Abstract
The complement system is a tightly controlled proteolytic cascade in the innate immune system, which tags intruding pathogens and dying host cells for clearance. An essential protein in this process is complement component C3. Uncontrolled complement activation has been implicated in several human diseases and disorders and has spurred the development of therapeutic approaches that modulate the complement system. Here, using purified proteins and several biochemical assays and surface plasmon resonance, we report that our nanobody, hC3Nb2, inhibits C3 deposition by all complement pathways. We observe that the hC3Nb2 nanobody binds human native C3 and its degradation products with low nanomolar affinity and does not interfere with the endogenous regulation of C3b deposition mediated by Factors H and I. Using negative stain EM analysis and functional assays, we demonstrate that hC3Nb2 inhibits the substrate–convertase interaction by binding to the MG3 and MG4 domains of C3 and C3b. Furthermore, we notice that hC3Nb2 is cross-reactive and inhibits the lectin and alternative pathway in murine serum. We conclude that hC3Nb2 is a potent, general, and versatile inhibitor of the human and murine complement cascades. Its cross-reactivity suggests that this nanobody may be valuable for analysis of complement activation within animal models of both acute and chronic diseases.
Keywords: single-domain antibody, complement system, inhibitor, innate immunity, antibody, structural biology, nanobody, C3 convertase, single-domain antibody (sdAb nanobody)
The complement system is a prominent part of our innate immune system and serves as a first line of defense through opsonization and lysis of intruding pathogens. Likewise, complement contributes to the clearance of dying host cells, which is crucial in homeostasis and minimizes the development of autoimmune diseases (1). The system also plays a role in development and aids the sculpting of the early developing brain by promoting the pruning of excess synapses (2). Complement is activated through three pathways, the classical pathway (CP), the lectin pathway (LP), and the alternative pathway (AP). The complement cascade may be triggered upon binding of pattern-recognizing molecules (PRMs) to pathogen-associated molecular patterns and danger-associated molecular patterns presented on the activating surface. In the CP, the PRM C1q detects antibody-antigen complexes and multiple other patterns (3), whereas five different PRMs of the LP recognize specific carbohydrate structures or patterns of acetyl groups (4). Serine proteases associated with the PRMs cleave complement factor C4 (C4) and C2, leading to the appearance of the classical pathway C3 convertase C4b2a formed by C4b and the active serine protease C2a on the activator surface (5) (see Fig. 1A). This C3 convertase cleaves C3 into C3b and the anaphylatoxin C3a. The C3b deposited on the activator associates with Factor B (FB) to form the AP proconvertase C3bB that, upon activation by Factor D (FD), gives rise to the active AP C3 convertase, C3bBb (Fig. 1A). The alternative pathway strongly amplifies the outcome of the CP and LP (6, 7), but the AP may also initiate independently through the tick-over of C3. Here, the thioester of C3 hydrolyzes, which induces structural rearrangements in C3 giving rise to C3(H2O), a functional homologue of C3b. This allows for the association with FB and ultimately the formation of a fluid-phase AP C3 convertase, C3(H2O)Bb (8).
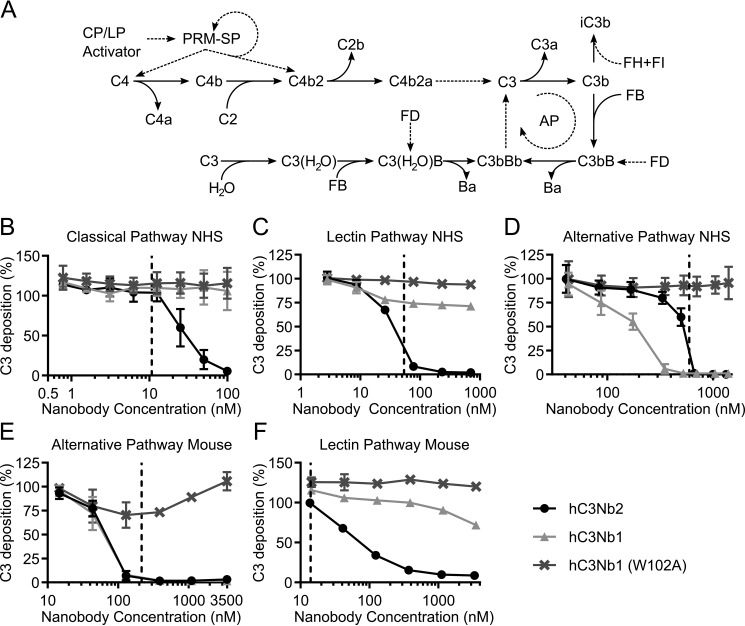
Figure 1.
The hC3Nb2 nanobody inhibits all complement activation pathways in human and mouse serum. A, the activation pathways of complement. PRMs of the CP and LP bind the activating surface. This binding activates the PRM-associated serine proteases (PRM-SP) that cleave C4 and the C2 in the proconvertase C4b2. This results in generation of the CP C3 convertase, C4b2a. This convertase cleaves C3 into C3a and C3b, and C3b may then associate with FB. FD cleaves the C3b-bound FB, resulting in the formation of the AP C3 convertase, C3bBb. C3 can spontaneously hydrolyze, and the resulting hydrolysis product C3(H2O) is a functional homolog of C3b and can associate with FB to, upon activation by FD, form the fluid-phase AP C3 convertase, C3(H2O)Bb. Dashed lines, proteolytic activation. Solid lines, conversions. B, hC3Nb2 is a potent classical pathway inhibitor of C3 fragment deposition onto a surface of heat-aggregated IgG in 0.2% NHS. C, as in B but on a mannan surface, showing that hC3Nb2 also inhibits C3 fragment deposition through the lectin pathway in 5% NHS. D, hC3Nb2 likewise suppresses C3 fragment deposition by the alternative pathway onto a surface of zymosan in 11% NHS. E, inhibition by nanobodies of the alternative pathway in 5% mouse serum (i.e. C3 fragment deposition onto zymosan-coated surfaces). F, inhibition of the LP (i.e. deposition onto a mannan surface) in 0.3% mouse serum. B–F display the C3 deposition at the indicated nanobody concentrations. The C3 deposition was normalized to the C3 deposition obtained without added nanobodies (100% deposition). The effect of hC3Nb2 (black) was compared with the strong alternative pathway inhibitor hC3Nb1 (light gray) and the inactive hC3Nb1 (W102A) mutant (dark gray). The data for hC3Nb1 and hC3Nb1 (W102A) in C and E were published previously (20). Dashed lines, final C3 concentration at the given serum dilution, assuming a C3 concentration of 5.3 µm in undiluted human serum (55) and 4.2 µm in undiluted mouse serum (56). Average and S.D. (error bars) are shown for n = 3 experiments in B and D and n = 2 experiments in C, E, and F.
If left uncontrolled, complement activation proceeds to the terminal pathway. Once C3b reaches a threshold surface density on the activator, the C3 convertases switch substrate specificity to C5. The appearance of these C5 convertases, which cleave C5 into C5b and the potent anaphylatoxin C5a, demarcates the initiation of the terminal pathway. C5b nucleates the assembly of the membrane attack complex that perforates and lyses pathogens (9). On healthy host cells, the terminal pathway is not activated because deposited C3b is rapidly degraded to iC3b by Factor I (FI) with the help of the co-factors factor H (FH), membrane cofactor protein (MCP/CD46), and complement receptor 1 (CR1). The resulting iC3b is incapable of forming a C3 convertase with FB. Additionally, decay acceleration factor (DAF/CD55), CR1, and FH irreversibly dissociate the AP C3 convertase (5). In contrast, properdin, also known as Factor P, is the only positive regulator of the AP C3 and C5 convertases and acts by stimulating AP proconvertase assembly, inhibiting AP convertase dissociation, and competing with FI for binding to C3b (10, 11).
Mutations in AP components are associated with the development of a number of diseases, including the chronic renal diseases C3 glomerulopathies (C3G) and atypical hemolytic uraemic syndrome (12, 13) as well as age-related macular degeneration, which leads to loss of vision (14). In the chronic disease paroxysmal nocturnal hemoglobinuria (PNH), dysregulation of the terminal pathway due to mutations in genes required for glycosylphosphatidylinositol anchor synthesis leads to erythrocyte lysis (15). Furthermore, complement activation has also been linked to multiple neurological diseases, including schizophrenia (16) and early stages of Alzheimer's disease (17). In addition to these chronic diseases, the complement system plays a cardinal role in the progression of various acute diseases, such as ischemia-reperfusion injury (18).
With the aim of controlling such dysregulated complement in patients, a plethora of different molecules, targeting receptors, proteases, and their substrates, have been developed (19). We have recently described the first nanobody (Nb)-based complement modulator, called hC3Nb1, that inhibits the AP in both human and murine serum (20). Nbs originate from the VHH domain of the heavy chain–only antibodies present in camelids. However, hC3Nb1 only exerts subtle effects on C3 cleavage by the CP C3 convertase, thus limiting the use of the Nb in the assessment of CP- and LP-driven diseases. Furthermore, hC3Nb1 prevents FI degradation of C3b, which may be undesirable for in vivo applications.
We thus set out to develop a C3-specific Nb that broadly inhibits complement through complete shutdown of the cascade, while still allowing negative regulation of C3b on host cells. Here we describe hC3Nb2, which binds C3, C3b, and a C3(H2O) mimic with low nanomolar affinity. The Nb inhibits both the AP and CP C3 convertase in human serum without interfering with the degradation of C3b to iC3b and is also cross-reactive with mouse C3 and is functional in murine serum. Through a combination of biochemical assays and negative stain EM (nsEM), we rationalize that hC3Nb2 exerts its function through inhibition of substrate binding to C3 convertases. We thus present a potent complement-specific nanobody, suitable for studies of the complement pathway in human diseases and murine models that may develop into a candidate for therapeutic control of complement-driven pathogenesis.
