ABSTRACT
Stressed cells downregulate translation initiation and assemble membrane-less foci termed stress granules (SGs). Although SGs have been extensively characterized in cultured cells, the existence of such structures in stressed adult stem cell pools remains poorly characterized. Here, we report that the Drosophila orthologs of the mammalian SG components AGO1, ATX2, CAPRIN, eIF4E, FMRP, G3BP, LIN-28, PABP and TIAR are enriched in adult fly intestinal progenitor cells, where they accumulate in small cytoplasmic messenger ribonucleoprotein complexes (mRNPs). Treatment with sodium arsenite or rapamycin reorganized these mRNPs into large cytoplasmic granules. Formation of these intestinal progenitor stress granules (IPSGs) depended on polysome disassembly, led to translational downregulation and was reversible. Although the canonical SG nucleators ATX2 and G3BP were sufficient for IPSG formation in the absence of stress, neither of them, nor TIAR, either individually or collectively, were required for stress-induced IPSG formation. This work therefore finds that IPSGs do not assemble via a canonical mechanism, raising the possibility that other stem cell populations employ a similar stress-response mechanism.
KEY WORDS: Drosophila; Intestinal progenitor; Stress granule; Intestinal stem cell, Messenger ribonucleoprotein particle; ROX8; ATX2; RIN
Highlighted Article: Previously defined canonical stress granule factors are not required for SG assembly in adult Drosophila progenitor cells, suggesting that other cell populations employ a similar noncanonical assembly mechanism in vivo.
INTRODUCTION
Eukaryotic cells form stress granules (SGs) in response to environmental stresses such as heat, oxidative stress, viral infections and UV irradiation (Anderson and Kedersha, 2006, 2009; van Leeuwen and Rabouille, 2019; Protter and Parker, 2016). The formation of these cytoplasmic assemblies, which contain both mRNAs and RNA-binding proteins (RBPs), is associated with a global reduction in protein translation. The connection between SG formation and protein translation is direct: pharmacological interventions that specifically affect polysome stability can both induce SGs in the absence of stress and also prevent SG formation in response to stress (Kedersha et al., 2000; Buchan et al., 2008; Buchan and Parker, 2009). While SGs allow cells to limit global translation during times of stress, they are also transient and dissolve as stress is relieved, allowing cells to return to normal programs of protein translation (Buchan and Parker, 2009; Wheeler et al., 2016). Although SG assembly involves the physical association between non-translating pre-initiation complexes and RBPs that contain ‘sticky’ stretches of internally disorganized regions (IDRs) (Wheeler et al., 2016), the precise mechanism underlying SG-mediated control of protein levels remains unresolved.
Based on biochemical analyses, SGs contain a solid core that is surrounded by a liquid-like shell that facilitates the rapid exchange of materials with the cytoplasm (Jain et al., 2016; Niewidok et al., 2018; Wheeler et al., 2016). Components of SGs identified by immunostaining of cultured mammalian cells treated with various stressors include poly(A)+ mRNAs, eukaryotic translation initiation factors, 40S ribosomes and a variety of RBPs (reviewed in Buchan and Parker, 2009). While the components and requirements for SG formation can differ depending on stress treatment (Aulas et al., 2017), a few of these RBPs are considered integral to SG assembly. These include ataxin-2 (ATX2, also known as ATXN2), the Ras GTPase-activating protein-binding proteins 1/2 (G3BP1/2) family proteins (Tourrière et al., 2003), and the T-cell internal antigen-1 (TIA1) and TIA1-related (TIAR, also known as TIAL1) family proteins (Kedersha et al., 1999). Forced expression of these integral RBPs can induce SG-like structures in the absence of stress (Kedersha et al., 2000; Tourrière et al., 2003), while their knockdown or elimination interferes with SG formation in a variety of stressed mammalian cells (Gilks et al., 2004; Kedersha et al., 2016; Nonhoff et al., 2007). Their requirement is also evolutionarily conserved; loss of the yeast orthologs of ATX2 and TIA1 interferes with SG assembly after glucose deprivation (Buchan et al., 2008), while knockdown of the fly ortholog of G3BP prevents arsenite-induced SGs in cultured Drosophila embryonic cells (Aguilera-Gomez et al., 2017). Other SG components, like caprin (CAPR, CAPRIN1 and CAPRIN2 in mammals) and Fragile X mental retardation protein (FMRP, also known as FMR1), can induce SG-like assemblies but are not absolutely required for SG formation (Didiot et al., 2009; Gareau et al., 2013; Kedersha et al., 2016). While functional analysis has focused on a relatively small subset of proteins, more recent biochemical analyses have identified as many as ∼300 mammalian and ∼200 yeast SG proteins (Jain et al., 2016; Markmiller et al., 2018; Youn et al., 2018). Key current questions include the molecular mechanisms that drive SG assembly and whether such mechanisms identified in cultured cells are also active across diverse cell type in vivo.
In this study, we identify and characterize the formation requirements of a class of SGs that form in progenitor cells in the adult Drosophila intestine. SG-like cytoplasmic foci have been previously identified in this model organism, including in injured brains and heat-stressed imaginal discs (Anderson et al., 2018; Jevtov et al., 2015), but neither the composition nor formation requirements of these structures have been investigated in detail. We previously found that the Drosophila orthologs of two mammalian SG components, LIN-28 (LIN28A and LIN28B in mammals) and FMRP, are abundantly expressed specifically in the cytoplasm of intestinal progenitor cells (Chen et al., 2015; Luhur et al., 2017). This cell population sustains the intestinal epithelium, an actively regenerating tissue that is constantly exposed to various types of stressors (Jiang et al., 2016), and is composed of two main cell types, mitotic intestinal stem cells (ISCs) and their non-mitotic and transient daughters, known as enteroblasts (EBs), which are destined for terminal differentiation (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006) (see Fig. S1A for a schematic of the ISC lineage as well as markers used to label lineage cell types). Here, we report that LIN-28 and FMRP are members of a much larger class of RNA-associated proteins, including the Drosophila orthologs of ATX2, G3BP1/2 and TIA1/TIAR, which are enriched in intestinal progenitor cells and reorganize into SG-like structures after intestines are exposed to stress treatments ex vivo. Importantly, while progenitor cells form SGs after stress treatment, their neighboring terminally differentiated cells do not under the conditions tested. Finally we show that, while these structures display key features of SGs, their formation does not require Drosophila ATX2, G3BP1/2 or TIA1/TIAR, either alone or in combination.
RESULTS
Stress granule components are enriched in intestinal progenitor cells
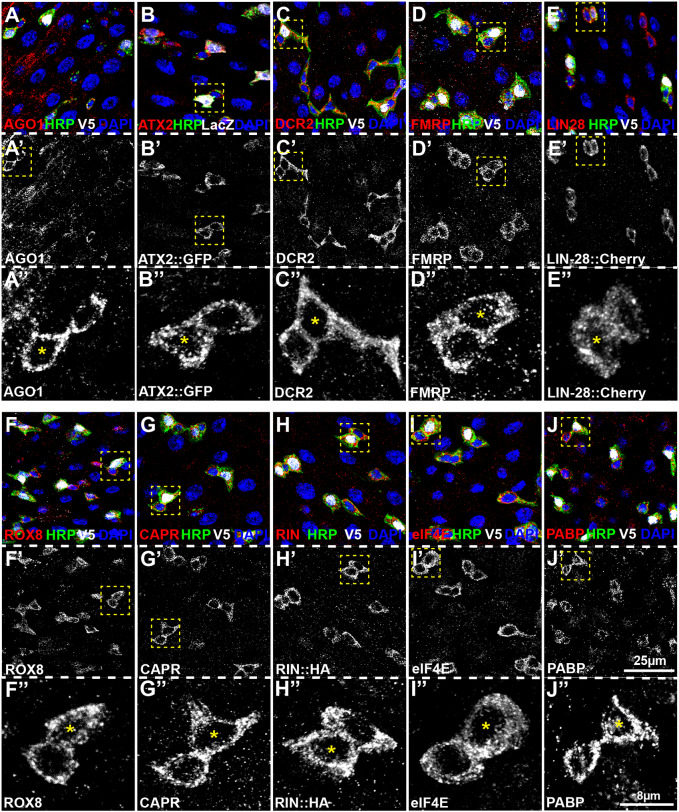
The enrichment of LIN-28 and FMRP in adult Drosophila intestinal progenitor cells (Chen et al., 2015; Luhur et al., 2017) raised the possibility that the fly orthologs of other known mammalian SG proteins might be similarly enriched in intestinal progenitors. To test this possibility, we stained intestines of 7-day-old wild-type adult female Drosophila reared under standard conditions using a series of antibodies or translational reporters. In addition to LIN-28 and FMRP, this series included representative examples of three main classes of SG proteins; RBPs linked to translational regulation or decay [Argonaute-1 (AGO1), ATX2, Dicer-2 (DCR2), and TIA1/TIAR, known in Drosophila as ROX8)], RBPs that regulate aspects of RNA metabolism other than translation or decay [CAPR and G3BP1/2, known in Drosophila as Rasputin (RIN)], and components of stalled initiation complexes [eukaryotic translation initiation factor 4E (eIF4E) and poly(A)-binding protein (PABP)] (Anderson and Kedersha, 2008; Balzer and Moss, 2007; Kedersha et al., 1999, 2002, 2005; Leung et al., 2006; Mazroui et al., 2002; Nonhoff et al., 2007; Solomon et al., 2007; Tourrière et al., 2003). Although there is a second TIA1-related protein encoded in the Drosophila genome, we did not include it in our study because it has less similarity to TIA1/TIAR than ROX8 and is not expressed in intestinal cells based on transcriptome data (Fig. S1B). We used previously published and verified reagents to detect all of these proteins except for RIN, for which we generated a homozygous viable rin::HA strain via CRISPR/Cas9 genome editing that contained an HA epitope immediately downstream of the start methionine common to all known rin transcripts (Fig. S1C), and ROX8, for which we generated and validated a rat polyclonal antibody against full-length ROX8 (see Fig. S1D–H). Strikingly, all ten of these markers displayed elevated signal in intestinal progenitor cells (Fig. 1A–J,A′–J′), identified in this experiment via horseradish peroxidase (HRP) staining (O'Brien et al., 2011). The progenitor cell staining was similar in all cases, appearing almost exclusively cytoplasmic and displaying no apparent difference in level or organization between ISCs and EBs, which were distinguished by the expression of EB markers gbe-LacZ (Micchelli and Perrimon, 2006) or 3Xgbe-smGFP::V5::nls. The distribution of all ten proteins within individual progenitor cells appeared non-uniform and somewhat granular as detected by conventional confocal microscopy (Fig. 1A″–J″). Thus, these staining patterns indicated the presence of a dense web of messenger ribonucleoprotein (mRNP) particles distributed throughout the cytoplasm of intestinal progenitor cells under homeostatic conditions.
Fig. 1.
