Abstract
Long noncoding RNA (lncRNA) plays crucial roles in various biological processes of different cancers, especially acting as a competing endogenous RNA (ceRNA). However, the role of lncRNA-mediated ceRNA in Wilms tumor (WT), which is the most common malignant kidney cancer in children, remains unknown. In present study, RNA sequence profiles and clinical data of 125 patients with WT consisting of 119 tumor and 6 normal tissues from Therapeutically Applicable Research To Generate Effective Treatments database were analyzed. A total of 1833 lncRNAs, 156 microRNAs (miRNAs), and 3443 messenger RNAs (mRNAs) were identified as differentially expressed (DE) using “DESeq2” package. The lncRNA-miRNA-mRNA ceRNA regulatory network involving 748 DElncRNAs, 33 DEmiRNAs, and 189 DEmRNAs was constructed based on miRcode, Targetscan, miRTarBase, and miRDB database. Gene Ontology term and Kyoto Encyclopedia of Genes and Genomes pathway analyses revealed that DEmRNAs were mainly enriched in cell proliferation-related processes and tumor-related pathways, respectively, and 13 hub genes were identified by a protein–protein interaction network. Survival analysis detected 48 lncRNAs, 7 miRNAs, and 16 mRNAs to have significant impact on the overall survival of patients with WT. Additionally, we found that 6 DElncRNAs with potential prognostic value were correlated with tumor stage (DENND5B-AS1) and histologic classification (TMPO-AS1, RP3-523K23.2, RP11-598F7.3, LAMP5-AS1, and AC013275.2) of patients with WT. Our research provides a great insight into understanding the molecular mechanism underlying occurrence and progression of WT, as well as the potential to develop targeted therapies and prognostic biomarkers.
Keywords: Wilms tumor, lncRNA, competing endogenous RNA, survival analysis, therapeutically applicable research to generate effective treatments
Introduction
Wilms tumor (WT) or nephroblastoma is the most common pediatric kidney cancer in children of age below 5 years.1 It accounts for nearly 95% of all pediatric renal malignancies, with an incidence of approximately 7 cases per million children.2 The overall 5-year survival rate for WT in the United States is 92%, whereas the proportion plummets to just 78% in poor parts of the world with fewer medical resources.3 Although a great improvement in survival rate has been achieved over past few decades through combination therapy including surgery, radiotherapy, and chemotherapy,4 the prognoses of patients with WT with higher risks of treatment failure or a more advanced stage are still poor. In addition, the exposure to toxic therapies has potential side effects,5 such as occurrence of renal dysfunction,6 cardiotoxicity,6 musculoskeletal problems,7 and development of secondary malignant neoplasms.8 It is thus urgent to improve outcomes for children with high risks of tumor characteristics and develop tailored therapy to reduce treatment burden for children with low risk of WT. Therefore, investigating the molecular mechanisms involved in WT for developing more effective targeted therapies and identifying biomarkers with highly sensitive and specific values for risk stratification are very essential.
Currently, the available information on the molecular background of WT pathogenesis largely implicates mutations in multiple genes, including WT1,9,10 CTNNB1,11 AMER1 (WTX),12 and TP53,13 amplification of MYCN gene,14 genomic imprinting of the IGF2 and H19,15 aberrations of 11p15 methylation,16 loss of heterozygosity at 1p and 16q,17 and dysregulation several microRNAs (miRNAs) including miR-17,18 miR-185,19 miR-204,20 and miR-483. 21 However, the critical roles of long noncoding RNA (lncRNA), a class of noncoding RNAs > 200 nt in length, in WT are largely unknown. Numerous studies have demonstrated that lncRNA participates in several biological processes, especially as regulatory factors in various types of malignant tumors.22,23 Notably, lncRNA could regulate messenger RNA (mRNA) expression by acting as a competing endogenous RNA (ceRNA) to affect tumorigenesis and progression. The ceRNA hypothesis was firstly proposed by Salmena et al24 in 2011, which establishes a new regulatory mechanism among different RNA transcripts including lncRNAs, pseudogene transcripts, miRNAs, and mRNAs. According to this hypothesis, lncRNA can interact with mRNA by sharing miRNA response elements (MREs). For instance, lncRNA FER1L4 can regulate PTEN expression by acting as a sponge for miR-106a-5p in gastric cancer and thus accelerate gastric cell proliferation.25 H19 can act as a ceRNA to regulate epithelial–mesenchymal transition and mesenchymal-to-epithelial transition in bladder cancer by targeting miR-29b-3p. 26 Recently, lncRNAs-associated ceRNA network have been constructed in colorectal cancer,27 gastric cancer,25 cholangiocarcinoma,28 head and neck squamous cell carcinoma,29 bladder cancer,26 and others. Nevertheless, there is still a lack of comprehensively analysis of an lncRNA-mediated ceRNA network in WT, especially based on whole transcriptome profile and large-scale sample sizes.
Therefore, the aims of this study were to identify key differentially expressed RNAs and construct an lncRNA-related ceRNA network to reveal the underlying mechanisms in carcinogenesis and development of WT, and detect novel biomarkers with prognostic value through a large-scale high-throughput sequence database. The results are expected to help further develop more personalized treatment strategies to minimize toxicity exposure for patients with WT.
Materials and Methods
Data Collection and Processing
All data in this study were available and freely downloaded from Therapeutically Applicable Research To Generate Effective Treatments (TARGET) database (https://ocg.cancer.gov/programs/target) using the TCGA data portal (https://www.cancer.gov/tcga, accessed September 5, 2018). Our research was in accordance with the publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines); therefore, further approval by an ethics committee was not required.
The high-throughput sequencing data (level 3) including miRNA count data for 137 WT samples and expression level of mRNA and lncRNA for 132 individuals, and the related clinical information of 652 patients with WT were downloaded. Data were filtered for samples with recurrent solid tumor and metastatic tumor, as well as samples without 3 different RNAs expression profiles simultaneously and complete clinical data information including tumor stage, survival status, and survival time. A total of 125 filtered samples were divided into 2 cohorts: 119 primary solid tumor and 6 normal tissues. The clinical characteristics of patients with WT are summarized in Table 1. Briefly, mean age of these people at initial pathologic diagnosis was 4.7 years. The number of patients with pathological WT stages I, II, III, and IV were 15 (12.60%), 48 (40.34%), 44 (36.97%), and 12 (10.08%), respectively. The majority of patients were FHWT (favorable histology WT; 67.22%) and the rest were DAWT (diffuse anaplastic WT; 32.77%). At the time of the last follow-up, 70 (58.82%) patients were still alive, while 49 (41.18%) individuals had died.
Table 1.
Corresponding Clinical Features of 119 Patients With Wilms Tumor.
| Items | Patients, N = 119 | |
|---|---|---|
| N | % | |
| Tumor stage | ||
| Stage I | 15 | 12.6 |
| Stage II | 48 | 40.34 |
| Stage III | 44 | 36.97 |
| Stage IV | 12 | 10.08 |
| Histologic classification | ||
| FHWT | 80 | 67.22 |
| DAWT | 39 | 32.77 |
| Gender | ||
| Female | 67 | 56.3 |
| Male | 52 | 43.7 |
| Race | ||
| Black or African American | 18 | 15.13 |
| White | 89 | 74.79 |
| Other | 5 | 4.2 |
| Not reported | 7 | 5.88 |
| Ethnicity | ||
| Hispanic or Latino | 11 | 9.24 |
| Not Hispanic or Latino | 80 | 67.23 |
| Not reported | 28 | 23.53 |
| Age (years) | ||
| <5 | 74 | 62.18 |
| ≥5 | 45 | 37.82 |
| Survival status | ||
| Alive | 70 | 58.82 |
| Dead | 49 | 41.18 |
Abbreviations: DAWT, diffuse anaplastic Wilms tumor; FHWT, favorable histology Wilms tumor.
