Abstract
The recent pandemic of SARS-CoV-2 infection has affected more than 3.0 million people worldwide with more than 200 thousand reported deaths. The SARS-CoV-2 genome has the capability of gaining rapid mutations as the virus spreads. Whole-genome sequencing data offers a wide range of opportunities to study mutation dynamics. The advantage of an increasing amount of whole-genome sequence data of SARS-CoV-2 intrigued us to explore the mutation profile across the genome, to check the genome diversity, and to investigate the implications of those mutations in protein stability and viral transmission. We have identified frequently mutated residues by aligning ~660 SARS-CoV-2 genomes and validated in 10,000 datasets available in GISAID Nextstrain. We further evaluated the potential of these frequently mutated residues in protein structure stability of spike glycoprotein and their possible functional consequences in other proteins. Among the 11 genes, surface glycoprotein, nucleocapsid, ORF1ab, and ORF8 showed frequent mutations, while envelop, membrane, ORF6, ORF7a and ORF7b showed conservation in terms of amino acid substitutions. Combined analysis with the frequently mutated residues identified 20 viral variants, among which 12 specific combinations comprised more than 97% of the isolates considered for the analysis. Some of the mutations across different proteins showed co-occurrences, suggesting their structural and/or functional interaction among different SARS-COV-2 proteins, and their involvement in adaptability and viral transmission. Analysis of protein structure stability of surface glycoprotein mutants indicated the viability of specific variants and are more prone to be temporally and spatially distributed across the globe. A similar empirical analysis of other proteins indicated the existence of important functional implications of several variants. Identification of frequently mutated variants among COVID-19 patients might be useful for better clinical management, contact tracing, and containment of the disease.
Keywords: SARS-CoV-2, Hot-spot mutations, Spike glycoprotein, Frequent mutation, Protein stability
Abbreviations: ACE2, Angiotensin-converting enzyme 2; CTD, C- terminal domain; MERS-CoV, Middle East respiratory syndrome coronavirus; MUSCLE, Multiple Sequence Comparison by Log- Expectation; NTD, N- terminal domain; ORF, Open Reading Frame; RBD, Receptor binding domain; SARS-Cov 2, Severe acute respiratory syndrome coronavirus 2
1. Introduction
In December 2019, the Chinese government reported several human pneumonia cases in Wuhan city and designated the disease as coronavirus disease 2019 (COVID 19) (Wang et al., 2020a). Major symptoms of COVID-19 include fever, cough, dyspnoea, and muscular soreness. There were some patients with COVID 19, where atypical symptoms like diarrhea and vomiting were also found (Ding, 2020; Wang, 2020; Zhu et al., 2020). Whole-genome sequencing showed that the causative agent is a novel coronavirus, initially termed as 2019-nCoV (Zhu et al., 2020; Wu et al., 2020). Later on, the International Committee on Taxonomy of Viruses (ICTV) officially designated the virus as SARS-CoV-2. WHO on March 11, 2020, has declared the COVID-19 outbreak as a global pandemic (Cucinotta and Vanelli, 2020).
Corona-viruses are a class of genetically diverse viruses found in a wide range of host species like mammals and birds (Drosten et al., 2003; Resta et al., 1985). SARS-CoV-2 is an enveloped virus and comprises a positive sense single-strand RNA genome of ~30 kb (Kim et al., 2020). This SARS-CoV-2 also belongs to the genus betacoronavirus like SARS-CoV and MERS-CoV. Primarily, it was thought to cause infections in birds and other mammals but recent outbreaks have revealed the ability of coronaviruses to cross species barriers and human transmission (Menachery et al., 2017). Coronaviruses carry the largest genome among all RNA viruses and each viral transcript consists of a 5′-cap structure and a 3′ poly-A tail (Lai and Stohlman, 1981). After entry to the host cell, the genomic RNA is translated to produce non-structural proteins (nsps) from two open reading frames (ORFs). On the other hand, the viral genome is also used as a template for replication and transcription via RNA-dependent RNA polymerase activity. In the intermediate stage, negative-strand RNA intermediates are produced to serve as a template for positive-sense genomic RNA and sub-genomic RNA synthesis. These shorter sub-genomic RNAs encode the structural proteins i.e. Spike, Envelope, Membrane and Nucleocapsid protein, and several other accessory proteins (Snijder et al., 2016; Sola et al., 2015; Dongwan Kim et al., 2020).
The mutation rate for RNA viruses is drastically high and this higher mutation rate is correlated with a virulence which is beneficial for viral adaptation (Duffy, 2018). The SARS-CoV-2 genome has the capability of gaining rapid mutations as the virus spreads (Lu et al., 2020). The advantage of the increasing amount of whole-genome sequence data of SARS-CoV-2 intrigued us to explore the mutation profile across the genome, to check the genome diversity and to investigate the consequences of those mutations on stability and transmission.
In this present study, we used ~660 complete SARS-CoV-2 genome data from NCBI virus database (16th April 2020) for in-silico analysis. We performed gene and protein sequence alignment and characterized the mutation status of all genes. The most conserved and variable regions were recognized for all genes along with synonymous and non-synonymous changes. As non-synonymous changes dictate the altered amino acid composition, a collection of all mutations for each protein has been determined. We cataloged these substitutions for all proteins and identified different variants that are prevalent in nature. We also evaluated the impact of mutating spike glycoprotein in protein stability, viral transmission, adaptability, and diversification. This brief characterization of SARS-CoV-2 variants and functional impact analysis of those variants could lead to better clinical management of the COVID-19 pandemic.
2. Materials and methods
2.1. Collection of SARS-CoV-2 genomic sequences
Total 664 SARS-CoV-2whole genome sequences were downloaded from NCBI Virus repository (https://www.ncbi.nlm.nih.gov/labs/virus) as of April 16, 2020. The repository provided the option of excluding the partially sequenced genomes by selecting only the complete sequences under the category ‘Nucleotide Completeness’. Additionally, sequences with one or more ambiguous sites denoted by ‘N’ were filtered out before further analysis. Number of sequences from each country is as follows: Australia:1, Brazil: 1, China: 57, Colombia: 1, France: 1, Greece: 4, India: 2, Iran: 1, Israel: 2, Italy: 2, Nepal: 1, Pakistan: 2, Peru: 1, South Africa: 5, Spain: 11, Sweden: 1, Taiwan: 3, Turkey: 1, USA: 565 and Vietnam: 2.
2.2. Identification of variable sites
We have aligned nucleotide and amino acid sequences of ORF1ab, ORF3a, ORF6, ORF7a, ORF8, ORF10, envelop (E), membrane (M), nucleocapsid (N) and surface glycoprotein (S)using MUSCLE multiple sequence alignment algorithms in MEGA-X (Kumar et al., 2018). The alignment files both at nucleotide and protein levels, generated in MEGA-X (in .meg format) are zipped and provided as Supplementary File 1. The SARS-CoV-2 isolate with GenBank accession ID ‘NC_045512’, as downloaded from NCBI (https://www.ncbi.nlm.nih.gov/nuccore/NC_045512) was used as the reference strain in this study. We tabulated the number of variable, singleton, parsimony informative sites at both gene and protein levels. After removing the ambiguous and deleted residues, we determined the amino acid substitutions in all ten proteins of the SARS-CoV-2 genome. Frequently mutated residues are those that showed a mutation in 1.5% of the strains. Co-occurring mutations are determined considering all frequently mutated residues for S, N, ORF3a, ORF8, and ORF1ab proteins.
Since it has been well established that bats as well as pangolins may be the sources of the original transmission of the virus in humans (Zhou et al., 2020; Zhang et al., 2020), we were interested to note the amino acid residues in the key mutational sites in Bat and pangolin coronavirus protein sequences. The protein sequences of Bat coronavirus RaTG13 (GenBank Accession ID MN996532.1), Pangolin coronavirus (MT072864.1), and two SARS-CoV strains TW11 (AY502924.1) and GD01 (AY278489.2) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/protein). These were aligned with the analogous protein sequences of SARS-Cov2 in MEGA-X, and the residues at the frequently mutated sites of the respective viral proteins were observed.