SG proteins accumulate in adult Drosophila intestinal progenitor cells. (A–J) Confocal micrographs of posterior midguts stained for (A) AGO1, (B) ATX2::GFP, (C) DCR2, (D) FMRP, (E) LIN-28::Cherry, (F) ROX8, (G) CAPR, (H) RIN, (I) eIF4E and (J) PABP. 3Xgbe-smGFP::V5::nls (green in A,C–J) and gbe-LacZ (green in B) label enteroblast cells, HRP (white) labels both EBs and ISCs, and the DNA-dye DAPI (blue) labels all cells. Grayscale images of each stained protein are also shown in A′–J′, with a higher magnification image of progenitor pairs marked with yellow dotted outlines shown in A″–J″. Cells labeled with a yellow asterisk (*) in A″–J″ are gbe+, HRP+ enteroblasts while neighboring gbe−, HRP+ cells are ISCs.
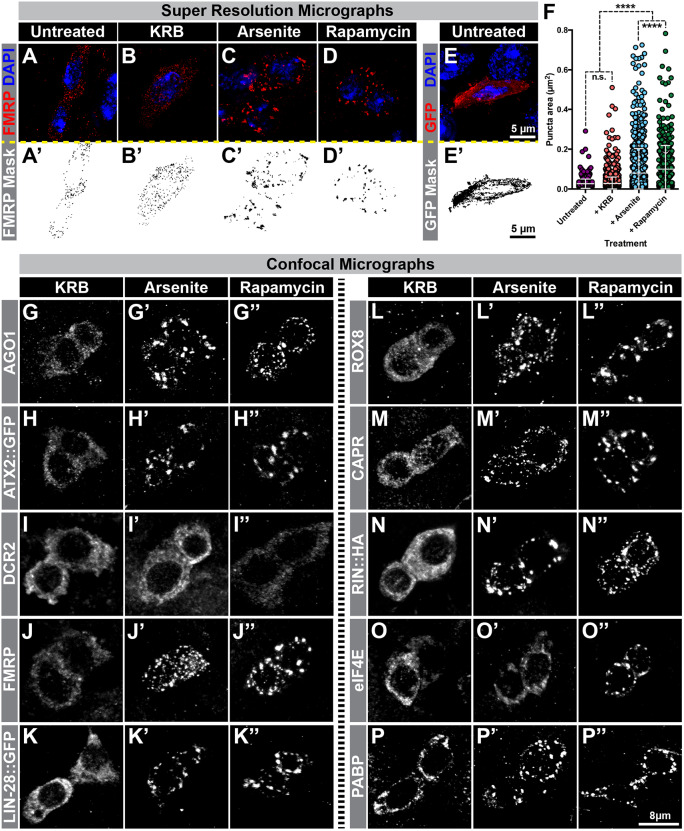
To rigorously assess the apparent granularity of the progenitor cytoplasm, we turned to structured illumination super-resolution microscopy and focused on FMRP distribution as a proxy for this collection of progenitor-enriched SG proteins. Under normal untreated conditions, FMRP appeared as a constellation of distinct small dots distributed throughout the cytoplasm (Fig. 2A,A′). These cytoplasmic puncta had an average area of 0.027±0.023 µm2 (mean±s.d., n=563 puncta, seven cells) but were present in a broad range of sizes, with ∼2% being at least 5-fold larger than average (Fig. 2F). As a control, we compared the FMRP distribution to GFP and found that the GFP pattern was not punctate but rather appeared as a meshwork present throughout the cell (Fig. 2E,E′). Because there were no distinct signal intensity maxima, we did not quantify the GFP distribution. This analysis suggested that the cytoplasm of adult intestinal progenitor cells contained a set of FMRP+ mRNP complexes that were present during normal homeostatic conditions.
Fig. 2.
Arsenite and rapamycin treatment induce IPSGs. (A–E) Super resolution micrographs of intestinal progenitor cells that are either (A,E) untreated or treated with (B) KRB (control), (C) 1 mM sodium arsenite in KRB, or (D) 1 mM rapamycin in KRB and stained for FMRP (red in A–D, black in A′–D′) or GFP (red in E, black in E′) and DAPI (blue in A–E). (F) Scatter dot plots of FMRP puncta area in untreated progenitor cells (n=563 puncta, 7 cells) or following KRB (n=2039 puncta, 15 cells), 1 mM sodium arsenite (n=1096 puncta, 20 cells), or 1 mM rapamycin (n=476 puncta, 18 cells). Graph shows mean±s.d. ****P<0.0001; n.s., not significant (Kruskal–Wallis test). (G–P) Grayscale confocal micrographs of progenitor cell pairs from intestines treated with KRB (G–P), 1 mM sodium arsenite in KRB (G′–P′), or 1 mM rapamycin in KRB (G″–P″) and stained for (G–G″) AGO1, (H–H″) ATX2::GFP, (I–I″) DCR2, (J–J″) FMRP, (K–K″) LIN-28::GFP, (L–L″) ROX8, (M–M″) CAPR, (N–N″) RIN::HA, (O–O″) eIF4E and (P–P″) PABP.
The enrichment of SG proteins in progenitor cells relative to surrounding cells suggested that these proteins were downregulated during differentiation. To evaluate this idea, we examined the distribution of RIN, another SG protein identified above, in progenitor cells as they differentiated into hormone-producing enteroendocrine cells (EEs). To track this process, we counterstained for piezo-Gal4-driven GFP, which labeled ISCs destined to differentiate into EEs as well as newborn EEs (He et al., 2018), and Prospero (Pros), which marked both newborn and mature EEs (Micchelli and Perrimon, 2006). We used RIN::HA in this case as a proxy for progenitor-enriched SG proteins due to compatibility with antibodies needed to detect both piezo-Gal4-driven GFP and Pros. Cytoplasmic RIN::HA staining was abundantly detected in piezo+, Pros− ISCs but almost entirely absent from piezo−, Pros+ mature EEs, indicating a reduction in its level during EE differentiation (Fig. S2A–A″). This observation, along with the expression patterns shown in Fig. 1, suggested the presence of an active post-transcriptional control program in progenitor cells that is cleared during differentiation.
mRNPs form large cytoplasmic granules after exposure to stress
As their name suggests, SG proteins can coalesce to form dense aggregates in some cell types experiencing environmental stress (Buchan and Parker, 2009; Kedersha et al., 1999). The prevalence of SG proteins in intestinal progenitor cells prompted us to evaluate whether SG formation is also a feature of this cell type. To induce stress, we treated intestines ex vivo and prior to fixation with either sodium arsenite or rapamycin prepared in Krebs Ringer bicarbonate buffer (KRB) at concentrations known to induce SGs in other cell types (McEwen et al., 2005; Panas et al., 2016). Control intestines were prepared in parallel and treated with KRB alone for the same length of time. For this analysis, we focused on FMRP, since FMRP is considered among the most reliable stress granule markers in mouse and human tissue culture cells (Kedersha and Anderson, 2007). Arsenite or rapamycin treatment significantly altered FMRP granule appearance: while the average size of FMRP granules increased ∼2.9-fold from 0.028±0.032 µm2 (mean±s.d., n=2039 puncta, 15 cells) after KRB treatment to 0.080±0.12 µm2 (n=1096 puncta, 20 cells) after arsenite or ∼3.6-fold to 0.100±0.12 µm2 (n=476 puncta, 18 cells) after rapamycin treatment (Fig. 2B–D,F), the total number of granules decreased from 138±58 puncta per cell (n=14) in KRB to only 50±17 puncta or 32±12 puncta per cell (n=16 or n=13) in arsenite or rapamycin treatment, respectively. This redistribution occurred in greater than 90% of progenitor cells after either arsenite or rapamycin treatment, in contrast to only 5% of cells after KRB-only treatment, based on quantification of cells that displayed fewer but visibly larger FMRP punctae (Fig. S3A–C). These and subsequent experiments were performed with 1 mM sodium arsenite or rapamycin, which we found was the lowest concentration to consistently induce stress granules. Consistent with the low level of SG protein expression in cells surrounding progenitor cells (Fig. 1A), arsenite or rapamycin treatment failed to induce the formation of punctae in labeled Pros+ EEs (Fig. S2B–D) or in non-progenitor cells (Fig. S3B,C). Collectively, these alterations suggested that pre-existing mRNPs in progenitor cells reorganized into distinctively larger stress-induced particles after stress. Because these stress-triggered granules seemed to be limited to progenitor cells in the intestinal epithelium, we refer to them as intestinal progenitor stress granules (IPSGs). IPSGs formed in progenitor cells throughout the intestine but because progenitor cells are most abundant and highly active in the posterior portion of the midgut (Marianes and Spradling, 2013; O'Brien et al., 2011), we focused on this region for all of our subsequent analyses.
To evaluate whether other proteins in addition to FMRP were recruited to IPSGs, we immunostained control and treated intestines for the protein series described above. After either arsenite or rapamycin treatment, AGO1, ATX2::GFP, LIN-28::GFP, ROX8, CAPR, RIN::HA and PABP were detected in large cytoplasmic punctae, indicating recruitment to IPSGs (Fig. 2G,H,J–N,P). In contrast, DCR2 did not show a distribution change after either treatment (Fig. 2I). Interestingly, eIF4E localized to IPSGs only after rapamycin treatment but not after arsenite treatment (Fig. 2O), suggesting that IPSGs induced by different stressors have different protein composition. Consistent with this, we noted that rapamycin treatment resulted in fewer and larger punctae relative to arsenite treatment (compare Fig. 2L′ and 2L″ or 2M′ and 2M″, quantified in Fig. 2F). Taken together, these data show that the SGs that form in intestinal progenitor cells after exposure to environmental stress are similar in protein composition to the SGs known to form in other cell types.
SG proteins colocalize in IPSGs after arsenite and rapamycin treatments
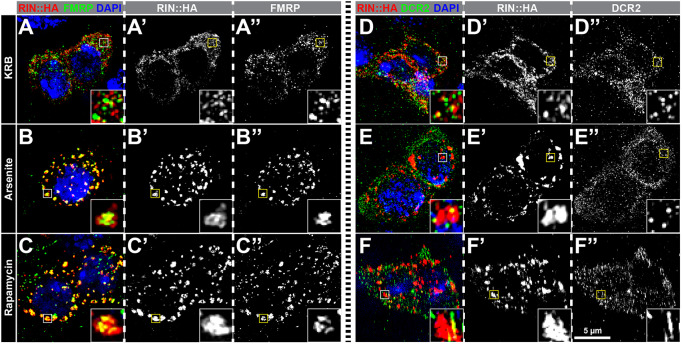
To determine whether individual SG proteins are recruited to the same IPSGs, we simultaneously analyzed FMRP and RIN::HA distribution in progenitor cells using super-resolution microscopy. After KRB treatment, FMRP and RIN::HA displayed distinct although partially overlapping patterns of small dots (Pearson's R value=0.26) distributed throughout the cytoplasm (Fig. 3A). These patterns suggested that mRNP distribution in the absence of acute stress is heterogenous, with a combination of FMRP+ RIN+, FMRP+ RIN−, and FMRP− RIN+ complexes. In contrast, FMRP and RIN::HA signals were almost entirely overlapping after arsenite (Pearson's R value=0.67) and rapamycin (Pearson's R value=0.65) treatments (Fig. 3B,C). This redistribution suggests that these treatments caused RIN::HA and FMRP granules to jointly aggregate into larger IPSGs. Careful inspection of the overlap found that RIN::HA staining was more broadly distributed while FMRP was more centrally distributed, suggesting an organized arrangement of components within IPSGs. We used DCR2, which was not recruited to IPSGs (Fig. 2I) as a negative control, and found that DCR2 signal does not overlap with RIN::HA signal after either KRB (Pearson's R value=0.24), arsenite (Pearson's R value=0.14) or rapamycin (Pearson's R value=0.13) treatment (Fig. 3D–F). These results, therefore, indicate that IPSGs contain an organized mixture of SG components.