Identification of Differentially Expressed RNAs
A total of 14 852 lncRNAs, 20 271 mRNAs, and 1881 miRNAs were annotated based on Gencode version 22 (https://www.gencodegenes.org/) and miRBase version 21 (http://www.mirbase.org/). The RNAs with a mean expression less than 1 and a median read counts equal to 0 were removed. The differentially expression analysis then was performed between normal or adjacent tissues and tumor tissues using DESeq2,30 a Bioconductor package in R, to detect potential RNAs involved in development of WT. The false discovery rate (FDR), as proposed by the Benjamini-Hochberg,31 was utilized for each RNA to correct statistical significance of multiple testing. Therefore, lncRNAs, mRNAs, and miRNAs were deemed as differentially expressed (DE), that is, DElncRNAs, DEmiRNAs, and DEmRNAs, when they passed the threshold of |log2 fold-change (FC)| ≥ 2 and FDR < 0.01. Finally, the cluster heat maps and volcano plots of above DERNAs were visualized using pheatmap (version 1.0.10, Raivo Kolde, 2019, pheatmap: Pretty Heatmaps) and ggplot232 packages in R.
Construction of a ceRNA Network
To construct a ceRNA regulation network in terms of ceRNA hypothesis, the DElncRNA–DEmiRNA intersections were firstly predicted according to the miRcode database.33 These paired DEmiRNAs were used to identify targeted mRNAs based on miRTarBase,34 miRDB,35 and TargetScan36 databases. Differentially expressed mRNAs only retrieved in all 3 databases were defined as candidate mRNAs. The co-expression network of DERNAs was built based on DElncRNA–DEmiRNA and DEmiRNA–DEmRNA intersections, which were visualized using Cytoscape version 3.7.0 software.37
Functional Enrichment Analysis and Protein–Protein Interaction Network Analysis of ceRNA
Functional enrichment analyses including Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for targeted DEmRNAs in the ceRNA network were conducted by clusterProfiler R package.38 The cutoff of q value < 0.05 for both GO and KEGG pathways was considered as statistically significant. Moreover, a protein–protein interaction (PPI) network was constructed using STRINGdb39 R package to investigate the molecular and cellular mechanism underlying WT tumor. The protein intersection with over 700 scores was used to detect highly interacted hub proteins by cytoHhbba28 plugin in the Cytoscape 3.7.0 software.
Survival Analysis
To identify potential prognostic signature in patients with WT, all RNAs in the ceRNA network were performed survival analysis using survival package40 in R platform. The WT children were divided into a high expression group and a low expression group based on median expression level of the specific RNA. Log-rank test was used to compare differences of overall survival rates between subgroups, and Kaplan-Meier method was applied to plot survival curves. Generally, P value less than .05 was considered statistically significant.
Correlation Analysis Between Key lncRNAs and Clinical Features
After screening out key RNAs according to the above-described bioinformatics analyses, χ2 test was performed to determine whether RNAs expression was correlated with different clinical features. These features included gender (female vs male), tumor stage (I+II vs III+IV), histologic classification (FHWT vs DAWT), race (black or African American vs white), ethnicity (Hispanic or Latino vs not Hispanic or Latino), and age (≥5 years vs <5 years). Notably, the histological classification followed protocol from the Children’s Oncology Group (COG).41 A P value <.05 was used as a significant cutoff value.
Results
Identification of DEmRNAs, DEmiRNAs, and DElncRNAs in WT
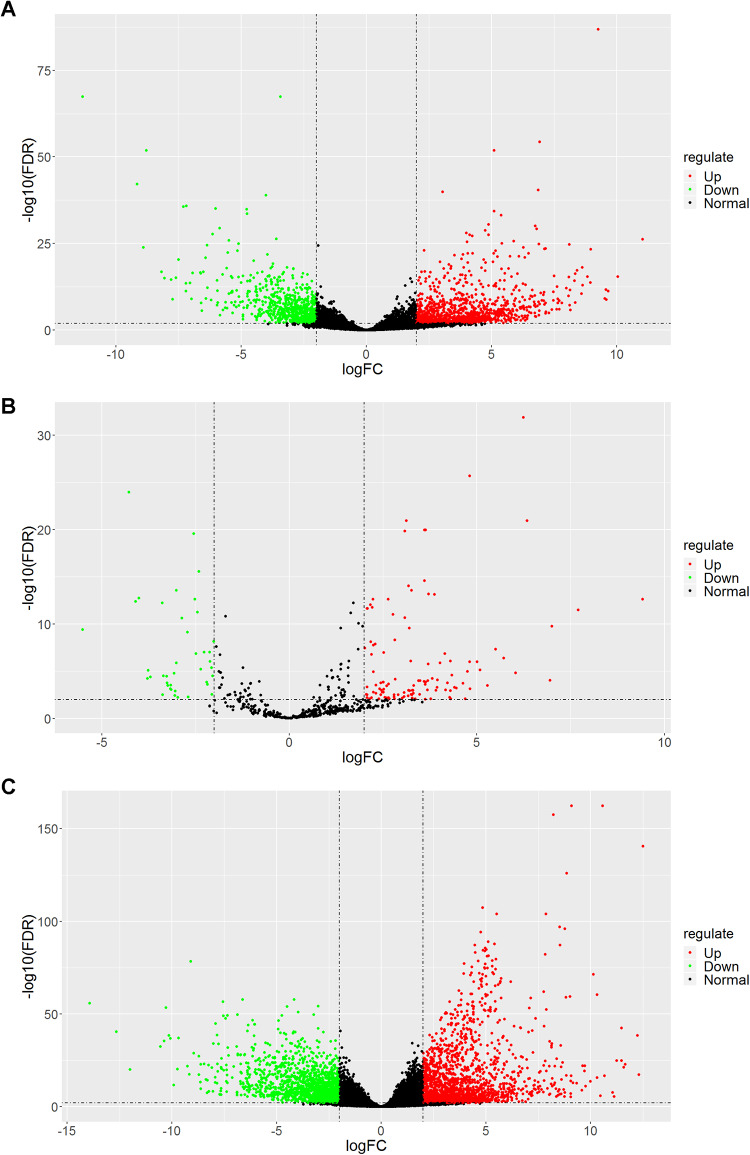
The expression level of RNAs between 119 primary solid tumor and 6 normal samples was compared in this study. A total of 1833 DElncRNAs (1115 upregulated and 718 downregulated), 156 DEmiRNAs (116 upregulated and 40 downregulated), and 3443 DEmRNAs (1777 upregulated and 1666 downregulated) were identified. The distribution of all DEmRNAs, DElncRNAs, and DEmiRNAs on −log10(FDR) and log2 FC are shown in volcano plot (Figure 1). Furthermore, heatmap in Figure S1 shows that the WT samples are clearly distinguishable from normal samples with respect to expression profiles of these DERNAs. The top 15 upregulated and 15 downregulated DElncRNAs, DEmiRNAs, and DEmRNAs in WT samples are presented in Tables S1 to S3.
