Abstract
Myelin protein P2 is a peripheral membrane protein of the fatty acid–binding protein family that functions in the formation and maintenance of the peripheral nerve myelin sheath. Several P2 gene mutations cause human Charcot-Marie-Tooth neuropathy, but the mature myelin sheath assembly mechanism is unclear. Here, cryo-EM of myelin-like proteolipid multilayers revealed an ordered three-dimensional (3D) lattice of P2 molecules between stacked lipid bilayers, visualizing supramolecular assembly at the myelin major dense line. The data disclosed that a single P2 layer is inserted between two bilayers in a tight intermembrane space of ∼3 nm, implying direct interactions between P2 and two membrane surfaces. X-ray diffraction from P2-stacked bicelle multilayers revealed lateral protein organization, and surface mutagenesis of P2 coupled with structure-function experiments revealed a role for both the portal region of P2 and its opposite face in membrane interactions. Atomistic molecular dynamics simulations of P2 on model membrane surfaces suggested that Arg-88 is critical for P2-membrane interactions, in addition to the helical lid domain. Negatively charged lipid headgroups stably anchored P2 on the myelin-like bilayer surface. Membrane binding may be accompanied by opening of the P2 β-barrel structure and ligand exchange with the apposing bilayer. Our results provide an unprecedented view into an ordered, multilayered biomolecular membrane system induced by the presence of a peripheral membrane protein from human myelin. This is an important step toward deciphering the 3D assembly of a mature myelin sheath at the molecular level.
Keywords: membrane structure, protein structure, myelin, molecular dynamics, membrane biophysics, membrane protein, membrane bilayer, Charcot-Marie-Tooth disease, neuropathy, proteolipid
Introduction
A central question in myelin biology is the molecular mechanism of the tight packing of dozens of apposing lipid bilayers into a mature, multilayered myelin sheath. A major role in this process is played by myelin-specific proteins. The high degree of order within the myelin sheath has been known since early experiments using X-ray diffraction (1); however, the details of the molecular assembly have remained enigmatic.
The spontaneous formation of lipid membrane multilayers is a common functional property of different myelin-specific proteins, which are not genetically related. In peripheral nervous system (PNS)7 myelin, the compact multilamellar membrane contains only a few proteins. The intrinsically disordered myelin basic protein (MBP) is irreversibly embedded into a single leaflet of the lipid bilayer (2). The cytoplasmic domain of myelin protein zero (P0) behaves much like MBP, although it embeds deeper into the membrane (3). Full-length P0 promotes membrane stacking through both extra- and intracellular interactions (3–6). Peripheral myelin protein 22 (PMP22), another PNS integral membrane protein, forms myelin-like assemblies (7), much like those observed with MBP and P0. P2 adheres to the cytoplasmic leaflet of the bilayer and can be classified as a peripheral membrane protein (8).
Peripheral membrane proteins associate with cellular membranes via diverse mechanisms. Membrane binding may be either irreversible, mediated by post-translational modifications (palmitoylation, myristoylation, or prenylation), or reversible with variable binding affinities. The specificity of protein-membrane interactions is affected by the physical properties of the protein and the lipid bilayer, such as surface charge or membrane curvature. Many peripheral membrane proteins utilize amphipathic helices or hydrophobic amino acids that penetrate into the hydrophobic bilayer core to form stable interactions with membranes (9).
P2 is a Schwann cell–specific protein expressed in the PNS myelin of tetrapods (10). Intriguingly, P2 is expressed in a mosaic fashion, not being present in all myelin sheaths (11, 12). This small β-barrel protein belongs to the family of fatty acid-binding proteins (FABPs). The bound fatty acid is enclosed inside the β barrel by a lid formed by two adjacent α helices (13–15); the opening of the β barrel may be of importance in fatty acid entry and egress (13). In addition to fatty acid binding, P2 can transfer lipids from/to membranes using a collisional transfer mechanism (16), as seen with several other FABPs (17–21). Besides fatty acids, P2 may bind cholesterol (14), which is abundant in the myelin membrane and essential for myelination (22). The tip of the α-helical lid is hydrophobic, whereas both ends of the β barrel present positively charged surfaces (14, 15), and these properties are likely important, when P2 stacks between two phospholipid bilayers.
Studies on P2-deficient mice revealed temporarily reduced motor nerve conduction velocity and altered lipid composition in PNS myelin. However, the overall PNS myelin structure remained normal (16). Further analyses on the mutant mice revealed that P2 has a role in remyelination of an injured PNS (23) and melanoma cell invasion (24). Five Charcot-Marie-Tooth 1 (CMT1) disease point mutations in human P2 have been discovered (25–29). Three CMT1-associated P2 protein variants have been characterized at the molecular level, showing altered fatty acid and lipid membrane binding properties. The most drastic CMT1 mutation, T51P, also reduced the membrane stacking capability of P2 (30). Overall, the stability of the mutant proteins was decreased, even though crystal structures indicated only minor structural changes compared with WT P2 (30).
In the current study, we incorporated human P2 into a model membrane multilayer system and visualized the myelin-like proteolipid structures using cryo-EM. P2-bicelle complexes were used for additional structural insights. We produced mutated forms of P2 to establish determinants of lipid bilayer and fatty acid binding and used atomistic molecular dynamics (MD) simulations to visualize the intimate interaction between P2 and a myelin bilayer. We show the spontaneous formation of an ordered, crystal-like lattice of P2 bound inside membrane multilayers and highlight factors that are important in this process, which involves a conformational change in the protein. The results provide a glimpse into the self-assembling properties of myelin proteins and lipid membranes, which are likely to be crucial for correct myelination in the vertebrate nervous system.
Results
Whereas the molecular composition of compact myelin is relatively simple, the arrangement of proteins within the membrane multilayers is to a large extent unknown. Here, we used the peripheral membrane protein P2 from the PNS myelin major dense line as a model system to study myelin-like membrane stack formation and structure. P2 interacts with lipid bilayers with high affinity (13, 15, 31–33). We explored its membrane binding characteristics, determinants, and dynamics more closely. The results provide further information on the molecular details of the major dense line in PNS myelin, as well as on CMT disease mechanisms linked to mutations in P2.
Arrangement of P2 in multilayered membrane stacks
P2 spontaneously binds lipid membranes together, as reflected by earlier studies using turbidimetry, simulation, and X-ray diffraction (15, 32). However, the molecular details of this phenomenon and the resulting supramolecular structure have remained elusive. Cryo-EM was used to follow membrane stacking and ordering of proteolipid components in multilayers induced by P2.
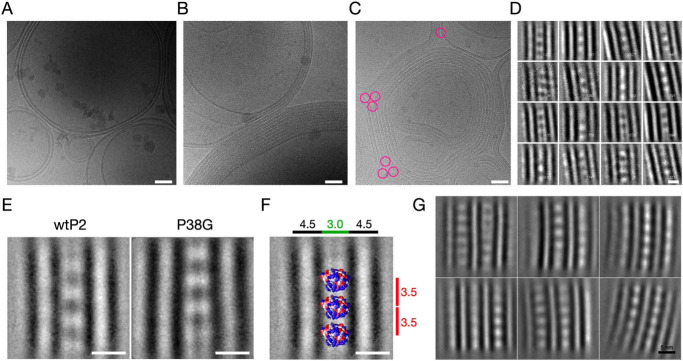
P2 induced the formation of highly ordered lipid bilayer stacks, whereas without P2, only unilamellar vesicles were observed (Fig. 1, A and B). The angle between two separating bilayers at the edge of a tight apposition is consistently <60° (Fig. 1C). Although P2 is only 15 kDa, it is visible in cryo-EM images as ordered rows of particles between two apposed membranes. Based on the calculated 2D class averages (Fig. 1, D–G), P2 evidently stabilizes the lipid membrane stacks and defines the spacing (∼3.0 nm) between two bilayer surfaces. This myelin-like spacing between two apposing lipid bilayers is constant throughout the membrane stacks. Based on the P2 crystal structure (14, 15), the longest diameter of P2 is 4.5 nm, indicating that either some parts of the protein are buried within the bilayer, or P2 is turned on its side on the membrane. The repeat distance in the multilayer, containing a 4.5-nm bilayer and a single layer of P2 molecules, is 7.5 nm. This is shorter than the distance measured in solution with X-ray diffraction under more hydrated conditions and close to the distance observed with MBP and the P0 cytoplasmic domain in diffraction experiments (2, 3, 15). P2 molecules are located between the bilayers with a lateral spacing of 3.5 nm between monomers (Fig. 1F), indicating lattice-like order between membranes. This order extends into neighboring membrane layers, and P2 molecules between the bilayers are at least to some extent in register between consecutive layers (Fig. 1G).
Figure 1.
