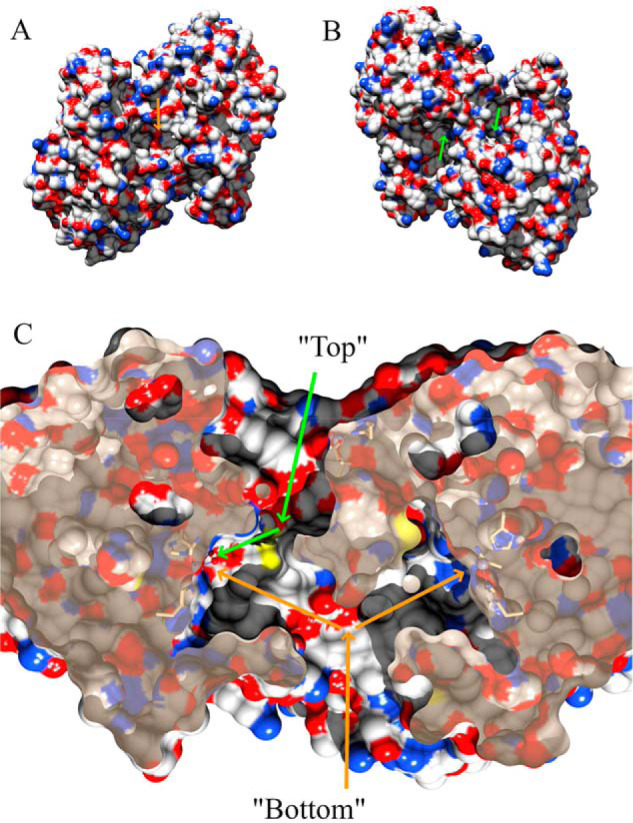
A, hydrophilicity surface map of the ChdT dimer from the bottom. The orange arrow indicates a single large opening. B, surface view of the ChdT dimer from the top. The green arrows indicate two separate openings. C, cross-sectional view of the ChdT dimer. The orange arrows indicate a Y-shaped channel with access to the catalytic Zn(II) ions from the bottom, whereas the green arrows indicate one branch of the Y-shaped channel that provides access to the catalytic Zn(II) from the top. The other Y-shaped channel from the top is on the backside of this view and is not visible.
The monomer protein fold is an αββα-sandwich
The overall protein fold of the ChdT monomer (Fig. 4A) was searched using the DALI server (30) for comparison against all known structures. Interestingly, known structures with any similarity to Chd were reported as either β-lactamases or alkylsulfatases (31–34). The most similar structures from each of these protein families, CphA and Pisa1, were selected for detailed comparison and structurally aligned with ChdT (Fig. 4, B and C). The overall fold of the ChdT monomer aligned well with the overall structure of the β-lactamase, CphA (PDB code 3IOG), except for an extra C-terminal motif in ChdT. The N-terminal portion (residues 1–277) of Chd forms an αββα-sandwich fold that is commonly observed in the β-lactamase superfamily; however, ChdT also possessed an additional C-terminal motif (residues 278–327) that originates in the middle of a long helix that is not found in the CphA structure. On the other hand, when ChdT was structurally aligned with one subunit of the alkylsulfatase, Pisa1 (PDB code 2YHE), the additional C-terminal motif observed in ChdT was present in Pisa1, which is much larger in size than either Chd or CphA.
Figure 4.
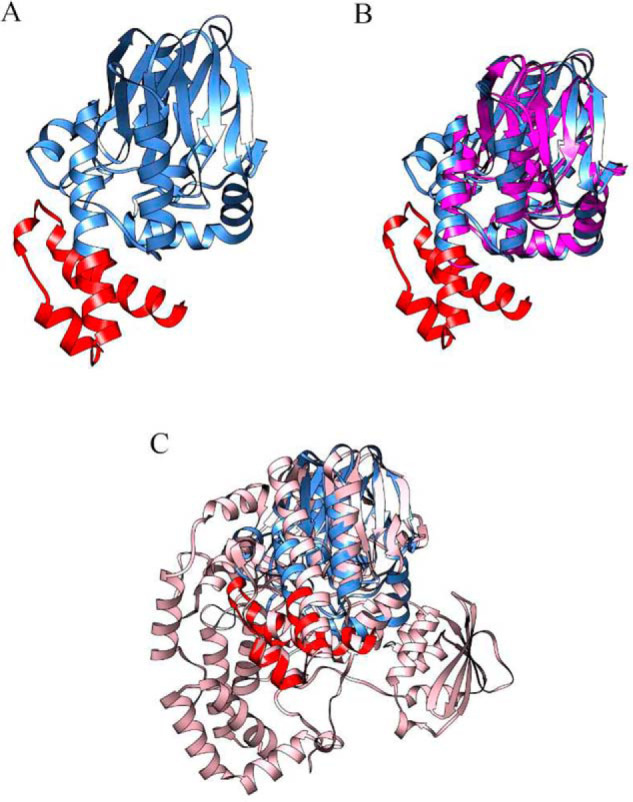
A, the overall protein fold of a ChdT monomer. The β-lactamase fold (residues 1–277) is in blue. A C-terminal motif (residues 278–327) is in red. B, ChdT is superimposed with the β-lactamase CphA (PDB code 3IOG), which is in magenta. C, ChdT is superimposed with an SEC alkyl sulfatase (PDB code 2YHE), which is in pink.
