Abstract
The spike protein (S) of SARS-CoV-2 mediates entry into human cells by interacting with human angiotensin-converting enzyme 2 (ACE2) through its receptor-binding domain (RBD). Here, we report identification of CD209L/L-SIGN and a related protein, CD209/DSIGN as alternative receptors capable of mediating SARS-CoV-2 entry into human cells. Immunofluorescence staining of human tissues revealed a prominent expression of CD209L in the lung and kidney epithelial and endothelial cells of small and medium-sized vessels, whereas CD209 was detected only in a limited number of cell types. Biochemical assays revealed that ectopically expressed CD209L and CD209 bind to S-RBD and mediate SARS-CoV-2 S-pseudotyped virus entry. Furthermore, we demonstrate that human endothelial cells endogenously express CD209L and are permissive to SARS-CoV-2 infection. Soluble CD209L-Fc neutralized virus entry. Our observations show that CD209L and CD209 serve as alternative receptors for SARS-CoV-2 in disease-relevant cell types, including the vascular system. This may have implications for antiviral drug development.
Introduction:
The outbreak of novel coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), has posed a serious threat to global public health with a major worldwide socio-economic impact1–3. Morbidity and mortality of SARS-CoV-2 is associated with acute respiratory distress syndrome (ARDS) and other complications such as coagulopathy, thrombosis and multi-organ failure in COVID-19 patients3–6. Although the role of pulmonary endothelial cells in pathogenesis of COVID-19 remains largely unknown, emerging evidence suggests that SARS-CoV-2 directly attacks the vascular system7–9. Severe endothelial injury, vascular thrombosis with micro-angiopathy and occlusion of alveolar capillaries, and angiogenesis distinctively were observed in the lung autopsies of COVID-19 patients10, underscoring the critical importance of the vasculature system in the pathogenesis of COVID-19.
Human angiotensin-converting enzyme 2 (ACE2) is known to interact with the surface spike (S) protein of SARS-CoV-2 and mediate viral entry11,12. To date, it is not known whether ACE2 is the only receptor utilized by SARS-CoV-2 for cellular entry. While it was previously reported that ACE2 is widely expressed in the lung and other organs13, a recent study demonstrated that ACE2 is expressed at low levels and only in a small subset of lung epithelial cells (Hikmet, et al., Bioxiv, 2020), suggesting that SARS-CoV-2 could infect human cells via alternative receptors. Consistent with this idea, Neuropilin receptor (Cantuti-Castelvetri et al., Bioxiv, 2020; Daly et al., Bioxiv 2020) that is highly expressed in endothelial and neuronal cells and CD147/Basigin (Wang et al., BioxiV 2020) are reported to facilitate SARS-CoV-2 entry. CD147 is expressed in erythrocytes14,15 and endothelial cells of brain and also known to act as a receptor for plasmodium14,16. Neuropilin receptors (Nrp 1 &2) play major roles in VEGF-dependent angiogenesis and semaphorin-dependent axon guidance17. Alternative entry receptors in addition to ACE2 have been reported for other, coronaviruses such as human NL-63 and SARS-CoV. These include CD209L (also known CLEC4M and L-SIGN) and CD209 (also known as DC-SIGN)18–21. Loss of CD209L in mice significantly reduced SARS-CoV infection22, underscoring the critical role of CD209L in SARS-CoV infection. CD209L and CD209 are members of the immunoglobulin-like cell adhesion molecule (IgSF CAM) superfamily receptors23. While CD209L is highly expressed in human type II alveolar cells and lung endothelial cells24,25, CD209 is largely expressed in dendritic cells and tissue resident macrophages, including dermal macrophages26, alveolar macrophages27 and peripheral blood mononuclear cells28,29. However, aside from their differential expression profiles, CD209L and CD209 are highly similar with 79.5% amino acid sequence homology. The most distinguished region of CD209L and CD209 is the C-type lectin domain, which functions as a calcium-dependent glycan-recognition domain30.
In this manuscript, we demonstrate that CD209L and CD209 bind to the receptor-binding domain (RBD) of SARS-CoV-2 S and mediate SARS-CoV-2 entry. CD209L and CD209 are differentially expressed in human lung and kidney epithelial and endothelial cells in a distinct manner. Our data suggests that in addition to ACE2, CD209L and CD209 can be used as alternative receptors to mediate SARS-CoV-2 infection. This has implications for antiviral drug design because CD209L and CD209 represent novel potential therapeutic targets against COVID19.