Fig. 3.
IPSG components colocalize in arsenite- and rapamycin-induced IPSGs. Super resolution micrographs of progenitor cells stained for (A–C) RIN::HA (red), FMRP (green) and DAPI (blue) or (D–F) RIN::HA (red), DCR2 (green) and DAPI (blue) after treatment with (A,D) KRB, (B,E) 1 mM sodium arsenite in KRB, or (C,F) 1 mM rapamycin in KRB. Insets are enlarged views of boxed regions in the main panels. RIN::HA and FMRP, and RIN::HA and DCR2 show only occasional colocalization under control conditions (A,D). However, after arsenite or rapamycin treatments, RIN::HA and FMRP colocalize with each other (B,C). In contrast, DCR2 does not colocalize with RIN::HA following stress induction (E,F).
IPSG and SG formation share key features
SG formation in other cell types has a number of well-characterized features, including the recruitment of mRNAs, a requirement for polysome disassembly, and reversibility after stress withdrawal (Anderson and Kedersha, 2002, 2006; Buchan and Parker, 2009; Kedersha et al., 2000). To compare IPSG formation with SG formation, we assessed whether IPSGs shared these properties. Since mammalian SGs are known to contain mRNAs as indicated by the accumulation of fluorescently tagged oligo-dT (Anderson and Kedersha, 2002; Jain et al., 2016; Kedersha et al., 1999), we first visualized poly-A+ mRNAs in progenitor cells following each treatment. Oligo-dT detected signals that were uniformly distributed throughout progenitor cytoplasm in the absence of stress (Fig. 4A). In contrast, oligo-dT staining was punctate after arsenite or rapamycin treatment, indicating the recruitment of poly-A+ mRNAs to IPSGs (Fig. 4B,C). Next, we assessed whether cycloheximide (CHX) treatment affected IPSG formation, since CHX stabilizes polysomes and is known to block SG formation in mammalian as well as in yeast cells (Buchan and Parker, 2009; Buchan et al., 2008; Kedersha and Anderson, 2009; Kedersha et al., 2000). Consistent with this, IPSGs were not detected after arsenite or rapamycin treatment in intestines pre-treated with CHX for 30 min as compared with samples pre-treated with KRB for the same length of time (Fig. 4D–G). Conversely, puromycin treatment readily induced IPSGs (Fig. 4H,I), consistent with previous results indicating that puromycin induces polysome disassembly and promotes SG formation in other cell types (Buchan and Parker, 2009; Buchan et al., 2008; Kedersha et al., 2000). Finally, because SGs are reversible cytoplasmic entities that get cleared away during recovery from stress (Anderson and Kedersha, 2002; Protter and Parker, 2016), we tested whether IPSGs were also cleared by treating intestines first with arsenite, rapamycin or puromycin for 45 min and then with Schneider's medium, a nutrient-rich medium, for 1 h to relieve stress. Similar to canonical SGs, IPSGs dissolved during recovery (Fig. 4J–O). Collectively, these results indicate that, similar to SG formation in other cell types, IPSG formation was part of an ordered process activated in intestinal progenitors to resist stress, rather than some indirect consequence of harsh tissue treatment whose effects persisted after the treatment was removed.
Fig. 4.
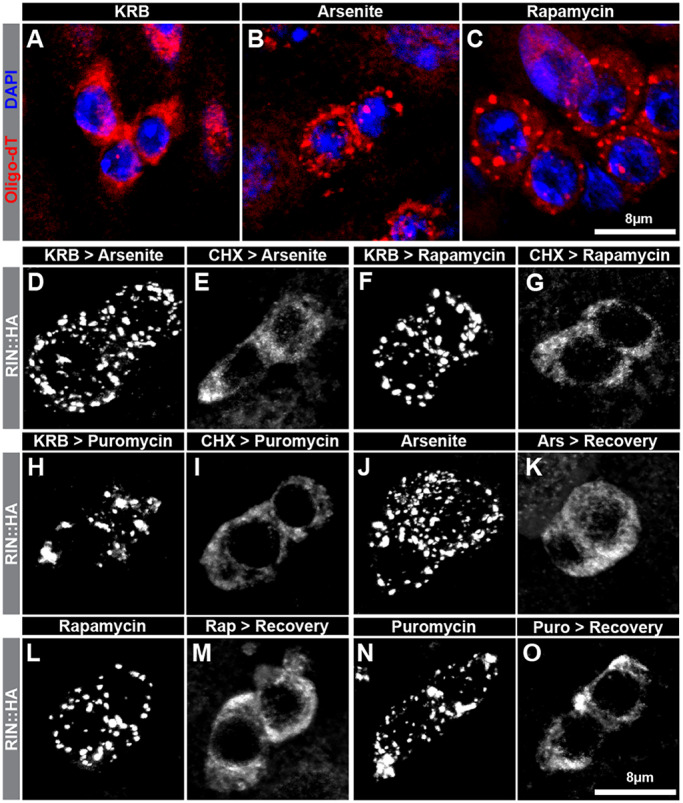
IPSGs contain mRNAs and their formation can be blocked and reversed. (A–C) Confocal micrographs of HRP+ progenitor cells treated ex vivo with (A) KRB, (B) 1 mM sodium arsenite in KRB, or (C) 1 mM rapamycin in KRB and stained with fluorescently tagged oligo-dT (red) and DAPI (blue). Note that HRP staining is not shown in these images. (D–I) Grayscale images of RIN::HA-stained progenitor cell pairs treated with either (D,F,H) KRB, or (E,G,I) cycloheximide (CHX) for 30 min followed by (D,E) 1 mM sodium arsenite for 60 min, (F,G) 1 mM rapamycin for 60 min, or (H,I) 250 µg/ml puromycin. (J–O) Grayscale images of RIN::HA-stained progenitor cell pairs treated with (J) 1 mM sodium arsenite in KRB for 45 min, (K) 1 mM sodium arsenite in KRB for 45 min followed by Schneider's medium for 60 min, (L) 1 mM rapamycin in KRB for 45 min, (M) 1 mM rapamycin in KRB for 45 min followed by Schneider's medium for 60 min, (N) 250 µg/ml puromycinin in KRB for 45 min, or (M) 250 µg/ml puromycin in KRB for 45 min followed by Schneider's medium for 60 min.
IPSG formation is associated with reduced protein synthesis
The above observations indicate that stress simultaneously induces the coalescence of mRNAs and the disassembly of polysomes, and suggest that IPSG formation might be associated with reduced protein production. To measure active protein synthesis in intestinal progenitors, we adapted an approach based on the modified puromycin analog O-propargyl-puromycin (OPP) (Strassburger et al., 2017). This method allows the fast and sensitive detection of protein synthesis using fluorescence microscopy after a highly specific Alexa-Fluor click reaction. Applying this method to intestines in which progenitor cells were labeled with GFP driven by the progenitor-specific escargot (esg) gene (Micchelli and Perrimon, 2006), high levels of OPP staining were detected in GFP+ cells after KRB treatment indicating robust mRNA translation within progenitors (Fig. 5A). However, following arsenite or rapamycin treatments, progenitor OPP signal was undetectable (Fig. 5B,D) and significantly lower than in the KRB control (Fig. 5K). As expected, this decrease in OPP level correlated with IPSG formation as detected by RIN::HA redistribution (Fig. 5F,G,I). To assess whether the subsequent loss of IPSGs was associated with a return to higher protein synthesis, we analyzed OPP levels in cells subjected to the same recovery regimen that had led to the disassembly of IPSGs (Fig. 4J–M). Progenitor cells one hour after recovery had a noticeably higher level of OPP staining than they did before recovery (compare Fig. 5B′ with C′, 5D′ with E′, 5G′ with H′ and 5I′ with J′; see Fig. 5L,M for quantification) and this higher OPP staining correlated with loss of IPSGs (compare 5G″ with H″, and I″ with J″). This inverse correlation between OPP level and IPSGs was particularly clear in neighboring progenitor cells that were at different stages of IPSG disassembly (Fig. 5N,O), and suggested a tight correlation between the restoration of translation and IPSG disassembly. Therefore, our data indicate that IPSG formation is associated with reduced mRNA translation in progenitor cells experiencing acute stress.
Fig. 5.