Results
hC3Nb2 inhibits all complement pathways
We previously described the generation of a phage library presenting C3-specific nanobodies after immunization of a llama with human C3b. From this library, we selected nanobodies against immobilized human C3b. Using this approach, we not only selected the AP inhibitor, hC3Nb1 (20), but also the hC3Nb2 nanobody presented here. First, we tested the effect of hC3Nb2 in classical and lectin pathway assays conducted in ELISA plates coated with either aggregated IgG or mannan. We compared the hC3Nb2 nanobody with our AP inhibitor hC3Nb1 and quantified the inhibition of the pathways by measuring the C3 fragments deposited on the surface. In our CP assay, we observed that, in contrast to the AP-specific hC3Nb1, the hC3Nb2 nanobody inhibits the C3 fragment deposition upon activation of the classical pathway when present in molar excess compared with C3 (Fig. 1B). Similarly, the nanobody inhibits C3 fragment deposition through the lectin pathway, indicating that hC3Nb2 inhibits C3 cleavage by the C4bC2a C3 convertase generated through the CP and the LP (Fig. 1C). In contrast, the AP-specific hC3Nb1 nanobody has only a very modest effect on this convertase. We proceeded to test the effect of the nanobody in an assay in which zymosan drives activation of the AP at conditions where the CP and LP are nonfunctional. This assay revealed that the nanobody also inhibits the alternative pathway in both human and mouse serum (Fig. 1, D and E). To confirm the cross-reactivity of the nanobody, we tested the effect of the nanobody in the lectin pathway in murine serum (Fig. 1F). Similar to the lectin pathway in human serum, hC3Nb2 inhibited the progression of the murine lectin pathway. Overall, these assays showed that hC3Nb2 is a broad inhibitor of the complement system capable of inhibiting C3 cleavage in all three pathways.
hC3Nb2 inhibits classical pathway–induced hemolysis
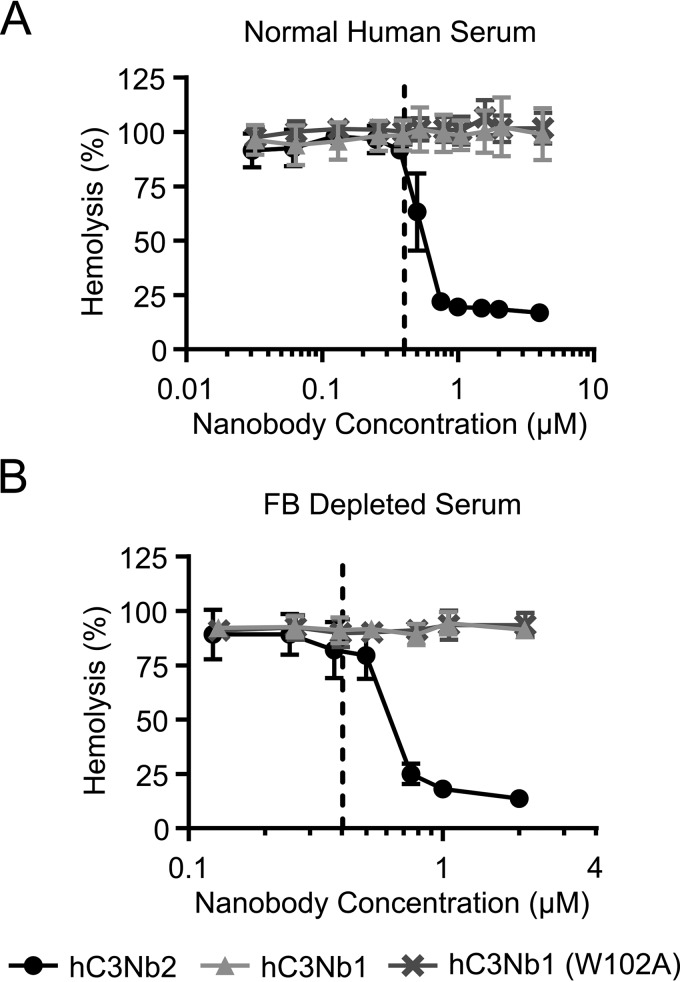
Next, we tested whether the inhibitory effect of the nanobody extends to hemolysis of sheep erythrocytes onto which classical pathway activity was induced with rabbit anti-sheep erythrocyte antibodies. We incubated the antibody-decorated cells in normal human serum (NHS) in the presence or absence of hC3Nb2. Again, we compared the effect to hC3Nb1 to confirm that the effect of hC3Nb2 was due to inhibition of the classical pathway, because hC3Nb1 effectively inhibits generation of C3b through the AP. In accordance with the observation that hC3Nb2 inhibits the classical pathway–mediated deposition of C3, the nanobody also inhibited the lysis of the sheep erythrocytes at concentrations exceeding that of C3 in the assay (Fig. 2A). The nanobody inhibited the hemolysis to ∼15% at a concentration of 1 µm in these assay conditions where C3 was present at 0.4 µm. When hC3Nb2 was replaced by the AP-specific inhibitor hC3Nb1 or its inactive derivative hC3Nb1 W102A, no inhibition of lysis was observed, indicating that AP amplification played an insignificant role in this hemolysis assay. To confirm that the CP was the dominating driver of hemolysis in this assay, we also conducted the hemolysis assay in FB-depleted serum in which AP amplification is absent (Fig. 2B). Again, hC3Nb2 inhibited the hemolysis when present in molar excess to C3, and the remaining lysis plateaued at ∼15%. These lysis experiments demonstrated that in addition to preventing C3 deposition, hC3Nb2 is a potent inhibitor of the progression of the complement cascade from the classical pathway to the terminal pathway.
Figure 2.
The hC3Nb2 inhibits classical pathway-mediated hemolysis. The classical pathway was activated on sheep erythrocytes by anti-sheep erythrocyte antibodies. The activated sheep erythrocytes were incubated in 7.5% NHS (A) or FB-depleted serum (B). The effects of hC3Nb2 (black) were compared with those of the alternative pathway inhibitor hC3Nb1 (light gray) and its inactive hC3Nb1 (W102A) mutant (dark gray). Lysis, as measured as absorption at 405 nm in the supernatants, was normalized to lysis by H2O (100%), whereas erythrocytes incubated in PBS were defined as 0% lysis. Dashed lines, putative C3 concentration in 7.5% serum. Average and S.D. (error bars) are shown for n = 3 experiments in A and n = 2 experiments in B.
hC3Nb2 binds C3, C3b, and C3MA with high affinity
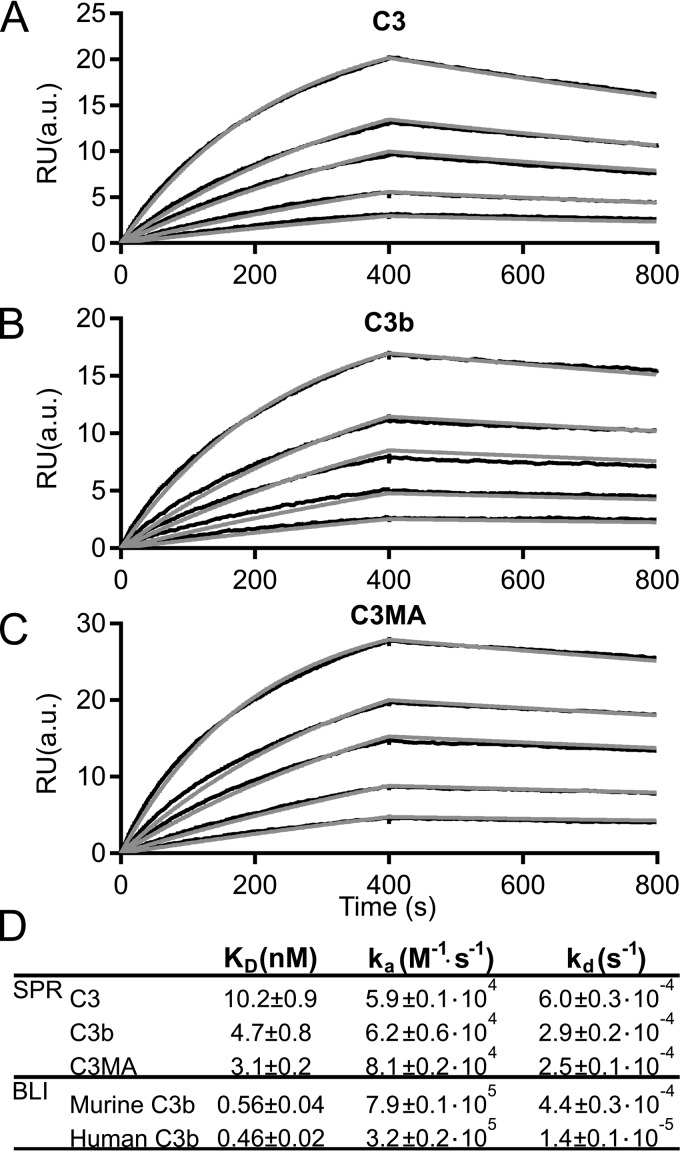
The C3b deposition and hemolysis assays in Figs. 1 and 2 indicate that a 1:1 molar ratio between hC3Nb2 and the C3 is needed for efficient inhibition of complement. However, the equilibrium dissociation constant (KD) as well as the rate constants for binding and dissociation are other key parameters for evaluating the potential of an inhibitory antibody. We assessed these constants by surface plasmon resonance (SPR). Here, hC3Nb2 was immobilized as ligand, utilizing a biotinylated C-terminal AviTag for binding to a streptavidin-coated chip. Increasing concentrations in the range of 5-60 nm of C3, C3b, and the C3(H2O) analog C3MA were injected as analytes over the chip. The rate and dissociation constants for complex formation with hC3Nb2 were determined by fitting the binding curves to a 1:1 binding mode (Fig. 3, A–D). The nanobody binds the three functional states of C3 with similar dissociation constants in the low nanomolar range (Fig. 3D). The off-rates were likewise similar between the three states and were on the order of 10−4 s−1. These SPR data showed that the nanobody binds with high affinity to a structurally conserved epitope present in C3, C3b, and the C3 tick-over mimic C3MA. We next analyzed the binding of hC3Nb2 to murine C3b. To this end, we coated bio-layer interferometry (BLI) sensors with biotinylated hC3Nb2 and transferred the sensors to wells containing murine C3b at concentrations ranging from 0.2 to 6.25 nm. Similar to the SPR data, we fitted the sensorgrams to a 1:1 binding mode (Fig. S1A). These data revealed that hC3Nb2 binds murine C3b with sub-nanomolar affinity (Fig. 3D). To allow direct comparison with binding to human C3b, we performed a parallel BLI experiment with human C3b and similarly fitted the data in a 1:1 model (Fig. S1B). Collectively, these data confirmed that the hC3Nb2 binds with similar affinity to murine and human C3b (Fig. 3D). Based on the similarity of dissociation constants for hC3Nb2 interaction with human C3 degradation fragments, we predict that the hC3Nb2 exhibits comparable affinities toward degradation products of murine C3.
Figure 3.