Materials and Methods:
Plasmids and antibodies:
CD209 cDNA (accession# BC110615), CD209L (accession# BC038851) and ACE2 (accession # BC039902.1) were cloned into retroviral pQCXIP vector. Retroviruses were produced in 293-GPG packaging cells as described31 and viruses were used to express CD209L, CD209 and ACE2 in HEK-293 cells. Chimeric Fc-CD209L, Fc-CoV-2-S-RBD, and CoV-2-S-RBD constructs was generated by PCR amplification and cloned in frame with human Fc fragment of IgE (accession # BC005912) and similarly soluble CD209L (amino acid 71–390) PCR amplified and cloned into pQCXIP-Myc vector. Lai-lucΔenv (Env-deficient HIV-1 containing a luciferase reporter gene in place of Nef), has been described previously32. The lentiviral vector pHAGE-Nanoluciferase was generated via PCR amplification of the nanoluc ORF with the following primers: 5’-ATTGCGGCCGCCATGGTCTTCACACTCGAAGATTTCG-3’ and 5’-TGAGGATCCTTACGCCAGAATGCGTTCGCAC-3’. The PCR amplified Nanoluc orf was subsequently cloned into the pHAGE-Zsgreen lentiviral vector (gracious gift of Dr. Darrell Kotton, BU CReM) using NcoI and BamHI restriction enzymes. The lentiviral packaging plasmid, psPAX2 has been described previously33. The SARS-CoV2 S/gp41 expression plasmid was a gracious gift of Dr. Nir Hachoen (Broad Institute) that expresses a codon-optimized version of CoV2 S and modified to include the eight most membrane-proximal residues of the HIV-1 envelope glycoprotein cytoplasmic domain after residue 1246 of the S protein. Anti-ACE2 antibody (cat# 4355) and anti-CD209 antibody (cat#13193) were purchased from Cell Signaling Technology (Danvers, MA). Anti-CD209L antibody (Cat# AV42396, Sigma-Aldrich) purchased from Sigma-Aldrich.
Cell culture and cell lines:
Vero E6 cells (ATCC CRL-1586), HEK293T cells, HEK-293 cells, HEK-293 cells expressing various constructs and HUVEC-TERT cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), L-glutamine (2mM), penicillin (50 units/ml) and streptomycin (50 mg/ml). Primary human umbilical vein endothelial cells were immortalized by pBABE-TERT-hgro (Plasmid #1773, Addgene) and hereafter named HUVEC-TERT.
SARS-CoV-2 virus propagation:
SARS-CoV-2 isolate USA_WA1/2020 was kindly provided by CDC’s Principal Investigator Natalie Thornburg and the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA). SARS-CoV-2 stocks were grown in Vero E6 cells and virus titers were determined by tissue culture infectious dose 50 (TCID50) assays. All work with SARS-COV-2 was performed in the BSL-4 facility of the National Emerging Infectious Diseases Laboratories (NEIDL) at Boston University following approved SOPs.
Infection of HUVEC-TERT cells and immunofluorescence analysis:
HUVEC-TERT cells (1x105) were seeded in 8-well chamber slides. The next day, cells were infected with SARS-CoV-2 at the indicated multiplicity of infection (MOI). One day post infection, the cells were fixed in 10% neutral buffered formalin for at least 6 hours at 4°C and removed from the BSL-4 laboratory for staining and imaging analysis. In brief, the cells were permeabilized with acetone-methanol solution (1:1) for 5 min at −20°C, incubated in 0.1 M glycine for 10 min at room temperature and subsequently incubated in blocking reagent (2% bovine serum albumin, 0.2% Tween 20, 3% glycerin, and 0.05% NaN3 in PBS) for 20 minutes at room temperature. After each step, the cells were washed three times in PBS. The cells were incubated for one hour at room temperature with a rabbit antibody directed against the SARS-CoV nucleoprotein (Rockland; 1:1000 dilution in blocking reagent), which also cross-reacts with the SARS-CoV-2 nucleoprotein34. The cells were washed four times in PBS and incubated with goat anti-rabbit antibody conjugated with AlexaFluor488 for 1 hour at room temperature (Invitrogen; 1:200 dilution in blocking reagent). 4’,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) was used at 200 ng/ml for nuclei staining. Images were acquired using a Nikon Eclipse Ti2 microscope with Photometrics Prime BSI camera and NIS Elements AR software.