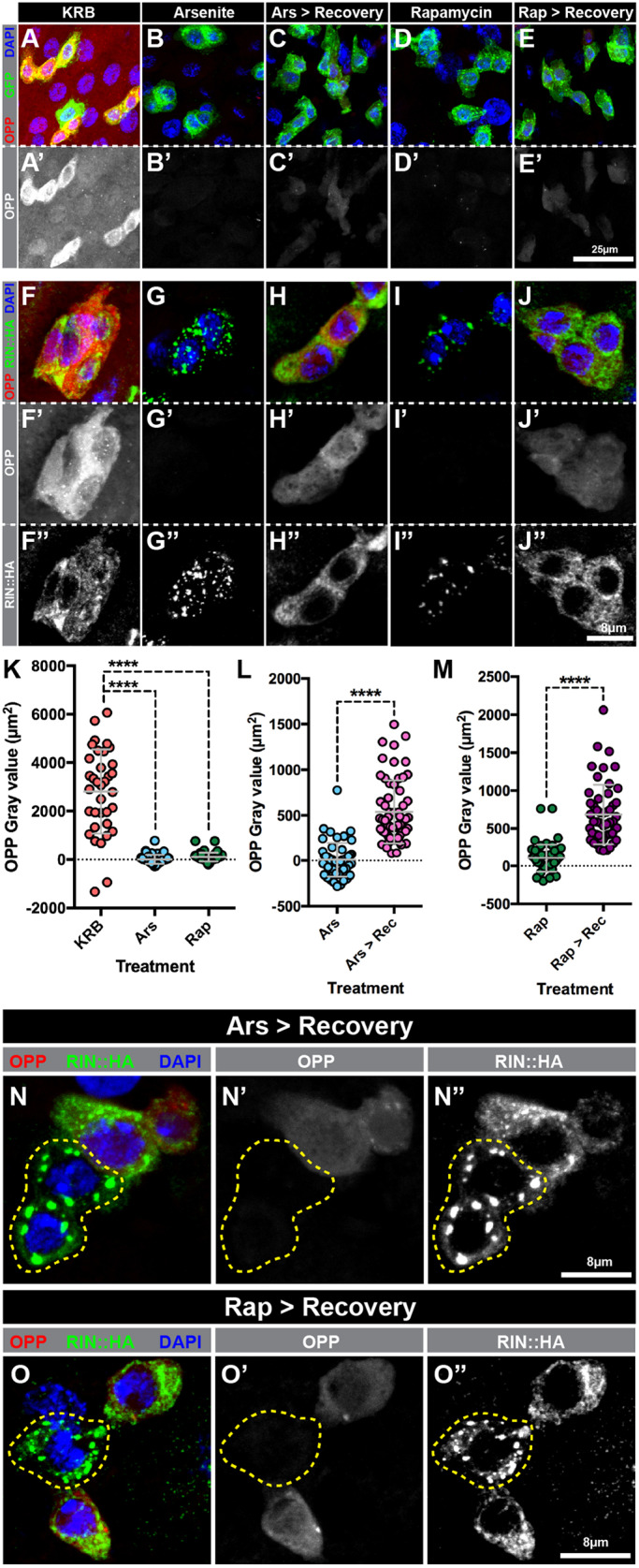
SG formation attenuates nascent protein synthesis in intestinal progenitor cells following acute stresses. Fluorescence micrographs of progenitor cells treated with (A,F) KRB for 45 min, (B,G) 1 mM sodium arsenite in KRB for 45 min, (C,H) 1 mM sodium arsenite in KRB for 45 min followed by Schneider's medium for 60 min, (D,I) 1 mM rapamycin in KRB for 45 min, or (E,J) 1 mM rapamycin in KRB for 45 min followed by Schneider's medium for 60 min and stained for (A–E) OPP (red in A–E, white in A′–E′), RIN::HA (green in A–E, white in A″–E″) and DAPI (blue), or (F–J) OPP (red in F–J, white in F′–J′), esg-Gal4>UAS-GFP (green) and DAPI (blue). (K) Scatter dot plot of normalized OPP fluorescence intensity of progenitor cells treated with KRB (n=35 cells, 6 intestines), 1 mM sodium arsenite in KRB (n=52 cells, 5 intestines) or 1 mM rapamycin in KRB (n=52 cells, 5 intestines). Normalized OPP fluorescence was calculated as the ratio of OPP fluorescence in progenitor cells relative to OPP fluorescence of neighboring cells. (L,M) Scatter dot plots of normalized OPP fluorescence intensity of progenitor cells treated with (L) 1 mM sodium arsenite in KRB (n=52 cells, 5 intestines) or 1 mM sodium arsenite in KRB followed by Schneider's medium (n=60 cells, 6 intestines), and (M) 1 mM rapamycin in KRB or 1 mM rapamycin in KRB followed by Schneider's medium (n=51 cells, 5 intestines). Graphs show mean±s.d. ****P<0.0001; n.s., not significant [Kruskal–Wallis test (K) and Mann–Whitney test (L,M)]. (N,O) Confocal micrographs of intestinal progenitors treated with (N) 1 mM sodium arsenite in KRB for 45 min followed by Schneider's medium for 60 min, or (O) 1 mM rapamycin in KRB for 45 min followed by Schneider's medium for 60 min and stained for OPP (red, N′ and O′), RIN::HA (green, N″ and O″) and DAPI (blue). Cells with persistent IPSGs are outlined by a yellow dotted line.
ATX2 and RIN are sufficient for IPSG formation
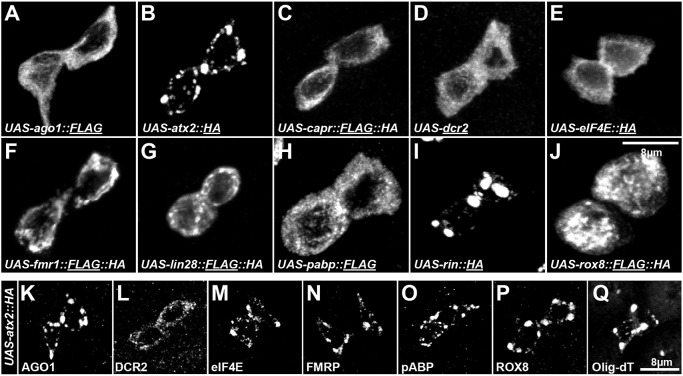
We next asked whether high levels of any IPSG components might be sufficient to induce stress granules even in the absence of acute stress. To identify such components, we overexpressed each of the ten proteins specifically in adult intestinal progenitor cells with a temperature sensitive version of esg-Gal4 (esgTS) for 24 h (Micchelli and Perrimon, 2006). ATX2 and RIN each coalesced into IPSG-like granules while the other eight components – AGO1, CAPR, DCR2, eIF4E, FMRP, LIN-28, ROX8 and PABP – failed to form cytoplasmic aggregates (Fig. 6A–J) and also failed to affect the cytoplasmic distribution of either FMRP (Fig. S4A–G) or ROX8 (Fig. S4H–N). All transgenes used in this experiment drove high expression of their respective protein products (Fig. 6A–J). However, since only the LIN-28 and FMRP transgenes have been previously verified (Luhur et al., 2017), one caveat with this experiment is that the transgenic versions of the proteins tested were not functional. To address this caveat, we tested whether sodium arsenite treatment caused transgenic AGO1, CAPR, DCR2, eIF4E, PABP and ROX8 proteins to aggregate. We observed that after such treatment, overexpressed AGO1, CAPR, PABP, ROX8 and eIF4E were able to aggregate (Fig. S4O,P,R–T), indicating that these transgenic proteins share at least some characteristics of their endogenous versions. As expected, overexpressed DCR2 failed to localize to IPSGs even after arsenite exposure (Fig. S4Q). Altogether, these observations suggest that AGO1, CAPR, DCR2, eIF4E, FMRP, LIN-28, PABP and ROX8 are not sufficient to form IPSGs in the absence of stress, although we cannot rule out the possibility that differences in transgenic protein levels may contribute to the effects we observed. To verify that ATX2 expression induced bona fide IPSGs, we analyzed the expression of other IPSG components and found that AGO1, eIF4E, FMRP, PABP and ROX8 as well as poly-A+ mRNAs localized to ATX2-induced IPSGs while, consistent with our previous observations, DCR2 failed to (Fig. 6K–Q). Furthermore, cells with ATX2-induced IPSGs showed low levels of OPP signal (Fig. S4U,V). Therefore, like stress-induced IPSGs, ATX2-induced IPSGs downregulated protein synthesis even in the absence of stress. These results indicate that ATX2 and RIN are sufficient for IPSG formation in a stress-independent manner. Interestingly, overexpression of RIN in Drosophila S2 cells failed to induce SGs (Aguilera-Gomez et al., 2017), indicating that SG formation following RIN overexpression is context dependent.
Fig. 6.
Overexpression of ATX2 or RIN induces IPSGs. (A–J) Grayscale confocal micrographs of progenitor cells with elevated expression of (A) AGO1::FLAG, (B) ATX2::HA, (C) CAPR::FLAG::HA, (D) DCR2, (E) eIF4E::HA, (F) FMRP::HA, (G) LIN-28::FLAG::HA, (H) pABP::FLAG, (I) RIN::HA or (J) ROX8:: FLAG::HA. The stained epitope is underlined in each panel. Note that only the elevated expression of ATX2 and RIN causes aggregation of cytoplasmic granules. (K–R) Grayscale confocal images of progenitor cells with elevated expression of ATX2::HA stained for (K) AGO1, (L) DCR2, (M) eIF4E, (N) FMRP, (O) PABP, (P) ROX8 and (Q) Oligo-dT. Similar to what is seen with acute stress-induced IPSGs, stress granules induced by ATX2 overexpression contain multiple RBPs as well as mRNAs.
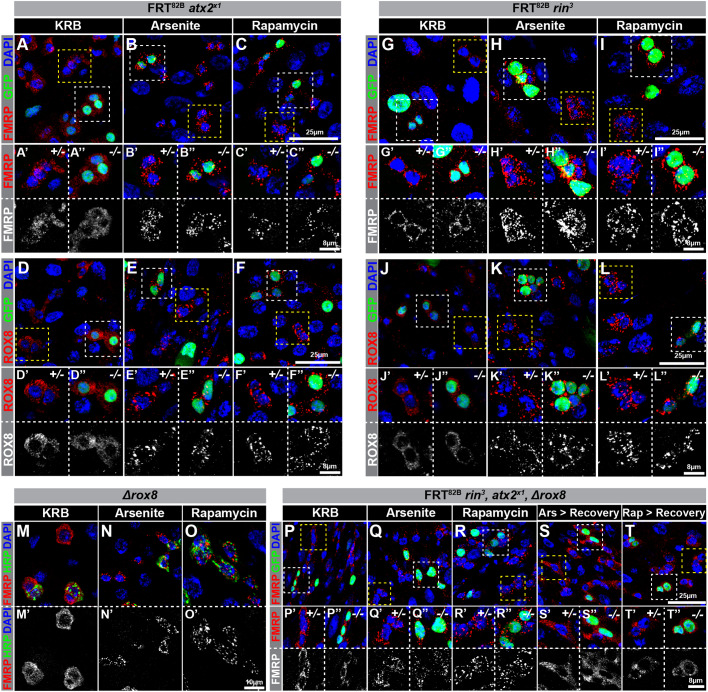
ATX2, RIN and/or ROX8 are not required for IPSG formation
To assess the genetic requirements of IPSGs, we analyzed whether depletion of individual IPSG components affected IPSG formation. We focused on atx2, rin and rox8 in particular since each of these genes or their orthologs has been shown to be required for SG formation in other cell types (Aguilera-Gomez et al., 2017; Bakthavachalu et al., 2018; Kedersha et al., 2016) and because we found that atx2 and rin expression were sufficient for IPSG formation as described above. Available and previously verified null alleles of atx2x1 and rin3 alleles were homozygous lethal (Bakthavachalu et al., 2018; Costa et al., 2013), so we used the mosaic analysis with a repressible cell marker (MARCM) technique (Lee and Luo, 1999) to generate and evaluate intestinal homozygous mutant progenitor cells. At 7–10 days after animals were subjected to clone induction, their intestines were dissected and treated with either arsenite or rapamycin, and scored for IPSGs based on FMRP and ROX8 distribution. Surprisingly, IPSGs were clearly detected in atx2x1 as well as in rin3 mutant progenitor cells (Fig. 7A–L). To rigorously assay for IPSG defects, we compared GFP+ homozygous mutant progenitor cells with surrounding, heterozygous progenitor cells identified based on their enrichment of IPSG proteins; careful observation revealed no obvious or consistent differences between these cell populations (compare neighboring ′ and ″ panels in Fig. 7). To corroborate this result and address whether protein perdurance might mask the clonal phenotypes of these two genes, we performed some additional experiments. In the case of rin3, we found that IPSGs formed normally after treatment of mutant intestines dissected from rare, viable ‘escapers’ that were trans-heterozygous for rin3 and an overlapping deficiency Df(3R)urd and that had been previously shown to be devoid of RIN protein (Costa et al., 2013). In the case of atx2, we analyzed adult atx2x1 progenitor cells derived from mutant cells induced in larvae, thereby allowing ∼2weeks of cell proliferation to occur between the time of clone induction and analysis, and found that IPSGs formed normally even under these conditions. Furthermore, progenitor-specific expression of three independent atx2 RNAi lines also did not affect IPSG formation (data not shown).
