Abstract
Bi-specific single chain antibody fragments (bi-scFv) represent an emerging class of biotherapeutics. We recently developed a fully human bi-scFv (EGFRvIII:CD3 bi-scFv) with the goal of redirecting CD3-expressing T cells to recognize and destroy malignant, EGFRvIII-expressing glioma. In mice, we showed that EGFRvIII:CD3 bi-scFv effectively treats orthotopic patient-derived malignant glioma and syngeneic glioblastoma. Here, we developed a targeted assay for pharmacokinetic (PK) analysis of EGFRvIII:CD3 bi-scFv, a necessary step in the drug development process. Using microflow liquid chromatography coupled to high resolution parallel reaction monitoring mass spectrometry, and data analysis in Skyline, we developed a bottom-up proteomic assay for quantification of EGFRvIII:CD3 bi-scFv in both plasma and whole blood. Importantly, a protein calibrator, along with stable isotope-labeled EGFRvIII:CD3 bi-scFv protein, were used for absolute quantification. A PK analysis in a CD3 humanized mouse revealed that EGFRvIII:CD3 bi-scFv in plasma and whole blood has an initial half-life of ~8 minutes and a terminal half-life of ~2.5 hours. Our results establish a sensitive, high-throughput assay for direct quantification of EGFRvIII:CD3 bi-scFv without the need for immunoaffinity enrichment. Moreover, these pharmacokinetic parameters will guide drug optimization and dosing regimens in future IND-enabling and Phase I studies of EGFRvIII:CD3 bi-scFv.
Keywords: bispecific antibody, pharmacokinetic analysis, targeted proteomics, biotherapeutic, glioma, mass spectrometry, absolute quantification, stable isotope labeled protein, parallel reaction monitoring
Graphical Abstract
Introduction
Over the past decade, immunotherapies have become a mainstay in the treatment of cancer, ranging from traditional monoclonal antibodies against tumor antigens or “checkpoint” receptors to multi-modal targeting strategies such as chimeric antigen receptors and bispecific antibodies. The recent approvals of blinatumomab, a CD19-CD3 bispecific antibody, and CD19-specific chimeric antigen receptor T cells by the Food and Drug Administration mark a significant milestone in cancer-specific treatment and have inspired a plethora of clinical trials testing novel immunotherapies.1
Glioblastoma (GBM) is a malignant brain tumor that remains virtually incurable in patients and has a median survival rate of <16 months.2 The gold-standard of care consists of surgery, radiation, and chemotherapy and is not tumor-specific, resulting in detrimental side effects. We developed a fully human bispecific antibody for the treatment of glioblastoma that leads to long term survival in our mouse models.3 Our antibody consists of two linked single-chain variable fragments that bind the epidermal growth factor receptor variant III (EGFRvIII), a tumor-specific antigen, and CD3 receptors on T cells (EGFRvIII:CD3 bi-scFv), which leads to T-cell cross-linking with tumor cells in the brain and subsequent tumor-specific cell lysis mediated by perforin and granzyme (Figure 1A).
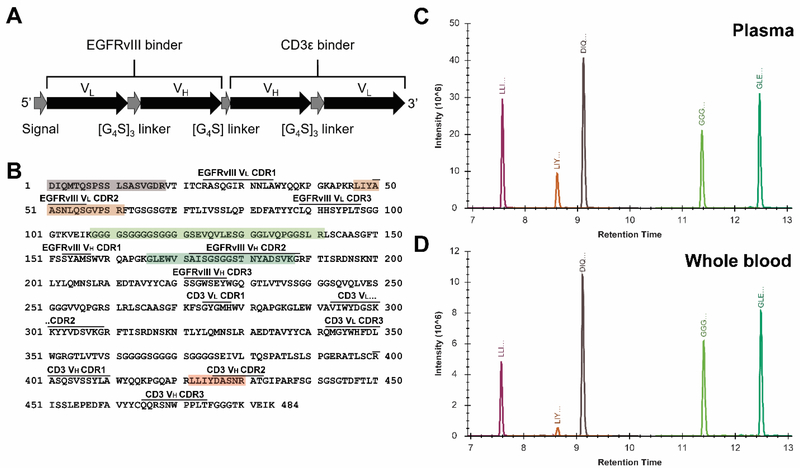
Figure 1: EGFRvIII:CD3 bi-scFv architecture, sequence, and peptide identification.
(A) Depiction of the sequence domains of EGFRvIII:CD3 bi-scFv, which consists primarily of two single chain Fv fragments joined by a [G4S]3 linker. (B) The protein sequence of EGFRvIII:CD3 bi-scFv. Complementarity determining regions (CDRs) corresponding to the normal and stable isotope labeled variable regions of the scFvs targeting EGFRvIII (clone 139) and CD3ε (clone 28F11) are overlined. Peptides targeted by our MS assay are highlighted. (C and D) 5.78 μg 12C14N EGFRvIII:CD3 bi-scFv was spiked into 100 μL of plasma and whole blood followed by reduction/alkylation and trypsinization. After quenching and precipitation of SDC, 5 μL of whole blood digests (out of a final digestion volume of 280 μL) and 10 μL of plasma tryptic digest (out of a final volume of 280 μL) were analyzed by microflow LC-MS/MS. Summed product ion (MS/MS) chromatograms (from four to nine product ions per peptide) were extracted for each of the 5 target peptides in Skyline for EGFRvIII:CD3 bi-scFv in (C) plasma and (D) whole blood. Individual peptides are colored according to (B).
Drug development of biologics such as bispecific antibodies requires formidable characterization alongside preclinical efficacy. To characterize EGFRvIII:CD3 bi-scFv we sought to establish the pharmacokinetics of the drug in circulation. Traditionally, ligand-binding assays (LBAs) are used for quantification of antibodies in biological samples. However, these assays have various limitations, including limited functionality for truncated antibodies, long development times, and variable selectivity.4–6 More recently, liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) has increasingly been used to characterize biotherapeutic proteoforms, identify host-cell contaminants and to quantify drug pharmacokinetics.7 The advantages of LC-MS/MS for antibody quantification include faster method development times and improved selectivity and coupled with stable isotope labeled internal peptide and protein standards, MS-based methods can achieve high accuracy and precision on par with or exceeding LBAs.8, 9 Quantification of biologics using LC-MS/MS has typically involved enrichment at the protein or peptide level using antibody-based resins. However, as the sensitivity of LC-MS/MS instrumentation has improved, the direct analysis in the presence of background matrix (e.g. blood or plasma) can be a feasible approach.6 It should be noted that quantification of drugs using MS in a matrix such as blood is done by measuring residual drug-derived peptides, allowing inference to the amount of bioavailable full-length protein. Importantly, the measurement of multiple peptides may reduce the possibility of masking of a single peptide or epitope by post-translational modification.
Here we have developed a streamlined, sensitive and high-throughput LC-MS/MS workflow for identifying and quantifying the level of EGFRvIII:CD3 bi-scFv in biological samples. For quantification of therapeutic doses of EGFRvIII:CD3 bi-scFv in plasma and whole-blood using tryptic digestion, we developed a minimal sample preparation workflow and coupled microflow ultraperformance liquid chromatography (UPLC) to parallel reaction monitoring (PRM) using a high-resolution benchtop orbitrap (Thermo Q-Exactive HF-X). Importantly, for accurate absolute quantification, we utilized an external calibration curve with authentic drug in plasma matrix, and we utilized a stable isotope-labeled (SIL) EGFRvIII:CD3 bi-scFv as an internal standard for normalization of matrix effects and other analytical variation between samples. Using our optimized workflow, we conducted an in vivo pharmacokinetic study in mice administered EGFRvIII:CD3 bi-scFv.