Electron cryomicroscopic analysis of lipid membrane stacking by P2. A, cryo-EM image of E. coli polar lipid liposomes without protein. The images in A and B are 480 × 480 nm in size. B, the same liposomes in the presence of human P2 make myelin-like multilayered stacks. Lipid/protein mass ratio is 2.0. C, the angle between stacked membranes at the edges (pink circles) is nearly constant at ∼60°. D, 2D class averages of a single P2-linked bilayer stack. Lipid headgroups and proteins are black. The size of the box is 140 × 140 pixels (18 × 18 nm). E, averaged structures of stacked membranes with WT and P38G mutant human P2. Based on the observed features and distances, a resolution of 1–2 nm can be estimated. F, the space between two membranes is enough to fit one layer of P2. The membrane diameter is 4.5 nm, the space between membranes is 3.0 nm, and the distance between individual P2 molecules is 3.5 nm. The crystal structure of a P2 monomer has been fitted into the assembly. The crystal structure is at the correct scale; it should be noted that the exact orientation is unknown, and the view is for visual evaluation. G, averaging of larger segments of P2-stacked multilayers, including two layers in the analysis, indicates the lattice-like arrangement of P2 throughout the myelin-like multilayer. Scale bars, 50 nm (A–C) and 5 nm (D–G).
Cryo-EM was similarly carried out with the “hyperactive” P38G variant (34) mixed with lipids (Fig. 1E). Neither the bilayer spacing nor protein-lipid organization altered in the presence of the mutant. During sample preparation, P38G induced membrane aggregation/stacking faster than human WT P2 (wtP2), and the turbidity effect was visible within 2–3 min (not shown), in line with earlier experiments (34).
Three-dimensional order in P2-bicelle complexes
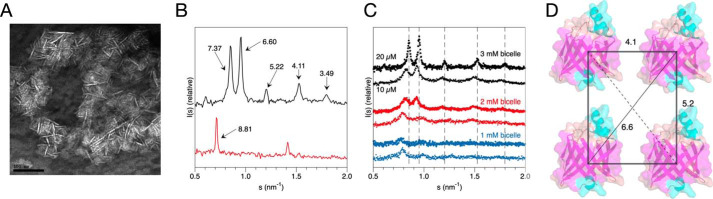
To obtain additional structural insight into P2-membrane complexes, P2 was studied in a bicelle environment. P2 induced turbidity in protein-bicelle suspensions, and EM imaging revealed stacked arrangements of bicelles in these samples (Fig. 2A). Thus, X-ray diffraction was used to gain more information on repetitive structures. In addition to the Bragg peaks originating from membrane stacking repeats of ∼7–8 nm, additional diffraction peaks were observed (Fig. 2, B and C) in samples with the highest lipid and protein concentrations. The corresponding distances are close to those expected from a lattice-like setup of P2 molecules between two membranes, as observed in cryo-EM. The distances can be used to deduce a possible lateral organization of P2 molecules in the membrane plane (Fig. 2D).
Figure 2.
Insights into P2 structure between membranes from bicelle complexes. A, negative staining EM micrograph of P2-stacked bicelles. Being flat lipid discs, the bicelles are here mainly seen as thin lines, corresponding to a side view. Scale bar, 100 nm. B, Bragg X-ray diffraction peaks from P2-stacked bicelles (black) and vesicles (red). The corresponding repeat distances are marked. C, titration of protein and lipid concentration in the bicelle samples indicates shorter distances and higher order when both protein and lipid concentrations increase. D, a model of P2 arrangement on the plane of the membrane, based on the peak positions in B. Note that the 7.37-nm peak corresponds to the bicelle membrane stack repeat distance, and the 3.49-nm distance could be a sign of an additional tighter arrangement of P2 on the membrane, because a 3.5-nm distance was seen in cryo-EM. For the vesicle sample in B, the 8.81-nm peak corresponds to the stack repeat between consecutive membrane layers, and no information is obtained on the lateral protein arrangement.
The distances observed in the experiment changed as a function of protein/lipid ratio. This behavior is similar, but not identical, to that observed for the P0 cytoplasmic domain, which caused tighter membrane packing at high protein/lipid ratios (3). For P2, both the lipid and protein concentration affect the repeat distance in a concerted fashion (Fig. 2C). The distances get shorter when lipid concentration increases, indicating an overall increase in order and tighter packing. On the other hand, at the same lipid concentration, shorter distances are observed with higher protein amounts. Hence, the protein and lipid components synergistically assemble into a compact, ordered, three-dimensional proteolipid structure.
Design of point mutants
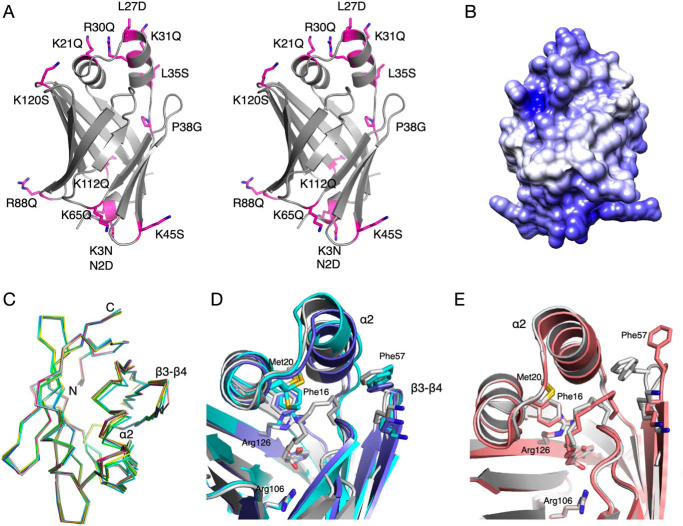
To elucidate structure-function relationships in P2, as a general model for a FABP with a collisional mechanism and tight interaction with membranes, we used the crystal structure of human P2 to design mutations that might affect membrane binding (Fig. 3A). The electrostatic surface of wtP2 shows two positively charged faces, at the helical lid domain and the bottom of the barrel structure (Fig. 3B). The mutations can be roughly divided into three classes: those removing positive surface charge, those affecting the hydrophobic surface of helix α2, and other mutations possibly affecting the portal region.
Figure 3.
Crystal structure analysis of selected P2 mutant variants. A, stereo view of all mutants analyzed. B, surface electrostatics of human P2 reveal two positively charged faces at opposite ends of the molecule. C, view from the top on the Cα traces of all P2 variant crystal structures indicates flexibility of helix α2 and the β3-β4 loop. D, conformational differences between liganded and unliganded P38G. P38G with palmitate (PDB entry 4D6B (34)) (gray) is superimposed with the two monomers of unliganded P38G (light and dark blue). E, partial opening of the portal in the K65Q variant (pink), superimposed on the wtP2 structure (PDB entry 4BVM (15)).
Crystal structures of P2 variants
For a high-resolution insight into the structure-function differences in the P2 variants, their respective crystal structures were solved (Table S1 and Fig. 3C). None of the mutations affected folding or secondary structure elements in the crystal state. With respect to this observation, it is important to note that the three studied CMT disease variants of P2 crystallized like wtP2, even though their stability and function were impaired (30). The root mean square deviations of the mutant structures compared with wtP2 vary between 0.08 and 0.36 Å, P38G being the most divergent.
Prior to the current work, all crystal structures for wtP2 or mutant P2 contained a bound ligand inside the β barrel. The P38G structure refined here is the first exception: its internal cavity is clearly empty; no electron density for a bound fatty acid is present. This allows comparison of details between liganded and unliganded P2 (Fig. 3D). In our earlier study, the P38G mutant contained bound palmitate (34). In the unliganded crystal structure of P38G, the amino acid side chains pointing inward mainly retain their conformation. The main-chain hydrogen bond between residue 38 and Leu-10 also exists in both P38G structures. However, helix α2 at the portal region has slightly shifted outward from helix α1 in both chains; a similar change is observed in the R30Q mutant, which could be linked to altered membrane-binding properties (see below). In addition, P38G electron density is poor for residues 33–37 at the end of helix α2 of chain B, supporting an increased flexibility/partial unfolding of the portal region in the P38G mutant in the absence of bound ligand, as seen in earlier simulations (34). Phe-57, as well as the whole β4-β5 loop of chain A, has somewhat tilted away from the α2 helix.
All P2 structures excluding P38G have a fatty acid, modeled as a mixture of palmitate or cis-vaccinate in the atomic-resolution structures (15, 35), bound inside the β barrel. The conformation and position of the fatty acid are similar in most structures. In the K65Q mutant, the conformation of the palmitate is different, and Phe-57 points outward in all four chains in the asymmetric unit (Fig. 3E). This supports the proposed role for Phe-57 as a gatekeeper residue in the FABP family (13, 36).
All human P2 crystal structures published thus far have an anionic group bound in proximity of the hinge region; the identity of the ligand depends on crystallization conditions and crystal contacts. In wtP2, either chloride or citrate interacts with Thr-56 and Lys-37 (14, 15). P38G and F57A contain chloride and sulfate, respectively (13, 34). In the CMT-associated P2 mutant structures, there is a malate located in the anion-binding site (30). In line with these data, all P2 mutant structures solved here have an anionic group bound in the same pocket. These observations lend further support to the hypothesis that this pocket may be involved in recognizing phospholipid headgroups and initiating membrane binding and/or conformational change (15).