Fig. 7.
atx2, rin and rox8 are not required for IPSG formation. (A–L) Confocal micrographs of intestinal sections harboring (A–F) atx2x1 or (G–L) rin3 homozygous mutant (white boxes) and heterozygous (yellow boxes) progenitor cells exposed to (A,D,G,J) KRB, (B,E,H,K) 1 mM sodium arsenite in KRB, or (C,F,I,L) 1 mM rapamycin in KRB and stained for (A–C,G–I) FMRP (red) or (D–F,J–L) ROX8 (red), (A–L) GFP (green) and (A–L) DAPI (blue). (M–O) Confocal micrographs of Δrox8 mutant progenitor cells treated with (M) KRB, (N) 1 mM sodium arsenite or (O) 1 mM rapamycin and stained for (M–O) FMRP (red, M′–O′ white), HRP (green) and DAPI (blue). (P–T) Confocal micrographs of intestinal sections harboring rin3, atx2x1, Δrox8 homozygous triple mutant (white boxes) and heterozygous (yellow boxes) progenitor cells exposed to (P) KRB, (Q) 1 mM sodium arsenite in KRB, (R) 1 mM rapamycin in KRB, (S) 1 mM sodium arsenite in KRB followed by Schneider's medium or (T) 1 mM rapamycin in KRB followed by Schneider's medium and stained for (P–T) FMRP (red), (P–T) GFP (green) and (P–T) DAPI (blue). A′–L′, A″–L″, P′–T′ and P″–T″ show insets of progenitor cells that are heterozygous (GFP negative, outlined with the yellow box) or homozygous (GFP positive, outlined with the white box) for null alleles of (A–F) atx2x1, (G–L) rin3 or (P–T) rin3, atx2x1, Δrox8. Note that IPSGs are formed in progenitor cells that are exposed to acute stress even in the absence of ATX2, RIN and ROX8 alone or in the absence of all three.
Because no rox8 alleles were available, we used CRISPR/Cas9 to generate a null allele, Δrox8 (Fig. S1D,E). This mutation removed 920 bp that encoded the first 250 amino acids of the 470 amino acid ROX8-coding sequence, disrupted the coding frame of the remaining 220 amino acids, completely or partially eliminated all three of the ROX8 RNA recognition motifs and eliminated ROX8 expression as detected by tissue staining and western blotting (Fig. S1F–H). Δrox8 mutants were homozygous viable, so we used homozygous mutant intestines for stress treatments. Following stress treatments, IPSGs were formed in Δrox8 mutant progenitor cells, indicating that ROX8, like ATX2 and RIN, was not necessary for IPSG formation (Fig. 7M–O).
Because these results indicate that atx2, rin, and rox8 were individually dispensable for IPSG formation, we considered whether two of the three genes might be redundant with one another, given that at least atx2 and rin were sufficient for IPSG formation. To test this possibility, we recombined atx2x1, rin3 and Δrox8 in each pairwise combination, and used these recombinant chromosomes to analyze double-mutant intestinal progenitor cells (Fig. S1I). Like the single mutants, double-mutant progenitors showed no defect in IPSG formation, indicating that atx2, rin and rox8 were not functionally redundant with one another, but leaving open the possibility that one of the three genes was sufficient in the absence of the other two. To test this possibility, we prepared an atx2x1, rin3, Δrox8 triple-mutant chromosome and generated triple-mutant intestinal progenitor cells (Fig. S1I). These cells, like wild-type cells, formed normal IPSGs (Fig. 7P–R) that reversed normally during recovery (Fig. 7S,T), indicating that adult intestinal progenitor cells can rely on a robust, non-canonical pathway to form stress granules in response to environmental perturbation.
To round out our genetic analysis, we analyzed intestinal progenitor cells that were homozygous mutant for previously verified null alleles in ago1 (ago1Q127X) (Pressman et al., 2012), caprin (capr2) (Papoulas et al., 2010) or lin-28 (Δlin-28) (Chen et al., 2015) or expressed an fmr1 RNAi line that we previously found matched the intestinal phenotypes of fmr1 null alleles (Luhur et al., 2017). In all cases, IPSGs were detected after arsenite or rapamycin treatment (Fig. S5). We opted not to include eif4e or pabp in this genetic analysis since both genes encode general factors known to be required for cell viability (Hernández et al., 2005; Zeng et al., 2015). Nevertheless, this comprehensive and systematic analysis indicated that individual IPSG components are dispensable for IPSG formation.
DISCUSSION
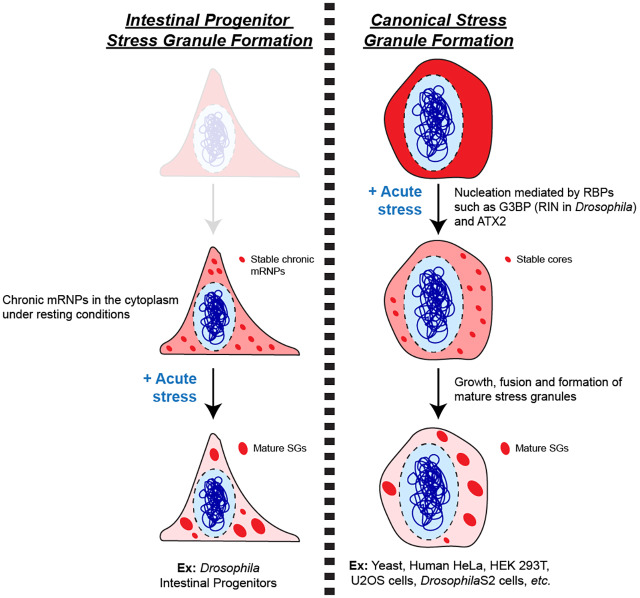
In this study, we characterized a population of SGs in Drosophila intestinal progenitor cells whose formation does not require the canonical stress granule nucleators needed in other cell types. We propose a model describing IPSG formation that is based on three main observations: (1) IPSGs are bona fide SGs because they contain mRNAs and their formation can be blocked and reversed, (2) IPSGs are composed of at least nine conserved proteins that are highly expressed and distributed throughout the cytoplasm of progenitor cells prior to stress and that are known to associate with SGs in other cell types, and (3) IPSGs form even in the absence of three components, the Drosophila orthologs of mammalian ATX2, G3BP and TIA1, that are considered to be integral to SG formation in other cells. We therefore propose that following acute stresses, pre-existing mRNP particles aggregate together to form mature SGs (Fig. 8, left side), bypassing the role of these IDR-rich proteins in nucleating stable cores needed during SG assembly in other cell types (Fig. 8, right side) (Bakthavachalu et al., 2018; Han et al., 2012; Kato et al., 2012; Kedersha et al., 1999, 2000, 2016; Panas et al., 2016; Tourrière et al., 2003). This model indicates that the initial steps of SG assembly are variable and depend upon the cytoplasmic constituency of cells at resting state. Subsequent steps of IPSG assembly might follow the same progression proposed for SGs, including microtubule-dependent fusion of core mRNPs (Wheeler et al., 2016); our super-resolution images of IPSGs indicate that they are not uniform but are rather likely composites of fused mRNPs (Fig. 3C). Since we observed a complex and partially overlapping pattern of IPSG proteins in the absence of stress, we hypothesize that mRNPs in unstressed cells are highly dynamic in nature. The pre-existence of such mRNPs may be an adaptation to the harsh intestinal environment and critical for proper epithelial homeostasis, allowing intestinal progenitor cells to rapidly respond to and recover from constant insult. The conservation of the proteins analyzed in this study raises the possibility that SGs in other cell types, including intestinal stem cells in other animals, may form via a pathway similar to Drosophila IPSGs.
Fig. 8.
Chronic mRNPs coalesce to form mature SGs in intestinal progenitors. Left, in Drosophila intestinal progenitors, SG proteins accumulate in chronic mRNPs even under basal, non-stressed conditions. Following acute stress, these pre-existing granules function as seeds for the formation of mature stress granules. Right, during canonical stress granule formation, proteins with internally disordered regions (IDRs) such as G3BP1/2 (Drosophila RIN) and ATX2 mediate the initial nucleation step following acute stress. These nascent granules subsequently grow and fuse together to form mature stress granules. The existence of chronic mRNPs in progenitor cells eliminates the requirement for ATX2 and G3BP1/2 in the initial steps of stress granule formation.
In contrast to IPSGs, other known SGs share a common need for three different IDR-containing proteins: ATX2, G3BP1/2 or TIA1/TIAR (Bakthavachalu et al., 2018; Kedersha et al., 1999, 2000, 2016; Tourrière et al., 2003). These IDR-containing proteins play a critical role in establishing the core structures of nascent SGs that fuse during mature SG formation (Fig. 8, right side) (Wheeler et al., 2016). Examples of the requirement of these proteins for SG formation include Drosophila G3BP in S2 cells, human G3BP1, either alone or in combination with its paralog G3BP2, in HEK293T, HeLa and U2OS cell types (Aguilera-Gomez et al., 2017; Kedersha et al., 2016; Matsuki et al., 2013; Reineke and Neilson, 2019; Somasekharan et al., 2015; Tsai et al., 2016), Drosophila ATX2 and specifically its C-terminal IDR in S2 cells (Bakthavachalu et al., 2018), and the prion-like domains of mammalian TIA1 in cultured COS7 cells (Gilks et al., 2004). Despite this general requirement, there is also evidence suggesting possible redundancy between these proteins in some contexts. For example, while G3BP1/2 is required for arsenite-induced SG formation, it is not necessary for SG formation following osmotic stress (i.e. after treatment with NaCl or sorbitol) (Kedersha et al., 2016; Protter and Parker, 2016). Furthermore, while loss of ATX2 or TIA1 severely compromises SG assembly in some mammalian and yeast cells, SGs are not completely eliminated (Buchan et al., 2008; Gilks et al., 2004; Nonhoff et al., 2007; Protter and Parker, 2016). While these data suggest some redundancy between SG nucleators in some contexts, this possibility has not been previously investigated. Since we observed that ATX2, RIN and ROX8 are co-expressed in intestinal progenitors, our study directly evaluated this potential functional redundancy using combinations of double and triple mutants of atx2, rin and rox8. This rigorous characterization showed that elimination of these integral SG nucleators, either alone or in combination, had little effect on either the size or number of SGs that form after either arsenite- or rapamycin-induced stress. While we cannot completely rule out subtle defects, the grossly normal appearance of SGs in triple-mutant intestinal progenitors suggests the existence of a non-canonical mechanism for SG formation in these and potentially other cell types.