Experimental procedures
Chemicals and reagents
The EGFRvIII peptide resin was made using a 14-mer peptide (LEEKKGNYVVTDHC; JPT Peptide Technologies) that spans the EGFRvIII fusion junction and corresponds to the extracellular epitope for EGFRvIII-specific antibodies. Briefly, the peptide was immobilized to Irreversible Thiol-coupling SepFast 4HF (BioToolomics) by incubation in coupling buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA, pH 8.5) at room temperature for 1 h in the dark. After three more washes in coupling buffer to remove unbound peptide, non-reacted sites were blocked by incubating the resin in coupling buffer containing an additional 50 mM cysteine for 30 min at room temperature. Subsequently, the resin was washed 4 times alternating between a high pH (100 mM Tris, 500 mM NaCl, pH 8) and low-pH (100 mM sodium acetate, 500 mM NaCl, pH 4) wash buffer. The resin was then packed into an HR 16/5 column (GE Healthcare) and stored in 20% ethanol.
The EGFRvIII peptide tetramer was made using the same 14-mer peptide conjugated to biotin. Briefly, the biotinylated peptide was incubated with a 5-fold molar excess with AlexaFluor 647-labeled streptavidin at room temperature for 3 h, and free biotinylated peptide was removed using a 30 kDa molecular weight cut-off filter.
Synthesis and purification of 12C14N (normal) and 13C15N (stable isotope labeled [SIL]) EGFRvIII:CD3 bi-scFv
EGFRvIII:CD3 bi-scFv (molecular weight of 50.9 kDa) was produced using a stably transfected, suspension adapted Chinese Hamster Ovary cell line (CHO). A CHO cell codon-optimized EGFRvIII:CD3 bi-scFv cDNA transfection-grade plasmid was linearized and used to transfect CHO-S cells. Transfected cells underwent two rounds of single cell cloning to obtain a high-yield, high-viability working clone. From this clone, a master cell bank was generated under the current Good Manufacturing Protocol (cGMP) standard and certified by BioReliance.
To produce EGFRvIII:CD3 bi-scFv for in vitro and in vivo studies, transfected CHO-S cells were cultured in 2 L shaker flasks containing 1 L of FortiCHO media each for 12 days. After centrifugation at 1500 xg for 15 min, the supernatant containing EGFRvIII:CD3 bi-scFv was subsequently purified using EGFRvIII peptide resin column on an AKTA FPLC system (GE Healthcare). Briefly, the column was equilibrated with wash buffer (50 mM Tris; 150 mM NaCl; pH 7.2), and the supernatant was loaded at 3 mL/min followed by washing with 100 mL of wash buffer. EGFRvIII:CD3 bi-scFv was eluted with elution buffer (0.5 M glycine; 150 mM NaCl; pH 2.75) into collection tubes containing neutralization solution (1 M Tris; 150 mM NaCl; pH 8.0) Resulting purified EGFRvIII:CD3 bi-scFv was concentrated to around 1 mg/mL using tangential flow filtration and dialysis (Spectrum Labs) and formulated in PBS.
To produce stable isotope-labeled 13C15N EGFRvIII:CD3 bi-scFv, MCB cells were grown in custom CD FortiCHO media (Thermo Fisher Scientific) containing 13C615N4 L-arginine and 13C615N2 L-lysine (Sigma Isotec) for >6 doublings to ensure >99% incorporation of stable isotope-labeled amino acids. To confirm stable isotope incorporation, protein was recovered from 1 ml of cell culture supernantant and analyzed by LC-MS/MS (Supplemental Methods). After verification of stable isotope incorporation, the stable isotope-cultured CHO cells were then expanded in shaker flasks to produce 13C15N EGFRvIII:CD3 bi-scFv. 13C15N EGFRvIII:CD3 bi-scFv was purified in an identical manner as the natural isotope 12C14N EGFRvIII:CD3 bi-scFv and was analyzed by LC-MS/MS as described in Supplemental Information.
In vitro binding studies
A flow cytometry assay was utilized to confirm that purified EGFRvIII:CD3 bi-scFv retained binding to both target epitopes. The assay consisted of a two-step binding protocol that ensured both arms of the bispecific antibody were functional at the same time. Specifically, 1 μg of EGFRvIII:CD3 bi-scFv was incubated with 1 × 106 human peripheral blood mononuclear cells (PBMCs) for 30 minutes, which resulted in binding of CD3ε. The samples were washed and incubated for 30 minutes with a custom-made fluorescent EGFRvIII petide tetramer, which binds the other arm of the bispecific antibody demonstrating bi-functionality of the antibody. After another wash step, the cells were analyzed in a BD Fortessa flow cytometer. Fluorescence data was analyzed using Flowjo.
In vivo pharmacokinetic (PK) studies
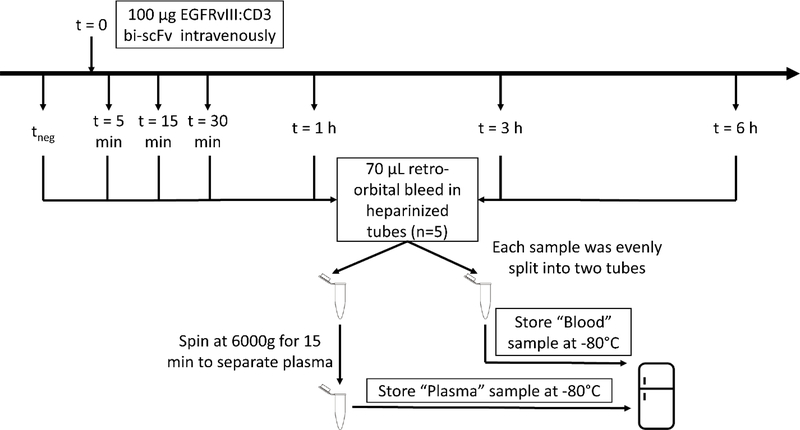
PK studies used transgenic mice with heterologous expression human CD3ε (hCD3ε−/+) on a C57B/6 background (tgε600, Jackson Laboratory, strain number 20456 crossed with wild-type C57/BL6). Mice (16 weeks old; 23.5 ± 1.6 (s.d.) g) were administered 100 μg of 12C14N EGFRvIII:CD3 bi-scFv (100 μl of 1 mg/mL drug) via tail vein injection. At each designated time point, 70 μL of blood was collected from n=5 mice via retro-orbital bleeds using heparinized microhematocrit capillary tubes (VWR) and transferred to 1.5 mL Eppendorf tubes (Figure 3). To minimize adverse effects of the retro-orbital technique and conform to IUCAC standards, only 2 time-points were collected per animal (i.e. 15 mice were dosed for collection of blood at 6 timepoints). As a control, blood was collected from an additional group of mice that did not receive EGFRvIII:CD3 bi-scFv injection (tneg). Blood from the remaining dosed animals was collected 5 min, 15 min, 30 min, 1 hour, 3 hours, and 6 hours after injection. After collection, blood was stored on ice and separated into 2 aliquots for whole blood and plasma. Whole blood samples were immediately frozen, and plasma samples were isolated by centrifugation of whole blood at 6000 xg for 15 min at 4 °C followed by collection of the cell-free supernatant. All samples were stored at −80 °C until analysis.
Figure 3. Pharmacokinetic experiment workflow.
