Abstract
The COVID-19 pandemic has created a public health crisis. Because SARS-CoV-2 can spread from individuals with pre-symptomatic, symptomatic, and asymptomatic infections, the re-opening of societies and the control of virus spread will be facilitated by robust surveillance, for which virus testing will often be central. After infection, individuals undergo a period of incubation during which viral titers are usually too low to detect, followed by an exponential viral growth, leading to a peak viral load and infectiousness, and ending with declining viral levels and clearance. Given the pattern of viral load kinetics, we model surveillance effectiveness considering test sensitivities, frequency, and sample-to-answer reporting time. These results demonstrate that effective surveillance depends largely on frequency of testing and the speed of reporting, and is only marginally improved by high test sensitivity. We therefore conclude that surveillance should prioritize accessibility, frequency, and sample-to-answer time; analytical limits of detection should be secondary.
Introduction
Successful surveillance testing of SARS-CoV-2 depends on understanding both the dynamics of spread between individuals and the dynamics of the virus within the human body. Critically, the ability of SARS-CoV-2 to spread from individuals who are pre-symptomatic, symptomatic, or essentially asymptomatic [1, 2, 3] means that diagnosis and isolation based on symptoms alone will be unable to prevent ongoing spread [4, 5]. As a consequence, the use of surveillance testing to identify infectious individuals presents one possible means to break enough transmission chains to suppress the ongoing pandemic and reopen societies, with or without a vaccine.
The reliance on testing as a means to safely reopen societies has placed a microscope on the analytical sensitivity of virus assays, with a gold-standard of quantitative real-time polymerase chain reaction (qPCR). These assays have analytical limits of detection that are usually within around 103 viral RNA copies per ml (cp/ml) [6]. However, qPCR remains expensive and as a laboratory based assay often have sample-to-result times of 24–48 hours. New developments in SARS-CoV-2 diagnostics have the potential to reduce cost significantly, allowing for expanded testing or greater frequency of testing and can reduce turnaround time to minutes [7, 8, 9]. These assays however largely do not meet the gold standard for analytical sensitivity, which has encumbered the widespread use of these assays [10].
Three features of the viral increase, infectivity, and decline during SARS-CoV-2 infection led us to hypothesize that there might be minimal differences in effective surveillance using viral detection tests of different sensitivities, such as RT-qPCR with a limit of detection (LOD) at 103 cp/ml [6] compared to often cheaper or faster assays with higher limits of detection (i.e., around 105 cp/ml [7, 8, 9]) such as point-of-care nucleic acid LAMP and rapid antigen tests (Figure 1A). First, since filtered samples collected from patients displaying less than 106 N or E RNA cp/ml contain minimal or no measurable infectious virus [11, 12, 13], either class of test should detect individuals who are currently infectious. The absence of infectious particles at viral RNA concentrations < 106 cp/ml is likely due to (i) the fact that the N and E RNAs are also present in abundant subgenomic mRNAs, leading to overestimation of the number of actual viral genomes by ~100–1000X [14], (ii) technical artifacts of RT-PCR at Ct values > 35 due to limited template [15, 16], and (iii) the production of non-infectious viral particles as is commonly seen with a variety of RNA viruses [17]. Second, during the exponential growth of the virus, the time difference between 103 and 105 cp/ml is short, allowing only a limited window in which only the more sensitive test could diagnose individuals. For qPCR, this corresponds to the time required during viral growth to go from Ct values of 40 to ~34. While this time window for SARS-CoV-2 is not yet rigorously defined [18], for other respiratory viruses such as influenza, and in ferret models of SARS-CoV-2 transmission, it is on the order of a day [19, 20]. Finally, high-sensitivity screening tests, when applied during the viral decline accompanying recovery, are unlikely to substantially impact transmission because such individuals detected have low, if any, infectiousness [14]. Indeed, a recent review by Cevik et al [18] notes that no study to date has successfully cultured live virus more than 9 days after the onset of symptoms.
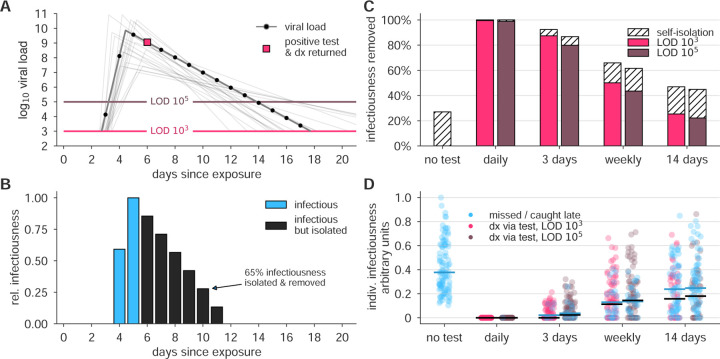
Figure 1: Surveillance testing effectiveness depends on frequency.
(A) An example viral load trajectory is shown with LOD thresholds of two tests, and a hypothetical positive test on day 6, two days after peak viral load. 20 other stochastically generated viral loads are shown to highlight trajectory diversity (light grey; see Methods). (B) Relative infectiousness for the viral load shown in panel A pre-test, totaling 35% (blue) and post-isolation, totaling 65% (black). (C) Surveillance programs using tests at LODs of 103 and 105 at frequencies indicated were applied to 10, 000 individuals’ trajectories of whom 35% would undergo symptomatic isolation near their peak viral load if they had not been tested and isolated first. Total infectiousness removed during surveillance (colors) and self isolation (hatch) are shown for surveillance as indicated, relative to total infectiousness with no surveillance or self-isolation. (D) The impact of surveillance on the infectiousness of 100 individuals is shown for each surveillance program and no testing, as indicated, with each individual colored by test if their infection was detected during infectiousness (medians, black lines) or colored blue if their infection was missed by surveillance or detected positive after their infectious period (medians, blue lines). Units are arbitrary and scaled to the maximum infectiousness of sampled individuals.