We also find that in a complex heterogenous tissue, such as the adult intestine, SGs selectively form in only a subset of cells. This observation may have been previously overlooked because much of the work on SG assembly has been conducted in homogenous cell culture systems rather than in intact tissues. Even the few previous studies on SG formation in Drosophila tissues has found that SG formation occurs uniformly by most cells throughout stressed tissues including, for example, heat-stressed ovarian follicular epithelia (Gareau et al., 2013) and larval imaginal discs (Jevtov et al., 2015), as well as mechanically stressed adult brain (Anderson et al., 2018). Unlike the adult intestine, however, which is populated by both an active stem cell contingent as well as terminally differentiated cells, these tissues are all relatively homogenous with respect to the differentiation state of resident cell types. Our analysis suggests that terminally differentiated intestinal cell types are refractory to SG formation because key RBPs are downregulated during their differentiation. Future studies investigating the molecular basis for this refractory state are medically relevant, since limiting SG formation could prevent the ectopic formation of pathogenic mRNP aggregates thought to underlie neurodegenerative disease (Wolozin and Ivanov, 2019).
Embryonic and some somatic stem cell populations are known to maintain low levels of translational activity (Baser et al., 2019; Blanco et al., 2016; Llorens-Bobadilla et al., 2015; Signer et al., 2014; Tahmasebi et al., 2019). This reduced translational activity helps these stem cells maintain an undifferentiated state, while increased translation drives differentiation (Baser et al., 2019; Tahmasebi et al., 2019). For instance, murine embryonic stem cells maintain global low translation during self-renewal, while differentiation proceeds with increased transcript abundance, ribosome loading, and protein synthesis and content (Sampath et al., 2008). In addition, inhibition of translation by phosphorylation of the eukaryotic translation initiation factor 2α helps maintain low translation in mouse skeletal muscle stem cells; failure to maintain low translation result in loss of quiescence, initiation of the myogenic process and consequent differentiation (Zismanov et al., 2016). Using a microscopy-based OPP-staining approach, we found that adult Drosophila intestinal progenitors, in contrast, display high levels of protein synthesis even under resting conditions. We suggest that since increased translation in stem cells is a hallmark of differentiation, intestinal progenitor cells are in a state primed for differentiation that allows ISCs to rapidly proliferate and differentiate in order to replenish cells when needed. Since translation is an energy expensive process (Holcik and Sonenberg, 2005), stress responses may divert this energy to more immediate needs. We propose that SG formation provides this layer of regulation in intestinal progenitors during episodes of cellular stress. Furthermore, the characteristics of ISCs we identified here may also be shared by stem cell populations that support other high-turnover adult tissues.
While IPSGs were easily detectable in ex vivo-treated intestines, additional treatments in which adults were either fed chemical stressors or starved for various lengths of time with various vehicles (water, sugar-water or nothing) failed to induce SGs. We hypothesize that ex vivo treatment induces IPSGs while feeding does not because orally fed chemicals are absorbed by enterocytes and fail to reach basally located and well protected progenitor cells. In addition, it is possible that endogenous IPSGs form transiently or require a dosage of proper duration that we did not test. We did find one other condition that induced small IPSG-like assemblies, namely heat shock. However, the induction was variable, and the treatments led to immediate death, precluding the ability to study them.
There is considerable interest in identifying stem cell-specific factors, and here we show that ten different RBPs are enriched in intestinal progenitors relative to surrounding differentiated cells. Previous transcriptional profiling analyses identified only one of these, lin-28, as being enriched in stem cells (Doupé et al., 2018). Consistent with this, the other nine genes display relatively uniform transcript levels in non-differentiated versus differentiated cells (Dutta et al., 2013) (see Fig. S1B). This apparent discrepancy between protein and transcript profiles in differentiated versus progenitor cells suggests active post-transcriptional regulatory mechanisms in intestinal cells. These mechanisms remain largely unexplored due to the lack of tools to profile the translatome relative to the transcriptome specifically in subsets of intestinal cells. Future work focused on developing such tools will likely identify post-transcriptional mechanisms that control stem cell behavior during resting and stressed conditions.
MATERIALS AND METHODS
Drosophila strains and husbandry
Female flies were used in all experiments and aged as indicated. All fly strains were cultured on standard Bloomington Drosophila stock center medium (https://bdsc.indiana.edu/information/recipes/bloomfood.html). Flies were reared in 18°C, 25°C and 29°C incubators set for a 12-h-light–12-h-dark schedule and 65% humidity. Fly strains were cultured in groups of 15–20 (typically 5 males and up to 15 females). All strains used in this study are listed in Table S1.
Temperature
For temporal and regional gene expression-targeting (TARGET) experiments, flies were grown at 18°C, collected over 2 days, and reared in 29°C for up to 10 days before being dissected. For clonal analysis using the mosaic analysis with repressible cell marker (MARCM) method, animals were reared at 25°C until eclosion, collected over 2 days and heat-shocked immediately at 37°C for 30 min in a Lauda circulating water bath. Subsequently, flies were reared at 25°C for 7–10 days.
Construction of new strains
{3Xgbe-smGFP::V5::nls}attP40
The 3xgbe-smGFP::V5::nls plasmid contains three main parts that were assembled as follows. First, a pair of annealed oligonucleotides (3202 and 3203; see Table S2 for all oligonucleotide sequences) encoding three copies of the Grainyhead binding element (GBE) (5′-CTTGGAAACCGGTTATGCGAG-3′) and two copies of Su(h)m8 binding sites (5′-AAACTTACTTTCAGCTCGGTTCCCACGCCAC-3′) were concatemerized three times and subcloned into a plasmid containing attB and the white transformation marker. Then, the smGFP::V5 open reading frame that contains ten copies of the V5 epitope embedded within an inactive version of ‘spaghetti monster’ GFP was PCR amplified with high-fidelity Q5 polymerase (NEB) from pJFRC206 (Addgene plasmid 63168) with oligonucleotides 3416 and 3412. Finally, the nuclear localization sequence from the transformer gene was PCR-amplified from genomic DNA with oligonucleotides 3402 and 3421. The two PCR products were subcloned into the XhoI and XbaI sites of the 3xgbe-containing plasmid using the HiFi DNA assembly master mix (NEB), and all three pieces were sequenced to verify the absence of errors. The transgene was subsequently inserted into the attP40 landing site by Rainbow Genetics (Camarillo, CA).
CRISPR/Cas9-mediated generation of RIN::HA
A 2×HA epitope tag (YPYDVPDYAYPYDVPDYA) was introduced at the rin locus, immediately after the RIN start methionine, using a single-stranded oligonucleotide (ssODN) homology directed repair CRISPR/Cas9 approach (Gratz et al., 2013). The gRNA plasmid was generated by subcloning annealed oligonucleotides 4153 and 4154 (see Table S2 for all oligonucleotide sequences) encoding gRNA 5′-CGGCGAAGGCTGCTGCGATT-3′ into the BbsI site of pU6-BbsI-chiRNA (Gratz et al., 2013) and co-injected along with ssODN repair template oligonucleotide 4158 into strain y1 M{vas-Cas9}ZH-2A w1118/FM7c (BL51323) by Rainbow Genetics (Camarillo CA). Stocks were established from individual F0 founder males, which were subsequently genotyped by PCR using forward oligonucleotide 4184, complementary to the HA tag, and reverse oligonucleotide 4179 that lay downstream of the ssODN sequence. Stocks that were derived from males positive for this PCR analysis were retained and further confirmed by sequencing of rin genomic DNA.
CRISPR/Cas9-mediated generation of Δrox8
The Δrox8 mutation, which removes 920 bp of sequence immediately downstream of the ROX8 start methionine, was generated with two guide RNA (gRNA) plasmids and a repair plasmid. The gRNA plasmids were generated by subcloning annealed oligonucleotides 4151 and 4152 encoding gRNA 5′-CTTCGGTTGCGACTCGTCCATGG-3′ as well as annealed oligonucleotides 4309 and 4310 encoding gRNA 5′-GAAGACCCGCACGTCCTGGATGG-3′ into the BbsI site of pU6-BbsI-chiRNA (Gratz et al., 2013). The repair plasmid was generated by PCR amplifying a left homology arm using oligonucleotides 4397 and 4398 (see Table S2 for oligonucleotide sequences) and a right homology arm using oligonucleotides 4399 and 4400 and subcloning both into pHD-DsRed-attP [Drosophila Genomics Resource Center (DGRC) plasmid 1361] using HiFi DNA Assembly Master Mix (NEB). These three plasmids were co-injected into strain y1 M{vas-Cas9}ZH-2A w1118/FM7c (BL51323) by Rainbow Genetics (Camarillo CA) and dsRed+ male progeny were used to establish stocks. Insertion of the repair plasmid into the rox8 locus was confirmed by PCR using dsRed oligonucleotides 3144 and 4420 paired with respective oligonucleotides 4417 and 4384, which lie beyond the homology arm sequences. In addition to the Δ deletion, four additional alleles were generated by injection of single gRNAs without any repair plasmid. These mutations disrupt the predicted ROX8 reading frame after the first, second, fourth and nineteenth amino acids. However, none of these mutations affected ROX8 accumulation as detected by western blotting, suggesting that the currently annotated start methionine may not be used (even though the next downstream methionine is located 20 amino acids into the RRM1 domain).
UAS transgenic lines
Second chromosome insertions of UAS-capr::3xFlag-HA, UAS-rox8::3xFlag-HA and UAS-ago1::TEV-Flag transgene were generated by injection of either pNIK1372 (UAS-capr::3xFlag-HA), pNIK1345 (UAS-rox8::3xFlag-HA) or pNIK1354 (UAS-ago1::TEV-Flag) into a strain containing the attP40 landing site by Rainbow Genetics (Camarillo, CA). pNIK1372 and pNIK1345 were generated by PCR amplifying with Q5 high-fidelity polymerase (NEB) the caprin-coding region from cDNA LP14942 (DGRC) using oligonucleotides 4266 and 4267, or the rox8-coding region from cDNA FM011201 (DGRC) using oligonucleotides 4189 and 4190, subcloning the resulting PCR product into a homemade attB plasmid containing 20 UAS sites, a multicloning site, the 3×FLAG-HA open reading frame, and SV40 3′UTR sequence using HiFi DNA assembly master mix (NEB), and sequence verifying the resulting plasmids to confirm the absence of any PCR-induced errors. pNIK1354 was similarly generated by PCR amplifying the ago1 coding region from cDNA SD07515 (DGRC) using oligonucleotides 3887 and 3888, and high-fidelity Q5 polymerase (NEB), subcloning the resulting PCR product into an attB plasmid containing the 3×FLAG open reading frame using HiFi DNA Assembly Master Mix (NEB), and sequence verifying the resulting plasmid to confirm the absence of any PCR-induced errors.
Antibody generation
ROX8 antibodies were generated in rats (Cocalico Biologicals) against a 6×HIS-tagged version of full-length ROX8 that was expressed and purified according to standard methods. The ROX8-encoding plasmid (pNIK1379) was generated by PCR amplifying the rox8 coding region from cDNA FM011201 (DGRC) using oligonucleotides 4285, 4286, and high-fidelity Q5 polymerase (NEB), subcloning the resulting PCR product into the NcoI and EcoRI sites of pHIS.parallel using HiFi DNA assembly master mix (NEB), and sequence verifying the resulting plasmid to confirm the absence of any PCR-induced errors.
