Abstract
Clinical diagnosis of hepatitis E viral (HEV) infection mainly relies on serological assays, and the current status of misdiagnoses regarding HEV infection is uncertain. In this study, patients with acute HEV infection were tested for anti-HEV IgM and IgG, a HEV antigen (Ag), and viral loads (HEV RNA). Serology was performed using four commercial HEV ELISA kits: Wantai, Kehua, Lizhu, and Genelabs IgM and IgG. The HEV RNA was detected using RT-PCR assays. The sensitivities of different kits for anti-HEV IgM ranged from 82.6% to 86%. Each kit for anti-HEV IgM was highly specific (97.8–100%). The sensitivities of all kits to detect anti-HEV IgG with (87.2–91.9%) had a substantial agreement, but the Kehua and Genelabs tests were more specific than the Wantai and Lizhu tests. The Wantai tests for the HEV Ag and HEV RNA were also important for acute HEV infections (Kappa = 0.787). Furthermore, a total of 6.98% of HEV infections were positive for HEV RNA but negative for both the HEV Ag and anti-HEV antibodies of IgM and IgG classes. Our findings demonstrate that the diagnosis of hepatitis E may be missed if only serological assays are used. Thus, a combination of serological and nucleic acid testing provides the optimal sensitivity and specificity to the diagnostic process.
Key words: hepatitis E virus, serological markers, diagnostic performance, enzyme-linked immunosorbent assay, misdiagnosis
Introduction
Hepatitis E is the infection of the liver caused by a virus known as the hepatitis E virus (HEV) and has posed severe public health hazards around the world. HEV has four major genotypes (1–4) that are globally distributed into different epidemiological patterns based on socioeconomic factors and ecology (Lu et al. 2006). HEV genotypes 1 and 2 infect humans solely (Ahmad et al. 2011). Generally, genotype 1 accounts for the epidemics in some parts of Asia, while genotype 2 is more prevalent in Africa, Mexico, and other developing countries (Colson et al. 2012). Genotypes 3 and 4 are zoonotic with an expanded host range (Okamoto 2007), while there have been noted chronic HEV infections in immunosuppressed patients (Honer zu Siederdissen et al. 2014). Genotype 3 is prevalent worldwide, while genotype 4 is mainly present in Asia. Besides, genotypes 5 and 6, which primarily infect wild boar, have been found in Japan (Sato et al. 2011; Takahashi et al. 2011). Recently, new genotypes, known as HEV-7 and HEV-8, were also found to infect camels and humans (Al-Sadeq et al. 2017).
Currently, HEV’s diagnosis depends on specific serological and nucleic acid tests, as the clinical manifestations and routine laboratory measures of HEV are similar to those of other acute hepatitis (Zhang et al. 2019). There are four major methods for diagnosing hepatitis E, including the detection of anti-HEV IgM and IgG antibodies, the antigen (Ag), and HEV RNA. Presently, the clinical diagnosis of acute hepatitis E cases mainly depends on the serological detection of anti-HEV antibodies (Dreier and Juhl 2014). However, equivalence, sensitivity, and specificity in the results of the HEV Enzyme-linked Immunosorbent Assay (ELISA) kits tend to differ between manufacturers, leading to discrepancies in the rates of anti-HEV antibodies among different populations (Herremans et al. 2007; Drobeniuc et al. 2010), together with the HEV genome heterogeneity, and the different antigenic structure of HEV proteins. Moreover, cross-reactions of anti-HEV IgM with the Epstein-Barr virus (EBV) and cytomegalovirus (CMV) antibodies have been reported, which cause false-positive results (Hyams et al. 2014). Currently, the development of the HEV RNA assay kits is in the early stages in China and has not yet been widespread. Thus, the clinical diagnosis of HEV infection still mainly relies on serological assays with a few reports of hepatitis E misdiagnoses occurring in China.
In the present study, the performance of four commercial serological assays and PCR assay for the detection of HEV infection was evaluated, and the possibility of misdiagnosing of this infection using serological detection alone was determined.
