Abstract
Aim
The aim of this study was to explore the expression of exosomal non-coding RNAs (ncRNAs) in the sera of patients with HCC versus control.
Methods
Firstly, Bioinformatics analysis was conducted to retrieve ncRNAs specific to HCC (hsa-miRNA-1298 and lncRNA-RP11-583F2.2). Afterwards, extraction and characterization of exosomes were performed. We measured the expression of the chosen exosomal RNAs by reverse transcriptase quantitative real-time PCR in sera of 60 patients with HCC, 42 patients with chronic hepatitis C (CHC) infection and 18 healthy normal volunteers.
Results
The exosomal ncRNAs [hsa-miRNA-1298, lncRNA-RP11-583F2.2] had better sensitivity and specificity than alpha-fetoprotein (AFP) in HCC diagnosis.
Conclusion
The exosomal hsa-miRNA-1298, lncRNA-RP11-583F2.2 can be potential biomarkers for HCC diagnosis.
Keywords: Hepatocellular carcinoma, exosome, miRNA, lncRNA, diagnosis, chronic hepatitis C
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world, with high morbidity and mortality [1]. According to the American Cancer Society, more than 700,000 people throughout the world are diagnosed with HCC each year and more than 600,000 deaths that occur each year are due to liver cancer [2].
In Egypt, cancer of the liver in males is the first most common cancer (33.63%) and is the second common cancer in females (13.5%). HCC represents the most common primary liver malignant tumor and it is the 2nd cause of mortality due to cancer in both sexes [3]. HCC occurs in a number of preexisting conditions, e.g. Hepatitis C virus (HCV) and hepatitis B virus (HBV), alcoholic and nonalcoholic cirrhosis. This is highly related to the HCV epidemic that involved about 10-15% of the Egyptian population through the last three decades, and was stated as the highest HCV prevalence in the world [4]. Egypt has the highest prevalence of HCV infection in the world with 13.8% of the population infected and seven million have chronic HCV liver disease [5].
Most of the screening tools used for HCC diagnosis, either serum alpha-fetoprotein (AFP) or ultrasound lack
adequate sensitivity and specificity [6]. Great efforts have been made towards the study of genomics and epigenomics to demonstrate the mechanisms of HCC and consequently identify novel therapeutic strategies and improve the clinical outcome of patients [7]. The development of cancer is a complex process in which exosomes have a major role. Exosomes are minute vesicles budded from the endosomal network in the form of vesicular bodies. The exosomes have a major role in the transport of important molecules between cells with concern in the targeted cells. The important molecules such as messenger RNAs (mRNAs), microRNAs (miRNAs), long noncoding RNAs (lncRNAs), ribosomal RNAs (rRNAs), small-nuclear RNAs (snRNAs) and transfer RNAs(tRNAs) [8, 9]. The exosomes derived from tumor liberated into the circulation and locally to react with the diversity of target cells, such as other cancer cells, immune and endothelial cells [10]. As the content of exosomes like that of the original cell, they can be considered as potential biomarkers for diagnosis and therapeutic strategies of cancer [11].
Sohn et al., (2015) reported that exosomal microRNAs, such as miRNA-18a, miRNA-221, miRNA-222, and miRNA-224 are elevated in patients of hepatocellular carcinoma as compared to patients with chronic hepatitis B or cirrhosis of the liver. Meantime, the levels of serum exosomal miRNA-101, miRNA-106b, miRNA-122, and miRNA-195 were lower in patients of hepatocellular carcinoma as compared to chronic hepatitis B patients [12]. Moreover, Takahashi et al., (2014) reported that the revelation of hepatocellular carcinoma cells to different agents of anticancer such as camptothecin, induce the lncRNA-VLDLR expression in transformed liver cells in addition to recruitment of these cells into exosomes derived from it. This result elucidates that this lncRNA-VLDLR could involve in chemo-resistance in HCC cells.
The same research group stated other lncRNA mediated in chemo-resistance of hepatocellular carcinoma is the regulator of reprogramming (ROR). LncRNA-ROR plays a role in inducing the maintenance of cancer stem cells and the advancement of HCC cells chemo-resistance, whereas knockdown of this lncRNA enhanced the chemo-sensitivity [13].
In the current study, ncRNAs specific to HCC were retrieved and the expression of the chosen ncRNAs in HCC was verified through in silico data analysis. Then the expression of serum exosomal ncRNAs was done to evaluate their usefulness as diagnostic biomarkers and the relationship between the selected RNA biomarkers and pathological changes of patients was explored.
2. PATIENTS AND METHODS
2.1. Patients and Samples
In the current study, 60 HCC patients were diagnosed based on the American Association for the Study of Liver Diseases (AASLD) practice guidelines. However, the clinical stage was determined by the Barcelona Clinic Liver Cancer (BCLC) classification, the clinical stages of HCC of patients classified as 54 (90%) early-stage (A and B) and 6 (C) (10%) late-stage HCC. All blood samples assembled before any surgical, chemotherapeutic or radiotherapeutic procedures. For each patient, complete follow-up data was available. Forty-two patients with chronic viral hepatitis C(CHC) were recruited at the tropical department Ain Shams University Hospital. As well as, blood samples were collected from 18 healthy normal volunteers during their routine medical checkup.