Results
Impact of surveillance on individuals
To examine how surveillance testing would reduce the average infectiousness of individuals, we first modeled the viral loads and infectiousness curves of 10,000 simulated individuals using the predicted viral trajectories of SARS-CoV-2 infections based on key features of latency, growth, peak, and decline identified in the literature (Figure 1A; see Methods). Accounting for these within-host viral kinetics, we calculated what percentage of their total infectiousness would be removed by surveillance and isolation (Figure 1B) with tests at LOD of 103 and 105, and at different frequencies. Here, infectiousness was taken to be proportional to the logarithm of viral load in excess of 106 cp/ml (with alternative assumptions addressed in sensitivity analyses; see Supplemental Materials), consistent with the observation that pre-symptomatic patients are most infectious just prior to the onset of symptoms [21], and evidence that the efficiency of viral transmission coincides with peak viral loads, which was also identified during the related 2003 SARS outbreak [22, 23]. We considered that 35% of patients would undergo symptomatic isolation within three days of their peak viral load if they had not been tested and isolated first, and 65% would have sufficiently mild or no symptoms such that they would not isolate unless they were detected by surveillance testing. Based on recent results, we modeled asymptomatic and symptomatic infections as having the same initial viral loads [1, 24, 25, 26], but with faster clearance among asymptomatics [24, 26, 27, 28, 29] (see Methods). This analysis demonstrated that there was little difference in averting infectiousness between the two classes of test. Dramatic reductions in total infectiousness of the individuals were observed by testing daily or every third day, ~65% reduction when testing weekly, and < 50% under biweekly testing (Figure 1C). Because viral loads and infectiousness vary across individuals, we also analyzed the impact of different surveillance regimens on the distribution of individuals’ infectiousness, revealing that more sporadic testing leads to an increased likelihood that individuals will test positive after they are no longer infectious or be missed by testing entirely (Figure 1D).
Impact of surveillance on a population
Above, we assumed that each infection was independent. To investigate the effects of surveillance testing strategies at the population level, we used simulations to monitor whether epidemics were contained or became uncontrolled, while varying the frequencies at which the test was administered, ranging from daily testing to testing every 14 days, and considering tests with LOD of 103 and 105, analogous to RT-qPCR and RT-LAMP / rapid antigen tests, respectively. We integrated individual viral load trajectories into two different epidemiological models to ensure that important observations were independent of the specific modeling approach. The first model is a simple fully mixed model representing a population of 20,000, similar to a large university setting, with a constant rate of external infection approximately equal to one new import per day. The second model is a previously described agent-based model with both within-household and age-stratified contact structure based on census microdata in a city representative of New York City [30], which we initialized with 100 cases without additional external infections. Individual viral loads were simulated for each infection, and individuals who received a positive test result were isolated, but contact tracing and monitoring was not included to more conservatively estimate the impacts of surveillance alone [31, 32]. Model details and parameters are fully described in Methods.
We observed that a surveillance program administering either test with high frequency limited viral spread, measured by both a reduction in the reproductive number R (Figures 2A and B; see Methods for calculation procedure) and by the total infections that persisted in spite of different surveillance programs, expressed relative to no surveillance (Figures 2C and D). Testing frequency was found to be the primary driver of population-level epidemic control, with only a small margin of improvement provided by using a more sensitive test. Direct examination of simulations showed that with no surveillance or biweekly testing, infections were uncontrolled, whereas surveillance testing weekly with either LOD = 103 or 105 effectively attenuated surges of infections (examples shown in Figure S1).
Figure 2: Surveillance testing affects disease dynamics.
Both the fully-mixed compartmental model (top row) and agent based model (bottom row) are affected by surveillance programs. (A, B) More frequent testing reduces the effective reproductive number R, shown as the percentage by which R0 is reduced, 100 × (R0 – R)/R0. Values of R were estimated from 50 independent simulations of dynamics (see Methods). (C, D) Relative to no testing (grey bars), surveillance suppresses the total number of infections in both models when testing every day or every three days, but only partially mitigates total cases for weekly or bi-weekly testing. Error bars indicate inner 95% quantiles of 50 independent simulations each.
The relationship between test sensitivity and the frequency of testing required to control outbreaks in both the fully mixed model and the agent-based model generalize beyond the examples shown in Figure 2 and are also seen at other testing frequencies, sensitivities, and asymptomatic fractions. We simulated both models at LODs of 103, 105, and 106, and for testing ranging from daily to every 14 days. For those, we measured each surveillance policy’s impact on total infections (Figure S2A and B) and on R (Figure S2C and D). In Figure 2, we modeled infectiousness as proportional to log10 of viral load. To address whether these finding are sensitive to this modeled relationship, we performed similar simulations with infectiousness proportional to viral load (Figure S3), or uniform above 106/ml (Figure S4). We found that results were robust to these large variations in the modeled relationship between infectiousness and viral load. To further address whether our results depended on the exact 35% fraction of individuals assumed to be behaviorally symptomatic, we performed sensitivity analyses with fewer (20%) or more (50%) symptomatic individuals and found no meaningful difference in results (Figure S5).
Impact of delayed test results
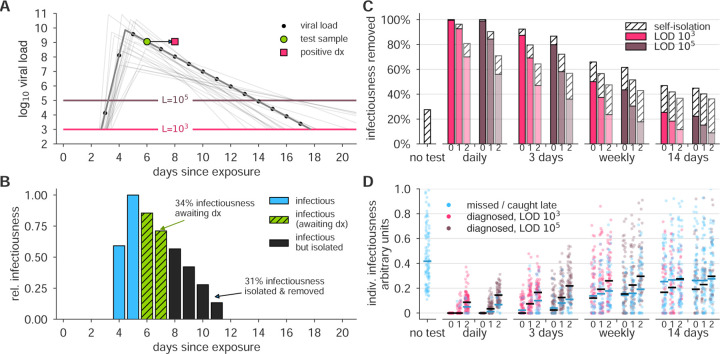
An important variable in surveillance testing is the time between a test’s sample collection and the reporting of a diagnosis. To examine how time to reporting affected epidemic control, we re-analyzed both the reduction in individuals’ infectiousness, as well as the epidemiological simulations, comparing the results of instantaneous reporting (reflecting a rapid point-of-care assay), one day delay, and two day delay (Figure 3A and B). Delays in reporting dramatically decreased the reduction in infectiousness in individuals as seen by the total infectiousness removed (Figure 3C), the distribution of infectiousness in individuals (Figure 3D), or the dynamics of the epidemiological models (Figure 4). This result was robust to the modeled relationship between infectiousness and viral load in both simulation models and for various test sensitivities and frequencies (Figure S6). These results highlight that delays in reporting lead to dramatically less effective control of viral spread and emphasize that fast reporting of results is critical in any surveillance testing. These results also reinforce the relatively smaller benefits of improved limits of detection.
