Abstract
Vestigial-like 3 (VGLL3) is a member of the VGLL family, whose members serve as cofactors for TEA domain–containing transcription factors (TEADs). TEADs promote tissue and tumor development together with the cofactors Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ). Although VGLL3 is involved in tumor cell proliferation, its relationship with TEADs and YAP/TAZ remains largely unknown. To close this research gap, here we established tumor cells stably expressing VGLL3 and found that they exhibit enhanced proliferation. Notably, YAP and TAZ were inactivated in the VGLL3-expressing cells, coinciding with activation of the Hippo pathway, which suppresses YAP/TAZ activities. VGLL3 in combination with TEADs promoted expression of the Hippo pathway components large tumor suppressor kinase (LATS2) and angiomotin-like 2 (AMOTL2). VGLL3 was highly expressed in malignant breast tumor cells and osteosarcoma cells, and VGLL3 knockdown increased nuclear localization of YAP and TAZ. Knockdown of LATS2 or AMOTL2, as well as VGLL3 knockdown, repressed proliferation of breast tumor cells. Together, these results suggest that VGLL3 together with TEADs promotes cell proliferation by activating the Hippo pathway through LATS2 and AMOTL2, leading to YAP/TAZ inactivation.
Keywords: cell proliferation, cell signaling, Hippo pathway, transcription coactivator, Yes-associated protein (YAP), TEA domain–containing transcription factor (TEAD), vestigial-like 3 (VGLL3), transcriptional coactivator with PDZ-binding motif (TAZ), cancer, cell proliferation, cell signaling
VGLL3 is one of the four members of the vestigial-like (VGLL) family of proteins. VGLL3 is thought to bind to TEA domain–containing transcription factors (TEADs) through the TONDU domain and function as a transcriptional cofactor for TEADs (1, 2). Although the physiological role of VGLL3 remains largely unknown, there is evidence suggesting that VGLL3 is involved in tumor development. For example, gene amplification and overexpression of VGLL3 were observed in several types of human sarcomas, such as myxoinflammatory fibroblastic sarcoma and soft-tissue sarcoma (3–5). In a cell line derived from a soft-tissue sarcoma patient, VGLL3 was shown to be required for proliferation (4). VGLL3 was also overexpressed in gastric cancer patients and was associated with a poor prognosis (6).
Mammalian cells have four TEADs, TEAD1–TEAD4, which are broadly expressed and involved in tissue development and tumorigenesis (7). TEAD activity is controlled by the Hippo pathway. The Hippo pathway primarily targets the transcriptional cofactor YAP (Yes-associated protein) and its paralog TAZ (transcriptional coactivator with PDZ-binding motif) (7–11). YAP/TAZ bind to TEADs in the nucleus and drive expression of target genes that are crucial for cell proliferation and survival. Hippo pathway activation induces phosphorylation of YAP/TAZ through the serine/threonine kinases LATS1/2 (large tumor suppressor kinase 1/2), promoting cytoplasmic retention and protein degradation of YAP/TAZ (12). The Hippo pathway is the key negative regulator of TEADs, and dysregulation of this pathway causes tissue overgrowth and tumor development (13).
Given the involvement of VGLL3 in tumor development, it is possible that VGLL3 promotes cell growth by regulating TEADs and YAP/TAZ. However, the relationship between these proteins is not clear. Therefore, this study was conducted to examine the interaction between VGLL3, YAP/TAZ, TEADs, and the Hippo pathway.
Results
Stable expression of VGLL3 enhances cell proliferation
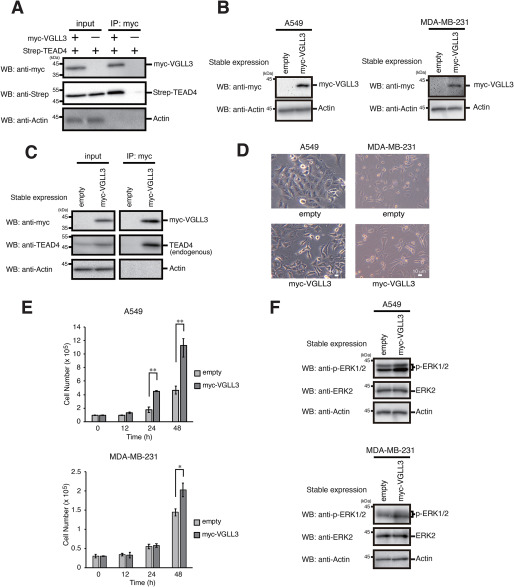
Because VGLL3 is thought to bind to TEADs, we first confirmed association between VGLL3 and TEADs using TEAD4 as a representative member of TEADs. Co-expression and a subsequent immunoprecipitation assay showed that TEAD4 co-immunoprecipitated with VGLL3, supporting the notion that VGLL3 binds to TEADs (Fig. 1A). To examine the pro-proliferative effect of VGLL3, we established cell pools stably expressing VGLL3 in human lung tumor A549 cells and breast tumor MDA-MB-231 cells (Fig. 1B). Stably expressed VGLL3 bound endogenous TEAD4 proteins (Fig. 1C). Stable expression of VGLL3 caused a spindle-like morphological change in A549 cells (Fig. 1D). Notably, VGLL3 expression significantly enhanced cell proliferation with activation of mitogenic extracellular signal–regulated kinases (ERKs) (Fig. 1, E and F). Consistent with previous data (4), these results showed that VGLL3 has a pro-proliferative effect in tumor cells probably through activation of ERK signaling.
Figure 1.