Venous blood samples from each participant were collected in plain collection tubes without clot activator and centrifuged at 1300xg at 4°C for 20 min. to obtain the serum. Then all sera samples were stored at -80°C until assayed. From all the participants of this study, written informed consent was obtained. The study was performed according to the Declaration of Helsinki, and approved by the Research Ethical Committee at the Faculty of Medicine, Ain Shams University, Egypt (ethical approval number; FWA 000017585). The clinical and demographic data of all the participants have been summarized (Table 1).
Table 1. Shows the clinicopathological factors in different groups of the study.
| - |
Malignant (HCC)
N (%) |
CHC
(HCV) N (%) |
Healthy Control
N (%) |
P | χ2(a) |
|---|---|---|---|---|---|
|
Age:
≥ 57.8 < 57.8 |
35 (58.3%) 25 (41.7%) |
21 (50%) 21 (50%) |
9 (50%) 9 (50%) |
.657 NS | 0.839 |
|
Sex:
Male (88) Female (32) |
43 (71.7%) 17 (28.3%) |
33 (78.6%) 9 (21.4%) |
12 (66.7%) 6 (33.3%) |
.582 NS | 1.084 |
|
Smoking:
Smoker (53) Non-Smoker (67) |
23 (38.3%) 37 (61.7%) |
21 (50%) 21 (50%) |
9 (50%) 9 (50%) |
.437 NS | 1.656 |
|
HCV-antibodies:
Positive (94) Negative (26) |
52 (86.7%) 8 (13.3%) |
42 (100%) 0 (0%) |
0 (0%) 18 (100%) |
.000** | 79.149 |
|
HBV-sAg:
Positive(6) Negative(114) |
6 (10%) 54 (90%) |
0 (0%) 42 (100%) |
0 (0%) 18 (100%) |
.043* | 6.316 |
|
Cirrhosis:
Cirrhotic(70) Non-cirrhotic(50) |
49(81.7%) 11(18.3%) |
9(21.4%) 33(78.6%) |
0(0%) 18(100%) |
.000** | 39.840 |
2.2. Viral Markers and Serum AFP Detection
Serum anti-HCV antibodies and hepatitis B surface antigen (HBsAg) were investigated by enzyme-linked immunosorbent assay (ELISA) using commercial kits. Serum AFP was measured using ELISA commercial kits (AbCam, Cambridge, MA).
2.3. Bioinformatics Based Selection of RNA Based Biomarker Network
The identification of the RNA based biomarker network was done in the following steps:
(i). We used scanning algorithms to search miRNA specific to HCC through several miR public databases, namely, miRWalk database, and miR2Disease. The 2 databases confirmed the correlation between selected hsa-miR-1298 dysregulation and HCC development depending on, fold change ≥ 2.0, P-value < 0.05, higher ranking score and novelty.
(ii). Pathway enrichment analysis revealed that hsa-miR-1298 has a high number of target genes related to carcinogenesis, for example, Mitogen Activated Kinase-Like Protein, exosomes secretion, apoptosis and cell adhesion through Diana database.
(iii). We chose LncRNA-RP11-583F2.2 targeting hsa-miR-1298 through the database of lncRBA acting as competing endogenous RNA (inCeDB) (http://gyanxet-beta.com/lncedb/index.php. LncRNA-RP11-583F2.2 has a higher number of target genes linked to HCC by bioinformatics tools, namely; Database of Cancer Gene Networks from Public Gene Expression Data(TCNG) (Available at http://tcng.hgc.jp/index.html) and lncRNA Expression database NRED database (Available at http://nred.matticklab.com/cgi-bin/ncrnadb.pl/) (Supplementary Table 1s (412.3KB, pdf) ).
2.4. Exosomal RNA Extraction and Characterization of Exosomes
Exosomal RNA was extracted from sera samples of all participants using exoRNeasy Serum/Plasma MidiKit (Cat. no. 77044, Qiagen, USA) according to manufacturer's instructions, the exoRNeasy Serum/Plasma Kits use a membrane-based affinity binding step to isolate exosomes. Purified exosomes suspensions were examined by A JEOL 1010 transmission electron microscopy (TEM) at various magnifications (Fig. 1). The extracted total RNA from exosomes was reverse transcribed into cDNA as soon as possible with a miScript II RT Kit (Cat. no. 218161, Qiagen, USA), following the manufacturer’s protocol for sera samples using Thermo Hybaid PCR express (Thermo Scientific, USA).
Fig. (1).