Figure 3: Effectiveness of surveillance testing is compromised by delays in reporting.
(A) An example viral load trajectory is shown with LOD thresholds of two tests, and a hypothetical positive test on day 6, but with results reported on day 8. 20 other stochastically generated viral loads are shown to highlight trajectory diversity (light grey; see Methods). (B) Relative infectiousness for the viral load shown in panel A pre-test (totaling 35%; blue) and post-test but pre-diagnosis (totaling 34%; green), and post-isolation (totaling 31%; black). (C) Surveillance programs using tests at LODs of 103 and 105 at frequencies indicated, and with results returned after 0, 1, or 2 days (indicated by small text beneath bars) were applied to 10, 000 individuals trajectories of whom 35% were symptomatic and self-isolated after peak viral load if they had not been tested and isolated first. Total infectiousness removed during surveillance (colors) and self isolation (hatch) are shown, relative to total infectiousness with no surveillance or self-isolation. Delays substantially impact the fraction of infectiousness removed. (D) The impact of surveillance with delays in returning diagnosis of 0, 1, or 2 days (small text beneath axis) on the infectiousness of 100 individuals is shown for each surveillance program and no testing, as indicated, with each individual colored by test if their infection was detected during infectiousness (medians, black lines) or colored blue if their infection was missed by surveillance or diagnosed positive after their infectious period (medians, blue lines). Units are arbitrary and scaled to the maximum infectiousness of sampled individuals.
Figure 4: Delays in reporting decrease the epidemiological impact of surveillance-driven isolation.
The effectiveness of surveillance programs are dramatically diminished by delays in reporting in both the fully-mixed compartmental model (top row) and agent based model (bottom row). (A, B) The impact of surveillance every day, 3 days, weekly, or biweekly, on the reproductive number R, calculated as 100 × (R0 – R)/R0, is shown for LODs 103 and 105 and delays of 0, 1, or 2 days (small text below axis). Values of R were estimated from 50 independent simulations of dynamics (see Methods). (C, D) Relative to no testing (grey bars), surveillance suppresses the total number of infections in both models when testing every day or every three days, but delayed results lead to only partial mitigation of total cases, even for testing every day or 3 days. Error bars indicate inner 95% quantiles of 50 independent simulations each.
Generality of findings to changes in modeling assumptions
Communities vary in their transmission dynamics, due to difference in rates of imported infections and in the basic reproductive number R0, both of which will influence the frequency and sensitivity with which surveillance testing must occur. We performed two analyses to illustrate this point. First, we varied the rate of external infection in our fully mixed model, and confirmed that when the external rate of infection is higher, more frequent surveillance is required to prevent outbreaks (Figure S7A). Second, we varied the reproductive number R0 between infected individuals in both models, and confirmed that at higher R0, more frequent surveillance is also required (Figure S7B and C). This may be relevant to institutions like college campuses or military bases wherein frequent classroom setting or dormitory living are likely to increase contact rates. Thus, the specific strategy for successful surveillance will depend on the current community infection prevalence and transmission rate.
The generality of our findings to different epidemiological parameters (Figure S7), relationships between viral load and infectiousness (Figures S3 and S4), and proportion of symptomatic individuals (Figure S5) led us to ask whether a more general mathematical formula could predict R without requiring epidemiological simulation. We derived such a formula (Supplemental Text S1) and found that its predicted values of R were nearly perfectly correlated with simulation-estimated values (Pearson’s r = 0.998, p < 10−6; Figure S8), providing a mathematical alternative to simulation-based sensitivity analyses.
Surveillance testing to mitigate an ongoing epidemic
The impact of surveillance testing on transmission dynamics led us to hypothesize that surveillance testing could be used as an active tool to mitigate an ongoing epidemic. To test this idea, we simulated an outbreak situation using both the fully-mixed and agent-based models but with three additional conditions. First, we assumed that in an ongoing pandemic, other mitigating interventions would cause the reproductive number to be lower, though nevertheless larger than one. Second, we considered that not all individuals would want to or be able to participate in a SARS-CoV-2 surveillance program. Third, we assumed that the collection of samples for testing, if performed on a large scale, could result in imperfect sample collection, causing an increase in the false negative rate, independent of an assay’s analytical sensitivity. These modifications are fully described in Methods.
We simulated epidemics in which surveillance testing began only at the point when uncontrolled infections reached 4% prevalence. Based on results from our previous analyses, we considered a less sensitive but rapid test with LOD 105 cp/ml and a zero-day delay in results, and further assumed that 10% of would-be positive samples would be negative due to improper sample collection. We then examined scenarios of testing every 3 days and every 7 days, with either 50% or 75% of individuals participating, starting from a partially mitigated R0 = 1.5. We found that surveillance testing of 75% of individuals every 3 days was sufficient to drive the epidemic toward extinction within 6 weeks and reduce cumulative incidence by 88%, and that other combinations also had successful but less rapid mitigating impacts, particularly when compared with no intervention (Figure 5). Notably, even weekly testing with 50% participation was able to reduce the peak and length of the outbreak, illustrating how even partial surveillance testing using a test with 100X lower molecular sensitivity than PCR can have public health benefits when used frequently (Figure 5). Repeating these simulations using a test with LOD 106 led to similar results (Figure S9). To further generalize these results, we modified our mathematical formula to predict the impacts of per-individual test refusal and pertest sampling-related sensitivity on the reproductive number R (see Supplemental Text S1).
Figure 5: Surveillance testing suppresses an ongoing epidemic.
Widespread testing and isolation of infected individuals drives prevalence downward for both (A) the fully-mixed compartmental model and (B) the agent based model. Time-series of prevalence, measured as the total number of infectious individuals, are shown for no intervention (solid) and surveillance testing scenarios (various dashed; see legend). Surveillance testing began only when prevalence reached 4% (box), and time series are shifted such that testing begins at t = 0. Scenarios show the impact of a test with LOD 105, no delay in results, and with 10% of samples assumed to be incorrectly collected (and therefore negative) to reflect decreased sensitivity incurred at sample collection in a mass testing scenario. Annotations show total number of post-intervention infections, as a percentage of the no-intervention scenario, labeled as 100%. See Fig. S9 for identical simulations using a test with LOD 106.