Human CD3 transgenic mice were administered 100 μg of 12C14N EGFRvIII:CD3 bi-scFv via tail vein injection. At each designated time point, 70 μL of blood was collected from n=5 mice via retro-orbital bleeds using heparinized microhematocrit capillary tubes and transferred to 1.5 mL Eppendorf tubes. As a control, blood was collected from an additional group that did not receive 12C14N EGFRvIII:CD3 bi-scFv injection (tneg). Blood from the remaining dosed animals was collected 5 min, 15 min, 30 min, 1 hour, 3 hours, and 6 hours after injection. After collection, blood was stored on ice and separated into 2 aliquots labeled whole blood and plasma. Whole blood samples were immediately frozen, and plasma samples were isolated by centrifugation of whole blood at 6000 xg for 15 min at 4°C followed by collection of the cell-free supernatant. All samples were stored at −80°C until analysis.
Sample preparation for plasma PK
For plasma PK analyses, 10 μL of heparin-plasma was diluted with 10 μL of 50 mM ammonium bicarbonate, pH 8 (AmBic) to account for addition of the same volume of calibrator in AmBic in the standard curve and QC samples (see below). To this we added 95 μL of master mix containing 5% w/v sodium deoxycholate (SDC), 10 mm dithiothreitol (DTT) and 57 ng (~1 pmol) of 13C15N EGFRvIII:CD3 bi-scFv followed by heating at 80 °C for 10 min. After cooling, 1:10 w/w TPCK trypsin (10 μl of 5 mg/mL trypsin in AmBic) was added and incubated at 37 °C for 4 h on a Thermomixer. Finally, 12.5 μL of 10% (v/v) trifluoroacetic acid (TFA) and 20% (v/v) acetonitrile (MeCN) were added (to yield final concentrations of 1% TFA and 2% MeCN). After vortexing, the resulting precipitate was removed by centrifugation at 15,000 xg for 10 min at 4 ºC, and the supernatant was transferred to Maximum Recovery LC vials (Waters).
For preparation of standard curve and QC samples, 10 μL of mouse EDTA-plasma was spiked with 10 μL of 12C14N EGFRvIII:CD3 bi-scFv in AmBic (as above) to yield an equivalent of 0.5, 1, 5, 10, 50 and 100 ng of EGFRvIII:CD3 bi-scFv per μL of plasma. QC samples were prepared similarly with 2.5, 7.5 and 75 ng 12C14N EGFRvIII:CD3 bi-scFv per μL of plasma. Finally, a blank sample was prepared by addition of 10 μL AmBic to 10 μL plasma. Master mix was added, and all samples were processed as described above.
Sample preparation for whole blood PK
Whole blood (WB; 10 μL) was processed as with plasma with the following modifications: 100 ng 13C15N EGFRvIII:CD3 bi-scFv (internal standard) was added per sample as part of the mastermix; digestions used 10 μL of 12.5 mg/mL trypsin; and a final concentration of 1.5% (v/v) TFA and 2% (v/v) MeCN were added to quench trypsin and precipitate the SDC. Calibration curves were prepared as described for plasma except that the QC samples were prepared by addition of 0.25, 2.5 and 25 ng 12C14N EGFRvIII:CD3 bi-scFv per μL of WB.
Microflow LC-MS/MS analysis
EGFRvIII:CD3 bi-scFv in plasma was analyzed by LC-MS/MS using a Waters ACQUITY LC interfaced to a Thermo Q-Exactive HF-X via a heated electrospray ionization (HESI-II) source. For analysis of plasma, approximately 50 μg of tryptic digest (equivalent to 1 μL of plasma) was separated on a 1 mm x 15 mm 1.7 μm CSH C18 column using a flow rate of 100 μL/min, a column temperature of 55 °C and a gradient of 3–25% (v/v) MeCN:H2O containing 0.1% formic acid over 15 min. Tune parameters were: sheath gas, 35; aux gas, 10; sweep gas, 1; spray voltage, 3.5 kV; capillary temperature, 275 °C; funnel RF, 45. PRM analysis used the following settings: resolution, 60,000; AGC target of 1e5 maximum injection time (IT) of 300 ms; 0.7 m/z isolation window; and 27 V normalized collision energy (NCE). The method utilized an inclusion list as shown in Table S1A.
EGFRvIII:CD3 bi-scFv in whole blood was analyzed as in plasma with the following modifications. Approximately 150 μg of tryptic digests (equivalent to 1 μL of whole blood) were analyzed per sample. Finally, a solvent divert was used from 0–7 min and from 14–20 min of the LC method. During these times, 50:49.9:0.1 v/v/v MeCN:H2O:FA was delivered to the source at 100 μL/min using a binary pump (Shimadzu LC-600). The method utilized an inclusion list as shown in Table S1B.
Analysis of mass spectrometry data
PRM data was imported into Skyline, and interfering transitions were manually removed based on visual inspection as well as matching to a spectral library (dotp > 0.9) and r2 fitting of standard curves. For both plasma and whole blood quantification, replicate standard curves (0.5–100 μg/mL) were fit in Skyline using a quadratic regression fit and 1/x weighting. Limit of quantitation (LOQ) was defined as lowest calibration point with bias <20%; and the limit of detection was defined as 3 times the average background signal in non-dosed animals. Curated Skyline files “Plasma_PK_bi_scFv” and “WB_PK_bi_scFv” and associated raw data were uploaded to the Panorama Targeted Proteomics data repository (https://panoramaweb.org/31jFrQ.url) and can also be accessed via ProteomeXchange (PXD012472).
Analysis of pharmacokinetic (PK) data
Concentration/time data for each quantified peptide was exported from Skyline, and pharmacokinetic parameters were obtained using non-compartmental analysis in the WinNonlin software. The areas under the concentration-time curve (AUClast and AUCinf) were calculated by linear trapezoidal rule; subscript ‘last’ refers to the last experimental time-point and ‘inf’ refers to extrapolation to infinity. The apparent and terminal elimination half-lifes (t1/2) were determined by regression analysis of the linear initial (first 3 time-points) and terminal (last 2 time-points) portion of the logarithmic plasma concentration-time curve, respectively. Clearance (CL) = Dose/AUCinf and the volume of distribution at steady state (Vss) = mean residence time (MRT) x CL. The PK parameter values given are average (arithmetic mean) obtained from PK analysis of individual concentration/time traces for each of the 5 peptide fragments measured after digestion of plasma and blood samples that were collected from individual animals (n=5) at different timepoints. Given also is the single standard deviation of the PK parameters obtained from the peptides.
Results
Development of a targeted proteomic assay for EGFRvIII:CD3 bi-scFv
In order to develop a targeted protein assay for quantitation of EGFRvIII:CD3 bi-scFv in mice, we first sequenced the antibody preparation using standard reduction/alkylation, trypsin digestion and nanoflow LC-MS/MS. This approach yielded ~85% protein sequence coverage, identifying numerous peptides for targeted quantification of EGFRvIII:CD3 bi-scFv (Figure S1). Matching of a spectral library to a reference mouse proteome in Skyline showed that there was low identity between the tryptic peptides of the recombinant protein and mouse immunoglobulins (Figure S1); therefore, the endogenous immunoglobulins are less likely to contribute to interferences in targeted quantification of EGFRvIII:CD3 bi-scFv in mouse models.