Membrane binding and multilayer stacking
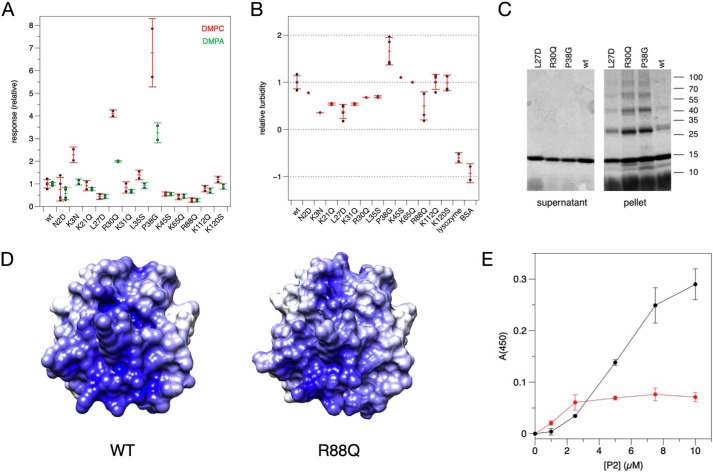
Surface plasmon resonance (SPR) was used to follow binding of the P2 variants onto immobilized lipid membranes, made of either dimyristoylphosphatidylcholine (DMPC) or dimyristoyl phospatidic acid (DMPA) (Fig. 4A). These membranes are net neutral and negatively charged, respectively. Whereas MBP essentially binds to lipids irreversibly on SPR (2), P2 dissociates from the membrane rapidly (15), suggesting different membrane interaction kinetics for these two proteins with overlapping function.
Figure 4.
Assays on P2 variant conformation and function. A, DMPC (red error bars) and DMPA (green error bars) membrane-binding assays by SPR. Each point represents the response at the highest concentration (1 μm) from a P2 concentration series, which was run 2–4 times for each variant. B, turbidity assay with 1:1 DMPC/DMPG vesicles. C, SDS-PAGE analysis of proteolipid pellets reveals SDS-resistant P2 multimers. D, surface electrostatics of the P2 bottom surface in wtP2 (left) and the R88Q mutant (right). E, turbidity assay of wtP2 with DMPC/DMPG vesicles (red) and bicelles (black). Error bars, S.D.
Four P2 mutants showed decreased binding to lipid membranes. One of these is L27D, which affects Leu-27 at the tip of the helical lid and reduces the hydrophobicity of the portal region. The other three mutations with reduced binding affinity toward lipid membranes are found in adjacent loops on the opposite face, at the bottom of the β barrel. All of these mutations (K45S, K65Q, and R88Q) affect surface residues and reduce the positive charge at the bottom of the β barrel. The locations of these mutations suggest two membrane-binding surfaces on opposite faces of P2, in line with its packing between two bilayers in vivo and in vitro.
Whereas some mutations caused diminished binding to the membrane surface, P38G and R30Q had increased levels of binding. These two mutations are located in the vicinity of the portal region and the helical lid domain. The difference in membrane binding of R30Q and P38G compared with wtP2 was more pronounced, when a DMPC membrane was studied.
Turbidimetry was used to assess the effectivity of P2 variants in aggregating DMPC/dimyristoylphosphatidylglycerol (DMPG) vesicles (Fig. 4B). When the turbid proteolipid suspensions were centrifuged and analyzed by SDS-PAGE, P2 co-sedimented with aggregated vesicles, and the strong proteolipid complex was only partially solubilized by SDS; P2 was present as a ladder of oligomeric forms (Fig. 4C). Again, P38G was the most effective variant, stacking vesicle membranes more than wtP2. Some mutations caused diminished turbidity compared with wtP2. The clearest of these were L27D and R88Q; the latter lies in the β6-β7 loop—in the middle of a large positively charged surface patch at the bottom of the β barrel (Fig. 4D). Hence, again, residues important for both membrane binding and stacking can be found on both positively charged faces of P2.
Another turbidimetric experiment was carried out to compare bicelles and vesicles in wtP2-induced multilayer formation. Like vesicles, bicelles are stacked by P2 into large structures causing turbidity (Fig. 4E). Such ordered complexes could be a step toward higher-resolution structure determination of myelin proteolipid complexes (e.g. due to restrained particle size and geometry).
Fatty acid and cholesterol binding
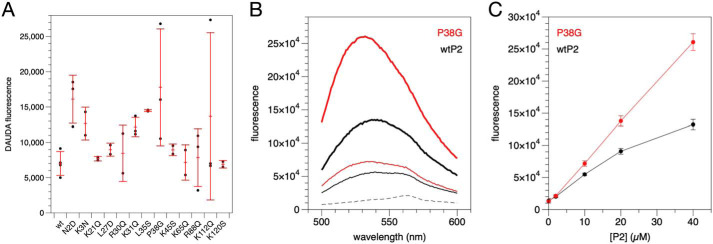
Using the fluorescent fatty acid analogue 11-dansylamino-undecanoic acid (DAUDA), we followed internal ligand binding to P2 variants (Fig. 5A). The situation is complicated by the fact that tightly bound fatty acids co-purify with P2 from the expression host. Thus, a quantitative analysis was not performed, as increased binding could reflect either higher affinity or lower amounts of co-purified ligand. However, the level of bound DAUDA should correlate with the opening of the portal region and/or the barrel, which is also required for removal of the bound fatty acid. Because bound fatty acid affects dynamics of P2 (13, 34), it is likely that some of the mutated variants have different affinities toward fatty acids. Most mutant variants showed slightly higher DAUDA signal than wtP2, and P38G was the strongest binder of all variants.
Figure 5.
Ligand binding by human P2. A, binding of the fluorescent fatty acid DAUDA. B, binding of NBD-cholesterol by wtP2 (black) and P38G (red). Dashed line, ligand alone; thin line, 10 μm P2; thick line, 40 μm P2. C, concentration dependence of fluorescence at 532 nm. Error bars, S.D.
We previously proposed cholesterol binding by P2 (14), in addition to fatty acids. Cholesterol binding was tested using wtP2 and P38G. wtP2 induced a clear, concentration-dependent change in the fluorescence spectrum of the environment-sensitive probe 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol (22-NBD-cholesterol); the fluorescence maximum shifted toward shorter wavelengths, and its intensity increased (Fig. 5, B and C). The spectral changes were more pronounced with P38G, which has a more flexible portal region (34). The experiment shows that cholesterol, which is very abundant in myelin, could be a physiologically relevant ligand for P2. These assays together indicate that protein flexibility is important when P2 binds to its biological ligands.
Folding and stability of point mutant variants
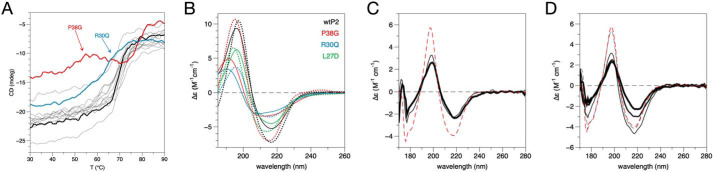
CD spectroscopy was used to analyze the folding and stability of the P2 variants. Whereas most mutations had little effect on thermal stability, P38G had two steps of unfolding, the first one appearing already at 50 °C and the second one only at >75 °C (Fig. 6A). Another outlier was R30Q, which had a slightly lowered stability compared with wtP2. Interestingly, these two mutations causing changes in stability are those with enhanced membrane binding and stacking properties. In the crystal structures, they present minor conformational differences in their helical lid, compared with wtP2.
Figure 6.
P2 stability and folding. A, melting curves for wtP2 and all studied mutants. The outliers are P38G (red) and R30Q (blue). Thick black line, wtP2. B, conformation of wtP2 and selected mutants in the presence (solid lines) and absence (dotted lines) of DMPC/DPC bicelles. C, WT P2 in water (red dashed line), 9:1 DMPC/DMPG (thin black line), and 1:1 DMPC/DMPG (thick black line). D, WT P2 in water (red dashed line) and lipid/DPC bicelles containing DMPC (black dashed line), 9:1 DMPC/DMPG (thin black line), 4:1 DMPC/DMPG (medium black line), and 1:1 DMPC/DMPG (thick black line).
To elucidate the conformational changes induced by membrane binding, synchrotron radiation CD (SRCD) spectra for wtP2 and some divergent mutants were measured in the presence and absence of DMPC/dodecylphosphocholine (DPC) bicelles (Fig. 6B). For wtP2, bicelle binding induced small changes in the SRCD spectrum. L27D exhibited less change in the spectrum in the presence of bicelles, supporting the reduced membrane binding of L27D observed in SPR and turbidity assays. On the other hand, P38G, having a higher propensity for membrane interactions, showed larger conformational changes in the bicelle environment. R30Q behaved much like P38G; both variants showed partial unfolding. These results highlight the importance of protein flexibility in membrane binding and indicate a role for the α-helical lid in P2-membrane interactions.
The bicelle system was used to deduce effects of lipid composition on wtP2 folding state, as well as to compare with vesicles with the same lipid composition. SRCD spectra showed that in vesicles, both 1:1 and 9:1 DMPC/DMPG gave the same conformational change of wtP2 compared with the protein in water (Fig. 6C). In bicelles, however, wtP2 behaved differently, in that very little change occurred in DMPC alone or in 9:1 DMPC/DMPG, and the spectrum changed more with 1:1 and 4:1 DMPC/DMPG, to resemble the one measured with vesicles (Fig. 6D). These differences with respect to lipid composition could be related to membrane curvature.