Experimental
Materials and Methods
Samples. From March 2014 to March 2018, 364 serum samples were collected from Tianjin Third Central Hospital and Tianjin Medical University General Hospital. A total of 86 cases were diagnosed with acute viral hepatitis E (Kamar et al. 2014; European Association for the Study of the Liver 2018), 91 cases with rheumatic diseases (RD) including systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA), and 91 cases with viral hepatitis by CMV or EBV according to the diagnostic guidelines of each disease. Meanwhile, 96 healthy volunteers were included in this study. Five milliliters of venous blood was collected and agglutinated for 10 min at 37°C, and subsequently centrifuged at 3,000 × g for 15 min at 4°C. The serum was taken and stored at –80°C before analysis. This study received approval from the Branch of Tianjin Third Central Hospital Ethics Committee (2019028). Our study was non-interventional and did not involve any specific sampling or addition to the usual procedures. An anonymized database provided analytical support. Therefore, the ethics committee waived the need for patient consent.
HEV serological assays. The commercially available HEV ELISA kits were selected with Wantai (Beijing, China), Kehua (Shanghai, China), Lizhu (Zhuhai, China), and Genelabs (Singapore, Singapore) for the detection of both IgM and IgG antibodies. For the HEV Ag assay, the Wantai was the only available commercial provider for the ELISA kit in China. All the experimental operations were performed according to the instructions recommended by manufacturers. Results of the ELISA tests were listed as ratios (s/co), and interpretations were made as advised in the instructions.
The HEV RNA assay. One-step reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay was set up by our research team and had been approved for clinical use. Primers and probes were designed based on a multiple sequence alignment of the HEV genome sequences in the ORF3 region, and synthesized by the Sangon Biotech Company (Shanghai, China): forward primer, 5’-GGTGGTTTCTGGGGT-GAC-3’ (Tm = 61.2°C); reverse primer, 5’-AGGGGTTG-GTTGGATGAA-3’ (Tm = 61.2°C); probe, 5’-TGATTC-TCAGCCCTTCGC-3’ (Tm = 62.5°C). NCBI-Primer-BLAST searches for the primers and probe showed that the genetic sequences of the different HEV genotypes 1–4 were highly conserved in the ORF3 region. Firstly, the target fragment of 70 bp was amplified from the strain of HEV genotype 4 (CHNXJ-SW13) by the forward and reverse primers mentioned above, and then inserted the fragment into Promega T-easy Vector (3105 bp) to obtain the standard plasmid. Serial dilutions plasmids of 5 × 100 ~ 5 × 109 copies/2 μl were obtained by 10-fold dilution, and the standard curve was established. Total RNA was extracted from 140 μl serum using TIANamp Virus RNA Kit (Tiangen, Beijing, China). The One-Step PrimeScript’ RT-PCR Kit (Takara, Dalian, China) enabled the performance of RT-qPCR in a total 20-μl reaction system, including 10 μl GoTaq Probe qPCR Master Mix (1×), 0.4 μl GoScript RT Mix for one-step RT-qPCR (1×), 1 μl forward primer (500 mM), 1 μl reverse primer (500 mM), 0.5 μl probe (250 nM), 5.1 μl RNase-Free H2O and 2 μl RNA template or standard plasmid of HEV. Subsequently, the reaction was conducted with the MX3000P Real-Time QPCR System (Aligent, California, USA). The conditions for PCR amplification involved the following: 1 cycle at 45°C for 30 min, 1 cycle at 95°C for 15 min, followed by 45 cycles at 95°C for 10 sec, at 55°C for 20 sec and at 72°C for 15 sec. The expression of the HEV RNA was calculated according to the standard curve established by plasmids with different dilutions.