The hC3Nb2 binds multiple functional states of C3 with nanomolar affinity. hC3Nb2 was immobilized through a biotinylated C-terminal AviTag on a streptavidin-coated surface plasmon resonance flow sensor. The analyte was injected over the hC3Nb2-coated sensor at concentrations of 5, 10, 20, 30, or 60 nm C3 (A), C3b (B), or C3MA (C). D, summary of the SPR and BLI binding and rate constants. Binding and rate constants from SPR were determined using the BiaCore T200 evaluation software; n = 3 for C3 and C3b, n = 2 for C3MA. BLI binding curves are presented in Fig. S1. Binding and rate constants from BLI-based experiments were determined as described under “Experimental procedures”; n = 2 for murine C3b, n = 2 for human C3b. RU, response units. a.u., arbitrary units.
hC3Nb2 inhibits C3 cleavage on the substrate level
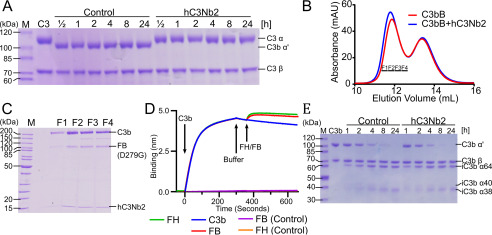
The hC3Nb2 nanobody was selected against human C3b and inhibits both CP mediated C3 deposition and hemolysis (Figs. 1 and 2). Together with the strong binding to native C3 observed by SPR, these data prompted the hypothesis that hC3Nb2 inhibits the C3 cleavage on the C3 substrate level rather than on the product level, because C3b has no function in the CP C3 convertase. To challenge this model, we tested whether the nanobody inhibits C3 cleavage by the alternative pathway analog, Cobra venom factor Bb (CVFBb). We assembled and activated CVFB with Factor D prior to mixing with C3 in the presence or absence of a 2-fold excess of hC3Nb2. Here, we observed that the nanobody potently inhibits the C3 cleavage, which was completed within the first 30 min in the absence of the nanobody but barely detectable after 24 h in the presence of hC3Nb2 (Fig. 4A). To preclude the possibility that this effect was due to hC3Nb2 interaction with CVF, we investigated their interaction by SPR. Again, hC3Nb2 was immobilized as ligand on the chip while CVF was applied as analyte. Sensorgrams obtained in the presence of CVF were indistinguishable from buffer control sensorgrams, indicating that the nanobody does not interact with CVF (Fig. S2). In summary, the data obtained with CVF confirmed the model that hC3Nb2 inhibits the C3 cleavage on the substrate level.
Figure 4.
The hC3Nb2 nanobody inhibits C3 cleavage but allows endogenous regulation by FI degradation. A, time course experiment where C3 degradation by CVFBb at 37 °C was monitored by SDS-PAGE in the presence or absence of hC3Nb2. Nanobody binding to C3 completely prevents cleavage revealing inhibition at the substrate level. B and C, proconvertase assembly assay. C3b was mixed with a 1.5-fold molar excess of the inactive, stabilized FB (D279G/S699A) and subjected to size-exclusion chromatography in either the presence or absence of a 2-fold molar excess of hC3Nb2. Nonreduced SDS-PAGE analysis of the fractions from B marked by bars reveals that FB and hC3Nb2 do not compete for binding to C3b. D, competition assay where immobilized hC3Nb2 on BLI sensors was dipped in 50 nm C3b. The sensors were subsequently transferred to FB (D279G) or FH. In addition, the binding of FB (D279G) and FH alone were analyzed for nonspecific background binding. Sensorgrams for FB (D279G) and FH are indistinguishable. E, a FH-mediated FI cleavage assay reveals that hC3Nb2 does not interfere with C3b degradation. The C3b was incubated with 0.2% (w/w) FH and 1% (w/w) FI for the indicated time at 37 °C in either the presence or absence of a 1.2-fold molar excess of hC3Nb2.
hC3Nb2 allows AP proconvertase formation
Prompted by the observation that the hC3Nb2 inhibits the C3 cleavage on substrate level by the C3 convertases, we turned to analyze its effect on the alternative pathway convertase. This is more difficult, because C3 is the substrate of the convertase and C3b is the noncatalytic subunit of the AP C3 convertase. We first tested whether the nanobody inhibits the assembly of the AP convertase because our hC3Nb1 has this effect (20). For the assembly of the AP C3 proconvertase, we used FB comprising the S699A and the D279G mutations; whereas the S699A renders the FB proteolytically inactive, the D279G mutation extends the t½ of the convertase (21). We incubated the FB S699A/D279G with C3b in the absence or presence of a 2-fold molar excess of hC3Nb2. A subsequent size-exclusion chromatography (SEC) analysis (Fig. 4B) indicated that the nanobody allows the assembly of the proconvertase, as indicated by a slightly earlier elution of the proconvertase in the presence of hC3Nb2. SDS-PAGE analysis confirmed the presence of a ternary complex of C3b, FB, and hC3Nb2 in the SEC peak fractions (Fig. 4C). To validate the SEC results in an orthogonal approach, we performed a BLI-based assessment of the effect of hC3Nb2 on proconvertase assembly. To this end, we immobilized hC3Nb2 on BLI sensors and loaded C3b from the well. Next, we applied FB (D279G) to the nanobody-bound C3b, which resulted in a marked increase in the signal compared with C3b alone (Fig. 4D). This confirms that the FB- and hC3Nb2-binding sites do not compete and underscores that the nanobody does not prevent the AP C3 convertase assembly.
The hC3Nb2 nanobody allows FH-assisted FI degradation of C3b
The data presented thus far show that hC3Nb2 is a potent inhibitor of C3 cleavage through the three activation pathways. However, a potential inhibition of the endogenous regulatory function undertaken by FI could result in an accumulation of C3b on activating surfaces. In an in vivo setting, such an accumulation may result in a burst of AP activation under conditions where the hC3Nb2 concentration becomes too low and the activity of the endogenous complement regulators is insufficient for FI degradation of host cell bound C3b to iC3b. Our AP-specific hC3Nb1 potently inhibits FI degradation (20) and may for this reason be a nonideal complement regulator in vivo. Maximal inhibition of complement at the C3 level would occur if the C3 cleavage was inhibited while the degradation of C3b to iC3b could still proceed. To test whether hC3Nb2 allows such C3b degradation, we mixed C3b with Factor H and Factor I and monitored the C3b cleavage in a time course experiment by SDS-PAGE. This showed that hC3Nb2 allows the FH-assisted cleavage by FI (Fig. 4E), as evident by the similar progress of C3b degradation in the absence and presence of hC3Nb2. However, if FI cleavage itself is the rate-limiting step in this assay, this approach may not be sufficiently sensitive to detect a partial interference by hC3Nb2 with FH binding to C3b that may have undesirable consequences in vivo. To investigate in an FI-independent manner whether the nanobody interferes with the FH-C3b interaction, we used a BLI competition assay to evaluate whether hC3Nb2 and FH can bind simultaneously to C3b. Similar to our BLI-based FB competition assay, we immobilized hC3Nb2 on BLI sensors, dipped the sensors in C3b, and transferred the sensors to FH. The BLI binding curves displayed in Fig. 4D document that hC3Nb2 and FH do not compete for binding to C3b. This indicates that hC3Nb2 does not interfere with the function of FH with respect to FI degradation.
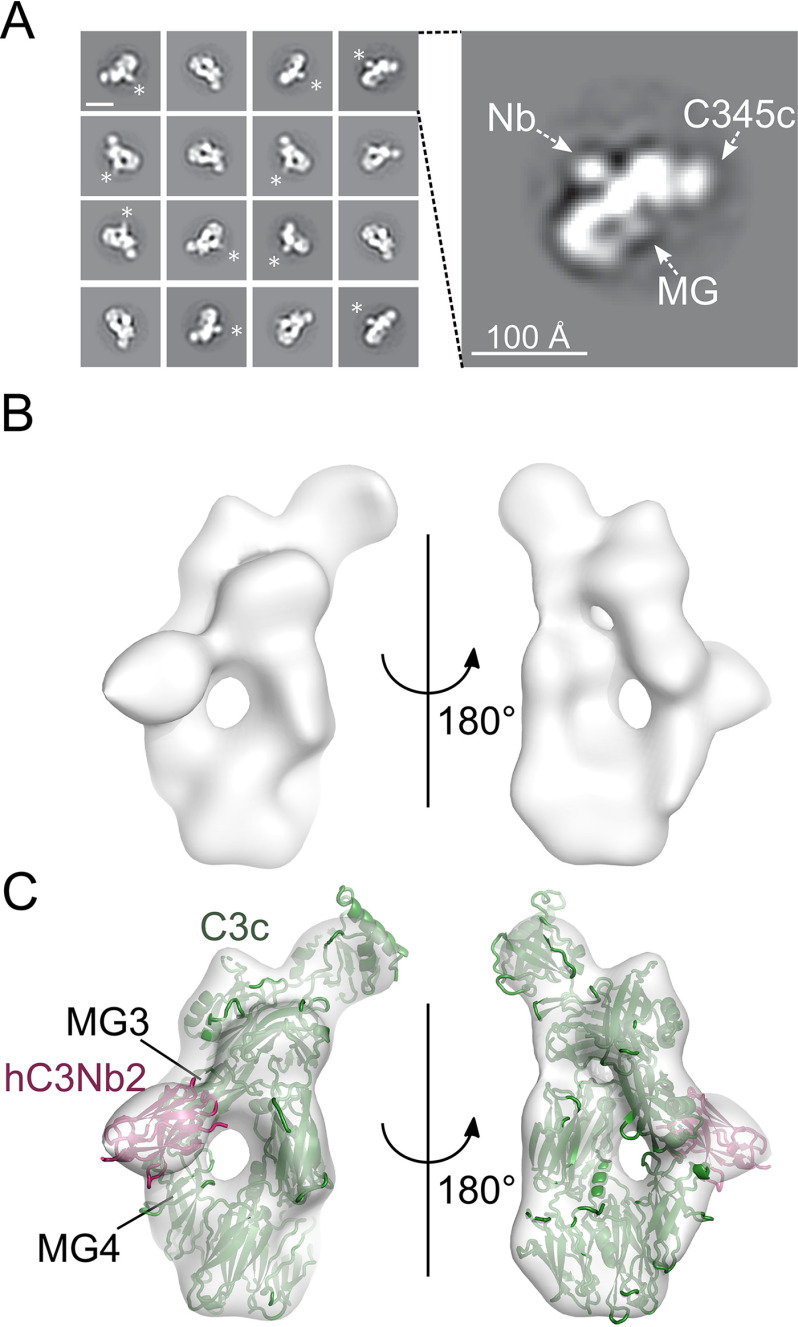
The hC3Nb2 nanobody binds the MG3-MG4 interface
To gain further insight into the inhibitory mechanism of hC3Nb2, we performed nsEM. To this end, we first isolated the hC3Nb2:C3c complex by SEC and obtained the complex from the early peak fractions. We applied the complex to EM grids and stained with uranyl formate. The 2D class averages obtained from micrographs recorded from these grids displayed recognizable features of C3c, including the C345c domain and the macroglobulin (MG) ring (Fig. 5A). Furthermore, hC3Nb2 could clearly be recognized as a density protruding from the MG ring on the “back” of the C3c MG ring opposite the C345c domain (Fig. 5A). To determine the domains involved in the interaction, stochastic gradient descent was used to generate an initial reference model, which was used for 3D classification and subsequent refinement. This gave rise to a high-quality 3D reconstruction with a good distribution of views (Fig. S3, A and B). Similar to the 2D class averages, the 3D reconstruction revealed clear density for the C345c domain and the MG ring (Fig. 5B). After rigid-body fitting of the crystal structure of C3c (22) into the 3D reconstruction, a clear density remained that was not occupied by C3c but matches the dimension of a nanobody (Fig. 5C). The density was present on the back of the MG ring, in agreement with the 2D class averages. The 3D reconstruction further revealed that hC3Nb2 binds at the interface between MG3 and MG4, with the major part of the epitope being present in the MG3 domain (Fig. 5C). To assess whether the hC3Nb2 epitope is conserved between different C3 fragments, the nsEM experiment was repeated using C3b instead of C3c. The data gave rise to 2D class averages where the MG ring, C345c, TE domain, and hC3Nb2 could easily be recognized (Fig. S4A). However, the 2D class averages also show that the hC3Nb2:C3b complex has a high tendency toward preferred orientations on the carbon-coated grid. This is also evident in the resulting 3D reconstruction, which severely limits the final resolution (Fig. S4, B–E). C3b can still be reliably rigid body–fitted into the 3D reconstruction, and comparison with the hC3Nb2:C3c complex reveals that hC3Nb2 binds the same epitope in both C3 fragments (Fig. S4, F and G).