Stable expression of VGLL3 promotes cell proliferation. A, A549 cells were transfected with Strep-TEAD4, with or without Myc-VGLL3. After incubation for 24 h, the cells were subjected to immunoprecipitation (IP) with the anti-Myc antibody. Input fractions and immunoprecipitations were analyzed by Western blotting with the indicated antibodies. B, A549 and MDA-MB-231 cells stably expressing the empty vector of Myc-VGLL3 were analyzed by Western blotting (WB) with the indicated antibodies. C, A549 cells stably expressing the empty vector of Myc-VGLL3 were subjected to immunoprecipitation with the anti-Myc antibody. Input fractions and immunoprecipitations were analyzed by Western blotting with the indicated antibodies. D, A549 cells stably expressing the empty vector of Myc-VGLL3 were visualized by phase-contrast microscopy. E, A549 and MDA-MB-231 cells stably expressing the empty vector of Myc-VGLL3 were seeded at 1 × 105 and 0.3 × 105 cells/well, respectively, and cultured for the indicated times. After the incubation, cell numbers were counted, and viable cells numbers were plotted. Results represent the mean ± S.D. (error bars) (n =3). Asterisks indicate the statistical significance (**, p < 0.005; *, p < 0.05) as calculated by Student's t test. F, A549 and MDA-MB-231 cells stably expressing the empty vector of Myc-VGLL3 were analyzed by Western blotting with the indicated antibodies.
VGLL3 target genes are distinct from those of Yap/TAZ
TEADs bind to the GTIIC motif (5′-ACATTCCA-3′) in promoter regions and drive gene expression in association with YAP/TAZ (14–16). We examined the effect of VGLL3 on the binding of TEADs to the GTIIC motif using reporter assays. YAP expression stimulated GTIIC-driven reporter activity (EGFP expression), whereas VGLL3 expression repressed, rather than induced, reporter activity (Fig. 2A). We also examined expression of CTGF (connective tissue growth factor), CYR61 (cysteine-rich angiogenic inducer 61), and ANKRD1 (ankyrin repeat domain 1), well-known targets of TEADs in association with YAP/TAZ (17–19). CTGF and ANKRD1 expression was almost completely repressed, and only slight induction of CYR61 expression was observed in VGLL3-expressing cells (Fig. 2B). These results suggest that the VGLL3-TEAD complex does not bind to the GTIIC motif and that target genes of this complex are distinct from those of YAP/TAZ-TEAD.
Figure 2.
VGLL3 target genes are distinct from those of YAP/TAZ. A, A549 cells were co-transfected with the GTIIC-driven reporter plasmid and pCAG/TetR, with or without Myc-VGLL3 or YAP. After incubation for 24 h, cells were analyzed by Western blotting (WB) with the indicated antibodies. Quantitative ratios of EGFP to TetR are shown as relative values. B, A549 cells stably expressing the empty vector of Myc-VGLL3 were analyzed for CTGF mRNA expression by qPCR. GAPDH mRNA expression was used to normalize the data. The expression level of CTGF mRNA in control cells was set to 1. Results represent the mean ± S.D. (n =3). N.D., not detected. Asterisks indicate statistical significance (***, p < 0.001; *, p < 0.05) as calculated by Student's t test.
VGLL3 inactivates Yap/TAZ via hippo pathway activation
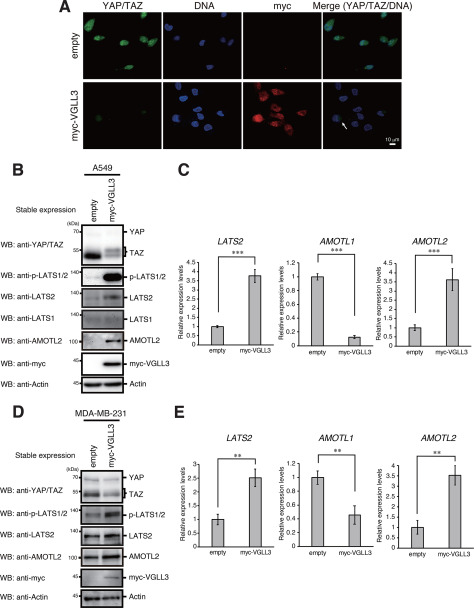
To investigate the involvement of YAP/TAZ in VGLL3-dependent cell proliferation, we examined activation of the Hippo pathway and subcellular localization of YAP/TAZ in VGLL3-expressing A549 cells. Notably, immunofluorescence revealed that YAP/TAZ levels were significantly reduced in these cells, and residual staining was detected in the cytoplasm but not in the nucleus (Fig. 3A). Western blotting showed that TAZ is the main component of YAP/TAZ proteins in A549 cells. In VGLL3-expressing A549 cells, the band of TAZ was weakened and showed increased molecular weight, likely caused by phosphorylation (Fig. 3B). LATS2, the key kinase in the Hippo pathway, showed elevated expression and phosphorylation levels following VGLL3 expression (Fig. 3, B and C). Expression of the angiomotin (AMOT) family, which inhibits nuclear translocation of YAP/TAZ (20), was also examined. Among this family, AMOTL2 expression was elevated in VGLL3-expressing cells (Fig. 3, B and C). Meanwhile, AMOTL1 expression was reduced, and AMOT expression was not detectable (data not shown). Similar phenomena were observed in VGLL3-expressing MDA-MB-231 cells (Fig. 3, D and E). These results suggest that VGLL3 inactivates YAP/TAZ via LATS2- and AMOTL2-mediated activation of the Hippo pathway.
Figure 3.