Data analysis. Commercially available software was used for all the statistical analyses (MedCalc, version 18.2, Belgium). The receiver operating characteristic (ROC) curve was used to analyze the diagnostic performances for anti-HEV IgG and IgM assays. Agreements between different kits were assessed by the Kappa statistic. The level of agreement was defined by the Kappa coefficient as excellent (> 0.8), substantial (0.6–0.8), moderate (0.4–0.6), fair (0.2–0.4), and poor (< 0.2) (Nogues-Sabate et al. 2018). A p-value of less than 0.05 was considered statistically significant. Additionally, two-sided 95% confidence intervals (CI) were calculated.
Results
Performance of anti-HEV IgM assays. The diagnostic performance characteristic of each anti-HEV IgM kit was determined by the ROC curve. As shown in Table I, the area under the curve (AUC) value of the ELISA kits ranged from 0.909 to 0.93. The sensitivity among the 86 acute HEV infections ranged from 82.6% to 86% (Table I), suggesting an excellent agreement (Kappa: 0.819–1) in anti-HEV IgM antibody detection between the ELISA kits (Table II). All the anti-HEV IgM assays evaluated demonstrated good specificities (97.8–100%), except for two false-positive results obtained from the RD cases detected by the Genelabs ELISA kit. No positive results were shown in CMV/EBV infected patients by all the assays (Table I), which demonstrated that there were no cross-reactions of IgM against CMV and EBV.
Table I.
Diagnostic performance of anti-HEV IgM assays.
| Commercial tests | % Sensitivity (95% CI) | % Specificity (95% CI) | % PPV (95% CI) | % NPV (95% CI) | AUC (95% CI) | p | % Specificity with RD (95% CI) | % Specificity with CMV/EBV (95% CI) | % Specificity with healthy (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| Wantai | 84.9 (75.5–91.7) | 100 (98.7–100) | 100 (100–100) | 95.5 (92.8–97.2) | 0.924 (0.892–0.949) | < 0.01 | 100 (100–100) | 100 (100–100) | 100 (100–100) |
| Kehua | 86.0 (76.9–92.6) | 100 (98.7–100) | 100 (100–100) | 95.9 (93.2–97.5) | 0.930 (0.899–0.954) | < 0.01 | 100 (100–100) | 100 (100–100) | 100 (100–100) |
| Lizhu | 83.7 (74.2–90.8) | 100 (98.7–100) | 100 (100–100) | 95.2 (92.5–97.0) | 0.919 (0.886–0.945) | < 0.01 | 100 (100–100) | 100 (100–100) | 100 (100–100) |
| Genelabs | 82.6 (72.9–89.9) | 99.3 (97.4–99.9) | 97.3 (89.9–99.3) | 94.8 (92.1–96.7) | 0.909 (0.875–0.937) | < 0.01 | 97.8 (94.8–100) | 100 (100–100) | 100 (100–100) |
Table II.
Concordance for anti-HEV IgM assays in the diagnosis of the acute HEV infections.
| Commercial tests | % Concordance | Kappa (95% CI) |
|---|---|---|
| Wantai | ||
| Kehua | 98.8 | 0.950 (0.852–1.000) |
| Lizhu | 98.8 | 0.953 (0.862–1.000) |
| Genelabs | 95.3 | 0.819 (0.648–0.990) |
| Kehua | ||
| Lizhu | 97.7 | 0.903 (0.771–1.000) |
| Genelabs | 96.5 | 0.860 (0.706–1.000) |
| Lizhu | ||
| Genelabs | 96.5 | 0.868 (0.722–1.000) |
Performance of anti-HEV IgG assays. The AUC value (0.827–0.929) of the four distinct anti-HEV IgG kits are shown in Table III. The sensitivity among the acute HEV infection patients ranged from 87.2% to 91.9% and showed a substantial agreement with Kappa coefficients from 0.752 to 0.927 between each different ELISA kit (Table III and IV). However, the specificities of anti-HEV IgG among the non-HEV population showed significant inconsistency between each ELISA kit (Table IV). The specificity of the Kehua (96.9–98.9%) and Genelab (96.7–98.9%) tests were significantly higher when compared with the Wantai (64.6–81.3%) and Lizhu assays (69.8–83.5%) (Table III).