Atomistic simulations on P2-membrane interactions
To combine aspects of high-resolution structural data and membrane binding, we studied the interactions of WT and P38G P2 with membrane surfaces using atomistic MD simulations (Fig. 7). Two membrane systems were built: a 1:1 mixture of DMPC/DMPG, which corresponds to compositions often used in the laboratory, and a myelin-like membrane based on literature values (37).
Figure 7.
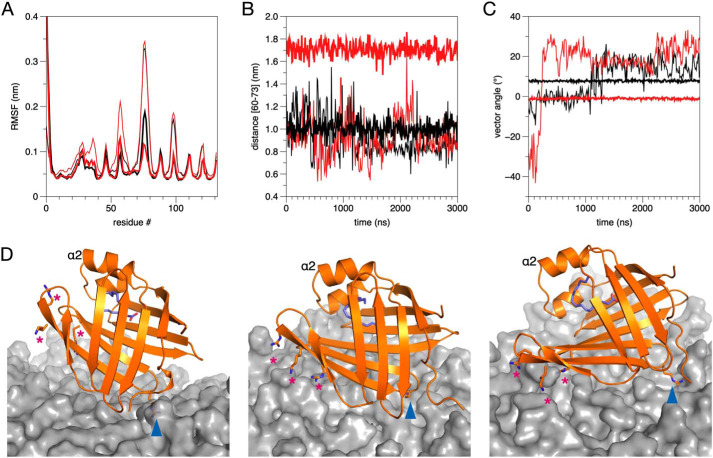
MD simulations on P2 binding to lipid membrane surface. A, root mean square fluctuation (RMSF) for wtP2 (black) and P38G (red) in 1:1 DMPC/DMPG (thin lines) and myelin lipid composition (thick lines). B, distance of the β4-β5 opening of the β barrel during the simulation. Coloring is as in A. C, angle of the P2 β barrel axis with respect to the membrane surface. Note how both wtP2 and P38G are rigidly anchored to the same orientation immediately after the equilibration period. D, snapshots from the simulations. Left, wtP2 on DMPC/DMPG at 1135 ns. Middle, wtP2 on myelin at 2060 ns. Right, P38G on myelin at 800 ns. Locations of the two membrane anchors, Arg-88 (blue arrowhead) and Arg-78/Lys-79/Arg-96 (magenta asterisks) are indicated.
During the attachment of wtP2 onto the membrane surface, a similar orientation was always observed: this involved the positively charged surface close to the bottom of the β barrel. Arg-88 is a central residue in initial P2-membrane interactions. Whereas it was expected that initial membrane binding would involve the portal region and the helical lid, this orientation, with the bottom face of the β barrel first approaching the membrane, is reproducible. The protein was further turned on its side in this arrangement in the myelin lipid composition, indicating that the rows of P2 molecules observed in cryo-EM images do not embed deep into the bilayers. The 3-nm spacing between membranes can accommodate one layer of P2 in this orientation.
A difference in orientation was observed between the DMPC/DMPG and myelin membranes; P2 remains more upright and dynamic in DMPC/DMPG, whereas it falls rigidly on its side on the myelin-like membrane (Fig. 7C). Differences in wtP2 dynamics were additionally observed between lipid compositions. The protein was more rigid when bound to the myelin membrane (Fig. 7A); on DMPC/DMPG, it had higher dynamics and hung on the membrane with the Arg-88 anchor (Fig. 7C). During the simulation with the myelin-like bilayer, the phosphatidylinositol 4,5-bisphosphate (PIP2) molecules within the myelin bilayer bound to the tip of the β5-β6 and β7-β8 loops, promoting opening of the β barrel, whereas Arg-88 at the other end of the protein, in the β6-β7 loop, interacted strongly with palmitoyloleoylphosphatidylserine (POPS) headgroups (Fig. S1 and Fig. 7D). The PIP2-binding site is formed of the side chains of Arg-78, Lys-79, and Arg-96. These results could reflect an important difference between a biological membrane composition and simplistic membrane models.
The electrostatic interactions of wtP2 and P38G were very similar with the membrane lipids during the simulations (Fig. S1). The P38G variant similarly attached to the myelin-like membrane surface, being anchored sideways, and opened up even more than wtP2 (Fig. 7, B and D). The portal region and the expected opening during ligand exchange (13) face upward in this setting, and upon the approach of another membrane, they could closely interact with its surface.
Discussion
Myelin protein P2 is a unique member of the FABP family, able to stack lipid bilayers together, in addition to being a member of the FABP subgroup carrying out collisional transfer. Lipid membrane binding by P2 involves the hydrophobic tip of the helical lid, electrostatic interactions, and dynamics of the portal region (13, 15, 34). Here, we have revealed details of the assembly of the P2-membrane stacks and the surprising role of the bottom region of the P2 β barrel in membrane binding. The data provide much-needed information on the assembly of the myelin membrane at the molecular level.
Structure of P2-induced proteolipid multilayers
Our cryo-EM experiments illustrate an organized lattice-like supramolecular three-dimensional arrangement of P2-membrane stacks. Surprisingly, P2, a 15-kDa protein, which has dimensions of 4.5 × 3.6 nm, is visible between the lipid bilayers as a lateral network. Both the cryo-EM images and calculated 2D class averages of P2-membrane stacks show a constant distance (3 nm) between the apposing lipid membranes and a repeat distance (containing a single bilayer and intermembrane space) of 7.5 nm. Earlier, a repeat distance of ∼9 nm for P2-membrane stacks was measured by X-ray diffraction using DMPC/DMPG in suspension (15). Bragg peaks in X-ray diffraction experiments support the highly organized arrangement of P2-membrane stacks seen in cryo-EM, and the conditions for preparing cryo-EM samples, with less hydration, might be more relevant to myelin in vivo. Indeed, using the bicelle model system, we measured repeat distances of 7.5 nm in stacks of bicelles induced by P2, and the distance evolved as a function of protein and lipid concentration. Thus, P2 may have a function in defining the membrane spacing in PNS compact myelin, together with MBP and P0. All three of these proteins produce membrane stacks in vitro (2, 3), with intermembrane spacing very close to that seen in the mature myelin major dense line.
The spacing between the neighboring P2 molecules between membrane bilayers is constant (∼3.5 nm), and there appears to be a relationship between the positioning of P2 molecules between consecutive membrane layers. The results suggest the presence of a near-crystalline lattice of P2 between membranes; this is also supported by our X-ray diffraction experiment using stacked bicelles, in which—unlike in earlier similar experiments using lipid vesicles—we see new distances much shorter than those coming from bilayer stacking per se. As these distances depend on protein concentration, they correspond to distances between proteins arranged as a lateral layer between two membranes. Whether such packing occurs in vivo depends on the local P2 concentration in myelin as well as the presence and organization of other highly abundant myelin proteins, such as P0 and MBP. The quantity of P2 has been reported to vary between different regions of PNS as well as from nerve fiber to nerve fiber (12). It is possible that altering levels of one compact myelin protein during development are linked to changes in one or more of the other proteins able to compensate for its function. Furthermore, P2 might be more important in thick myelin sheaths, as suggested by the very mild phenotype of P2 mouse mutants and the link to human CMT via P2 mutations.
We recently reported, using similar cryo-EM approaches, the arrangement of the extracellular domains of P0 as a zipper-like assembly between the membranes (3). The assembly of P2 at the cytoplasmic face shown here completes the picture of PNS myelin molecular assembly. Importantly, whereas P0 extracellular domains interact with each other as two layers between membranes, only a single layer of P2 is observed, and each protein molecule must interact with two cytoplasmic leaflets simultaneously. The details of this aspect were further characterized here through mutagenesis, functional experiments, and high-end computer simulations.
Functional residues revealed by point mutations
The unique ability of P2 to stack lipid membranes requires two membrane-binding sites on opposite faces of the protein; P2 has two positively charged surfaces. Membrane binding experiments for surface-mutated P2 gave information about crucial regions and mechanisms of protein-membrane interaction. The L27D mutation at the tip of the α-helical portal region reduces membrane stacking and binding as well as diminishes the changes in CD spectrum upon introducing membrane-mimetic bicelles. Thus, Leu-27 may be inserted into the hydrophobic core of a lipid bilayer. This insertion is presumably facilitated by a conformational change in the portal region (15). In addition, other portal region mutations (K21Q and K31Q), which remove a positive charge, also decreased membrane binding and stacking. These residues probably interact with negatively charged lipid head groups and, together with Leu-27, form a membrane anchor of the P2 portal region. We earlier showed that the L27D mutation impairs the formation of stacked membrane systems in a cell culture system (15).
On the other hand, the removal of a positive charge at the opposite end of the β barrel (mutations K45Q, K65S, R88Q, and K112Q) caused reduced membrane binding and stacking. In MD simulations, R88Q protrudes into the lipid membrane and forms tight interactions with lipid headgroups, especially PS in the myelin-like bilayer. However, there are no hydrophobic residues at the bottom face of P2, and the barrel bottom interaction with the lipid membrane is facilitated by electrostatic interactions. The bottom region of P2 is unlikely to be deeply inserted into membranes; nor will it undergo large conformational changes.