Figure 5.
Mapping of the hC3Nb2 binding site on C3c. A, negative stain EM 2D class averages of the hC3Nb2:C3c complex. *, extra density, which was ascribed to the hC3Nb2 in the 2D class averages. In the expanded view of one 2D class to the right, the MG ring and the C-terminal C345c domain are marked by arrows, whereas the likely location of hC3Nb2 is marked Nb. The scale bar in the top left 2D class is 100 Å. B, a 3D EM reconstruction based on the hC3Nb2:C3c particles. C, fitting of the C3c (PDB entry 2A74) and the hC3Nb1 (PDB entry 6EHG) into the 3D reconstruction. The locations of the MG3 and MG4 domains are marked.
hC3Nb2 blocks the C3:C3 convertase association through steric hindrance
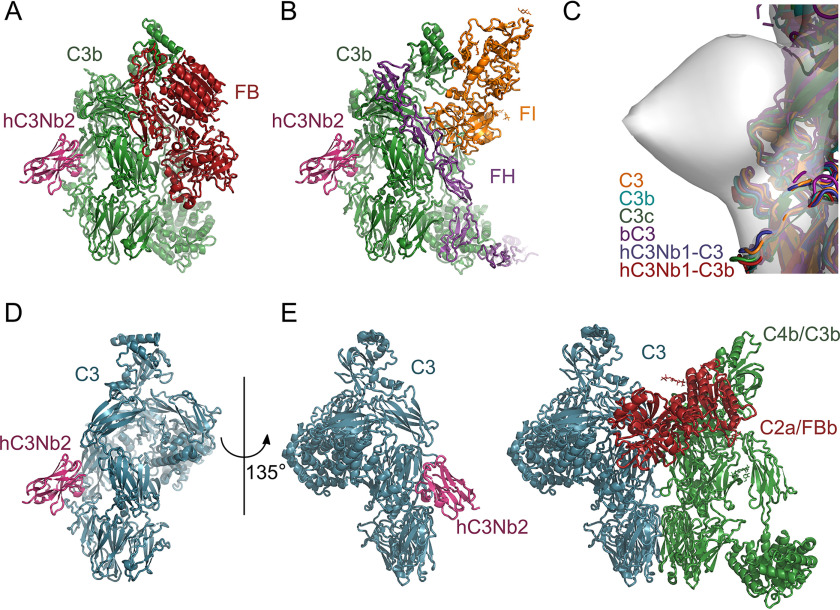
We next investigated whether an hC3Nb2 epitope on the back of C3b with contributions from the MG3 and MG4 domains could explain our functional data. A comparison with the structure of the C3bB complex (23) rationalized why hC3Nb2 allows C3bB proconvertase assembly (Fig. 4, B and C), because FB interacts with C3b through the C345c and the CUB domains on the opposite side of C3b compared with the Nb (Fig. 6A). In the same manner, the suggested epitope agrees with the observation that the hC3Nb2 does not interfere with FH association and FI degradation (Fig. 4, D and E). A superposition of C3b:FH:FI structure (24) with the hC3Nb2:C3c model reveals that hC3Nb2 binds somewhat distant from the FH- and FI-binding sites (Fig. 6B). Similarly, the suggested epitope places hC3Nb2 distant from the binding site of hC3Nb1, which interacts with MG6 and MG7 domains of C3b (Fig. S5A) (20). To test this experimentally, we coated BLI sensors with hC3Nb2 and transferred the sensors to C3b or C3b with a 10-fold molar excess of either hC3Nb1 or hC3Nb2. The binding curves revealed no competition between the two nanobodies for binding to C3b (Fig. S5B). Importantly, the suggested hC3Nb2 epitope also offers a straightforward explanation for how the nanobody inhibits C3 cleavage by preventing the association between the substrate C3 and the C3 convertases. We earlier proposed a general model for the convertase-substrate complexes stating that the noncatalytic subunit of a convertase, C4b in CP convertases and C3b in AP convertases, contacts the substrates C3 and C5 through a two-point interface (25, 26). One component of this interface involves recognition of the substrate MG7 domain by the MG6 and MG7 domains in the noncatalytic convertase subunit, whereas at the second part of the convertase-substrate interface, the MG4 and MG5 domains in both the substrate and the noncatalytic subunit convertase form contacts. A comparison of known structures of C3, C3b, and C3c (10, 20, 22, 27, 28) shows that the proposed epitope of hC3Nb2 is structurally conserved in these three functional states of C3 (Fig. 6C). Based on this structural conservation and on the finding that the dissociation constants for C3b and C3 are almost identical (Fig. 3D), we suggest that hC3Nb2 binds to native C3 in the same manner as to C3b and C3c (Fig. 6D). We may now compare our model of substrate–convertase complexes with a model of the C3 substrate from which hC3Nb2 protrudes out from the MG3 domain (Fig. 6E). This comparison argues that the nanobody sterically hinders association between substrate and C3 convertase at the MG4-MG5 interface. This agrees with the inhibition of C3 cleavage by CVFBb and C4b2a (Figs. 1, B and C and 4A), where the effect of hC3Nb2 cannot be due to its strong binding to C3b. Hence, our mapping of the hC3Nb2 to the MG3 and MG4 domains of C3 and C3b corroborates the functional data. In summary, our data demonstrate that the ability of hC3Nb2 to efficiently block all three activation pathways is due to its binding to the substrate C3. When it comes to the AP C3 convertase, hC3Nb2 will bind to both the C3 substrate and the convertase C3b in agreement with its strong inhibition of the alternative pathway (Fig. 1, D and E).
Figure 6.
A structure-based model for the inhibitory mechanism. A docking of hC3Nb2 onto the structures of C3bB (PDB entry 2XWJ) (A) and C3b:miniFH:FI (PDB entry 5O32) (B) using the hC3Nb2:C3c envelope is consistent with the interpretation that hC3Nb2 does not compete with either FB or FH. C, a superposition of the known structures at the MG3-MG4 interface suggests that the epitope of hC3Nb2 is conserved in C3 (PDB entries 2A73 and 2B39), C3b (PDB entry 5FO7), C3c (PDB entry 2A74), hC3Nb1:C3 (PDB entry 6RU5), and hC3Nb1:C3b (PDB entry 6EHG). D, mapping of hC3Nb2 onto native C3 (PDB entry 2A73) by comparison with C3c. E, a comparison between the model of the hC3Nb2:C3 complex in D and the general model of the convertase-substrate complexes implies that hC3Nb2 inhibits C3 cleavage by all convertases by preventing substrate recognition.
Discussion
Here we describe the hC3Nb2 nanobody, which binds C3 with high affinity and efficiently inhibits the activation of complement activation through both the alternative and the classical pathway C3 convertases. The nanobody is cross-species–reactive, as it inhibits the complement progression in both human and murine serum. Using a combination of biochemistry, biophysics, and structural information, we have unraveled the mechanism of inhibition and shown that hC3Nb2 blocks all three activation pathways by preventing the binding of substrate by the C3 convertases.
Several complement regulators surveil the complement system to prevent activation on host cells, and C3b constitutes a central regulation target recognized by a number of such regulators. One of the most appealing properties of hC3Nb2 is that it does not interfere with the cofactor function of FH, the most important fluid-phase complement regulator of C3b. In contrast to the cofactor function of FH, the effect of hC3Nb2 on the decay-accelerating activity of FH has not been directly addressed. However, both functions rely on the binding of FH to C3b, and, given the absence of interference with the cofactor activity, we expect that hC3Nb2 does not interfere with the decay-accelerating activity. Related to decay is the influence of hC3Nb2 on the t½ of C3bBb. The epitope is located far from the C-terminal C345c domain of C3b that holds the catalytic Bb; hence, we predict that the nanobody does not change the rate of Bb dissociation from C3b. Other negative regulators of C3b are membrane cofactor protein (MCP/CD46), complement receptor 1 (CR1/CD35), and decay-accelerating factor (DAF/CD55). MCP is ubiquitously expressed and serves as a cofactor for FI in the cleavage of C3b (29). CR1 expressed on peripheral blood cells has a dual function. It acts as an FI cofactor and on the erythrocyte surface; it serves as a carrier of C3b- and C4b-opsonized objects to the spleen and liver (30). Finally, DAF promotes the decay of the C3bBb complex (5). All of these regulators comprise a number of complement control protein domains that interact with C3b in a mode similar to FH. The main interaction surface of these regulators lies in an L-shaped binding site on C3b in the region from the MG7 domain to the MG1-TE domain interface, crossing MG2 (27). The binding site of hC3Nb2 is in the MG3-MG4 region of C3b and hence lies far from this general regulator surface, and we expect therefore that hC3Nb2 does not interfere with any of the C3b regulators DAF, CR1, or MCP.
