Abstract
Polyamines regulate gene expression in Escherichia coli by translationally stimulating mRNAs encoding global transcription factors. In this study, we focused on histone acetylation, one of the mechanisms of epigenetic regulation of gene expression, to attempt to clarify the role of polyamines in the regulation of gene expression in eukaryotes. We found that activities of histone acetyltransferases in both the nucleus and cytoplasm decreased significantly in polyamine-reduced mouse mammary carcinoma FM3A cells. Although protein levels of histones H3 and H4 did not change in control and polyamine-reduced cells, acetylation of histones H3 and H4 was greatly decreased in the polyamine-reduced cells. Next, we used control and polyamine-reduced cells to identify histone acetyltransferases whose synthesis is stimulated by polyamines. We found that polyamines stimulate the translation of histone acetyltransferases GCN5 and HAT1. Accordingly, GCN5- and HAT1-catalyzed acetylation of specific lysine residues on histones H3 and H4 was stimulated by polyamines. Consistent with these findings, transcription of genes required for cell proliferation was enhanced by polyamines. These results indicate that polyamines regulate gene expression by enhancing the expression of the histone acetyltransferases GCN5 and HAT1 at the level of translation. Mechanistically, polyamines enhanced the interaction of microRNA-7648-5p (miR-7648-5p) with the 5′-UTR of GCN5 mRNA, resulting in stimulation of translation due to the destabilization of the double-stranded RNA (dsRNA) between the 5′-UTR and the ORF of GCN5 mRNA. Because HAT1 mRNA has a short 5′-UTR, polyamines may enhance initiation complex formation directly on this mRNA.
Keywords: polyamine, spermidine, gene expression, histone modification, histone acetyltransferase, polyamine modulon, translational regulation, histone acetyltransferase GCN5 (GCN5), histone acetyltransferase 1 (HAT1), miR-7648-5p, histone acetylase, translation regulation, miR-7648-5p
Gene expression independent of DNA sequence is regulated by epigenetic mechanisms. Epigenetic control of gene expression comprises three pathways: histone modifications, DNA methylation, and RNA-based pathways. A number of different posttranslational modifications are known to occur on histone tails, including acetylation, methylation, ubiquitination, phosphorylation, and sumoylation (1, 2). Acetylation of lysine residues on histones, in particular, has been shown to play important roles in the regulation of chromatin structure and nucleosome dynamics. The dynamic structural change of chromatin by histone acetylation is linked to cellular processes, including transcriptional activation, DNA damage repair, and cell cycle. Reversible lysine acetylation is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), which add or remove acetyl groups to or from target histones, respectively. HATs and HDACs are also known as transcription coactivators and corepressors. These enzymes are highly conserved from yeast to human. Aberrations of HATs and HDACs have been reported in many diseases, such as cancer. However, the cause of the collapse of the balance between HATs and HDACs remains unknown (3–5).
Polyamines (putrescine, spermidine, and spermine) are present at millimolar concentrations in prokaryotic and eukaryotic cells and play regulatory roles in cell growth and viability (6–8). We have proposed that a set of genes whose expression is enhanced by polyamines at the level of translation can be classified as a “polyamine modulon” (9) and thus far have identified 20 different genes in E. coli and 7 different genes in eukaryotes as components of a polyamine modulon (10, 11). In E. coli, using a polyamine-requiring mutant, MA261, it was found that polyamines regulate gene expression because many proteins encoded by the polyamine modulon are transcription factors.
In mammalian cells, to identify members of the polyamine modulon, polyamine-reduced cells were prepared using α-difluoromethylornithine (DFMO), an inhibitor of ornithine decarboxylase, and proteins in cells were compared between control and polyamine-reduced cells. Seven proteins stimulated by polyamines thus far identified were Cct2, Hnrpl, and Pgam1 (12), eEF1A (13), p16 (7), EXT2 (14) and CHSY1 (15). The mechanisms of polyamine stimulation are as follows. First, polyamines enhance ribosome shunting, which involves discontinuous scanning by 40S ribosomal subunits during the scanning of the 5′-UTR of mRNA. This was observed in Cct2 (T-complex protein 1, β subunit) synthesis. Second, in the 5′-UTR of mRNA, a complementary sequence, consisting of more than five nucleotides, to the nucleotide sequences at the 3′-end of 18S rRNA, i.e. a CR sequence (complementary sequence to 18S rRNA), is normally present at -17 to -32 upstream from the initiation codon AUG. When the CR sequence is located more distally from the initiation codon AUG, e.g. –33 to –39, polyamines stimulated protein synthesis. This was observed in eEF1A (a translation elongation factor). Third, when a microRNA inhibits protein synthesis through the interaction with the complementary sequence in some mRNAs, polyamines release the microRNA from mRNA and stimulate protein synthesis. This was observed in EXT2 (a protein involved in extension of polysaccharide chains) synthesis. Fourth, when an RNA G-quadruplex exists at the 5′-UTR of mRNA, polyamines stimulate protein synthesis through the unfolding of the G-quadruplex. This was observed in CHSY1 (chondroitin synthase 1) synthesis (15). The polyamine modulon involved in gene expression has not been identified, and the role of polyamines in gene expression is unknown in eukaryotes.
In this study, we have found another two members of the polyamine modulon, Gcn5 and Hat1 histone acetyltransferases. We have studied the physiological functions of polyamines through stimulation of histone acetyltransferase synthesis and the mechanisms by which polyamines stimulate synthesis of these two proteins.
Results
Polyamines enhance histone acetylation and HAT activity
We looked for transcription factors in mammalian cells whose synthesis is stimulated by polyamines, because synthesis of many transcription factors in Escherichia coli are stimulated by polyamines at the translational level (6, 7, 10). However, synthesis of none of the 10 transcription factors was stimulated by polyamines (Fig. S1).
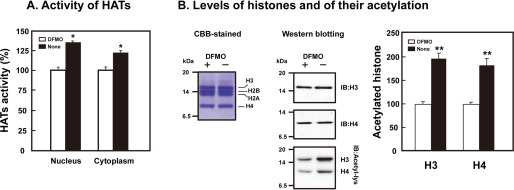
We then focused on epigenetic regulation of gene expression and looked for a polyamine modulon among genes encoding histone modification enzymes such as histone acetyltransferases (HATs) (16). For this purpose, HAT activities were first compared in control FM3A cells and DFMO-treated, polyamine-reduced FM3A cells, in which putrescine and spermidine became negligible and spermine decreased to about half of control cells (13). As shown in Fig. 1A, HAT activities in both nuclei and cytoplasm were significantly higher in control cells than in polyamine-reduced cells, when the activity was measured using the mixture of histones as substrates. Under these conditions, the level of histone acetylation (H3 and H4) was higher in control cells than polyamine-reduced cells, although protein levels of H3 and H4 were nearly equal in both cells (Fig. 1B).
Figure 1.