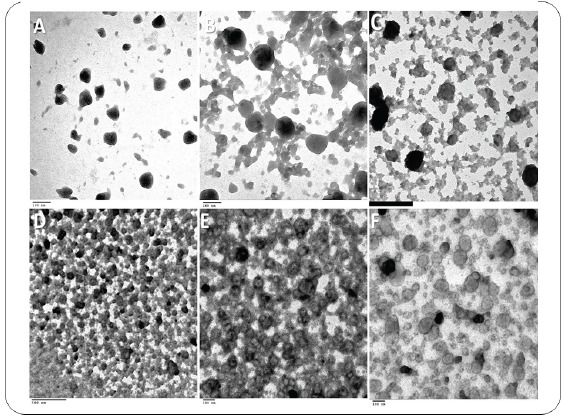
Transmission electron microscopy image showing the morphology of Exosomes isolated from serum of control group (A&B), HCV serum-derived Exosomes (C) and HCC serum-derived Exosomes (D, E & F). The isolated Exosomes are intact small round shaped nanovesicles, morphologically homogeneous and of different sizes ranging from 30 to 100 nm in size, with a typical round or cup shape appearance (scale bar, 100 nm). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2.5. Real Time-PCR (qPCR) Quantification of RNA Based Biomarker
Exosomal hsa-miR-1298 expression in sera samples was assessed by mixing the total cDNAs with miScript SYBR Green PCR Kit (Cat. no. 218076, Qiagen, Helman Germany), according to the manufacturer’s protocol; along with the miScript Universal primer provided by the manufacturer and the forward primer specific to miRNA (Hs_miR-1298_1 miScript Primer Assay, MIMAT0005800, Cat. no. MS00014574).
Exosomal LncRNA-RP11-583F2.2 expression in sera samples was estimated using RT2 SYBR Green ROX qPCR Master mix Kit (Cat. no. 330520, Qiagen, Germany) and RT2 lncRNA qPCR Primer Assay for Human RP11-583F2.2 (ENST00000583416, Cat. no. LPH24879A).
All the PCR primers were purchased from Qiagen, (Hs_ACTB_1_SG QuantiTect Primer Assay, NM_001101, Cat. no. QT00095431) and both and small nucleolar RNA, C/D box 68 [SNORD-68]) were used as housekeeping genes to normalize our raw data for lncRNA and miRNA respectively. The PCR program for the Rotor gene 5Plex (Qiagen, Hilden, Germany) based qPCR is as follow: firstly, denaturation at 95°C for 15 min; followed by 45 cycles of denaturation for 15 sec at 94°C; then annealing for 30 sec at 55°C. For relative lncRNA-RP11-583F2.2 quantification, the real-time cycler was programmed as follows: initial activation step for 10 min at 95ºC to activate HotStarTaq DNA Polymerase. Forty cycles of PCR performed under the following conditions; 15 sec at 95ºC, 30 sec at 55ºC and 30 sec at 72ºC for denaturation, annealing and extension, respectively. Each reaction was carried out in duplicate.
Using the Leviak method, the relative quantification of RNA based biomarker panel expression was calculated, where the RQ= 2-ΔΔCt method [14]. We compared 2 snoRNAs to identify the best reference gene for relative miR quantification. SNORD-68 was detected in all cases, with higher and more stable expression than RNU-6. Using the Rotor-Gene 5Plex real-time PCR detection system, the threshold cycle (Ct) value of each sample was calculated. Any Ct value more than 36 is considered negative. The results were analyzed by the plot curve analysis of Rotor-Gene software.
2.6. Statistics
All statistical analyses were performed by Statistical Package for the Social Sciences (SPSS software version 20). Comparisons were performed using Krausakul-Wallis, one-way analysis of variance (ANOVA test), and chi-square test, as appropriate. The receiver operating characteristic (ROC) curve was generated to explore the predictive value of selected RNA based biomarker network for HCC. The association between expressions of RNAs and clinicopathological data assessed with the Spearman rank correlation. Two-tailed P value of ≤ 0.05 was considered statistically significant.
3. RESULTS
3.1. Description of the Study Population
There was no statistically significant difference as regard age, sex, smoking and HBVsAg among the three study groups (p>0.05), details of the clinical data are presented in Table 1.
3.2. TEM Canning of Serum Exosomes Among the Studied Groups
The isolated exosomes are intact small round shaped nanovesicles, morphologically homogeneous and of different sizes ranging from 30 to 100 nm in size, with a typical round or cup shape appearance (scale bar, 100 nm). TEM images in (Fig. 1A and B) shows a low level of abundance of exosomes in healthy control samples compared with more abundance exosomes noted chronic hepatitis C (HCV) patient, as shown in the image in (Fig. 1C). However, the highest abundance of exosomes was found in the HCC patients as per images when compared with healthy control and chronic HCV induction patients (Fig. 1D, E and F).
3.3. Expression of Serum Exosomal ncRNAs Among the Study Groups
The exosomal ncRNAs biomarker levels based on RQ values in serum have been summarized (Table 2). The median (RQ) were, 2.77, 2.3, 0.411, for miRNA-1298 and 0.96, 0.643 and 29.9 for lncRNA-RP11-583F2.2 in healthy control, CHC group and malignant group (HCC) respectively. Compared with the non-malignant groups, the malignant group (HCC) had a higher expression of lncRNA-RP11-583F2.2 and lower expression of miRNA-1298 (p< 0.01) in the serum. The positivity rate of the serum exosomal (miRNA-1298 and lncRNA-RP11-583F2.2) was 95% and 96.7% respectively in the malignant group. However, they were 0% in normal individuals (p< 0.01), as shown in Table 2, Figs. (2A, B and 3).