Another aspect worth considering is the influence of hC3Nb2 on the function of complement receptors supporting phagocytosis of C3b-opsonized objects and mediating cross-talk with the adaptive immune system. By presentation of iC3b/C3dg-opsonized antigens, complement receptor 2 (CR2) stimulates B cell receptor signaling on B cells (31). Complement receptor 3 (CR3) and complement receptor 4 (CR4) are expressed on subsets of myeloid cells and some activated lymphoid cells and mediate binding and phagocytosis of iC3b-opsonized pathogens and immune complexes (32). A comparison of the C3b:hC3Nb2 complex with X-ray structures of CR2 and CR3 fragments bound to the thioester domain of the iC3b ligand (33, 34) suggests that hC3Nb2 will not interfere with the CR2 and CR3 recognition of C3-opsonized objects. In contrast, nsEM data indicate that CR4 may also interact with the MG3-MG4 domains of iC3b (35), and we cannot exclude the possibility that the hC3Nb2- and CR4-binding sites on iC3b overlap. hC3Nb2 likewise appears to overlap with the binding site of the complement receptor, CRIg, which contacts the MG3, MG4, MG5, and MG6 domains and the LNK region of C3b (36). CRIg is expressed on tissue-residing macrophages and plays a role in pathogen clearance (37). Studies in knockout mice show that, although the liver readily clears bacteria from circulation in a complement-independent manner, CRIg participates in the clearance of complement-opsonized bacteria that are bound on platelets (38). The potential interference with the function of CR4 and CRIg in phagocytosis should hence be kept in mind if hC3bNb2 is to be used in an in vivo setting.
Uncontrolled complement activation constitutes a cornerstone in multiple diseases, and taming this cascade is hence of the utmost interest. Mutations, especially in the regulators of the alternative pathway can lead to uncontrolled complement activation and disease progression. The dysregulation often results in tissue damage in the kidney. To date, the only complement-specific inhibitor approved for therapy is eculizumab, which binds C5 and inhibits the cleavage of C5 to C5b and C5a by the C5 convertase (39). Eculizumab is used to treat PNH, in which the lack of complement regulators CD59 and DAF allows for the formation of membrane attack complex on erythrocytes and thus lysis (15). The use of eculizumab has provided proof of evidence for the safety and efficacy of therapeutic control of the terminal pathway in the complement system. Similar to eculizumab, hC3Nb2 inhibits the progression of the complement system, unaffected by the route of initiation. However, the nanobody targets complement early in the cascade, thereby avoiding the build-up of C3b on a dysregulated complement activator as exemplified with PNH, where C3b opsonization of erythrocytes may lead to extravascular hemolysis (40).
In many ways, hC3Nb2 has functional properties resembling those of the well-established complement inhibitor compstatin, a cyclic peptide that prevents the cleavage of C3 both at the substrate and the convertase level (18). Compstatin binds in the MG4-MG5 interface of C3c (41) and is likely to bind to the same site in native C3. Based on our nsEM data, the binding sites of compstatin and hC3Nb2 do not directly overlap. Although compstatin is a promising drug, it does not inhibit murine C3 (18, 42), thus limiting its value in murine models of complement-associated disease. Like compstatin, hC3Nb2 binds native C3 and therefore requires high concentrations to saturate the circulating C3, which is present in the 1 mg/ml regime in plasma. However, local administration may benefit from broad inhibitors of complement, like hC3Nb2. Additionally, complete shutdown of the complement system is not only of therapeutic interest but may also allow the exploration of the complement diseases in disease models.
Numerous inhibitors of the complement system have emerged in recent years. Among alternative pathway inhibitors, the small molecule danicopan (Achillon) that targets FD is in phase II trials for the treatment of both C3G and PNH. Another small molecule targeting the alternative pathway is LNP023 from Novartis, which blocks the active site of FB and is currently in phase II evaluation targeting C3G, PNH, and IgA nephropathy. Whereas C3-targeting drugs are largely dominated by compstatin derivatives, mirococept, which is based on the C3 convertase–inhibiting properties of CR1, is currently in phase II testing of the effects on ischemia-reperfusion injury. For the development of C5 inhibitors, the pharmacokinetic properties of eculizumab have been improved with ravalizumab, but in addition, the peptide zilucoplan (RaPharma), the RNAi cemdisiran (Alnylam Pharmaceuticals Inc.), and the protein nomacopan (Akari) all target C5 and are in phase II clinical trials. Concerning the classical pathway and the lectin pathway, the antibodies sutimlimab and narsoplimab target C1s and mannose-binding lectin–associated serine protease (MASP)-2, respectively, and hence inhibit complement activation through these molecules (18). With the introduction of the hC3Nb2 nanobody, we expand the arsenal of complement inhibitors with a versatile reagent that can be easily prepared by recombinant protein expression in bacteria and mammalian cell culture.
hC3Nb2 offers broad inhibition of the complement cascade, whereas it allows the endogenous degradation mediated by the prime regulators of the system. Whereas hC3Nb2 allows investigations of whether the complement contributes to a given disease, it does not allow the pinpointing of which pathway drives the disease. Similar to hC3Nb2, our previously described nanobody hC3Nb1 binds C3 species with high affinity and shows reactivity with both human and murine C3. The hC3Nb1, however, primarily acts on C3b and inhibits the assembly of the AP C3 proconvertase, whereas it only exhibits modest effects on the CP C3 convertase–mediated cleavage. The introduction of hC3Nb2 hence supplements the hC3Nb1 nanobody. Together, these nanobodies allow for both the assessment of the involvement of complement and the identification of the driving pathways in disease development. The humanized nanobody caplacizumab was recently approved for treatment of acquired thrombotic thrombocytopenic purura (43). This represents a proof of principle for the application of nanobodies in the treatment of human diseases, and nanobody humanization strategies are well-established (44). Nanobodies may offer new therapeutic modalities as compared with conventional antibodies. Their small size offers high penetrance in tissue but also high clearance rates, but the latter problem can be addressed by, for example, fusion to IgG Fc and by coupling to serum albumin binders (45).
In conclusion, we have presented here the hC3Nb2 nanobody, which inhibits all complement pathways and additionally shows potent inhibition of the both the alternative and lectin pathways in murine serum. The nanobody inhibits the progression of the complement cascade by preventing the substrate–convertase interaction of the C3 convertases and binds both the substrate C3 and C3b in the AP convertase with low nanomolar affinity. Importantly, the nanobody does not interfere with endogenous regulation of C3b. We have thereby expanded the nanobody toolbox for studies of the complement system in vitro and murine in vivo models.
Experimental procedures
Purification of proteins and proconvertase assembly assay
The hC3Nb2 nanobody was selected as described (20). Briefly, a llama (Lama glama) was immunized with human C3b. RNA was purified from peripheral blood lymphocytes, and cDNA was generated. The Nbs were cloned into a phagemid vector, allowing a phage-display–based selection strategy. Nanobodies were selected against C3b by coating a microtiter plate with 1 µg of C3b in PBS for 12 h. PBS supplemented with BSA was added to the plate to block excess binding sites, and next 3 × 1012 nanobody-presenting M13 phages in PBS were added. Upon incubation for 1 h at room temperature, unbound phages were removed by 15 washes in PBS, 0.1% Tween 20 followed by 15 washes in PBS. The C3b-binding phages were eluted by incubation for 15 min in 0.2 m glycine (pH 2.2). The phages were subjected to a second round of selection, albeit with only 0.1 µg of C3b coated in each well. C3b-binding nanobodies were identified by ELISA as described (20). To this end, an ELISA plate was coated with 0.1 µg of C3b per well and subsequently blocked in PBS with 0.1% Tween and 2% BSA. Single phage-infected Escherichia coli colonies were picked and grown at 37 °C in lysogeny broth, and Nb expression was induced in 0.8 mm isopropyl β-d-1-thiogalactopyranoside in a 96-well plate setup. The cells were grown for 16 h at 30 °C, and the supernatant was applied to the C3b-coated ELISA plate. The plates were incubated for 1 h, washed six times in PBS-Tween 20, and then incubated in 1:10,000 diluted anti-E-tag-horseradish peroxidase antibody (Bethyl) for 1 h. The plates were washed, developed using 3,3′,5,5′-tetramethylbenzidine, and quenched in 1 m HCl, and the A450 was measured. Positive nanobodies were sequenced and cloned into pET22b (+) for bacterial expression.