Stimulation of acetylation of histones H3 and H4 by polyamines. FM3A cells were cultured in the presence and absence of 50 μm DFMO for 72 h. Nucleus and cytoplasm fractions were prepared, and activity of histone acetyltransferases (HATs) was measured as described under “Experimental Procedures.” (A) Activity of histone acetyltransferases. (B) Levels of histones and their acetylation. * p < 0.05; ** p < 0.01.
Stimulation of Gcn5 and Hat1 synthesis by polyamines
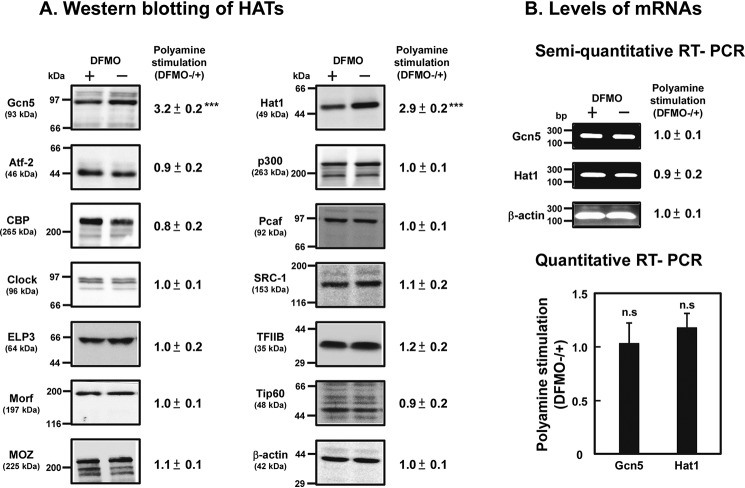
We searched 13 HATs for proteins whose synthesis is stimulated by polyamines using control and polyamine-reduced cells. As shown in Fig. 2, protein levels of Gcn5 (also known as Kat2a in mouse), located in the nucleus (type A), and Hat1 (also known as Kat1 in mouse), located in the cytoplasm (type B), were higher in control cells than polyamine-reduced cells, although mRNA levels of these proteins did not alter, suggesting that synthesis of Gcn5 and Hat1 was stimulated significantly by polyamines at the level of translation. The degree of polyamine stimulation of both Gcn5 and Hat1 was ∼3-fold.
Figure 2.
Western blotting of histone acetyltransferases (A), and the levels of mRNA of Gcn5 and Hat1 (B). (A) Protein levels of 13 histone acetyltransferases and of β-actin as a control were determined by Western blotting as described under “Experimental Procedures” using 20 μg of cell lysate, and the ratio of control/DFMO-treated cells was shown. *** p < 0.001. (B) mRNA levels of Gcn5, Hat1, and β-actin were determined as described under “Experimental Procedures.”
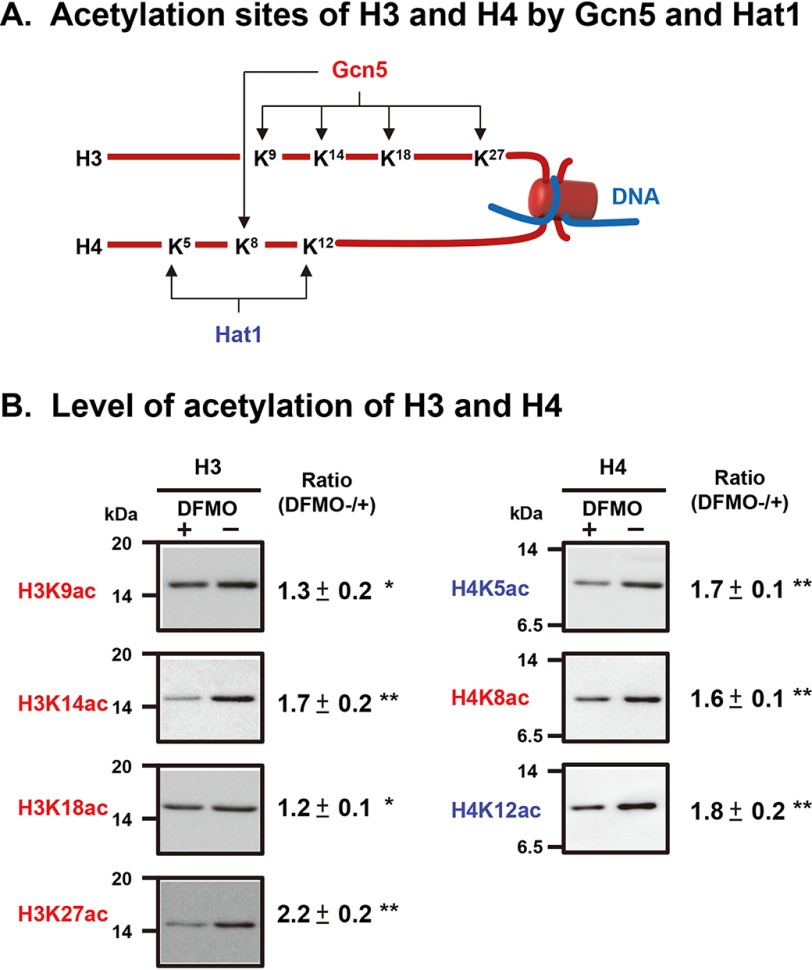
We then sought to determine which lysine was subject to more polyamine-stimulated acetylation. Gcn5 catalyzes acetylation of Lys-9, -14, -18, and -27 in histone H3 and Lys-8 in histone H4. Hat1 catalyzes acetylation of Lys-5 and -12 in histone H4 (Fig. 3A) (17). As shown in Fig. 3B, the acetylation of Lys-14 and -27 in histone H3 and that of Lys-5, -8, and -12 in histone H4 was stimulated by polyamines more than 1.5-fold. Stimulation of acetylation of Lys-9 and -18 in histone H3 by polyamines was also significant, although the degree of stimulation was less compared with the acetylation in other Lys residues. Protein levels of H3 and H4 were nearly equal in both cells as shown in Fig. 1B. These results indicate that acetylation by Gcn5 and Hat1 of all lysine residues located in the NH2-terminal region of histones H3 and H4 is stimulated by polyamines.
Figure 3.
Comparison of polyamine stimulation of histone acetylation at each lysine residue in histones H3 and H4. (A) Acetylation sites of histones H3 and H4 by Gcn5 and Hat1 (17). (B) Level of acetylation of H3 and H4. Histones were isolated from control and DFMO-treated, polyamine-reduced FM3A cells and analyzed by Western blotting using site-specific acetyl lysine antibodies as described under “Experimental Procedures.” * p < 0.05; ** p < 0.01.