Discussion
Our results lead us to conclude that surveillance testing of asymptomatic individuals can be used to limit the spread of SARS-CoV-2. However, our findings are subject to a number of limitations. First, the sensitivity of a test may depend on factors beyond LOD, including manufacturer variation and improper clinical sampling [33], though the latter may be ameliorated by different approaches to sample collection, such as saliva-based testing [34]. Second, the exact performance differences between testing schemes will depend on whether our model truly captures viral kinetics and infectiousness profiles [21], particularly during the acceleration phase between exposure and peak viral load. Continued clarification of these within-host dynamics would increase the impact and value of this, and other [31, 32, 35, 36] modeling studies. Finally, we modeled participation in surveillance testing regimens (or refusal thereof) as statistically independent between individuals, but health-related behaviors have been shown to be socially [37] and geographically [38, 39] correlated. Clustered refusal of surveillance testing, or refusal to isolate upon testing positive, could present challenging barriers to implementation.
Our findings show that the impact of surveillance testing can be expressed as a reduction of the reproductive number R. By mapping a given testing regimen to a reduction in R, the impact of testing regimen can be approximated and generalized without complicated simulations. For instance, one could estimate the maximum allowable turnaround time delays, or the minimum testing frequency required to bring R below one, based on user-specified and scenario-specific assumptions. To facilitate such generalizations and scenario planning, open-source calculation tools accompany this manuscript.
A critical point is that the requirements for surveillance testing are distinct from clinical testing. Clinical diagnoses target symptomatic individuals, need high accuracy and sensitivity, and are not limited by cost. Because they focus on symptomatic individuals, those individuals can isolate such that a diagnosis delay does not lead to additional infections. In contrast, results from the surveillance testing of asymptomatic individuals need to be returned quickly, since even a single day diagnosis delay compromises the surveillance program’s effectiveness. Indeed, at least for viruses with infection kinetics similar to SARS-CoV-2, we find that speed of reporting is much more important than sensitivity, although more sensitive tests are nevertheless somewhat more effective.
The difference between clinical and surveillance testing highlights the need for additional tests to be approved and utilized for surveillance. Such tests should not be held to the same degree of sensitivity as clinical tests, in particular if doing so encumbers rapid deployment of faster cheaper SARS-CoV-2 assays. We suggest that the FDA, other agencies, or state governments, encourage the development and use of alternative faster and lower cost tests for surveillance purposes, even if they have poorer limits of detection. If the availability of point-of-care or self-administered surveillance tests leads to faster turnaround time or more frequent testing, our results suggest that they would have high epidemiological value.
Our modeling suggests that some types of surveillance will subject some individuals to unnecessary quarantine days. For instance, the infrequent use of a sensitive test will not only identify (i) those with a low viral load in the beginning of the infection, who must be isolated to limit viral spread, but (ii) those in the recovery period, who still have detectable virus or RNA but are below the infectious threshold [13, 14]. Isolating this second group of patients will have no impact on viral spread but will incur costs of isolation, as would the isolation of individuals who received a false positive test result due to imperfect test specificity. The use of serology, repeat testing 24 or 48 hours apart, or some other test, to distinguish low viral load patients on the upslope of infection from those in the recovery phase could allow for more effective quarantine decisions.
Materials and Methods
Viral Loads
Viral loads were drawn from a simple viral kinetics model intended to capture (1) a variable latent period, (2) a rapid growth phase from the lower limit of PCR detectability to a peak viral load, (3) a slower decay phase, and (4) prolonged clearance for symptomatic infections vs asymptomatic infections. These dynamics were based on the following observations.
Latent periods prior to symptoms have been estimated to be around 5 days [40]. Latent periods prior to detection via virological tests at secondary sites of replication or shedding have been estimated to be up to 4 days [41], corresponding to a latent or eclipse phase observed with other viruses [42]. Viral load appears to peak prior to symptom onset [21], and peaks within 2 days of challenge in a macaque model [43, 44], though it should be noted that macaque challenge doses were high. Viral load decreases monotonically from the time of symptom onset [21, 45, 46, 47, 48], but may be high and detectable 3 or more days before symptom onset [1, 49]. Peak viral loads are difficult to measure due to lack of prospective sampling studies of individuals prior to exposure and infection, but viral loads have been reported in the range of to copies per ml [12, 47, 48]. Viral loads appear to become undetectable by PCR within 3 weeks of symptom onset [45, 48, 50], but detectability and timing may differ depending on the degree or presence of symptoms [50, 51]. The majority of studies reviewed by Cevik et al [18] found initial viral loads to be similar between symptomatic and asymptomatic infections [1, 24, 25, 26], but viral clearance was significantly and substantially faster among asymptomatic infections [24, 26, 27, 28, 29]. Finally, we note that the general understanding of viral kinetics may vary depending on the mode of sampling, as demonstrated via a comparison between sputum and swab samples [12]. For a comprehensive review of viral load dynamics, duration of shedding, and infectiousness, see Ref. [18].
To mimic growth and decay, log10 viral loads were specified by a continuous piecewise linear “hinge” function, specified uniquely with three control points: (t0, 3), (tpeak, Vpeak),(tf, 6) (Figure 6A; green squares). The first point represents the time at which an individual’s viral load first crosses 103, with t0 ~ unif[2.5, 3.5], measured in days since exposure. The second point represents the peak viral load. Peak height was drawn Vpeak ~ unif[7, 11], and peak timing was drawn with respect to the start of the exponential growth phase, tpeak – t0 ~ 0.5 + gamma(1.5) with a maximum of 3. The third point represents the time at which an individual’s viral load crosses beneath the 106 threshold, at which point viral loads no longer cause active cultures in laboratory experiments [11, 12, 13, 18]. For asymptomatic infections, this point was drawn with respect to peak timing, tf – tpeak ~ unif[4, 9]. For symptomatic infections, a symptom onset time was first drawn with respect to peak timing, tsymptoms – tpeak ~ unif[0, 3], and then the third control point was drawn with respect to symptom onset, tf – tsymptoms ~ unif[4, 9]. Thus, symptomatic trajectories are systematically longer, in both duration of infectiousness (see below) and duration of viral shedding, reflecting the documented prolonged clearance and relationship with viral culture experiments (Figure 6B; red circles). In simulations, each viral load’s parameters were drawn independently of others, and the continuous function described here was evaluated at 28 integer time points (Figure 6; black dots) representing a four week span of viral load values.