Stable expression of VGLL3 activates the Hippo pathway. A, A549 cells stably expressing the empty vector of Myc-VGLL3 were fixed with paraformaldehyde and doubly stained with anti-YAP/TAZ and anti-Myc antibodies. DNA was counterstained with propidium iodide. Cytoplasm-localized YAP/TAZ is indicated by an arrow. B and D, A549 (B) and MDA-MB-231 (D) cells stably expressing the empty vector of Myc-VGLL3 were analyzed by Western blotting (WB) with the indicated antibodies. C and E, A549 (C) and MDA-MB-231 (E) cells stably expressing the empty vector of Myc-VGLL3 were analyzed for LATS2, AMOTL1, and AMOTL2 mRNA expression by qPCR as in Fig. 2B. Asterisks indicate statistical significance (***, p < 0.001; **, p < 0.01) as calculated by Student's t test. Error bars, S.D.
VGLL3 activates the hippo pathway through TEADs
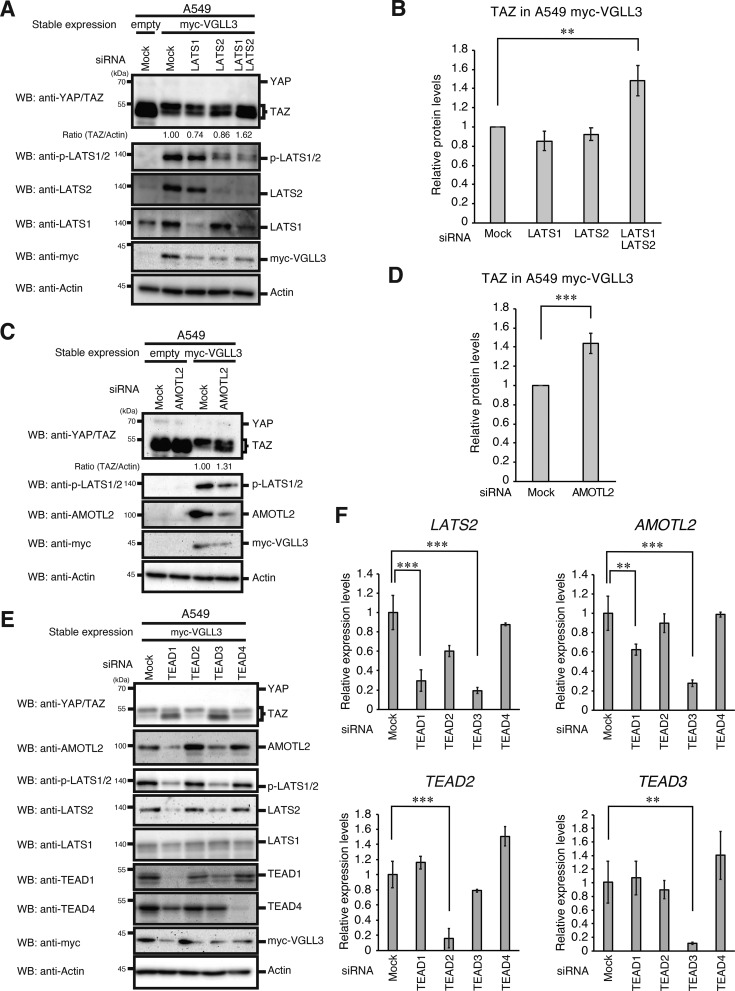
To investigate the involvement of the Hippo pathway in down-regulating YAP/TAZ in VGLL3-expressing cells, several knockdown experiments were performed. LATS1 plus LATS2 double knockdown recovered the reduction in TAZ levels, and LATS2 single knockdown increased the lower band of TAZ (unphosphorylated TAZ proteins) (Fig. 4, A and B). AMOTL2 knockdown also recovered TAZ reduction (Fig. 4, C and D), suggesting that elevated expression of LATS2 and AMOTL2 down-regulates TAZ in VGLL3-expressing cells. To determine the TEAD members that are crucial for induction of LATS2 and AMOTL2, each TEAD was knocked down. Knockdown of TEAD1 or TEAD3 reduced expression of LATS2 and AMOTL2 and recovered TAZ reduction in VGLL3-expressing cells (Fig. 4, E and F). These results suggest that VGLL3 induces LATS2 and AMOTL2 expression via TEAD1 and TEAD3, thereby activating the Hippo pathway.
Figure 4.
VGLL3 activates the Hippo pathway through TEADs. A, C, and E, A549 cells stably expressing the empty vector of Myc-VGLL3 were transfected with the indicated siRNAs for 48 h. After incubation, cells were lysed and subjected to Western blotting (WB) with the indicated antibodies. B and D, quantitative ratios of TAZ to actin, which were calculated based on the data in A and C, are shown as relative values and represent the mean ± S.D. (error bars) (n =3). Asterisks indicate statistical significance (***, p < 0.001; **, p < 0.01) as calculated by Student's t test. F, A549 cells stably expressing the empty vector of Myc-VGLL3 were transfected with the indicated siRNAs for 48 h. After incubation, cells were analyzed for LATS2, AMOTL2, TEAD2, and TEAD3 mRNA expression by qPCR as in Fig. 2B. Asterisks indicate statistical significance (***, p < 0.001; **, p < 0.01) as calculated by Student's t test.