Effect of histone acetylation by Gcn5 and Hat1 on gene expression
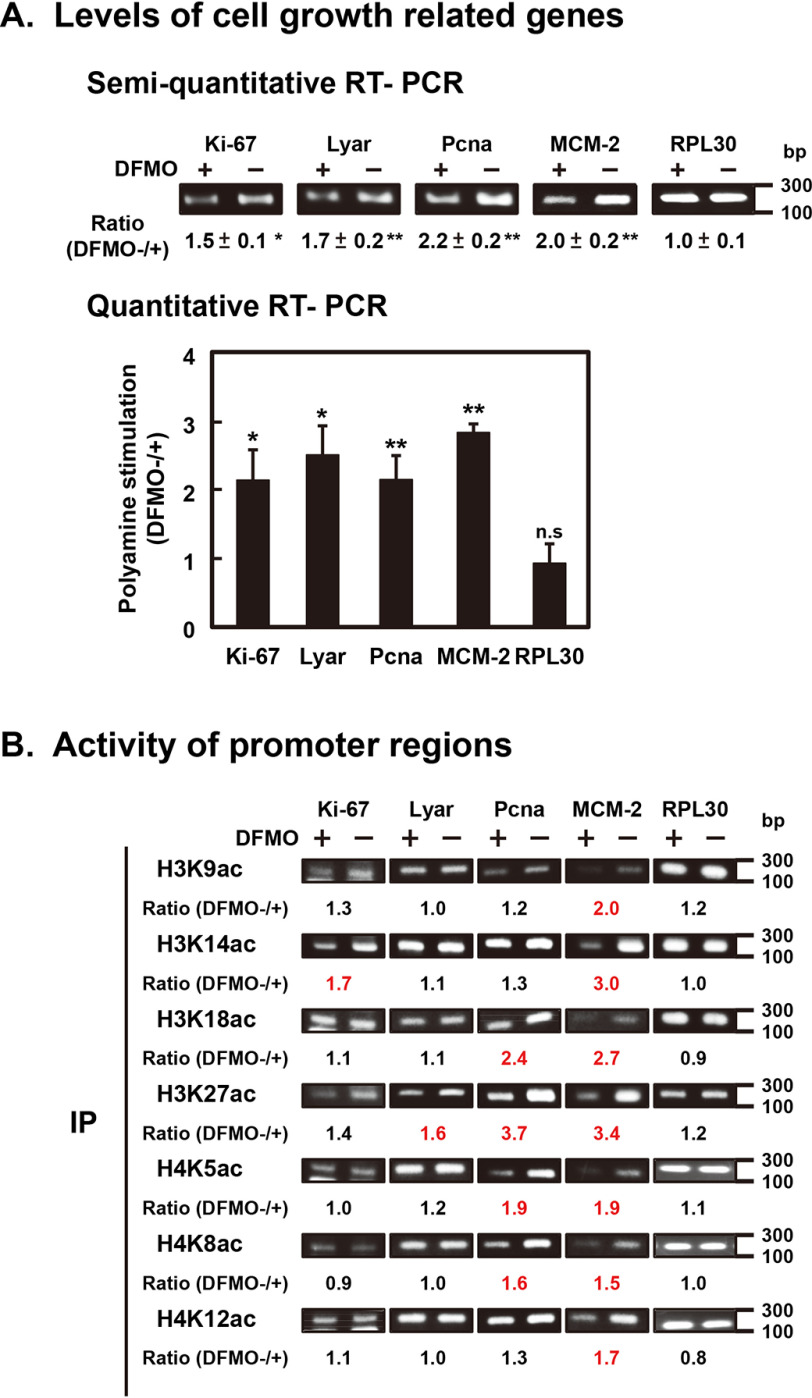
It is expected that synthesis of specific mRNAs which are involved in cell proliferation is stimulated by polyamines through stimulation of Gcn5 and Hat1 synthesis. Consistent with this idea, synthesis of mRNAs encoding Ki-67 protein associated with proliferation (18), Lyar (a nucleolar protein with zinc finger DNA binding motif) protein (19), Pcna (proliferating cell nuclear antigen) protein (20), and MCM-2 (minichromosome maintenance-2) protein (21) was stimulated by polyamines (Fig. 4A). The degree of polyamine stimulation of these mRNAs was 2- to 3-fold. As a control, the level of RPL30 (ribosomal protein L30) mRNA was measured. Synthesis of RPL30 mRNA was nearly equal in control and polyamine-reduced cells. The results indicate that synthesis of specific mRNAs, which are important for the functions of mouse mammary gland FM3A cells, is increased by polyamines through stimulation of Gcn5 and Hat1 synthesis.
Figure 4.
Stimulation of the synthesis of several mRNAs by polyamines through stimulation of acetylation of lysine residues in histones H3 and H4. (A) mRNA levels of cell growth-related genes, Ki-67, Lyar, Pcna, and MCM-2 and of PRL30 as a control. mRNA was isolated from control and DFMO-treated FM3A cells, and cDNA was synthesized and analyzed by quantitative real-time PCR as described under “Experimental Procedures.” * p < 0.05; ** p < 0.01. (B) Analysis of active promoter regions. The ChIP assay was carried out using anti-site-specific acetyl lysine of histones H3 and H4, and promoter regions of cell growth related genes, Ki-67, Lyar, Pcna, and MCM-2, and PRL30 as a control were amplified. Experiments were repeated twice, and the mean ratio (control/DFMO-treated) is shown. More than 1.5 is shown in red.
Hyperacetylation of histones can open compacted chromatin to induce transcription activation. The correlation between an increase in acetylation in histones H3 and H4 by polyamines and an increase in mRNA synthesis encoding factors involved in proliferation in FM3A cells was examined using a ChIP assay. Chromatin was isolated from control and polyamine-reduced cells, immunoprecipitated with anti-acetyl histone H3 and H4 antibodies, and analyzed with PCR amplification of promoter regions of the specific genes involved in proliferation in FM3A cells. The promoter regions of these genes were searched using the Eukaryotic Promoter Database (RRID:SCR_002485). As shown in Fig. 4B, the promoter regions of Ki-67, Lyar, Pcna, and MCM-2 genes in chromatin immunoprecipitated with antiacetyl histones H3 and H4 antibodies was increased significantly by polyamines. In particular, acetylation of Lys-27 in histone H3 was most strongly enhanced by polyamines (Fig. 3B), and promoters of genes for Lyar, Pcna, and MCM-2 were increased most strongly by polyamines in chromatin immunoprecipitated with anti-H3K27ac antibody (Fig. 4B). As a control, the level of RPL30 promoter in immunoprecipitated chromatin was examined, and it did not alter in control and polyamine-reduced cells. Therefore, synthesis of specific mRNAs that are important for the functions of mouse mammary gland FM3A cells is stimulated by polyamines. These results indicate that polyamines regulate gene expression through stimulation of histone acetylations.