Table III.
Diagnostic performance of anti-HEV IgG assays.
| Commercial tests | % Sensitivity (95% CI) | % Specificity (95% CI) | % PPV (95% CI) | % NPV (95% CI) | AUC (95% CI) | p | % Specificity with RD (95% CI) | % Specificity with CMV/EBV (95% CI) | % Specificity with healthy (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| Wantai | 91.9 (83.9–96.7) | 74.8 (69.3–79.8) | 53.0 (47.7–58.3) | 96.7 (93.6–98.4) | 0.833 (0.791–0.870) | < 0.01 | 79.2 (71.0–87.3) | 81.3 (73.3–89.3) | 64.6 (55.0–74.1) |
| Kehua | 87.2 (78.3–93.4) | 97.5 (94.9–99.0) | 91.5 (83.7–95.7) | 96.1 (93.4–97.7) | 0.923 (0.891–0.949) | < 0.01 | 98.9 (96.6–100) | 97.8 (94.8–100) | 96.9 (93.4–100) |
| Lizhu | 89.5 (81.1–95.1) | 75.9 (70.4–80.8) | 53.5 (48.0–58.9) | 95.9 (92.6–97.8) | 0.827 (0.784–0.865) | < 0.01 | 83.5 (75.9–91.1) | 74.7 (65.8–83.6) | 69.8 (60.6–79.0) |
| Genelabs | 88.4 (79.7–94.3) | 97.5 (94.9–99.0) | 91.6 (83.9–95.8) | 96.4 (93.8–98.0) | 0.929 (0.898–0.953) | < 0.01 | 98.9 (96.6–100) | 96.7 (93.0–100) | 97.9 (95.0–100) |
Table IV.
Concordance for anti-HEV IgG assays.
| Commercial tests | Concordance of HEV | Concordance of RD | Concordance of CMV/EBV | Concordance of healthy | ||||
|---|---|---|---|---|---|---|---|---|
| % | Kappa (95% CI) | % | Kappa (95% CI) | % | Kappa (95% CI) | % | Kappa (95% CI) | |
| Wantai | ||||||||
| Kehua | 95.3 | 0.753 (0.524–0.982) | 80.2 | 0.081 (–0.069–0.231) | 81.3 | 0.069 (–0.102–0.239) | 68.8 | 0.147 (0.015–0.279) |
| Lizhu | 98.8 | 0.927 (0.785–1.000) | 91.2 | 0.712 (0.525–0.898) | 88.5 | 0.809 (0.664–0.954) | 94.8 | 0.882 (0.782–0.982) |
| Genelabs | 96.5 | 0.805 (0.592–1.000) | 80.2 | 0.081 (–0.069–0.231) | 82.4 | 0.153 (–0.064–0.369) | 67.7 | 0.111 (–0.007–0.229) |
| Kehua | ||||||||
| Lizhu | 96.5 | 0.823 (0.629–1.000) | 84.6 | 0.107 (–0.087–0.300) | 76.9 | 0.125 (–0.034–0.283) | 74.0 | 0.183 (0.238–0.341) |
| Genelabs | 96.5 | 0.837 (0.658–1.000) | 100 | 1.000 (1.000–1.000) | 98.9 | 0.795 (0.403–1.000) | 99.0 | 0.852 (0.566–1.000) |
| Lizhu | ||||||||
| Genelabs | 95.3 | 0.752 (0.520–0.984) | 84.6 | 0.107 (–0.087–0.300) | 78.0 | 0.183 (0.001–0.366) | 72.9 | 0.139 (–0.005–0.282) |
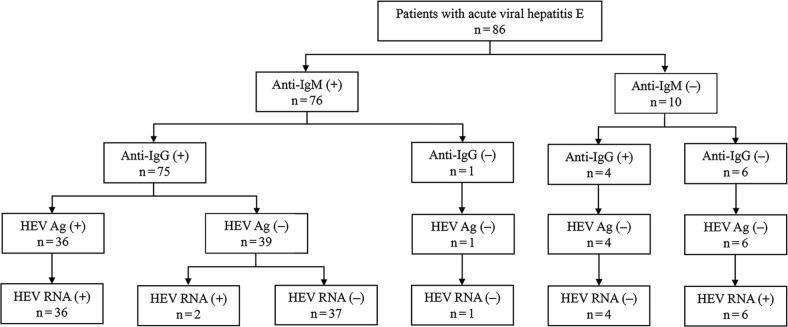
The HEV Ag and HEV RNA assays. A total of 36 acute HEV patients were positive for the HEV Ag using the Wantai ELISA kit, with no positive cases found among the non-HEV infection patients. A total of 44 acute HEV infection patients were positive for HEV RNA. The consistency rate between the HEV Ag and RNA was 90.7% (78/86), as shown in Table V. The two methods had a substantial agreement with a Kappa coefficient of 0.787 (0.656–0.918). Furthermore, six of the 86 samples were positive for HEV RNA but negative for anti-HEV IgM and IgG antibodies, and the HEV Ag by all ELISA kits (Fig. 1).