The hippo signaling pathway is involved in proliferation in breast cancer cells
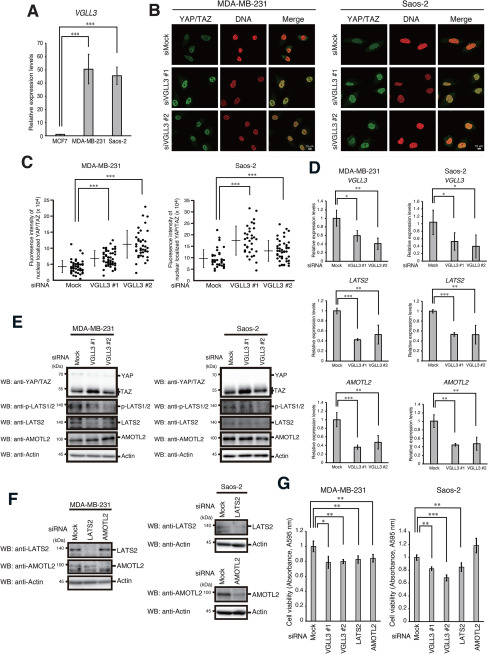
Because the VGLL family was suggested to be involved in breast cancer development (21), we examined VGLL3 expression in human breast cancer MCF7 cells and MDA-MB-231 cells. VGLL3 was highly expressed in MDA-MB-231 cells, a malignant type of breast cancer cells. High expression of VGLL3 was also observed in human osteosarcoma Saos-2 cells (Fig. 5A). To analyze the role of endogenous VGLL3 in activation of YAP/TAZ and the Hippo pathway, we depleted VGLL3 and analyzed the subcellular localization of YAP/TAZ in MDA-MB-231 and Saos-2 cells. VGLL3 depletion significantly increased the nuclear localization of YAP/TAZ (Fig. 5, B and C). VGLL3 depletion also caused a reduction in LATS2 expression and phosphorylation (Fig. 5, D and E). VGLL3 depletion repressed AMOTL2 mRNA expression but not its protein levels. Notably, knockdown of LATS2 or AMOTL2, as well as knockdown of VGLL3, significantly repressed proliferation in these cells. AMOTL2 depletion did not repress proliferation in Saos-2 cells. These results suggest that VGLL3-dependent induction of LATS2 is critically involved in tumor cell proliferation at the endogenous level (Fig. 5, F and G).
Figure 5.
The Hippo pathway is involved in tumor cell proliferation. A, MCF7, MDA-MB-231, and Saos-2 cells were analyzed for VGLL3 mRNA expression by qPCR as in Fig. 2B, except that the expression level of VGLL3 mRNA in MCF7 cells was set to 1. Asterisks indicate statistical significance (***, p < 0.001) as calculated by Student's t test. B and C, MDA-MB-231 and Saos-2 cells were transfected with the indicated siRNAs for 48 h. After incubation, cells were fixed with paraformaldehyde and stained with anti-YAP/TAZ antibody. DNA was counterstained with propidium iodide (B). The level of nuclear localized YAP/TAZ in each cell was plotted. Bars, mean ± S.D. (error bars). Asterisks indicate statistical significance (***, p < 0.001) as calculated by Student's t test (C). D, MDA-MB-231 and Saos-2 cells were transfected with the indicated siRNAs for 48 h. After incubation, cells were analyzed for VGLL3, LATS2, and AMOTL2 mRNA expression by qPCR as in Fig. 2B. Asterisks indicate statistical significance (**, p < 0.01; *, p < 0.05) as calculated by Student's t test. E and F, MDA-MB-231 and Saos-2 cells were transfected with the indicated siRNAs for 48 h. After incubation, cells were lysed and subjected to Western blotting (WB) with the indicated antibodies. G, MDA-MB-231 and Saos-2 cells were seeded and transfected with the indicated siRNAs for 48 h, and cell viability was analyzed by an MTT assay. Absorbance levels at 595 nm were plotted. Results represent the mean ± S.D. (n = 3). Asterisks indicate statistical significance (***, p < 0.001; **, p < 0.01; *, p < 0.05) as calculated by Student's t test.
Discussion
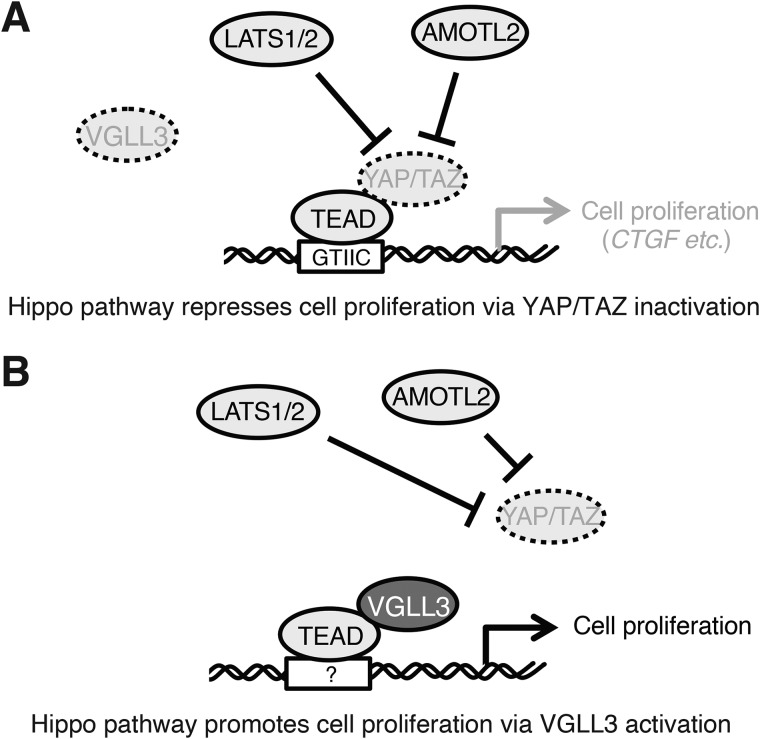
The Hippo pathway is an evolutionarily conserved regulator of tissue and cell growth. Involvement of the Hippo pathway in human tumors was initially recognized by the robust overgrowth of Drosophila melanogaster tissues that had loss-of-function mutations in components of this pathway. Subsequent studies revealed that Hippo pathway dysregulation promotes tumorigenesis in mammals and that several genes of this pathway have mutations and altered expression in human tumors. These observations established that the Hippo pathway is a tumor-suppressing signaling pathway (Fig. 6A) (7, 11, 13).