The active site of Chd
The coordination geometry of the active site Zn(II) ion is identical in both monomers and the simulated annealing composite omit map around each metal ion and their coordinating residues are consistent with the active-site Zn(II) ion residing in a distorted trigonal bipyramid or TBP geometry (Fig. 5A). The catalytic Zn(II) ion ligands include the Nϵ nitrogens of His117 (2.0 Å) and His257 (2.0 Å) along with a water/hydroxide oxygen atom (2.6 Å) making up the equatorial positions, whereas a carboxylate oxygen atom of Asp116 (2.1 Å) and a carbonyl oxygen atom of Asn216 (2.2 Å) (Table 2 and Fig. 5A) constitute the axial ligands. The coordinating water/hydroxide forms hydrogen bonds with the axial ligands Asp116 (2.6 Å) and Asn216 (2.9 Å) (Fig. 5A). Another active site residue, His114, is observed in two alternate conformations supported by simulated annealing composite omit maps (Fig. 5B). In one conformation (a), the Nϵ nitrogen atom of the imidazole ring is hydrogen bound to the coordinated water/hydroxide oxygen atom (2.9 Å) and the nitrogen atom (2.7 Å) of the axial ligand, Asn216. The second conformation (b) places the imidazole ring perpendicular to the imidazole ring in the (a) conformation, resulting in the loss of hydrogen-bonding interactions with both the coordinated water/hydroxide oxygen atom (5.4 Å) and possibly the nitrogen atom (3.1 Å) of the axial ligand Asn216. The structural Zn(II) ion is coordinated by the Nϵ nitrogen atoms of His143 (2.0 Å) and the carboxylate oxygen atom of Asp146 (1.9 Å) from each subunit (Fig. 5, C and D), providing a tetrahedral coordination geometry.
Figure 5.
A, ChdT active site. Zn(II)-ligand bonds are shown in gray sticks, and hydrogen bonds are shown as black dashed lines. The key distances are shown in angstroms. The two alternate conformations of His114 are labeled H114a and H114b. B, simulated annealing composite omit maps (2Fo − Fc) are shown in gray mesh at the 1.0 σ level for the coordinated residues, water, Zn(II), and His114. The residues are shown in the same orientations as that in A. C, the structural Zn(II) ion at the dimer interface. Zn(II)-ligand bonds are shown as gray sticks. One dimer is in tan, and the other is in cyan. His143 and Asp146 from different subunits are indicated with A or B. D, the simulated annealing composite omit map (2Fo − Fc) is contoured at 1.0 σ around the structural Zn(II) ion and its coordinated residues.
Table 2.
Selected bond lengths
| Atom 1 | Atom 2 | Length |
|---|---|---|
| Å | ||
| Active-site zinc | Carboxylate oxygen atom of Asp116 | 2.1 |
| Active-site zinc | Nϵ nitrogens of His117 | 2.0 |
| Active-site zinc | Nϵ nitrogens of His257 | 2.0 |
| Active-site zinc | Carbonyl oxygen atom of Asn216 | 2.2 |
| Active-site zinc | Metal-bound water/hydroxide | 2.6 |
| Nϵ nitrogen's of His114 | Metal-bound water/hydroxide | 2.9 |
| Nϵ nitrogen's of His114 | Nitrogen atom of Asn216 | 2.7 |
| Nϵ nitrogen's of His114 (perpendicular) | Metal-bound water/hydroxide | 5.4 |
| Nϵ nitrogen's of His114 (perpendicular) | Nitrogen atom of Asn216 | 3.1 |
| Structural zinc | Nϵ nitrogens of His143 | 2.0 |
| Structural zinc | Carboxylate oxygen atom of Asp146 | 1.9 |
Role of the structural Zn(II) site
The structural Zn(II) site ligands were eliminated by mutating both His143 and Asp146 to Ala. Removal of the structural Zn(II) site ligands had no apparent effect on ChdT activity, because the double mutant exhibited a kcat of 25 ± 1, which is identical to that of WT Chd (15, 26). Surprisingly, homodimer formation was also not affected based on gel-filtration results (Fig. S1). However, elimination of the structural Zn(II) site did increase the Km value 2-fold (Km = 0.24 ± 0.07 μm) and severely affected protein stability because the purified mutant protein precipitated within ∼2 h when incubated at 23–25 °C.
Chd biological assembly in solution
The intact molecular masses of both the truncated and structural Zn(II) Chd mutants were obtained by MALDI-TOF MS (Fig. 6). Masses consistent with homodimer formation were observed between 66 and 70 kDa. These MS data are consistent with the hydrodynamic volume for both samples determined by size-exclusion chromatography (Fig. S1), which indicate a molecular mass consistent with a homodimer in solution. It should also be noted that a small portion of ChdT fragmented (<8%) during MALDI-TOF MS analysis.
Figure 6.
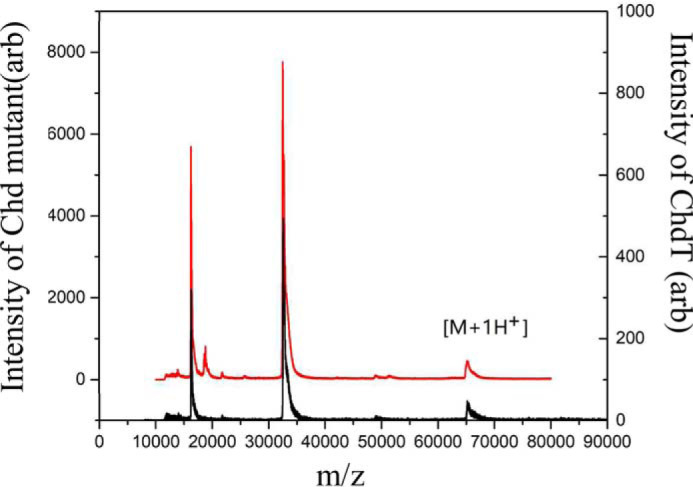
MALDI-TOF mass spectra of truncated ChdT (black) and the ChdT double mutant (red).