Figure 1.
Volcano plots of differentially expressed lncRNAs (A), miRNAs (B), and mRNAs (C). The red spots represent upregulated RNAs and the green spots represent downregulated RNAs with statistical significance in Wilms tumor samples. lncRNAs indicates long noncoding RNAs; mRNAs, messenger RNA; miRNAs, microRNAs.
Construction of a ceRNA Network in WT
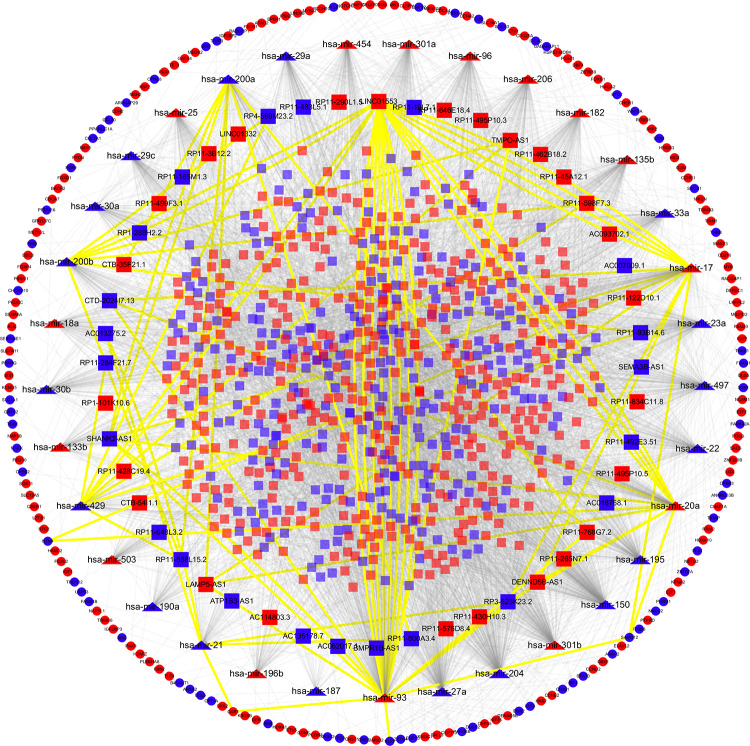
The ceRNA hypothesis suggests that lncRNAs could competitively bind with the MREs to communicate with mRNAs. To explore interactions among above DERNAs, a lncRNA-miRNA-mRNA-related ceRNA network of WT was built. First, the 1833 DElncRNAs were used to retrieve paired DEmiRNAs using miRcode database. As a result, 5032 pairs of DElncRNAs–DEmiRNAs involved in 759 DElncRNAs (eg, SEMA3B-AS1, TMPO-AS1, RP11-576D8.4, and ATP1B3-AS1) and 39 DEmiRNAs (eg, hsa-mir-20a, hsa-mir-17, and hsa-mir-21) were identified for further analysis. The top 10 DElncRNAs targeted by most DEmiRNAs are presented in Table S4. Subsequently, the DEmiRNAs–DEmRNAs intersection was constructed based on the 39 DEmiRNAs through Targetscan, miRTarBase, and miRDB databases. We detected 33 DEmiRNAs interacting with 189 DEmRNAs in all 3 databases (Table S5). The 6 remaining DEmiRNAs and their 11 paired DElncRNAs were discarded. In total, 748 DElncRNAs, 33 DEmiRNAs, and 189 DEmRNAs were incorporated into the ceRNA regulatory network of WT, as shown in Figure 2. Noticeably, the lncRNA RP11-205M3.3 with most connections could interact with 9 DEmiRNAs and 28 DEmRNAs.
Figure 2.
The lncRNA–miRNA–mRNA ceRNA regulatory network in Wilms tumor. LncRNAs, miRNAs, and mRNAs are denoted by rectangle, triangle, and ellipse, respectively. The nodes colored in red and blue, respectively, refer to upregulated and downregulated RNAs. The 62 lncRNAs–miRNAs interactions and 8 miRNAs–mRNAs interactions involved in RNAs with potential prognostic value were highlighted by yellow color. ceRNA indicates competing endogenous RNA; lncRNAs, long noncoding RNAs; mRNAs, messenger RNA; miRNAs, microRNAs.
Functional Enrichment Analysis and PPI Network Analysis
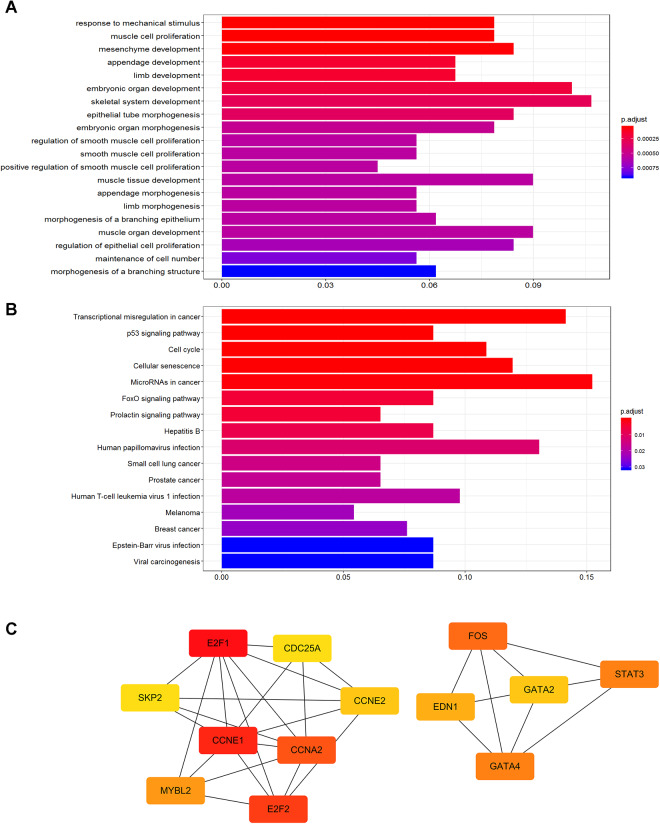
We subsequently explored the biological functions of 189 DEmRNAs in the ceRNA network by GO and KEGG analyses. The GO analysis revealed that these DEmRNAs significantly enriched in 440 GO terms where the top 20 GO biological processes are depicted in Figure 3A. The main biological processes implicated in WT contained in response to mechanical stimulus, muscle cell proliferation, mesenchyme development, and appendage development. The 16 significantly enriched KEGG pathways mainly included tumor-related pathways (Figure 3B), such as transcriptional misregulation in cancer, p53 signaling pathway, cell cycle, cellular senescence, and miRNAs in cancer. Finally, PPI network analysis involved 96 DEmRNAs with combined score >700 was conducted to understand crucial proteins involved in WT. Then, we used cytoHhbba app to screen out top 13 hub proteins (E2F1, CCNE1, E2F2, CCNA2, FOS, GATA4, STAT3, MYBL2, EDN1, CCNE2, GATA2, CDC25A, and SKP2) ranked by Matthew’s correlation coefficient method from the constructed PPI network (Figure 3C).
Figure 3.