Mechanism of polyamine stimulation of Gcn5 and Hat1 synthesis
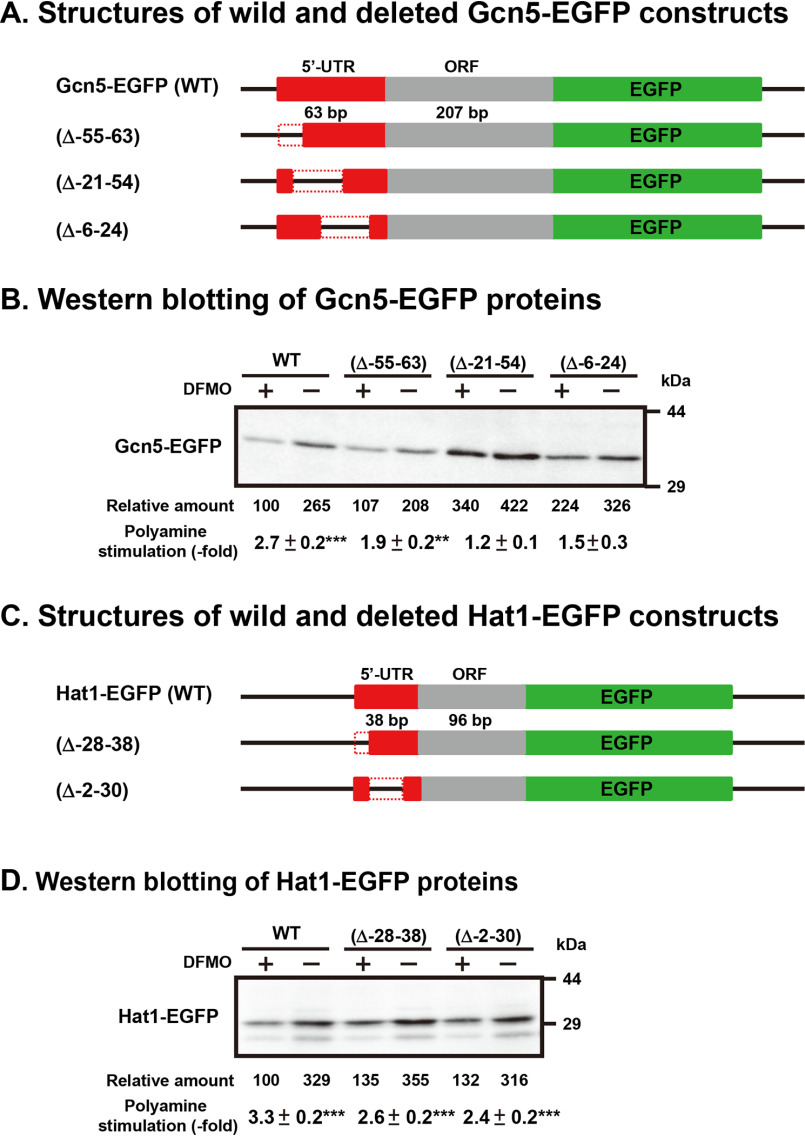
To analyze the mechanism of polyamine stimulation, we employed fusion mRNAs containing a 5′-UTR and part of the ORF of Gcn5 or Hat1 mRNA fused to EGFP (enhanced GFP) mRNA. Plasmids encoding fusion mRNAs were transfected into NIH3T3 cells, and the effect of polyamines was examined in control and 500 µm DFMO-treated, polyamine-reduced cells. As shown in Fig. 5, the size of the 5′-UTR of Gcn5 and Hat1 mRNA was 63 and 38 nucleotides, respectively. In the case of Hat1-EGFP, synthesis of the fusion protein from the wide-type (WT) construct was stimulated 3.3-fold by polyamines, similarly to FM3A cells, and the deletion of –28 to –38 or –2 to –30 nucleotides of the 5′-UTR of Hat1 mRNA did not influence the degree of polyamine stimulation of Hat1-EGFP synthesis (Figs. 5C and 5D). These results suggest that the 5′-UTR of Hat1 mRNA is not involved in the translational regulation by polyamines. Since it was previously reported that formation of the initiation complex consisting of mRNA, Met-tRNAi, and 40S ribosomal subunits was enhanced by spermidine, Hat1 synthesis may be stimulated due to the enhancement of initiation complex formation by polyamines (22).
Figure 5.
Identification of polyamine acting sites on Gcn5 and Hat1 mRNAs. WT and 5′-UTR-deleted Gcn5-EGFP and Hat1-EGFP fusion plasmids were transfected into NIH3T3 cells and cultured in the presence and absence of 500 μm DFMO for 72 h. Protein levels were analyzed by Western blotting using anti-EGFP antibody (Clontech). ** p < 0.01; *** p < 0.001.
In the case of Gcn5-EGFP (Figs. 5A and 5B), synthesis of fusion protein from the WT construct was stimulated 2.7-fold by polyamines, similarly to FM3A cells, and the deletion of –55 to –63, –21 to –54 and –6 to 24 nucleotides of the 5′-UTR of mRNA decreased the degree of stimulation from 2.7-fold to 1.9-, 1.2-, and 1.5-fold, respectively. These results suggest that –21 to –54 nucleotides in the 5′-UTR of Gcn5 mRNA are involved in the polyamine stimulation of Gcn5 synthesis. When this region was removed from Gcn5 mRNA, protein synthetic activity in polyamine-reduced cells increased more than 3-fold, whereas polyamine stimulation almost disappeared.