Table V.
Consistency for HEV Ag and HEV RNA assays in the diagnosis of the acute HEV infections.
| HEV RNA +, n (%) | HEV RNA −, n (%) | Total, n (%) | |
|---|---|---|---|
| HEV Ag +, n (%) | 36 (41.86) | 0 (0) | 36 (41.86) |
| HEV Ag −, n (%) | 8 (9.3) | 42 (48.84) | 50 (58.14) |
| Total, n (%) | 44 (51.16) | 42 (48.84) | 86 (100) |
Fig. 1.
Flow diagram for patients with acute viral hepatitis E.
Discussion
To date, the identification of serological markers in HEV infections using accurate diagnostic assays remain a challenge. There are a plethora of issues regarding the specificity and sensitivity of HEV serological assays in epidemiological and clinical settings that require urgent attention. In this present study, we evaluated four dominant, commercially available anti-HEV IgM and IgG assays, as well as the HEV Ag and HEV RNA to investigate the misdiagnosis’s current status rely on the current measurements.
Anti-HEV IgM appears in the early phase of acute hepatitis E. The antibodies can be detected as early as four days after the onset of jaundice and last up to five months (Kuniholm et al. 2009). There are two main methods in anti-HEV IgM serological assays: the capture method with anti-human IgM μ chain (Wantai, Kehua and Lizhu), and the indirect method (Genelabs). The sensitivity and specificity of different methods present variations in anti-HEV IgM assays with a recent study demonstrating a high cross-reactivity of HEV IgM compared to EBV and CMV (Drobeniuc et al. 2010). However, in the present study, no false-positive results due to cross-reaction with EBV- or CMV-infected patients were observed, indicating the highly specific nature of the anti-HEV IgM assays. Moreover, these findings were found to be consistent with those of the other groups, including immunocompromised patients (Abravanel et al. 2013) and infections with HEV genotype 3 (Legrand-Abravanel et al. 2009). The Genelabs ELISA kit detected two false-positive results obtained from patients with RD. This finding supports the opinion that the capture method using the antihuman IgM μ chain is more specific than the indirect method using the anti-HEV IgM assay.
In general, the detection of anti-HEV IgG is usually used as an indicator of past infection. However, the appearance of the anti-HEV IgG antibody is early, which could be used in the clinical diagnosis of acute HEV infection (Aggarwal and Jameel 2011). The Qatar research group found that Wantai HEV-IgG assays revealed high sensitivity and specificity with excellent Kappa concordance using different enzyme immunoassays in assessing seroprevalence of HEV antibodies (Al-Absi et al. 2018). However, a significant discrepancy in anti-HEV IgG results between different assay kits in the non-HEV population was found in our study. The positive rates were significantly higher by the Wantai and Lizhu kits than those of Kehua and Genelabs. A Korean research compared anti-HEV IgG antibody results using the Genelabs and Wantai ELISA kits to estimate HEV serum prevalence in the Korean population (Park et al. 2012). They found a significant inconsistency in the results between the two assays, which was also observed in our study. Therefore, epidemiological investigations of HEV in the population may lead to significant inconsistencies when different kits are used. The Kehua and Genelabs IgG assays had high specificities in the non-HEV population and could be used in the clinical diagnosis of HEV. On the other hand, the Wantai and Lizhu IgG assays were more suitable for epidemiological investigations because the positive rates in the non-HEV population were too high to distinguish the acute HEV infection from the previous disease.