Table 2. Shows the median (RQ) and the positivity rate of serum exosomal RNA based biomarkers among the study groups.
| - | Malignant (HCC) |
CHC
(HCV) |
Healthy Control | P | χ2(a) |
|---|---|---|---|---|---|
| RQ of miRNA-1298 | 0.41 | 2.3 | 2.77 | .000** | 81.344 |
| RQ of Lnc-RNA-RP11-583F2.2 | 29.9 | 0.643 | .96 | .000** | 71.162 |
| Positivity rate of miRNA-1298 | 57 (95%) | 1 (2.4%) | 0 (0%) | .000 ** | 104.678 |
| Positivity rate of Lnc-RNA-RP11-583F2.2 | 58 (96.7%) | 5 (11.9%) | 0 (0%) | .000 ** | 86.022 |
Fig. (2).
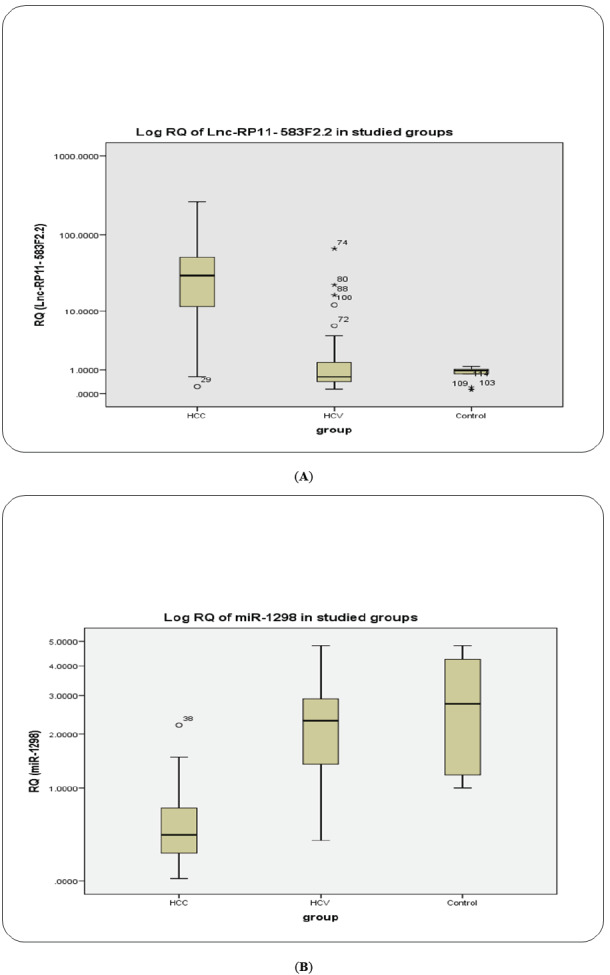
BOXPLOT: Serum exosomal lncRNA-RP11-583F2.2 and exosomal RAB11A mRNA as determined by qRT‐PCR among the HCC, CHC, and healthy control groups. (A) LncRNA‐RP11‐513I15.6; (B) Has‐miR‐1298; The data are presented as the median fold changes (P < .05). CHC, chronic hepatitis C virus; HCC, hepatocellular carcinoma; qRT‐PCR, quantitative reverse‐transcriptase polymerase chain reaction. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig. (3).
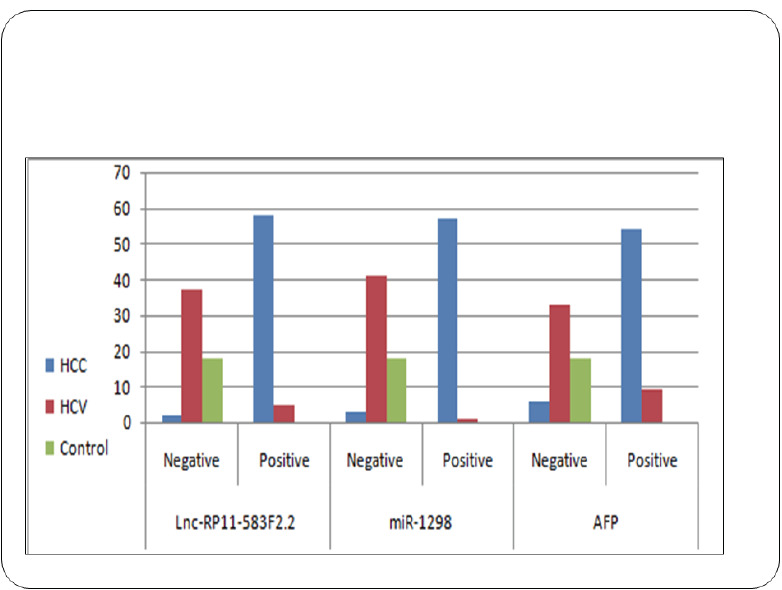
Bar chart showing the positivity rates for serum exosomal lncRNA-RP11-583F2.2, and exosomal Hsa‐miR‐1298 expression among the HCC, the CHC, and healthy control groups. The data are presented as percentages, and * indicates P < .05 compared with the control group (chi‐square test). CHC, chronic hepatitis C virus; HCC, hepatocellular carcinoma. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.4. Correlation of the Serum Exosomal with Demographic and Clinical Factors
There was no statistically significant difference between fold change (RQ) value of exosomal miRNA-1298 and lncRNA-RP11-583F2.2 and the different clinicopathological factors within the malignant group (HCC) (P >0.05) as shown in Table 3A. There was no statistically significant correlation between fold change (RQ) value of exosomal miR-1298 and lncRNA-RP11-583F2.2 and the different laboratory parameters within the malignant group (HCC) (P>0.05) as shown in Table 3B. Interestingly, there was a highly significant negative correlation between exosomal miR-1298 and lncRNA-RP11-583F2.2 based on fold change (RQ) among the three study groups. While, there was no significant correlation between exosomal miR-1298 and lncRNA-RP11-583F2.2 based on fold change (RQ) in malignant group as shown in Table 4.