2.3. Protein structural analyses
Cryo-EM three-dimensional structures of SARS-CoV-2 spike glycoprotein have been recently made available in RCSB Protein Data Bank (https://www.rcsb.org/) (Berman et al., 2000). Three PDB structures i.e. 6VXX (Closed state) (Walls et al., 2020), 6VYB (Open state) (Walls et al., 2020) and 6VSB (Prefusion) (Wrapp et al., 2020) were downloaded. We used FoldXBuildModel function to construct the mutant 3D protein structures (Schymkowitz et al., 2005). The differences in total energy, electrostatics, solvation energy etc.were calculated for all the closed, open state as well as prefusion spike protein mutant 3D models with FoldX empirical force field (Schymkowitz et al., 2005; Kiel et al., 2004). All three cryo-EM structures have some missing residues in the available PDBs, and are not considered for stability calculation or further structural analysis. However missing residues and loops were generated for each conformation to have a proper environment of the mutated residues to increase the accuracy of energy calculations. For this purpose, we have used SWISS-MODEL (https://swissmodel.expasy.org/) homology modelling online server with Spike glycoprotein sequence (uniprt_ID: P0DTC2) and three individual PDBs were used as a template to generate three conformations. The models were refined by OpenMM7 (Eastman et al., 2017) with CHARMM27 force-field (Mackerell Jr. et al., 2004) implemented in the SWISS-MODEL server. The models were further validated by RAMPAGE (Lovell et al., 2003) which shows 91–92% residues are in the favoured region and 6–7% are in the allowed region in the phi-psi map of all the three models. Structural analyses and figures have been generated in Discovery studio 2020 (Dassault System BIOVIA Corp) (Dassault Systèmes BIOVIA, 2016).
2.4. Phylogeny, and spatial and temporal distribution of SARS-CoV-2 strains
To study the impact of non-synonymous mutations in the transmission and viability of the newly emerging SARS-CoV-2 sequences across the world, we looked into Nextstrain tool, a powerful visualization tool comprising >10,000 SARS-CoV-2 samples to study the evolution of various pathogens (https://nextstrain.org/) (Hadfield et al., 2018). The frequency of occurrence, the date of collection, and the corresponding geographical location of the mutant strains was noted on 27th April 2020. Please note that the genotype information on Nextstrain are not consistent on different dates, at least for some residues. Major amino acid substitutions were mapped and visualized in the phylogeny from Nextstrain Next hCoV-19 App. Minor (less frequently) mutated residues that showed variations in Nextstrain data compared to NCBI virus database were excluded from the analysis. Amino acid residues of ORF1ab were split as ORF1a from 1 to 4400 and the rest as ORF1b in Nextstrain Next hCoV-19 App.
2.5. Empirical analysis of the functional implication of mutations
To understand the implications of the amino acid substitutions in the mutants, charge state (at neutral pH), Kyte-Doolittle hydropathy indices (Kyte and Doolittle, 1982), and Grantham's mean chemical difference indices, which takes into account side-chain chemical structure, volume, and polarity (Grantham, 1974), were compared. Stabilities of their side-chains between exposed and buried forms were compared using apparent partition energies as reported by Wertz and Scheraga (Wertz and Scheraga, 1978). The typical contributions of a putative H-bond and salt-bridge towards protein stabilities were assumed to be in the range of 0.5–1.5 kcal/mol (Pace et al., 2014; Sheu et al., 2003; Pace, 1995) and 3 kcal/mol (Nayek et al., 2014) based on previous studies on protein folding.
3. Results
3.1. Synonymous and non-synonymous mutations
After aligning the individual ORFs of the SARS-CoV-2 genome, we identified mutations both at gene and protein levels (Table 1 ). At the gene level, the number of mutations per 100 bases was found to be relatively high in N, ORF10, ORF6, ORF7a, ORF8, and ORF3a, suggesting that these genes may be more prone to mutations as compared to others. Among the structural proteins, M and E proteins contained the least variability, which indicated that these proteins may be associated with housekeeping functions and consequently have a greater resistance to mutations. Looking into the changes per 100 amino acids for each of the proteins (Table 1), we observed that ORF3a exhibited the highest mutability, closely followed by N and ORF8. While looking into the synonymous and non-synonymous changes, we have found that the ORF1ab and spike protein contained the largest number of non-synonymous mutations (Table 1). When we normalized with respect to the length, ORF3a, ORF8, and N exhibited a relatively high number of non-synonymous changes.
Table 1.
Nucleotide and protein alignment of SARS-CoV-2 genes.
| Name | Total No of Samples | No. of nucleotides | Variable sites | Variable Sites per 100 bp | Synonymous | Non Synonymous | Singleton Informative Site per 100 bp | Parsimony Informative Site per 100 bp | No. of Amino acids | Variable Site | Variable Sites per 100 aa | Singleton Informative Site per 100 aa | Parsimony Informative Site per 100 aa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Envelope Protein | 664 | 228 | 4 | 1.75 | 2 | 2 | 1.32 | 0.44 | 76 | 2 | 0.88 | 0.88 | 0.00 |
| Membrane Protein | 649 | 669 | 7 | 1.20 | 2 | 5 | 0.60 | 0.60 | 223 | 5 | 0.75 | 0.45 | 0.30 |
| Nucleocapsid Protein | 664 | 1260 | 50 | 3.97 | 24 | 26 | 2.54 | 1.43 | 420 | 26 | 2.06 | 1.43 | 0.63 |
| ORF10 | 660 | 117 | 4 | 3.42 | 2 | 2 | 2.56 | 0.85 | 39 | 2 | 1.71 | 1.71 | 0.00 |
| ORF1ab | 639 | 21,291 | 383 | 1.80 | 148 | 235 | 1.34 | 0.46 | 7097 | 235 | 1.10 | 0.83 | 0.28 |
| ORF3a | 660 | 828 | 28 | 3.38 | 8 | 20 | 2.05 | 1.33 | 276 | 20 | 2.42 | 1.33 | 1.09 |
| ORF6 | 657 | 186 | 6 | 3.23 | 3 | 3 | 1.61 | 1.61 | 62 | 3 | 1.61 | 1.08 | 0.54 |
| ORF7a | 658 | 366 | 9 | 2.73 | 3 | 6 | 1.91 | 0.82 | 122 | 6 | 1.64 | 1.09 | 0.55 |
| ORF7b | 446 | 132 | 2 | 1.51 | 2 | 0 | 0.76 | 0.76 | 44 | 0 | 0 | 0 | 0 |
| ORF8 | 661 | 366 | 9 | 2.46 | 1 | 8 | 1.37 | 1.09 | 122 | 8 | 2.19 | 1.09 | 1.09 |
| Spike Protein | 643 | 3822 | 74 | 1.96 | 30 | 44 | 1.52 | 0.44 | 1274 | 44 | 1.13 | 0.92 | 0.21 |
3.2. Frequently mutated amino acids in SARS-CoV-2 proteins
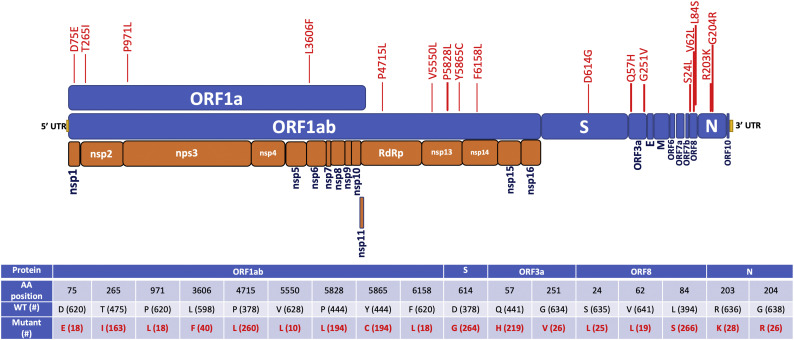
Considering the altered protein sequences due to non-synonymous changes, we next focused on amino acid substitutions among all proteins of the SARS-CoV-2 genome (Supplementary Table 1). An abridged version of the table containing only those substitutions that have been observed in a minimum of 10 isolates (~1.5% of the total isolates) is provided in Fig. 1. Among the structural proteins, E and M showed most conserved structures across all the viral genomes under consideration, with substitutions at two sites of each E and M proteins only in 1and5isolatesrespectively. Upon examining the S proteins, we have found several mutations; nevertheless, most of these substitutions are perceived only in a single isolate, with notable exceptions being the D614G. There have been 264 (41%) instances of D614G, suggesting its pivotal role in regards to the protein stability and other key characteristics. Among the other changes, V483A, L5F, Q675H, H655Y, and S939F occurred in 6, 5, 3, 2, and 2 isolates respectively. The N protein also depicted substitutions R203K and G204R.