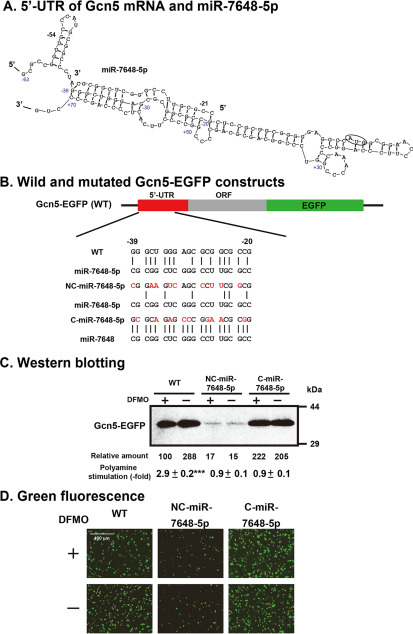
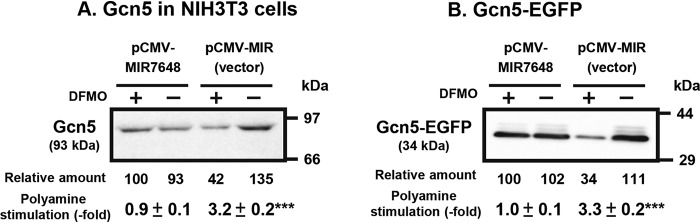
The mechanism of polyamine stimulation of Gcn5 synthesis was studied further. As shown in Fig. 6A, the –21 to –39 nucleotides of the 5′-UTR can interact with the +50 to +70 nucleotides in the ORF of Gcn5 mRNA. This suggests that the scanning of the initiation complex consisting of Met-tRNAi-40S ribosomal subunits may be perturbed. Among microRNAs in mammalian cells, there is a miR-7648-5p, which interacts with the –20 to –39 nucleotides of the 5′-UTR of Gcn5 mRNA (Fig. 6A and 6B). To study the role of miR-7648-5p, two forms of mutated 5′-UTR of Gcn5-EGFP mRNAs were constructed: one interacts more strongly with miR-7648-5p, i.e. C-miR-7648-5p-Gcn5-EGFP mRNA, and the other interacts more weakly with miR-7648-5p, i.e. NC-miR-7648-5p-Gcn5-EGFP mRNA (Fig. 6B). When the NC-miR-7648-5p-Gcn5-EGFP construct was transfected into NIH3T3 cells, Gcn5-EGFP synthesis was greatly reduced with or without polyamines (Fig. 6C and 6D). In contrast, Gcn5-EGFP synthesis was increased ∼2.2-fold in polyamine-reduced cells compared with the WT construct when the C-miR-7648-5p-Gcn5-EGFP construct was transfected into NIH3T3 cells. Accordingly, polyamine stimulation disappeared. From these results, it was hypothesized that miR-7648-5p binds to the 5′-UTR of Gcn5 mRNA and stimulates its translation. To test this hypothesis, microRNA expression plasmid pCMV-MIR7648 or its vector was transfected into NIH3T3 cells for overexpression of miR-7648-5p. As shown in Fig. 7A, polyamine stimulation of Gcn5 synthesis disappeared, although Gcn5 synthesis was enhanced by miR-7648-5p in DFMO-treated, polyamine-reduced cells. In contrast, when NIH3T3 cells were transfected with the vector, Gcn5 synthesis was stimulated 3.2-fold by polyamines, similarly to FM3A cells. In addition, when the pGcn5-EGFP fusion plasmid and pCMV-MIR7648 were cotransfected into NIH3T3 cells, the same results were obtained (Fig. 7B). These results indicate that scanning of the initiation complex was stimulated by weakening the intramolecular interaction between the –21 to –39 and the +50 to +70 nucleotides of Gcn5 mRNA, due to the interaction between the –20 to –39 nucleotides of Gcn5 mRNA and miR-7648-5p and that polyamines enhance the interaction between the –20 to –39 segment of Gcn5 mRNA and miR-7648-5p.
Figure 6.
Involvement of miR-7648-5p on the polyamine stimulation of Gcn5 synthesis. (A) Initiation region of Gcn5 mRNA (–63 to +70) and miR-7648-5p sequences are shown. (B) WT and 5′-UTR mutated Gcn5-EGFP constructs. (C, D) Expression levels of WT and mutated Gcn5-EGFP fusion proteins were analyzed by fluorescence microscopy and by Western blotting using lysates of NIH3T3 cells cultured in the presence and absence of 500 μm DFMO. *** p < 0.001.
Figure 7.
Effect of miR-7648-5p on polyamine stimulation of Gcn5 synthesis. (A) MicroRNA expression plasmid pCMV-MIR7648 and its vector were transfected into NIH3T3, and the effect of polyamines was examined using DFMO-treated and untreated cells. (B) MicroRNA expression plasmid pCMV-MIR7648 and its vector were cotransfected with pGcn5-EGFP into NIH3T3 cells. Protein levels of Gcn5 and Gcn5-EGFP were determined by Western blotting as described under “Experimental Procedures” using 20 μg of cell lysate, and the ratio of control/DFMO-treated cells was shown. *** p < 0.001.
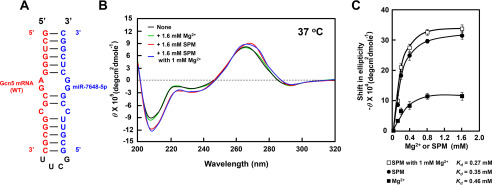
The effect of polyamines on the interaction between the –20 to –39 nucleotides of Gcn5 mRNA and miR-7648-5p was examined by CD using a synthetic RNA consisting of the –37 to –22 segment of the 5′-UTR of Gcn5 mRNA connected with the 3 to 18 nucleotides of miR-7648-5p in between four nucleotides. This RNA can make a dsRNA with a bulged-out region, which can be recognized by polyamines (Fig. 8A) (23). A substantial increase in the relative intensity of the negative band at 208 nm in CD reflects stabilization (or an increase) of the A-form dsRNA (24). Although this interaction was not enhanced by Mg2+, it was strongly enhanced by spermine in the presence and absence of Mg2+ (Fig. 8C). Similar results were obtained with spermidine. The results suggest that polyamines enhance the interaction between the 5′-UTR of Gcn5 mRNA with miR-7648-5p, resulting in stimulation of translation of Gcn5 mRNA.
Figure 8.
CD analysis of the complex between the 5′-UTR of Gcn5 mRNA and miR-7648-5p in the presence and absence of spermine. (A) Structures of Gcn5-miR-7648-5p fusion RNA. (B) CD spectra were recorded as described under “Experimental Procedures.” (C) Concentration-dependent shifts induced by Mg2+ (■), spermine (●), and spermine with 1 mm Mg2+ (□) at 37 °C in magnitude at 208 nm are shown. Values are means ± S.E. of triplicate determinations. The Kd values were determined according to the double reciprocal equation plot.
Discussion
In this study, to elucidate a mechanism of regulation of gene expression by polyamines in eukaryotes, we looked for new members of the polyamine modulon, which are involved in transcription, because many members of the polyamine modulon are transcription factors in E. coli. However, there was no polyamine modulon among the transcription factors tested. Thus, we started to study the effects of polyamines on the epigenetic control of gene expression. It was found that synthesis of two histone acetyltransferases, Gcn5 and Hat1, was stimulated by polyamines at the level of translation. HATs play an important role in chromatin remodeling and gene expression in eukaryotes and perform both global and specific histone acetylation, as well as acetylation of nonhistone proteins such as transcriptional factors (25, 26). Gcn5 was originally characterized as a transcriptional coactivator, which is thought to be recruited to regulatory gene regions where it contributes to gene activation by acetylation of histones. We found that the acetylation of histone lysine residues (H3 K9, K14, K18, and K27 and H4 K8) that can be acetylated by Gcn5 was increased by polyamines (Fig. 3). In addition, the synthesis of several mRNAs which are important for cell proliferation is stimulated by polyamines (Fig. 4). Hat1 plays an important role in chromatin assembly and DNA damage repair through acetylation of histones (27, 28). Furthermore, it has been reported that polyamines promote DNA damage repair (29) and that activation of p53 by spermine mediates induction of autophagy (30). Our results also strongly suggest that polyamines are involved in cellular processes such as DNA damage repair and transcription activation by stimulation of Gcn5 and Hat1 synthesis.