In this study, all four anti-HEV IgG serological assays used the indirect method. There are two major types of antigens coated on the plates for binding of anti-HEV IgG antibodies, including synthetic peptides and recombinant proteins (Innis et al. 2002; Ulanova et al. 2008). The use of the recombinant ORF2-encoded protein in numerous serological studies has revealed its significant efficacy in the identification of antibodies against various HEV strains (Christensen et al. 2008; Kuniholm et al. 2009). Since recombinant proteins can replicate the HEV neutralizing epitope better than the synthetic peptides, the results in the Wantai anti-HEV IgG assay were more sensitive. A French research group also substantiated that the Wantai IgG assay was the most sensitive amongst all other eight commercial ELISA kits used to detect HEV of genotypes 1 and 3 (Abravanel et al. 2013). The results suggested that the anti-HEV IgM assay was superior in the diagnosis of acute HEV infection due to its good specificity when paired with the Wantai anti-HEV IgG assay, which could improve the accuracy of diagnosis.
The latest reports have indicated that this novel HEV Ag is a resourceful serum marker to detect the acute HEV infection and has a good consistency with HEV RNA (Zhang et al. 2006; Zhao et al. 2015; Fraga et al. 2018; Zhang et al. 2019). Our findings in this study also supported this view. However, all the HEV Ag positive samples showed positive anti-HEV IgM results (Fig. 1), which provided no direct evidence to support that Ag detection could improve diagnostic efficiency. Furthermore, six samples were only positive for HEV RNA but negative for anti-HEV IgM and IgG, as well as Ag in all ELISA kits employed, which showed that 6.98% of acute HEV infection patients have a chance to be misdiagnosed if reliant on serological assays detection alone. It indirectly indicates that HEV RNA detection can improve diagnostic efficiency. However, despite the highly specific and sensitive capability of some PCR assays for the detection of HEV RNA, their utility has been restricted due to the short period of HEV viremia detection. Therefore, the incidence of acute HEV infection cannot be completely ruled out by a negative HEV PCR result.
In conclusion, for the successful diagnosis of acute viral hepatitis E, a combination of nucleic acid and serological tests is imperative to provide excellent specificity and sensitivity to the diagnosis. However, we also observed significant inconsistencies between the serological and HEV RNA assays; thereby, caution is warranted while interpreting the results of both serological and molecular tests in HEV diagnosis.
Supplementary materials are available on the journal’s website.
Acknowledgments
This project was supported by the Tianjin Science and Technology Program of China (No. 17ZXHLSY00020).