Pseudovirus production and viral entry assay:
Single round nanoluc-expressing lentivirus vectors were generated via transient co-transfection of HEK293T cells with pHAGE-Nanoluc, psPAX2 and SARS COV2 S/gp41 plasmids using calcium chloride and BES buffered saline33. Alternatively, HEK293T cells were co-transfected with Lai-lucΔenv and SARS-CoV2 S/gp41 plasmids for generation of luciferase-expressing single-cycle lentiviruses. Virus containing cell supernatants were harvested 2 days post transfection and filtered using a 0.45µm syringe filters, aliquoted and stored at −80°C until further use33. The p24gag content of the virus stocks was quantified using a p24gag ELISA, as described before33.
Pseudovirus Infections:
HEK293, HEK293-CD209L, HEK293-CD209 and ACE2/HEK-293 cells (2x104/well) were infected by spinoculation, as described before35. Cells were lysed 48 hours post infection, and cell lysates were quantified for firefly luciferase or nanoluciferase activity.
Immunofluorescence staining of human tissues:
Paraffin embedded sections of normal human organs were obtained from the Department of Pathology achieve without any personal identifiers. Tissues were sectioned into 5 micrometer sections and stained were processed for immunofluorescence assay using the following antibody. Rabbit polyclonal anti-CD209L (1:100) dilution, Rabbit polyclonal anti-CD209 (1:100) dilution), anti-mouse CD31 (Abcam, ab9498, 1:100 dilution), anti-mouse MUC1 (Abcam, ab70745, 1:50 dilution), Aquaporin 1 anti-mouse (Abcam, ab9566, 1:50 dilution) were used. Rabbit polyclonal antibody (Abcam, ab37415), and appropriate isotype mouse IgG1 and IgG2 control antibodies were used.
Results:
CD209L is expressed in human lung epithelial and endothelial cells:
To investigate the potential role of CD209L and CD209 in COVID-19, we first examined expression of CD209L in human tissues from SARS-CoV-2 target organs including lung endothelial and epithelial cells, renal vessels, tubules and glomeruli and temporal artery in central nervous system via immunofluorescence staining. Lung tissue was co-stained with CD209L and MUC1, which served as a marker for type II alveolar cells36,37 and examined using laser microscopy. Isotope antibody served as a control and showed negligible staining of the lung tissue (S. Figure 1). Our results showed prominent expression of CD209L in the alveolar cells, which in part co-localized with MUC1 (Figure 1A). We also observed expression of CD209L in pulmonary capillaries (Figure 1B), endothelium of the small and medium sized temporal artery (Figure 1C) and renal arterioles (Figure 1D). CD31 served as a marker of endothelial cells (Figure 1B, C, D). Moreover, we show that CD209L is expressed in the renal proximal tubular epithelial cells marked by aquaporin1 (S. Figure 2A). However, CD209L was not observed in the glomerular capillaries and minimal expression was noted in the mesangial area probably in the infiltrating immune cells (S. Figure 2B). The results demonstrate a prominent expression of CD209L in the type II alveolar cells, pulmonary endothelium as well as renal vessels and renal tubular cells which are also potential cells targeted by SARS-CoV-2.
