Abstract
Introduction
N6-methyladenosine (m6A) is one of the most widely studied epigenetic modifications. It plays important roles in various biological processes, such as splicing, RNA localization and degradation, many of which are related to the functions of introns. Although a number of computational approaches have been proposed to predict the m6A sites in different species, none of them were optimized for intronic m6A sites. As existing experimental data overwhelmingly relied on polyA selection in sample preparation and the intronic RNAs are usually underrepresented in the captured RNA library, the accuracy of general m6A sites prediction approaches is limited for intronic m6A sites prediction task.
Methodology
A computational framework, WITMSG, dedicated to the large-scale prediction of intronic m6A RNA methylation sites in humans has been proposed here for the first time. Based on the random forest algorithm and using only known intronic m6A sites as the training data, WITMSG takes advantage of both conventional sequence features and a variety of genomic characteristics for improved prediction performance of intron-specific m6A sites.
Results and Conclusion
It has been observed that WITMSG outperformed competing approaches (trained with all the m6A sites or intronic m6A sites only) in 10-fold cross-validation (AUC: 0.940) and when tested on independent datasets (AUC: 0.946). WITMSG was also applied intronome-wide in humans to predict all possible intronic m6A sites, and the prediction results are freely accessible at http://rnamd.com/intron/.
Keywords: m6A, intron, site prediction, sequence features, genomic features, RNA methylation
1. INTRODUCTION
Recent advances have shown that, among 150 known RNA modifications, N6-methyladenosine (m6A) has attracted the most extensive attention due to its prevalence and various biological functions [1-4]. The m6A RNA methylation usually occurs in the conserved sequence DRACH (D = G, A; H = A, C or U) or GGAC [5]. Studies showed that m6A appears in almost all the RNA transcripts, including coding and non-coding transcripts [6, 7], and is enriched near the stop codon, 3’ untranslated regions and the last exon region of mRNA [8, 9]. Moreover, increasing evidences suggest that pre-mRNA contains a large number of m6A modification sites, and more than 2,000 m6A sites were detected in introns, which may have important functions [10]. Recent studies have found that [11], as a common molecular tag, m6A modification involves in many important biological processes, including RNA localization and degradation [12, 13], RNA structural dynamics [11], variable splicing [12], primary microRNA process [14, 15], cell differentiation and adaptation, and clock regulation [16]. It is also associated with protein translation, obesity, abnormal brain development and other diseases [17]. Therefore, accurate localization of m6A is particularly important for understanding the function of RNA methylation in biology. In addition, there is evidence that methylation modification in introns can affect alternative splicing in three ways. First, RNA modification in introns can affect the interaction between snRNA and pre-mRNA. Secondly, the modification sites in introns can directly regulate the binding of RNA-binding proteins by strengthening the relationship between binding factors and their binding proteins, thus affecting variable splicing. Thirdly, RNA modification indirectly affects splicing sites by altering the secondary structure of RNA [18].
With the rapid development of high-throughput sequencing technology, the appearance of MeRIP-Seq opened the prelude to the global and unbiased analysis of RNA methylation in 2012 [5]. MeRIP-Seq high-throughput sequencing is the first technique to detect the m6A spectrum in the whole transcriptome, in which, the RNA fragments containing m6A are precipitated, purified, sequenced and then further analyzed. It is expected that there are more m6A-containing RNA fragments enriched near true m6A sites in immunoprecipitation samples (IP samples) compared with the input control samples (control samples), and people can use exomePeak [19] or other detection methods to detect the m6A peak (site) with a resolution of around 100nt. As the MeRIP-Seq data depends on both IP and input control samples simultaneously, the process is similar to the peak calling procedure widely used in ChIP-Seq [20] to predict histone modification or transcription factor binding sites. It is possible to further determine the exact location of m6A sites by searching for the m6A conforming to DRACH motif in the peak detected by exomePeak and other methods. However, the main disadvantage of this method is that it is often difficult to distinguish the random DRACH motifs from the real m6A-containing motifs nearby. If all the DRACH motifs (including random ones) located at the m6A peak are reported as the m6A sites, positive predictions will be made. Currently, both MeTDB [21] and RMBase [22] databases report a large number (more than 300,000) of m6A sites in the transcriptome, many of which should be false-positive sites due to the randomly DRACH motifs located within m6A peaks.