The mechanism of polyamine stimulation of synthesis of the two proteins was then studied. In the case of Hat1 synthesis, the size of the 5′-UTR of mRNA was short, i.e. 38 nucleotides, and the 5′-UTR of Hat1 mRNA was not involved in polyamine stimulation of Hat1 synthesis. In the case of Gcn5 synthesis, a microRNA was involved in the polyamine stimulation. Usually, most microRNAs are involved in the inhibition of protein synthesis (31–33). Thus far, there are several reports in which microRNAs are involved in the stimulation of protein synthesis (34–36). Among these reports, both miR-122 (34) and microRNA-10a (36) function at the 5′-UTR of mRNAs, and other microRNAs (miR369-3 and micro RNAs-Let7 (35)) function at the 3′-UTR of mRNAs. MicroRNA-122 stimulates HCV (hepatitis C virus) translation by enhancing the association of ribosomes with the viral RNA at an early initiation stage (34). MicroRNA-10a also functions at the 5′-end of mRNAs (36). There is no report on miR-7648-5p, but we suggest that the function of miR-7648-5p is unique. This microRNA interacted with the 5′-UTR of Gcn5 mRNA and disturbed the intramolecular interaction between the 5′-UTR and ORF of the Gcn5 mRNA. As a result, microRNA miR-7648-5p enhanced translation of Gcn5 mRNA. Polyamines, especially spermine, strongly contribute to the enhancement of the interaction between miR-7648-5p with the 5′-UTR of Gcn5 mRNA, because the interaction of the bulged-out region of dsRNA is enhanced by spermine (Fig. 8) (23).
Thus far, we have identified nine members of the mammalian polyamine modulon. To clarify the function of polyamines in eukaryotes, further identification of members of the polyamine modulon are necessary. We expect that more than 20 members of the polyamine modulon exist, since the gene numbers of eukaryotic cells are much greater than those in E. coli. Through the identification of the polyamine modulon, the function of polyamines in cell proliferation and viability will be clarified like with E. coli (10).
We demonstrated that polyamines regulate gene expression through stimulation of the synthesis of Gcn5 and Hat1. Since our results concern only one form of regulation of gene expression by polyamines, further study is necessary to assess other histone modifications.
Experimental Procedures
Cell culture of mouse mammary carcinoma FM3A cells and preparation of cell lysate
FM3A cells (Japan Health Science Foundation) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Wako) supplemented with 2% heat-inactivated fetal bovine serum (FBS) at 37 °C for 72 h in an atmosphere of 5% CO2 in air. To make polyamine-reduced FM3A cells, 50 µm DFMO was added to the medium. Control and polyamine-reduced FM3A cells (2 × 106 cells) were treated with 200 μl of 5% (w/v) TCA, and centrifuged at 12,000 × g for 10 min. The precipitate was dissolved with 100 μl of a buffer containing 25 mm Tris-HCl, pH 6.8, 1% 2-mercaptoethanol, 5% glycerol, and 1% SDS, and the pH of the homogenate was adjusted to pH 6.8 with 1 M Tris-HCl, pH 8.0. After standing at room temperature overnight, supernatant was obtained by centrifugation at 17,000 × g for 15 min and used as cell lysate. Protein content was determined using the method of Bradford (37).
Western blotting analysis
Western blotting analysis was performed with the method of Nielsen et al. (38), using horseradish peroxidase conjugated anti-rabbit IgG (GE Healthcare Bio-Sciences) as the secondary antibody and ECL Western blotting reagents (GE Healthcare Bio-Sciences). Antibodies against Gcn5, Hat1, p300, Tip60, MOZ, and β-actin were obtained from Santa Cruz Biotechnology; those against Atf-2, CBP, Pcaf, Clock, SRC-1, ELP3, TFIIB, and acetylated-lysine, the acetyl-histone H3 antibody sampler kit (H3, H3K9ac, H3K14ac, H3K18ac, and H3K27ac), and the acetyl-histone H4 antibody sampler kit (H4, H4K5ac, H4K8ac, and H4K12ac) were from Cell Signaling Technology; and that against Morf was from Abcam. The level of histone acetyltransferase was analyzed using 20 μg cell lysate and the above antibodies. Histones were extracted using a total histone extraction kit (EpiGentek) according to the accompanying manual, and 2 μg histones were separated by 15% SDS-PAGE. The level of acetylation of histones was analyzed using the above-mentioned acetyl-histone H3 and H4 antibody sampler kits. The level of protein on the blot was quantified with a LAS-3000 luminescent image analyzer (Fujifilm).
Assays for histone acetyltransferases
Nuclear and cytosol fractions of FM3A cells were prepared using Exsubcell extract (ATTO), and the activities of histone acetyltransferases were measured using these fractions and EpiQuickTM HAT activity/inhibition assay kit (EpiGentek) according to the accompanying manuals.
RNA isolation and measurement of the level of mRNA by semiquantitative or quantitative real-time PCR
Total RNA was extracted from control and DFMO-treated FM3A cells cultured for 72 h with TRIzol reagent (Thermo Fisher Scientific). Complementary DNA was synthesized using a cDNA synthesis kit (ReverTra-PlusTM; Toyobo), and a semiquantitative real-time PCR was performed using EmeraldAmp®Max PCR master mix (TaKaRa) at 95 °C for 5 min; 25 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s; and 72 °C for 5 min according to the accompanying manuals. The PCR product was separated by agarose-gel electrophoresis and stained with ethidium bromide, and band intensity was quantified using a LAS-3000 luminescent image analyzer (Fujifilm). The PCR primers used are P1 and P2 for Gcn5 mRNA (Gene ID: 107435), P3 and P4 for Hat1 mRNA (Gene ID: 14534), P5 and P6 for β-actin, P7 and P8 for Ki-67 mRNA, P9 and P10 for Lyar mRNA, P11 and P12 for Pcna mRNA, P13 and P14 for MCM-2 mRNA, and P15 and P16 for PRL30 mRNA (ribosomal protein L30 mRNA) (Table S1). Quantitative real-time PCR was performed using these primers, the SYBR green master mix, and an Applied Biosystems® 7500 real-time PCR system. mRNA levels were normalized using β-actin mRNA as a control.
ChIP assay
A ChIP assay was performed using a SimpleChIP enzymatic chromatin IP kit (magnetic beads) (Cell Signaling Technology) (39, 40). FM3A cells were cultured in the presence and absence of 50 µm DFMO for 72 h. Cells were fixed with formaldehyde, and chromatin was digested with micrococcal nuclease into 150–900-bp DNA/protein fragments. Then, complexes of histones H3 and H4 and bound DNA were immunoprecipitated with the above-mentioned acetyl-histone H3 antibody sampler kit (H3, H3K9ac, H3K14ac, H3K18ac, and H3K27ac) and acetyl-histone H4 antibody sampler kit (H4, H4K5ac, H4K8ac, and H4K12ac). The acetylated histone-DNA complexes were collected using protein G magnetic beads. PCR was carried out for amplification of promoter regions of genes for Ki-67 (primers P17 and P18), Lyar (primers P19 and P20), Pcna (primers P21 and P22), MCM-2 (primers P23 and P24), and RPL30 (primers provided with the ChIP kit) as a control. PCR products were analyzed as described above.