The functional enrichment analysis of 189 DEmRNAs in the ceRNA network and PPI network of top 13 hub DEmRNAs identified by MCC method. A, Top 20 significantly enriched GO terms (q value <0.05). B, Sixteen significantly KEGG pathways (q value <0.05). C, The node color in PPI network changes gradually from red to yellow in decreasing order according to the intersection scores. DEmRNAs indicates differentially expressed messenger RNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MCC, Matthew’s correlation coefficient; PPI, protein–protein interaction.
Important RNAs Relevant to the Prognosis of Patients With WT
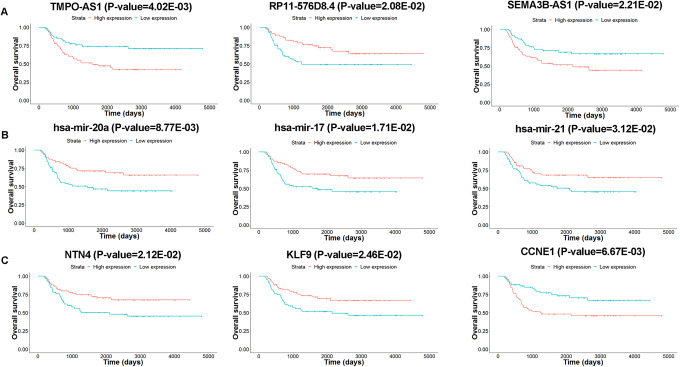
In order to identify potentially significant RNAs associated with the survival of patients with WT, Kaplan-Meier survival curve and log-rank test analysis were conducted for each RNA in the ceRNA network. Consequently, 48 lncRNAs, 7 miRNAs, and 16 mRNAs were considered as potential prognosis-related RNAs (P value <.05, Table 2). Among these, 17 of the 48 DElncRNAs were positively correlated with overall survival of the patients with WT, while the remaining 31 DElncRNAs were negatively associated with overall survival (Table 2). As shown in Figure 4A, survival curves for RP11-576D8.4 appeared to be protective, as patients with high expression levels of RP11-576D8.4 tended to have longer survival time, whereas TMPO-AS1 and SEMA3B-AS1 were considered as risk factors, as their high expression were associated with shorter overall survival time. The high expression of 7 DEmiRNAs (hsa-mir-93, hsa-mir-20a, hsa-mir-17, hsa-mir-429, hsa-mir-21, hsa-mir-200a, and hsa-mir-200b) and 5 DEmRNAs (BCL6, NTN4, KLF9, ARC, and FGF9) were associated with prolonged survival time, on the contrary, high expression level of the rest of 11 DEmRNAs were related to poor prognosis (Figure 4B and C).
Table 2.
The 48 DElncRNAs, 7 DEmiRNAs, and 16 DEmRNAs in the ceRNA Network Significantly Associated With Overall Survival of Patients With Wilms Tumor
| Node | RNAa | Ensembl ID | Regulate | P value | Functionb |
|---|---|---|---|---|---|
| DElncRNA | LINC01332* | ENSG00000250446 | Up | 1.01E-03 | Risky |
| RP11-430H10.3* | ENSG00000254651 | Up | 1.98E-03 | Risky | |
| RP11-93B14.6* | ENSG00000167046 | Down | 2.79E-03 | Risky | |
| AC062017.1 | ENSG00000222020 | Down | 3.15E-03 | Risky | |
| TMPO-AS1* | ENSG00000257167 | Up | 4.02E-03 | Risky | |
| BMPR1B-AS1* | ENSG00000249599 | Down | 4.64E-03 | Protective | |
| RP11-495P10.5* | ENSG00000238107 | Up | 5.47E-03 | Risky | |
| CTB-54I1.1 | ENSG00000254299 | Up | 6.74E-03 | Risky | |
| RP11-834C11.8 | ENSG00000248576 | Up | 7.12E-03 | Risky | |
| RP11-45A12.1* | ENSG00000255079 | Up | 7.24E-03 | Protective | |
| RP1-101K10.6* | ENSG00000227627 | Up | 8.42E-03 | Risky | |
| AC135178.7 | ENSG00000226871 | Down | 8.57E-03 | Risky | |
| RP11-165M1.3* | ENSG00000261394 | Down | 9.33E-03 | Protective | |
| AC013275.2* | ENSG00000231013 | Down | 1.20E-02 | Protective | |
| RP11-483L5.1 | ENSG00000255462 | Down | 1.23E-02 | Risky | |
| LAMP5-AS1* | ENSG00000225988 | Up | 1.37E-02 | Protective | |
| RP11-462B18.2* | ENSG00000231193 | Up | 1.47E-02 | Risky | |
| RP11-800A3.4* | ENSG00000260401 | Down | 1.62E-02 | Protective | |
| DENND5B-AS1* | ENSG00000255867 | Up | 1.77E-02 | Risky | |
| RP11-10L7.1* | ENSG00000246375 | Down | 1.81E-02 | Risky | |
| AC093702.1 | ENSG00000231156 | Up | 2.01E-02 | Risky | |
| CTB-35F21.1* | ENSG00000249526 | Up | 2.02E-02 | Risky | |
| RP11-576D8.4* | ENSG00000224717 | Up | 2.08E-02 | Protective | |
| RP11-428C19.4 | ENSG00000255308 | Up | 2.12E-02 | Risky | |
| SEMA3B-AS1 | ENSG00000232352 | Down | 2.21E-02 | Risky | |
| RP11-265N7.1* | ENSG00000259450 | Up | 2.34E-02 | Risky | |
| RP11-646E18.4* | ENSG00000260377 | Up | 2.47E-02 | Risky | |
| RP11-598F7.3* | ENSG00000256948 | Up | 2.49E-02 | Risky | |
| RP11-122D10.1* | ENSG00000259280 | Up | 2.73E-02 | Risky | |
| RP11-768G7.2* | ENSG00000241213 | Up | 2.88E-02 | Risky | |
| LINC01553* | ENSG00000235931 | Up | 3.15E-02 | Risky | |
| RP11-499F3.1 | ENSG00000259610 | Up | 3.20E-02 | Risky | |
| RP1-288H2.2 | ENSG00000257989 | Down | 3.57E-02 | Protective | |
| CTD-2024I7.13* | ENSG00000246422 | Down | 3.58E-02 | Protective | |
| AC016768.1* | ENSG00000232451 | Down | 3.68E-02 | Protective | |
| RP11-588L15.2* | ENSG00000250893 | Down | 3.70E-02 | Protective | |
| ATP1B3-AS1* | ENSG00000244124 | Down | 3.74E-02 | Protective | |
| AC007009.1 | ENSG00000244239 | Down | 3.76E-02 | Risky | |
| RP11-290L1.5 | ENSG00000257329 | Up | 4.08E-02 | Risky | |
| RP11-284F21.7 | ENSG00000229953 | Down | 4.15E-02 | Protective | |
| SHANK2-AS1* | ENSG00000226627 | Down | 4.21E-02 | Protective | |