Dissections and immunostaining
Gastrointestinal tracts of adult female flies were dissected in ice cold 1× PBS and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Cat. No. 15714) in PBS for 45 min. These samples were washed with 1× PBT (1× PBS, 0.1% Triton X-100) and then blocked (1× PBT, 0.5% bovine serum albumin) for at least 45 min. Subsequently, samples were incubated at 4°C overnight with primary antibodies, including rabbit anti-GFP (A11122, Life Technologies, 1:1000), mouse anti-V5 (MCA1360GA, Bio-Rad, 1:250), chicken anti-V5 (NB600-379, Novus Biologicals, 1:1000), chicken anti-LacZ (ab9361, Abcam, 1:2000), mouse anti-FLAG (F3165, Sigma, 1:1000), mouse anti-HA (2367S, Cell Signaling Technology, 1:100), rabbit anti-HA (3724S, Cell Signaling Technology, 1:1000), rabbit anti-mCherry (5993-100, BioVision, 1:1000), mouse anti-FMR1 (5A11, Developmental Studies Hybridoma Bank, 1:100), mouse anti-Prospero (MR1A, Developmental Studies Hybridoma Bank, 1:100), rabbit anti-RIN (Aguilera-Gomez et al., 2017, 1:500), rat anti-ROX8 (this study, 1:1500), rabbit anti-Caprin (Papoulas et al., 2010) (1:500), rabbit anti-pABP (Duncan et al., 2009) (1:2000), rabbit anti-AGO1 (ab5070, Abcam, 1:500), rabbit anti-eIF4E (a gift from Akira Nakamura, Department of Germline Development, Kumamoto University, Japan; 1:1500), and mouse anti-DCR2 (Miyoshi et al., 2009) (1:500). Samples were washed and incubated for 2–3 h with secondary antibodies, including Alexa-Fluor-488 and -568-conjugated goat anti-rabbit, -mouse, -rat and -chicken IgG antibodies (Life Technologies, 1:1000). Alexa-Fluor-647-conjugated goat anti-HRP antibodies were used in the secondary antibody solution whenever required. Samples were washed and treated with DAPI (1:10,000) and mounted in Vectashield mounting medium (Vecta Laboratories).
Microscopy and image processing
Images were collected on either a Leica SP8 Scanning Confocal microscope (Leica DMi8 inverted microscope platform; equipped with WLL 470–670 nm, 405 nm and 440 nm lasers, Huygens deconvolution software and controlled by Leica LAS-X software) or an OMX 3D-SIM Super-Resolution microscope (DeltaVision OMX system; equipped with 405, 488, 561 and 642 nm lasers, and controlled by AquireSR software with image processing by SoftWorx imaging software) housed at the Light Microscopy Imaging Center, Indiana University, Bloomington. Images captured on the SP8 confocal microscope were acquired using a Leica HC PL APO CS2 63×/ NA 1.40 lens with Leica Type F immersion liquid (N=1.518). Super resolution images collected using the OMX system were acquired using an Olympus PL APO N 60×/ NA 1.42 lens with Applied Precision immersion oil (N=1.516). Samples to be compared were collected under identical settings on the same day, image files were adjusted simultaneously using Adobe Photoshop CC, and figures were assembled using Adobe Illustrator CC. ImageJ FIJI (https://fiji.sc/). The Analyze Particles plugin was used to quantify the number of puncta and average area using 0.01 µm2 as the lower cut off of puncta size.
Oligo-dT fluorescent in situ hybridization
RNase-free solutions were used throughout the in situ protocol. First, gastrointestinal tracts of adult female flies were dissected in ice cold 1× PBS and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, cat. no. 15714) in PBS for 45 min. Subsequently, samples were washed three times 5 min each in 0.3% PBT (1× PBS and 0.3% Triton X-100). Intestines were then dehydrated by washing in 7:3, 1:1 and 3:7 0.3% PBT:MeOH for 5 min each. Next, samples were washed in 100% MeOH for 10 min. Then samples were rehydrated by washing them in 3:7, 1:1 and 7:3 0.3% PBT:MeOH for 5 min each. Following the series of washes, samples were washed in 0.3% PBT for 10 min. Subsequently, samples were first rinsed and then washed in H-wash (20% formaldehyde, 20× SSC, DEPC-treated water) for 10 min. Then, H-wash was completely removed and oligo-dT probes were added with Hyb-buffer (Ambion, cat. no. AM8670). Samples were incubated with probes overnight at 37°C. The following day, Hyb-buffer was removed, and samples were washed in H-wash two times for 30 min each. After the second wash, samples were mounted using ProLong Diamond mounting solution (Invitrogen, Cat. No. P36971).
Ex vivo treatments
For ex vivo arsenite and rapamycin treatments, intestines from females aged 8–10 days on normal diet were incubated in KRB (Alfa Aesar, cat. no. J67591), or KRB supplemented with 1 mM arsenite (Sigma) or 1 mM rapamycin (LC Laboratories, cat. no. r-5000) for 60 min and then fixed. Note that the 1 mM concentration was determined following testing a series of arsenite and rapamycin dilutions (250 µM, 500 µM, 1 mM, 1.5 mM and 2 mM in KRB) for consistent stress granule formation in intestinal cells. To assess the effects of polysome stabilization on stress granule formation, dissected intestines were incubated with KRB supplemented with 0.1 µg/ml cycloheximide (Sigma, cat. no. C7698) for 30 min prior to arsenite or rapamycin treatments. To examine the effects of polysome destabilization on stress granule formation, dissected intestines were incubated with KRB supplemented with 250 µg/ml puromycin (Sigma, cat. no. P8833) for 60 min and then fixed. For stress granule recovery, arsenite-, rapamycin- or puromycin-treated intestines (for 45 min) were incubated with Schneider's medium (Sigma, cat. no. S0146) for 1 h before fixation. All these ex vivo experiments were performed with solutions pre-warmed to room temperature.
Ex vivo OPP assay to detect active translation
Adult female Drosophila intestines were dissected in ice-cold 1× PBS. Subsequently, dissected intestines were treated with chemicals as described above. Treated intestines were washed with KRB and the O-propargyl-puromycin (OPP) assay was performed based on the kit (Invitrogen, Cat. No. C10457) manual with modifications. Briefly, washed intestines were treated with Click-iT® OPP reagent (1000× diluted in KRB) for 30 min and washed three times with 1× PBS. Subsequently, tissues were fixed in 4% paraformaldehyde in 1× PBS for 45 min. Samples were washed in 1× PBT and incubated with OPP reaction cocktail for 30 min protected from light. Samples were kept in the dark for all subsequent steps. After the incubation, the reaction cocktail was removed and tissues were washed once with Click-iT® reaction rinse buffer. Subsequently, intestines were washed overnight with 1× PBS. Whenever antibody staining was necessary, the regular immunostaining protocol described above was followed.
Protein isolation and western blot analysis
Female Drosophila flies were used for protein isolation. Females aged 8–10 days on a normal diet were lysed in protein lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM KCl, 5 mM MgCl2, 1% NP-40, 0.5 mM DTT and 1× protease inhibitor cocktail), protein extracts were resolved on a 4–20% gradient polyacrylamide gel (Bio-Rad, Cat. No. 456-1093), transferred to Immobilon®-P membrane (Millipore, cat. no. IPVH00010) and probed with rat anti-ROX8 (1:500) or mouse anti-α-tubulin (12G10, Developmental Studies Hybridoma Bank, 1:1000) antibodies. Subsequently, blots were washed extensively with 1× TBST (1× TBS and 0.1% Tween-20) and incubated with HRP-conjugated anti-rat or -mouse secondary antibodies. After extensive secondary washes with 1× TBST, blots were treated with ECL detection reagent 1 and 2 (Thermo Scientific, cat. no. 1859701 and 1859698) and finally exposed to chemiluminescence films (GE Healthcare, cat. no. 28906839) and the signal was developed.
Statistical analysis
All statistical analyses were performed using Prism (GraphPad, Version 7.0). First datasets were tested for normality using D'Agostino-Pearson test. For comparisons of two datasets, if datasets followed (1) a parametric distribution, an unpaired t-test or (2) a non-parametric distribution, a Mann–Whitney test was performed. An ordinary one-way ANOVA test was used when comparing three or more datasets following a parametric distribution. Multiple comparisons of three or more datasets following a non-parametric distribution were analyzed using Kruskal–Wallis test. Unless otherwise noted, significance is indicated as follows: n.s., not significant; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Graphs show mean±s.d.
Supplementary Material
Acknowledgements
We thank Richard Carthew, Matthias Hentze, Craig Micchelli, Akira Nakamura, Ophelia Papoulas, Norbert Perrimon, Catherine Rabouille, Mani Ramaswami, Mikiko Siomi, Ernst Hafen, the Bloomington Drosophila Stock Center (supported by grant NIHP4OOD018537), the Drosophila Genome Resource Center (supported by grant NIH2P40OD010949), and the Developmental Studies Hybridoma Bank (created by the NICHD of the NIH) for reagents, and the Light Microscopy Imaging Center (supported by grant NIH1S10OD024988-01) for access to the SP8 confocal and OMX super-resolution microscopes.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.B., N.S.S.; Methodology: K.B., I.S.A.; Formal analysis: K.B.; Investigation: K.B., I.S.A., M.A.H., D.R.; Resources: N.S.S.; Writing - original draft: K.B., N.S.S.; Writing - review & editing: K.B., N.S.S.; Visualization: K.B.; Supervision: K.B., N.S.S.; Project administration: K.B., N.S.S.; Funding acquisition: N.S.S.