Substrate docking
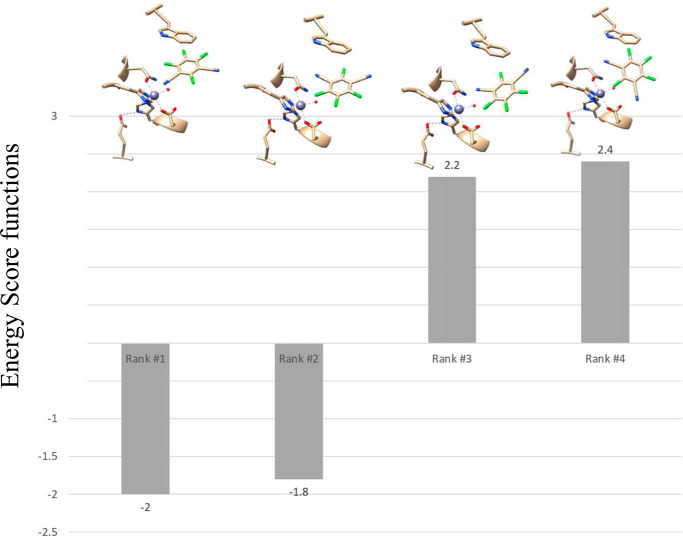
Attempts to co-crystallize or soak crystals of ChdT with the substrate TPN or recently identified inhibitors were unsuccessful; therefore, substrate docking was utilized to examine possible substrate-binding poses. Substrate docking generated four different poses ranked by the docking scoring function, where a lower value indicates lower total energy after docking (Fig. 7) (35–37). The top ranked model (model 1) suggested a binding interaction between the nitrogen atom of a cyano group of TPN and the active site zinc ion (2.3 Å), along with an energetically favorable π–π interaction between the TPN aromatic ring and that of the active-site residue Trp227. Interestingly, this pose preorganizes a possible transition state of TPN hydrolysis by placing the ortho carbon of TPN within ∼3.0 Å of the O atom of the bound water/hydroxide. In each of the other poses, all possible TPN ligating groups are too far from the catalytic Zn(II) ion to be considered a bonding interaction (>3.5 Å). However, model 3 revealed a hydrogen bond between the nitrogen atom of a TPN cyano group and the amide nitrogen of the Zn(II) ligands Asn216 (2.3 Å). The resulting substrate position placed the ortho carbon of TPN within 3.1 Å of the O atom of the bound water/hydroxide. In contrast to model 1, TPN is rotated in this pose, which abolished the energetically favorable π–π interaction between the TPN aromatic ring and that of Trp227.
Figure 7.
Four TPN docking poses for the active site of ChdT. The best pose (pose 1) indicates ligation of a CN group from TPN to the active site Zn(II) ion.
Discussion
Chd is a protein with an unusual sequence: the highest sequence similarity to a structurally characterized protein, glyoxylase II from Arabidopsis thaliana (PDB code 1XM8), is 14.7%. Here, a 1.96 Å resolution structure of ChdT, a truncated construct of Chd, is reported that reveals an αββα-sandwich fold that is also observed in the MBL superfamily (38). Chd is a “head-to-tail” homodimer, formed between two α-helices from each monomer (Fig. 4). Each monomer contains a mono-Zn(II) active site, and a third Zn(II) ion anchors an intersubunit structural site. Otherwise, and uniquely for enzymes with the same functionality, no coenzyme is associated with active Chd.
The mononuclear Zn(II) active site in each monomer of Chd is a characteristic shared with the B2 metallo-β-lactamases. The third Zn(II) ion in the Chd homodimer, which is coordinated by the Nϵ nitrogen atom of a His143 and a carboxylate oxygen atom of Asp146 from each subunit, resides in a tetrahedral coordination geometry that is buried at the dimer interface and likely functions as a structural site. A ChdT(H143A, D146A) construct predictably lacked the ability to bind this third zinc ion but, surprisingly, exhibited full catalytic activity, albeit with a 2-fold elevated Km. Additionally, the ChdT(H143A, D146A) variant retained the homodimeric architecture of the native protein in solution. The most striking effect of the double mutation was markedly impaired stability in solution at ambient temperatures.
The X-ray structure of Chd revealed two previously unknown solvent-accessible channels in each monomer that likely provide different routes for substrate and products to access/leave the catalytic Zn(II) site (Fig. 3). Because TPN is hydrophobic in nature, hydrolysis of an ortho-chlorine produces more hydrophilic products: 4-OH-TPN, a chloride ion and a proton. Because the products of TPN hydrolysis vary greatly in solubility and size, it would be reasonable to have two sets of solvent channels capable of shuttling different reactants and products to and from the active site. The Y-shaped channel that originates on the bottom of the Chd homodimer contains a large opening (8.5 × 17.8 Å) with a channel that is largely hydrophobic. This Y-shaped channel likely serves to recognize substrate and guide TPN to the active site. On the other hand, water and H+ and/or chloride ions can migrate through the second active-site channel, which is more hydrophilic and resides on the top of the dimer. It has been reported that dimerization of SidA, which is an alkylsulfatase similar to Pisa1, ensures resistance to biocidic SDS at toxic concentrations (39). Given that TPN is toxic, Chd may have evolved as a dimer with two active-site channels to elicit a similar response.