Fig. 1.
Frequently mutated residues, i.e. those observed in a minimum of 10 SARS-Cov-2 isolates are plotted on the respective proteins of the SARS-CoV-2 genome. The tabular presentation depicted the number of occurrences (#) of wildtype and mutant residues among the sequences of SARS-CoV-2 ORFs.
Among the non-structural proteins, ORFs 6, 7a, and 10 shared similar behavior to E and M proteins with them being mostly conserved. In contrast, ORF3a exhibited non-synonymous mutations, the majority of which had mostly been distributed at 2residues (Q57H and G251V). Mutations in ORF8 showed a major substitution at L84S and an accompanied change of V62L. Another substitution, S24L was observed in 25 samples out of the 660 sequences analysed. We moved on to the largest encoded SARS-CoV-2 protein, ORF1ab that encodes replicase polyproteins required for viral RNA replication and transcription (Ziebuhr, 2005). ORF1a and ORF1b encode two polypeptides, pp1a and pp1ab, and finally processed into 16 nsps (Fig. 1) (Ziebuhr, 2005; Chen et al., 2020). Majority of the non-synonymous mutations at ORF1ab led to amino acid changes at the 265th, 4725th, 5828th, and 5865thresidues (T265I, P4715L, P5828L, and Y5865C). Notably, L5828P and C5865Y occurred simultaneously in all strains, suggesting a possible functional relationship between these two residues.
3.3. Identification of prevalent SARS-CoV-2 variants
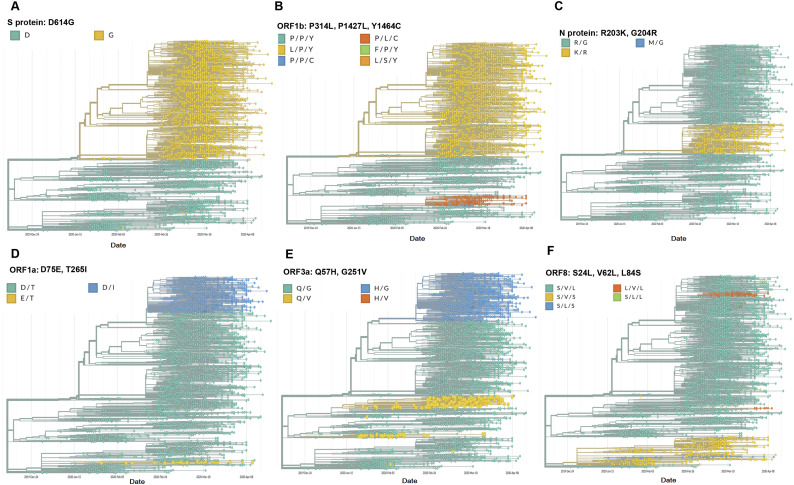
To identify the variants that are prevalent over time, we next determined the frequently mutated residues that occurred at least 1.5% of the samples. The combined analysis of all proteins with these frequently mutated residues identified 20 possible SARS-CoV-2 variants, among which 15variants comprised more than 97% of the analysed sequences having frequency > 1% (Table 2 ). Apart from the wild type variant (13.3%), other frequent SARS-CoV-2 variants are ORF8:L84S/ORF1ab:P5828L/Y5865C (30.6%), S:D614G/ORF3a:Q57H/ORF1ab:T265I/P4715L (20.4%), S:D614G/ORF3a:Q57H/ORF1ab:P4715L (7.2%), ORF8:L84S (4.6%) and S:D614G/ORF1ab:P4715L(4.6%). We noted that G251V of ORF3a, V62L of ORF8, D75E, P971L, P5828L, Y5865C, and F6158L of ORF1ab substitutions occurred only with the D614 wild type variant of the S protein. While, substitutions R203K and G204R of N protein, Q57H of ORF3a, S24L of ORF8, T265I, P4715L, and V5550L of ORF1ab occurred only with the D614G mutant of S protein. Co-occurrences of these mutations might have implications in direct structural interactions or indirect regulations of these proteins on the survivability of the virus. To further validate these findings, we visualized these major substitutions by observing the phylogeny of SARS-CoV-2 in Nextstrain, which contains a curated database of more than 10,000 SARS-CoV-2 sequences, depicted in the form of phylogenetic trees (Fig. 2 ).
Table 2.
Strains with co-occurring major mutations in N, S, ORF3a, ORF8 and ORF1ab proteins.
| Proteins (Positions)⁎ | N(203,204) | S(614) | orf3a(57,251) | orf8(24,62,84) | orf1ab (75,265,971,3606,4715,5550,5828,5865,6158) | # of isolates | Freq. (%) | ||||||||||||
| Amino acid position | 203 | 204 | 614 | 57 | 251 | 24 | 62 | 84 | 75 | 265 | 971 | 3606 | 4715 | 5550 | 5828 | 5865 | 6158 | ||
| SARS-CoV-2 | R | G | D | Q | G | S | V | S⁎ | D | T | P | L | P | V | L⁎ | C⁎ | F | 186 | 30.6 |
| R | G | G⁎ | H⁎ | G | S | V | L | D | I⁎ | P | L | L⁎ | V | P | Y | F | 124 | 20.4 | |
| R | G | D | Q | G | S | V | L | D | T | P | L | P | V | P | Y | F | 81 | 13.3 | |
| R | G | G⁎ | H⁎ | G | S | V | L | D | T | P | L | L⁎ | V | P | Y | F | 44 | 7.2 | |
| R | G | D | Q | G | S | V | S⁎ | D | T | P | L | P | V | P | Y | F | 28 | 4.6 | |
| R | G | G⁎ | Q | G | S | V | L | D | T | P | L | L⁎ | V | P | Y | F | 28 | 4.6 | |
| K⁎ | R⁎ | G⁎ | Q | G | S | V | L | D | T | P | L | L⁎ | V | P | Y | F | 23 | 3.8 | |
| R | G | D | Q | V⁎ | S | V | L | D | T | P | F⁎ | P | V | P | Y | F | 20 | 3.3 | |
| R | G | G⁎ | H⁎ | G | L⁎ | V | L | D | I⁎ | P | L | L⁎ | V | P | Y | F | 20 | 3.3 | |
| R | G | D | Q | G | S | L⁎ | S⁎ | E⁎ | T | L⁎ | L | P | V | P | Y | L⁎ | 17 | 2.8 | |
| R | G | D | Q | G | S | V | L | D | T | P | F⁎ | P | V | P | Y | F | 11 | 1.8 | |
| R | G | G⁎ | H⁎ | G | S | V | L | D | I⁎ | P | L | L⁎ | L⁎ | P | Y | F | 8 | 1.3 | |
| R | G | D | Q | V⁎ | S | V | L | D | T | P | L | P | V | P | Y | F | 5 | 0.8 | |
| R | G | D | Q | G | S | V | S⁎ | D | T | P | F⁎ | P | V | L⁎ | C⁎ | F | 4 | 0.7 | |
| R | G | D | Q | G | S | V | S⁎ | D | T | P | F⁎ | P | V | P | Y | F | 3 | 0.5 | |
| R | G | D | Q | G | S | L⁎ | S⁎ | E⁎ | T | L⁎ | L | P | V | P | Y | F | 1 | 0.2 | |
| R | G | D | Q | G | S | L⁎ | S⁎ | D | T | P | L | P | V | P | Y | F | 1 | 0.2 | |
| R | G | G⁎ | H⁎ | G | S | V | L | D | I⁎ | P | F⁎ | L⁎ | V | P | Y | F | 1 | 0.2 | |
| K⁎ | G | G⁎ | H⁎ | G | S | V | L | D | I⁎ | P | L | L⁎ | V | P | Y | F | 1 | 0.2 | |
| R | G | G⁎ | H⁎ | G | S | V | S⁎ | D | I⁎ | P | L | L⁎ | V | P | Y | F | 1 | 0.2 | |
| Bat coronavirus RaTG13 | R | G | D | Q | G | S | V | S | D | T | P | V | P | V | P | Y | F | ||
| Pangolin coronavirus | R | G | D | Q | G | S | V | S | D | N | Q | V | P | V | P | Y | F | ||
| SARS coronavirus TW11 | R | G | D | Q | G | E | F | Y | D | T | V | V | P | V | P | Y | F | ||
| SARS coronavirus GD01 | R | G | D | Q | G | E | F | Y | D | T | V | V | P | V | P | Y | F | ||
Mutated residues.