An exception within all P2 mutants concerns P38G. In line with earlier data (38), it is more active in most of the experiments, including membrane stacking as well as membrane–, fatty acid–, and cholesterol–binding assays. The P38G mutation, however, does not alter the organization or repeat distance of the P2-membrane stacks in cryo-EM. In the crystal structure of P38G, there is no fatty acid bound, and this mutant is more flexible and has altered dynamics compered with wtP2 (38). In MD simulations, the fatty acid was observed nearly escaping from the barrel (13). The weak electron density of the portal region in the P38G mutant crystal structure supports the idea of a flexible lid in this mutant, making it more dynamic (34, 39) and prone to opening. The other mutation, R30Q, which increased the flexibility of the portal region, causes smaller but similar effects on the activity of P2 in several assays, confirming the importance of the dynamics of the portal region in the function of P2. For other FABPs, the Arg residue corresponding to P2 Arg-30 has been suggested to attract negatively charged fatty acids (40, 41); while this could be happening in P2 as well, the R30Q mutation clearly has larger-scale effects on membrane interactions and local folding or dynamics.
Phe-57 is a conserved residue within the FABP family, suggested to be a general gatekeeper for ligand binding (13, 36). It controls ligand entry into the β barrel and can flip between two conformations (13, 42). Phe-57 points outward in the K65Q crystal structure, and the fatty acid shifts toward the opening cleft. It is unclear how a mutation located at the other end of the β strand might induce the flipping of Phe-57. However, the Phe-57 flip may be one initial step in P2 opening, which was observed in MD simulations and structural studies (13, 30).
Arg-88 appears central to the initial P2-membrane contact, functioning as an anchor. Within the human FABP family, Arg-88 is conserved in P2, but not other family members (15). This indicates its possible importance for the membrane-stacking function, because other collision-type FABPs bind transiently to single membrane surfaces.
A specific case, not within the scope of the current study, is the effect on the observed supramolecular assembly by the several P2 mutations linked to CMT (25–29). In our earlier study, we showed that three of these mutations were linked to both lowered protein stability and altered membrane binding properties (30). It will be of interest to find out whether the highly ordered assemblies of P2 between membranes are disorganized when CMT mutations are present.
Conformational changes and dynamics upon membrane binding
The binding of P2 onto a myelin-like membrane is accompanied by a conformational change, opening the likely entry/egress site of the bound fatty acid. This change can be observed both experimentally and in computer simulations. The change is similar to that observed in solution for the CMT disease variants and in extended computer simulations of P2 (13, 30, 34). Similar conformational changes were observed for H-FABP during long simulations (40). The bound ligand could be exchanged with the apposing membrane in a multilayer, when this conformational change occurs.
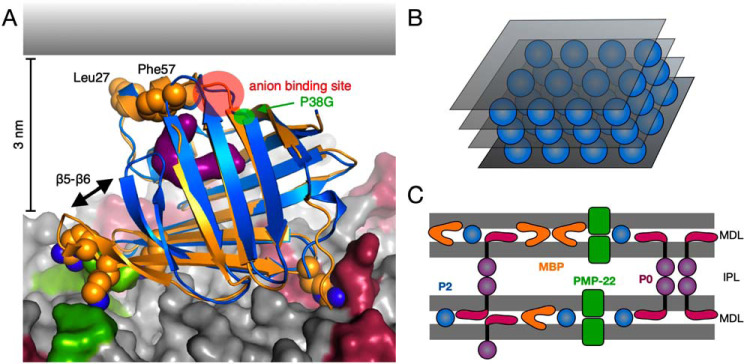
Atomistic simulations of P2 on a membrane surface revealed different behavior on a simplistic model membrane compared with a myelin composition. Importantly, some of the lipids concentrated on the myelin membrane formed specific interactions with P2 during the long atomistic simulations, contributing to the conformational change. The presence of two anchor points for P2-membrane binding enabled the membrane to contribute to P2 barrel opening, unraveling the bound ligand. The negatively charged lipids, PS and PIP2, might also affect other myelin proteins in a specific fashion, and further experiments will be required to grasp the full scope of intertwined interactions between myelin proteins and specific lipids. A model combining current data on P2 bound to the cytoplasmic leaflet of myelin is shown in Fig. 8A.
Figure 8.
Model for P2-membrane interactions in myelin based on current data. A, membrane binding based on MD simulations. Shown is a superposition of wtP2 crystal structure (blue) and the membrane-bound conformation of P38G (orange). Upon membrane binding, Arg-88 gets anchored by POPS molecules (red), and the basic residues around the β5-β6 loop interact strongly with PIP2 (green). The rest of the membrane lipids are colored gray. The opening of the β5-β6 flap exposes the fatty acid ligand (purple). Leu-27, Phe-57, the anion-binding site (red circle), and the location of the P38G mutation (green circle) are facing the apposing membrane surface (gray) in this setting. B, schematic model of lattice-like P2 (blue spheres) arrangement between consecutive membranes (gray). C, integral (P0 and PMP-22) and peripheral (P2 and MBP) membrane proteins in PNS compact myelin. Although each individual protein has been shown to promote membrane stacking, thus far no data exist for their arrangement in compact myelin together. MDL, major dense line (cytosolic apposition); IPL, intraperiod line (extracellular apposition).
Upon the formation of a P2-membrane complex (Fig. 8B), the dynamics of both the protein and lipid components are altered. When bound to P2, the dynamics of the lipid membrane are decreased (31), whereas P2 becomes extremely heat-stable when bound to membranes (15). These observations are likely to be linked to the synergistic effects of P2 and the lipids in the tightening of the 3D molecular assembly, as shown here by X-ray diffraction from myelin-mimicking bicelle complexes. Furthermore, they are in line with the decreased dynamics of P2 on a myelin-like membrane in the simulations.
Conclusions
We have shown that, similarly to P0, MBP, and PMP22, myelin protein P2 is able to spontaneously induce the formation of myelin-like membrane multilayers. We have for the first time visualized the arrangement of P2 between membranes, providing an unprecedented view into the structure of the major dense line in peripheral nerves. Furthermore, our observations provide a lipid composition-dependent mechanism for the opening of the P2 structure for ligand entry and egress; in the case of a multilayered membrane, the ligand could be exchanged with the apposing membrane. How myelin proteins act together in forming native myelin multilayers through interactions at both extracellular and intracellular surfaces of the bilayer (Fig. 8C) is a major question in myelin biology; the tools and materials exist for solving this question in the coming years.
Experimental procedures
Protein production
The expression and purification of wtP2 was done as described (14). Mutagenesis and the expression and purification of P2 variants were described earlier (43).
Electron cryomicroscopy and image processing
0.6 mg/ml purified wtP2 or the P38G variant was mixed with Escherichia coli polar lipids (Avanti Polar Lipids) using a lipid/protein ratio of 2 (w/w), corresponding to a molar ratio of ∼40, and incubated for 1–2 h at +23 °C. For grid preparation, samples were applied to glow-discharged, holey carbon grids (QUANTIFOIL R 1.2/1.3, R 2/2, or R 3.5/1). 3-μl samples were adsorbed for 1 min at +20 °C, 90% humidity. Grids were then blotted for 2 s and vitrified by plunging into liquid nitrogen–cooled liquid ethane using an FEI Vitrobot MK4 (Vitrobot, Maastricht Instruments). The frozen grids were imaged using an FEI Titan Krios transmission electron microscope operated at 300 keV. Images were recorded using a Gatan K2 Summit direct electron detector, in counting mode (0.2 s/frame, 8 s in total, 6–7 e/pixels/s). Movie frames were aligned with MotionCorr (44) and preprocessed by 2dx_automator (45). The effective pixel size of the images was 1.3 Å/pixel. Particles were boxed with EMAN2 (Helixboxer) (46) and further processed by Spring (47) with helical reconstruction. In total, 25,000 overlapping and CTF-corrected segments with a size of 240 × 240 pixels were used with a binning value of 2 to calculate 2D class averages.
Structural analysis of P2-stacked bicelles
0.5 mg/ml P2 was mixed with 0.5 mg/ml bicelles (phospholipid/DPC ratio 2.85, phospholipids 1:1 DMPC/DMPG) and incubated for 1 h at room temperature. 4-μl samples were then pipetted onto glow-discharged carbon-coated copper grids before incubating for 1 min. Excess solution was removed with filter paper (Whatman), and the samples were washed with four drops of Milli-Q water. Samples were stained with two drops of 2% uranyl acetate for 12 s in each drop and air-dried. Transmission EM was performed using a Jeol JEM-1230 (MedWOW) instrument.
To examine repetitive structures in turbid samples, 2, 10, and 20 μm P2 was mixed with 1, 2, or 3 mm bicelles in 20 mm HEPES (pH 7.5), 150 mm NaCl. Samples were prepared at ambient temperature right before the measurements and measured at +25 °C. Synchrotron SAXS data from the suspensions were collected at the PETRA III storage ring (DESY, Hamburg, Germany) on EMBL beamline P12 (48). Data were processed and analyzed using ATSAS (49). Repeat distances in the sample were deduced from Bragg peak positions.