As predicted by Wang et al. (15), the first His in the B2 β-lactamase MBL (His-Xaa-His-Asp-His) is replaced by a Ser (Ser112 in Chd). This minor change in the B2 β-lactamase MBL active site may explain why a dinuclear Zn(II) site does not occur in both B2 enzymes and Chd. The single active-site Zn(II) ion in Chd resides in a distorted TBP geometry with His117, His257, Asp116, Asn216, and water/hydroxide as ligands (Fig. 5). Several potentially important hydrogen-bonding interactions also exist in the active site of Chd. Perhaps most notably, there is at least one interaction between the metal bound water/hydroxide oxygen atom and the Nϵ nitrogen atom of His114. His114 forms a strong hydrogen bond (∼2.9 Å) with the equatorial metal bound water/hydroxide in Chd and also with the nitrogen atom of the axial Asn216 ligand (∼2.7 Å). Interestingly, His114 clearly occupies two orientations in the structure, the second of which is perpendicular to the hydrogen bound conformation, which places the Nϵ nitrogen atom out of hydrogen-bonding range of the metal-bound water/hydroxide (∼5.4. Å) but close enough to the axial ligand Asn216 (∼3.1 Å) to possibly form a hydrogen bond. The functional role of His114 and its flexibility is currently unknown but may play a catalytic role as a general acid/base. This is consistent with both ChpA and Pisa1, because His114 is strictly conserved in both enzymes, and in ChpA, it was also proposed to be the general acid to protonate the leaving group (40).
Other potentially important hydrogen-bonding interactions occur between the Asn216 carboxamide nitrogen proton and the metal bound water/hydroxide (2.9 Å) and the second axial Asp116 ligand carboxylate oxygen, also with the metal bound water/hydroxide (2.6 Å). These hydrogen-bonding interactions likely assist in the deprotonation of the bound water to a nucleophilic hydroxide. Finally, a potentially important hydrogen-bonding interaction occurs between the Nδ proton of the His117 ligand with a side chain oxygen atom of Glu81 (∼2.7 Å), forming a Glu-His-Zn triad that has been postulated to decrease the Lewis acidity of zinc ions (41) and may further assist in facilitating the coordination of substrate or water to the active site Zn(II) ion. Similar arrangements and fully conserved Asp-His-zinc triads are observed for other hydrolases (42).
The discovery of a large hydrophobic channel on the bottom of the Chd homodimer that provides access to the catalytic Zn(II) ion suggests that it may serve as the initial substrate recognition site. To properly position an aromatic substrate, such as TPN, for catalysis the substrate would need to either bind to or near the active site Zn(II) ion. Interestingly, Trp227 resides in the hydrophobic channel ∼9.1 Å from the active site metal ion. Previous mutagenesis studies indicated that Trp227 is catalytically essential (15), suggesting that it may be involved in recognizing and positioning TPN near the active site Zn(II) ion via a π–π interaction. To test this hypothesis, TPN was docked into the hydrophobic channel with the most energetically favorable pose occurring for a TPN orientation that maximizes the π–π interaction with Trp227, places a CN nitrogen atom within a bonding distance to the active site Zn(II) (∼2.3 Å), and expands its coordination geometry to octahedral (Fig. 7; pose 1). Because the C4 and C6 chlorines in TPN are chemically equivalent (homotopic) given the symmetry arising from the positions of the two CN substitutes, it does not matter which CN group binds to the active-site Zn(II) ion. Chd catalyzes the dehalogenation of only one chlorine, and the resulting product cannot serve as a substrate for Chd. In pose 1, the C4/C6 chlorine would be positioned so that once the hydrolysis reaction occurs, the newly formed chloride ion would be oriented toward the small backend channel for expulsion to the bulk solvent. Although a second pose was reasonably energetically favorable (Fig. 7; pose 2), TPN was rotated, thus abolishing the energetically favorable π–π interaction between the TPN aromatic ring and that of Trp227. However, this pose does reveal a potential hydrogen bond between the nitrogen atom of a TPN cyano group and the amide nitrogen Asn216, which places the ortho carbon of TPN within 3.1 Å of the O atom of the bound water/hydroxide.
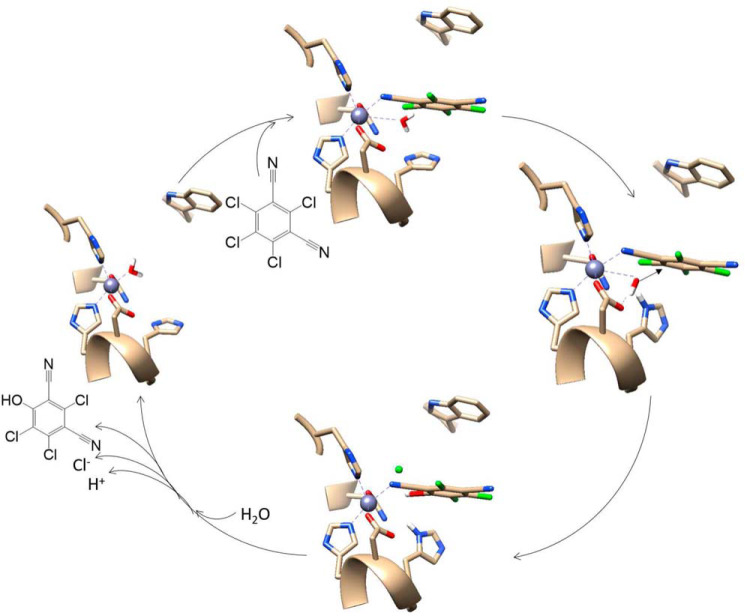
In conclusion, the X-ray crystal structure of ChdT, in combination with previously reported kinetic data, allows us to refine our proposed catalytic mechanism for Chd (Fig. 8) (26). The first step in catalysis is likely recognition of TPN by the large hydrophobic channel that leads to the Zn(II) active site. In this scenario, TPN is positioned by a π–π interaction with Trp227 that was shown to be important for catalytic turnover (15). Next, a CN nitrile nitrogen atom of TPN coordinates to the active-site Zn(II), although no direct evidence exists to support this assumption at this time, expanding its coordination number from five to six. Binding of the substrate nitrile nitrogen has a 2-fold impact. First, it removes electron density from the aromatic ring activating the ortho carbon for nucleophilic attack (27, 43), and second, it activates the coordinated water/hydroxide to function as a nucleophile. Deprotonation of the metal-bound water molecule by His114, with assistance from the Zn(II) ligands Asp116 and Asn227, forms a nucleophilic hydroxide moiety that is consistent with the postulated pKa of the zinc-bound water molecule (44). Once the zinc-bound hydroxide is formed, it can attack the activated ortho carbon of TPN, forming an η-1-μ-transition state complex (45). Solvent kinetic isotope effect studies indicate that one proton is transferred in the transition state, likely because of the breaking of a water O–H bond and the formation of a protonated His114 (26). The observed conformational flexibility of His114 may facilitate the formation of products and enable their release. Once the products are released, the Zn(II)-bound water molecule is replaced. Although a mechanism could also be proposed where a nitrile nitrogen atom of TPN does not bind to the active site Zn(II) ion, there is no residue other than the Zn(II) ligand Asn216 that could reasonably form a hydrogen bond to the nitrile nitrogen of TPN, which would be important for activating the substrate for hydrolysis.