Besides MeRIP-seq, a base-resolution technique such as miCLIP-Seq [23] has been proposed for the identification of precise m6A sites at base-resolution. However, due to the technical difficulty and the cost of the experiments, it has not been widely used to study the m6A epitranscriptome under different biological contexts, instead it provides necessary information for computational prediction of methylation sites. A number of computational methods have been developed so far for m6A sites prediction. iRNA-Methyl [24] combined the dinucleotide components with the enthalpy, entropy and free energy to form a pseudo dinucleotide composition (PseDNC) that represents the RNA sequence, then used the SVM classifier to predict the m6A methylation sites of Saccharomyces cerevisiae. Zhou et al. [25] proposed an m6A predictor called SRAMP, which takes advantage of sequence coding feature, K-nearest base-pair similarity feature and base-pair frequency feature and the random forest (RF) classifier respectively, and then integrated classification results by weighted sum method for mammalian m6A sites prediction. MethyRNA [26] encoded RNA sequences using the chemical characteristics of nucleotides and accumulated frequency information of nucleotides and predicted the m6A modification sites in Saccharomyces cerevisiae based on SVM classifier. PRNAm-PC [27] extracted 10 physicochemical characteristics of dinucleotides and combined them with their autocovariance and cross-covariance transformation features to form the PseDNC feature representing RNA sequence, and input into the SVM classifier to predict the m6A methylation sites of Saccharomyces cerevisiae. RAM-ESVM [28] uses PseDNC, Transcription Starting Sites (TSS) and Transcription Factor Binding Sites (TFBS) and their k-mer features to build three SVM classifiers, respectively. Then the classification results were integrated with the voting rules to detect the m6A methylation sites of Saccharomyces cerevisiae. BERMP [29] method used two classifiers to predict m6A methylation modification sites. Firstly, the base coding and the frequency of each base in a sliding window of a certain length were input into the Gated Recurrent Unit (GRU) classifier and the random forest classifier, respectively, and the final prediction results were obtained by the logistic regression model based on the results of the two classifiers. Gene2vec [30] employed the Convolutional Neural Network (CNN) for m6A prediction, which represented mRNA sequences with word embedding. Deep-m6A [31] took the product of one-hot coding of sequence characteristics and the reads count of sites in the IP samples as input to predict m6A sites by CNN. In addition, AthMethPre [32] and other methods [33-39] also extract features based on sequence information for the prediction of RNA methylation sites. WHISTLE [40] combined sequence features and 35 genomic features to predict m6A sites, and drafted the entire predicted m6A epitranscriptome. Although there are already many methods proposed for predicting RNA methylation sites, to our best knowledge, all of them focus on the prediction of methylation sites in full transcripts (including both exons and introns) or mature mRNA (including only exons), none is dedicated to predict methylation sites in introns. None of them considered the impact of polyA selection in RNA library preparation and the under-representation of intronic RNAs in the data and the detected results, which should induce strong bias when these approaches were used to apply for m6A sites located in the introns.
In this paper, a framework which is based on the whole-intronome m6A methylation sites prediction by combining sequence features with genomic features (WITMSG) dedicated to the prediction of m6A sites in the introns. WITSMG extracted physicochemical characteristics and frequency accumulation characteristics of bases based on the sequence information and multiple genomic features and predicted whole-intronome m6A methylation sites with the random forest classifier.
2. MATERIALS and METHODS
2.1. Datasets Construction
The single based m6A sites used are the same as the raw data in WHISTLE project [40], including six single-base resolution m6A experiments from six datasets of five cell types (Table 1), including HEK293T, MOLM13, A549, CD8T and HeLa, where HEK293T has two samples. The positive m6A sites are defined as m6A sites conforming to the DRACH consensus motifs and supported on at least 2 of the 6 datasets. The negative m6A sites were randomly selected on the same transcripts containing the positive sites. There are an equal number of negative and positive sites for each of the training data, and the underlying motifs are restricted on DRACH. The exons and introns are defined by the primary transcript (longest transcript) of each gene. The regions that can be mapped to multiple genes are masked, and no sites are reported from those regions.
Table 1. Single-base resolution datasets in intronic m6A prediction.
In the end, 5258 intronic m6A sites were collected, including 2629 positive sites and 2629 negative m6A sites. For testing purposes, a total of 108952 sites in exons were also collected, with 54476 positive m6A sites and 54476 negative m6A sites. Among the total 57105 m6A sites, 95.4% (54476) were from exons, which reflected the bias induced by the RNA library protocol. Four-fifths of the sites were retained for training and the other one-fifth of the sites were retained for testing purposes. We also combine the intronic and exonic sites to mimic the real scenario, in which both the exonic and intronic m6A sites were simultaneously considered for both training and testing (Fig. 1).
Fig. (1).
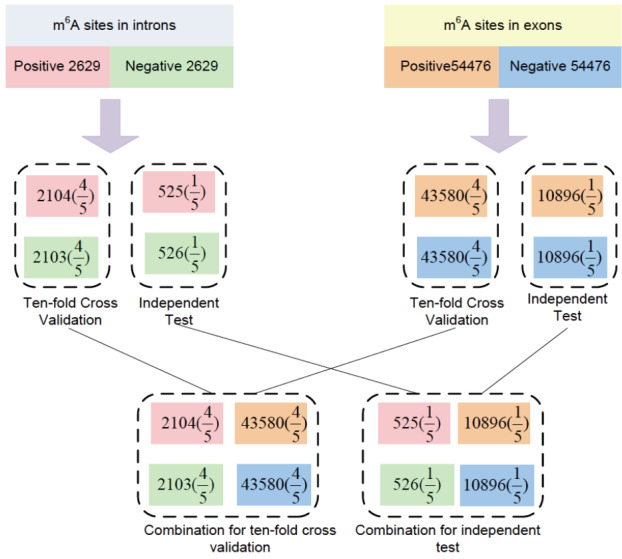
The training and testing data. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2.2. Feature Representation
In this work, an m6A site is represented by two groups of features, i.e., the sequence features and the genomic features.