Table 3A. Shows correlation of the serum level of RNA based biomarkers with different clinicopathological factors within the malignant group.
| Clinicopathological Factors | RQ of miR-1298 | RQ of Lnc-RNA-RP11-583F2.2 |
|---|---|---|
|
Mean age:
≥57.8 years <57.8 years |
.33
.52 |
33.12
29.85 |
| P, χ2(a) | 133 NS, 2.256 | .808 NS, .059 |
|
Sex:
Male Female |
.40
.51 |
21.25
41.35 |
| P, χ2(a) | .131 NS, 2.285 | .489 NS, .478 |
|
Smoking:
Smoker Non-smoker |
.41
.40 |
29.85
30.06 |
| P, χ2(a) | .30 NS, 1.072 | .257 NS, 1.286 |
|
HCV-Ab:
Positive Negative |
.41
.46 |
29.96
26.82 |
| P, χ2(a) | .296 NS, 1.093 | .573 NS, .318 |
|
HBV-sAg:
Positive Negative |
.39
.41 |
41.09
28.76 |
| P, χ2(a) | .554 NS, .351 | .632 NS, .230 |
|
Cirrhosis:
Cirrhotic Non-cirrhotic |
.40
.42 |
27.66
35.01 |
| P, χ2(a) | .491 NS, .475 | .496 NS, .464 |
|
Child-Pugh score:
A2 A5 A6 B6 B7 B8 C10 |
.236
.524 .408 .459 .476 .414 .407 |
5.61
11.31 44.94 25.63 49.40 29.85 19.40 |
| P, χ2(a) | .575 NS, 4.762 | .651NS, 4.19 |
|
BCLC stage:
Early Late |
.41
.40 |
30.92
19.40 |
| P, χ2(a) | .167 NS, 1.91 | .632 NS, .230 |
|
Average Tumor size:
< 3 cm ≥ 3 cm |
.408
.903 |
28.76
57.81 |
| P, χ2(a) | .741NS, .109 | .789 NS, .071 |
Table 3B. Shows correlation of RQ of miR-1298 and RQ of Lnc-RNA RP11-583F2.2 with different laboratory parameters within the malignant group.
| Spearman's rho | ||||||||
|---|---|---|---|---|---|---|---|---|
| - | - | AST | AST | Albumin | total bilirubin | direct bilirubin | INR | AFP |
| RQ (miR-1298) | Correlation Coefficient | -.017- | .175 | -.127- | -.021- | .155 | .107 | .021 |
| Sig. (2-tailed) | .896 | .181 | .333 | .871 | .236 | .417 | .873 | |
| RQ (Lnc-RNA RP11-583F2.2) | Correlation Coefficient | -.117- | .186 | -.005- | -.049- | -.115- | .088 | -.244- |
| Sig. (2-tailed) | .373 | .154 | .967 | .710 | .380 | .503 | .060 | |
Table 4. Shows the correlation between RQ of miR-1298 and RQ of Lnc-RNA RP11-583F2.2 among all the study group and malignant group.
| Group Type | - | RQ (miR-1298) | RQ (Lnc-RNA RP11-583F2.2) | |
|---|---|---|---|---|
| All groups | RQ (miR-1298) | Correlation Coefficient | 1.000 | -.602** |
| Sig. | . | .000 | ||
| RQ (Lnc-RNA RP11-583F2.2) | Correlation Coefficient | -.602** | 1.000 | |
| Sig. | .000 | . | ||
| RQ (miR-1298) | Correlation Coefficient | 1.000 | .138 | |
| Malignant | Sig. | . | .293 | |
| RQ (Lnc-RNA RP11-583F2.2) | Correlation Coefficient | .138 | 1.000 | |
| Sig. | .293 | |||
3.5. Accuracy of Serum Parameters for Predicting HCC by ROC Analysis
The use of ROC curves analysis and values of the area under the curve (AUC) evaluate the ncRNAs diagnostic value as shown in Supplementary Figs. (1s (412.3KB, pdf) a-d).