Funding
This work was funded by the National Institute of General Medical Sciences (award R01GM124220). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.243451.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.243451.reviewer-comments.pdf
References
- Aguilera-Gomez A., Zacharogianni M., van Oorschot M. M., Genau H., Grond R., Veenendaal T., Sinsimer K. S., Gavis E. R., Behrends C. and Rabouille C. (2017). Phospho-rasputin stabilization by Sec16 is required for stress granule formation upon amino acid starvation. Cell Rep. 20, 935-948. 10.1016/j.celrep.2017.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. and Kedersha N. (2002). Stressful initiations. J. Cell Sci. 115, 3227-3234. [DOI] [PubMed] [Google Scholar]
- Anderson P. and Kedersha N. (2006). RNA granules. J. Cell Biol. 172, 803-808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. and Kedersha N. (2008). Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141-150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Anderson P. and Kedersha N. (2009). Stress granules. Curr. Biol. 19, R397-R398. 10.1016/j.cub.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Anderson E. N., Gochenaur L., Singh A., Grant R., Patel K., Watkins S., Wu J. Y. and Pandey U. B. (2018). Traumatic injury induces stress granule formation and enhances motor dysfunctions in ALS/FTD models. Hum. Mol. Genet. 27, 1366-1381. 10.1093/hmg/ddy047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A., Fay M. M., Lyons S. M., Achorn C. A., Kedersha N., Anderson P. and Ivanov P. (2017). Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 130, 927-937. 10.1242/jcs.199240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavachalu B., Huelsmeier J., Sudhakaran I. P., Hillebrand J., Singh A., Petrauskas A., Thiagarajan D., Sankaranarayanan M., Mizoue L., Anderson E. N. et al. (2018). RNP-granule assembly via Ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98, 754-766.e4. 10.1016/j.neuron.2018.04.032 [DOI] [PubMed] [Google Scholar]
- Balzer E. and Moss E. G. (2007). Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 4, 16-25. 10.4161/rna.4.1.4364 [DOI] [PubMed] [Google Scholar]
- Baser A., Skabkin M., Kleber S., Dang Y., Gülcüler Balta G. S., Kalamakis G., Göpferich M., Ibañez D. C., Schefzik R., Lopez A. S. et al. (2019). Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature 566, 100-104. 10.1038/s41586-019-0888-x [DOI] [PubMed] [Google Scholar]
- Blanco S., Bandiera R., Popis M., Hussain S., Lombard P., Aleksic J., Sajini A., Tanna H., Cortés-Garrido R., Gkatza N. et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335-340. 10.1038/nature18282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R. and Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932-941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Muhlrad D. and Parker R. (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441-455. 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Luhur A. and Sokol N. (2015). Lin-28 promotes symmetric stem cell division and drives adaptive growth in the adult Drosophila intestine. Development 142, 3478-3487. 10.1242/dev.127951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A., Pazman C., Sinsimer K. S., Wong L. C., McLeod I., Yates J., Haynes S. and Schedl P. (2013). Rasputin functions as a positive regulator of Orb in Drosophila oogenesis. PLoS ONE 8, e72864 10.1371/journal.pone.0072864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot M.-C., Subramanian M., Flatter E., Mandel J.-L. and Moine H. (2009). Cells lacking the Fragile X Mental Retardation Protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol. Biol. Cell 20, 428-437. 10.1091/mbc.e08-07-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupé D. P., Marshall O. J., Dayton H., Brand A. H. and Perrimon N. (2018). Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. Proc. Natl. Acad. Sci. USA 115, 12218-12223. 10.1073/pnas.1719169115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K. E., Strein C. and Hentze M. W. (2009). The SXL-UNR corepressor complex uses a PABP-mediated mechanism to inhibit ribosome recruitment to msl-2 mRNA. Mol. Cell 36, 571-582. 10.1016/j.molcel.2009.09.042 [DOI] [PubMed] [Google Scholar]
- Dutta D., Xiang J. and Edgar B. A. (2013). RNA expression profiling from FACS-isolated cells of the Drosophila intestine. Curr. Protoc. Stem Cell Biol. 27, 2F.2.1-2F.2.12. 10.1002/9780470151808.sc02f02s27 [DOI] [PubMed] [Google Scholar]
- Gareau C., Houssin E., Martel D., Coudert L., Mellaoui S., Huot M.-E., Laprise P. and Mazroui R. (2013). Characterization of fragile X mental retardation protein recruitment and dynamics in Drosophila stress granules. PLoS ONE 8, e55342 10.1371/journal.pone.0055342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M. and Anderson P. (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383-5398. 10.1091/mbc.e04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., Harrison M. M., Wildonger J. and O'Connor-Giles K. M. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029-1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G. et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768-779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- He L., Si G., Huang J., Samuel A. D. T. and Perrimon N. (2018). Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel. Nature 555, 103-106. 10.1038/nature25744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G., Altmann M., Sierra J. M., Urlaub H., Diez del Corral R., Schwartz P. and Rivera-Pomar R. (2005). Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech. Dev. 122, 529-543. 10.1016/j.mod.2004.11.011 [DOI] [PubMed] [Google Scholar]
- Holcik M. and Sonenberg N. (2005). Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318-327. 10.1038/nrm1618 [DOI] [PubMed] [Google Scholar]
- Jain S., Wheeler J. R., Walters R. W., Agrawal A., Barsic A. and Parker R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487-498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtov I., Zacharogianni M., van Oorschot M. M., van Zadelhoff G., Aguilera-Gomez A., Vuillez I., Braakman I., Hafen E., Stocker H. and Rabouille C. (2015). TORC2 mediates the heat stress response in Drosophila by promoting the formation of stress granules. J. Cell Sci. 128, 2497-2508. 10.1242/jcs.168724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Tian A. and Jiang J. (2016). Intestinal stem cell response to injury: lessons from Drosophila. Cell. Mol. Life Sci. 73, 3337-3349. 10.1007/s00018-016-2235-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J. et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753-767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N. and Anderson P. (2007). Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61-81. 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N. and Anderson P. (2009). Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90, 155-185. 10.1016/S1877-1173(09)90004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M., Li W., Miller I. and Anderson P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431-1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E. and Anderson P. (2000). Dynamic shuttling of Tia-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257-1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I. J., Stahl J. and Anderson P. (2002). Evidence that ternary complex (eIF2-GTP-tRNAi Met)–deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195-210. 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E. and Anderson P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871-884. 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Panas M. D., Achorn C. A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G. M., Ivanov P. et al. (2016). G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845-860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. and Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451-461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Leung A. K. L., Calabrese J. M. and Sharp P. A. (2006). Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA 103, 18125-18130. 10.1073/pnas.0608845103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K. and Martin-Villalba A. (2015). Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329-340. 10.1016/j.stem.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Luhur A., Buddika K., Ariyapala I. S., Chen S. and Sokol N. S. (2017). Opposing post-transcriptional control of InR by FMRP and LIN-28 adjusts stem cell-based tissue growth. Cell Rep. 21, 2671-2677. 10.1016/j.celrep.2017.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marianes A. and Spradling A. C. (2013). Physiological and stem cell compartmentalization within the Drosophila midgut. eLife 2, e00886 10.7554/eLife.00886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S., Soltanieh S., Server K. L., Mak R., Jin W., Fang M. Y., Luo E.-C., Krach F., Yang D., Sen A. et al. (2018). Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590-604.e13. 10.1016/j.cell.2017.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki H., Takahashi M., Higuchi M., Makokha G. N., Oie M. and Fujii M. (2013). Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 18, 135-146. 10.1111/gtc.12023 [DOI] [PubMed] [Google Scholar]
- Mazroui R., Huot M.-E., Tremblay S., Filion C., Labelle Y. and Khandjian E. W. (2002). Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11, 3007-3017. 10.1093/hmg/11.24.3007 [DOI] [PubMed] [Google Scholar]
- McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J.-J., Anderson P. and Kaufman R. J. (2005). Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925-16933. 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- Micchelli C. A. and Perrimon N. (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475-479. 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Okada T. N., Siomi H. and Siomi M. C. (2009). Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA 15, 1282-1291. 10.1261/rna.1541209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewidok B., Igaev M., Pereira da Graca A., Strassner A., Lenzen C., Richter C. P., Piehler J., Kurre R. and Brandt R. (2018). Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J. Cell Biol. 217, 1303-1318. 10.1083/jcb.201709007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U., Ralser M., Welzel F., Piccini I., Balzereit D., Yaspo M.-L., Lehrach H. and Krobitsch S. (2007). Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 18, 1385-1396. 10.1091/mbc.e06-12-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien L. E., Soliman S. S., Li X. and Bilder D. (2011). Altered modes of stem cell division drive adaptive intestinal growth. Cell 147, 603-614. 10.1016/j.cell.2011.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B. and Spradling A. (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470-474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Panas M. D., Ivanov P. and Anderson P. (2016). Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215, 313-323. 10.1083/jcb.201609081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulas O., Monzo K. F., Cantin G. T., Ruse C., Yates J. R., Ryu Y. H. and Sisson J. C. (2010). dFMRP and Caprin, translational regulators of synaptic plasticity, control the cell cycle at the Drosophila mid-blastula transition. Development 137, 4201-4209. 10.1242/dev.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressman S., Reinke C. A., Wang X. and Carthew R. W. (2012). A systematic genetic screen to dissect the microRNA pathway in Drosophila. G3 2, 437-448. 10.1534/g3.112.002030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter D. S. W. and Parker R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668-679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L. C. and Neilson J. R. (2019). Differences between acute and chronic stress granules, and how these differences may impact function in human disease. Biochem. Pharmacol. 162, 123-131. 10.1016/j.bcp.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P., Pritchard D. K., Pabon L., Reinecke H., Schwartz S. M., Morris D. R. and Murry C. E. (2008). A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2, 448-460. 10.1016/j.stem.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Signer R. A. J., Magee J. A., Salic A. and Morrison S. J. (2014). Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49-54. 10.1038/nature13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S., Xu Y., Wang B., David M. D., Schubert P., Kennedy D. and Schrader J. W. (2007). Distinct structural features of Caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27, 2324-2342. 10.1128/MCB.02300-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan S. P., El-Naggar A., Leprivier G., Cheng H., Hajee S., Grunewald T. G. P., Zhang F., Ng T., Delattre O., Evdokimova V. et al. (2015). YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J. Cell Biol. 208, 913-929. 10.1083/jcb.201411047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassburger K., Lorbeer F. K., Lutz M., Graf F., Boutros M. and Teleman A. A. (2017). Oxygenation and adenosine deaminase support growth and proliferation of ex vivo cultured Drosophila wing imaginal discs. Development 144, 2529-2538. 10.1242/dev.147538 [DOI] [PubMed] [Google Scholar]
- Tahmasebi S., Amiri M. and Sonenberg N. (2019). Translational control in stem cells. Front. Genet. 9, 709 10.3389/fgene.2018.00709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E. and Tazi J. (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823-831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai W.-C., Gayatri S., Reineke L. C., Sbardella G., Bedford M. T. and Lloyd R. E. (2016). Arginine Demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 291, 22671-22685. 10.1074/jbc.M116.739573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W. and Rabouille C. (2019). Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic 20, 623-638. 10.1111/tra.12669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler J. R., Matheny T., Jain S., Abrisch R. and Parker R. (2016). Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. and Ivanov P. (2019). Stress granules and neurodegeneration. Nat. Rev. Neurosci. 20, 649-666. 10.1038/s41583-019-0222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J.-Y., Dunham W. H., Hong S. J., Knight J. D. R., Bashkurov M., Chen G. I., Bagci H., Rathod B., MacLeod G., Eng S. W. M. et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517-532.e11. 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Zeng X., Han L., Singh S. R., Liu H., Neumüller R. A., Yan D., Hu Y., Liu Y., Liu W., Lin X. et al. (2015). Genome-wide RNAi screen identifies networks involved in intestinal stem cell regulation in Drosophila. Cell Rep. 10, 1226-1238. 10.1016/j.celrep.2015.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V., Chichkov V., Colangelo V., Jamet S., Wang S., Syme A., Koromilas A. E. and Crist C. (2016). Phosphorylation of eIF2α is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 18, 79-90. 10.1016/j.stem.2015.09.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.