2.3. Sequence Features
A 21nt sequence around the DRACH motif was described using the same method of MethyRNA. There are four kinds of nucleotides in RNA, including adenine (A), guanine (G), cytosine (C) and uracil (U). According to the different structural properties, a nucleotide was depicted by three features: ring number, chemical functions and hydrogen bonds. For example, cytosine and uracil have only one ring structure, while adenine and guanine have two rings; adenine and cytosine both contain amino, divided into the amino group, while guanine and uracil both contain keto, divided into keto group. In addition, guanine and cytosine have strong hydrogen bonds when forming secondary structure, while adenine and uracil have weak hydrogen bonds. Therefore, following the above features, each nucleotide could be encoded by a three- dimensional vector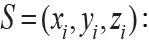 :
:
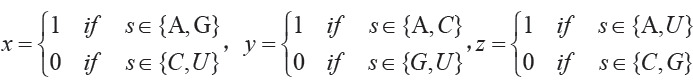
Thus, based on chemical properties defined, A can be encoded by a vector (1,1,1), C can be encoded by a vector (0,1,0), G can be encoded by a vector (1,0,0), and U can be encoded by a vector (0,0,1).
In addition, base frequency information and the distribution of each base in the sequence were also considered, i.e., the base accumulation frequency feature is the frequency of the i -th base in the previous bases. The density  of the i -th base is defined as the frequency of the occurrence of the i -th base before position, that is,, where is defined as the sum number of occurrences of the i -th base in the previous i bases. For example, for the sequence “CUGGAUCGUU”, cytosine appears at the first and seventh positions with cumulative frequencies of 1.00(1/1) and 0.29(2/7), respectively, while uracil frequencies are 0.5(1/2), 0.33(2/6), 0.33(3/9) and 0.4(4/10), respectively.
of the i -th base is defined as the frequency of the occurrence of the i -th base before position, that is,, where is defined as the sum number of occurrences of the i -th base in the previous i bases. For example, for the sequence “CUGGAUCGUU”, cytosine appears at the first and seventh positions with cumulative frequencies of 1.00(1/1) and 0.29(2/7), respectively, while uracil frequencies are 0.5(1/2), 0.33(2/6), 0.33(3/9) and 0.4(4/10), respectively.
2.4. Genomic Features
Sequence features are often used alone in current RNA methylation sites prediction methods, but the sequence features cannot represent the topological information of RNA methylation sites; therefore 57 additional genomic features were generated that may contribute to the RNA methylation sites prediction. Specifically, genomic features 1-4 stand for the dummy variable features, which represent whether the site is overlapped to the topological region on the major RNA transcript. In order to prevent the influence of transcript isoforms, the primary transcripts (longest transcripts) were selected to extract genomic features. All features were extracted by using the transcriptional annotations of the hg19 TxDb package [22]. Genomic Features 5-6 represent the length of the transcript region containing the methylation site. If the region did not contain the methylation site, the value is set to 0. Feature 7-24 indicate that the adenosine site belongs to which consensus motif it is. Feature 25-28 capture the distances toward the splicing junctions or the nearest neighboring sites. Feature 29-34 represent clustering indicators or motif clustering. They are used to measure the clustering effect of the RNA methylation sites. Feature 35-38 are the scores related to evolutionary conservation, including two Phast-Cons scores and two fitness consequences score, to measure the conservation level. Feature 39 and feature 40 indicate the RNA secondary structures predicted by RNAfold [43]. Feature 41-52 are the RNA annotation related to m6A biology. Supplementary Table (S1 (484.7KB, pdf) ) contains the detailed information of the genomic features we considered in the prediction.
3. RESULTS and DISCUSSION
3.1. Evaluation Metrics
In order to measure the performance of the model, we used four kinds of performance metrics, including Sn (sensitivity), Sp (specificity), ACC (accuracy) and MCC (Matthews's correlation coefficient). The sensitivity reflects the success rate of positive sample prediction. The specificity reflects the success rate of negative sample prediction. MCC is a comprehensive performance evaluation index considering unbalanced data sets. The four indicators are defined as follows:
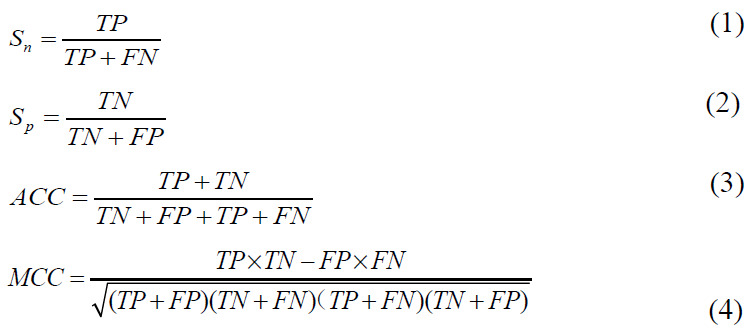
where TP is true positive, TN is true negative, FP is false positive, and FN is false negative. In addition, Receiver Operating Characteristic (ROC) curves were plotted and the areas (as called “AUC”) under the curves were calculated and used as the primary evaluation metrics.
