Abstract
Background
Thyroid nodules are extremely common and typically diagnosed with ultrasound whether benign or malignant. Imaging diagnosis assisted by Artificial Intelligence has attracted much attention in recent years. The aim of our study was to build an ensemble deep learning classification model to accurately differentiate benign and malignant thyroid nodules.
Material/Methods
Based on current advanced methods of image segmentation and classification algorithms, we proposed an ensemble deep learning classification model for thyroid nodules (EDLC-TN) after precise localization. We compared diagnostic performance with four other state-of-the-art deep learning algorithms and three ultrasound radiologists according to ACR TI-RADS criteria. Finally, we demonstrated the general applicability of EDLC-TN for diagnosing thyroid cancer using ultrasound images from multi medical centers.
Results
The method proposed in this paper has been trained and tested on a thyroid ultrasound image dataset containing 26 541 images and the accuracy of this method could reach 98.51%. EDLC-TN demonstrated the highest value for area under the curve, sensitivity, specificity, and accuracy among five state-of-the-art algorithms. Combining EDLC-TN with models and radiologists could improve diagnostic accuracy. EDLC-TN achieved excellent diagnostic performance when applied to ultrasound images from another independent hospital.
Conclusions
Based on ensemble deep learning, the proposed approach in this paper is superior to other similar existing methods of thyroid classification, as well as ultrasound radiologists. Moreover, our network represents a generalized platform that potentially can be applied to medical images from multiple medical centers.
MeSH Keywords: Artificial Intelligence; Image Processing, Computer-Assisted; Thyroid Nodule; Ultrasonography
Background
Thyroid nodules are common clinically, and with application of high-frequency ultrasound, their incidence has increased. Ultrasound diagnosis of benign and malignant nodules is mainly performed under guidelines from the American College of Radiology (ACR) [1] and the ultrasound section of the American Thyroid Association (ATA) [2], both of which have been increasingly improved in recent years. But there still remain some defects, and diagnostic accuracy is not consistent due to differing levels of experience among radiologists performing ultrasound [3]. With gradual development of machine learning in recent years, intelligent medical image diagnosis has become available. Deep learning can reveal subtler and more abstract information embedded in images along with the deepening of the network layers. In addition, use of artificial intelligence (AI) for medical or auxiliary medical care can lighten the burden of doctors and optimize medical treatments. Medical image processing is one of the breakthroughs in this field. Both deep learning and AI have achieved high accuracy for classification of skin cancer and detection of pneumonia [4,5], even exceeding that of physicians. This is also true for diagnosis of thyroid nodules [6–9].
In 2008, Lim KJ et al. [10] were the first to apply a neural network to differentiation of benign and malignant thyroid nodules. Ma J et al. [11] were the first to use a convolutional neural network in this field in 2017. They separately trained two networks in the ImageNet database. Then, by concatenating feature images, they used the softmax classifier to diagnose thyroid nodules with an accuracy of 83.02%±0.72%. Imaging diagnosis assisted by AI has attracted much attention in the past several years. If the diagnostic effectiveness of AI – including accuracy, sensitivity, and specificity – is found to be comparable to that of an experienced radiologist performing ultrasound, it will have a tremendous impact on the imaging diagnosis.
However, if ultrasound images are directly used as inputs to a neural network, the shape information from thyroid nodules may be lost. Thus, two different AI models were trained on the basis of ensemble learning [12]. To accurately diagnose thyroid nodules, we calculated the mean output of these two types of models and determined whether the thyroid nodules were benign or malignant using a new model: EDLC-TN (ensemble deep learning-based classifier for thyroid nodules). The aim of our research was to use the deep learning method to differentiate benign and malignant thyroid nodules, thereby improving the accuracy of lesion identification.