Figure 6: Example asymptomatic and symptomatic viral loads with model control points.

Examples of model viral loads (lines) and corresponding stochastically drawn control points (squares, circles) are shown for (A) an asymptomatic viral load trajectory and (B) a symptomatic viral load trajectory. Because simulations took place in discrete time, black dots show points at which this example viral load would have been sampled. Light grey lines show 20 alternative trajectories in each panel to illustrate the diversity of viral loads drawn from the simple model. Red circles indicate the control points which are modified in symptomatic trajectories to account for symptom onset and prolonged time till clearance.
Infectiousness
Infectiousness F was assumed to be directly related to viral load V in one of three ways. In the main text, each individual’s relative infectiousness was proportional log10 of viral load’s excess beyond 106, i.e. . In the supplementary sensitivity analyses, we investigated two opposing extremes. To capture a more extreme relationship between infectiousness and viral load, we considered F to be directly proportional to viral load’s excess above 106, i.e. , and to capture a more extreme relationship, but in the opposing direction, we considered F to simply be a constant when viral load exceeded 106, i.e. . We call these three functions log-proportional, proportional, and threshold throughout the text and supplemental materials.
We note that a comprehensive review of viral loads, shedding, and infectiousness [18] found that across the surveyed literature, no virus was able to be cultured beyond 9 days post-symptoms. Thus, the choice of the final control point in our symptomatic viral load model (Figure 6B), which corresponds to the latest time at which an individual is infectious, is at most 9 days post-symptom onset.
Recently, He et al [21] published an analysis of infectiousness relative to symptom onset which was corrected by Bonhoeffer et al (see [21] for details). Among our infectiousness functions, this inferred relationship bears the greatest similarity, over time, to the log-proportional infectiousness function, as visualized in Figs. 1 and 3. The proportional and threshold models therefore represent one of many types of sensitivity analysis. Results for those models can be found in Figures S3, S4, and S6.
In all simulations, the value of the proportionality constant implied by the infectiousness functions above was chosen to achieve the targeted value of R0 for that simulation, and confirmed via simulation as described below.
Disease Transmission Models
Overview
Two models were used to simulate SARS-CoV-2 dynamics, both based on a typical compartmental framework. The first model was a fully-mixed model of N = 20, 000 individuals with all-to-all contact structure, zero initial infections, and a constant 1/N per-person probability of becoming infected from an external source. This model could represent, for instance, a large college campus with high mixing, situated within a larger community with low-level disease prevalence. The second model was an agent-based model of N = 8.4 million agents representing the population and contact structure of New York City, as previously described [30]. Contact patterns were based on a combination of individual-level household contacts drawn from census microdata and age-stratified contact matrices which describe outside of household contacts. This model was initialized with 100 initial infections and no external sources of infection.
Both the fully-mixed and agent-based models tracked discrete individuals who were Susceptible (S), Infected (I), Recovered (R), Isolated (Q), and Self-Isolated (SQ) at each discrete one day timestep. Upon becoming infected (S → I), a viral load trajectory V (t) was drawn which included a latent period, growth, and decay. Each day, an individual’s viral load trajectory was used to determine whether their diagnostic test would be positive if administered, as well as their infectiousness to susceptible individuals. Based on a schedule of testing each person every D days, if an individual happened to be tested on a day when their viral load exceeded the limit of detection L of the test, their positive result would cause them to isolate (I → Q), but with the possibility of a delay in turnaround time. A fraction 1 – f of individuals self-isolate on the day of symptom onset, which occurs 0 to 3 days after peak viral load, to mimic symptom-driven isolation (I → SQ), with f = 0.65 for both models, with f = 0.8 and f = 0.5 explored in sensitivity analyses (Figure S5). Presymptomatic individuals were isolated prior to symptom onset only if they received positive test results. When an individual’s viral load dropped below 103, that individual recovered (I, Q, SQ → R). Details follow.
Testing, Isolation, and Sample-to-Answer Turnaround Times
All individuals were tested every D days, so that they could be moved into isolation if their viral load exceeded the test’s limit of detection V (t) > L. Each person was deterministically tested exactly every D days, but testing days were drawn uniformly at random such that not all individuals were tested on the same day. To account for delays in returning test results, we included a sample-to-answer turnaround time T, meaning that an individual with a positive test on day t would isolate on day t + T.
Transmission, Population Structure, and Mixing Patterns: Fully-mixed model
Simulations were initialized with all individuals susceptible, S = N. Each individual was chosen to be symptomatic independently with probability f, and each individual’s first test day (e.g. the day of the week that their weekly test would occur) was chosen uniformly at random between 1 and D. Relative infectiousness was scaled up or down to achieve the specified R0 in the absence of any testing policy, but inclusive of any assumed self-isolation of symptomatics.
In each timestep, those individuals who were marked for testing that day were tested, and a counter was initialized to T, specifying the number of days until that individual received their results. Next, individuals whose test results counters were zero were isolated, I → Q. Then, symptomatic individuals whose viral load had declined relative to the previous day were self-isolated, I → SQ. Next, each susceptible individual was spontaneously (externally) infected independently with probability 1/N, S → I. Then, all infected individuals contacted all susceptible individuals, with the probability of transmission based on that day’s viral load V (t) for each person and the particular infectiousness function, described above, S → I.
To conclude each time step, individuals’ viral loads and test results counters were advanced, with those whose infectious period had completely passed moved to recovery, I, Q, SQ → R.
Transmission, Population Structure, and Mixing Patterns: Agent-based model
The agent-based model added viral kinetics and testing policies (as described above) to an existing model for SARS-CoV-2 transmission in New York City. A full description of the agent-based model is available [30]; here we provide an overview of the relevant transmission dynamics.
Simulations were initialized with all individuals susceptible, except for 100 initially infected individuals, S = N – 100. As in the fully-mixed model, each individual’s test day was chosen uniformly at random and relative infectiousness was scaled to achieve the specified R0.
In each timestep, those individuals who were marked for testing that day were tested, and a counter was initialized to T, specifying the number of days until that individual received their results. Next, individuals whose test results counters were zero were isolated, I → Q. There was no self-isolation in this model (and accordingly, the model did not label individuals as symptomatic or asymptomatic).