3.2. Comparison of RF and Other Classifiers in Cross-validation
Four classifiers were used for m6A sites prediction, including random forest (RF) [44, 45], support vector machine (SVM) [46], K-nearest neighbor (KNN) [47] and logistic regression (LR) [48], respectively. RF is one of the most widely used machine learning algorithms for biological data, based on which, SRAMP was developed for predicting the mammalian m6A sites. SVM is also one of the most widely used machine learning algorithms in computational biology. iRNA-Methyl and RAM-ESVM predicted m6A sites using SVM. KNN is one of the simplest methods in data mining classification technology, and LR is a classification model in machine learning, which has the characteristics of simple algorithms and high efficiency. For comparing the four classifiers, 10-fold cross-validation was employed on the training datasets, and the classifier with the best results was used in independent test data. Besides, the data of the introns, exons, and introns merged with exons (as called “combines”) were also tested, respectively. The performance of different classifiers were summarized. As shown in Table 2, RF, SVM and LR achieved very similar performance under all the 3 modes tested. Notably, the performance achieved on exon (AUC = 0.9133) or intron (AUC = 0.9403) is better than that on combined data (AUC = 0.9095). Although more training sites were available under the combined mode, mixing intronic and exonic m6A sites actually negatively affect the prediction performance, suggesting that the exonic and intronic m6A sites exhibited different characteristics in our data.
Table 2. Performance under cross-validation.
| Data | Method | Evaluation Metrics | ||||
|---|---|---|---|---|---|---|
| Sn | Sp | ACC | MCC | AUC | ||
| Introns | RF | 0.8184 | 0.9334 | 0.8759 | 0.7573 | 0.9403 |
| SVM | 0.8242 | 0.8949 | 0.8595 | 0.7217 | 0.9292 | |
| KNN | 0.4988 | 0.5021 | 0.5005 | 0.0010 | 0.8142 | |
| LR | 0.7809 | 0.9496 | 0.8652 | 0.7413 | 0.9352 | |
| Exons | RF | 0.8600 | 0.813 | 0.8396 | 0.6798 | 0.9133 |
| SVM | 0.8383 | 0.8385 | 0.8384 | 0.6769 | 0.9131 | |
| KNN | 0.4993 | 0.5011 | 0.5002 | 0.0004 | 0.7984 | |
| LR | 0.7486 | 0.8922 | 0.8204 | 0.6476 | 0.9073 | |
| Combined | RF | 0.8462 | 0.8253 | 0.8357 | 0.6716 | 0.9095 |
| SVM | 0.8291 | 0.8341 | 0.8316 | 0.6632 | 0.9065 | |
| KNN | 0.4995 | 0.5010 | 0.5003 | 0.0005 | 0.7954 | |
| LR | 0.7250 | 0.8938 | 0.8094 | 0.6279 | 0.8977 | |
3.3. Independent Tests
We considered using sites from different regions for training and testing. Specifically, we used m6A sites from intron, exon and combined regions as training and then testing on sites from the 3 categories as well (Fig. 2), and the results were summarized in Table 3, where the red font indicates the maximum value of AUC in the current category.
Fig. (2).
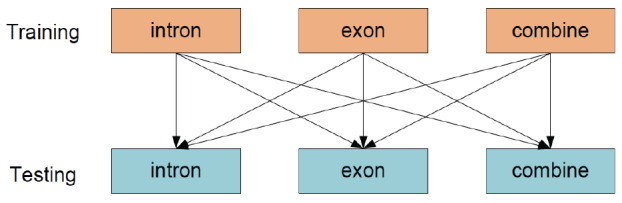
Independent tests. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Table 3. Results on independent tests.
| Testing | Training | Method | Evaluation Metrics | ||||
|---|---|---|---|---|---|---|---|
| Sn | Sp | ACC | MCC | AUC | |||
| Intron | intron | RF | 0.8229 | 0.9544 | 0.8887 | 0.7841 | 0.9458 |
| SVM | 0.8362 | 0.9297 | 0.8830 | 0.7693 | 0.9333 | ||
| KNN | 0.4981 | 0.5067 | 0.5024 | 0.0047 | 0.8268 | ||
| LR | 0.7752 | 0.9562 | 0.8658 | 0.7439 | 0.9366 | ||
| exon | RF | 0.4667 | 0.9924 | 0.7298 | 0.5398 | 0.8794 | |
| SVM | 0.4133 | 0.9962 | 0.7050 | 0.5042 | 0.9021 | ||
| KNN | 0.4971 | 0.5010 | 0.4990 | -0.0019 | 0.6256 | ||
| LR | 0.2514 | 1 | 0.6261 | 0.3794 | 0.8934 | ||
| combine | RF | 0.6019 | 0.9848 | 0.7935 | 0.6353 | 0.9253 | |
| SVM | 0.8398 | 0.8328 | 0.8363 | 0.6726 | 0.9096 | ||
| KNN | 0.4981 | 0.5067 | 0.5024 | 0.0047 | 0.7977 | ||
| LR | 0.3505 | 1 | 0.6755 | 0.4611 | 0.8886 | ||
| Exon | intron | RF | 1 | 0.0012 | 0.5006 | 0.0244 | 0.8412 |
| SVM | 0.9990 | 0.0310 | 0.5150 | 0.1195 | 0.6938 | ||
| KNN | 0.4983 | 0.5027 | 0.5005 | 0.0010 | 0.5459 | ||
| LR | 0.9989 | 0.0258 | 0.5123 | 0.1072 | 0.8309 | ||
| exon | RF | 0.8584 | 0.8245 | 0.8414 | 0.6833 | 0.9165 | |
| SVM | 0.8401 | 0.8419 | 0.8410 | 0.6820 | 0.9151 | ||
| KNN | 0.4992 | 0.5014 | 0.5003 | 0.0006 | 0.8001 | ||
| LR | 0.7421 | 0.8951 | 0.8186 | 0.6448 | 0.9081 | ||
| combine | RF | 0.8568 | 0.8247 | 0.8407 | 0.6819 | 0.9141 | |