Material and Methods
Study cohort and datasets
We used four independent ultrasound datasets to develop and evaluate EDLC-TN in four different hospitals: Tianjin Medical University Cancer Institute and Hospital (Center 1), Jilin Integrated Traditional Chinese and Western Medicine Hospital (Center 2), Cangzhou Hospital of Integrated Traditional Chinese, Western Medicine of Hebei Province (Center 3), and Peking University BinHai Hospital (Center 4). Between January 2015 and December 2017, consecutive patients in these four medical centers who underwent diagnostic thyroid ultrasound examination and subsequent surgery were included in the study. Exclusion criteria were: (1) images from anatomical sites that were judged as not having tumor according to postoperative pathology; (2) nodules with incomplete or low-quality ultrasound images; and (3) cases with incomplete clinicopathological information. Finally, three datasets from Centers 1 to 3 including a total of 25 509 thyroid ultrasound images were used to train and test the model, of which 15 255 were malignant and 10 254 were benign (confirmed by postoperative pathological diagnosis). Images (n=1,032) from Center 4 differed greatly from the other three in terms of style, clarity, and machine types. Therefore, the dataset from Center 4 was only used as an external validation set for verifying the generalizability of the model. Data from each medical Centers 1 to 3 were randomly divided into training and testing sets at a ratio of approximately 7: 3 (Table 1). In all settings, testing data did not include any images used in training.
Table 1.
Number of training and testing images from four datasets.
| Center 1 (23504) | Center 2 (530) | Center 3 (1475) | Center 4 (1032) | Total of all (26541) | |
|---|---|---|---|---|---|
| Training dataset | |||||
| Benign | 6464 | 205 | 522 | – | 7191 |
| Malignant | 10090 | 164 | 414 | – | 10668 |
| Total for training | 16554 | 369 | 936 | – | 17859 |
| Testing dataset | |||||
| Benign | 2620 | 84 | 359 | 502 | 3565 |
| Malignant | 4330 | 77 | 180 | 530 | 5117 |
| Total for testing | 6950 | 161 | 539 | 1032 | 8682 |
This study was approved by the Tianjin Medical University Cancer Institute and Hospital ethics committee. Informed consent from patients was waived due to the retrospective nature. In training and test datasets, ultrasound images were collected and stored by various brands of ultrasonic equipment, such as PHILIPS, GE, Siemens, Mindray, and TOSHIBA. In addition, the images were acquired with superficial probes.
Experimental pathways
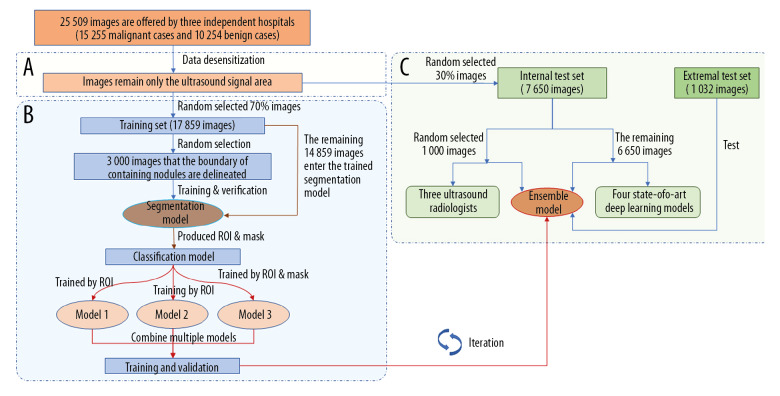
Our experimental pathways mainly included three parts (Figure 1): segmentation of nodules, ensemble learning for classification, and testing the diagnostic performance of the model. The purpose of the training segmentation model was only to find the nodule automatically. To verify whether the algorithm was effective or not, we manually performed a test check of 500 images, reaching a relevance ratio of more than 98%. Using the segmentation model, the region of interest (ROI) containing the nodule was first segmented and then classification was modeled. Results of the classification were calculated quantitatively as the comprehensive evaluation of the two processes. The classification model was improved based on DenseNet [13] and adopted as a multistep cascade experiment pathway, as shown in Figure 2. The classification result was determined according to the voting of three weak models by the average method and the voting method. Finally, we compared diagnostic performance of the EDLC-TN with that of ultrasonographers and four advanced deep learning models, and conducted an external test.
Figure 1.