Figure 8.
A revised catalytic mechanism for TPN hydrolysis by Chd.
Experimental procedures
Materials
NEB restriction enzymes, T4 DNA ligase, Agilent QuikChange II site-directed mutagenesis kit, nickel–nitrilotriacetic acid columns, Wizard® SV gel and PCR clean-up systems, size exclusion columns 4-(2-hydroxyethyl)-1-piperazineethane-sulfonic acid, potassium bromide, potassium sodium tartrate, and TPN were purchased commercially (Fisher Scientific) at the highest purity available. TEV protease (EC 3.4.22.44) was obtained via previously published methods (46).
N-terminal sequence modification of Chd
The first 15 residues of the N terminus of WT Chd were removed by PCR (sense primer, ACTCCATGGATGCCGTGAAATTTAG, and antisense primer, CATAAGCTTTTAATGGTGATGGTGATGGTGATG). The modified Chd gene containing NcoI and HindIII restriction sites was amplified under standard PCR conditions for gene cloning (47). The PCR products were purified by agarose gel electrophoresis and Wizard SV gel and subcloned into pET28a+, creating the new expression vector pET28a+–ChdT. The modified sequence was verified by Functional Biosciences (Madison, WI, USA).
Expression and purification of ChdT
The plasmid containing ChdT was freshly transformed into BL21(DE3) competent cells (Stratagene), and a single colony was used to inoculate 50 ml of LB-Miller culture containing 50 μg/ml kanamycin with shaking overnight at 37 °C as previously described (26). Briefly, this culture was used to inoculate a 1-liter culture, and the cells were grown at 37 °C until an A600 nm of 0.8–1.0 was reached. The culture was cooled on ice, induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside supplemented with 0.05 mm ZnCl2, and expressed at 25 °C for 16 h. The cells were harvested by centrifugation at 6370 × g and 4 °C for 10 min in a Beckman Coulter Avanti JA-10 rotor and resuspended in 20 mm Tris-HCl buffer, pH 7.0, containing 50 mm NaCl, 10 mm tris(2-carboxyethyl)phosphine, and 25 mm imidazole at a ratio of 5 ml/g of cells. The cells were lysed by ultrasonication (Misonix Sonicator 3000) for 4 min (30 s on, 45 s off) at 21 W. The crude extract was obtained after centrifugation in a JA-20 rotor at 31,000 × g and 4 °C for 20 min.
Crude extracts of ChdT (100 mg) were loaded onto a 5-ml nickel–nitrilotriacetic acid Superflow Cartridge (Qiagen) using an ÄKTA FPLC P-960. The column was washed with 50 ml of 20 mm Tris-HCl buffer, pH 7.0, containing 50 mm NaCl and 25 mm imidazole, followed by 50 ml of 20 mm Tris-HCl buffer, pH 7.0, containing 50 mm NaCl and 75 mm imidazole. The protein was eluted using a linear imidazole gradient (75–500 mm) at a flow rate of 2 ml/min. Active protein fractions were pooled and concentrated using 50 mm Tris buffer, pH 7.0, containing 1 mm EDTA with an Amicon Ultra-15 10,000 molecular weight cutoff centrifugal filter unit (Millipore) resulting in ∼12 mg/liter of soluble ChdT-His6.
The His6 tag was removed by treating His6-tagged ChdT with His6-tagged TEV protease (EC 3.4.22.44) for 16 h at 4 °C in 50 mm Tris, pH 8.0. Cleaved protein was concentrated with a Centricon (15,000 molecular weight cutoff; Amicon) to 3 ml and loaded on immobilized metal affinity chromatography to remove the remaining cleaved His6 tag, uncut protein, and the His6-tagged TEV protease, whereas the flow-through containing ChdT was collected and washed with 50 mm HEPES buffer, pH 7.0, containing 10% glycerol. Purified protein was analyzed by SDS-PAGE with a 12.5% polyacrylamide SPRINT NEXT GELTM (Amresco). The gels were stained with Gel Code Blue (Thermo Fisher Scientific). Protein concentration of crude extracts was determined using a Coomassie (Bradford) protein assay kit (Pierce) and pure protein by measuring the absorbance at 280 nm with a Shimadzu UV-2450 spectrophotometer equipped with a TCC-240A temperature-controlled cell holder. Theoretical molecular masses and protein extinction coefficients were calculated with the ExPASy compute pI/Mw tool.
Activity assay of ChdT
The enzymatic activity of ChdT toward TPN was measured using a Shimadzu UV-2450 spectrophotometer equipped with a TCC-240A temperature-controlled cell holder in 1-ml quartz cuvettes. A 1-ml reaction consisted of 50 mm HEPES buffer, pH 7.0, at 25 °C and various concentrations of TPN. The rate of TPN dehalogenation was determined by continuously monitoring the formation of 4-OH-TPN at 345 nm (Δϵ345 = 3.5 mm−1 cm−1). Data analysis was performed using OriginPro 9.0 (OriginLab, Northampton, MA, USA). The kinetic constants Vmax and Km were calculated by fitting to the Michaelis–Menten equation. One unit of enzyme activity was defined as the amount of enzyme that catalyzed the production of 1 μmol of the TPN minute at 25 °C.