Figure 1. CD209L is expressed in lung, and renal epithelial and endothelial cells:
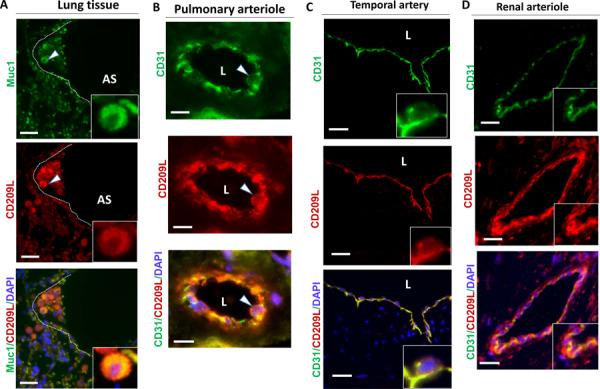
PFA fixed human lung, renal and temporal arteriole tissues were subjected to immunofluorescence staining. Lung tissue stained with anti-MUC1, anti-CD31, and anti-CD209L antibodies. (A) Type II alveoli epithelial cells of alveoli were positive for CD209L (red) and MUC1 (green). (B) Pulmonary arteriole endothelial cells were positive for CD31 (green) and CD209L (red). (C) Endothelial cells of temporal arteriole were stained with CD209L (green) and CD31 (red). (D) Renal endothelial cells were positive for CD209L (red) and CD31 (green). White arrowhead pointing to the alveolar cell and AS= alveolar space and white dotted line corresponds to alveolar septa.
Interestingly, CD209 was expressed only in a subset of type II alveolar cells marked by MUC1 (S. Figure 2A). Furthermore, unlike CD209L which is highly expressed in endothelial cells (S. Figure 1B, C), we did not observe expression of CD209 in the pulmonary or renal arterioles (data not shown). However, a distinct expression of CD209 was observed in the limited proximal tubular epithelial cells in kidneys (S. Figure 2B). Furthermore, glomerular capillaries were also mostly negative for CD209 (S. Figure 2C). Overall, these results suggest that CD209 has a limited expression profile compared to CD209L in the organs examined.
Endothelial cells express CD209L and are infected by SARS-CoV-2:
It has been proposed that the vascular system might be a direct target of SARS-CoV-2 infection8. To explore if human endothelial cells are permissive to SARS-CoV-2, we infected HUVEC-TERT cells (human umbilical vein endothelial cells immortalized with TERT) with SARS-CoV-2 at various multiplicities of infection (MOIs). A robust infection was observed at 1 day post infection, when the cells were infected with SARS-CoV-2 at a high MOI (MOI=15). Even a low MOI of 0.15 led to detectable infection levels at 1 day post infection, indicating that HUVEC-TERT cells are highly permissive to SARS-CoV-2 infection (Figure 2A). Next, we analyzed expression levels of CD209L and ACE2 in HUVEC-TERT, PAE (porcine aortic endothelial cells), and COS-1 (non-endothelial cells transformed green monkey kidney cells). Among the three cell lines tested, the highest levels of CD209L were detected in HUVEC-TERT cells followed by PAE cells (Figure 2B). CD209L was also detected in COS-1 cells, albeit at the significantly low levels (Figure 2B). Curiously, expression of ACE2 in these cells compared to CD209L was relatively low and only overnight incubation with anti-ACE2 antibody detected faint protein bands likely correspond to ACE2 (Figure 2B). Next, we asked whether expression of CD209L in HUVEC-TERT cells can mediate SARS-CoV-2 entry. To this end, we generated SARS-CoV-2 S-pseudotyped lentiviral particles and infected HUVEC-TERT cells. SARS-CoV-2 S-pseudotyped lentiviral particles infected HUVEC-TERT cells in a concentration-dependent manner (Figure 2C). To examine if CD209L promotes SARS-CoV-2 S-mediated entry, we carried out neutralization assays using soluble CD209L encompassing the extracellular domain of CD209L fused to Fc. Soluble CD209L-Fc reduced viral entry by 48% (Figure 2D). Taken together the data demonstrate that endothelial cells express CD209L and soluble CD209L neutralizes SARS-CoV-2 S-pseudotyped viral entry.