Regarding the HCC patients versus CHC patients and healthy control, the best discriminating cutoff values of miR-1298 and lncRNA-RP11-583F2.2 were ≤ 0.965 and ≥ 5.02 respectively. Accordingly, the sensitivities were 95% and 96.7% respectively and specificity were 98.3% and 91.7% respectively, which indicated that these threshold values could be used to distinguish malignant group (HCC) from non-malignant groups (CHC patients and healthy subjects) as shown in Supplementary Tables 2s (412.3KB, pdf) and 3s (412.3KB, pdf) .
4. DISCUSSION
The incidence of liver cancer has increased more than triple since 1980. Since 2000, liver cancer death rates have increased by almost 3% per year [2].
In Egypt, HCC represents an important public health problem, the estimated number of liver cancer cases in Egypt 2013 was 27,991 for both sexes and expected to be 85,471 for both sexes in 2050 due to population growth [15].
Therefore, the aim of bioinformatics in the cancer biomarker discovery is to give priority lists of marker candidates with the preferred sensitivity and specificity [16].
Currently, tumor-derived exosomes have shown potential in the field of cancer [17]. The contents of tumor-derived exosomes, such as miRNAs, lncRNAs and oncoproteins reflect pathophysiological status of their endosomal origin [18, 19]. Some studies reported that exchange of exosomal RNAs and proteins not only plays a major role in onset and progression of HCC but also it may be considered as potential noninvasive markers in addition to targets of therapy [20]. The cells of hepatocellular carcinoma excrete high amounts of exosomes whose participation in pathogenesis is actually under investigation. Though the use of HCC derived exosomes as potential diagnostic and prognostic biomarkers is still at the primary stage, but there are supporting observations that exosomes released from HCC cells are different in RNA and protein content that released from untransformed cells [21]. Tumor-derived exosomes are protected against degrading enzymes (e.g., RNAses), as they are enclosed in a lipid bilayer membrane [22]. Interestingly, these enriched mRNAs, miRNAs, lncRNAs and oncoproteins in tumor-derived exosomes are selected, suggesting that the exosomal content may provide novel serological biomarkers for various types of cancer [23-26].
In light of these findings, bioinformatics analysis was used to choose hsa-miR-1298 and lncRNA-RP11-583F2.2 as promising non-coding RNAs relevant to hepatocellular carcinoma based on previous microarray studies followed by clinical validation in HCC patients versus control.
Fornari et al., (2015) stated the exosomal secretion of miRNA-21, miRNA-221a, miRNA-519d and miRNA-1228 from hepatocellular carcinoma patients and showed a relation between serum and tissue levels of miRNA-21, miRNA-494, and miRNA-519d [27]. Furthermore, Sugimachi et al. (2015) explored serum exosomal biomarkers that may predict HCC recurrence after surgery [28]. Kogure et al., (2013) reported that the certain miRNAs enriched in exosomes released from HCC cells in vitro e.g., miR-133b, miR142-5p, miR-215, miR-367, miR-376a, miR-378, miR-451, miR-517c, miR-518d, miR- 520f and miR-584 [29]. In addition, Liu et al., (2015) stated that miR-10b and miR-21 were enriched in exosomes released from HCC cells in vivo [30]. Wei et al., (2015) found miR-10b-5p and miR-486-5p were highly abundant in exosomes in the SMMC-7721 HCC cell line but less abundant in cellular RNAs. While, let-7b-5p, let-7d-5p, and let-7c-5p were highly abundant in cellular miRNAs, but occurred only in very low numbers in exosomal RNAs. These results revealed selective and specific enrichment of exosomes with specific miRNAs in HCC cells [31]. Zhou et al., (2016) stated that miR-1298 inhibits mutant KRAS-driven tumor growth by repressing tyrosine kinase FAK and the laminin subunit LAMB3 [32].
Hsa-miR-1298 is a microRNA gene, and is affiliated with undefined RNA class, located on the X chromosome (Xq23), (114,715,233bp - 114,715,344bp) with the length of 112 bases. Fornari et al., stated the excretion of circulating miRNA-21, miRNA-221a, miRNA-519d and miRNA-1228 from HCC patients [33]. The present study reports that exosomal miR-1298 expression down-regulated in patients of hepatocellular carcinoma compared with patients of hepatitis C virus and normal good health control.
Increasing evidence also pointed out lncRNA’s role as transmissible molecules in hepatocellular carcinoma, in addition to use exosomes to carry them. Kogure et al., (2013) reported that the long noncoding RNA TUC339 was significantly up-regulated in HCC derived exosomes and this lncRNA TUC339 was mediated in the growth of the tumor, adhesion of cell and progression of cell cycle [28]. Liu et al., (2017) stated that lncRNA-RP11-62F24.2 was found significantly expressed in tissues of gastric cancer in comparison with normal gastric tissue (p< 0.05) and its expression level was significantly correlated with invasion and tumor size [34].
Meanwhile, Jingxu et al., (2015) reported that lncRNA-RP11-119F7.4 expression was down-regulated in gastric cancer [35].