Crystallization
Purified ChdT was exchanged into 50 mm HEPES buffer, pH 7.5, containing 10% glycerol for crystallization and concentrated to a final protein concentration of 10 mg/ml. Initial crystallization screens using a Gryphon Crystallization Robot (Art Robins Inc.) and Emerald Biosystems Wizard III/IV screen sets, produced crystals of ChdT in 10% PEG 6000 and 100 mm HEPES buffer, pH 7.0. Crystallization optimization leads revealed the best quality ChdT crystals via sitting-drop vapor diffusion occurred by mixing 6 μl of (10 mg/ml) ChdT with 6 μl of well solution (10% PEG 6000, 100 mm HEPES, pH 7.0). The crystals were allowed to grow at room temperature and reached optimal size and morphology for data collection after 7–10 days. The crystals were manually transferred to well solution supplemented with 30% glycerol (cryo-protectant) and flash frozen in liquid N2.
X-ray diffraction data collection
SAD and monochromatic data were collected at the LS-CAT 21-ID Advanced Photon Source (APS) at Argonne National Laboratory. SAD data sets were collected at a wavelength of 1.28 Å, whereas another monochromatic data set was collected at 0.98 Å at 100 K using a Dectris Eiger 9M detector. The data sets were indexed and integrated using the HKL2000 suite.
Model building and refinement
The structure of ChdT was solved by SAD using autosol in the Phenix software suite (48). The model building was first conducted in autobuild followed by COOT (49), refined using Phenix, and analyzed in COOT and USCF Chimera (50). Final refinement statistics are reported in Table 1, and structural figures were made using USCF Chimera.
Double mutation at the structural zinc-binding site
His143 and Asp146, structural Zn(II) site ligands, were mutated to alanine (Ala143 and Ala146) by a single mutagenic primer as follows: sense primer, TTGATGGTGCCCTGGCGGCCGATAAGGCGTTCATGGT; and antisense primer: ACCATGAACGCCTTATCGGCCGCCAGGGCACCATCAA (mutated residues are shown in bold type). The sense primer had a completely complementary sequence to the corresponding antisense primer. The first PCR was performed using sense primer and antisense primer in separate tubes (25 μl). Following denaturation at 98 °C for 1 min, five PCR cycles were carried out with each cycle comprised of incubation at 98 °C for 15 s, followed by a 30-s annealing step at 65 °C, and a 1-min extension at 71 °C. A 50-μl solution was prepared by combining two tubes for a second PCR, which was carried out under the same conditions as the first PCR but for 55 cycles. The final products were transformed directly into XL10-gold ultracompetent cells. The mutated ChdT sequence was verified by DNA sequencing by Functional Biosciences.
MALDI-TOF MS
Saturated α-CHCA thin layer solution was prepared on a MALDI target (51). A 2-μl protein sample (10 μm) was mixed with a DHB-CHCA matrix on a MALDI target by the “sandwich method” (21). Mass spectra of each sample were obtained on a Bruker microflex LRFTM MALDI-TOF MS.
Substrate docking
TPN poses were created and optimized by Molecular Operating Environment (Chemical Computing Group, Montreal, Canada). The TPN substrate was docked into the ChdT active site pocket using Autodock Vina, which is a structural analysis panel of Chimera 1.13.1rc (UCSF Chimera, San Francisco, CA, USA). The optimized TPN molecule was deemed a ligand, whereas the active site of Chd was assigned as a receptor. The oxygen position of the active-site Zn(II)-bound water/hydroxide was fixed as were the protein residue ligands. The best pose, based on the docking scoring function versus binding energy (37), was considered the final model.
Data availability
All data described in the manuscript are contained within the manuscript or the supporting information. The final refined model was deposited at the Protein Data Bank under code 6UXU.
Supplementary Material
Acknowledgments
This work used resources of the Advanced Proton Source, a U.S. Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and Michigan Technology Tri-Corridor Grant 085P1000817.
This article contains supporting information.
Author contributions—D. S. C. and D. L. data curation; D. S. C., X. Y., B. B., R. C. H., and D. L. formal analysis; D. S. C. and X. Y. writing-original draft; X. Y. investigation; X. Y. methodology; B. B., R. C. H., and D. L. conceptualization; B. B., R. C. H., and D. L. funding acquisition; B. B., R. C. H., and D. L. project administration; B. B., R. C. H., and D. L. writing-review and editing.
Funding and additional information—This work was supported by National Science Foundation Grant CHE-1808711 (to B. B. and R. C. H.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- Chd
- chlorothalonil dehalogenase
- ChdT
- N-terminally truncated Chd
- TPN
- 2,4,5,6-tetrachloroiso-phtalonitrile
- SAD
- single-wavelength anomalous dispersion
- 4-OH-TPN
- 4-hydroxytrichloro-isophthalonitrile
- MBL
- metallo-β-lactamase
- TBP
- trigonal bipyramidal.