Fig. 2.
Phylogeny (generated from GISAID Next hCoV-19 App) with colour coding for the wild type and substituted residues of A. Spike glycoprotein (S) at the 614th amino acid position, B. ORF1b at 314, 1427 and 1464th amino acid positions, C. Nucleocapsid (N) at 203 and 204th amino acid positions, D. ORF1a at 75 and 265th, E. ORF3a at 57 and 251st, and F. ORF8 at 24, 62 and 84th amino acid positions. All possible combinations of residues at the frequently mutated sites have been represented by a distinct colour for each ORF/protein, the strains are colour-coded as per the combination of residues at the reference sites. The horizontal axis depicts the dates around which the isolates were sequenced and submitted.
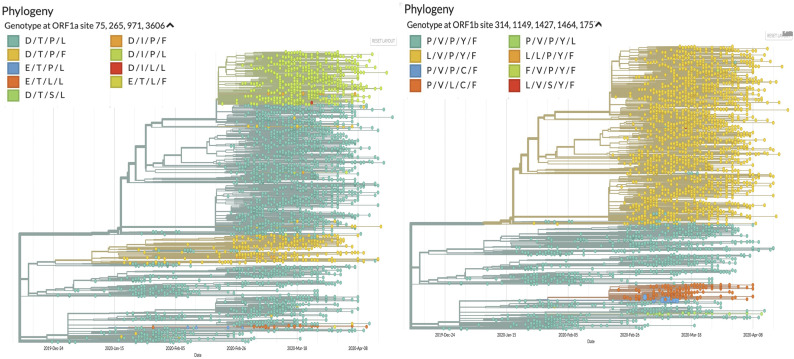
We observed the variants that contained these specific substitutions are mostly clustered together. For S protein, the proportion of the samples with D614G substitutions was roughly equal to that of wild type variant, which showed the adaptability of this substitution. For ORF1b, we assessed the substitutions at positions 314, 1427, and 1464, which corresponded to 4715th, 5828th, and 5865thresidues of ORF1ab. Mapping of all frequently mutated residues of ORF1a and ORF1b on the phylogeny of SARS-CoV-2 is presented in Supplementary Fig. 1. As observed previously, substitutions at 5828th and 5865th position co-occurred even in this large sample set of Nextstrain data. Viral variants with residues L/P/Y, P/P/Y and P/L/C dominated the bulk of the sequences, which showed that the conjoined mutations (either P/Y or L/C) at 5828th and 5865th residues were linked with increasing survivability (Fig. 2). Interestingly, we could not overlook the fact that the two representations of the mutations for both S protein and ORF1b were remarkably co-occurring. We have seen that those variants that have D614G substitutions in S also have L/P/Y residues at 314, 1427, and 1464 positions of ORF1b. It suggested that these residues in S and ORF1b, irrespective of whether they have occurred simultaneously, show good viability (Fig. 2). When focusing on the D614 residue in S protein, the most prevalent variant was with P/P/Y at all the three positions of ORF1b, with exception of few variants with P/L/C residues. Looking into the mutation profile of N, we have found that the wild type variants (R/G at 203 and 204 positions) appeared to be predominant. Comparison with mutation profile of S protein identified co-occurrence of 614G variant with K/R at 203rd and 204th positions of N protein (Fig. 2). We checked the mutation status at positions 75 and 265 of ORF1a protein. We withheld the inclusion of the 971stsite here, as we had seen that substitutions at this site and the 75th residue were co-linked, i.e. they were mostly identical (Fig. 2). We noticed that the wild type variants formed the majority with few isolates of D75E mutants. A fairly good number of samples with T265I substitutions of ORF1a were also observed having D614G substitutions of S protein. Comparing the mutational profile with the ORF3a at 57th and 251st positions, we again found a stark resemblance to both these profiles. We saw that the D/I variant in ORF1a mostly went hand-in-hand with H/G variants in ORF3a (Fig. 2). Viewing the mutational profile of ORF8 with respect to positions V62L and L84S occurred mostly with the D614 of S protein, while S24L occurred with the G variants, also observed in our analysis with ~660 samples. Overall, the mutation profile that we identified with 664 samples showed excellent concordance with the Nextstrain data comprising ~10,000 samples.
The following are the supplementary data related to this article.
Supplementary Fig. 1: Phylogeny (generated from GISAID Next hCoV-19 App) with colour coding for the wild type and all substituted residues of ORF1a and ORF1b.
We compared these frequently mutated residues with the corresponding protein sequences of Bat coronavirus RaTG13, Pangolin coronavirus, SARS coronavirus TW11, and GD01 (Table 2). All these frequently mutated residues completely matched with Bat coronavirus RaTG13. The notable exceptions in here being mismatches at positions 265, 971, and 3606 of ORF1a in case of pangolin coronavirus and mismatches in all three residues of ORF8 in both the SARS-CoV isolates.
3.4. Empirical analysis of changes in structure and stability parameters
Parameters for 17 frequently mutated residues and V483A substitution at the receptor-binding domain of S protein that may affect the protein structures are presented in Table 3 . Only 3 out of these 18 substitutions were associated with any change in the charge of the side chain. The change in apparent side-chain partitioning energy varied from −2.79 to 3.13 kcal/mol. The maximum expected difference in the number of H-bond and salt bridges associated with mutations were 4 and 1, respectively. While only 3 variants could have changed in salt-bridge interactions, 8 of them could potentially have a difference in H-bonding reflecting the fact that average energy associated with a salt-bridge interaction is much higher.
Table 3.