| RP11-492E3.51 | ENSG00000239353 | Down | 4.57E-02 | Protective | |
| RP11-3B12.2 | ENSG00000237764 | Up | 4.60E-02 | Risky | |
| AC114803.3 | ENSG00000230432 | Up | 4.67E-02 | Risky | |
| RP11-648L3.2 | ENSG00000253116 | Down | 4.74E-02 | Protective | |
| RP3-523K23.2* | ENSG00000261116 | Down | 4.75E-02 | Protective | |
| RP11-495P10.3* | ENSG00000224481 | Up | 4.84E-02 | Risky | |
| RP4-569M23.2 | ENSG00000231119 | Down | 4.91E-02 | Risky | |
| DEmRNA | CDCA4 | ENSG00000170779 | Up | 4.14E-03 | Risky |
| CCNE1 | ENSG00000105173 | Up | 6.67E-03 | Risky | |
| OSR1 | ENSG00000143867 | Up | 7.81E-03 | Risky | |
| BCL6 | ENSG00000113916 | Down | 1.67E-02 | Protective | |
| NTN4 | ENSG00000074527 | Down | 2.12E-02 | Protective | |
| KLF9 | ENSG00000119138 | Down | 2.46E-02 | Protective | |
| SAMD12 | ENSG00000177570 | Down | 2.74E-02 | Risky | |
| DEPDC1 | ENSG00000024526 | Up | 2.88E-02 | Risky | |
| ARC | ENSG00000198576 | Down | 3.47E-02 | Protective | |
| FOXP4 | ENSG00000137166 | Up | 3.90E-02 | Risky | |
| QSER1 | ENSG00000060749 | Up | 4.14E-02 | Risky | |
| DBF4 | ENSG00000006634 | Up | 4.37E-02 | Risky | |
| MYCN | ENSG00000134323 | Up | 4.38E-02 | Risky | |
| IGF2BP2 | ENSG00000073792 | Up | 4.66E-02 | Risky | |
| CHEK1 | ENSG00000149554 | Up | 4.68E-02 | Risky | |
| FGF9 | ENSG00000102678 | Down | 4.97E-02 | Protective | |
| DEmiRNA | hsa-mir-93 | Up | 1.71E-03 | Protective | |
| hsa-mir-20a | Up | 8.77E-03 | Protective | ||
| hsa-mir-17 | Up | 1.71E-02 | Protective | ||
| hsa-mir-429 | Down | 2.89E-02 | Protective | ||
| hsa-mir-21 | Down | 3.12E-02 | Protective | ||
| hsa-mir-200a | Down | 3.63E-02 | Protective | ||
| hsa-mir-200b | Down | 4.46E-02 | Protective |
Abbreviations: DE, differentially expressed; lncRNAs, long noncoding RNAs; miRNAs, microRNAs; mRNAs, messenger RNA.
a Star highlights DElncRNA that has a positive association with DEmRNAs through linear regression analysis.
b The relationship between expression level of RNAs and survival of patients with Wilms tumor. “Protective” refers to patient with high expression levels of RNA tended to have longer survival time, whereas “risky” represents their high expression were associated with shorter overall survival time.
Figure 4.
The Kaplan-Meier curves for RNAs associated with overall survival in Wilms tumor. A, lncRNAs for TMPO-AS1, RP11-576D8.4, and SEMA3B-AS1. B, miRNAs for hsa-mir-20a, hsa-mir-17, and hsa-mir-21. C, mRNAs for NTN4, KLF9, and CCNE1. Horizontal axis represents overall survival time (days) and vertical axis shows survival probability. lncRNAs indicates long noncoding RNAs; mRNAs, messenger RNA; miRNAs, microRNAs.
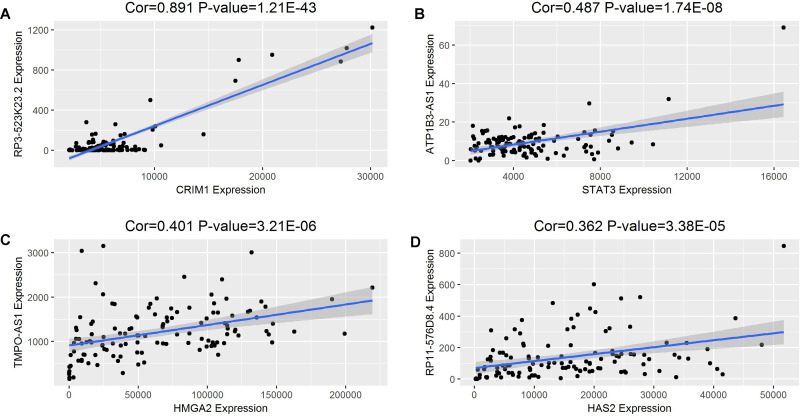
To confirm the speculation that lncRNAs may indirectly interacted with mRNAs in the constructed ceRNA regulatory network of WT, a linear regression analysis was conducted to analyze the relationships between expression levels of 48 prognostic DElncRNAs and 189 DEmRNAs. Consequently, 30 of 48 lncRNAs were positively correlated with 76 DEmRNAs mediated by 28 DEmiRNAs. The P value and correlation coefficient of 4 DElncRNAs against DEmRNAs expression level are depicted in Figure 5. The relatively high positive correlations suggested a possibility of indirect interactions between lncRNA and mRNA expression. For example, RP3-523K23.2 and ATP1B3-AS1 may regulate the expression of CRIM1 (cor = 0.891 and P value = 1.21E-43) and STAT3 (cor = 0.487, P value = 1.74E-08), respectively.
Figure 5.
The linear regression of 4 representative DElncRNAs against DEmRNAs expression level including RP3-523K23.2 versus CRIM1 (A), ATP1B3-AS1 versus STAT3 (B), TMPO-AS1 versus HMGA2 (C), and RP11-576D8.4 versus HAS2 (D). The blue line represents the linear model fitted by the dots in each figure, while gray region refers to the 95% CI. “Cor” is the correlation coefficient between the expressions of lncRNA and mRNA. DE indicates differentially expressed; lncRNAs, long noncoding RNAs; mRNAs, messenger RNA; miRNAs, microRNAs.
Correlation Analysis Between Key lncRNAs and Clinical Features
To further investigate relationship between expression level of 748 lncRNAs involved in the ceRNA network and clinical features of the patients, a comprehensive analysis was performed by comparing lncRNAs expression profiles with gender, tumor stage, histologic classification, race, ethnicity, and age. A total of 133 lncRNAs were found to be associated with clinical features as summarized in Table 3. Notably, DENND5B-AS1 and 5 DElncRNAs (TMPO-AS1, RP3-523K23.2, RP11-598F7.3, LAMP5-AS1, and AC013275.2) were correlated with tumor stage and histologic classification, respectively, which suggested their great value in diagnosis, therapy, and prognosis for patients with WT.
Table 3.