Crystal structure determination
All P2 variants were crystallized, and X-ray diffraction data were collected as described (43). Data were processed with XDS (50, 51), and molecular replacement was done using Phaser (52) using human wtP2 (PDB code 2WUT) (14) as a search model. Structures were refined with phenix.refine (53), and rebuilding was done in Coot (54). The structures were validated using MolProbity (55). The refined coordinates and structure factors were deposited in the PDB (see Table S1 for statistics and entry codes).
Proteolipid vesicle aggregation
A 5 μm concentration of each P2 mutant was mixed with DMPC/DMPG (1:1) vesicles in a buffer containing 10 mm HEPES (pH 7.4), 150 mm NaCl and incubated for 10 min at room temperature. Lysozyme and BSA were used as negative controls. Turbidity was measured on a Tecan Infinite M200 plate reader at 450 nm. The turbidity values were plotted as relative turbidity compared with wtP2 from the same measurement series. For further characterization, the turbid samples were centrifuged, and the supernatant and pellet fractions were analyzed by SDS-PAGE to detect co-sedimentation of P2 with aggregated vesicles.
Turbidity was also studied with protein-bicelle complexes, using wtP2. For this purpose, bicelles (phospholipid/detergent ratio 2.85) were prepared as above, but the phospholipid composition was varied. Bicelles were at 5 mm and P2 at 33 μm. wtP2 was simultaneously used to examine the effect of phospholipid vesicle composition on protein-induced turbidity; the lipid concentration was 0.5 mm. Turbidity was measured using a Tecan Spark 20M microplate reader at +30 °C.
Surface plasmon resonance
SPR was used to determine the effect of mutations in P2 on the binding of the protein to lipid monolayers using the Biacore T100 SPR instrument. Lipid monolayers consisting of either DMPC or DMPA were immobilized on an HPA chip (GE Healthcare) according to the manufacturer's instructions. P2 at 0.05–1.0 μm was injected onto the chip at +25 °C using 10 mm HEPES (pH 7.4), 150 mm NaCl as running buffer.
CD spectroscopy
CD spectra were measured in 10 mm sodium phosphate (pH 7.0) at a protein concentration of 0.1 mg/ml, using quartz cuvettes with 0.1-cm pathlength and a Jasco J-715 spectropolarimeter. Melting curves were measured using 0.2 mg/ml protein at 217 nm. The temperature was increased 1 °C/min from +20 to +90 °C.
SRCD measurements for selected variants were performed on the CD1 beamline of the ASTRID storage ring at the ISA synchrotron (Aarhus, Denmark). Scans from 280 to 165 nm were performed in 1-nm steps at +20 °C in H2O. Three mutants with large effects on membrane binding (L27D, R30Q, and P38G), were studied in a bicelle environment (4:1 DMPC/DPC).
Bicelles and vesicles with varying phospholipid compositions were used for further SRCD experiments with wtP2. 0.4 mg/ml wtP2 was mixed with 5 mm bicelles or 2.65 mm vesicles. Spectra were recorded from 280 to 170 nm at +30 °C, using a 100-μm cuvette. These experiments were performed on the AU-CD beamline of the ASTRID2 storage ring at the ISA synchrotron (Aarhus, Denmark).
Fluorescence spectroscopy
The fluorescent fatty acid DAUDA was used to study fatty acid binding by P2. DAUDA has been used to study ligand binding in FABPs before (56, 57). DAUDA was dissolved in DMSO, and the final DMSO concentration in the samples was 1%. 20 μm DAUDA was mixed with 0, 1, 5, or 10 μm protein. Samples were incubated for 2 h at +23 °C. Fluorescence excitation at 280 nm was used, and emission was recorded at 530 nm using a Tecan Infinite M200 plate reader.
The binding of wtP2 and the P38G mutant to cholesterol was studied using the environment-sensitive fluorescent cholesterol analogue 22-NBD-cholesterol. The fluorescence intensity of 22-NBD-cholesterol increases and the fluorescence emission maximum shifts if the probe is moved to a nonpolar environment. 100 μm 22-NBD-cholesterol stock solution was prepared in 100% ethanol, and the maximum ethanol concentration in the sample was 2%. All experiments were carried out in 10 mm HEPES (pH 7.5). 2 μm 22-NBD-cholesterol was incubated for 16 h at +23 °C with varying amounts of P2. Fluorescence spectra were recorded on a Horiba Fluoromax-4 instrument, using excitation at 473 nm and emission between 500 and 600 nm, with a bandwidth of 5 nm.
Atomic scale molecular dynamics simulations
Structures of wtP2 and P38G were prepared for the simulations essentially as described elsewhere (34). Briefly, the P2 structure with bound palmitate was taken from the PDB entry 4BVM (15) and converted to match an all-atom representation consistent with the CHARMM36 force field (58), which was used for simulating the system components, unless mentioned otherwise. The topology for wtP2 was directly obtained from this conversion. The P38G mutation was made in silico and equilibrated in a water environment. Both protein-palmitate complexes had a total charge of +10.
Lipid bilayers were constructed using the CHARMM-GUI membrane builder (59). Two different membrane systems were considered: a 1:1 DMPC/DMPG bilayer as a general reference with a net negative surface charge and a myelin bilayer mimicking the cytoplasmic leaflet of the myelin membrane. The composition of the myelin-like bilayer was 44 mol % cholesterol, 27 mol % palmitoyloleoylphosphatidylethanolamine, 2 mol % PIP2, 11 mol % palmitoyloleoylphosphatidylcholine, 13 mol % POPS, and 3 mol % sphingomyelin (37). The bilayers were symmetrical, comprised of a total of 200 lipid molecules each.
Ten Cl− ions were included to neutralize the total charge of each protein-palmitate complex. The systems were solvated with a total of 15,000 water molecules each, with 0.1 m KCl. Water was modeled using the TIP3P model (60). Additional counterions (90 K+ in the DMPC/DMPG and 32 K+ in the myelin membrane system) were included to neutralize the system total charge. The total system volume was approximately (7.5 × 7.5 × 12) nm3 for the DMPC/DMPG and (6.5 × 6.5 × 13.5) nm3 for the myelin membrane systems. Periodic boundary conditions were used to make the bilayer structure continuous.
MD simulations were carried out under NpT conditions. Temperature coupling was performed with the velocity-rescale method (61), using separate thermostats for the protein, the bilayer, and the solvent. Reference temperatures were set at 310 K, with coupling time constants of 2.0 ps. Pressure coupling was done semi-isotropically with the Parrinello–Rahman barostat (62), using reference pressures of 1.0 bar with coupling time constants of 2.0 ps and compressibility constants of 4.5 × 10−5 bar−1.
All bonds were constrained with the LINCS algorithm (63). Cut-off radii of 1.0 nm were introduced for the Coulombic and Lennard–Jones interactions, including the neighbor list. Long-range electrostatics were calculated using the particle-mesh Ewald method (64) with cubic interpolation and a spacing of 0.16 nm for the Fourier grid.
The simulation systems were built by adding the protein structure near the bilayer and solvating the system thereafter. After a short steepest-descent equilibration, the systems were simulated long enough for the protein to spontaneously come into close contact with the bilayer. This was used as the starting structure, after which the systems were simulated for 3 μs each. A total of four full simulations were run (one for each protein-membrane combination), in addition to several shorter simulations on the P2 membrane attachment phase. The first 500 ns of each simulation were removed as an equilibration period, and the final 2.5 μs were used for analyses. All simulations were conducted with GROMACS 4.6.7 (65), using the CHARMM36 all-atom representation and a time step of 2 fs, saving the trajectory coordinates every 50 ps.
Data availability
Refined crystal structure coordinates and experimental structure factors for the P2 variants are available at the Protein Data Bank with accession codes 6XU5 (variant N2D), 6XU9 (K3N), 6XUA (K21Q), 6XUW (L27D), 6STS (R30Q), 6XVQ (K31Q), 6XVR (L35S), 6XVS (P38G), 4A1H (K45S), 4A1Y (K65Q), 6XVY (R88Q), 4A8Z (K112Q), and 6XW9 (K120S). All other data are either presented in the manuscript or available from the corresponding author (Petri Kursula, University of Bergen, Petri.kursula@uib.no) upon request.
Author contributions
S. R., M. L., I. V., H. S., and P. K. conceptualization; S. R., O. C. K., J. K., T. N., M. L., A. R., V. P. D., and P. K. investigation; S. R., J. K., T. N., M. L., and P. K. visualization; S. R., O. C. K., J. K., T. N., M. L., V. P. D., I. V., and H. S. methodology; S. R. and P. K. writing-original draft; S. R., O. C. K., J. K., T. N., M. L., A. R., I. V., H. S., and P. K. writing-review and editing; J. K., A. R., I. V., H. S., and P. K. supervision; I. V. and P. K. funding acquisition; P. K. project administration.
Supplementary Material
Acknowledgments
Beamtime and user support at EMBL/DESY and ISA are gratefully acknowledged. Travel to synchrotrons was supported by the Norwegian Research Council (SYNKNØYT Project 247669) and the European Union Horizon 2020 programs iNEXT (Grant 653706) and CALIPSOplus (Grant 730872).