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Literature
- Abravanel F, Chapuy-Regaud S, Lhomme S, Miedougé M, Peron JM, Alric L, Rostaing L, Kamar N, Izopet J.. Performance of anti-HEV assays for diagnosing acute hepatitis E in immunocompromised patients. J Clin Virol. 2013. Dec;58(4):624–628. 10.1016/j.jcv.2013.10.003 [DOI] [PubMed] [Google Scholar]
- Aggarwal R, Jameel S.. Hepatitis E. Hepatology. 2011. Dec;54(6): 2218–2226. 10.1002/hep.24674 [DOI] [PubMed] [Google Scholar]
- Ahmad I, Holla RP, Jameel S.. Molecular virology of hepatitis E virus. Virus Res. 2011. Oct;161(1):47–58. 10.1016/j.virusres.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Absi ES, Al-Sadeq DW, Younis MH, Yassine HM, Abdalla OM, Mesleh AG, Hadwan TA, Amimo JO, Thalib L, Nasrallah GK.. Performance evaluation of five commercial assays in assessing seroprevalence of HEV antibodies among blood donors. J Med Microbiol. 2018. Sep 01;67(9):1302–1309. 10.1099/jmm.0.000807 [DOI] [PubMed] [Google Scholar]
- Al-Sadeq DW, Majdalawieh AF, Nasrallah GK.. Seroprevalence and incidence of hepatitis E virus among blood donors: A review. Rev Med Virol. 2017. Sep;27(5):e1937 10.1002/rmv.1937 [DOI] [PubMed] [Google Scholar]
- Christensen PB, Engle RE, Hjort C, Homburg KM, Vach W, Georgsen J, Purcell RH.. Time trend of the prevalence of hepatitis E antibodies among farmers and blood donors: a potential zoonosis in Denmark. Clin Infect Dis. 2008. Oct 15;47(8):1026–1031. 10.1086/591970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, Romanet P, Moal V, Borentain P, Purgus R, Benezech A, Motte A, Gérolami R.. Autochthonous infections with hepatitis E virus genotype 4, France. Emerg Infect Dis. 2012. Aug;18(8): 1361–1364. 10.3201/eid1808.111827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier J, Juhl D.. Autochthonous hepatitis e virus infections: a new transfusion-associated risk? Transfus Med Hemother. 2014; 41(1):29–39. 10.1159/000357098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobeniuc J, Meng J, Reuter G, Greene-Montfort T, Khudyakova N, Dimitrova Z, Kamili S, Teo CG.. Serologic assays specific to immunoglobulin M antibodies against hepatitis E virus: pangenotypic evaluation of performances. Clin Infect Dis. 2010. Aug; 51(3):e24–e27. 10.1086/654801 [DOI] [PubMed] [Google Scholar]
- European Association for the Study of the Liver EASL Clinical Practice Guidelines on hepatitis E virus infection. J Hepatol. 2018. Jun;68(6):1256–1271. 10.1016/j.jhep.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Fraga M, Doerig C, Moulin H, Bihl F, Brunner F, Müllhaupt B, Ripellino P, Semela D, Stickel F, Terziroli Beretta-Piccoli B, et al. Hepatitis E virus as a cause of acute hepatitis acquired in Switzerland. Liver Int. 2018. Apr;38(4):619–626. 10.1111/liv.13557 [DOI] [PubMed] [Google Scholar]
- Herremans M, Bakker J, Duizer E, Vennema H, Koopmans MPG.. Use of serological assays for diagnosis of hepatitis E virus genotype 1 and 3 infections in a setting of low endemicity. Clin Vaccine Immunol. 2007. May;14(5):562–568. 10.1128/CVI.00231-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höner zu Siederdissen C, Pischke S, Schlue J, Deterding K, Hellms T, Schuler-Lüttmann S, Schwarz A, Manns MP, Cornberg M, Wedemeyer H.. Chronic hepatitis E virus infection beyond transplantation or human immunodeficiency virus infection. Hepatology. 2014. Sep;60(3):1112–1113. 10.1002/hep.26987 [DOI] [PubMed] [Google Scholar]
- Hyams C, Mabayoje DA, Copping R, Maranao D, Patel M, Labbett W, Haque T, Webster DP.. Serological cross reactivity to CMV and EBV causes problems in the diagnosis of acute hepatitis E virus infection. J Med Virol. 2014. Mar;86(3):478–483. 10.1002/jmv.23827 [DOI] [PubMed] [Google Scholar]