References
- 1. Sims J. L., Suflita J. M., and Russell H. H. (1991) Reductive Dehalogenation of Organic Contaminants in Soils and Ground Water, U.S. Environmental Protection Agency, Superfund Technology Support Center for Ground Water [Google Scholar]
- 2. Arora P. K., and Bae H. (2014) Role of dehalogenases in aerobic bacterial degradation of chlorinated aromatic compounds. J. Chem. Dalton Trans. 2014, 1–11 10.1155/2014/157974 [DOI] [Google Scholar]
- 3. Baxter J., and Cummings S. P. (2006) The current and future applications of microorganism in the bioremediation of cyanide contamination. Antonie Van Leeuwenhoek 90, 1–17 10.1007/s10482-006-9057-y [DOI] [PubMed] [Google Scholar]
- 4. Copley S. D. (1998) Microbial dehalogenases: enzymes recruited to convert xenobiotic substrates. Curr. Opin. Chem. Biol. 2, 613–617 10.1016/S1367-5931(98)80092-6 [DOI] [PubMed] [Google Scholar]
- 5. de Jong R. M., and Dijkstra B. W. (2003) Structure and mechanism of bacterial dehalogenases: different ways to cleave a carbon-halogen bond. Curr. Opin. Struct. Biol. 13, 722–730 10.1016/j.sbi.2003.10.009 [DOI] [PubMed] [Google Scholar]
- 6. Janssen D. B., Oppentocht J. E., and Poelarends G. J. (2001) Microbial dehalogenation. Curr. Opin. Biotechnol. 12, 254–258 10.1016/S0958-1669(00)00208-1 [DOI] [PubMed] [Google Scholar]
- 7. Reddy G. V. B., and Gold M. H. (2000) Degradation of pentachlorophenol by Phanerochaete chrysosporium: intermediates and reactions involved. Microbiology 146, 405–413 10.1099/00221287-146-2-405 [DOI] [PubMed] [Google Scholar]
- 8. Beil S., Mason J. R., Timmis K. N., and Pieper D. H. (1998) Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J. Bacteriol. 180, 5520–5528 10.1128/JB.180.21.5520-5528.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xun L., Topp E., and Orser C. S. (1992) Purification and characterization of a tetrachloro-p-hydroquinone reductive dehalogenase from a Flavobacterium sp. J. Bacteriol. 174, 8003–8007 10.1128/JB.174.24.8003-8007.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Orser C. S., Dutton J., Lange C., Jablonski P., Xun L., and Hargis M. (1993) Characterization of a Flavobacterium glutathione S-transferase gene involved reductive dechlorination. J. Bacteriol. 175, 2640–2644 10.1128/JB.175.9.2640-2644.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yokota T., Fuse H., Omori T., and Minora Y. (1986) Microbial dehalogenation of haloalkanes mediated byoxy genases or halidohydrolase. Agric. Biol. Chem. 50, 453–460 10.1271/bbb1961.50.453 [DOI] [Google Scholar]
- 12. Bunge M., Adrian L., Kraus A., Opel M., Lorenz W. G., Andreesen J. R., Görisch H., and Lechner U. (2003) Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421, 357–360 10.1038/nature01237 [DOI] [PubMed] [Google Scholar]
- 13. Leisinger T., Bader R., Hermann R., Schmid-Appert M., and Vuilleumier S. (1994) Microbes, enzymes and genes involved in dichloromethane utilization. Biodegradation 5, 237–248 10.1007/BF00696462 [DOI] [PubMed] [Google Scholar]
- 14. Ang T. F., Maiangwa J., Salleh A. B., Normi Y. M., and Leow T. C. (2018) Dehalogenases: from improved performance to potential microbial dehalogenation applications. Molecules 23, E1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang G., Li R., Li S., and Jiang J. (2010) A novel hydrolytic dehalogenase for the chlorinated aromatic compound chlorothalonil. J. Bacteriol. 192, 2737–2745 10.1128/JB.01547-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caux P. Y., Kent R. A., Fan G. T., and Stephenson G. L. (1996) Environmental fate and effects of chlorothalonil: a Canadian perspective. Crit. Rev. Environ. Sci. Technol. 26, 45–93 10.1080/10643389609388486 [DOI] [Google Scholar]
- 17. Sakkas V. A., Lambropoulou D. A., and Albanis T. A. (2002) Study of chlorothalonil photodegradation in natural waters and in the presence of humic substances. Chemosphere 48, 939–945 10.1016/S0045-6535(02)00121-2 [DOI] [PubMed] [Google Scholar]
- 18. Mozzachio A. M., Rusiecki J. A., Hoppin J. A., Mahajan R., Patel R., Beane-Freeman L., and Alavanja M. C. (2008) Chlorothalonil exposure and cancer incidence among pesticide applicator participants in the agricultural health study. Environ. Res. 108, 400–403 10.1016/j.envres.2008.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carlo-Rojas Z., Bello-Mendoza R., Figueroa M. S., and Sokolov M. (2004) Chlorothalonil degradation under anaerobic conditions in an agricultural tropical soil. Water Air Soil Pollution 151, 397–409 10.1023/B:WATE.0000009918.70113.c5 [DOI] [Google Scholar]
- 20. Vickers A. E., Sloop T. C., and Lucier G. W. (1985) Mechanism of action of toxic halogenated aromatics. Environ. Health Perspect. 59, 121–128 10.1289/ehp.59-1568090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kussmann M., and Roepstorff P. (2000) Sample preparation techniques for peptides and proteins analyzed by MALDI-MS. Methods Mol. Biol. 146, 405–424 [DOI] [PubMed] [Google Scholar]
- 22. Viciu M. S., Grasa G. A., and Nolan S. P. (2001) Catalytic dehalogenation of aryl halides mediated by a palladium/imidazolium salt system. Organometallics 20, 3607–3612 10.1021/om010332s [DOI] [Google Scholar]
- 23. Liang B., Wang G., Zhao Y., Chen K., Li S., and Jiang J. (2011) Facilitation of bacterial adaptation to chlorothalonil-contaminated sites by horizontal transfer of the chlorothalonil hydrolytic dehalogenase gene. Appl. Environ. Microbiol. 77, 4268–4272 10.1128/AEM.02457-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ren X., Li H., and Chen S. (2011) Cloning of the chlorothalonil-degrading gene cluster and evidence of its horizontal transfer. Curr. Microbiol. 62, 1068–1073 10.1007/s00284-010-9824-x [DOI] [PubMed] [Google Scholar]