This work was supported by the Academy of Finland, the Emil Aaltonen Foundation (Finland), the Jane and Aatos Erkko Foundation (Finland), the Helsinki Institute of Life Science Fellow program (Finland), the Science and Research Foundation of the City of Hamburg (Germany), and the Sigrid Jusélius Foundation (Finland). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains Table S1 and Fig. S1.
- PNS
- peripheral nervous system
- MBP
- myelin basic protein
- PMP22
- peripheral myelin protein 22
- FABP
- fatty acid-binding protein
- CMT
- Charcot-Marie-Tooth
- MD
- molecular dynamics
- 2D and 3D
- two- and three-dimensional, respectively
- DMPC
- dimyristoylphosphatidylcholine
- DMPG
- dimyristoylphosphatidylglycerol
- DMPA
- dimyristoyl phospatidic acid
- 22-NBD-cholesterol
- 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol
- DPC
- dodecylphosphocholine
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- wtP2
- human WT P2
- PDB
- Protein Data Bank
- SRCD
- synchrotron radiation CD
- DAUDA
- 11-dansylamino-undecanoic acid
- POPS
- palmitoyloleoylphosphatidylserine.
References
- 1. Schmitt F. O., Bear R. S., and Clark G. L. (1935) The role of lipoids in the X-ray diffraction patterns of nerve. Science 82, 44–45 10.1126/science.82.2115.44 [DOI] [PubMed] [Google Scholar]
- 2. Raasakka A., Ruskamo S., Kowal J., Barker R., Baumann A., Martel A., Tuusa J., Myllykoski M., Bürck J., Ulrich A. S., Stahlberg H., and Kursula P. (2017) Membrane association landscape of myelin basic protein portrays formation of the myelin major dense line. Sci. Rep. 7, 4974 10.1038/s41598-017-05364-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Raasakka A., Ruskamo S., Kowal J., Han H., Baumann A., Myllykoski M., Fasano A., Rossano R., Riccio P., Bürck J., Ulrich A. S., Stahlberg H., and Kursula P. (2019) Molecular structure and function of myelin protein P0 in membrane stacking. Sci. Rep. 9, 642 10.1038/s41598-018-37009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Filbin M. T., Walsh F. S., Trapp B. D., Pizzey J. A., and Tennekoon G. I. (1990) Role of myelin P0 protein as a homophilic adhesion molecule. Nature 344, 871–872 10.1038/344871a0 [DOI] [PubMed] [Google Scholar]
- 5. Wong M. H., and Filbin M. T. (1994) The cytoplasmic domain of the myelin P0 protein influences the adhesive interactions of its extracellular domain. J. Cell Biol. 126, 1089–1097 10.1083/jcb.126.4.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wong M. H., and Filbin M. T. (1996) Dominant-negative effect on adhesion by myelin Po protein truncated in its cytoplasmic domain. J. Cell Biol. 134, 1531–1541 10.1083/jcb.134.6.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mittendorf K. F., Marinko J. T., Hampton C. M., Ke Z., Hadziselimovic A., Schlebach J. P., Law C. L., Li J., Wright E. R., Sanders C. R., and Ohi M. D. (2017) Peripheral myelin protein 22 alters membrane architecture. Sci. Adv. 3, e1700220 10.1126/sciadv.1700220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trapp B. D., Dubois-Dalcq M., and Quarles R. H. (1984) Ultrastructural localization of P2 protein in actively myelinating rat Schwann cells. J. Neurochem. 43, 944–948 10.1111/j.1471-4159.1984.tb12828.x [DOI] [PubMed] [Google Scholar]
- 9. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- 10. Gould R. M., Oakley T., Goldstone J. V., Dugas J. C., Brady S. T., and Gow A. (2008) Myelin sheaths are formed with proteins that originated in vertebrate lineages. Neuron Glia Biol. 4, 137–152 10.1017/S1740925X09990238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belin S., Ornaghi F., Shackleford G., Wang J., Scapin C., Lopez-Anido C., Silvestri N., Robertson N., Williamson C., Ishii A., Taveggia C., Svaren J., Bansal R., Schwab M. H., Nave K., et al. (2019) Neuregulin 1 type III improves peripheral nerve myelination in a mouse model of congenital hypomyelinating neuropathy. Hum. Mol. Genet. 28, 1260–1273 10.1093/hmg/ddy420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trapp B. D., McIntyre L. J., Quarles R. H., Sternberger N. H., and Webster H. D. (1979) Immunocytochemical localization of rat peripheral nervous system myelin proteins: P2 protein is not a component of all peripheral nervous system myelin sheaths. Proc. Natl. Acad. Sci. U.S.A. 76, 3552–3556 10.1073/pnas.76.7.3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laulumaa S., Nieminen T., Raasakka A., Krokengen O. C., Safaryan A., Hallin E. I., Brysbaert G., Lensink M. F., Ruskamo S., Vattulainen I., and Kursula P. (2018) Structure and dynamics of a human myelin protein P2 portal region mutant indicate opening of the β barrel in fatty acid binding proteins. BMC Struct. Biol. 18, 8 10.1186/s12900-018-0087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Majava V., Polverini E., Mazzini A., Nanekar R., Knoll W., Peters J., Natali F., Baumgärtel P., Kursula I., and Kursula P. (2010) Structural and functional characterization of human peripheral nervous system myelin protein P2. PLoS ONE 5, e10300 10.1371/journal.pone.0010300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruskamo S., Yadav R. P., Sharma S., Lehtimäki M., Laulumaa S., Aggarwal S., Simons M., Bürck J., Ulrich A. S., Juffer A. H., Kursula I., and Kursula P. (2014) Atomic resolution view into the structure-function relationships of the human myelin peripheral membrane protein P2. Acta Crystallogr. D Biol. Crystallogr. 70, 165–176 10.1107/S1399004713027910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zenker J., Stettner M., Ruskamo S., Domènech-Estévez E., Baloui H., Médard J. J., Verheijen M. H., Brouwers J. F., Kursula P., Kieseier B. C., and Chrast R. (2014) A role of peripheral myelin protein 2 in lipid homeostasis of myelinating Schwann cells. Glia 62, 1502–1512 10.1002/glia.22696 [DOI] [PubMed] [Google Scholar]
- 17. Kim H. K., and Storch J. (1992) Mechanism of free fatty acid transfer from rat heart fatty acid-binding protein to phospholipid membranes: evidence for a collisional process. J. Biol. Chem. 267, 20051–20056 [PubMed] [Google Scholar]
- 18. Storch J. (1993) Diversity of fatty acid-binding protein structure and function: studies with fluorescent ligands. Mol. Cell Biochem. 123, 45–53 10.1007/BF01076474 [DOI] [PubMed] [Google Scholar]
- 19. Thumser A. E., and Storch J. (2000) Liver and intestinal fatty acid-binding proteins obtain fatty acids from phospholipid membranes by different mechanisms. J. Lipid Res. 41, 647–656 [PubMed] [Google Scholar]
- 20. Thumser A. E., Tsai J., and Storch J. (2001) Collision-mediated transfer of long-chain fatty acids by neural tissue fatty acid-binding proteins (FABP): studies with fluorescent analogs. J. Mol. Neurosci. 16, 143–150; discussion 151–157 10.1385/JMN:16:2-3:143 [DOI] [PubMed] [Google Scholar]
- 21. Zamarreño F., Giorgetti A., Amundarain M. J., Viso J. F., Córsico B., and Costabel M. D. (2018) Conserved charged amino acids are key determinants for fatty acid binding proteins (FABPs)-membrane interactions: a multi-methodological computational approach. J. Biomol. Struct. Dyn. 36, 861–877 10.1080/07391102.2017.1301271 [DOI] [PubMed] [Google Scholar]
- 22. Saher G., and Stumpf S. K. (2015) Cholesterol in myelin biogenesis and hypomyelinating disorders. Biochim. Biophys. Acta 1851, 1083–1094 10.1016/j.bbalip.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 23. Stettner M., Zenker J., Klingler F., Szepanowski F., Hartung H. P., Mausberg A. K., Kleinschnitz C., Chrast R., and Kieseier B. C. (2018) The role of peripheral myelin protein 2 in remyelination. Cell Mol. Neurobiol. 38, 487–496 10.1007/s10571-017-0494-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graf S. A., Heppt M. V., Wessely A., Krebs S., Kammerbauer C., Hornig E., Strieder A., Blum H., Bosserhoff A. K., and Berking C. (2019) The myelin protein PMP2 is regulated by SOX10 and drives melanoma cell invasion. Pigment Cell Melanoma Res. 32, 424–434 10.1111/pcmr.12760 [DOI] [PubMed] [Google Scholar]
