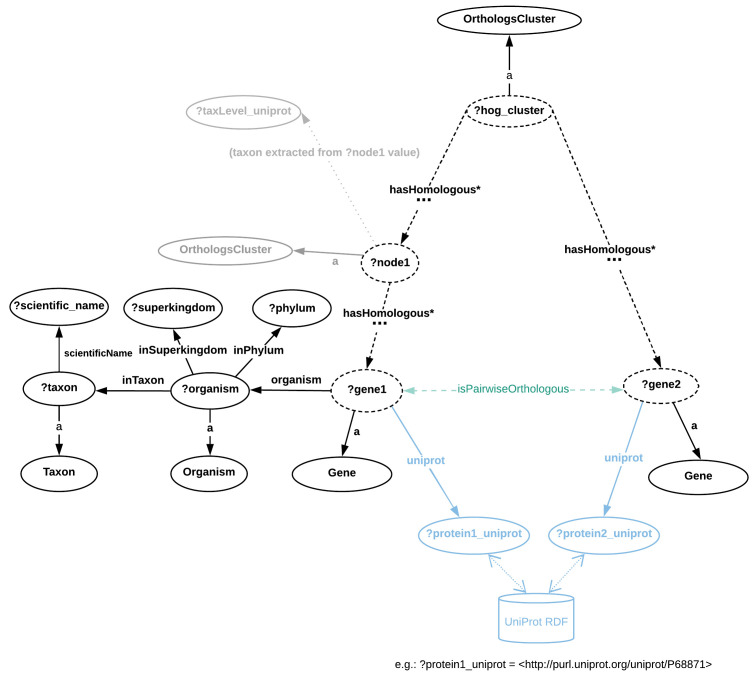
Figure 3. Directed graph abstraction of a portion of the MBGD RDF graph related to hierarchical orthologous groups.
In Figure 3, nodes are either classes or variables, and edges are RDF properties. The terms preceded by a question mark (e.g. ?gene1) represent variables assigned with either zero or more literals or URIs. Dashed edges illustrate the orth:hasHomologous property that can be stated zero or more times, recursively. URI prefixes were omitted. MBGD is gene-centric and contains taxonomic ranges where HOGs are built are not directly available in RDF - in some cases these can be extracted from the cluster URI (e.g. http://mbgd.genome.ad.jp/rdf/resource/cluster/2018-01_tax32_8537 corresponds to taxonomic identifier 32, Myxococcus). By contrast, the taxonomic information per gene entry is richer in MBGD than in OMA, including explicit Superkingdom and Phylum information. Example SPARQL queries based on this graph abstraction are provided in the “Protocols” section, as well as in the accompanying Jupyter notebook. The pairwise orthology information is not directly available (e.g. through an RDF property), but can be extracted from the Orthologs Cluster (to highlight this, the “isPairwiseOrthologous” is shown in green with a dashed arrow).