We next investigated whether EGFRvIII:CD3 bi-scFv could be detected at therapeutic levels in plasma and whole blood without an affinity enrichment step. One hundred microliters of plasma and whole blood were spiked with ~5 μg/mL 12C14N EGFRvIII:CD3 bi-scFv, which was in the concentration range of prior dosing studies.6, 10 Plasma digestions employed sodium deoxycholate (SDC) as a denaturant, reduction/alkylation with DTT and iodoacetamide and a 4 h digestion with 1:10 (w/w) TPCK-trypsin.11 Digestions were stopped, and SDC precipitated by addition of 1% (v/v) TFA; and ~50 μg of digests (equivalent to 1 μL plasma) were analyzed by using microflow LC-MS/MS. Specifically, a 1 mm x 15 cm, charged surface hybrid (CSH) C18 column was selected for its high peptide loading capacity, reproducibility and throughput. MS acquisition used parallel reaction monitoring (PRM) because of its high sensitivity and specificity.12 Digestion of whole blood was done with 5 times more trypsin than in plasma (based on a protein concentration in blood of 250 μg/mL). In addition, because of the higher protein concentrations in blood versus plasma, we found that increasing the TFA concentration to 1.5% was needed to reach the desired pH for optimal precipitation of SDC and heme cofactor (evidenced by color of supernatant versus pellet). Finally, analyses were scaled to ~150 μg (or ~0.6 μL) of whole blood, which was still under the estimated maximum column loading capacity of 200 μg.
We focused the PRM analysis on peptides lacking cysteine residues in order to avoid peptides with variable modifications and allow simplification of the sample prep. We identified five peptides that eluted over a relatively narrow range (~5 min) of the 20 min LC-MS/MS method and had excellent signal-to-noise. These peptides spanned both the N-terminal and C-terminal domains containing scFvs targeting EGFR-VIII and CD3ε and overlapped several of the complementarity-determining regions (CDRs) of the antibody variable regions (Figure 1B). There was little or no difference in the retention times of these peptides between the plasma and whole blood samples despite significant potential matrix differences (Figure 1C and 1D). In addition, the elution times of these peptides were non-overlapping, which could allow for maximum utilization of MS duty cycle. On the other hand, observed peptide responses were lower in whole blood (even when accounting for injection of half as much samples versus plasma), suggesting that other factors, such as digestion bias or ion suppression, could affect quantification of the protein between the two matrices. Nonetheless, these data suggested the feasibility of the microflow LC-MS/MS approach for the direct detection of therapeutically relevant quantities of EGFRvIII:CD3 bi-scFv in plasma and whole blood.
Synthesis and characterization of stable isotope-labeled 13C15N EGFRvIII:CD3 bi-scFv
Targeted proteomic assays often utilize isotope-labeled internal standards for cross-run normalization, confident peptide identification and accurate peptide/protein quantification.13 Stable isotope-labeled (SIL) peptides are often utilized as single-point internal calibrators via stable-isotope dilution; however, external protein calibrators and corresponding SIL protein internal standards may afford better quantitative precision and accuracy.14, 15 To take advantage of these benefits, and to try and prevent any biases due to structure or function differences between drug and internal standard (e.g. deficient post-translational modifications in bacterially-expressed peptides), we sought to produce a SIL protein that was identical to the EGFRvIII:CD3 bi-scFv drug molecule used in our preclinical studies.3
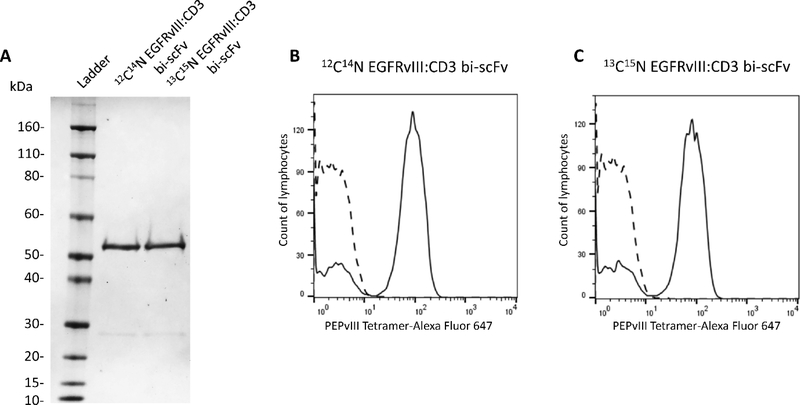
The 13C15N stable isotope-labeled EGFRvIII:CD3 bi-scFv was produced in suspension-adapted CHO-S cells using a GMP-certified master cell bank and custom media containing 13C615N2-Lys and 13C615N4-Arg. Approximately 3.8 mg 13C15N protein was purified from 1 L of conditioned media by affinity chromatography and >99% 13C15N incorporation was confirmed by PRM (Figure S2). SDS-PAGE also revealed that the SIL protein and the EGFRvIII:CD3 bi-scFv drug both migrated at predicted molecular weights (~51 kDa) and had a similar proportion of full-length protein to minor low molecular weight (~26 kDa) impurity (Figure 2A). Furthermore, both antibodies bound both EGFRvIII and CD3, as shown by flow cytometry (Figure 2B and 2C); thus, the SIL protein appeared to retain the properties of the antibody drug used in pre-clinical studies. Based on these data, we expected that the 13C15N EGFRvIII:CD3 bi-scFv would be effective as an internal standard for normalization and in controlling for analytical variability.
Figure 2. Analysis of 2C14N and 13C15N EGFRvIII:CD3 bi-scFv.
(A) SDS-PAGE analysis revealed that the 2C14N [position 2] and 13C15N [position 3] EGFRvIII:CD3 bi-scFv were identical in size at around 51 kDa [Novex Sharp Pre-stained Protein Standard in position 1]. (B and C) Flow cytometry analysis showed that (B) 12C14N and (C) 13C15N EGFRvIII:CD3 bi-scFv display functional EGFRvIII and CD3 binding.
Targeted pharmacokinetic analysis of EGFRvIII:CD3 bi-scFv in mouse plasma and whole blood
Since EGFRvIII:CD3 bi-scFv is a fully humanized antibody that does not bind mouse CD3ε, we utilized a transgenic mouse expressing human CD3ε (tg-hCD3) in order to better model drug biodistribution. For PK analysis, blood was collected by retro-orbital sampling from non-dosed tg-hCD3 mice versus tg-hCD3 mice receiving a single i.v. injection of 12C14N EGFRvIII:CD3 bi-scFv with blood sampling at 5 min, 15 min, 30 min, 1 h, 3 h and 6 h post-injection (n=5 per timepoint; Figure 3). Time points were chosen based on the pharmacokinetic profile of related antibody fragments.16 After collection of whole blood (WB) and plasma using heparinized microhematocrit capillary tubes, samples were stored at −80 °C. For absolute quantification in plasma, an external standard curve was generated using 12C14N EGFRvIII:CD3 bi-scFv spiked into 10 μL plasma; in addition, low, mid and high-quality control (QC) samples were prepared by addition of known amounts of 12C14N EGFRvIII:CD3 bi-scFv to plasma. Samples were denatured and reduced by dilution of the plasma with a master mix containing 13C15N EGFRvIII:CD3 bi-scFv, sodium deoxycholate (SDC) and DTT, followed by heating. Since no cysteine-containing peptides were targeted, an alkylation step was omitted. Finally, samples were trypsinized as described previously.