- 25. Yue W., Xiong M., Li F., and Wang G. (2015) The isolation and characterization of the novel chlorothalonil-degrading strain Paracoccus sp. XF-3 and the cloning of the chd gene. J. Biosci. Bioeng. 120, 544–548 10.1016/j.jbiosc.2015.03.013 [DOI] [PubMed] [Google Scholar]
- 26. Yang X., Bennett B., and Holz R. C. (2019) Insights into the catalytic mechanism of a bacterial hydrolytic dehalogenase that degrades the fungicide chlorothalonil. J. Biol. Chem. 264, 13411–13420 10.1074/jbc.RA119.009094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peat T. S., Newman J., Balotra S., Lucent D., Warden A. C., and Scott C. (2015) The structure of the hexameric atrazine chlorohydrolase AtzA. Acta Crystallogr. D Biol. Crystallogr. 71, 710–720 10.1107/S1399004715000619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krissinel E. (2010) Crystal contacts as nature's docking solutions. J. Comput. Chem. 31, 133–143 10.1002/jcc.21303 [DOI] [PubMed] [Google Scholar]
- 29. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 30. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crowder M. W., Spencer J., and Vila A. J. (2006) Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res. 39, 721–728 10.1021/ar0400241 [DOI] [PubMed] [Google Scholar]
- 32. Fryszczyn B. G., Adamski C. J., Brown N. G., Rice K., Huang W., and Palzkill T. (2014) Role of β-lactamase residues in a common interface for binding the structurally unrelated inhibitory proteins BLIP and BLIP-II. Protein Sci. 23, 1235–1246 10.1002/pro.2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Knaus T., Schober M., Kepplinger B., Faccinelli M., Pitzer J., Faber K., Macheroux P., and Wagner U. (2012) Structure and mechanism of an inverting alkylsulfatase from Pseudomonas sp. DSM6611 specific for secondary alkyl sulfates. FEBS J. 279, 4374–4384 10.1111/febs.12027 [DOI] [PubMed] [Google Scholar]
- 34. Walsh T. R., Toleman M. A., Poirel L., and Nordmann P. (2005) Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18, 306–325 10.1128/CMR.18.2.306-325.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quiroga R., and Villarreal M. A. (2016) Vinardo: a scoring function based on Autodock Vina improves scoring, docking, and virtual screening. PLoS One 11, e0155183 10.1371/journal.pone.0155183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seeliger D., and de Groot B. L. (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 24, 417–422 10.1007/s10822-010-9352-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baier F., and Tokuriki N. (2014) Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 426, 2442–2456 10.1016/j.jmb.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 39. Hagelueken G., Adams T. M., Wiehlmann L., Widow U., Kolmar H., Tümmler B., Heinz D. W., and Schubert W. D. (2006) The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc. Natl. Acad. Sci. U.S.A. 103, 7631–7636 10.1073/pnas.0510501103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garau G., Bebrone C., Anne C., Galleni M., Frère J. M., and Dideberg O. (2005) A metallo-β-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J. Mol. Biol. 345, 785–795 10.1016/j.jmb.2004.10.070 [DOI] [PubMed] [Google Scholar]
- 41. Christianson D. W., and Cox J. D. (1999) Catalysis by metal-activated hydroxide in zinc and manganese metalloenzymes. Annu. Rev. Biochem. 68, 33–57 10.1146/annurev.biochem.68.1.33 [DOI] [PubMed] [Google Scholar]
- 42. Desmarais W., Bienvenue D. L., Bzymek K. P., Petsko G. A., Ringe D., and Holz R. C. (2006) The high-resolution structures of the neutral and the low pH crystals of the aminopeptidase from Aeromonas proteolytica. J. Biol. Inorg. Chem. 11, 398–408 10.1007/s00775-006-0093-x [DOI] [PubMed] [Google Scholar]
- 43. Hu J., Su Q., Schlessman J. L., and Rokita S. E. (2019) Redox control of iodotyrosine deiodinase. Protein Sci. 28, 68–78 10.1002/pro.3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sakurai M., Furuki T., and Inoue Y. (1995) The pKa of the zinc-bound water in carbonic anhydrase and its model compounds as studied by the AM1 calculation coupled with a reaction field theory. J. Phys. Chem. 99, 17789–17794 10.1021/j100050a019 [DOI] [Google Scholar]
- 45. Cosper N. J., Bienvenue D. L., Shokes J. E., Gilner D. M., Tsukamoto T., Scott R. A., and Holz R. C. (2003) The dapE-encoded N-succinyl-l,l-diaminopimelic acid desuccinylase from Haemophilus influenzae is a dinuclear metallohydrolase. J. Am. Chem. Soc. 125, 14654–14655 10.1021/ja036650v [DOI] [PubMed] [Google Scholar]
- 46. Tropea J. E., Cherry S., and Waugh D. S. (2009) Expression and purification of soluble His6-tagged TEV protease. Methods Mol. Biol. 498, 297–307 10.1007/978-1-59745-196-3_19 [DOI] [PubMed] [Google Scholar]
- 47. Yang X., Bennett B., and Holz R. C. (2018) Analyzing the function of the insert region found between the α- and β-subunits in the eukaryotic nitrile hydratase from Monosiga brevicollis. Arch. Biochem. Biophys. 657, 1–7 10.1016/j.abb.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 51. Signor L., and Boeri Erba E. (2013) Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometric analysis of intact proteins larger than 100 kDa. J. Vis. Exp. 10.3791/50635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the manuscript are contained within the manuscript or the supporting information. The final refined model was deposited at the Protein Data Bank under code 6UXU.