Figure 6.
Model of the roles of Hippo pathway in VGLL3-low or -high cells. A, Hippo pathway–mediated inactivation of YAP/TAZ represses cell proliferation through inhibition of TEAD-dependent gene expression (CTGF etc.) in VGLL3-low cells. B, Hippo pathway-mediated inactivation of YAP/TAZ facilitates association of VGLL3 with TEAD and then promotes cell proliferation through TEAD-dependent gene expression in VGLL3-high cells.
In this study, we found that stable expression of VGLL3 promotes cell proliferation. We detected association between VGLL3 and TEAD4, supporting the notion that VGLL3 is a cofactor for TEADs. Because Hippo pathway activation represses TEAD activity, the cell growth–promoting effect of VGLL3 led us to speculate that VGLL3 inhibits the Hippo pathway and in turn induces nuclear translocation and activation of YAP/TAZ. However, contrary to this speculation, components of the Hippo pathway, LATS2 and AMOTL2, were increased, and YAP/TAZ were inactivated in VGLL3-expressing cells. VGLL3 was highly expressed in the malignant breast tumor cell line MDA-MB-231, and its knockdown caused a reduction in LATS2 and AMOTL2 expression and induction of YAP/TAZ nuclear localization. Notably, knockdown of LATS2 or AMOTL2, as well as knockdown of VGLL3, repressed proliferation in MDA-MB-231 cells, suggesting that VGLL3-dependent up-regulation of Hippo pathway components and the following down-regulation of YAP/TAZ are required for VGLL3-dependent cell growth. Previous studies showed that VGLL1 and VGLL4 have binding sites on TEADs that overlap with YAP/TAZ-binding sites and thus compete with YAP/TAZ for TEAD binding (10, 22). Therefore, VGLL3 is likely to inactivate YAP/TAZ through the Hippo pathway to enhance binding of itself to TEADs to facilitate gene expression for proliferation. In this context, it can be concluded that the Hippo pathway has the potential to act as a tumor-promoting signaling pathway via VGLL3 activation (Fig. 6B).
Although VGLL3 was found to have a cell growth–promoting effect, probably through activation of mitogenic ERK signaling, target genes underlying this effect have not been identified. Whereas CTGF is a well-known YAP/TAZ-dependent TEAD target involved in cell proliferation (19), CTGF expression was almost completely repressed in VGLL3-expressing cells. Reporter assays showed that VGLL3 did not activate GTIIC TEAD-binding site–dependent gene expression. Therefore, we hypothesize that the DNA-binding site and target genes of the VGLL3-TEAD complex are different from those of the YAP/TAZ-TEAD complex. Given that VGLL1 was shown to facilitate anchorage-independent cell growth via expression of insulin-like growth factor-binding protein 5 (IGFBP5) (10), a cell proliferation-related gene, we analyzed IGFBP5 expression in VGLL3-expressing cells. However, we could not detect IGFBP5 expression in these cells, suggesting that IGFBP5 is not a target of the VGLL3-TEAD complex (data not shown). To determine the genes responsible for VGLL3-induced cell proliferation, analysis of gene expression profiles of a VGLL3-expressing cell is required.
In contrast to our findings, VGLL3 was hypothesized to have a tumor-suppressive role in ovarian tumor (23). This hypothesis was derived from the chromosome transfer experiment: transfer of a chromosome fragment containing the VGLL3 gene suppressed tumor phenotypes in the ovarian tumor cell line OV90. Although transfer of this fragment recovered VGLL3 expression (24), VGLL3 single-gene transfer failed to repress proliferation in OV90 cells (25). Therefore, we speculate that the hypothesis of a tumor-suppressive role of VGLL3 may be still disputable.
In conclusion, we found that VGLL3 promotes cell growth through activation of the Hippo pathway and subsequent inactivation of YAP/TAZ. Our findings suggest that the Hippo pathway can function as a tumor-promoting signaling pathway via VGLL3 activation. In VGLL3-expressing tumors, such as sarcomas and malignant breast tumors, the Hippo pathway may be a therapeutic target to inhibit VGLL3 activation.
Experimental procedures
Plasmids
cDNA fragments encoding VGLL3, YAP, or TEAD4 were generated by PCR from the reverse-transcribed product of A549 total RNA and subcloned into pIRESpuro3-CAG vector (26). VGLL3 and TEAD4 cDNAs were fused with an N-terminal Myc tag and Strep tag, respectively. The following primers were used for PCR: VGLL3, 5′-AAA AGA ATT CAT GAG TTG TGC GGA GGT GAT GTA TCA CCC CCA G-3′ (sense) and 5′-AAA AGC GGC CGC TCA GTA CCA CGG TGA TTC CTT ACT CTT GTC TTG-3′ (antisense); YAP, 5′-AAA GAA TTC ATG GAT CCC GGG CAG CAG CCG CCG CCT CAA C-3′ (sense) and 5′-TTT GCG GCC GCC TAT AAC CAT GTA AGA AAG CTT TCT TTA TCT AG-3′ (antisense); and TEAD4, 5′-AAA GGA TCC GAG GGC ACG GCC GGC ACC ATT ACC TC-3′ (sense) and 5′-TTT GAA TTC TCA TTC TTT CAC CAG CCT GTA GAT GTG-3′ (antisense). The 8× GTIIC (5′-ACATTCCA-3′) reporter plasmid was constructed as follows. The enhanced GFP (EGFP) expression vector (pcDNA/EGFP) (26) was digested with MluI and BamHI, and the cytomegalovirus promoter was removed. The synthesized DNA fragment containing 8× GTIIC sites and a minimal promoter was inserted into the MluI and BamHI sites of pcDNA/EGFP vector. The sequence of the synthesized DNA fragment is shown in Table S1. pCAG/TetR vector (26), which constitutively expresses tetracycline repressor (TetR), was used to assess transfection efficiency.