| SVM | 0.8398 | 0.8328 | 0.8363 | 0.6726 | 0.9096 | ||
| KNN | 0.4994 | 0.5009 | 0.5001 | 0.0003 | 0.7980 | ||
| LR | 0.7349 | 0.8906 | 0.8128 | 0.6333 | 0.9015 | ||
| Combine | intron | RF | 0.9921 | 0.0444 | 0.5182 | 0.1144 | 0.8270 |
| SVM | 0.4983 | 0.5028 | 0.5006 | 0.0012 | 0.5555 | ||
| KNN | 0.9884 | 0.0686 | 0.5285 | 0.1455 | 0.8165 | ||
| LR | 0.9914 | 0.0723 | 0.5318 | 0.1618 | 0.6955 | ||
| exon | RF | 0.8320 | 0.8390 | 0.8355 | 0.6710 | 0.9110 | |
| SVM | 0.8142 | 0.8478 | 0.8310 | 0.6624 | 0.9055 | ||
| KNN | 0.4993 | 0.5009 | 0.5009 | 0.0002 | 0.7908 | ||
| LR | 0.7045 | 0.9024 | 0.8034 | 0.6191 | 0.8968 | ||
| combine | RF | 0.8463 | 0.8326 | 0.8395 | 0.6790 | 0.9126 | |
| SVM | 0.8294 | 0.8394 | 0.8344 | 0.6689 | 0.9077 | ||
| KNN | 0.4994 | 0.5011 | 0.5004 | 0.0005 | 0.7979 | ||
| LR | 0.7173 | 0.8956 | 0.8065 | 0.6229 | 0.8979 | ||
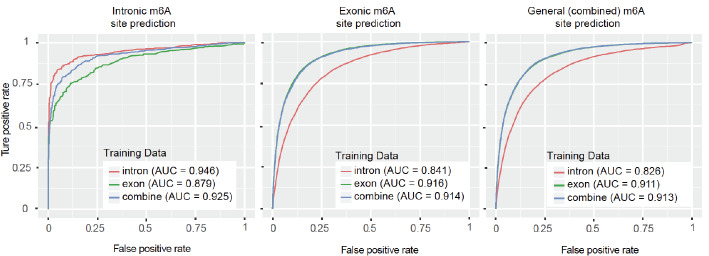
Interestingly, the best prediction performance was achieved when the testing data and training data were from the same category. For intronic m6A sites prediction, substantially better performance was achieved when intronic sites were used (AUC = 0.9458) compared with when exonic sites (AUC = 0.9021) or combined sites were used (AUC = 0.9253). A similar trend is also observed for exonic or general (or combined) m6A sites prediction. In addition, it can be seen that RF gets the best performance among the four methods tested in intronic sites prediction in both cross-validation and independent test. Therefore, RF is chosen as the classifier for predicting whole intronome RNA methylation sites later.
Additionally, the ROC curves of these 9 tests were shown in Fig. 3. We can see that the highest AUC for intronic m6A sites prediction was achieved when intronic sites were used as training. There is little difference in the performance of exonic or general m6A sites prediction between using exonic or general sites as the training data. This is because of the over-representation of exonic sites (95.4%) in the gold standard data from the WHISTLE project, which is likely due to the widely adopted polyA selection RNA library preparation protocol.
Fig. (3).
The ROC curve of independent tests. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.4. Feature Selection for Intronic and Exonic m6A Sites Prediction
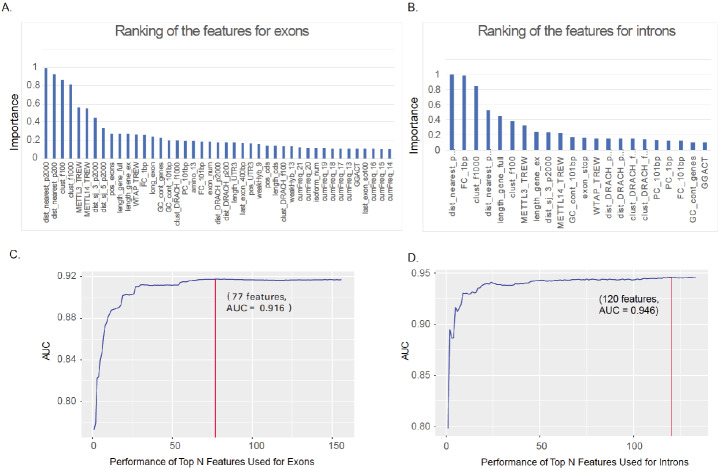
To further optimize the prediction performance and differentiate the characteristics of intronic and exonic m6A sites, we used feature selection to identify the most effective features for m6A sites prediction in introns and exons, respectively. Firstly, the importance of features which are calculated by the random-forest R package is ranked as their respective AUCs in ten-fold cross-validation. Then, one feature is added into the training set at each time from the ordered feature set, and the prediction performance was evaluated using ten-fold cross-validation. The feature set returned highest AUC was selected as the optimal feature subset. As shown in Fig. (4A and 4B), the top 5 most important predictive features for exons are the distance to the nearest neighbors (peaked at 2000bp), the distance to the nearest neighbors (peaked at 200bp), the number of neighbors within 100bp flanking regions, the number of neighbors within 1000bp flanking regions and the TREW data of METTL3 RNA binding sites, while the top 5 features for introns are the distance to the nearest neighbors (peaked at 2000bp), the fitness consequences scores 1bp z score, the number of neighbors within 1000bp flanking regions, the distance to the nearest neighbors (peaked at 200bp) and the full transcript length. While clearly indicates that RNA methylation sites exhibit certain clustering characteristics, the difference in top features also suggests the intrinsic difference in m6A sites located in exons and introns. Fig. (4C and 4D) shows the results of the feature selection. The feature set with the highest AUC was selected. In the prediction of methylation sites in exons and introns, the highest AUC can be obtained from the top 77 and 120 features, respectively. Therefore, the top 77 features make up the optimal subset when predicting m6A methylation sites in exons, while the top 120 features for intronic sites prediction.