To prepare recombinant hC3Nb2, nanobody encoding pET22b (+) plasmid was transformed into chemically competent E. coli low-background strain (LOBSTR) cells (46). A single colony was inoculated in 2xYT + 100 µg/ml ampicillin and grown for 16 h at 37 °C. Large cultures of 2xYT with 100 µg/ml ampicillin were inoculated with 1.25% of the culture. The culture was grown to an A600 of 0.6-0.8, induced by 1 mm isopropyl β-d-1-thiogalactopyranoside, and grown overnight at 18 °C. Then cells were harvested by centrifugation at 7,800 × g for 20 min at 4 °C, and the hC3Nb2 was purified from the pelleted cells as described (20). In short, pelleted cells were resuspended and sonicated, and debris was pelleted by centrifugation. The cleared lysate was loaded onto a HisTrap Crude FF (GE Healthcare), and the nanobody was eluted using 400 mm imidazole. The nanobody was dialyzed against 20 mm sodium acetate, 50 mm NaCl and then applied to a Source S column (GE Healthcare) equilibrated in the dialysis buffer. hC3Nb2 was eluted by a linear gradient from 20 to 500 mm NaCl over 35 ml. The nanobody was finally purified on a Superdex 75 (GE Healthcare) column equilibrated in 20 mm HEPES (pH 7.5), 150 mm NaCl. Complement FB was purchased from Complement Technology, and CVF was purified as described (47). Active FD was purified from HEK293 F-cells by co-transfection of FD and MASP-3. To this end, plasmids encoding FD and MASP-3 were mixed and incubated with a 2-fold excess of polyethyleneimine prior to transfection. 5 days after the transfection, the conditioned medium was harvested, and the FD was captured on a 5-ml His-Trap Excel (GE Healthcare) column equilibrated in 20 mm HEPES (pH 7.5), 500 mm NaCl, 30 mm imidazole. FD was eluted by application of 300 mm imidazole and subsequently dialyzed against 20 mm MES (pH 6.0). The dialyzed protein was applied to a 1-ml Mono S (GE Healthcare) column equilibrated in 20 mm MES (pH 6.0) to separate pro-FD from activated FD. The protein was eluted by a linear gradient from 0 to 500 mm NaCl over 10 ml. Human native C3 was purified from outdated plasma as described (20). To generate C3b, C3 was mixed with 1% (w/w) trypsin, and the mix was incubated for 4 min at 37 °C. Cleavage progression was stopped by the addition of 1 mm phenylmethylsulfonyl fluoride and a 10-fold (w/w) excess of pancreatic trypsin inhibitor relative to trypsin. To separate C3 from the C3b, the mix was diluted 3-fold and applied to a Source 15S (GE Healthcare) column equilibrated in 20 mm MES (pH 6.0), 20 mm NaCl. The protein was eluted by a linear gradient from 20 to 250 mm NaCl over 60 ml. C3b-containing fractions were added 100 mm Tris (pH 8.5), 10 mm iodoacetamide. The C3b was applied to a 24-ml Superdex 200 Increase (GE Healthcare) equilibrated in 20 mm HEPES (pH 7.5), 150 mm NaCl. To generate C3MA, C3 was incubated for 2 h at room temperature in 100 mm Tris (pH 8.5), 100 mm methylamine (pH 8.7), and 10 mm iodoacetamide and then dialyzed against 20 mm MES (pH 6.0), 250 mm NaCl for 3 h at 4 °C. The dialysate was incubated at 37 °C for 16 h and then diluted 3-fold in 20 mm MES (pH 6.0). The diluted protein was applied to a 9-ml Source 15S column equilibrated in 20 mm MES (pH 6.0), 20 mm NaCl and subsequently eluted by a linear gradient from 200 to 500 mm NaCl over 80 ml. C3c was prepared from C3b, and to this end, C3b was incubated with 0.2% (w/w) Factor I (Complement Technologies) and 1% (w/w) Factor H (Complement Technologies) for 24 h at 37 °C. The cleaved product was diluted 2-fold in 20 mm HEPES (pH 7.5) and applied to a 1-ml Mono Q (GE Healthcare) column equilibrated in 20 mm HEPES (pH 7.5), 130 mm NaCl. The C3c was eluted by a 25-ml linear gradient from 130 to 320 mm NaCl. The FB (D279G/S699A) was purified from HEK293F cell supernatants as described (11). The alternative pathway C3 proconvertase was assembled by mixing C3b with a 1.5-fold molar excess of inactive FB (D279G/S699A) in Running Buffer (20 mm HEPES (pH 7.5), 150 mm NaCl, 2 mm MgCl2). The proconvertase was incubated in either the presence or absence of a 2-fold molar excess of hC3Nb2 for 15 min on ice. Next, the mix was loaded on a 24-ml Superdex 200 increase column equilibrated in Running Buffer and operated at 4 °C.
C3 deposition assays
For the human classical pathway assay, each well in a 96-well Maxisorp plate (Thermo, catalog no. 446612) was coated with 100 µl of 15 µg/ml heat-aggregated IgG diluted in 50 mm sodium carbonate (pH 9.6) (AMPLIQON Laboratory Reagents) and incubated overnight. Residual binding sites in the wells were blocked by TBS (10 mm Tris (pH 7.4), 140 mm NaCl) supplemented with 5 mm CaCl2 and 1 mg/ml HSA for 1 h at room temperature and then washed three times with TBS-T (TBS, 0.05% (v/v) Tween 20). Nanobodies were diluted in veronal buffer (5 mm barbital (pH 7.4), 145 mm NaCl, 0.25 mm CaCl2, 0.8 mm MgCl2) (LONZA, catalog no. 12-624E) supplemented with 0.2% NHS, and 100 µl was added to each well. The wells were incubated for 1 h at 37 °C in a humidity box and then washed three times with TBS-T. Deposited C3 was detected using 100 µl of biotinylated rabbit anti-C3d antibody (Dako, catalog no. A006302-2) diluted to 0.5 µg/ml in TBS-T and incubation for 2 h at room temperature followed by three washes in TBS-T. Wells were then supplemented with 100 µl of 1 µg/ml europium-labeled streptavidin (PerkinElmer Life Sciences, catalog no. 1244-360) diluted in TBS-T supplemented with 25 µm EDTA and incubated for 1 h at room temperature. The wells were washed three times in TBS-T, and then 200 µl of enhancement buffer (Ampliqon laboratory reagent, catalog no. Q99800) was added to each well. The fluorescence signal, read as time-resolved fluorometry, was measured using a VICTOR3 multilabel plate counter (PerkinElmer Life Sciences). A test for the influence of the nanobodies on the lectin pathway assay was performed in a similar manner; in this case, the surface was coated with mannan instead of IgG, as described (20).
For the alternative pathway assay, a 96-well Maxisorp plate was coated overnight with 100 µl of 20 µg/ml zymosan (Sigma, catalog no. Z4250) in 50 mm sodium carbonate (pH 9.6) (AMPLIQON Laboratory Reagents) per well. Next, the wells were blocked for 1 h at room temperature in 200 µl of TBS buffer containing 1 mg/ml human serum albumin. The wells were washed three times in 300 µl TBS-T. 100 µl of nanobody was diluted in VBS/EGTA/Mg2+ buffer (veronal buffer, 10 mm EGTA, 4.2 mm MgCl2) containing 11% (v/v) serum. We further included wells containing only buffer and wells containing serum without nanobody. The plate was incubated in a humidity box at 37 °C for 1.5 h. The effect of nanobodies on the murine AP was tested in murine serum diluted to 5%. The serum was from male C57Bl6 mice, and the deposited C3 was quantified using a rat anti-mouse C3 antibody (Cederlane, catalog no. CL7503NA) as described (20).
Hemolysis assays
Classical pathway assay was essentially performed as described (48). Sheep red blood cells (RBCs) from sheep blood in Alsevers solution (SSI Diagnostics) were washed three times and then resuspended to 6% in BI buffer (Veronal buffer supplemented with 2 mm CaCl2) supplemented with 1 mg/ml gelatin. The erythrocytes were mixed at 1:1 (v/v) with anti-sheep RBC antibodies (Sigma, catalog no. S1389-1VL) diluted 1:200 in BI buffer supplemented with 1 mg/ml gelatin. The mix was incubated for 30 min at room temperature followed by harvesting of RBCs at 1,000 × g for 6 min. Half of the supernatant was decanted, and the RBCs were resuspended. 15 µl of NHS was mixed with 65 µl of PBS containing the indicated concentration of nanobody. Next, 120 µl of BI buffer was added, and the mix was incubated for 2 h at 37 °C. Next, 60 µl of the mix was transferred to each well (in at least duplicates) in a conical C-bottom 96-well microplate, and 30 µl of the RBCs was added. The plate was incubated for 2 h at 37 °C, and the lysis was stopped by the addition of 100 µl of cold stop solution containing 0.9% (w/v) NaCl, 5 mm EDTA. The plate was centrifuged at 2,000 rpm for 10 min at 4 °C to pellet surviving cells. The supernatant was transferred to a 96-well microplate (Costar, catalog no. 3897), and the absorbance at 405 nm was measured using a VICTOR3 multilabel plate counter. The experiment was performed three times. Experiments with FB-depleted serum (Complement Technologies) were performed similarly, but this experiment was only repeated twice.
Surface plasmon resonance
Nanobodies with a C-terminal AviTag were mixed in a 20-fold excess (w/w) with BirA-ligase in 20 mm HEPES (pH 7.5), 150 mm NaCl, 5 mm MgCl2, 2 mm ATP, 0.15 mm d-biotin. The mix was incubated overnight at room temperature and then applied to a 1-ml Source 15S (GE Healthcare) column equilibrated in 20 mm sodium acetate (pH 5.5), 20 mm NaCl. The protein was eluted by a linear gradient from 20 to 500 mm NaCl. All SPR experiments were performed on a BiaCore Instrument T200 operated at 25 °C. The biotinylated Avi-tagged nanobody was immobilized via streptavidin immobilized on CMD500M chips (XanTec Bioanalytics) sensor chips. A reference flow cell, without immobilized nanobody, was used for background subtraction. All experiments were performed with a flow rate of 30 µl/min in a running buffer (150 mm NaCl, 20 mm HEPES (pH 7.5), 0.05% Tween 20, 3 mm MgCl2) unless otherwise stated. In the beginning of each experiment, 30 µg/ml biotinylated hC3Nb2 was applied to the flow cell for 10 s before sample was applied. C3, C3b, and C3MA were applied to the flow cells in concentrations of 60, 30, 20, 10, and 5 nm. Sample association lasted 400 s and was immediately followed by a 500-s dissociation period. Following each sample application, three 10-s pulses of 100 mm glycine (pH 2.7) followed by a 120-s stabilization period, were used to regenerate the surface. The CVF-binding assay was performed similarly. Buffer or 50 nm CVF was applied to the flow cells for 200 s, followed by a 600-s dissociation period on the flow cells equilibrated in 150 mm NaCl, 20 mm HEPES, pH 7.5, 3 mm MgCl2. The assay was performed in duplicates.
Bio-layer interferometry
All bio-layer interferometry experiments were performed on an Octet Red96 (ForteBio) at 30 °C and shaking at 1,000 rpm, using the buffer 20 mm HEPES, pH 7.5, 150 mm NaCl, 0.05% Tween 20 unless otherwise stated. The streptavidin biosensors (ForteBio) were washed for 12 min in buffer before 5 µg/ml biotinylated hC3Nb2-avi was loaded on the sensors for 10 min. The sensors were then washed for 2 min before starting any assay. In the binding kinetics assays, the sensors were first baselined for 2 min before being dipped in a well containing either human C3b (5, 2.5, 1.25, 0.625, 0.31, 0.16, and 0 nm) or mouse C3b (6.25, 3.13, 1.56, 0.78, 0.39, 0.20, and 0 nm), and association was followed for 5 min. Thereafter, the sensors were dipped in the same well that was used for baselining, and the dissociation was followed for 5 min. The 0 nm measurements were subtracted from all experiments before fitting the curves to a 1:1 Langmuir binding model, where the association is modeled as
and the dissociation is modeled as a first-order exponential decay.