Pathways of experiments. Our experimental pathways mainly included three parts. (A) Data desensitization, removal of the sections of the patient’s personal information in the images. (B) Training and validation of ensemble learning for classification of thyroid nodules. In the segmentation part, the nodule area was manually marked and used to train the segmentation model. ROI and mask were extracted by the segmentation model. Then, three weak models were trained and combined to obtain an advanced classification model. (C) Comparison experiments with radiologists and other deep learning models, and external validation experiment. We then compared performance of the classification model with that of three ultrasound radiologists and four state-of-the-art deep learning models. Finally, we conducted an external validation using an independent dataset.
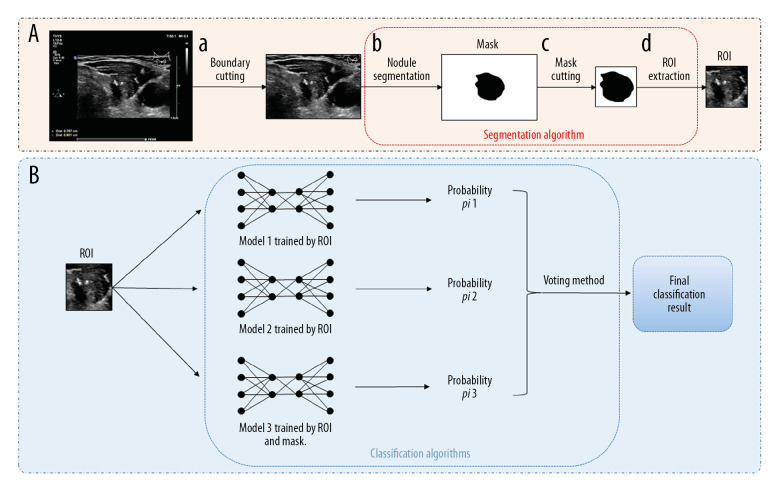
Figure 2.
The multistep cascade experiment pathway of EDLC-TN. (A) The process of extracting ROI and mask. First, the boundary was cut off (a). Second, the nodule area was segregated (b). Then, the mask image of the thyroid nodule was depicted (c). Finally, ROI was segmented (d). (B) The process of classifying images by ensemble learning model. After obtaining the ROI and its corresponding mask, three classification models were trained and combined to obtain an advanced classification model. ROI was put into models and got the final classification result through the voting method.
EDLC-TN model
A multistep cascade experiment pathway was adopted, as shown in Figure 2.
First, the image boundary with annotation was cut off (Supplementary Table 1) for data cleaning. Then, the nodule and the surrounding area of the image (region of interest, ROI) was extracted. We used a semiautomatic method to achieve this goal, that is, carefully annotating the boundaries of thyroid nodules in 3000 images by hand, and training a nodule segmentation model with these marked images to segment all of the rest images. The structure of segmentation model is shown in Supplementary Table 2, and the method of converting the segmentation results to ROI is shown in Supplementary Table 1.
Through the above process, each image generated a three-channel ROI R, and a one-channel mask M. We used these data to train nodule classification models based on the structure shown in Supplementary Table 3. For better performance, we trained multiple models and combined them through two ensemble learning methods, namely the average and voting methods. The average method calculates the mean value of all base model results. For the voting method, each base model votes on the category of the image, and the final result is the category with more votes.
The Adam optimizer was used during the training. The learning rate was initialized as 0.1. After 60 epoch iterations, it was decreased to 0.01, and then reduced by 10 times after every 200 epochs. The batch size was adjusted to the maximum within the limits of the computer memory. We trained our models on NVIDIA TITAN XP GPU based on the TensorFlow framework.