Then, transmission from infected individuals to susceptible individuals was simulated both within and outside households. To model within-household transmission, each individual had a set of other individuals comprising their household. Household structures, along with the age of each individual, were sampled from census microdata for New York City [52]. The probability for an infectious individual to infect each of their household members each day was determined by scaling the relative infectiousness values to match the estimated secondary attack rate for close household contacts previously reported in case cluster studies [53].
Outside of household transmission was simulated using age-stratified contact matrices, which describe the expected number of daily contacts between an individual in a given age group and those in each other age group. Each infectious individual of age i drew Poisson(Mij) contacts with individuals in age group j, where M is the contact matrix. The contacted individuals were sampled uniformly at random from age group j. We use a contact matrix for the United States estimated by [54]. Each contact resulted in infection, S → I, with probability proportional to the relative infectiousness of the infected individual on that day, scaled to obtain the specified value of R0.
To conclude each time step, individuals’ viral loads and test results counters were advanced, with those whose infectious period had completely passed moved to recovery, I, Q → R.
Calibration to achieve targeted R0 and estimation of R
As a consistency check, each simulation’s R0 was estimated as follows, to ensure that simulations were properly calibrated to their intended values. Note that to vary R0, the proportionality constant in the function that maps viral load to infectiousness need only be adjusted up or down. In a typical SEIR model, this would correspond to changing the infectiousness parameter which governs the rate at which I-to-S contacts cause new infections β.
For the fully-mixed, the value of R0 was numerically estimated by running single-generation simulations in which a 50 infected individual were placed in a population of N – 50 others. The number of secondary infections from those initially infected was recorded and used to directly estimate R0.
For the agent-based model, the value of R0 depends on the distribution of infected agents due to stratification by age and household. We numerically estimate R0 by averaging over the number of secondary infections caused by each agent who was infected in the first 15 days of the simulation (at which point the population is still more than 99.99% susceptible).
Estimations of R proceeded exactly as estimations of R0 for both models, except with interventions applied to the the viral loads and therefore the dynamics. Prediction of R without direct simulation is described in Supplemental Text S1.
Supplementary Material
Acknowledgements
The authors wish to thank the BioFrontiers Institute IT HPC group. This work was supported by grants NIH F32 AI145112 (James Burke), NIH F30 AG063468 (Evan Lester), MURI W911NF1810208 (Bryan Wilder, Milind Tambe), an NIH directors DP5 award 1DP5OD028145-01 (Michael Mina), and the Howard Hughes Medial Institute (Roy Parker).
References
- [1].Arons Melissa M, Hatfield Kelly M, Reddy Sujan C, Kimball Anne, James Allison, Jacobs Jesica R, Taylor Joanne, Spicer Kevin, Bardossy Ana C, Oakley Lisa P, et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. New England Journal of Medicine, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sutton Desmond, Fuchs Karin, D’alton Mary, and Goffman Dena. Universal screening for SARS-CoV-2 in women admitted for delivery. New England Journal of Medicine, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Oran Daniel P and Topol Eric J. Prevalence of asymptomatic SARS-CoV-2 infection: A narrative review. Annals of Internal Medicine, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moghadas Seyed M, Fitzpatrick Meagan C, Sah Pratha, Pandey Abhishek, Shoukat Affan, Singer Burton H, and Galvani Alison P. The implications of silent transmission for the control of covid-19 outbreaks. Proceedings of the National Academy of Sciences, 117(30):17513–17515, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grassly Nicholas C, Margarita Pons-Salort, Parker Edward P K, White Peter J, Ferguson Neil M, and “The Imperial College COVID-19 Response Team”. Comparison of molecular testing strategies for covid-19 control: a mathematical modelling study. The Lancet Infectious Diseases, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vogels Chantal BF, Brito Anderson F, Wyllie Anne Louise, Fauver Joseph R, Ott Isabel M, Kalinich Chaney C, Petrone Mary E, Landry Marie-Louise, Foxman Ellen F, and Grubaugh Nathan D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 qRT-PCR assays. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Butler Daniel J, Meydan Christopher Mozsary Cem, Danko David, Foox Jonathan, Rosiene Joel, Shaiber Alon, Afshinnekoo Ebrahim, MacKay Matthew, Sedlazeck Fritz J, et al. Shotgun transcriptome and isothermal profiling of sars-cov-2 infection reveals unique host responses, viral diversification, and drug interactions. bioRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dao Thi Viet Loan, Herbst Konrad, Boerner Kathleen, Meurer Matthias, Kremer Lukas PM, Kirrmaier Daniel, Freistaedter Andrew, Papagiannidis Dimitrios, Galmozzi Carla, Stanifer Megan L, et al. A colorimetric rt-lamp assay and lamp-sequencing for detecting sars-cov-2 rna in clinical samples. Science Translational Medicine, 12(556), 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Meyerson Nicholas R, Yang Qing, Clark Stephen Kyle, Paige Camille L, Fattor Will T, Gilchrist Alison R, Barbachano-Guerrero Arturo, and Sawyer Sara L. A community-deployable sars-cov-2 screening test using raw saliva with 45 minutes sample-to-results turnaround. medRxiv, 2020. [Google Scholar]
- [10].Coronavirus (COVID-19) update: FDA informs public about possible accuracy concerns with Abbott ID NOW Point-of-Care Test. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-informs-public-about-possible-accuracy-concerns-abbott-id-now-point, May 14, 2020.