For assessing the ability of the hC3Nb2:C3b complex to bind FH or FB (D279G), the sensors were first baselined for 2 min before being dipped in a well containing 50 nm C3b. The sensors were then washed for 1 min in 20 mm HEPES, pH 7.5, 150 mm NaCl, 0.05% Tween 20, 2 mm MgCl2, before assessing complex formation with 100 nm FB (D279G) or FH for 300 s. For the nanobody competition assay, the sensors were first baselined for 2 min before being dipped in 20 nm C3b or 20 nm C3b pre-incubated for 30 min with either 200 nm hC3Nb1 or 200 nm hC3Nb2. The complex formation was assessed for 5 min, after which the dissociation rate was assessed for 5 min.
CVFBb cleavage of C3 and FI cleavage of C3b
The CVFBb was assembled by mixing FB (D279A) with a 2-fold molar excess of CVF and 10% (w/w) FD in 150 mm NaCl, 20 mm HEPES (pH 7.5), 2 mm MgCl2. The mix was incubated for 15 min at room temperature to allow convertase assembly and activation, followed by a 10-min incubation on ice. Meanwhile, C3 was mixed with a 2-fold excess of hC3Nb2. Nb-treated or -untreated C3, in a 10-fold excess to FB, was added to the CVFBb, and the reaction was incubated at 37 °C. Samples were obtained after 0.5, 1, 2, 4, 8, and 24 h, mixed with reducing SDS-loading dye, and boiled for 30 s. C3b, in 20 mm HEPES (pH 7.5), 150 mm NaCl, was incubated for 5 min on ice in the presence or absence of a 1.2-fold molar excess of hC3Nb2, followed by the addition of FI and FH (Complement Technology) at 1% (w/w) and 0.2% (w/w), respectively. The reaction mix was incubated at 37 °C. Samples were obtained after 1, 2, 4, and 8 h, mixed with reducing SDS-PAGE loading dye, and boiled for 30 s.
Negative stain EM
For negative stain EM, hC3Nb2 was added in a 2-fold molar excess to C3c or C3b, followed by 5-min incubation on ice. The complex was purified using a 24-ml Superdex 200 Increase (GE Healthcare) column equilibrated in 20 mm HEPES (pH 7.5), 150 mm NaCl. A sample of 3 µl at ∼20 µg/ml complex taken from the early peak fractions was applied to a carbon-coated copper grid (Gilder 400-C3) glow-discharged at 25 mA for 45 s on an easiGlow glow-discharge system (PELCO). After the sample was blotted away, it was washed in 3 µl of 2% (w/v) uranyl formate, followed by a staining step where a new 3-µl drop of 2% (w/v) uranyl formate was incubated on the grid for 45 s. Image acquisition was performed on FEI Technai G2 Spirit transmission electron microscope operated at 120 kV with a magnification of ×67,000 and a defocus range from −0.7 to −1.7 µm. Images were acquired using Leginon (49). For the hC3Nb2:C3b complex, particles were picked from the collected micrograph images by the automated particle picker (DoG picker) in the Appion Framework (50, 51), whereas cisTEM (52) was used for the hC3Nb2:C3c complex . 2D and 3D classification was performed using RELION (53). An initial input model for 3D classification was generated by stochastic gradient descent in RELION for the hC3Nb2:C3c complex, whereas the initial model used for the hC3Nb2:C3b complex was C3b (PDB entry 5FO7) filtered to 80 Å using EMAN (54).
Data availability
The EM 3D reconstructions presented here are available upon request from Gregers Rom Andersen (gra@mbg.au.dk). All other data are contained within the article.
Supplementary Material
Acknowledgments
We thank Christine Schar for assistance with SPR experiments, Thomas Boesen for help with electron microscopy, and Karen Margrethe Nielsen for technical support.
This article contains supporting information.
Author contributions—H. P., R. K. J., and G. R. A. conceptualization; H. P., R. K. J., S. T., and G. R. A. data curation; H. P., R. K. J., A. G. H., S. T., and G. R. A. formal analysis; R. K. J., A. G. H., S. T., N. S. L., and G. R. A. supervision; G. R. A. funding acquisition; H. P., R. K. J., and S. T. validation; H. P., R. K. J., A. G. H., and S. T. investigation; H. P., R. K. J., and S. T. visualization; H. P., R. K. J., T. A. G., S. T., and N. S. L. methodology; H. P., R. K. J., S. T., and G. R. A. writing-original draft; G. R. A. project administration; H. P., R. K. J., S. T., and G. R. A. writing-review and editing; T. A. G. resources.
Funding and additional information—This work was supported by the Lundbeck Foundation (BRAINSTRUC, R155-2015-2666) and the Aarhus University Science and Technology Ph.D. School GSST.
Conflict of interest—G. R. A. has a collaboration with Alexion pharmaceuticals.
- CP
- classical pathway
- AP
- alternative pathway
- BLI
- bio-layer interferometry
- C3G
- C3 glomerulopathies
- C3MA
- complement component C3 methylamine
- CR
- complement receptor
- CVF
- cobra venom factor
- DAF
- decay acceleration factor
- FB
- Factor B
- FD
- Factor D
- FH
- Factor H
- FI
- Factor I
- LP
- lectin pathway
- MASP
- mannose-binding lectin–associated serine protease
- MCP
- membrane cofactor protein
- MG
- macroglobulin
- Nb
- nanobody
- NHS
- normal human serum
- nsEM
- negative stain EM
- PDB
- Protein Data Bank
- PNH
- paroxysmal nocturnal hemoglobinuria
- PRM
- pattern-recognizing molecule
- SEC
- size-exclusion chromatography
- SPR
- surface plasmon resonance
- TE
- thioester
- RBC
- red blood cell
- 2D
- two-dimensional
- 3D
- three-dimensional.
References
- 1. Merle N. S., Noe R., Halbwachs-Mecarelli L., Fremeaux-Bacchi V., and Roumenina L. T. (2015) Complement system part II: role in immunity. Front. Immunol. 6, 257 10.3389/fimmu.2015.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stephan A. H., Barres B. A., and Stevens B. (2012) The complement system: an unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 35, 369–389 10.1146/annurev-neuro-061010-113810 [DOI] [PubMed] [Google Scholar]
- 3. Kojouharova M., Reid K., and Gadjeva M. (2010) New insights into the molecular mechanisms of classical complement activation. Mol. Immunol. 47, 2154–2160 10.1016/j.molimm.2010.05.011 [DOI] [PubMed] [Google Scholar]
- 4. Kjaer T. R., Thiel S., and Andersen G. R. (2013) Toward a structure-based comprehension of the lectin pathway of complement. Mol. Immunol. 56, 222–231 10.1016/j.molimm.2013.05.220 [DOI] [PubMed] [Google Scholar]
- 5. Bajic G., Degn S. E., Thiel S., and Andersen G. R. (2015) Complement activation, regulation, and molecular basis for complement-related diseases. EMBO J. 34, 2735–2757 10.15252/embj.201591881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harboe M., Garred P., Karlstrøm E., Lindstad J. K., Stahl G. L., and Mollnes T. E. (2009) The down-stream effects of mannan-induced lectin complement pathway activation depend quantitatively on alternative pathway amplification. Mol. Immunol. 47, 373–380 10.1016/j.molimm.2009.09.005 [DOI] [PubMed] [Google Scholar]
- 7. Harboe M., Ulvund G., Vien L., Fung M., and Mollnes T. E. (2004) The quantitative role of alternative pathway amplification in classical pathway induced terminal complement activation. Clin. Exp. Immunol. 138, 439–446 10.1111/j.1365-2249.2004.02627.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bexborn F., Andersson P. O., Chen H., Nilsson B., and Ekdahl K. N. (2008) The tick-over theory revisited: formation and regulation of the soluble alternative complement C3 convertase (C3(H2O)Bb). Mol. Immunol. 45, 2370–2379 10.1016/j.molimm.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heesterbeek D. A. C., Angelier M. L., Harrison R. A., and Rooijakkers S. H. M. (2018) Complement and bacterial infections: from molecular mechanisms to therapeutic applications. J. Innate Immun. 10, 455–464 10.1159/000491439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pedersen D. V., Gadeberg T. A. F., Thomas C., Wang Y., Joram N., Jensen R. K., Mazarakis S. M. M., Revel M., El Sissy C., Petersen S. V., Lindorff-Larsen K., Thiel S., Laursen N. S., Fremeaux-Bacchi V., and Andersen G. R. (2019) Structural basis for properdin oligomerization and convertase stimulation in the human complement system. Front. Immunol. 10, 2007 10.3389/fimmu.2019.02007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pedersen D. V., Roumenina L., Jensen R. K., Gadeberg T. A., Marinozzi C., Picard C., Rybkine T., Thiel S., Sørensen U. B., Stover C., Fremeaux-Bacchi V., and Andersen G. R. (2017) Functional and structural insight into properdin control of complement alternative pathway amplification. EMBO J. 36, 1084–1099 10.15252/embj.201696173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith R. J. H., Appel G. B., Blom A. M., Cook H. T., D'Agati V. D., Fakhouri F., Fremeaux-Bacchi V., Józsi M., Kavanagh D., Lambris J. D., Noris M., Pickering M. C., Remuzzi G., de Córdoba S. R., Sethi S., et al. (2019) C3 glomerulopathy—understanding a rare complement-driven renal disease. Nat. Rev. Nephrol. 15, 129–143 10.1038/s41581-018-0107-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodríguez de Córdoba S., Hidalgo M. S., Pinto S., and Tortajada A. (2014) Genetics of atypical hemolytic uremic syndrome (aHUS). Semin. Thromb. Hemost. 40, 422–430 10.1055/s-0034-1375296 [DOI] [PubMed] [Google Scholar]
- 14. van Lookeren Campagne M., Strauss E. C., and Yaspan B. L. (2016) Age-related macular degeneration: complement in action. Immunobiology 221, 733–739 10.1016/j.imbio.2015.11.007 [DOI] [PubMed] [Google Scholar]
- 15. Hill A., DeZern A. E., Kinoshita T., and Brodsky R. A. (2017) Paroxysmal nocturnal haemoglobinuria. Nat. Rev. Dis. Primers 3, 17028 10.1038/nrdp.2017.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sekar A., Bialas A. R., de Rivera H., Davis A., Hammond T. R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V., Genovese G., Rose S. A., Handsaker R. E., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Daly M. J., et al. (2016) Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hong S., Beja-Glasser V. F., Nfonoyim B. M., Frouin A., Li S., Ramakrishnan S., Merry K. M., Shi Q., Rosenthal A., Barres B. A., Lemere C. A., Selkoe D. J., and Stevens B. (2016) Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 10.1126/science.aad8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mastellos D. C., Ricklin D., and Lambris J. D. (2019) Clinical promise of next-generation complement therapeutics. Nat. Rev. Drug Discov. 18, 707–729 10.1038/s41573-019-0031-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zelek W. M., Xie L., Morgan B. P., and Harris C. L. (2019) Compendium of current complement therapeutics. Mol. Immunol. 114, 341–352 10.1016/j.molimm.2019.07.030 [DOI] [PubMed] [Google Scholar]