Fig. (4).
Feature selection results. A. The ranking of the features for exonic m6A sites prediction. B. The ranking of the features for intronic m6A sites prediction. C. Top 77 features were selected for exonic m6A sites prediction, and achieved AUC of 0.916. D. Top 120 features were selected for intronic m6A sites prediction, and achieved AUC of 0.946. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.5. Comparison with Existing Methods
To further verify the effectiveness of the proposed algorithm in predicting m6A RNA methylation sites in introns, we compared WITMSG with SRAMP, MethyRNA and M6AMRFS. The results were summarized in Table 4 and the ROC curves were shown in Fig. (5). It can be seen that the proposed approach substantially outperformed competing approaches in intron-specific m6A sites prediction.
Table 4. Performance comparison for intronic m6A sites prediction.
| Method | Sn | Sp | ACC | MCC | AUC |
|---|---|---|---|---|---|
| SRAMP | 0.7333 | 0.8213 | 0.7774 | 0.5568 | 0.8425 |
| MethyRNA | 0.6419 | 0.6996 | 0.6708 | 0.3421 | 0.7249 |
| M6AMRFS | 0.5352 | 0.2281 | 0.3815 | -0.2487 | 0.6171 |
| WITMSG | 0.8152 | 0.9506 | 0.8830 | 0.7730 | 0.9458 |
Fig. (5).
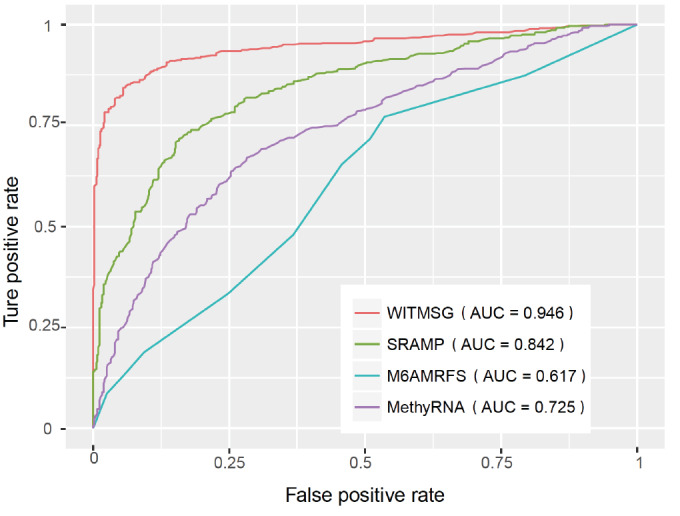
ROC for intronic m6A sites prediction. The proposed approach substantially outperformed competing approaches. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.6. Intronome-wide m6A Sites Prediction
To generate a complete map of all the intronic m6A sites in humans, we searched the entire intronome (collection of all the introns) for the m6A DRACH motifs as the candidate
m6A sites and then used the proposed approach to evaluate the probability of m6A methylation at these locations. In the end, 1,841,962 out of the total 20,156,510 intronic DRACH motifs were predicted to contain m6A RNA methylation sites, and the complete prediction results are freely accessible at http://rnamd.com/intron/.
CONCLUSION
With the rapid development of high throughput sequencing technology and bioinformatics efforts, people can now predict transcriptome m6A RNA modification sites with reasonable accuracy. However, till this day, efforts have not been made to address the bias induced in the RNA library preparation, which led to limited accuracy in intron-specific m6A sites prediction. We showed, for the first time, the different characteristics exhibited in intronic and exonic m6A sites, and then presented here WITMSG, a method to predict m6A epitranscriptome in introns. Unlike most of the other methods that relied on sequence information only, WITMSG extracted the physicochemical, frequency accumulation characteristics of bases, and 57 additional genomic characteristics to predict the m6A methylation modification sites in introns based on random forest. To the best of our knowledge, WITMSG is the first m6A sites predictor optimized for introns. By using only intronic m6A sites as the training data and integrating multiple genomic features besides conventional sequence features, WITMSG substantially outperformed competing approaches (SRAMP, M6AMRFS and MethyRNA) in intronic m6A sites prediction. In the end, we scanned the entire intronome for possible intronic m6A sites and made results of 1,841,962. The predicted intronic m6A sites publically accessible to share with researchers of the field, especially those who are interested in the function of m6A related to pre-mRNA. Notably, the proposed WITMSG framework can be easily extended to study the intronic RNA modification sites of other RNA modification types such as PSI, m1A and m5C as well as in other species such as mouse and yeast.