- [11].Quicke Kendra, Gallichote Emily, Sexton Nicole, Young Michael, Janich Ashley, Gahm Gregory, Carlton Elizabeth J, Ehrhart Nicole, and Ebel Gregory D. Longitudinal surveillance for SARS-CoV-2 RNA among asymptomatic staff in five colorado skilled nursing facilities: Epidemiologic, virologic and sequence analysis. medRxiv, 2020. [Google Scholar]
- [12].Wӧlfel Roman, Corman Victor M, Guggemos Wolfgang, Seilmaier Michael, Zange Sabine, Müller Marcel A, Niemeyer Daniela, Jones Terry C, Vollmar Patrick, Rothe Camilla, et al. Virological assessment of hospitalized patients with COVID-2019. Nature, 581(7809):465–469, 2020. [DOI] [PubMed] [Google Scholar]
- [13].Scola Bernard La, Bideau Marion Le, Andreani Julien, Van Thuan Hoang Clio Grimaldier, Colson Philippe, Gautret Philippe, and Raoult Didier. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. European Journal of Clinical Microbiology & Infectious Diseases, 39(6):1059, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alexandersen Soren, Chamings Anthony, and Bhatta Tarka Raj. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Caraguel Charles GB, Stryhn Henrik, Gagné Nellie, Dohoo Ian R, and Hammell K Larry. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: analytical and epidemiologic approaches. Journal of Veterinary Diagnostic Investigation, 23(1):2–15, 2011. [DOI] [PubMed] [Google Scholar]
- [16].Ruiz-Villalba Adrián, van Pelt-Verkuil Elizabeth, Gunst Quinn D, Ruijter Jan M, and van den Hoff Maurice JB. Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR). Biomolecular detection and quantification, 14:7–18, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Klasse PJ. Molecular determinants of the ratio of inert to infectious virus particles. In Progress in molecular biology and translational science, volume 129, pages 285–326. Elsevier, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cevik Muge, Tate Matthew, Lloyd Oliver, Maraolo Alberto Enrico, Schafers Jenna, and Ho Antonia. Sars-cov-2, sars-cov-1 and mers-cov viral load dynamics, duration of viral shedding and infectiousness: a living systematic review and meta-analysis. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Smith Amber M and Perelson Alan S. Influenza A virus infection kinetics: quantitative data and models. Wiley Interdisciplinary Reviews: Systems Biology and Medicine, 3(4):429–445, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Richard Mathilde, Kok Adinda, de Meulder Dennis, Bestebroer Theo M, Lamers Mart M, Okba Nisreen MA, van Vlissingen Martje Fentener, Rockx Barry, Haagmans Bart L, Koopmans Marion PG, et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. bioRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].He Xi, Lau Eric HY, Wu Peng, Deng Xilong, Wang Jian, Hao Xinxin, Lau Yiu Chung, Wong Jessica Y, Guan Yujuan, Tan Xinghua, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nature Medicine, 26(5):672–675, 2020. [DOI] [PubMed] [Google Scholar]
- [22].Shen Zhuang, Ning Fang, Zhou Weigong, He Xiong, Lin Changying, Chin Daniel P, Zhu Zonghan, and Schuchat Anne. Superspreading sars events, beijing, 2003. Emerging Infectious Diseases, 10(2):256, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Malik Peiris Joseph Sriyal, Chu Chung-Ming, Cheng Vincent Chi-Chung, Chan KS, Hung IFN, Poon Leo LM, Law Kin-Ip, Tang BSF, Hon TYW, Chan CS, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated sars pneumonia: a prospective study. The Lancet, 361(9371):1767–1772, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang Zheng, Xiao Tongyang, Wang Yanrong, Yuan Jing, Ye Haocheng, Wei Lanlan, Wang Haiyan, Liao Xuejiao, Qian Shen, Wang Zhaoqin, et al. Early viral clearance and antibody kinetics of COVID-19 among asymptomatic carriers. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lavezzo Enrico, Franchin Elisa, Ciavarella Constanze, Cuomo-Dannenburg Gina, Barzon Luisa, Claudia Del Vecchio Lucia Rossi, Manganelli Riccardo, Loregian Arianna, Navarin Nicolò, et al. Suppression of a sars-cov-2 outbreak in the italian municipality of vo’. Nature, pages 1–5, 2020. [DOI] [PubMed] [Google Scholar]
- [26].Vinh Chau Nguyen Van, Lam Vo Thanh, Dung Nguyen Thanh, Yen Lam Minh, Quang Minh Ngo Ngoc, Ngoc Nghiem My, Dung Nguyen Tri, Huy Man Dinh Nguyen, Nguyet Lam Anh, Han Ny Nguyen Thi, et al. The natural history and transmission potential of asymptomatic sars-cov-2 infection. medRxiv, 2020. [Google Scholar]
- [27].Chen Xudan, Zhang Yang, Zhu Baoyi, Zeng Jianwen, Hong Wenxin, He Xi, Chen Jingfeng, Zheng Haipeng, Qiu Shuang, Deng Ying, et al. Associations of clinical characteristics and antiviral drugs with viral rna clearance in patients with covid-19 in guangzhou, china: a retrospective cohort study. medRxiv, 2020. [Google Scholar]
- [28].Hu Zhiliang, Song Ci, Xu Chuanjun, Jin Guangfu, Chen Yaling, Xu Xin, Ma Hongxia, Chen Wei, Lin Yuan, Zheng Yishan, et al. Clinical characteristics of 24 asymptomatic infections with covid-19 screened among close contacts in nanjing, china. Science China Life Sciences, 63(5):706–711, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yang Rongrong, Gui Xien, and Xiong Yong. Comparison of clinical characteristics of patients with asymptomatic vs symptomatic coronavirus disease 2019 in wuhan, china. JAMA Network Open, 3(5):e2010182–e2010182, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wilder Bryan, Charpignon Marie, Killian Jackson A, Ou Han-Ching, Mate Aditya, Jabbari Shahin, Perrault Andrew, Desai Angel, Tambe Milind, and Majumder Maimuna S. Modeling between-population variation in COVID-19 dynamics in hubei, lombardy, and new york city. Available at SSRN 3564800, 2020. [DOI] [PMC free article] [PubMed]
- [31].Peak Corey M, Kahn Rebecca, Grad Yonatan H, Childs Lauren M, Li Ruoran, Lipsitch Marc, and Buckee Caroline O. Individual quarantine versus active monitoring of contacts for the mitigation of COVID-19: a modelling study. The Lancet Infectious Diseases, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kucharski Adam J, Klepac Petra, Conlan Andrew, Kissler Stephen M, Tang Maria, Fry Hannah, Gog Julia, Edmunds John, CMMID COVID-19 Working Group, et al. Effectiveness of isolation, testing, contact tracing and physical distancing on reducing transmission of SARS-CoV-2 in different settings. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fang Yicheng, Zhang Huangqi, Xie Jicheng, Lin Minjie, Ying Lingjun, Pang Peipei, and Ji Wenbin. Sensitivity of chest ct for COVID-19: comparison to RT-PCR. Radiology, page 200432, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wyllie Anne Louise, Fournier John, Casanovas-Massana Arnau, Campbell Melissa, Tokuyama Maria, Vijayakumar Pavithra, Geng Bertie, Muenker M Catherine, Moore Adam J, Vogels Chantal BF, et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. Medrxiv, 2020. [Google Scholar]