- 20. Jensen R. K., Pihl R., Gadeberg T. A. F., Jensen J. K., Andersen K. R., Thiel S., Laursen N. S., and Andersen G. R. (2018) A potent complement factor C3 specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement. J. Biol. Chem. 293, 6269–6281 10.1074/jbc.RA117.001179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roumenina L. T., Jablonski M., Hue C., Blouin J., Dimitrov J. D., Dragon-Durey M. A., Cayla M., Fridman W. H., Macher M. A., Ribes D., Moulonguet L., Rostaing L., Satchell S. C., Mathieson P. W., Sautes-Fridman C., et al. (2009) Hyperfunctional C3 convertase leads to complement deposition on endothelial cells and contributes to atypical hemolytic uremic syndrome. Blood 114, 2837–2845 10.1182/blood-2009-01-197640 [DOI] [PubMed] [Google Scholar]
- 22. Janssen B. J., Huizinga E. G., Raaijmakers H. C., Roos A., Daha M. R., Nilsson-Ekdahl K., Nilsson B., and Gros P. (2005) Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 437, 505–511 10.1038/nature04005 [DOI] [PubMed] [Google Scholar]
- 23. Forneris F., Ricklin D., Wu J., Tzekou A., Wallace R. S., Lambris J. D., and Gros P. (2010) Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 330, 1816–1820 10.1126/science.1195821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xue X., Wu J., Ricklin D., Forneris F., Di Crescenzio P., Schmidt C. Q., Granneman J., Sharp T. H., Lambris J. D., and Gros P. (2017) Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses. Nat. Struct. Mol. Biol. 24, 643–651 10.1038/nsmb.3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laursen N. S., Andersen K. R., Braren I., Spillner E., Sottrup-Jensen L., and Andersen G. R. (2011) Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex. EMBO J. 30, 606–616 10.1038/emboj.2010.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schatz-Jakobsen J. A., Pedersen D. V., and Andersen G. R. (2016) Structural insight into proteolytic activation and regulation of the complement system. Immunol. Rev. 274, 59–73 10.1111/imr.12465 [DOI] [PubMed] [Google Scholar]
- 27. Forneris F., Wu J., Xue X., Ricklin D., Lin Z., Sfyroera G., Tzekou A., Volokhina E., Granneman J. C., Hauhart R., Bertram P., Liszewski M. K., Atkinson J. P., Lambris J. D., and Gros P. (2016) Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode. EMBO J. 35, 1133–1149 10.15252/embj.201593673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fredslund F., Jenner L., Husted L. B., Nyborg J., Andersen G. R., and Sottrup-Jensen L. (2006) The structure of bovine complement component 3 reveals the basis for thioester function. J. Mol. Biol. 361, 115–127 10.1016/j.jmb.2006.06.009 [DOI] [PubMed] [Google Scholar]
- 29. Liszewski M. K., and Atkinson J. P. (2015) Complement regulator CD46: genetic variants and disease associations. Hum. Genomics 9, 7 10.1186/s40246-015-0029-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krych-Goldberg M., and Atkinson J. P. (2001) Structure-function relationships of complement receptor type 1. Immunol. Rev. 180, 112–122 10.1034/j.1600-065X.2001.1800110.x [DOI] [PubMed] [Google Scholar]
- 31. Carroll M. C., and Isenman D. E. (2012) Regulation of humoral immunity by complement. Immunity 37, 199–207 10.1016/j.immuni.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lukácsi S., Nagy-Baló Z., Erdei A., Sándor N., and Bajtay Z. (2017) The role of CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in complement-mediated phagocytosis and podosome formation by human phagocytes. Immunol. Lett. 189, 64–72 10.1016/j.imlet.2017.05.014 [DOI] [PubMed] [Google Scholar]
- 33. van den Elsen J. M., and Isenman D. E. (2011) A crystal structure of the complex between human complement receptor 2 and its ligand C3d. Science 332, 608–611 10.1126/science.1201954 [DOI] [PubMed] [Google Scholar]
- 34. Bajic G., Yatime L., Sim R. B., Vorup-Jensen T., and Andersen G. R. (2013) Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3. Proc. Natl. Acad. Sci. U.S.A. 110, 16426–16431 10.1073/pnas.1311261110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen X., Yu Y., Mi L. Z., Walz T., and Springer T. A. (2012) Molecular basis for complement recognition by integrin αXβ2. Proc. Natl. Acad. Sci. U.S.A. 109, 4586–4591 10.1073/pnas.1202051109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiesmann C., Katschke K. J., Yin J., Helmy K. Y., Steffek M., Fairbrother W. J., McCallum S. A., Embuscado L., DeForge L., Hass P. E., and van Lookeren Campagne M. (2006) Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature 444, 217–220 10.1038/nature05263 [DOI] [PubMed] [Google Scholar]
- 37. Helmy K. Y., Katschke K. J. Jr., Gorgani N. N., Kljavin N. M., Elliott J. M., Diehl L., Scales S. J., Ghilardi N., and van Lookeren Campagne M. (2006) CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell 124, 915–927 10.1016/j.cell.2005.12.039 [DOI] [PubMed] [Google Scholar]
- 38. Broadley S. P., Plaumann A., Coletti R., Lehmann C., Wanisch A., Seidlmeier A., Esser K., Luo S., Rämer P. C., Massberg S., Busch D. H., van Lookeren Campagne M., and Verschoor A. (2016) Dual-track clearance of circulating bacteria balances rapid restoration of blood sterility with induction of adaptive immunity. Cell Host Microbe 20, 36–48 10.1016/j.chom.2016.05.023 [DOI] [PubMed] [Google Scholar]
- 39. Rother R. P., Rollins S. A., Mojcik C. F., Brodsky R. A., and Bell L. (2007) Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat. Biotechnol. 25, 1256–1264 10.1038/nbt1344 [DOI] [PubMed] [Google Scholar]
- 40. Risitano A. M., Marotta S., Ricci P., Marano L., Frieri C., Cacace F., Sica M., Kulasekararaj A., Calado R. T., Scheinberg P., Notaro R., and Peffault de Latour R. (2019) Anti-complement treatment for paroxysmal nocturnal hemoglobinuria: time for proximal complement inhibition? A position paper from the SAAWP of the EBMT. Front. Immunol. 10, 1157 10.3389/fimmu.2019.01157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Janssen B. J., Halff E. F., Lambris J. D., and Gros P. (2007) Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J. Biol. Chem. 282, 29241–29247 10.1074/jbc.M704587200 [DOI] [PubMed] [Google Scholar]
- 42. Mastellos D. C., Yancopoulou D., Kokkinos P., Huber-Lang M., Hajishengallis G., Biglarnia A. R., Lupu F., Nilsson B., Risitano A. M., Ricklin D., and Lambris J. D. (2015) Compstatin: a C3-targeted complement inhibitor reaching its prime for bedside intervention. Eur. J. Clin. Invest. 45, 423–440 10.1111/eci.12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elverdi T., and Eskazan A. E. (2019) Caplacizumab as an emerging treatment option for acquired thrombotic thrombocytopenic purpura. Drug Des. Devel. Ther. 13, 1251–1258 10.2147/DDDT.S134470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vincke C., Loris R., Saerens D., Martinez-Rodriguez S., Muyldermans S., and Conrath K. (2009) General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J. Biol. Chem. 284, 3273–3284 10.1074/jbc.M806889200 [DOI] [PubMed] [Google Scholar]
- 45. Coppieters K., Dreier T., Silence K., Haard H. D., Lauwereys M., Casteels P., Beirnaert E., Jonckheere H., Wiele C. V. D., Staelens L., Hostens J., Revets H., Remaut E., Elewaut D., and Rottiers P. (2006) Formatted anti–tumor necrosis factor α VHH proteins derived from camelids show superior potency and targeting to inflamed joints in a murine model of collagen-induced arthritis. Arthritis Rheum. 54, 1856–1866 10.1002/art.21827 [DOI] [PubMed] [Google Scholar]
- 46. Andersen K. R., Leksa N. C., and Schwartz T. U. (2013) Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins 81, 1857–1861 10.1002/prot.24364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laursen N. S., Gordon N., Hermans S., Lorenz N., Jackson N., Wines B., Spillner E., Christensen J. B., Jensen M., Fredslund F., Bjerre M., Sottrup-Jensen L., Fraser J. D., and Andersen G. R. (2010) Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 107, 3681–3686 10.1073/pnas.0910565107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yatime L., Merle N. S., Hansen A. G., Friis N. A., Ostergaard J. A., Bjerre M., Roumenina L. T., Thiel S., Kristensen P., and Andersen G. R. (2018) A single-domain antibody targeting complement component C5 acts as a selective inhibitor of the terminal pathway of the complement system and thus functionally mimicks the C-terminal domain of the Staphylococcus aureus SSL7 protein. Front. Immunol. 9, 2822 10.3389/fimmu.2018.02822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C. S., and Carragher B. (2005) Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 10.1016/j.jsb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- 50. Voss N. R., Yoshioka C. K., Radermacher M., Potter C. S., and Carragher B. (2009) DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 10.1016/j.jsb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lander G. C., Stagg S. M., Voss N. R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P. W., Lyumkis D., Potter C. S., and Carragher B. (2009) Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 10.1016/j.jsb.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Grant T., Rohou A., and Grigorieff N. (2018) cisTEM, user-friendly software for single-particle image processing. Elife 7, e35383 10.7554/eLife.35383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scheres S. H. (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ludtke S. J., Baldwin P. R., and Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 55. Engströom G., Hedblad B., Berglund G., Janzon L., and Lindgärde F. (2007) Plasma levels of complement C3 is associated with development of hypertension: a longitudinal cohort study. J. Hum. Hypertens. 21, 276–282 10.1038/sj.jhh.1002129 [DOI] [PubMed] [Google Scholar]
- 56. Kotimaa J., Klar-Mohammad N., Gueler F., Schilders G., Jansen A., Rutjes H., Daha M. R., and van Kooten C. (2016) Sex matters: systemic complement activity of female C57BL/6J and BALB/cJ mice is limited by serum terminal pathway components. Mol. Immunol. 76, 13–21 10.1016/j.molimm.2016.06.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EM 3D reconstructions presented here are available upon request from Gregers Rom Andersen (gra@mbg.au.dk). All other data are contained within the article.