Potential implications of SARS-COV2 mutations on viral protein structure and stability.
| Protein | Mutant | Relative abundance of the mutation (%) | Expected change in charge state | Mean chemical difference index | Difference in Kyte-Doolittle hydropathy index | Apparent partition energy (parent residue) | Apparent partition energy (mutant residue) | Difference in apparent partition energy1 | Potential difference in number of H-bonds | Potential difference in stability due to H-bond2 | Presumptive difference due to salt bridge interactions3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S | D614G | 69.8 | 1 | 94 | 3.1 | 0.41 | 0.31 | 0.1 | -2 | 1–3 | 3 |
| V483A | 0.9 | 0 | 64 | −2.4 | −0.46 | 0.05 | −0.51 | 0 | 0 | 0 | |
| N | R203K | 4.4 | 0 | 26 | 0.6 | 0.12 | 0.57 | −0.45 | −2 | 1–3 | 0 |
| G204R | 4.1 | −1 | 125 | −4.1 | 0.31 | 0.12 | 0.19 | 4 | -(2–6) | −3 | |
| ORF1ab | D75E | 2.9 | 0 | 45 | 0 | 0.41 | 0.46 | −0.05 | 0 | 0 | 0 |
| T265I | 34.3 | 0 | 89 | 5.2 | 0.38 | −0.69 | 1.07 | −1 | 0.5–1.5 | 0 | |
| P971L | 2.9 | 0 | 98 | 5.4 | 0.46 | −2.67 | 3.13 | 0 | 0 | 0 | |
| L3606F | 6.7 | 0 | 22 | −1 | −2.67 | −1.03 | −1.64 | 0 | 0 | 0 | |
| P4715L | 68.8 | 0 | 98 | 5.4 | 0.46 | −2.67 | 3.13 | 0 | 0 | 0 | |
| V5550L | 1.6 | 0 | 32 | −0.4 | −0.46 | −2.67 | 2.21 | 0 | 0 | 0 | |
| P5828L | 43.7 | 0 | 98 | 5.4 | −2.67 | 0.46 | 3.13 | 0 | 0 | 0 | |
| Y5865C | 43.7 | 0 | 194 | 3.8 | −0.84 | −0.25 | 0.59 | −1 | 0.5–1.5 | 0 | |
| F6158L | 2.9 | 0 | 22 | 1 | −1.03 | −2.67 | 1.64 | 0 | 0 | 0 | |
| ORF3a | Q57H | 49.7 | −1 | 24 | 0.3 | 0.38 | −0.41 | 0.79 | 1 | -(0.5–1.5) | −3 |
| G251V | 4.1 | 0 | 109 | 4.6 | 0.31 | −0.46 | 0.77 | 0 | 0 | 0 | |
| ORF8 | S24L | 3.9 | 0 | 145 | 4.6 | 0.12 | −2.67 | 2.79 | −1 | 0.5–1.5 | 0 |
| V62L | 3.0 | 0 | 32 | −0.4 | −0.46 | −2.67 | 2.21 | 0 | 0 | 0 | |
| L84S | 67.5 | 0 | 145 | −4.6 | −2.67 | 0.12 | −2.79 | 1 | -(0.5–1.5) | 0 |
In (kcal/mol). More positive values indicate mutant is relatively less comfortable to be exposed in water and negative value the otherwise.
In (kcal/mol). Negative values indicate the mutation may to contribute towards stability through H-bonding and positive value the otherwise.
In (kcal/mol) Negative value indicates the mutation may to contribute towards stability through salt-bridge and positive value the otherwise. It should be noted that a residue cannot engage in all probable H-bond interactions and salt-bridge at the same time.
The relative abundance of a mutation can be taken as a surrogate of viral viability, which would be dictated by the effect of the mutation on protein stability and its function with respect to specific biomolecular events during host-pathogen interaction. The relative abundance of the D614G mutant (69.8%) was the highest. The partitioning energy difference was minimal (0.1 kcal/mol) indicating that unless the sidechain of D614 was involved in any consequential H-bond, this mutant could be as viable as the parent strain. The analysis of the larger Nextstrain dataset indicated that D614G mutation is significantly more prevalent, indeed. The high relative abundance (67.5%) of the L84S mutant (ORF8) could be due to additional H-bond or favourable partitioning energy. T265I and Y5865C mutants of ORF1ab associated with removal of an alcoholic -OH group, which is often associated with a modest contribution to protein stability (Pace et al., 2014; Sheu et al., 2003; Pace, 1995) showed a similar relative abundance of 34.3% and 43.7%, respectively. However, the S24L mutant (ORF8) showed a significant decrease in abundance. This may be attributed to a more unfavorable change in apparent partitioning energy (by ~2 kcal/mol) due to a significant difference in chain length of serine and leucine. The significant decrease in abundance of G204R (N protein) in spite of the potential for additional H-bonding may also be attributed to significantly larger chain length. However, the V483A (S protein), V62L (ORF8), V5550L, and D75E (ORF1ab) mutants with relatively low values of differences in Grantham's index or Kyte-Doolittle index and comparable H-bonding or salt-bridge capacity showed a dramatic decrease in relative abundance. While P4715L and P5828L mutants showed relatively high abundance (39.2 and 29.2%, respectively), P971L showed only 2.7% abundance. Interestingly, both F6158L and L3606F with very low differences in Grantham's index and reversal in the difference in apparent partitioning energy showed low abundance.
3.5. Stability of mutant spike glycoproteins
We have encountered several different variants pertaining to S protein of SARS-CoV-2. Apart from D614G and some co-occurring mutations, other changes have been observed in a few cases, e.g., L5F occurring in 5 strains, V483A in 6 strains, while among others, most of these substitutions were observed in a single strain. By performing the stability analysis of spike glycoprotein for mutating residues that are available in all three pdb file, we found that some of the variants are stable in nature corresponding to negative total energies, calculated for both open, closed as well as prefusion models (Table 4, Supplementary Table 2). Among 22 analysed substitutions in S protein, 9 structures showed a reduction in total free energy in all three conformations. Mutants S50L and H49Y showed the most reduction in total free energy, while all mutants with D614G substitutions showed stabilizing structure, suggesting its prevalent role in spike protein evolution. Interestingly, reduction in solvation polar energy was found in only 5 structures, including the D614G mutant. Detailed information for the differences in energy for all residues are presented in Supplementary Table 2.
Table 4.
Total free energy and solvation polar energy changes of Spike glycoprotein mutants (Protein Stability analysis using FoldX).
| Substitution in S protein⁎ | Change in total energy (ΔΔG) in kcal/mol |
Change in Solvation Polar energy |
||||
|---|---|---|---|---|---|---|
| PDB ID: 6VXX (Closed) | PDB ID: 6VYB (Open) | PDB ID: 6VSB (Prefusion) | PDB ID: 6VXX (Closed) | PDB ID: 6VYB (Open) | PDB ID: 6VSB (Prefusion) | |
| A27V | 1.86108 | 1.25673 | NA | 2.56145 | 2.29815 | NA |
| Y28N | 3.65923 | 3.87214 | 2.27761 | −3.09106 | −1.99917 | −2.83006 |
| T29I | 5.84563 | 1.66361 | −3.83215 | 1.39819 | 0.408085 | −0.427407 |
| H49Y⁎ | −2.45444 | -5.2875 | −2.34688 | 1.21591 | −0.583689 | 0.228903 |
| S50L⁎ | −6.67437 | −7.34299 | −6.67805 | 1.31686 | 1.07831 | 1.53239 |
| L54F, D614G⁎ | −1.96221 | −0.511008 | −1.80246 | −3.71794 | −6.41299 | −3.86936 |
| D111N | −1.06165 | 2.71599 | −1.01473 | 0.267207 | 0.175352 | −1.32065 |
| S221W | 2.40534 | 2.09354 | 31.1189 | 4.01364 | 3.33787 | 9.05937 |
| T240I⁎ | −4.13551 | −4.27492 | 9.3503 | −1.21194 | −1.08835 | −0.641467 |
| A348T | 2.07674 | 9.89322 | 1.00384 | 4.89654 | 4.78981 | 3.54288 |
| R408I | 1.12775 | 3.10298 | 1.50848 | −0.474226 | −2.09272 | −0.452398 |
| H519Q | −2.43624 | −0.545803 | −4.62908 | −0.642438 | −2.29236 | −0.688364 |
| A520S⁎ | −0.139498 | 1.19999 | 0.939507 | 1.80759 | 1.36561 | 1.62258 |
| A570V | 3.62668 | 6.71797 | 8.03373 | 4.26631 | 1.79105 | 2.23977 |
| D614G⁎ | −0.765945 | −0.743935 | −1.40123 | −2.93183 | −3.13725 | −3.21621 |
| V772I⁎ | −1.07838 | -1.1456 | −1.21835 | 8.89E-01 | 0.867421 | 0.992304 |
| F797C | 15.5152 | 10.927 | 12.8012 | −3.28608 | −5.36719 | −2.97664 |
| A930V | 8.81798 | 9.24991 | 3.19247 | 3.04437 | 2.96865 | 3.47189 |
| D614G, D936Y⁎ | −3.281 | −2.58379 | −2.39667 | −2.49131 | −2.19201 | −1.81321 |
| D614G, S939F⁎ | −1.19889 | −1.87823 | −4.61428 | −3.23393 | −5.71654 | −2.00416 |
| A1078V | 3.60391 | 2.84238 | 2.42529 | 2.56687 | 2.67595 | 3.10323 |
Mutants that show reduction in total free energy in all three conformations are presented in bold.