Antibodies
The following antibodies were used: anti-Myc (9E10; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-ERK2 (sc-154; Santa Cruz Biotechnology), anti-Strep (GT661; Genetex, Irvine, CA, USA), anti-YAP/TAZ (8418; Cell Signaling Technology, Beverly, MA, USA), anti-LATS1 (3477; Cell Signaling Technology), anti-LATS2 (5888; Cell Signaling Technology), anti-phospho-LATS1/2 (9157; Cell Signaling Technology), anti-phospho-ERK (4377; Cell Signaling Technology), anti-AMOTL2 (AP8860c; ABGENT, San Diego, CA, USA), anti-GFP (mFX75; Fujifilm Wako, Tokyo, Japan), anti-actin (clone C4; Merck Millipore, Darmstadt, Germany), anti-TEAD1 (GT13112; Genetex), anti-TEAD4 (ab58310; Abcam), and anti-TetR (TET01; MoBiTech). Horseradish peroxidase-F(ab′)2 secondary antibodies were purchased from GE Healthcare.
Cells and transfection
A549, MDA-MB-231, MCF7, and Saos-2 cells were cultured in Dulbecco's modified Eagle's medium (Nissui, Tokyo, Japan) containing 5% fetal bovine serum at 37 °C. To establish stable cell pools, A549 and MDA-MB-231 cells were transfected with pIRESpuro3-CAG/Myc-VGLL3 vector using Lipofectamine2000 (Thermo Fisher Scientific) following the manufacturer's instructions. Transfected cells were then selected for puromycin resistance. siRNAs (5 nm) were transfected by the reverse-transfection method with Lipofectamine RNAiMax (Thermo Fisher Scientific) following the manufacturer's instructions. Silencer select siRNAs for VGLL3 (s52380 (#1), s52378 (#2)), LATS1 (s17393), LATS2 (s25505), TEAD1 (s13961), TEAD2 (s16075), TEAD3 (s13967), TEAD4 (s13964), AMOTL2 (s28109), and mock siRNA (4390843) were purchased from Thermo Fisher Scientific.
Western blotting and immunoprecipitation
Western blotting and immunoprecipitation were performed as described previously (26). Whole-cell lysates prepared in SDS-sample buffer were subjected to SDS-PAGE and electrotransferred onto polyvinylidene difluoride membranes. Protein bands were detected with the appropriate antibodies and analyzed with a ChemiDoc XRS+ image analyzer (Bio-Rad). The intensity of bands was measured by Quantity One software (Bio-Rad).
Quantitative real-time PCR (qPCR) analysis
Total RNA was isolated from cells with the RNAiso Plus reagent (Takara, Shiga, Japan), and cDNAs were synthesized from 0.5 µg of each RNA preparation using the ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan), following the manufacturer's instructions. The following primers were used for PCR: GAPDH, 5′-GGA GCG AGA TCC CTC CAA AAT-3′ (sense) and 5′-GGC TGT TGT CAT ACT TCT CAT GG-3′ (antisense); LATS2, 5′-ACC CCA AAG TTC GGA CCT TAT-3′ (sense) and 5′-CAT TTG CCG GTT CAC TTC TGC-3′ (antisense); AMOTL1, 5′-GAA ACA TCT GCT TTG ACG GTG G-3′ (sense) and 5′-GAA GTT TGG GGA GTG GAA GTT AC-3′ (antisense); AMOTL2, 5′-ACC ATG CGG AAC AAG ATG GAC-3′ (sense) and 5′-GGC GGC GAT TTG CAG ATT C-3′ (antisense); AMOT, 5′-ACC TCG TGA AGT CAT CCT CCA-3′ (sense) and 5′-CCT CCG AAT CTC GCC CTC TA-3′ (antisense); CTGF, 5′-CAG CAT GGA CGT TCG TCT G-3′ (sense) and 5′-AAC CAC GGT TTG GTC CTT GG-3′ (antisense); CYR61, 5′-ACC GCT CTG AAG GGG ATC T-3′ (sense) and 5′-ACT GAT GTT TAC AGT TGG GCT G-3′ (antisense); ANKRD1, 5′-AGT AGA GGA ACT GGT CAC TGG-3′ (sense) and 5′-TGT TTC TCG CTT TTC CAC TGT T-3′ (antisense); VGLL3, 5′-TAT GGA GCG TCC CAG TAT CTG-3′ (sense) and 5′-TGA ATA CCG CTA ACT TCT TCT GC-3′ (antisense); TEAD2, 5′-CTT CGT GGA ACC GCC AGA T-3′ (sense) and 5′-GGA GGC CAC CCT TTT TCT CA-3′ (antisense); TEAD3, 5′-TCA TCC TGT CAG ACG AGG G-3′ (sense) and 5′-TCT TCC GAG CTA GAA CCT GTA TG-3′ (antisense); and IGFBP5, 5′-ACC TGA GAT GAG ACA GGA GTC-3′ (sense) and 5′-GTA GAA TCC TTT GCG GTC ACA A-3′ (antisense). After initial denaturation at 95 °C for 1 min, PCR was performed for 40 cycles (15 s at 95 °C and 45 s at 60 °C) using the Thunderbird SYBR Green Polymerase Kit (Toyobo) and Eco Real-Time PCR System (Illumina, San Diego, CA, USA).