Plasmids
To make plasmid pGcn5-EGFP(WT), PCR was performed using primers P25 and P26 (Table S1) and the above-described synthesized cDNA as templates. The amplified Gcn5 cDNA (a 63-nucleotide 5′-UTR and a 207-nucleotide ORF) was digested with EcoRI and BamHI and inserted into the same restriction sites of plasmid pEGFP-N1 (Clontech). To make plasmid pGcn5-EGFP(Δ-55-63), PCR was performed using primers P26 and P27 and pGcn5-EGFP(WT) as templates, and PCR product was digested with EcoRI and BamHI and inserted into the same restriction sites of plasmid pEGFP-N1. Plasmids pGcn5-EGFP(Δ-21-54), pGcn5-EGFP(Δ-6-24), pGcn5-EGFP(NC-miR-7648-5p), and pGcn5-EGFP(C-miR-7648-5p) were prepared by site-directed mutagenesis employing overlap extension using PCR (41), and the first PCR primer sets are P25 and P29:28 and P26 for pGcn5-EGFP(Δ-21-54), P25 and P31: P30 and P26 for pGcn5-EGFP(Δ-6-24), P25 and P33:P32 and P26 for pGcn5-EGFP(NC-miR-7648-5p), and P25 and P35:P34 and P26 for pGcn5-EGFP(C-miR-7648-5p). pGcn5-EGFP(WT) was used as a template. Primers P25 and P26 were used for second PCR. The second PCR product was digested with EcoRI and BamHI and inserted into the same restriction sites of plasmid pEGFP-N1. Plasmids pHat1-EGFP(WT) (a 38-nucleotide 5′-UTR and a 96-nucleotide ORF) and pHat1-EGFP(Δ-28-38) were similarly constructed using primers P36 and P37, and P38 and P37, respectively. Primers used for pHat1-EGFP(Δ-2-30) are P36 and P40:P39 and P37 for the first PCR and P36 and P37 for the second PCR. The nucleotide sequence of the plasmids was confirmed with the 3130 genetic analyzer (Applied Biosystems) using P41 as a primer.
MicroRNA Expression Plasmid pCMV-MIR7648 and its vector were purchased from ORIGENE.
Transient transfection of fusion plasmids into NIH3T3 cells and measurement of their protein levels
NIH3T3 cells (3 × 105/10 ml) were cultured in D-MEM supplemented with 50 units/ml streptomycin, 100 units/ml penicillin G, and 10% FBS at 37 °C in an atmosphere of 5% CO2 in air for 36 h. Then, cells were cultured in the presence and absence of 500 µm DFMO for 12 h. After changing the medium with a fresh one without FBS, cells were transfected with 4 µg of various fusion plasmids using LipofectamineTM 2000 reagents (Thermo Fisher Scientific) according to the manufacturer's instructions and cultured for 3 h. After changing the medium with a fresh one containing FBS, cells were cultured in the presence and absence of 500 µm DFMO for a further 24 h. Prior to harvesting of NIH3T3 cells, images were captured with an EVOS®FL cell imaging system (Thermo Fisher Scientific) containing a GFP filter. NIH3T3 cells attached to the culture dish were washed twice with 5 ml of PBS, and incubated with 0.4 ml of 0.25% trypsine-0.02% EDTA-4Na solution at 37 °C for 3 min, and 5 ml of D-MEM containing 10% FBS was added to the culture dish. Dispersed cells were collected by centrifugation at 300 × g for 5 min, washed twice with PBS, and used for Western blotting as described above.
Search for miR
MicroRNA was searched using miRBase (RRID:SCR_017497).
CD measurement of RNA
The 36 nucleotides containing interaction sites of Gcn5 mRNA and miR-7648-5p (5′-GCUGGGAGCGCGGCGCUUCGGCGUUCCGGGCUCGGC-3′) were obtained from Hokkaido System Science, Japan. CD spectra were recorded over 200–300 nm on a Jasco J-820 spectropolarimeter (Jasco International Co.) using a 0.1-cm path-length cuvette at 37 °C (24). Scan speed was 100 nm/min, and CD samples contained 10 mm Tris-HCl (pH 7.5), 50 mm KCl, and 50 µm RNA. Where indicated, Mg2+ and/or spermine were added to the CD samples. Typical spectra at 37 °C corresponded to the average of three scans.
Statistics
Values are indicated as the mean ± S.E. of triplicate determinations. Data of control and DFMO-treated groups were analyzed by Student's t test, and a statistically significant difference was shown by probability values.
Data availability
All data are contained within the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. A. J. Michael for his help in preparing the manuscript.
This article contains supporting information.
Author contributions—A. S. formal analysis; A. S. funding acquisition; A. S. and Y. T. investigation; A. S. writing-original draft; Y. T., T. U., K. I., and K. K. supervision; Y. T. and T. U. validation; K. I. and K. K. writing—review and editing.
Funding and additional information—This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (19K16357).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- HAT
- histone acetyltransferase
- ChIP
- chromatin immunoprecipitation
- DFMO
- α-difluoromethylornithine
- HDAC
- histone deacetylase
- 5′-UTR
- 5′-untranslated region.