Correlations Between DElncRNAs and Clinical Features of Patients With Wilms Tumor.
| Clinical parameters | Upregulate | Downregulate |
|---|---|---|
| Tumor stage (I+II vs III+IV) | CASC6, DENND5B-AS1, AC012462.1, AC011288.2, AC114808.2, AC007285.7, RP11-650L12.2, LINC00574, RP11-524C21.2, AC010729.2, AC010974.3, RP11-119F7.5, RP11-626G11.4, RP11-573J24.1 | RP11-438D8.2, SHANK2-AS3, GRM5-AS1, CTC-321K16.1 |
| Histologic classification (FHWT vs DAWT) | RP11-472M19.2, CTA-254O6.1, BARX1-AS1, AC079135.1, CTD-3007L5.1, RP11-429By14.4, RP11-881M11.4, TMPO-AS1, RP11-626H12.2, RP1-182D15.2, CTD-2320G14.2, LINC01280, RP11-626G11.4, RP11-847H18.3, LINC01284, LINC00337, RP11-348F1.2, AC068057.1, RP11-443B7.2, RP11-553A10.1, AP000462.1, CTD-2298J14.2, HCG23, RP1-170O19.14, RP11-343B18.2, RP11-384P7.7, LINC00595, RP1-45N11.1, CTD-2591A6.2, GPR50-AS1, RP11-3B12.5, RP11-434D9.2, AC005592.1, RP11-1084I9.1, RP11-152L7.2, AC092159.3, RP11-219B4.3, RP11-314D7.2, AC011288.2, RP11-343J3.2, RP11-379B8.1, LINC01231, RP11-560A15.3, RP11-567G11.1, AP000432.2, RP11-608O21.1, RP11-67H24.2, LAMP5-AS1, RP3-522D1.1, RP11-598F7.3, RP3-510O8.4, AC133633.2, LINC01059, DEPDC1-AS1, CTD-2587M23.1, RP11-524C21.2 | LINC01484, RP4-781K5.4, RP11-386B13.4, RP11-22C11.2, AC104794.4, RP3-523K23.2, RP11-327J17.2, WWTR1-AS1, RP11-311H10.4, LINC00505, RP11-106M7.1, RP5-884G6.2, RP1-18D14.7,RP1-187B23.1, KCNC4-AS1, RP11-710F7.2, GSN-AS1, AC013275.2, RP11-67L3.5, |
| Gender (female vs male) | AC012442.5, AC079150.3, RP11-907D1.1, RP11-54O7.1, TMPO-AS1, AC003092.2, RP11-9L18.3, RP11-430H10.3 | CTC-498J12.1, DDX39B-AS1, RP11-165M1.3, RP11-403P17.2 |
| Race (white vs black or African American) | RP11-1042B17.3, CTD-2147F2.1 | AP001627.1, RP1-187B23.1, AC012485.2, LINC01123 |
| Ethnicity (not Hispanic or Latino vs Hispanic or Latino) | RP11-626G11.4, RP11-13E5.2, RP11-847H18.3, CTD-2130F23.2, RP11-897M7.1, RP11-495P10.6, CTD-2544H17.1, LINC01096 | KCNC4-AS1, GLIS3-AS1, RP11-21A7A.4, RP5-1021I20.2 |
| Age (≥5 vs <5 years) | FAM225A, CTD-2383M3.1, CTD-2116N20.1, RP11-67H24.2, RP11-474D1.4, AC005592.1, RP11-266N13.2, LINC01310, AC068057.1, RP11-575H3.1, RP11-316I3.1, STARD4-AS1, RP11-64P14.7, RP11-893F2.5, RP11-307C19.1, RP11-292D4.1, RP11-58A18.1, LINC00390 | RP11-800A3.7, WWTR1-AS1, GATA2-AS1, RP11-386B13.4 |
| RP11-16O9.2 |
Discussion
Although numerous studies have endeavored to elucidate on the molecular mechanisms underlying WT, the exact pathogenesis remains elusive, particularly the roles of related lncRNAs. Recently, the ceRNA hypotheses have been proposed that different RNAs including lncRNAs and mRNAs could interact with each other through shared MREs. To comprehensively illustrate how lncRNA-associated ceRNA network involved in pathogenesis and progression of WT, we analyzed the large cohort of patients with WT with high-throughput sequencing data from TARGET database in this study. The aberrantly expressed RNAs (DElncRNAs, DEmiRNAs, and DEmRNAs) were identified and incorporated into construction of ceRNA network. To evaluate biological roles of dysregulated RNAs in the ceRNA network, we investigated their associations with the overall survival and clinical features of patients with WT. In addition, the functional enrichment analysis and PPI network of DEmRNAs were performed to detect hub genes.
Recently, accumulative studies have been reported that lncRNA acting as a ceRNA plays critical roles in different cancers, for instance, UCA1 42 in renal cell carcinoma, FER1L4 in gastric cancer,25 and H19 in bladder cancer.26 Despite considerable literature revealing key lncRNA roles in various cancers, a few studies have reported crucial association of dysregulated lncRNA as a ceRNA with progression and development of WT. To our knowledge, only 1 lncRNA (LINC00473) has been reported as a potential oncogene for WT through sponging miR-195 and activating IKKα to promote tumor proliferation.43 In our current study, 1833 DElncRNAs including LINC00473 were detected. Additionally, 48 DElncRNAs in the ceRNA network were significantly correlated with overall survival, indicating their potential prognostic value for WT. Guo et al44 detected that overexpression level of SEMA3B-AS1 inhibited gastric cancer cell proliferation, migration, and invasion in vitro and had a prognostic value in gastric cardia adenocarcinoma. TMPO-AS1 has been found to be a diagnostic and prognostic marker for prostate cancer45 and could be a prognostic signature for stages I and II lung adenocarcinoma.46 The linear regression of expression of TMPO-AS1 against mRNAs in the present study indicated that TMPO-AS1 may interact with 5 genes (FSCN1, HMGA2, MYB, DENND5B, and ETV5) mediated by hsa-mir-200b, hsa-mir-429, hsa-mir-33a, and hsa-mir-150 (Figure 5). In addition, we observed that RP11-576D8.4 had a potential to indirectly regulate 3 genes (HAS2, JARID2, and CDH2) through hsa-mir-204 (Figure 5) in this study. Yuan et al47 reported that RP11-576D8.4 was upregulated in triple negative breast cancer. Moreover, ATP1B3-AS1 has been recently reported to be aberrantly expressed in uninfected infants of HIV-positive mothers and HIV-negative mothers.48 The linear regression results showed that ATP1B3-AS1 interacted with 24 genes mediated by hsa-mir-96, hsa-mir-93, and hsa-mir-182. To our best knowledge, the other 44 lncRNAs identified in our study are novel potential biomarkers in survival prediction, which provide new clues to further investigate the molecular mechanism underling initiation and progression of WT.
It is widely acknowledged that miRNA, a bridge of the ceRNA network, binds with lncRNA and mRNA in different cancers, including WT. In our study, 7 miRNAs in the ceRNA were associated with overall survival rate, 3 (hsa-mir-20a, hsa-mir-17, and hsa-mir-21) of which agreed with reports by other literature. He et al49 identified that the hsa-mir-20a and hsa-mir-17 were involved in the DE regulatory network of WT, which were targeted by transcription factor E2F3. In addition, hsa-mir-20a and hsa-mir-17-5p were upregulated in WT and acted as an oncogene.18,49 Upregulated miR-21 indicated a poor prognosis of WT, and the miR-21 expression level was associated with clinicopathological parameters.50 However, the roles of hsa-mir-93, hsa-mir-429, hsa-mir-200a, and hsa-mir-200b in WT have not been reported.