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay
Cells were plated with 5,000 cells/well in a 96-well plate and transfected with the indicated siRNAs for 2 days. After incubation, the solution in each well was replaced with Dulbecco's modified Eagle's medium containing 0.5 mg/ml MTT (Dojindo, Kumamoto, Japan) and incubated for an additional 1 h. The resulting crystalized product was dissolved in 100 µl of 100% DMSO and measured at 595 nm using a Filter Max F5 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Immunofluorescence
Images were obtained using an LSM 780 confocal laser-scanning microscope with a ×63 1.40 numerical aperture oil-immersion objective (Zeiss, Jena, Germany) as described previously (27, 28). One planar (xy) slice (1.0-μm thickness) was shown in all experiments. Briefly, cells were fixed in 4% paraformaldehyde for 20 min at room temperature and permeabilized in PBS containing 0.2% Triton X-100 and 3% BSA at room temperature. Cells were subsequently reacted with an appropriate primary antibody for 1 h, washed with PBS containing 0.1% saponin, and stained with an Alexa Fluor 488– or Alexa Fluor 546–conjugated secondary antibody for 1 h. DNA was stained with a mixture of 20 µg/ml propidium iodide and 200 µg/ml RNase A for 30 min Stained cells were mounted with Prolong Antifade reagent (Thermo Fisher Scientific). Intensities of nuclear localized YAP/TAZ were quantified by ZEN software (Zeiss).
Data availability
All data are contained within the article.
Supplementary Material
This article contains supporting information.
Author contributions— N. H., K. O., Y. T., H. T., Naoto Yamaguchi, and Noritaka Yamaguchi data curation; N. H., K. O., and Y. T. investigation; K. O., Y. T., H. T., and Naoto Yamaguchi formal analysis; H. T., Naoto Yamaguchi, and Noritaka Yamaguchi resources; Naoto Yamaguchi and Noritaka Yamaguchi funding acquisition; Noritaka Yamaguchi conceptualization; Noritaka Yamaguchi supervision; Noritaka Yamaguchi validation; Noritaka Yamaguchi writing-original draft; Noritaka Yamaguchi project administration.
Funding and additional information—This work was supported in part by Grant-in-aid for Challenging Research (Exploratory) 19K22482 (to Noritaka Yamaguchi) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; grants from the Takeda Science Foundation (to Noritaka Yamaguchi), the Foundation for Promotion of Cancer Research in Japan (to Noritaka Yamaguchi), and the Chiba Foundation for Health Promotion and Disease Prevention (to Naoto Yamaguchi); and a scholarship donation from the Daiichi Sankyo Co., Ltd. (to Naoto Yamaguchi).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- VGLL
- vestigial-like 3
- TEAD
- TEA domain–containing transcription factor
- YAP
- Yes-associated protein
- TAZ
- transcriptional coactivator with PDZ-binding motif
- ERK
- extracellular signal–regulated kinase
- CTGF
- connective tissue growth factor
- CYR61
- cysteine-rich angiogenic inducer 61
- ANKRD1
- ankyrin repeat domain 1
- AMOT
- angiomotin
- IGFBP5
- insulin-like growth factor-binding protein 5
- EGFP
- enhanced GFP
- TetR
- tetracycline repressor
- qPCR
- quantitative real-time PCR
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MTT
- 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide.
References
- 1. Figeac N., Mohamed A. D., Sun C., Schönfelder M., Matallanas D., Garcia-Munoz A., Missiaglia E., Collie-Duguid E., De Mello V., Pobbati A. V., Pruller J., Jaka O., Harridge S. D. R., Hong W., Shipley J., et al. (2019) VGLL3 operates via TEAD1, TEAD3 and TEAD4 to influence myogenesis in skeletal muscle. J. Cell Sci. 132, jcs225946 10.1242/jcs.225946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simon E., Faucheux C., Zider A., Thézé N., and Thiebaud P. (2016) From vestigial to vestigial-like: the Drosophila gene that has taken wing. Dev. Genes Evol. 226, 297–315 10.1007/s00427-016-0546-3 [DOI] [PubMed] [Google Scholar]
- 3. Hallor K. H., Sciot R., Staaf J., Heidenblad M., Rydholm A., Bauer H. C., Aström K., Domanski H. A., Meis J. M., Kindblom L. G., Panagopoulos I., Mandahl N., and Mertens F. (2009) Two genetic pathways, t(1;10) and amplification of 3p11-12, in myxoinflammatory fibroblastic sarcoma, haemosiderotic fibrolipomatous tumour, and morphologically similar lesions. J. Pathol. 217, 716–727 10.1002/path.2513 [DOI] [PubMed] [Google Scholar]
- 4. Hélias-Rodzewicz Z., Pérot G., Chibon F., Ferreira C., Lagarde P., Terrier P., Coindre J. M., and Aurias A. (2010) YAP1 and VGLL3, encoding two cofactors of TEAD transcription factors, are amplified and overexpressed in a subset of soft tissue sarcomas. Genes Chromosomes Cancer 49, 1161–1171 10.1002/gcc.20825 [DOI] [PubMed] [Google Scholar]
- 5. Antonescu C. R., Zhang L., Nielsen G. P., Rosenberg A. E., Dal Cin P., and Fletcher C. D. (2011) Consistent t(1;10) with rearrangements of TGFBR3 and MGEA5 in both myxoinflammatory fibroblastic sarcoma and hemosiderotic fibrolipomatous tumor. Genes Chromosomes Cancer 50, 757–764 10.1002/gcc.20897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang L. H., Wang Z., Li L. H., Liu Y. K., Jin L. F., Qi X. W., Zhang C., Wang T., and Hua D. (2019) Vestigial like family member 3 is a novel prognostic biomarker for gastric cancer. World J. Clin. Cases 7, 1954–1963 10.12998/wjcc.v7.i15.1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin K. C., Park H. W., and Guan K. L. (2017) Regulation of the Hippo pathway transcription factor TEAD. Trends Biochem. Sci. 42, 862–872 10.1016/j.tibs.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng Y., and Pan D. (2019) The Hippo signaling pathway in development and disease. Dev. Cell 50, 264–282 10.1016/j.devcel.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calses P. C., Crawford J. J., Lill J. R., and Dey A. (2019) Hippo pathway in cancer: aberrant regulation and therapeutic opportunities. Trends Cancer 5, 297–307 10.1016/j.trecan.2019.04.001 [DOI] [PubMed] [Google Scholar]