3.6. Structural analysis of the wild type and mutant spike glycoproteins
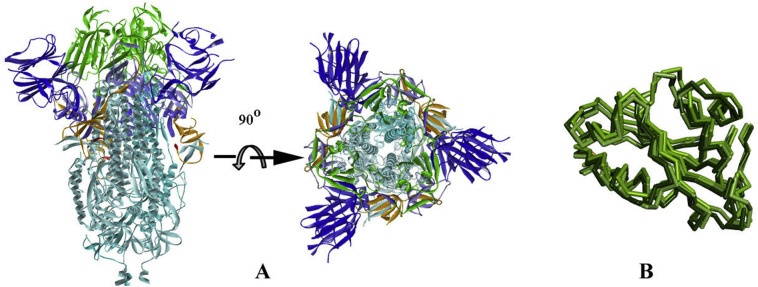
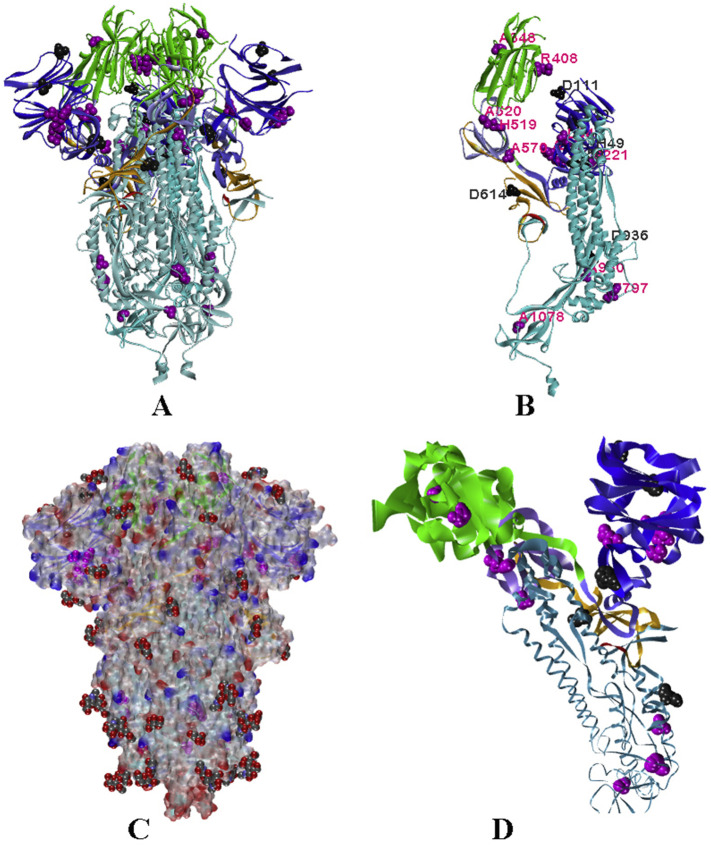
To further understand the implication of these mutants, we have analysed all three structures of S (Walls et al., 2020; Wrapp et al., 2020; Gui et al., 2017). The spike glycoprotein is a homo-trimeric protein (Walls et al., 2020; Wrapp et al., 2020; Gui et al., 2017) having two subunits S1 and S2 in each monomer protruding from the viral surface. S1 subunit forms a budding head responsible for host–receptor binding, while S2 is mainly a stalk-like structure that helps in the fusion of viral and host membranes (Fig. 3A). S proteins are cleaved at the S1/S2 interface but remain non-covalently linked with each other in the prefusion state (Gui et al., 2017). S1 subunit can further be divided into sub-domains namely N-terminal domain (NTD: residues 15–261), C-terminal domains 1, 2 and 3 (CTD1: residues 320–516; CTD2: residues 517–579; CTD3: residues 580–663) (Fig. 3A).
Fig. 3.
Ribbon diagram of Spike Protein (PDB_ID 6VXX), A. In two views; colour code: NTD blue, RBD green, CTD2 light blue, CTD3 orange, S1/S2 linker red and S2 sky blue. B. Superposition of RBD of DOWN/closed conformation (6VXX) with UP/open (6VYB) and pre-fusion state (6VSB) in faded green colour ribbons.
CTD1, which is the main region of S protein for host-receptor interaction, is also termed as the receptor-binding domain (RBD) (Walls et al., 2020; Wrapp et al., 2020; Gui et al., 2017). RBD undergoes conformational changes during receptor binding (human ACE2) that leads to the blossom of the S1 bud in an open or ‘UP’ conformation conducive for S-ACE2 interaction. Comparing the inert ‘DOWN' and active ‘UP’ conformations (PDB_ID s: 6VXX and 6VYB respectively), it is found that RBD moves as a rigid-body in a hinge bending motion around its linker region with NTD and CTD2 with all-atom rmsd for 198 residues is around 2.8 Å (Fig. 3B). A similar change of conformation is also observed in prefusion state (Wrapp et al., 2020).
The miss-sense mutations in S protein are mainly single point mutations with few double mutations. All these mutations can be classified as stabilizing and destabilizing based on the free-energy changes (Table 4) of the in-silico generated mutant structures w.r.t the wild type variant (Li et al., 2020) (Fig. 1). Our study depicts that there are no stabilizing mutations in the receptor-binding domain RBD (Fig. 4A, B). This observation indicates that the mechanism of S protein for a high affinity human ACE2 binding is unique in nature and any mutation (found to date) leads to an unstable structure and this could be correlated with lower viability of these mutations containing isolates.
Fig. 4.
Location of mutations. A. Ribbon diagram of trimeric S protein with colour code as described in Fig. 3. The amino acids undergone to mutations are represented by Vander-wall radii with purple for destabilizing/neutral mutation points and grey for stabilizing ones. B. The monomeric unit of S-protein with a label for the same amino acids as in A. C. A semi-transparent electro-static surface presentation of the S-protein with glycans as Van der Waal presentation. D. Mutations in the monomeric unit of S protein, where ribbon size is proportional to the average isotropic displacement of amino acid residue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
There are 42 miss-sense mutations found in S protein, we have considered 21 of them that are available in every monomer structure of S protein as well as in three pdbs. Out of these, 32 are in the S1 subunit and only 10 are found in the S2 subunit. The cryo-EM structure of the protein (6VXX) shows a high thermal parameter for the NTD and RBD (Fig. 4D). The high-temperature factor of RBD could be correlated with its dynamic nature leading to the conformational switch between close and open states. It is also observed that most of the stabilizing mutations are in the NTD, which is an inherently unstable domain as depicted from the high thermal parameter. This observation puts an open question, whether the virus adopts viability through mutations, stabilizing the flexible NTD.