- Innis BL, Seriwatana J, Robinson RA, Shrestha MP, Yarbough PO, Longer CF, Scott RM, Vaughn DW, Myint KS.. Quantitation of immunoglobulin to hepatitis E virus by enzyme immunoassay. Clin Diagn Lab Immunol. 2002. May;9(3):639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamar N, Dalton HR, Abravanel F, Izopet J.. Hepatitis E virus infection. Clin Microbiol Rev. 2014. Jan 01;27(1):116–138. 10.1128/CMR.00057-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuniholm MH, Purcell RH, McQuillan GM, Engle RE, Wasley A, Nelson KE.. Epidemiology of hepatitis E virus in the united states: results from the third national health and nutrition examination survey, 1988–1994. J Infect Dis. 2009. Jul;200(1):48–56. 10.1086/599319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand-Abravanel F, Thevenet I, Mansuy JM, Saune K, Vischi F, Peron JM, Kamar N, Rostaing L, Izopet J.. Good performance of immunoglobulin M assays in diagnosing genotype 3 hepatitis E virus infections. Clin Vaccine Immunol. 2009. May;16(5):772–774. 10.1128/CVI.00438-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li C, Hagedorn CH.. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006. Jan;16(1):5–36. 10.1002/rmv.482 [DOI] [PubMed] [Google Scholar]
- Nogués-Sabaté A, Aviles-Jurado FX, Ruiz-Sevilla L, Lehrer E, Santamaría-Gadea A, Valls-Mateus M, Vilaseca I.. Intra and interobserver agreement of narrow band imaging for the detection of head and neck tumors. Eur Arch Otorhinolaryngol. 2018. Sep; 275(9):2349–2354. 10.1007/s00405-018-5063-8 [DOI] [PubMed] [Google Scholar]
- Okamoto H. Genetic variability and evolution of hepatitis E virus. Virus Res. 2007. Aug;127(2):216–228. 10.1016/j.virusres.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Park HK, Jeong SH, Kim JW, Woo BH, Lee DH, Kim HY, Ahn S.. Seroprevalence of anti-hepatitis E virus (HEV) in a Korean population: comparison of two commercial anti-HEV assays. BMC Infect Dis. 2012. Dec;12(1):142 10.1186/1471-2334-12-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Sato H, Naka K, Furuya S, Tsukiji H, Kitagawa K, Sonoda Y, Usui T, Sakamoto H, Yoshino S, et al. A nationwide survey of hepatitis E virus (HEV) infection in wild boars in Japan: identification of boar HEV strains of genotypes 3 and 4 and unrecognized genotypes. Arch Virol. 2011. Aug;156(8):1345–1358. 10.1007/s00705-011-0988-x [DOI] [PubMed] [Google Scholar]
- Takahashi M, Nishizawa T, Sato H, Sato Y, Jirintai, Nagashima S, Okamoto H.. Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J Gen Virol. 2011. Apr 01;92(4):902–908. 10.1099/vir.0.029470-0 [DOI] [PubMed] [Google Scholar]
- Ulanova TI, Obriadina AP, Talekar G, Burkov AN, Fields HA, Khudyakov YE.. A new artificial antigen of the hepatitis E virus. J Immunoassay Immunochem. 2008. Dec 31;30(1):18–39. 10.1080/15321810802570269 [DOI] [PubMed] [Google Scholar]
- Zhang F, Li X, Li Z, Harrison TJ, Chong H, Qiao S, Huang W, Zhang H, Zhuang H, Wang Y.. Detection of HEV antigen as a novel marker for the diagnosis of hepatitis E. J Med Virol. 2006. Nov; 78(11):1441–1448. 10.1002/jmv.20717 [DOI] [PubMed] [Google Scholar]
- Zhang H, Rao H, Wang Y, Wang J, Kong X, Ji Y, Zhu L, Liu Y, Fang J, Yang M, et al. Evaluation of an antigen assay for diagnosing acute and chronic hepatitis E genotype 4 infection. J Gastroenterol Hepatol. 2019. Feb;34(2):458–465. 10.1111/jgh.14405 [DOI] [PubMed] [Google Scholar]
- Zhao C, Geng Y, Harrison TJ, Huang W, Song A, Wang Y.. Evaluation of an antigen-capture EIA for the diagnosis of hepatitis E virus infection. J Viral Hepat. 2015. Nov;22(11):957–963. 10.1111/jvh.12397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials are available on the journal’s website.