The LC-MS/MS analysis of EGFRvIII:CD3 bi-scFv in plasma used a scheduled PRM assay targeting the 12C14N and 13C15N forms of the five EGFRvIII:CD3 bi-scFv peptides (as in Figure 1B). Because of the narrow chromatographic peaks, we were able to schedule the assay using 0.6 min wide, non-overlapping retention time windows. At an MS2 resolution of 60,000 and a 300 ms maximum ion injection time (Table 1), we achieved 20 or more data points (i.e. points-across-the-peak) for each quantified peptide. Standard curve and QC samples were analyzed in duplicate (from low to high; at the beginning and end of the analysis) and the time-course samples were analyzed once each, beginning with the blank (un-dosed) samples and continuing in the order that samples were collected. Interestingly, we did note that the intensities of three out of five peptides from the SIL protein internal standard were lower in the standard curve and QC samples versus the PK samples (Skyline Data). This demonstrates that in some cases even a full length protein drug/calibrator and internal standard may not behave like a metabolized protein drug in all cases; notably, while such effects may have an impact on the absolute quantity of the drug measured, it will not impact the half-life calculation. The standards and QCs were created from a pool of plasma obtained by cardiac puncture and collected in EDTA-microcentrifuge tubes, whereas the PK samples were obtained via retro-orbital bleeding using heparinized capillary tubes. Variations in sample collection, such as the small effect of EDTA on trypsinization, might explain these differences that should nonetheless be controlled for by normalization to the SIL peptides across samples.
Table 1.
Summary of LC-MS/MS method
| LC Instrument | Waters ACQUITY UPLC |
| LC column | Waters 1 mm × 15 cm, 1.8 μm CSH (Part #186006935) |
| Mobile phase (MP) | MPA, 0.1% formic acid/99.9% H2O MPB, 0.1% formic acid/99.9% MeCN |
| Gradient | 0–15 min, 3–25% MPB 15–15.5 min, 25–90% MBP 15.5–17.5 min, 90% MBP 17.5–18 min, 90%–3% MPB 18–20 min, 3% MPB |
| Flow rate | 100 μL/min |
| Injection volume | 14 μL (plasma); 8 μL (whole blood) |
| Divert solvent (whole blood only) | 0.1% FA/49.9% H2O/50% MeCN, 100 μL/min; 0–7 min and |
| MS Instrument | Thermo Q-Exactive HF-X |
| MS method | PRM, 0–20 min |
| MS2 resolution | 60,000 |
| AGC target | 1E5 |
| Max injection time | 300 ms |
| Isolation width | 0.7 m/z |
| Normalized collision energy | 27 |
Since EGFRvIII:CD3 bi-scFv binds to circulating hCD3ε-expressing cells, the molecule may partition between plasma and blood cells. Thus, to measure total circulating levels of EGFRvIII:CD3 bi-scFv, we performed a parallel analysis of whole blood from identical experimental groups using the same experimental design and MS method as employed on plasma. In addition to changes in the digestion protocol and analysis of ~40% lower volume of blood versus plasma (equivalent to ~150 μg tryptic digests; see experimental procedures), we used a solvent divert to minimize the potential effects of high protein load and residual heme cofactor on MS system performance. Briefly, a divert valve was used to deliver 50/50 (v/v) H2O:MeCN containing 0.1% FA to the source via an auxiliary pump, during the first 7 min and the last 6 min of the LC-MS method; during this time, the UPLC column eluent was diverted to waste (Figure S3). Peak broadening was not observed with the longer flow path, nor was there evidence of carryover from the divert valve setup based on analysis of a double blank analyzed after the highest QC standard (Skyline Data). Notably, equivalent response of all SIL peptides between the standard curve and the samples was observed in whole blood.
Raw data was imported into Skyline and analyzed using the built-in quantitation feature (see Methods).17 First, data was matched to the 10 most intense product ions based on library spectra, and interfering product ions were removed by manual inspection of the data. Additional refinement was performed based on the dot product (>0.9 for samples with high abundant peptides) and normal/SIL product ion ratios, and identical transitions were selected for the normal and SIL pairs. Note that we did not place any limitation on the number of transitions used for quantification, which ranged between four and nine (Skyline Data). Following the initial transition selection and normalization to SIL peptides, the data was fit to the standard curves using the calibration feature in Skyline.17 In addition to goodness of fit (r2), we evaluated several other figures of merit, including lower limit of quantitation (LOQ) and accuracy of QC samples above LOQ (Table 2). LOQ was defined as the lowest calibration point with calculated concentration within ±20% of expected (nominal). Accuracy of our QC samples above LOQ was 100% ± 25% in all cases with the exception of peptide LIYA in whole blood. Accuracy of 100% ± 20% is the typical acceptance range based on FDA guidance for bioanalytical method validation; the peptide LLIYDASNR meets this criteria in both matrices at all QC levels, therefore this peptide would be the top candidate should the assay be carried forward for full validation.18 Nonetheless, in this study we were interested in understanding pharmacokinetics of EGFRvIII:CD3 bi-scFv processing and degradation using peptides from all protein regions, so all peptides were used in both matrices with the exception of LIYA in whole blood.
Table 2.
Peptide quantification figures of merit
| Plasma | Whole blood | |||||
|---|---|---|---|---|---|---|
| Peptide | r2 | LOQ (μg/mL) | QC accuracy, low, mid, high1 (average %) | r2 | LOQ (μg/mL) | QC accuracy, low, mid2 (average %) |
| DIQMTQSPSSLS ASVGDR | 0.999 | 0.5 | 107, 120, 112 | 0.999 | 0.5 | 78, 87 |
| LIYAASNLQSGVPSR | 0.999 | 0.5 | 108, 121, 109 | 0.999 | 5 | 67, 92 |
| GGGGSGGGGSGGGGSEVQVLE SGGGLVQPGGSLR | 0.999 | 0.5 | 113, 124, 109 | 0.999 | 0.5 | 78, 90 |
| GLEWVSAISGSGGSTNYADSVK | 0.998 | 0.5 | 116, 118, 122 | 0.999 | 0.5 | 79, 89 |
| LLIYDASNR | 0.996 | 0.5 | 102, 112, 107 | 0.998 | 0.5 | 91, 100 |
low, mid, and high QC samples were 2.5 μg/mL, 7.5 μg/mL and 75 μg/mL EGFRvIII:CD3 bi-scFv in plasma matrix
average low and mid QC samples were 2.5 μg/mL and 25 μg/mL EGFRvIII:CD3 bi-scFv in whole blood matrix
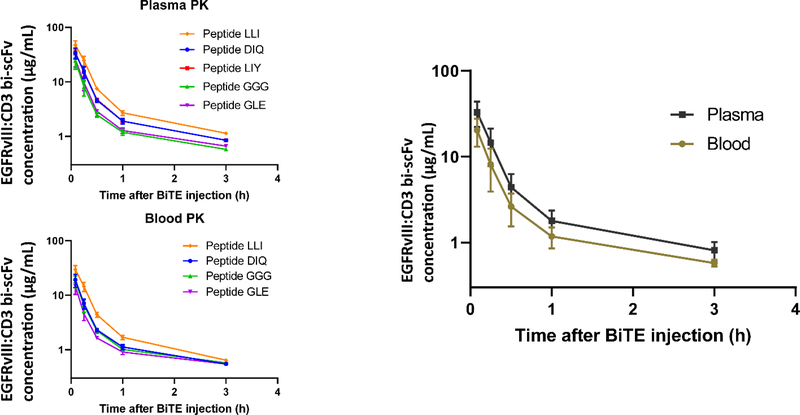
In plasma, the quantified levels of all peptides in plasma were >LOQ (0.5 μg/mL) at the 3 h time-point, but at 6 h, levels dropped below LOQ; thus,6 h data were excluded from the PK analysis. Still, the bulk of the decay in plasma EGFRvIII:CD3 bi-scFv occurred during the first hour, so we were able to capture the most important phase of the protein clearance (Figure 4).