Abd El Gwad et al., (2018) reported that the panel of 3 exosomal RNA-based biomarkers (lncRNA-RP11-513I15.6, miR-1262, and RAB11A) showed excellent sensitivity and specificity in discriminating HCC patients from CHC patients and healthy controls [36].
There was a statistically negative correlation between miR-1298 and lncRNA RP11-583F2.2, mRNA statistical significance (P<0.01). It seemed that lncRNA RP11-583F2.2 may act as a sponge for miR-1298 inhibiting its action suggesting the hypothesis of acting as competing endogenous RNA alongside possible participation in HCC pathogenesis [12]. These statistical associations agree with insilco data analysis for retrieval of ncRNAs specific to HCC. It is worth noting that the identification of the aforementioned statistical correlation where HCC cells can be targeted with knocking out lncRNA activity, to turn off specific target genes may have significant therapeutic potential in HCC.
To the best of our knowledge, the present study is the first to report that exosomal lncRNA-RP11-583F2.2 expression up-regulated in the serum of hepatocellular carcinoma patients as compared with hepatitis C virus patients and normal good health control so exosomal lncRNA-RP11-583F2.2 may be used as a potential novel biomarker in the diagnosis of HCC.
Our results indicated that miR-1298 and lncRNA-RP11-583F2.2 were superior to the AFP in sensitivity and specificity (95%, 96.7%) and (98.3%, 91.7%) respectively as compared to AFP(90%,85%). The selected miR-1298 and lncRNA-RP11-583F2.2 reduce false negative results as compared to AFP.
The findings of this study demonstrated that the estimation of serum exosomal miR-1298 and lncRNA-RP11-583F2.2 could have a potential diagnostic and therapeutic target in patients with HCC in the future especially when used in combination.
The study limitations include a relatively small sample size. Therefore, larger multicenter studies are needed for validation. Although, this study addressed the statistical association of the lncRNA-RP11-583F2.2 by linking it to miR-1298 but more mechanistic studies are needed.
CONCLUSION
In conclusion, the presented strategy enables us to identify a significant differential exosomal miRNA-1298 and lncRNA-RP11-583F2.2 expression between HCC and control with high accuracy through in silico data analysis followed by clinical validation.
ACKNOWLEDGEMENTS
Declared none.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Ethics Committee of Faculty of Medicine, Ain Shams University, Egypt (ethical approval number; FWA 000017585).
HUMAN AND ANIMAL RIGHTS
No animals were used for studies that are the basis of this research. All human research procedures followed were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
Written informed consent was taken from all the participants of this study.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
Ain Shams Faculty of Medicine, Grants Office and Grant No. 2016-36 supported this work.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
REFERENCES
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Cancer facts and figures Atlanta. Atlanta, GA: American Cancer Society; 2019. pp. 1–71. [Google Scholar]
- 3.National Cancer Registry Program of Egypt. Magnitude of hepatocellular carcinoma in Egypt. 2011 [Google Scholar]
- 4.Elghazaly H., Gaballah A., Bahie Eldin N. Clinic-pathological pattern of hepatocellular carcinoma (HCC) in Egypt. Ann. Oncol. 2018;29(5):v5–v6. doi: 10.1093/annonc/mdy151.018. [DOI] [Google Scholar]
- 5.Amer N.A., Gemaay M.A., Mohamed A.E., Hussein M.M., Shehad I. Prevalence of viral hepatitis in Egyptian patients with hepatocellular carcinoma. Egyptian Liver J. 2013;3(1):6–9. doi: 10.1097/01.ELX.0000424247.25858.79. [DOI] [Google Scholar]
- 6.Papandreou I., Lim A.L., Laderoute K., Denko N.C. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15(10):1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 7.Jin S., White E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008;4:563–566. doi: 10.4161/auto.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christianson H.C., Svensson K.J., van Kuppevelt T.H., Li J.P., Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA. 2013;110(43):17380–17385. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gajos-Michniewicz A., Duechler M., Czyz M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014;347(1):29–37. doi: 10.1016/j.canlet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Stoorvogel W., Kleijmeer M.J., Geuze H.J., Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 11.Pant S., Hilton H., Burczynski M.E. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem. Pharmacol. 2012;83(11):1484–1494. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohn W., Kim J., Kang S.H., Yang S.R., Cho J-Y., Cho H.C., Shim S.G., Paik Y-H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 2015;47:e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi K., Yan I.K., Kogure T., Haga H., Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Amal S.I., Hussein M.K., Nabiel N.H., Hoda B., Hossam K. Cancer incidence in Egypt: results of the national population- based cancer registry program. J. Cancer Epidemiol. 2014;437971:18. doi: 10.1155/2014/437971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair M., Sandhu S.S., Sharma A.K. Prognostic and predictive biomarkers in cancer. Curr. Cancer Drug Targets. 2014;14(5):477–504. doi: 10.2174/1568009614666140506111118. [DOI] [PubMed] [Google Scholar]