Figure 2: Endothelial cells express CD209L and are infected by SARS-CoV-2:
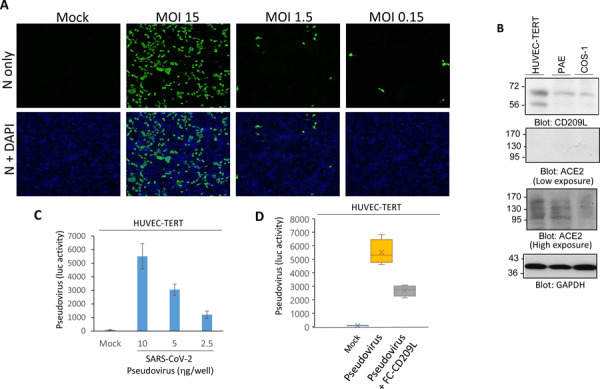
(A) HUVEC-TERT cells seeded in chamber slides were mock-infected or infected with SARS-CoV-2 at the indicated MOIs. Cells were fixed at 1 day post infection, and stained with an antibody directed against the viral nucleoprotein, N (green). Cell nuclei were stained with DAPI (blue). (B) Western blot analysis of HUVEC-TERT, PAE and COS-1 cells for CD209L, ACE2 and loading control, GAPDH. (C) HUVEC-TERT cells (2X104/well, 96-well plate) were infected with SARS-CoV-2 pseudovirus with different concentrations (6 wells/group). After 24hours, cells were processed and subjected to luciferase activity.
To directly examine the role of CD209L and CD209 in mediating SARS-CoV-2 entry, we over-expressed CD209L, CD209 or ACE2 in HEK-293 cells and comparatively analyzed the individual roles of these receptors in SARS-CoV-2 S mediated entry using SARS-CoV-2 S-pseudotyped lentiviral particles. The results show that both CD209L and CD209 were able to facilitate SARS-CoV-2 S-pseudotyped virus entry, albeit to a lesser extent than ACE2 (Figure 3A). Expression of CD209L rendered HEK-293 cells permissive to SARS-CoV-2 infection as shown by immunofluorescence analysis (Figure 3B). Active viral replication was indicated by the presence of double-stranded RNA in the infected cells (Figure 3C).
Figure 3: CD209L and CD209 mediate SARS-CoV-2 spike-pseudotyped cell entry.
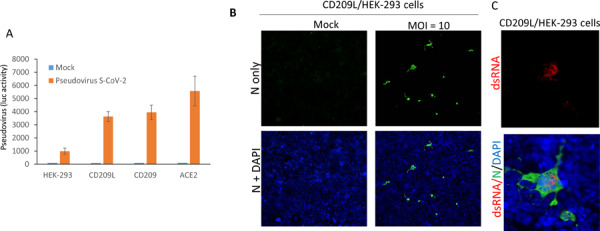
(A) HEK-293 cells expressing CD209L, CD209 or ACE2 (2X104/well, 96-well plate and 6 wells/group) were infected with pseudotyped virus (10ng/ml) or mock virus. After 24hours, cells were analyzed for luciferase activity. (B, C) CD209L/HEK-293 cells seeded in chamber slides were mock-infected or infected with SARS-CoV-2 at the indicated MOI. Cells were fixed at 1 day post infection, and stained with an antibody directed against the viral nucleoprotein, N (green). Cell nuclei were stained with DAPI (blue) or dsRNA.
SARS-CoV-2 spike via its RBD domain binds to CD209L and CD209:
Given that CD209L is able to facilitate SARS-CoV-2 entry, we decided to investigate the physical interaction of CD209L with SARS-CoV-2-S. Specifically, we asked whether the RBD domain of SARS-CoV-2-S binds to CD209L. To this end, we generated a chimeric soluble Fc-S-RBD-protein and used it for immunoprecipitation assays to test its binding with CD209L. The result showed that FC-S-RBD domain binds to CD209L (Figure 4A and B). Next, we comparatively examined the binding of soluble SARS-CoV-2-S-RBD with CD209L, CD209 and ACE2 in a dot blot assay. SARS-CoV-2-S-RBD interacted with CD209L, CD209 and ACE2 in a concentration dependent manner with ACE2 showing the highest biding affinity to RBD (Figure 4E).
Figure 4: CD209L and CD209 bind to SARS-CoV-2-S-RBD.