ACKNOWLEDGEMENTS
Declared none.
AUTHORS’ CONTRIBUTIONS
ZW and LL initialized the project; LL, XL, ZW and JM designed the research plan; ZW constructed the genomic features considered in site prediction; LL performed the site prediction; LL drafted the manuscript. All authors read, critically revised and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
FUNDING
This work has been supported by the National Natural Science Foundation of China [61902230, 61972451, 31671373]; China Postdoctoral Science Foundation [2018 M640949]; Fundamental Research Funds for the Central Universities [GK201903083, GK201901010]; XJTLU Key Program Special Fund [KSF-T-01].
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
REFERENCES
- 1.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 2.Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15(5):313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J., Jia G. Methylation modifications in eukaryotic messenger RNA. J. Genet. Genomics. 2014;41(1):21–33. doi: 10.1016/j.jgg.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Liu L. LITHOPHONE: improving lncRNA methylation site prediction using an ensemble predictor. Front. Genet. 2020 doi: 10.3389/fgene.2020.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 7.Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F.N. 6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T.N. (6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., Dai Q., Chen W., He C.A. METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke S., Pandya-Jones A., Saito Y., Fak J.J., Vågbø C.B., Geula S., Hanna J.H., Black D.L., Darnell J.E., Jr, Darnell R.B. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31(10):990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roost C., Lynch S.R., Batista P.J., Qu K., Chang H.Y., Kool E.T. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 2015;137(5):2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., Ren B., Pan T., He C.N. 6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue H. Prediction of RNA methylation status from gene expression data using classification and regression methods. Evol. Bioinform. Online. 2020 doi: 10.1177/1176934320915707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T., Hao Y.J., Zhang Y., Li M.M., Wang M., Han W., Wu Y., Lv Y., Hao J., Wang L., Li A., Yang Y., Jin K.X., Zhao X., Li Y., Ping X.L., Lai W.Y., Wu L.G., Jiang G., Wang H.L., Sang L., Wang X.J., Yang Y.G., Zhou Q. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16(3):289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S., Ben-Haim M.S., Eyal E., Yunger S., Pinto Y., Jaitin D.A., Viukov S., Rais Y., Krupalnik V., Chomsky E., Zerbib M., Maza I., Rechavi Y., Massarwa R., Hanna S., Amit I., Levanon E.Y., Amariglio N., Stern-Ginossar N., Novershtern N., Rechavi G., Hanna J.H. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 16.Fustin J.M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka M.S., Kakeya H., Manabe I., Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 17.Peng L., Yuan X., Jiang B., Tang Z., Li G.C. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 2016;37(3):2779–2788. doi: 10.1007/s13277-015-4663-9. [DOI] [PubMed] [Google Scholar]
- 18.Martinez N.M., Gilbert W.V. Pre-mRNA modifications and their role in nuclear processing. Quant. Biol. 2018;6(3):210–227. doi: 10.1007/s40484-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng J., Cui X., Rao M.K., Chen Y., Huang Y. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics. 2013;29(12):1565–1567. doi: 10.1093/bioinformatics/btt171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valouev A., Johnson D.S., Sundquist A., Medina C., Anton E., Batzoglou S., Myers R.M., Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5(9):829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H., Wang H., Wei Z., Zhang S., Hua G., Zhang S.W., Zhang L., Gao S.J., Meng J., Chen X., Huang Y. MeT-DB V2.0: elucidating context-specific functions of N6-methyl-adenosine methyltranscriptome. Nucleic Acids Res. 2018;46(D1):D281–D287. doi: 10.1093/nar/gkx1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xuan J.J., Sun W.J., Lin P.H., Zhou K.R., Liu S., Zheng L.L., Qu L.H., Yang J.H. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46(D1):D327–D334. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12(8):767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W., Feng P., Ding H., Lin H., Chou K.C. iRNA-Methyl: Identifying N(6)-methyladenosine sites using pseudo nucleotide composition. Anal. Biochem. 2015;490:26–33. doi: 10.1016/j.ab.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Y., Zeng P., Li Y.H., Zhang Z., Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44(10):e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W., Tang H., Lin H. MethyRNA: a web server for identification of N6-methyladenosine sites. J. Biomol. Struct. Dyn. 2017;35(3):683–687. doi: 10.1080/07391102.2016.1157761. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z., Xiao X., Yu D.J., Jia J., Qiu W.R., Chou K.C. pRNAm-PC: Predicting N(6)-methyladenosine sites in RNA sequences via physical-chemical properties. Anal. Biochem. 