Figure 4. Pharmacokinetics of EGFRvIII:CD3 bi-scFv.
Antibody concentration versus time data from plasma and whole blood samples of our murine PK experiment are presented in log-linear plot. The data shows a complex two-step decay, with a short initial half-life of ~8 min and a longer terminal half-life of ~2.5 h.
In whole blood, we achieved the same LOQ (0.5 μg/mL) for four of the peptides despite the lower volume analyzed. However, the peptide LIYAASNLQSGVPSR had an LOQ of 5 μg/mL and was excluded from the analysis because it fell below LOQ after 60 min. For each of the other peptides, data quality was suitable and quantified levels were ≥LOQ out to the 3 h time-point. However, the 6 h time-point was again <LOQ and excluded from the PK analysis. Nonetheless, the data demonstrated feasibility of EGFRvIII:CD3 bi-scFv quantification in whole blood without an enrichment step. In both plasma and blood, we observed a rapid decay of EGFRvIII:CD3 bi-scFv, which suggested that this was not due to sequestration by CD3ε-expressing T-cells but rather clearance from blood (Figure 4).
Plasma and blood pharmacokinetics
Absolute concentration of the individual peptides (in μg/mL) was exported and independently analyzed using a non-compartmental approach in the WinNonlin software to obtain relevant pharmacokinetic parameters for whole blood and plasma (Table 3). Within the first 30 minutes, around 90% of the drug was lost from both whole blood and plasma (t1/2, initial = 8.5 min). Extrapolation to time zero (curve-stripping) of this process gives the initial drug concentration, C0. Terminal half-life was estimated to be between 1 h and 3 h (t1/2, terminal = 1.8 h). Additional values and single standard deviation of the main PK parameters obtained from the non-compartmental analysis are listed in Table 3.
Table 3.
Non-compartmental pharmacokinetic analysis
| PK parameter | Blood [SD] | Plasma [SD] | Blood/Plasma |
|---|---|---|---|
| C0 [μg/mL] | 32.3 [6.0] | 50.0 [9.6] | 0.65 |
| Cmax [μg/mL] | 20.3 [5.6] | 33.0 [9.1] | 0.62 |
| Clast [μg/mL] | 0.59 [0.04] | 0.81 [0.22] | 0.72 |
| AUClast [μg · h/mL] | 8.5 [2.6] | 13.9 [4.6] | 0.61 |
| t1/2 initial [min] | 8.3 [0.41] | 8.5 [0.7] | 0.98 |
| t1/2 terminal [h] | 2.8 [1.6] | 1.8 [0.2] | 1.56 |
| AUCinfinity [μg · h/mL] | 8.6 [2.6] | 14.1 [4.7] | 0.61 |
| Clearance [L · kg/h] | 0.52 [0.13] | 0.33 [0.10] | 1.60 |
| MRTlast [h] | 0.51[0.04] | 0.47 [0.01] | 1.08 |
| Vss [L/kg] | 0.29 [0.09] | 0.16 [0.05] | 1.76 |
C0, extrapolated concentration at time 0; Cmax, concentration observed 5 min post-injection; Clast, concentration observed 3 h post-injection; AUClast, area under the curve between 0–3 h; t1/2 initial, half-life during first 30 minutes; t1/2 terminal, half-life observed between 1 h and 3 h; AUCinfinity area under the curve extrapolated to infinity; MRTlast, mean residence time observed between 0–3 h; Vss, steady state volume of distribution
Discussion
While ligand-binding assays (e.g. ELISA) or affinity purification coupled to mass spectrometry (AP-MS)) have been mainstays for the quantification of biotherapeutics, these assays may only detect quantities of “free” protein and suffer from epitope masking by autoantibodies or other interacting proteins. In the case of EGFRvIII:CD3 bi-scFv, the presence of a target epitope (CD3ε) on numerous circulating T-cell subtypes at steady state raises the question of how best to quantify this and related proteins.19 Our goal here was to develop a targeted MS assay for the pharmacokinetic analysis of free and cell-bound EGFRvIII:CD3 bi-scFv that did not require antibody-based enrichment and could utilize whole blood, thus allowing the quantification of total drug in the sample. The result is an assay that combines the LC-MS/MS approaches which are common in the fields of clinical MS and proteomics but not often used in combination, namely high flow chromatography and parallel reaction monitoring using high resolution accurate mass MS, respectively, enabling high throughput and system robustness, while achieving high sensitivity and specificity. In addition, we have shown the feasibility of this approach for targeted quantification in whole blood, which importantly, by using denaturing conditions, enabled us to simultaneously detect both free and cell-bound drug and allowed us to infer that decay of protein from plasma was not simply due to binding to blood cells. The analysis of whole blood was performed with minimal loss in sensitivity and required relatively few changes to the method: quantities of trypsin were scaled 5-fold; 50% more TFA was used in quenching the reaction; and injection volumes were scaled back to ~0.6 μl of whole blood per injection. The whole blood analysis was further aided by our selection of peptides that eluted over a narrow retention time window, allowing us to employ a solvent divert in order to minimize the potential for dirtying of the MS and avoid off-line fractionation for these high peptide loads.
While there has been considerable debate around the best approaches for absolute quantitation by mass spectrometry, there is general agreement that the use of protein calibrators and use of stable isotope-labeled (SIL) protein internal standards added to samples early in the workflow can afford high accuracy and precision, because this approach accounts for the most sources of potential variance.8 For quantification of bi-specific scFVs, we show here that it can be relatively easy to produce an identical SIL internal standard protein, which can then be used in both standards and samples (Figure 2). In addition, this approach allows for immense flexibility in target peptide selection. For example, the 34-mer peptide we chose as one of the targets for EGFRvIII:CD3 bi-scFv quantification would have been difficult to obtain by routine peptide synthesis. Furthermore, given the high homology between EGFRvIII:CD3 bi-scFv and the human proteome, the potential ease in expanding the assay to different tryptic peptides or alternative proteases should help to ease the translation of the assay to PK analysis in human subjects. Still, there remain challenges and limitations with this approach. For example, we did not perform absolute quantitation of either forms of the protein by the gold-standard approach of amino acid analysis (AAA) or by gravimetric analysis; thus, the absolute quantitation is dependent on the accuracy of a Bradford assay versus a BSA calibrator.
Our pharmacokinetic analysis of EGFRvIII:CD3 bi-scFv whole blood and plasma concentration over 3 hours suggest a bi-phasic disposition profile. The major process, responsible for ~90% of drug loss from plasma, occurs within the first 30 min (t1/2 initial = 8.5 min, Table 3). Given that the size of our 50.9 kDa antibody is below the molecular weight cutoff of the glomeruli, the initial drug loss is assumed to be urinary elimination.20, 21 After the first 30 minutes, the rate of drug loss slows significantly, resulting in the apparent terminal half-life of ~2.5 hours. Data collected beyond 3 h suggest another (slower) elimination process but the accurate calculation and interpretation is precluded due to assay limitations (LOQ). In the scenario where the drug has no interaction with the blood cells, based on the expected 41% mouse hematocrit we would expect a blood:plasma PK parameter ratio of 0.59.22, 23 Indeed, Cmax, Clast, and AUClast blood:plasma ratio of 0.62, 0.72, and 0.61, respectively, suggest a very weak interaction of EGFRvIII:CD3 bi-scFv with blood cells (Table 3). This also explains the identical corresponding half-life values observed in plasma and blood, suggesting that the elimination processes are solely associated with plasma. This further suggests that plasma can be substituted with whole blood in future PK studies of EGFRvIII:CD3 bi-scFv. We observed low plasma clearance (0.33 L/h per kg BW) as compared to mouse kidney plasma flow (2.3 L/h per kg) and plasma cardiac output (14.2 L/h per kg) indicating low first-pass clearance by the liver and kidneys. As expected for a protein, our antibody exhibits a low volume of distribution at steady state, Vss (0.16 L/kg), as compared to total body water (0.73 L/kg).24 The mean residence time (MRT) of the drug in plasma was estimated to be 0.47 h.