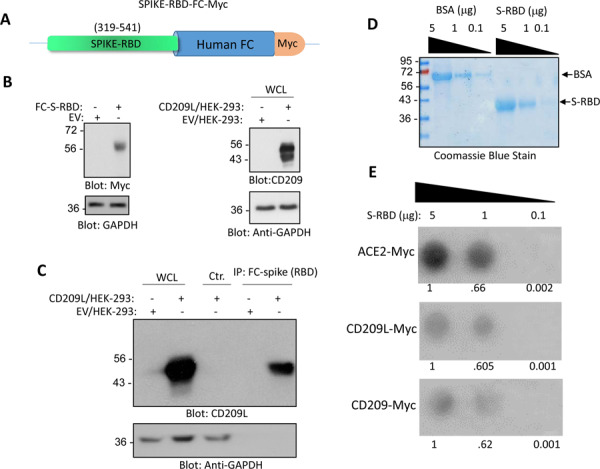
(A) Schematic of Fc-CoV-2-S protein is shown. (B) Expression of Fc-CoV-2-S and CD209L in HEK-293 cells. (C) Immunoprecipitation assay demonstrates the binding of CD209L with Fc-CoV-2-S protein. (D) Coomassie blue stain of CoV-2-S-RBD is shown. (E) CoV-2-S-RBD applied onto PFVD membrane with varying concentrations via Dot blot apparatus. The membranes after blocking with 5%BSA were incubated with cell lysates derived from HEK-293 cells expressing ACE-2-Myc, CD209L or CD209 and binding of ACE2, CD209L and CD209 to CoV-2-S-RBD was detected anti-Myc antibody.
Discussion:
The mechanisms of SARS-CoV-2 infection of human cells remain largely unknown. We demonstrate that CD209L and CD209 serve as attachment factors for SARS-CoV-2 by interacting with the RBD domain of the SARS-CoV-2 spike protein. Recognition of CD209L and CD209 by RBD domain of SARS-CoV-2 S can contribute to virus entry and infection. HUVEC-TERT cells endogenously express CD209L and are permissive to SARS-CoV-2 infection. Furthermore, SARS-CoV-2 S-pseudotyped lentiviral particles infected HUVEC-TERT cells and soluble CD209L-Fc neutralized virus cell entry by nearly 50%, suggesting that SARS-CoV-2 can target human cells via alternative receptors such as CD209L. Consistent with this idea, previous studies demonstrated that CD209L interacts with Ebola virus glycoprotein and mediate endothelial cell infection38,39.
Although CD209L and CD209 can physically interact with SARS-CoV-2 S-RBD and facilitate SARS-CoV-2 entry, ACE2 appears to have a higher affinity to SARS-CoV-2. However, unlike ACE2 which is present at relatively low levels in lung and other organs (Hikmet, et al., Bioxiv, 2020), CD209L is expressed in alveoli type II epithelial cells of lung, renal proximal epithelial cells and blood vessels. Interestingly, unlike ACE2 which is present at relatively low levels in lung and other organs (Hikmet, et al., Bioxiv, 2020), CD209L is highly expressed in alveoli type II epithelial cells of lung, renal proximal epithelial cells and blood vessels. However, we did not observe CD209 expression in blood vessels or lung epithelial cells. Previous studies have shown that CD209 is mostly expressed in dendritic cells, and tissue-resident macrophages, and B cells40, suggesting that these cell types could be targeted by SARS-CoV-2 via recognition of CD209.
Emerging evidence now suggest that the vascular system is a major site of assault by SARS-CoV-27,8 and COVID-19 patients suffer from distinct endothelial injury and altered angiogenesis10. Furthermore, widespread microvascular thrombosis in patients with SARS infection involving kidneys, lungs and other organs were reported3,4. Therefore, future research should investigate the molecular and cellular effects of SARS-CoV-2 in vascular system and explore the selective roles of CD209L versus ACE2 in the vascular system and explore therapeutic potential of CD209L against COVID-19.
Supplementary Material
Acknowledgement:
This work was supported in part through grants from the National institute of health NIH/NCI (R21CA191970, R21CA193958, CTSI grant 1UL1TR001430 and Malory Fund, Department of Pathology, Boston University to N.R.), R01 AI064099 (SG) and R01 AG060890 (SG), the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01-AI133486 (EM) and Evergrande MassCPR sub-award 280870.5116795.0025 (EM) and R01 HL132325 (VCC).
Footnotes
Conflict of interest: Authors declare no conflict of interest.