In our constructed ceRNA network, 16 of 189 DEmRNAs were detected to be significantly correlated with survival time of patients with WT. Five DEmRNAs (BCL6, NTN4, KLF9, ARC, and FGF9) were protective and the other 11 DEmRNAs (CDCA4, CCNE1, OSR1, SAMD12, DEPDC1, FOXP4, QSER1, DBF4, MYCN, IGF2BP2, and CHEK1) were unfavorable. These results suggested that these 16 genes might play important roles in progression of WT and could serve as potential prognostic factors for patients with WT. Some of the 16 mRNAs have been reported to have a substantial influence on several diseases. Paschou et al51 revealed that NTN4 directed axon outgrowth and guidance as an extracellular protein and could be a candidate gene for Tourette syndrome susceptibility. The KLF9 was a valuable prognostic factor and its high expression level meant longer survival time in human pancreatic ductal adenocarcinoma.52 Huang et al53 showed that DEPDC1 promoted cell proliferation and might be a potential therapeutic target of prostate cancer. Furthermore, GO and KEGG analyses revealed that DEmRNAs in the ceRNA network might be involved in cell proliferation and tumor-related biological processes, such as p53 signaling pathway, a well-known cancer-related pathway. Lahoti et al54 detected that p53 expression associated with the development of WT, and p53 immunopositivity indicated negative outcome. Besides, most of the hub DEmRNAs identified by PPI network have been well documented as closely correlated with carcinogenesis and progression of WT. The CCNE1 gene, which is an essential gene for controlling cell cycle, was detected to be negatively associated with WWOX (a tumor suppressor gene) in WT using real-time reverse transcript polymerase chain reaction experiment.55 STAT3 activation can promote WT proliferation and indicated unfavorable prognosis in WT.56 In addition, Cao et al57 found that STAT3 could inhibit WTX expression by regulating microRNA-370 in WT cells.
Due to the high correlation of 133 RNAs expression level with clinical features (Table 3), we performed multivariable Cox regression analyses for each RNA with potential prognostic value and 4 vital clinical variables including tumor stage (I+II vs III+IV), histologic classification (FAWT vs DAWT), gender (female vs male), and age (≥5 vs <5) to investigate the independence of RNAs in survival prediction. The regression coefficients of RNAs and clinical features are listed in Table S6, where red color represents significance of variables due to its P value less than .05. We found that tumor stage, gender, and age had large coefficients and were significant for most RNAs, whereas histologic classification derived from COG had slightly impact on survival prediction. Despite of the effect of clinical features, few RNAs still had significant prognostic value, which were 4 DElncRNAs (AC135178.7, AC093702.1, RP11-483L5.1, and RP3-523K23.2) and 3 DEmiRNA (hsa-mir-200a, hsa-mir-200b, and hsa-mir-429). These results suggested single RNA as a prognostic biomarker was largely dependent from clinical factors, and integrating multiple RNAs as a risk index would be promising as previously proposed by Golub et al.58
Conclusion
In the current study, we identified WT-specific DElncRNAs, DEmiRNAs, and DEmRNAs through multiple bioinformatics analyses of genome-wide transcriptome profiling. We successfully constructed an lncRNA-associated ceRNA network and found some key RNAs significantly associated with overall survival and clinical features. Some of these identified key RNAs were agreement with the previous reports; however, majority of them were identified to be associated with WT for the first time by our study. These results further provide a novel insight into better understanding of the molecular genetic mechanisms underlying pathogenesis of WT and enrich the database to develop targeted therapies and biomarkers that would minimize toxicity for patients. In addition, the mismatched data on WT and normal samples from TARGET database may lead to systematic bias in identification of DERNAs, and therefore in vitro and in vivo experiments for above candidate RNAs are needed for further validation.
Supplemental Material
supplemental_material_(2) for Comprehensive Analysis of a Long Noncoding RNA-Associated Competing Endogenous RNA Network in Wilms Tumor by Feng Zhang, Liping Zeng, Qinming Cai, Zihao Xu, Ruida Liu, Haicheng Zhong, Robert Mukiibi, Libin Deng, Xiaoli Tang and Hongbo Xin in Cancer Control
Acknowledgments
The authors thank TCGA for providing the RNA sequence and clinical data involved in this study.
Authors’ Note: XH and ZF conceived and designed the study. ZF, ZL, DL, and TX downloaded clinical and RNA sequence data and performed data analyses. CQ, XZ, LR, and ZH prepared figures and tables. ZF, ZL, and MR wrote the manuscript. All authors read, commented, and approved the final manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics: All data in this study were freely downloaded from Therapeutically Applicable Research To Generate Effective Treatments (TARGET) database (https://ocg.cancer.gov/programs/target) using the TCGA data portal (https://www.cancer.gov/tcga, accessed September 5, 2018). Our research was in accordance with the publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines); therefore, further approval by an ethics committee was not required.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Natural Science Foundation of Jiangxi Province (20181BAB205008 and 20133BCB23007 to Libin Deng and 20171BAB205109 to Xiaoli Tang).
ORCID iD: Feng Zhang  https://orcid.org/0000-0003-2723-4037
https://orcid.org/0000-0003-2723-4037
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Hohenstein P, Pritchard-Jones K, Charlton J. The yin and yang of kidney development and Wilms’ tumors. Genes Dev. 2015;29(5):467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pizzo PA, Poplack DG, Adamson PC, Blaney SM, Helman L. Principles and Practice of Pediatric Oncology. 7th ed Wolters Kluwer Health; 2016. [Google Scholar]
- 3. Leslie SW, Murphy PB. Cancer, Wilms (Nephroblastoma). Secondary Cancer, Wilms (Nephroblastoma). 2019. https://www.ncbi.nlm.nih.gov/books/NBK442004/
- 4. Niedzielska E, Bronowicki K, Pietras W, et al. Clinical factors in relapses of Wilms’ tumor – results for the Polish Pediatric Solid Tumors Study Group. Adv Clin Exp Med. 2014;23(6):925–931. [DOI] [PubMed] [Google Scholar]
- 5. Szychot E, Apps J, Pritchard-Jones K. Wilms’ tumor: biology, diagnosis and treatment. Transl Pediatr. 2014;3(1):12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ko EY, Ritchey ML. Current management of Wilms’ tumor in children. J Pediatr Urol. 2009;5(1):56–65. [DOI] [PubMed] [Google Scholar]
- 7. Davidoff AM. Wilms tumor. Adv Pediatr. 2012;59(1):247–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breslow NE, Lange JM, Friedman DL, et al. Secondary malignant neoplasms after Wilms tumor: an international collaborative study. Int J Cancer. 2010;127(3):657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haber DA, Buckler AJ, Glaser T, et al. An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms’ tumor. Cell. 1990;61(7):1257–1269. [DOI] [PubMed] [Google Scholar]
- 10. Huang A, Campbell CE, Bonetta L, et al. Tissue, developmental, and tumor-specific expression of divergent transcripts in Wilms tumor. Science. 1990;250(4983):991–994. [DOI] [PubMed] [Google Scholar]
- 11. Maiti S, Alam R, Amos CI, Huff V. Frequent association of beta-catenin and WT1 mutations in Wilms tumors. Cancer Res. 2000;60(22):6288–6292. [PubMed] [Google Scholar]