References
- 1. Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 10.1038/38664 [DOI] [PubMed] [Google Scholar]
- 2. Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 3. Di Martile M., Del Bufalo D., and Trisciuoglio D. (2016) The multifaceted role of lysine acetylation in cancer: prognostic biomarker and therapeutic target. Oncotarget 7, 55789–55810 10.18632/oncotarget.10048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Farria A., Li W., and Dent S. Y. (2015) KATs in cancer: functions and therapies. Oncogene 34, 4901–4913 10.1038/onc.2014.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trisciuoglio D., Di Martile M., and Del Bufalo D. (2018) Emerging role of histone acetyltransferase in stem cells and cancer. Stem Cells Int. 2018, 1–11 10.1155/2018/8908751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Igarashi K., and Kashiwagi K. (2010) Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 42, 39–51 10.1016/j.biocel.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 7. Igarashi K., and Kashiwagi K. (2015) Modulation of protein synthesis by polyamines. IUBMB Life 67, 160–169 10.1002/iub.1363 [DOI] [PubMed] [Google Scholar]
- 8. Pegg A. E. (2009) Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894 10.1002/iub.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Igarashi K., and Kashiwagi K. (2006) Polyamine modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J. Biochem. 139, 11–16 10.1093/jb/mvj020 [DOI] [PubMed] [Google Scholar]
- 10. Igarashi K., and Kashiwagi K. (2018) Effects of polyamines on protein synthesis and growth of Escherichia coli. J. Biol. Chem. 293, 18702–18709 10.1074/jbc.TM118.003465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Igarashi K., and Kashiwagi K. (2019) The functional role of polyamines in eukaryotic cells. Int. J. Biochem. Cell Biol. 107, 104–115 10.1016/j.biocel.2018.12.012 [DOI] [PubMed] [Google Scholar]
- 12. Nishimura K., Okudaira H., Ochiai E., Higashi K., Kaneko M., Ishii I., Nishimura T., Dohmae N., Kashiwagi K., and Igarashi K. (2009) Identification of proteins whose synthesis is preferentially enhanced by polyamines at the level of translation in mammalian cells. Int. J. Biochem. Cell Biol. 41, 2251–2261 10.1016/j.biocel.2009.04.021 [DOI] [PubMed] [Google Scholar]
- 13. Terui Y., Sakamoto A., Yoshida T., Kasahara T., Tomitori H., Higashi K., Igarashi K., and Kashiwagi K. (2015) Polyamine stimulation of eEF1A synthesis based on the unusual position of a complementary sequence to 18S rRNA in eEF1A mRNA. Amino Acids 47, 345–356 10.1007/s00726-014-1867-z [DOI] [PubMed] [Google Scholar]
- 14. Imamura M., Higashi K., Yamaguchi K., Asakura K., Furihata T., Terui Y., Satake T., Maegawa J., Yasumura K., Ibuki A., Akase T., Nishimura K., Kashiwagi K., Linhardt R. J., Igarashi K., et al. (2016) Polyamines release the let-7b-mediated suppression of initiation codon recognition during the protein synthesis of EXT2. Sci. Rep. 6, 33549 10.1038/srep33549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamaguchi K., Asakura K., Imamura M., Kawai G., Sakamoto T., Furihata T., Linhardt R. J., Igarashi K., Toida T., and Higashi K. (2018) Polyamines stimulate the CHSY1 synthesis through the unfolding of the RNA G-quadruplex at the 5′-untraslated region. Biochem. J. 475, 3797–3812 10.1042/BCJ20180672 [DOI] [PubMed] [Google Scholar]
- 16. Lee K. K., and Workman J. L. (2007) Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 10.1038/nrm2145 [DOI] [PubMed] [Google Scholar]
- 17. Peterson C. L., and Laniel M. A. (2004) Histones and histone modifications. Curr. Biol. 14, R546–R551 10.1016/j.cub.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 18. Scholzen T., and Gerdes J. (2000) The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 182, 311–322 [DOI] [PubMed] [Google Scholar]
- 19. Su L., Hershberger R. J., and Weissman I. L. (1993) LYAR, a novel nucleolar protein with zinc finger DNA-binding motifs, is involved in cell growth regulation. Genes Dev. 7, 735–748 10.1101/gad.7.5.735 [DOI] [PubMed] [Google Scholar]
- 20. Maga G., and Hubscher U. (2003) Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116, 3051–3060 10.1242/jcs.00653 [DOI] [PubMed] [Google Scholar]
- 21. Jurikova M., Danihel L., Polak S., and Varga I. (2016) Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 118, 544–552 10.1016/j.acthis.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 22. Shimogori T., Kashiwagi K., and Igarashi K. (1996) Spermidine regulation of protein synthesis at the level of initiation complex formation of Met-tRNAi, mRNA and ribosomes. Biochem. Biophys. Res. Commun. 223, 544–548 10.1006/bbrc.1996.0931 [DOI] [PubMed] [Google Scholar]
- 23. Higashi K., Terui Y., Inomata E., Katagiri D., Nomura Y., Someya T., Nishimura K., Kashiwagi K., Kawai G., and Igarashi K. (2008) Selective structural change of bulged-out region of double-stranded RNA containing bulged nucleotides by spermidine. Biochem. Biophys. Res. Commun. 370, 572–577 10.1016/j.bbrc.2008.03.137 [DOI] [PubMed] [Google Scholar]
- 24. Nakano S., Kanzaki T., and Sugimoto N. (2004) Influences of ribonucleotide on a duplex conformation and its thermal stability: study with the chimeric RNA-DNA strands. J. Am. Chem. Soc. 126, 1088–1095 10.1021/ja037314h [DOI] [PubMed] [Google Scholar]
- 25. Nagy Z., and Tora L. (2007) Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26, 5341–5357 10.1038/sj.onc.1210604 [DOI] [PubMed] [Google Scholar]
- 26. Narita T., Weinert B. T., and Choudhary C. (2019) Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174 10.1038/s41580-018-0081-3 [DOI] [PubMed] [Google Scholar]
- 27. Parthun M. R. (2007) Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene 26, 5319–5328 10.1038/sj.onc.1210602 [DOI] [PubMed] [Google Scholar]
- 28. Yang X., Li L., Liang J., Shi L., Yang J., Yi X., Zhang D., Han X., Yu N., and Shang Y. (2013) Histone acetyltransferase 1 promotes homologous recombination in DNA repair by facilitating histone turnover. J. Biol. Chem. 288, 18271–18282 10.1074/jbc.M113.473199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee C. Y., Su G. C., Huang W. Y., Ko M. Y., Yeh H. Y., Chang G. D., Lin S. J., and Chi P. (2019) Promotion of homology-directed DNA repair by polyamines. Nat. Commun. 10, 65 10.1038/s41467-018-08011-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chae Y. B., and Kim M. M. (2014) Activation of p53 by spermine mediates induction of autophagy in HT1080 cells. Int. J. Biol. Macromol. 63, 56–63 10.1016/j.ijbiomac.2013.10.041 [DOI] [PubMed] [Google Scholar]
- 31. Pillai R. S., Bhattacharyya S. N., and Filipowicz W. (2007) Repression of protein synthesis by miRNAs: how many mechanisms?. Trends Cell Biol. 17, 118–126 10.1016/j.tcb.2006.12.007 [DOI] [PubMed] [Google Scholar]
- 32. Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A., and Lund A. H. (2008) Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 283, 1026–1033 10.1074/jbc.M707224200 [DOI] [PubMed] [Google Scholar]
- 33. Gironella M., Seux M., Xie M. J., Cano C., Tomasini R., Gommeaux J., Garcia S., Nowak J., Yeung M. L., Jeang K. T., Chaix A., Fazli L., Motoo Y., Wang Q., Rocchi P., et al. (2007) Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. U. S. A. 104, 16170–16175 10.1073/pnas.0703942104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henke J. I., Goergen D., Zheng J., Song Y., Schuttler C. G., Fehr C., Junemann C., and Niepmann M. (2008) microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27, 3300–3310 10.1038/emboj.2008.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vasudevan S., Tong Y., and Steitz J. A. (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 36. Orom U. A., Nielsen F. C., and Lund A. H. (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- 37. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 38. Nielsen P. J., Manchester K. L., Towbin H., Gordon J., and Thomas G. (1982) The phosphorylation of ribosomal protein S6 in rat tissues following cycloheximide injection, in diabetes, and after denervation of diaphragm. A simple immunological determination of the extent of S6 phosphorylation on protein blots. J. Biol. Chem. 257, 12316–12321 [PubMed] [Google Scholar]
- 39. Orlando V. (2000) Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25, 99–104 10.1016/S0968-0004(99)01535-2 [DOI] [PubMed] [Google Scholar]
- 40. Kuo M. H., and Allis C. D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19, 425–433 10.1006/meth.1999.0879 [DOI] [PubMed] [Google Scholar]
- 41. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., and Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.