REFERENCES:
- 1.Bhatraju P.K., et al. Covid-19 in Critically Ill Patients in the Seattle Region - Case Series. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang-Hua Y., et al. Severe acute respiratory syndrome and venous thromboembolism in multiple organs. Am J Respir Crit Care Med 182, 436–437 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Ng K.H., et al. Pulmonary artery thrombosis in a patient with severe acute respiratory syndrome. Postgrad Med J 81, e3 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang N., Li D., Wang X. & Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 18, 844–847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y, W.X., Yang P, Zhang S. COVID-19 Complicated by Acute Pulmonary Embolism. Radiology: Cardiothoracic Imaging 2(2)(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varga Z., et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet 395, 1417–1418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ackermann M., et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monteil V., et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 181, 905–913 e907 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ackermann M., et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. The New England journal of medicine (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuba K., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nature medicine 11, 875–879 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamming I., et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203, 631–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanaguru M., Liu W., Hahn B.H., Rayner J.C. & Wright G.J. RH5-Basigin interaction plays a major role in the host tropism of Plasmodium falciparum. Proc Natl Acad Sci U S A 110, 20735–20740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aniweh Y., et al. P. falciparum RH5-Basigin interaction induces changes in the cytoskeleton of the host RBC. Cell Microbiol 19(2017). [DOI] [PubMed] [Google Scholar]
- 16.Zenonos Z.A., et al. Basigin is a druggable target for host-oriented antimalarial interventions. J Exp Med 212, 1145–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karpanen T., et al. Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J 20, 1462–1472 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Marzi A., et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J Virol 78, 12090–12095 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z.Y., et al. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol 78, 5642–5650 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffers S.A., et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 101, 15748–15753 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffers S.A., Hemmila E.M. & Holmes K.V. Human coronavirus 229E can use CD209L (L-SIGN) to enter cells. Advances in experimental medicine and biology 581, 265–269 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan V.S., et al. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat Genet 38, 38–46 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mateo M., Generous A., Sinn P.L. & Cattaneo R. Connections matter--how viruses use cell-cell adhesion components. J Cell Sci 128, 431–439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang D.M., et al. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod Pathol 18, 1–10 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.To K.F. & Lo A.W. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2). J Pathol 203, 740–743 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soilleux E.J., et al. Placental expression of DC-SIGN may mediate intrauterine vertical transmission of HIV. The Journal of pathology 195, 586–592 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Tailleux L., et al. DC-SIGN induction in alveolar macrophages defines privileged target host cells for mycobacteria in patients with tuberculosis. PLoS Med 2, e381 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bashirova A.A., et al. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. The Journal of experimental medicine 193, 671–678 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mummidi S., et al. Extensive repertoire of membrane-bound and soluble dendritic cell-specific ICAM-3-grabbing nonintegrin 1 (DC-SIGN1) and DC-SIGN2 isoforms. Inter-individual variation in expression of DC-SIGN transcripts. The Journal of biological chemistry 276, 33196–33212 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Zelensky A.N. & Gready J.E. The C-type lectin-like domain superfamily. FEBS J 272, 6179–6217 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Rahimi N., Dayanir V. & Lashkari K. Receptor chimeras indicate that the vascular endothelial growth factor receptor-1 (VEGFR-1) modulates mitogenic activity of VEGFR-2 in endothelial cells. The Journal of biological chemistry 275, 16986–16992 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Miller C.M., et al. Virion-Associated Vpr Alleviates a Postintegration Block to HIV-1 Infection of Dendritic Cells. J Virol 91(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akiyama H., et al. HIV-1 intron-containing RNA expression induces innate immune activation and T cell dysfunction. Nat Commun 9, 3450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thao T.T.N., et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature (2020). [DOI] [PubMed] [Google Scholar]
- 35.Akiyama H., et al. HIV-1 intron-containing RNA expression induces innate immune activation and T cell dysfunction. Nature communications 9, 3450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo S.H., et al. Expression patterns of markers for type II pneumocytes in pulmonary sclerosing hemangiomas and fetal lung tissues. Archives of pathology & laboratory medicine 129, 915–919 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Jarrard J.A., et al. MUC1 is a novel marker for the type II pneumocyte lineage during lung carcinogenesis. Cancer Res 58, 5582–5589 (1998). [PubMed] [Google Scholar]
- 38.Simmons G., et al. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305, 115–123 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Alvarez C.P., et al. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. Journal of virology 76, 6841–6844 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rappocciolo G., et al. DC-SIGN on B lymphocytes is required for transmission of HIV-1 to T lymphocytes. PLoS pathogens 2, e70 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