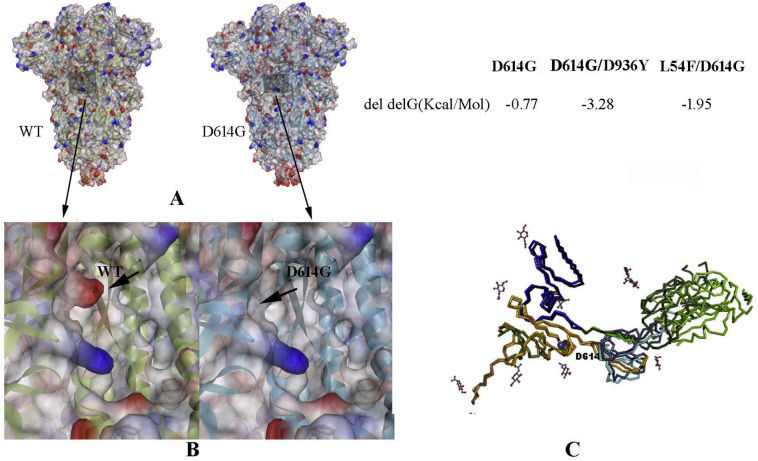
D614G substitution in CTD3 is found to be very stable and prevalent in nature. It occurs either as a single mutation or coupled with other mutations (L54F/D614G, D614G/D936Y, and D614G/S939F). Surprisingly, L54F is a sort of neutral (in terms of free energy change) mutation, however, when coupled with D614G, the double mutant becomes a stable one (Table 4). The structural comparison of wild-type and in-silico generated D614G mutant shows that a change from Aspartic acid to Glycine alters the electro-static potential of the surface of the protein (Fig. 5A). This change creates a favourable environment in a hydrophobic pocket of the S protein (Fig. 5B). Moreover, we have also observed that D614 is at the proximity of the hinge bending region (CTD2/NTD linker) of RBD (Fig. 5C), therefore mutation of D to a small residue G without any side-chain might increase the flexibility for a smooth switch over from inactive DOWN state to the active UP state, makes the mutant containing variants more virulent in terms of its smoother binding with ACE2.
Fig. 5.
The implication of D614G mutation, A. Wild-type (close conformation; pdb_id 6VXX) and D614G mutant structures have been represented as electrostatic potential surfaces. The position of mutation is highlighted. B. A close view near the position of the mutation. C. Superposition of a monomer of the DOWN/close wild-type (6VXX) and the UP/open (6VYB) of S protein showing a hinge-bending motion of RBD (green) around NTD linker (blue) and CTD2 (light blue). The location of D614 residue in CTD3 (orange) is indicated and represented as Van der Waal's presentation. The neighbouring glycans are represented by a stick model. Free energy change due to D614G single and D614G containing double mutants are given. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. Temporal and geographical distribution of wild type and mutant spike glycoproteins
Among these multiple variants, the ones that are occurring in a large fraction of the samples can be said to have adapted, while those strains which only existed with very few samples were likely to get eliminated in the way of selective process and are not generally perceived among the emerging variants. This implies that the favourable variants should be associated with greater stability and/or higher transmission rates of the SARS-CoV-2 proteins, while a decreased stability or transmission rate is expected in the case of the minor variants.
Looking into the spatial and temporal distribution of these variants of S protein in Nextstrain and noting down the number of occurrences of each variant along with the country it originated with the corresponding date (Table 5 ), we find that the variant with D614G substitution is characterized by greater viability across different countries as seen over a span of time, first originating on 24th December 2019 and prevailing since last recorded date. This mutation was also accompanied by L54F, D936Y, and S939Fin different isolates. However, all these variants are observed in multiple samples, which show that the change at the 614th residue is the impactful one which is imparting greater stability to the mutant protein. On the other hand, the less stable mutants that were found only in a few samples did not show such prevalence and were seen to be dwindling out with time.
Table 5.
Geographical and temporal distribution of Spike protein mutants.
| Mutations in spike protein⁎ | No. of sample | Country | First Collection Date | Last collection date | |
|---|---|---|---|---|---|
| A27V | 1 | USA | 23.03.2020 | NA | |
| Y28N | 1 | Australia | 28.02.2020 | NA | |
| T29I | 3 | Australia, USA, Netherlands | 21.03.2020 | 28.03.2020 | |
| H49Y⁎ | 13 | China, USA, Taiwan, Mexico, Australia | 17.01.2020 | 26.03.2020 | |
| S50L⁎ | 5 | China, Singapore, Australia | 28.02.2020 | 19.03.2020 | |
| R408I | 1 | India | 27.01.2020 | NA | |
| H519Q | 1 | Belgium | 29.02.2020 | NA | |
| A520S⁎ | 2 | USA | 13.03.2020 | 03.04.2020 | |
| A570V | 1 | China | 29.01.2020 | NA | |
| D614G⁎ | Innumerable | Across the globe | 24.01.2020 | Till date | |
| V772I | 1 | Turkey | 17.03.2020 | NA | |
| F797C | 1 | Sweden | 07.02.2020 | NA | |
| A930V | 1 | India | 31.01.2020 | NA | |
| D614G & L54F⁎ | D/L | Innumerable | Across the globe | 24.12.2019 | 15.04.2020 |
| G/L | Innumerable | Across the globe | 24.01.2020 | 20.04.2020 | |
| D/F | 1 | Netherlands | 31.03.2020 | NA | |
| G/F | 5 | France, USA, UK, Australia | 07.03.2020 | 02.04.2020 | |
| D614G &D936Y⁎ | D/D | Innumerable | Across the globe | 24.12.2019 | 15.04.2020 |
| G/D | Innumerable | Across the globe | 24.01.2020 | 20.04.2020 | |
| G/Y | 11 | Netherlands, Australia, Sweden, Wales | 12.03.2020 | 02.04.2020 | |
| D/H | 1 | Singapore | 05.01.2020 | NA | |
| D614G & S939F⁎ | D/S | Innumerable | Across the globe | 24.12.2019 | 15.04.2020 |
| G/S | Innumerable | Across the globe | 24.01.2020 | 20.04.2020 | |
| D/F | 1 | Switzerland | 26.02.2020 | NA | |
| G/F | 7 | France, Iceland, USA | 04.03.2020 | 02.04.2020 | |
Mutants with reduction in total free energies are presented in bold.
4. Discussion
We have performed a thorough mutational characterization encompassing the variations occurring in all ORFs of the SARS-CoV-2 genome. Among structural proteins, both the membrane and envelope proteins are more resilient to frequent mutations, while among non-structural proteins, ORFs 6, 7a, and 10 shared similar behavior to E and M proteins, with them being mostly conserved. This signifies that these proteins could have some essential functions, perhaps housekeeping roles that are critical to the virus, which is why these sequences cannot generally withstand any variations. In contrast, S, N, ORF3a, ORF8, and ORF1ab exhibits mutations. An intriguing feature for N protein that we noticed here was that both substitutions (R203K and G204R) were present simultaneously in 26 of the 28 samples, with only the 2 remaining samples lacking the G to R changes at the 204th residue. Mutations in ORF8 showed a major substitution L84S and an accompanied substitution of V62L with few isolates with S24L substitution. Moreover, all of these changes are not independent with respect to one another, which is established from the fact that V62L is also accompanied by a corresponding substitutions S84L. Two major amino acid substitutions D75E and P971L of ORF1ab occurred in the same eighteen strains that harboured both of these mutations and with no instance of any other strain having mutated at only one of these positions. These implicate that these two positions may have a linked relationship and may have some critical functions. Likewise, another clear-cut division of two variants was observed at the 4715th position which possessed L and P variants. We can discern a possible link between this mutation and the one discussed at the 265th position, both of which explicitly divided the isolates into two groups. Additionally, we also detected two mutations at 5828th (L to P) and 5865th (C to Y) positions, and those strains that contained any one of these variants was also forced to accommodate the other variation, with no exception to this event being observed in any sample. Combined analysis, only with the frequently mutated residues, identified at least 20 possible variants, among which 17 variants occurred at least more than one among the samples considered in this study. Frequent occurrences of some of the specific combinations of mutations at 5 genes indicated their direct or indirect interaction leading to stability, adaptability, viability, and transmission efficiency of the virus. Less frequently occurred variants might have eventually lost due to their low transmission efficiency or less adaptability in nature. Country specific under- and/or over-sampling could be a confounding factor for this variation. However, our observation with ~660 samples showed excellent concordance with the data generated from ~10,000 samples, suggesting the generality of this observation.