- 12. Rivera MN, Kim WJ, Wells J, et al. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science. 2007;315(5812):642–645. [DOI] [PubMed] [Google Scholar]
- 13. Bardeesy N, Falkoff D, Petruzzi MJ, et al. Anaplastic Wilms’ tumour, a subtype displaying poor prognosis, harbours p53 gene mutations. Nat Genet. 1994;7(1):91–97. [DOI] [PubMed] [Google Scholar]
- 14. Williams RD, Chagtai T, Alcaide-German M, et al. Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget. 2015;6(9):7232–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scott RH, Douglas J, Baskcomb L, et al. Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nat Genet. 2008;40(11):1329–1334. [DOI] [PubMed] [Google Scholar]
- 16. Gadd S, Huff V, Huang C-C, et al. Clinically relevant subsets identified by gene expression patterns support a revised ontogenic model of Wilms tumor: a Children’s Oncology Group Study. Neoplasia. 2012;14(8):742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grundy PE, Breslow NE, Li S, et al. Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: a report from the National Wilms Tumor Study Group. J Clin Oncol. 2005;23(29):7312–7321. [DOI] [PubMed] [Google Scholar]
- 18. Yu X, Li Z, Chan MT, Wu WK. The roles of microRNAs in Wilms’ tumors. Tumour Biol. 2016;37(2):1445–1450. [DOI] [PubMed] [Google Scholar]
- 19. Imam JS, Buddavarapu K, Lee-Chang JS, et al. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene. 2010;29(35):4971–4979. [DOI] [PubMed] [Google Scholar]
- 20. Koller K, Pichler M, Koch K, et al. Nephroblastomas show low expression of microR-204 and high expression of its target, the oncogenic transcription factor MEIS1. Pediatr Develop Pathol. 2014;17(3):169–175. [DOI] [PubMed] [Google Scholar]
- 21. Liu M, Roth A, Yu M, et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 2013;27(23):2543–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126(8):2775–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng B, Jeong S, Zhu Y, Chen L, Xia Q. miRNA and lncRNA as biomarkers in cholangiocarcinoma(CCA). Oncotarget. 2017;8(59):100819–100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xia T, Chen S, Jiang Z, et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lv M, Zhong Z, Huang M, et al. lncRNA H19 regulates epithelial–mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta Mol Cell Res. 2017;1864(10):1887–1899. [DOI] [PubMed] [Google Scholar]
- 27. Fan Q, Liu B. Comprehensive analysis of a long noncoding RNA-associated competing endogenous RNA network in colorectal cancer. Onco Targets Ther. 2018;11:2453–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song W, Miao D-l, Chen L. Comprehensive analysis of long noncoding RNA-associated competing endogenous RNA network in cholangiocarcinoma. Biochem Biophy Res Commun. 2018;506(4):1004–1012. [DOI] [PubMed] [Google Scholar]
- 29. Fang XN, Yin M, Li H, et al. Comprehensive analysis of competitive endogenous RNAs network associated with head and neck squamous cell carcinoma. Sci Rep. 2018;8(1):10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10): R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benjamini Y, Hochberg Y.Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 32. Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat. 2011;3(2):180–185. [Google Scholar]
- 33. Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2014;43(D1): D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with cytoscape 3. Curr Protoc Bioinformatics. 2014;47(1):8.13.11-18.13. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an r package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fox J, Carvalho MS. The RcmdrPlugin. survival package: extending the R Commander interface to survival analysis. J Stat Soft. 2012;49(7):1–32. [Google Scholar]
- 41. Mullen EA, Geller JI, Gratias EJ, Perlman EJ, Dome J. Real-time central review: a report of the first 3,000 patients enrolled on the Children’s Oncology Group Renal Tumor Biology and Risk Stratification protocol AREN03B2. J Clin Oncol. 2014;32(15 suppl):10000. [Google Scholar]
- 42. Lu Y, Liu W-G, Lu J-H, et al. LncRNA UCA1 promotes renal cell carcinoma proliferation through epigenetically repressing p21 expression and negatively regulating miR-495. Tumour Biol. 2017;39(5). [DOI] [PubMed] [Google Scholar]
- 43. Zhu S, Fu W, Zhang L, et al. LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of Wilms tumour via IKKalpha. Cell Prolifer 2018;51(1):e12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guo W, Liang X, Liu L, et al. MiR-6872 host gene SEMA3B and its antisense lncRNA SEMA3B-AS1 function synergistically to suppress gastric cardia adenocarcinoma progression. Gastric Cancer. 2019;22(4):705–722. [DOI] [PubMed] [Google Scholar]
- 45. Huang W, Su X, Yan W, et al. Overexpression of AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and poor prognosis in prostate cancer. Prostate. 2018;78(16):1248–1261. [DOI] [PubMed] [Google Scholar]
- 46. Peng F, Wang R, Zhang Y, et al. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol Cancer. 2017;16(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuan N, Zhang G, Bie F, et al. Integrative analysis of lncRNAs and miRNAs with coding RNAs associated with ceRNA crosstalk network in triple negative breast cancer. Onco Targets Ther. 2017;10:5883–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Musimbi KZ. Between Transcriptional Comparison of the Immune System of HIV-Exposed Uninfected and HIV-Uninfected Infants in Kilifi, Kenya. University of Nairobi; 2018. [Google Scholar]
- 49. He J, Guo X, Sun L, Wang K, Yao H. Networks analysis of genes and microRNAs in human Wilms’ tumors. Oncol Lett. 2016;12(5):3579–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cui M, Liu W, Zhang L, et al. Clinicopathological parameters and prognostic relevance of miR-21 and PTEN expression in Wilms’ tumor. J Pediatr Surg. 2017;52(8):1348–1354. [DOI] [PubMed] [Google Scholar]
- 51. Paschou P, Yu D, Gerber G, et al. Genetic association signal near NTN4 in Tourette syndrome. Ann Neurol. 2014;76(2):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mao Z, Fan X, Zhang J, et al. KLF9 is a prognostic indicator in human pancreatic ductal adenocarcinoma. Anticancer Res. 2017;37(7):3795–3799. [DOI] [PubMed] [Google Scholar]
- 53. Huang L, Chen K, Cai Z-p, et al. DEPDC1 promotes cell proliferation and tumor growth via activation of E2F signaling in prostate cancer. Biochem Biophys Res Commun. 2017;490(3):707–712. [DOI] [PubMed] [Google Scholar]
- 54. Lahoti C, Thorner P, Malkin D, Yeger H. Immunohistochemical detection of p53 in Wilms’ tumors correlates with unfavorable outcome. Am J Pathol. 1996;148(5):1577–1589. [PMC free article] [PubMed] [Google Scholar]
- 55. Płuciennik E, Nowakowska M, Wujcicka WI, et al. Genetic alterations of WWOX in Wilms’ tumor are involved in its carcinogenesis. Oncol Rep. 2012;28(4):1417–1422. [DOI] [PubMed] [Google Scholar]
- 56. Zhang LJ, Liu W, Gao YM, Qin YJ, Wu RD. The expression of IL-6 and STAT3 might predict progression and unfavorable prognosis in Wilms’ tumor. Biochem Biophys Res Commun. 2013;435(3):408–413. [DOI] [PubMed] [Google Scholar]
- 57. Cao X, Liu D, Yan X, et al. Stat3 inhibits WTX expression through up-regulation of microRNA-370 in Wilms tumor. FEBS Lett. 2013;587(6):639–644. [DOI] [PubMed] [Google Scholar]
- 58. Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplemental_material_(2) for Comprehensive Analysis of a Long Noncoding RNA-Associated Competing Endogenous RNA Network in Wilms Tumor by Feng Zhang, Liping Zeng, Qinming Cai, Zihao Xu, Ruida Liu, Haicheng Zhong, Robert Mukiibi, Libin Deng, Xiaoli Tang and Hongbo Xin in Cancer Control