Blinatumomab, a CD19/CD3 bispecific antibody fragment with similar architecture and molecular weight (55 kDa) as EGFRvIII:CD3 bi-scFv, is the only clinically approved bispecific antibody to date. As part of the drug approval process, blinatumomab was extensively characterized in multiple models.25 Across their mouse, rat, cynomolgus monkey, and chimpanzee pharmacokinetic studies, Amgen’s drug had terminal half-life’s that did not exceed 7 hours. Specifically, in mice the half-life did not exceed 2.5 hours and nephrectomy increased half-life significantly. In chimpanzees, the initial half-life was in the range of minutes and terminal half-life was ~2 hours.16 In Amgen’s human studies, a terminal half-life of 1.25±0.63 h was reported.26 Our pharmacokinetic study of EGFRvIII:CD3 bi-scFv in mice showed initial and terminal half-life’s that are very similar to blinatumomab and which are typical for various antibody fragments (Table 3). 27, 28 Further studies of EGFRvIII:CD3 bi-scFv in other models are in development and will be useful for determining an inter-species range of pharmacokinetic parameters and for doing allometric conversion to humans.29
The short half-life of our bispecific antibody will require careful clinical implementation and poses unique challenges and opportunities. On one hand, bispecific antibodies are extremely potent and have led to numerous serious adverse events, including death, in patients enrolled in clinical trials. 4, 30 In fact, blinatumomab, lists death as a potential side effect on its packaging insert.31 Thus, a short half-life can help control drug-concentration and allows rapid discontinuation of the drug in patients facing unmanageable side effect. On the other hand, EGFRvIII:CD3 bi-scFv will likely require continuous intravenous infusion to retain biological efficacy, especially when compared with full-length antibodies that maintain stable serum concentrations for several weeks following single bolus injections. Continuous infusion can be burdensome for both clinical trial operations and patient quality of life and the treatment must thus show a considerable clinical benefit to offset these limitations.
In conclusion, we have developed a sensitive and high-throughput LC-MS assay for the absolute quantification of EGFRvIII:CD3 bi-scFv in plasma and whole blood. Using this assay, we obtained pharmacokinetic parameters in mice that will be crucial for advancing this therapeutic into human clinical trials.
Supplementary Material
Acknowledgements
We thank Dr. Chris Shuford (Laboratory Corporation of America) for advice on assay development. We thank Darby Larson for his assistance with EGFRvIII:CD3 bi-scFv production. We thank David Snyder for his assistance with animal studies. This work was supported by R01NS085412 and U01-NS090284 grants to John H Sampson from the National Institute of Health. This work was also supported by grant 2P30-CA014236-41 (Comprehensive Cancer Center Core Grant – Pharmaceutical Research) to Ivan Spasojevic.
Abbreviations
- AP-MS
affinity purification coupled to mass spectrometry
- bi-scFv
Bi-specific single change antibody fragments
- CDR
complementarity-determining region
- cGMP
current Good Manufacturing Protocol
- CL
clearance
- GBM
glioblastoma
- IT
injection time
- LBA
ligand binding assay
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
- LOQ
limit of quantification
- MRT
mean residence time
- PK
pharmacokinetics
- PRM
parallel reaction monitoring
- PBMC
peripheral blood mononuclear cell
- QC
quality control
- SD
standard deviation
- SIL
stable isotope labeled
- UPLC
ultraperformance liquid chromatography
- Vss
volume of distribution at steady state
- v/v
volume per volume
- WB
whole blood
Footnotes
John Sampson has an equity interest in Annias Immunotherapeutics, which has licensed intellectual property from Duke related to the use of the pepCMV vaccine in the treatment of glioblastoma multiforme. J.H.S. has an equity interest in Istari Oncology, which has licensed intellectual property from Duke related to the use of poliovirus and D2C7 in the treatment of glioblastoma. John Sampson is an inventor on patents related to PEP-CMV DC vaccine with tetanus, as well as poliovirus vaccine and D2C7 in the treatment of glioblastoma.
Associated Content
Supporting Information
The following supporting information is available free of charge on the ACS Website at http://pubs.acs.org. Method of the LC-MS/MS analysis of 13C15N EGFRvIII:CD3 bi-scFv; Table S1A. Inclusion list for plasma PK assay; Table S1B. Inclusion list for whole blood PK assay; Figure S1. EGFRvIII:CD3 bi-scFv sequence coverage and peptides with homology to mouse proteome; Figure S2. PRM analysis of 13C15N EGFRvIII:CD3 bi-scFv incorporation; Figure S3. Divert Valve Setup; Curated Skyline files “Plasma_PK_bi_scFv” and “WB_PK_bi_scFv” and associated raw data were uploaded to the Panorama Targeted Proteomics data repository and can be accessed at https://panoramaweb.org/31jFrQ.url and via ProteomeXchange (PXD012472).