- 17.Azmi A.S., Bao B., Sarkar F.H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32(3-4):623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kogure T., Lin W-L., Yan I.K., Braconi C., Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54(4):1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji H., Greening D.W., Barnes T.W., Lim J.W., Tauro B.J., Rai A., Xu R., Adda C., Mathivanan S., Zhao W., Xue Y., Xu T., Zhu H.J., Simpson R.J. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13(10-11):1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 20.Laura S., Cecilia B., Claudia M., Franca C., Raffaele S., Carla C. Functional roles and therapeutic applications of exosomes in hepatocellular carcinoma. BioMed Res. Int. 2017;2017:1–8. doi: 10.1155/2017/2931813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He M., Qin H., Poon T.C., Sze S-C., Ding X., Co N.N., Ngai S-M., Chan T-F., Wong N. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis. 2015;36(9):1008–1018. doi: 10.1093/carcin/bgv081. [DOI] [PubMed] [Google Scholar]
- 22.Yuyama K., Sun H., Mitsutake S., Igarashi Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. J. Biol. Chem. 2012;287(14):10977–10989. doi: 10.1074/jbc.M111.324616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N., Reissfelder C., Pilarsky C., Fraga M.F., Piwnica-Worms D., Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avgeris M., Panoutsopoulou K., Papadimitriou M.A., Scorilas A. Circulating exosomal miRNAs: clinical significance in human cancers. Expert Rev. Mol. Diagn. 2019;19(11):979–995. doi: 10.1080/14737159.2019.1673732. [DOI] [PubMed] [Google Scholar]
- 25.Alegre E., Zubiri L., Perez-Gracia J.L., González-Cao M., Soria L., Martín-Algarra S., González A. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin. Chim. Acta. 2016;454:28–32. doi: 10.1016/j.cca.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 26.Suchorska W.M., Lach M.S. The role of exosomes in tumor progression and metastasis. Oncol. Rep. 2016;35(3):1237–1244. doi: 10.3892/or.2015.4507. [Review]. [DOI] [PubMed] [Google Scholar]
- 27.Fornari F., Ferracin M., Trerè D., Milazzo M., Marinelli S., Galassi M., Venerandi L., Pollutri D., Patrizi C., Borghi A., Foschi F.G., Stefanini G.F., Negrini M., Bolondi L., Gramantieri L. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One. 2015;10(10):e0141448. doi: 10.1371/journal.pone.0141448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugimachi K., Matsumura T., Hirata H., Uchi R., Ueda M., Ueo H., Shinden Y., Iguchi T., Eguchi H., Shirabe K., Ochiya T., Maehara Y., Mimori K. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer. 2015;112(3):532–538. doi: 10.1038/bjc.2014.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kogure T., Yan I.K., Lin W-L., Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer. 2013;4(7-8):261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W-H., Ren L-N., Wang X., Wang T., Zhang N., Gao Y., Luo H., Navarro-Alvarez N., Tang L.J. Combination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats. J. Cancer Res. Clin. Oncol. 2015;141(10):1767–1778. doi: 10.1007/s00432-015-1943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei J-X., Lv L-H., Wan Y.L., Cao Y., Li G.L., Lin H-M., Zhou R., Shang C-Z., Cao J., He H., Han Q-F., Liu P-Q., Zhou G., Min J. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61(4):1284–1294. doi: 10.1002/hep.27660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y., Dang J., Chang K.Y., Yau E., Aza-Blanc P., Moscat J., Rana T.M. miR-1298 inhibits mutant KRAS-driven tumor growth by repressing FAK and LAMB3. Cancer Res. 2016;76(19):5777–5787. doi: 10.1158/0008-5472.CAN-15-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fornari F., Pollutri D., Patrizi C., La Bella T., Marinelli S., Casadei Gardini A., Marisi G., Baron Toaldo M., Baglioni M., Salvatore V., Callegari E., Baldassarre M., Galassi M., Giovannini C., Cescon M., Ravaioli M., Negrini M., Bolondi L., Gramantieri L. In hepatocellular carcinoma miR-221 modulates sorafenib resistance through inhibition of caspase-3-mediated apoptosis. Clin. Cancer Res. 2017;23(14):3953–3965. doi: 10.1158/1078-0432.CCR-16-1464. [DOI] [PubMed] [Google Scholar]
- 34.Liu C., Cao B., Liu N., Zhou Z., Yang G., Zhou P. Increased expression of long noncoding RNA RP11-62F24.2 in gastric cancer and its clinical significance. Clin. Lab. 2017;63(9):1475–1479. doi: 10.7754/Clin.Lab.2017.170334. [DOI] [PubMed] [Google Scholar]
- 35.Sun J., Song Y., Chen X., Zhao J., Gao P., Huang X., Xu H., Wang Z. Novel long non-coding RNA RP11-119F7.4 as a potential biomarker for the development and progression of gastric cancer. Oncol. Lett. 2015;10(1):115–120. doi: 10.3892/ol.2015.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abd E.L., Gwad A., Matboli M., El-Tawdi A., Habib E.K., Shehata H., Ibrahim D., Tash F. Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J. Cell. Biochem. 2018;119(10):8600–8610. doi: 10.1002/jcb.27109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
Not applicable.