- [35].Chin Elizabeth T, Huynh Benjamin Q, Murrill Matthew, Basu Sanjay, and Lo Nathan C. Frequency of routine testing for covid-19 in high-risk environments to reduce workplace outbreaks. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Paltiel A David, Zheng Amy, and Walensky Rochelle P. Assessment of sars-cov-2 screening strategies to permit the safe reopening of college campuses in the united states. JAMA network open, 3(7):e2016818–e2016818, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Holtz David, Zhao Michael, Benzell Seth G., Cao Cathy Y., Rahimian Mohammad Amin, Yang Jeremy, Allen Jennifer, Collis Avinash, Moehring Alex, Sowrirajan Tara, Ghosh Dipayan, Zhang Yunhao, Dhillon Paramveer S., Nicolaides Christos, Eckles Dean, and Aral Sinan. Interdependence and the cost of uncoordinated responses to covid-19. Proceedings of the National Academy of Sciences, 117(33):19837–19843, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lieu Tracy A, Ray G Thomas, Klein Nicola P, Chung Cindy, and Kulldorff Martin. Geographic clusters in underimmunization and vaccine refusal. Pediatrics, 135(2):280–289, 2015. [DOI] [PubMed] [Google Scholar]
- [39].Kissler Stephen, Kishore Nishant, Prabhu Malavika, Goffman Dena, Beilin Yaakov, Landau Ruth, Gyamfi-Bannerman Cynthia, Bateman Brian, Katz Daniel, Gal Jonathan, Bianco Angela, Stone Joanne, Larremore Daniel, Buckee Caroline, and Grad Yonatan. Reductions in commuting mobility correlate with geographic differences in sars-cov-2 prevalence in new york city. Nature Communications (in press), 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lauer Stephen A, Grantz Kyra H, Bi Qifang, Jones Forrest K, Zheng Qulu, Meredith Hannah R, Azman Andrew S, Reich Nicholas G, and Lessler Justin. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Annals of internal medicine, 172(9):577–582, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hernandez Vargas Esteban Abelardo and Velasco-Hernandez Jorge X. In-host modelling of covid-19 kinetics in humans. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Burke James M, Bass Clovis R, Kincaid Rodney P, Ulug Emin T, and Sullivan Christopher S. The murine polyomavirus microrna locus is required to promote viruria during the acute phase of infection. Journal of virology, 92(16):e02131–17, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chandrashekar Abishek, Liu Jinyan, Martinot Amanda J, McMahan Katherine, Mercado Noe B, Peter Lauren, Tostanoski Lisa H, Yu Jingyou, Maliga Zoltan, Nekorchuk Michael, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yu Jingyou, Tostanoski Lisa H, Peter Lauren, Mercado Noe B, McMahan Katherine, Mahrokhian Shant H, Nkolola Joseph P, Liu Jinyan, Li Zhenfeng, Chandrashekar Abishek, et al. Dna vaccine protection against SARS-CoV-2 in rhesus macaques. Science, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Borremans Benny, Gamble Amandine, Prager KC, Helman Sarah K, McClain Abby M, Cox Caitlin, Savage Van, and Lloyd-Smith James O. Quantifying antibody kinetics and RNA shedding during early-phase SARS-CoV-2 infection. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].van Kampen Jeroen J.A., van de Vijver David A.M.C., Fraaij Pieter L.A., Haagmans Bart L., Lamers Mart M., Okba Nisreen, van den Akker Johannes P.C., Endeman Henrik, Gommers Diederik A.M.P.J., Cornelissen Jan J., Hoek Rogier A.S., van der Eerden Menno M., Hesselink Dennis A., Metselaar Herold J., Verbon Annelies, de Steenwinkel Jurriaan E.M., Aron Georgina I., van Gorp Eric C.M., van Boheemen Sander, Voermans Jolanda C., Boucher Charles A.B., Molenkamp Richard, Koopmans Marion P.G., Geurtsvankessel Corine, and van der Eijk Annemiek A. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (covid-19): duration and key determinants. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kawasuji Hitoshi, Takegoshi Yusuke, Kaneda Makito, Ueno Akitoshi, Miyajima Yuki, Kawago Koyomi, Fukui Yasutaka, Yoshida Yoshihiko, Kimura Miyuki, Yamada Hiroshi, et al. Viral load dynamics in transmissible symptomatic patients with COVID-19. medRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].To Kelvin Kai-Wang, Tsang Owen Tak-Yin, Leung Wai-Shing, Tam Anthony Raymond, Wu Tak-Chiu, Lung David Christopher, Yip Cyril Chik-Yan, Cai Jian-Piao, Chan Jacky Man-Chun, Chik Thomas Shiu-Hong, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. The Lancet Infectious Diseases, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kim Jin Yong, Ko Jae-Hoon, Kim Yeonjae, Kim Yae-Jean, Kim Jeong-Min, Chung Yoon-Seok, Kim Heui Man, Han Myung-Guk, So Yeon Kim, and Bum Sik Chin. Viral load kinetics of SARS-CoV-2 infection in first two patients in korea. Journal of Korean medical science, 35(7), 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Xiao Ai Tang, Tong Yi Xin, and Zhang Sheng. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clinical Infectious Diseases, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Liu Yang, Yan Li-Meng, Wan Lagen, Xiang Tian-Xin, Le Aiping, Liu Jia-Ming, Peiris Malik, Leo LM Poon, and Wei Zhang. Viral dynamics in mild and severe cases of COVID-19. The Lancet Infectious Diseases, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Minnesota Population Center. Integrated public use microdata series, international: Version 7.2 [dataset], 2019. 10.18128/D020.V7.2. [DOI] [Google Scholar]
- [53].Liu Yang, Eggo Rosalind, and Kucharski Adam. Secondary attack rate and superspreading events for SARS-CoV-2. The Lancet, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Prem Kiesha, Cook Alex, and Jit Mark. Projecting social contact matrices in 152 countries using contact surveys and demographic data. PLoS Computational Biology, 13(9):e1005697, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.