References
- 1.Morrissey KM; Yuraszeck TM; Li CC; Zhang Y; Kasichayanula S, Immunotherapy and Novel Combinations in Oncology: Current Landscape, Challenges, and Opportunities. Clin Transl Sci 2016, 9 (2), 89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT; Gittleman H; Fulop J; Liu M; Blanda R; Kromer C; Wolinsky Y; Kruchko C; Barnholtz-Sloan JS, CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol 2015, 17 Suppl 4, iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gedeon PC; Schaller TH; Chitneni SK; Choi BD; Kuan CT; Suryadevara CM; Snyder DJ; Schmittling RJ; Szafranski SE; Cui X; Healy PN; Herndon JE 2nd; McLendon RE; Keir ST; Archer GE; Reap EA; Sanchez-Perez L; Bigner DD; Sampson JH, A Rationally Designed Fully Human EGFRvIII:CD3-Targeted Bispecific Antibody Redirects Human T Cells to Treat Patient-derived Intracerebral Malignant Glioma. Clin Cancer Res 2018, 24 (15), 3611–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staatz CE; Taylor PJ; Tett SE, Comparison of an ELISA and an LC/MS/MS method for measuring tacrolimus concentrations and making dosage decisions in transplant recipients. Ther Drug Monit 2002, 24 (5), 607–15. [DOI] [PubMed] [Google Scholar]
- 5.de Jong LA; Uges DR; Franke JP; Bischoff R, Receptor-ligand binding assays: technologies and applications. J Chromatogr B Analyt Technol Biomed Life Sci 2005, 829 (1–2), 1–25. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen T; Mistarz UH; Costa N; Herbet A; Boquet D; Becher F; Rand KD, Investigating the utility of minimized sample preparation and high-resolution mass spectrometry for quantification of monoclonal antibody drugs. J Pharm Biomed Anal 2018, 159, 384–392. [DOI] [PubMed] [Google Scholar]
- 7.van den Broek I; Niessen WM; van Dongen WD, Bioanalytical LC-MS/MS of protein-based biopharmaceuticals. J Chromatogr B Analyt Technol Biomed Life Sci 2013, 929, 161–79. [DOI] [PubMed] [Google Scholar]
- 8.Lebert D; Dupuis A; Garin J; Bruley C; Brun V, Production and use of stable isotope-labeled proteins for absolute quantitative proteomics. Methods Mol Biol 2011, 753, 93–115. [DOI] [PubMed] [Google Scholar]
- 9.Vessman J; Stefan R; Van Staden JF; Danzer K; Lindner W; Burns DT; Fajgelj A; Mueller H, Selectivity in analytical chemistry (IUPAC Recommendations 2001). Pure and Applied Chemistry 2001, 73 (8). [Google Scholar]
- 10.Strop P; Liu SH; Dorywalska M; Delaria K; Dushin RG; Tran TT; Ho WH; Farias S; Casas MG; Abdiche Y; Zhou D; Chandrasekaran R; Samain C; Loo C; Rossi A; Rickert M; Krimm S; Wong T; Chin SM; Yu J; Dilley J; Chaparro-Riggers J; Filzen GF; O’Donnell CJ; Wang F; Myers JS; Pons J; Shelton DL; Rajpal A, Location matters: site of conjugation modulates stability and pharmacokinetics of antibody drug conjugates. Chem Biol 2013, 20 (2), 161–7. [DOI] [PubMed] [Google Scholar]
- 11.Leon IR; Schwammle V; Jensen ON; Sprenger RR, Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol Cell Proteomics 2013, 12 (10), 2992–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallien S; Duriez E; Crone C; Kellmann M; Moehring T; Domon B, Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol Cell Proteomics 2012, 11 (12), 1709–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carr SA; Abbatiello SE; Ackermann BL; Borchers C; Domon B; Deutsch EW; Grant RP; Hoofnagle AN; Huttenhain R; Koomen JM; Liebler DC; Liu T; MacLean B; Mani DR; Mansfield E; Neubert H; Paulovich AG; Reiter L; Vitek O; Aebersold R; Anderson L; Bethem R; Blonder J; Boja E; Botelho J; Boyne M; Bradshaw RA; Burlingame AL; Chan D; Keshishian H; Kuhn E; Kinsinger C; Lee JS; Lee SW; Moritz R; Oses-Prieto J; Rifai N; Ritchie J; Rodriguez H; Srinivas PR; Townsend RR; Van Eyk J; Whiteley G; Wiita A; Weintraub S, Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics 2014, 13 (3), 907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuford CM; Walters JJ; Holland PM; Sreenivasan U; Askari N; Ray K; Grant RP, Absolute Protein Quantification by Mass Spectrometry: Not as Simple as Advertised. Anal Chem 2017, 89 (14), 7406–7415. [DOI] [PubMed] [Google Scholar]
- 15.Nouri-Nigjeh E; Zhang M; Ji T; Yu H; An B; Duan X; Balthasar J; Johnson RW; Qu J, Effects of calibration approaches on the accuracy for LC-MS targeted quantification of therapeutic protein. Anal Chem 2014, 86 (7), 3575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlereth B; Quadt C; Dreier T; Kufer P; Lorenczewski G; Prang N; Brandl C; Lippold S; Cobb K; Brasky K; Leo E; Bargou R; Murthy K; Baeuerle PA, T-cell activation and B-cell depletion in chimpanzees treated with a bispecific anti-CD19/anti-CD3 single-chain antibody construct. Cancer Immunol Immunother 2006, 55 (5), 503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson CM; Shulman NJ; MacLean B; MacCoss MJ; Hoofnagle AN, Skyline Performs as Well as Vendor Software in the Quantitative Analysis of Serum 25-Hydroxy Vitamin D and Vitamin D Binding Globulin. Clin Chem 2018, 64 (2), 408–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bioanalytical Method Validation Guidance for Industry. FDA, Ed. May 24, 2018. [Google Scholar]
- 19.Rieckmann JC; Geiger R; Hornburg D; Wolf T; Kveler K; Jarrossay D; Sallusto F; Shen-Orr SS; Lanzavecchia A; Mann M; Meissner F, Social network architecture of human immune cells unveiled by quantitative proteomics. Nat Immunol 2017, 18 (5), 583–593. [DOI] [PubMed] [Google Scholar]
- 20.Jia L; Zhang L; Shao C; Song E; Sun W; Li M; Gao Y, An attempt to understand kidney’s protein handling function by comparing plasma and urine proteomes. PLoS One 2009, 4 (4), e5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobo ED; Hansen RJ; Balthasar JP, Antibody pharmacokinetics and pharmacodynamics. J Pharm Sci 2004, 93 (11), 2645–68. [DOI] [PubMed] [Google Scholar]
- 22.Physiological Data Summary – C57BL/6J (000664). Laboratory TJ, Ed. 2007. [Google Scholar]
- 23.O’Connell KE; Mikkola AM; Stepanek AM; Vernet A; Hall CD; Sun CC; Yildirim E; Staropoli JF; Lee JT; Brown DE, Practical murine hematopathology: a comparative review and implications for research. Comp Med 2015, 65 (2), 96–113. [PMC free article] [PubMed] [Google Scholar]
- 24.Davies B; Morris T, Physiological parameters in laboratory animals and humans. Pharm Res 1993, 10 (7), 1093–5. [DOI] [PubMed] [Google Scholar]
- 25.Blincyto Assessment Report. (CHMP), C. f. M. P. f. H. U., Ed. 2015.
- 26.Klinger M; Brandl C; Zugmaier G; Hijazi Y; Bargou RC; Topp MS; Gokbuget N; Neumann S; Goebeler M; Viardot A; Stelljes M; Bruggemann M; Hoelzer D; Degenhard E; Nagorsen D; Baeuerle PA; Wolf A; Kufer P, Immunopharmacologic response of patients with B-lineage acute lymphoblastic leukemia to continuous infusion of T cell-engaging CD19/CD3-bispecific BiTE antibody blinatumomab. Blood 2012, 119 (26), 6226–33. [DOI] [PubMed] [Google Scholar]
- 27.Schneider EL; Hearn BR; Pfaff SJ; Fontaine SD; Reid R; Ashley GW; Grabulovski S; Strassberger V; Vogt L; Jung T; Santi DV, Approach for Half-Life Extension of Small Antibody Fragments That Does Not Affect Tissue Uptake. Bioconjug Chem 2016, 27 (10), 2534–2539. [DOI] [PubMed] [Google Scholar]
- 28.Cumber AJ; Ward ES; Winter G; Parnell GD; Wawrzynczak EJ, Comparative stabilities in vitro and in vivo of a recombinant mouse antibody FvCys fragment and a bisFvCys conjugate. J Immunol 1992, 149 (1), 120–6. [PubMed] [Google Scholar]
- 29.Mahmood I, Pharmacokinetic allometric scaling of antibodies: application to the first-in-human dose estimation. J Pharm Sci 2009, 98 (10), 3850–61. [DOI] [PubMed] [Google Scholar]
- 30.Smith J Cancer Immunotherapy Phase I on Hold After Patient Death. https://labiotech.eu/medical/affimed-cancer-immunotherapy-death/ (accessed 2018/10/21).
- 31.Blincyto (blinatumomab) [package insert]. Amgen Inc., Thousand Oaks, CA: 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.