The contribution of mutations to the stability and function of the gene product, which depends on its location, including interaction with other viral or host molecules, may determine the viability of the mutant. Absence of any charge reversal, either among SARS-COV-2 mutants or other coronaviruses, and low frequency of mutations with a change in charge state underscore that it plays an important role in the viability of variants. The high viability of D614G mutant of S-protein seemed to be attributable to miniscule changes in partitioning energy as well as the exposed aspartate side chain located on a flexible loop in a relatively hydrophobic environment was not involved in any H-boding while it's substitution by glycine could facilitate the movement of the hinge. Compensatory effects of additional H-bond could be a plausible explanation for the relatively high frequency of L84S (ORF8) and Q57H (ORF3) mutants. However, these simple parameters could not explain low abundance of V483A (S-protein), V62L (ORF8), V5550L and D75E (ORF1ab), R203K (N) or discrepancies in an abundance of L3606F vs F6158L and P/L mutants (at 4715 and 5528 vs 971 positions) of Orf1ab. Unlike D614A, V483A mutant is a part of the crucial receptor binding domain. The tighter binding (4–10-fold compared to SARS-COV-1) of the S1-CTD to hACE2 receptor (Wrapp et al., 2020; Wang, 2020) has been attributed to the enhanced infectiousness of SARS-COV-2. Thus, low frequencies of the V483A, as well as other S1-CTD mutants, seem attributable to their role in interaction with the host receptor. It is possible that V62 (ORF8); R203 (N); V5550, D75, P971, L3606 and F6158 (ORF1ab) positions are also associated with crucial functional roles beyond stability.
Two variants with co-occurring mutants were more prevalent than the wild type variant. The most prevalent variant showed co-occurrence of P5828L and Y5865C in ORF1ab. The ability of proline to introduce kink in the structure - often in turns and loops close to surfaces and the tendency of the upstream cysteine to be modified if exposed or form S—S bond if buried may explain the co-occurrence. The next prevalent variant showed a co-occurrence of T265I and P4715L in ORF1ab. This might be indicative of these hydrophobic substitutions coming closer in the tertiary structure and stabilizing it through van der Waals interaction. The co-occurrence of these ORF1ab mutants with D614G (S-protein) and Q57H (ORF3a) is suggestive of functional interaction among these proteins. However, these interpretations are contingent upon the reported mutation frequencies being representative of the actual variant distribution and certainly begs more investigation and analysis.
Among the structural proteins which were mostly conserved, only the spike protein showed several mutations including a dominant mutational variant at the 614th position. We have investigated the thermodynamic stability of the variants to identify the variants which are correlated with greater stability and sustainability. Those strains that corresponded to structures with low stabilities were consequently found to have low transmission capabilities as verified in the Nextstrain data. We have identified several mutants with stable structures, including mutations at positions 49, 50, 54, 614, and 936 and have verified that these variants are enduring among the general population over time, with D614G be the most viable among them. Although some of the mutated residues of spike protein showed a greater reduction of total free energy compared to D614G substitution, their Spatio-temporal distribution and number of isolates are comparatively lower than the substitution at 614. It clearly suggests that spike protein alone is not the determining factor of the stability, adaptability, and transmission efficiency of the virus. The specific combination of all frequently mutated variants might be necessary for the prediction of the viability of the viral variants. However, considering only the disparity in the effectiveness of transmission among the different spike protein variants, we have two important suggestions to the different nations in tackling and curbing the spread of COVID-19 with greater efficacy. First and foremost, the mutational profile of a patient found to be COVID-19 positive needs to be analysed, specifically at these key sites of five proteins, either by Sanger sequencing or designing probes corresponding to these regions. Thereafter, a model can be predicted using the patients' severity and transmission of infection among the contacts for each combination of frequently mutated residues. Though one could argue that as the sequencing of the viral genome had been carried out at different time-points in different countries, with some countries like China imposing higher levels of quarantine measures at an earlier time compared to other countries (Cyranoski, 2020), our interpretations may not have 100% accuracy. However, our hypothesis and interpretation of the mutations show good concordance as evidenced by the Nextstrain data. Further research on the identification of mutational status SARS-CoV-2 infected individuals and determination of infection among their contacts might help to substantiate the idea of the correlation between genotypes with survivability and transmission of different strains. In conclusion, we maintain the belief that the propositions voiced here if followed adequately can work to curb the spread of the disease with much higher success.
Isolates with amino acid substitutions in each protein of SARS-CoV-2
Protein stability analysis of Spike glycoprotein substitutions
MEGA alignemnet files of SARS-CoV-2 proteins
Declaration of Competing Interest
There is no conflict of interest in this manuscript.
Acknowledgments
We would like to acknowledge Prof. Nitai P. Bhattacharya, retired professor, SINP Kolkata for his critical comments and evaluation of the manuscript.
References
- Berman H.M., The Protein Data Bank, et al. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D. What China’s coronavirus response can teach the rest of the world. Nature. 2020;579:479–480. doi: 10.1038/d41586-020-00741-x. [DOI] [PubMed] [Google Scholar]
- Dassault Systèmes BIOVIA . 2016. Discovery Studio Modeling Environment,Release 2017,San Diego: Dassault Systèmes. [Google Scholar]
- Ding Q., et al. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J. Med. Virol. 2020 doi: 10.1002/jmv.25781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongwan Kim J.-Y.L., Yang Jeong-Sun, Kim Jun Won, Kim V. Narry, Chang Hyeshik. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman P., et al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017;13(7) doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185(4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Gui M., et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C., Serrano L., Herrmann C. A detailed thermodynamic analysis of ras/effector complex interfaces. J. Mol. Biol. 2004;340(5):1039–1058. doi: 10.1016/j.jmb.2004.05.050. [DOI] [PubMed] [Google Scholar]
- Kim J.M., et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11(1):3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Stohlman S.A. Comparative analysis of RNA genomes of mouse hepatitis viruses. J. Virol. 1981;38(2):661–670. doi: 10.1128/jvi.38.2.661-670.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell S.C., et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell A.D., Jr., Feig M., Brooks C.L., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Graham R.L., Baric R.S. Jumping species-a mechanism for coronavirus persistence and survival. Curr. Opin. Virol. 2017;23:1–7. doi: 10.1016/j.coviro.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayek A., et al. Salt-bridge energetics in halophilic proteins. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0093862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C.N. Evaluating contribution of hydrogen bonding and hydrophobic bonding to protein folding. Methods Enzymol. 1995;259:538–554. doi: 10.1016/0076-6879(95)59060-9. [DOI] [PubMed] [Google Scholar]
- Pace C.N., et al. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014;23(5):652–661. doi: 10.1002/pro.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resta S., et al. Isolation and propagation of a human enteric coronavirus. Science. 1985;229(4717):978–981. doi: 10.1126/science.2992091. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J., et al. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu S.Y., et al. Energetics of hydrogen bonds in peptides. Proc. Natl. Acad. Sci. U. S. A. 2003;100(22):12683–12687. doi: 10.1073/pnas.2133366100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., et al. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2(1):265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. (e6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., et al. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz D.H., Scheraga H.A. Influence of water on protein structure. An analysis of the preferences of amino acid residues for the inside or outside and for specific conformations in a protein molecule. Macromolecules. 1978:11. doi: 10.1021/ma60061a002. [DOI] [PubMed] [Google Scholar]
- Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30(8):1578. doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolates with amino acid substitutions in each protein of SARS-CoV-2
Protein stability analysis of Spike glycoprotein substitutions
MEGA alignemnet files of SARS-CoV-2 proteins