- 25. Palaima P., Chamova T., Jander S., Mitev V., Van Broeckhoven C., Tournev I., Peeters K., and Jordanova A. (2019) Peripheral myelin protein 2: a novel cluster of mutations causing Charcot-Marie-Tooth neuropathy. Orphanet J. Rare Dis. 14, 197 10.1186/s13023-019-1162-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzaga-Jauregui C., Harel T., Gambin T., Kousi M., Griffin L. B., Francescatto L., Ozes B., Karaca E., Jhangiani S. N., Bainbridge M. N., Lawson K. S., Pehlivan D., Okamoto Y., Withers M., Mancias P., et al. (2015) Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep. 12, 1169–1183 10.1016/j.celrep.2015.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong Y. B., Joo J., Hyun Y. S., Kwak G., Choi Y. R., Yeo H. K., Jwa D. H., Kim E. J., Mo W. M., Nam S. H., Kim S. M., Yoo J. H., Koo H., Park H. T., Chung K. W., and Choi B. O. (2016) A mutation in PMP2 causes dominant demyelinating Charcot-Marie-Tooth neuropathy. PLoS Genet. 12, e1005829 10.1371/journal.pgen.1005829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Motley W. W., Palaima P., Yum S. W., Gonzalez M. A., Tao F., Wanschitz J. V., Strickland A. V., Löscher W. N., De Vriendt E., Koppi S., Medne L., Janecke A. R., Jordanova A., Zuchner S., and Scherer S. S. (2016) De novo PMP2 mutations in families with type 1 Charcot-Marie-Tooth disease. Brain 139, 1649–1656 10.1093/brain/aww055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Punetha J., Mackay-Loder L., Harel T., Coban-Akdemir Z., Jhangiani S. N., Gibbs R. A., Lee I., Terespolsky D., Lupski J. R., and Posey J. E. (2018) Identification of a pathogenic PMP2 variant in a multi-generational family with CMT type 1: clinical gene panels versus genome-wide approaches to molecular diagnosis. Mol. Genet. Metab. 125, 302–304 10.1016/j.ymgme.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruskamo S., Nieminen T., Kristiansen C. K., Vatne G. H., Baumann A., Hallin E. I., Raasakka A., Joensuu P., Bergmann U., Vattulainen I., and Kursula P. (2017) Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2. Sci. Rep. 7, 6510 10.1038/s41598-017-06781-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Knoll W., Natali F., Peters J., Nanekar R., Wang C., and Kursula P. (2010) Dynamic properties of a reconstituted myelin sheath. Spectroscopy 24, 585–592 10.1155/2010/317525 [DOI] [Google Scholar]
- 32. Sedzik J., Blaurock A. E., and Hoechli M. (1985) Reconstituted P2/myelin-lipid multilayers. J. Neurochem. 45, 844–852 10.1111/j.1471-4159.1985.tb04071.x [DOI] [PubMed] [Google Scholar]
- 33. Suresh S., Wang C., Nanekar R., Kursula P., and Edwardson J. M. (2010) Myelin basic protein and myelin protein 2 act synergistically to cause stacking of lipid bilayers. Biochemistry 49, 3456–3463 10.1021/bi100128h [DOI] [PubMed] [Google Scholar]
- 34. Laulumaa S., Nieminen T., Lehtimäki M., Aggarwal S., Simons M., Koza M. M., Vattulainen I., Kursula P., and Natali F. (2015) Dynamics of the peripheral membrane protein P2 from human myelin measured by neutron scattering—a comparison between wild-type protein and a hinge mutant. PLoS ONE 10, e0128954 10.1371/journal.pone.0128954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laulumaa S., and Kursula P. (2019) Sub-atomic resolution crystal structures reveal conserved geometric outliers at functional sites. Molecules 24, E3044 10.3390/molecules24173044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simpson M. A., and Bernlohr D. A. (1998) Analysis of a series of phenylalanine 57 mutants of the adipocyte lipid-binding protein. Biochemistry 37, 10980–10986 10.1021/bi980507a [DOI] [PubMed] [Google Scholar]
- 37. Inouye H., and Kirschner D. A. (1988) Membrane interactions in nerve myelin: II. Determination of surface charge from biochemical data. Biophys. J. 53, 247–260 10.1016/S0006-3495(88)83086-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laulumaa S., Blakeley M. P., Raasakka A., Moulin M., Härtlein M., and Kursula P. (2015) Production, crystallization and neutron diffraction of fully deuterated human myelin peripheral membrane protein P2. Acta Crystallogr. F Struct. Biol. Commun. 71, 1391–1395 10.1107/S2053230X15017902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laulumaa S., Koza M. M., Seydel T., Kursula P., and Natali F. (2019) A quasielastic neutron scattering investigation on the molecular self-dynamics of human myelin protein P2. J. Phys. Chem. B 123, 8178–8185 10.1021/acs.jpcb.9b05320 [DOI] [PubMed] [Google Scholar]
- 40. Guo Y., Duan M., and Yang M. (2019) The observation of ligand-binding-relevant open states of fatty acid binding protein by molecular dynamics simulations and a Markov state model. Int. J. Mol. Sci. 20, E3476 10.3390/ijms20143476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Young A. C., Scapin G., Kromminga A., Patel S. B., Veerkamp J. H., and Sacchettini J. C. (1994) Structural studies on human muscle fatty acid binding protein at 1.4 Å resolution: binding interactions with three C18 fatty acids. Structure 2, 523–534 10.1016/S0969-2126(00)00052-6 [DOI] [PubMed] [Google Scholar]
- 42. Jones T. A., Bergfors T., Sedzik J., and Unge T. (1988) The three-dimensional structure of P2 myelin protein. EMBO J. 7, 1597–1604 10.1002/j.1460-2075.1988.tb02985.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lehtimäki M., Laulumaa S., Ruskamo S., and Kursula P. (2012) Production and crystallization of a panel of structure-based mutants of the human myelin peripheral membrane protein P2. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 1359–1362 10.1107/S1744309112039036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li X., Mooney P., Zheng S., Booth C. R., Braunfeld M. B., Gubbens S., Agard D. A., and Cheng Y. (2013) Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 10.1038/nmeth.2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scherer S., Kowal J., Chami M., Dandey V., Arheit M., Ringler P., and Stahlberg H. (2014) 2dx_automator: implementation of a semiautomatic high-throughput high-resolution cryo-electron crystallography pipeline. J. Struct. Biol. 186, 302–307 10.1016/j.jsb.2014.03.016 [DOI] [PubMed] [Google Scholar]
- 46. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., and Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 47. Desfosses A., Ciuffa R., Gutsche I., and Sachse C. (2014) SPRING: an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J. Struct. Biol. 185, 15–26 10.1016/j.jsb.2013.11.003 [DOI] [PubMed] [Google Scholar]
- 48. Blanchet C. E., Spilotros A., Schwemmer F., Graewert M. A., Kikhney A., Jeffries C. M., Franke D., Mark D., Zengerle R., Cipriani F., Fiedler S., Roessle M., and Svergun D. I. (2015) Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 48, 431–443 10.1107/S160057671500254X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Franke D., Petoukhov M. V., Konarev P. V., Panjkovich A., Tuukkanen A., Mertens H. D. T., Kikhney A. G., Hajizadeh N. R., Franklin J. M., Jeffries C. M., and Svergun D. I. (2017) ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 50, 1212–1225 10.1107/S1600576717007786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kursula P. (2004) XDSi: a graphical interface for the data processing program XDS. J. Appl. Crystallogr. 37, 347–348 10.1107/S0021889804000858 [DOI] [Google Scholar]
- 52. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Davis I. W., Murray L. W., Richardson J. S., and Richardson D. C. (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 10.1093/nar/gkh398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Davies J. K., Thumser A. E., and Wilton D. C. (1999) Binding of recombinant rat liver fatty acid-binding protein to small anionic phospholipid vesicles results in ligand release: a model for interfacial binding and fatty acid targeting. Biochemistry 38, 16932–16940 10.1021/bi991926q [DOI] [PubMed] [Google Scholar]
- 57. Patil R., Laguerre A., Wielens J., Headey S. J., Williams M. L., Hughes M. L., Mohanty B., Porter C. J., and Scanlon M. J. (2014) Characterization of two distinct modes of drug binding to human intestinal fatty acid binding protein. ACS Chem. Biol. 9, 2526–2534 10.1021/cb5005178 [DOI] [PubMed] [Google Scholar]
- 58. Brooks B. R., Brooks C. L. 3rd, Mackerell A. D. Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., et al. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jo S., Kim T., Iyer V. G., and Im W. (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- 60. Jorgensen W. L., Chandrasekar J., and Madura J. D. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 10.1063/1.445869 [DOI] [Google Scholar]
- 61. Bussi G., Donadio D., and Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- 62. Parrinello M., and Rahman A. (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182 10.1063/1.328693 [DOI] [Google Scholar]
- 63. Hess B., Bekker H., Berendsen H. J. C., and Fraaije J. G. E. M. (1997) LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 [DOI] [Google Scholar]
- 64. Darden T., York D., and Pedersen L. (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 10.1063/1.464397 [DOI] [Google Scholar]
- 65. Hess B., Kutzner C., van der Spoel D., and Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Refined crystal structure coordinates and experimental structure factors for the P2 variants are available at the Protein Data Bank with accession codes 6XU5 (variant N2D), 6XU9 (K3N), 6XUA (K21Q), 6XUW (L27D), 6STS (R30Q), 6XVQ (K31Q), 6XVR (L35S), 6XVS (P38G), 4A1H (K45S), 4A1Y (K65Q), 6XVY (R88Q), 4A8Z (K112Q), and 6XW9 (K120S). All other data are either presented in the manuscript or available from the corresponding author (Petri Kursula, University of Bergen, Petri.kursula@uib.no) upon request.