- 10. Pobbati A. V., Han X., Hung A. W., Weiguang S., Huda N., Chen G. Y., Kang C., Chia C. S., Luo X., Hong W., and Poulsen A. (2015) Targeting the central pocket in human transcription factor TEAD as a potential cancer therapeutic strategy. Structure 23, 2076–2086 10.1016/j.str.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moroishi T., Hansen C. G., and Guan K. L. (2015) The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hao Y., Chun A., Cheung K., Rashidi B., and Yang X. (2008) Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 10.1074/jbc.M709037200 [DOI] [PubMed] [Google Scholar]
- 13. Harvey K. F., Zhang X., and Thomas D. M. (2013) The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 10.1038/nrc3458 [DOI] [PubMed] [Google Scholar]
- 14. Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., and Piccolo S. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 15. Mahoney W. M. Jr., Hong J. H., Yaffe M. B., and Farrance I. K. (2005) The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem. J. 388, 217–225 10.1042/BJ20041434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao J. H., Davidson I., Matthes H., Garnier J. M., and Chambon P. (1991) Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65, 551–568 10.1016/0092-8674(91)90088-G [DOI] [PubMed] [Google Scholar]
- 17. Li C., Srivastava R. K., Elmets C. A., Afaq F., and Athar M. (2013) Arsenic-induced cutaneous hyperplastic lesions are associated with the dysregulation of Yap, a Hippo signaling-related protein. Biochem. Biophys. Res. Commun. 438, 607–612 10.1016/j.bbrc.2013.08.008 [DOI] [PubMed] [Google Scholar]
- 18. Zhang H., Pasolli H. A., and Fuchs E. (2011) Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc. Natl. Acad. Sci. U.S.A. 108, 2270–2275 10.1073/pnas.1019603108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., and Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao B., Li L., Lu Q., Wang L. H., Liu C. Y., Lei Q., and Guan K. L. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 10.1101/gad.2000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Shen H., Withers H. G., Yang N., Denson K. E., Mussell A. L., Truskinovsky A., Fan Q., Gelman I. H., Frangou C., and Zhang J. (2017) VGLL4 selectively represses YAP-dependent gene induction and tumorigenic phenotypes in breast cancer. Sci. Rep. 7, 6190 10.1038/s41598-017-06227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiao S., Wang H., Shi Z., Dong A., Zhang W., Song X., He F., Wang Y., Zhang Z., Wang W., Wang X., Guo T., Li P., Zhao Y., Ji H., et al. (2014) A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180 10.1016/j.ccr.2014.01.010 [DOI] [PubMed] [Google Scholar]
- 23. Cody N. A., Ouellet V., Manderson E. N., Quinn M. C., Filali-Mouhim A., Tellis P., Zietarska M., Provencher D. M., Mes-Masson A. M., Chevrette M., and Tonin P. N. (2007) Transfer of chromosome 3 fragments suppresses tumorigenicity of an ovarian cancer cell line monoallelic for chromosome 3p. Oncogene 26, 618–632 10.1038/sj.onc.1209821 [DOI] [PubMed] [Google Scholar]
- 24. Cody N. A., Shen Z., Ripeau J. S., Provencher D. M., Mes-Masson A. M., Chevrette M., and Tonin P. N. (2009) Characterization of the 3p12.3-pcen region associated with tumor suppression in a novel ovarian cancer cell line model genetically modified by chromosome 3 fragment transfer. Mol. Carcinog. 48, 1077–1092 10.1002/mc.20535 [DOI] [PubMed] [Google Scholar]
- 25. Gambaro K., Quinn M. C., Wojnarowicz P. M., Arcand S. L., de Ladurantaye M., Barrès V., Ripeau J. S., Killary A. M., Davis E. C., Lavoie J., Provencher D. M., Mes-Masson A. M., Chevrette M., and Tonin P. N. (2013) VGLL3 expression is associated with a tumor suppressor phenotype in epithelial ovarian cancer. Mol. Oncol. 7, 513–530 10.1016/j.molonc.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamaguchi N., Nakayama Y., and Yamaguchi N. (2017) Down-regulation of Forkhead box protein A1 (FOXA1) leads to cancer stem cell-like properties in tamoxifen-resistant breast cancer cells through induction of interleukin-6. J. Biol. Chem. 292, 8136–8148 10.1074/jbc.M116.763276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuki R., Tatewaki T., Yamaguchi N., Aoyama K., Honda T., Kubota S., Morii M., Manabe I., Kuga T., Tomonaga T., and Yamaguchi N. (2019) Desuppression of TGF-β signaling via nuclear c-Abl-mediated phosphorylation of TIF1γ/TRIM33 at Tyr-524, -610, and -1048. Oncogene 38, 637–655 10.1038/s41388-018-0481-z [DOI] [PubMed] [Google Scholar]
- 28. Aoyama K., Fukumoto Y., Ishibashi K., Kubota S., Morinaga T., Horiike Y., Yuki R., Takahashi A., Nakayama Y., and Yamaguchi N. (2011) Nuclear c-Abl-mediated tyrosine phosphorylation induces chromatin structural changes through histone modifications that include H4K16 hypoacetylation. Exp. Cell Res. 317, 2874–2903 10.1016/j.yexcr.2011.09.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.