2016;497:60–67. doi: 10.1016/j.ab.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Chen W., Xing P., Zou Q. Detecting N6-methyladenosine sites from RNA transcriptomes using ensemble Support Vector Machines. Sci. Rep. 2017;7:40242. doi: 10.1038/srep40242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y., He N., Chen Y., Chen Z., Li L. BERMP: a cross-species classifier for predicting m6A sites by integrating a deep learning algorithm and a random forest approach. Int. J. Biol. Sci. 2018;14(12):1669–1677. doi: 10.7150/ijbs.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou Q.P.X., Leyi W., Bin L. Gene2vec: gene subsequence embedding for prediction of mammalian N6-methyladenosine sites from mRNA. RNA. 2018;25(2):205–218. doi: 10.1261/rna.069112.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S.Y., Zhang S.W., Fan X.N., Meng J., Chen Y., Gao S.J., Huang Y. Global analysis of N6-methyladenosine functions and its disease association using deep learning and network-based methods. PLOS Comput. Biol. 2019;15(1):e1006663. doi: 10.1371/journal.pcbi.1006663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang S., Yan Z., Liu K., Zhang Y., Sun Z. AthMethPre: a web server for the prediction and query of mRNA m6A sites in Arabidopsis thaliana. Mol. Biosyst. 2016;12(11):3333–3337. doi: 10.1039/C6MB00536E. [DOI] [PubMed] [Google Scholar]
- 33.Li G.Q. Methylation modifications in eukaryotic messenger RNA. J. Genet. Genomics. 2016;41(1):21–33.. doi: 10.1016/j.jgg.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Feng P., Ding H., Chen W., Lin H. Identifying RNA 5-methylcytosine sites via pseudo nucleotide compositions. Mol. Biosyst. 2016;12(11):3307–3311. doi: 10.1039/C6MB00471G. [DOI] [PubMed] [Google Scholar]
- 35.Chen W., Feng P., Tang H., Ding H., Lin H. Identifying 2′-O-methylationation sites by integrating nucleotide chemical properties and nucleotide compositions. Genomics. 2016;107(6):255–258. doi: 10.1016/j.ygeno.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Chen W. Identification and analysis of the N6-methyladenosine in the Saccharomyces cerevisiae transcriptome. Sci. Rep. 2015;13859:5. doi: 10.1038/srep13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Z., Peng H., Lan C., Zheng Y., Fang L., Li J. Imbalance learning for the prediction of N6-Methylation sites in mRNAs. BMC Genomics. 2018;19(1):574. doi: 10.1186/s12864-018-4928-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu W.R., Jiang S.Y., Sun B.Q., Xiao X., Cheng X., Chou K.C. iRNA-2methyl: identify RNA 2′-O-methylation sites by incorporating sequence-coupled effects into general PseKNC and ensemble classifier. Med. Chem. 2017;13(8):734–743. doi: 10.2174/1573406413666170623082245. [DOI] [PubMed] [Google Scholar]
- 39.Song B., Tang Y., Wei Z., Liu G., Su J., Meng J., Chen K. PIANO: a web server for pseudouridine site (Ψ) identification and functional annotation. Front. Genet. 2020;11:88. doi: 10.3389/fgene.2020.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q. Nucleic Acids Res. 2019;47(7):e41. doi: 10.1093/nar/gkz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vu L.P., Pickering B.F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., Chou T., Chow A., Saletore Y., MacKay M., Schulman J., Famulare C., Patel M., Klimek V.M., Garrett-Bakelman F.E., Melnick A., Carroll M., Mason C.E., Jaffrey S.R., Kharas M.G. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017;23(11):1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ke S., Alemu E.A., Mertens C., Gantman E.C., Fak J.J., Mele A., Haripal B., Zucker-Scharff I., Moore M.J., Park C.Y., Vågbø C.B., Kusśnierczyk A., Klungland A., Darnell J.E., Jr, Darnell R.B. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015;29(19):2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruber A.R., Bernhart S.H., Lorenz R. RNA bioinformatics. Springer; 2015. pp. 307–326. [Google Scholar]
- 44.Liu B. BioSeq-Analysis: a platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief. Bioinform. 2017;20(4):1280–1294. doi: 10.1093/bib/bbx165. [DOI] [PubMed] [Google Scholar]
- 45.Wei L., Xing P., Su R., Shi G., Ma Z.S., Zou Q. CPPred-RF: a sequence-based predictor for identifying cell-penetrating peptides and their uptake efficiency. J. Proteome Res. 2017;16(5):2044–2053. doi: 10.1021/acs.jproteome.7b00019. [DOI] [PubMed] [Google Scholar]
- 46.Song J. iProt-Sub: a comprehensive tool for accurately mapping and predicting protease-specific substrates and cleavage sites. Brief. Bioinform. 2019;20(2):638–658. doi: 10.1093/bib/bby028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia C.Z., Zhang J.J., Gu W.Z. RNA-MethylPred: A high-accuracy predictor to identify N6-methyladenosine in RNA. Anal. Biochem. 2016;510:72–75. doi: 10.1016/j.ab.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 48.Cha S., Yu H., Park A.Y., Oh S.A., Kim J.Y. The obesity-risk variant of FTO is inversely related with the So-Eum constitutional type: genome-wide association and replication analyses. BMC Complement. Altern. Med. 2015;15(1):120. doi: 10.1186/s12906-015-0609-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.