Radiologist evaluation and comparison
To assess the predictive effect of this deep learning algorithm, this paper reflects the performance of radiologists (W.X., Z.J.L. and Z.S.) on 1000 (11.52%, 1000/8,682) ultrasound images randomly selected from the test set and compares accuracy in differentiating between benign and malignant thyroid nodules on ultrasound images with the predictive results of deep learning models. The radiologists assessed nodules according to ACR TI-RADS criteria [1] and predicted whether a nodule was benign or malignant. After each individual independently judged and labeled each ultrasound image, in a kind of double-blind experiment, we used postoperative pathological analysis results (i.e., benign and malignant diagnoses that were completely correct) for statistical analysis. Finally, the average accuracy rate was calculated to assess each individual radiologist’s accuracy in evaluation of an ultrasound image of a thyroid nodule. The independent radiologists involved in the evaluation work were the attending doctor or associate professors. The first reader (W.X.) had 13 years of experience, the second reader (Z.J.L) had 8 years of experience, and the third reader (Z.S.) had more than 30 years of experience in diagnosing thyroid nodules.
Comparison with four state-of-the-art deep learning models
We compared the diagnostic performance of our model with the four machine learning algorithms which are currently most popular and advanced, including ResNeXt [14], SE_Inception_v4 [15], SE_Net [16] and Xception [17]. These models are widely used in the field of AI of medical images [18,19]. The 3000 ultrasound images randomly selected from the test set in Center 1 were used for this part of the study. The area under the receiver operating characteristic (ROC) curve with a 95% confidence interval (CI), accuracy, sensitivity, and specificity were calculated to compare capability for diagnosing thyroid cancer on ultrasound.
General applicability test
In this section, we aimed to investigate the general applicability of our AI system for diagnosing thyroid cancer. We did so by testing our network on a dataset of ultrasound images (n=1032) from Peking University BinHai Hospital, including 502 benign nodule images and 530 malignant nodule images (Table 1).
Statistical analysis
Data are shown as the means and standard deviations for continuous variables. The number of patients and images were analyzed for categorical variables. Diagnostic performance of the EDLC-TN and the radiologists was evaluated by calculating sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy. To determine whether the diagnostic performance of our models significantly differed, the AUCs between the EDLC-TN and the other four models were compared using the Z test. The intraclass correlation coefficient (ICC) and Kappa value were used to assess test-retest reliability and inter-reader agreement for different radiologists. All statistical results shown were calculated using MedCalc for Windows v15.8 (MedCalc Software, Ostend, Belgium), and P<0.05 was considered statistically significant.
Results
Four image datasets and study population
The total number of ultrasound images in this work was 26 541, including 10 756 benign nodule images and 15 785 malignant nodule images. Of the images, 17 859 (67.29%) images from Centers 1 to 3 were used for training. A total of 7 560 (28.82%) images from Centers 1 to 3 were used for internal testing. The dataset from Center 4 containing 1 032 (3.89%) images was only used as an external test set without training for verifying the generalizability of the model. Table 1 summarizes the number of images used in our training and testing datasets.
A total of 11 865 patients who underwent ultrasound examination and surgery between January 2015 and December 2017 at one of these four centers were included in this research. Demographic data and image information for all patients from four medical centers are shown in Table 2.
Table 2.
Demographic data and image information for all patients from four medical centers.
| Center 1 | Center 2 | Center 3 | Center 4 | |
|---|---|---|---|---|
| No. of patients | 10993 | 151 | 460 | 261 |
| Sex | ||||
| Female (n) | 8379 (76.22%) | 117 (77.48%) | 369 (80.22%) | 197 (75.48%) |
| Male (n) | 2614 (23.78%) | 34 (22.52%) | 91 (19.78%) | 64 (24.52%) |
| Age (years) | 46 (18–84) | 49 (21–67) | 51 (18–73) | 52 (23–70) |
| Position of nodules | ||||
| Left lobe | 5063 (46.06%) | 79 (52.31%) | 214 (46.52%) | 127 (48.66%) |
| Right lobe | 5652 (51.41) | 70 (46.36%) | 228 (49.57%) | 128 (27.77%) |
| Isthmus | 278 (2.53%) | 2 (1.32%) | 18 (3.91%) | 6 (2.61%) |
| Size (cm) | 1.25 (0.38–7.80) | 1.72 (0.58–6.25) | 1.53 (0.30–6.91) | 2.12 (0.49–7.21) |
| Postoperative pathology | ||||
| Benign nodules | 3996 (36.35%) | 82 (54.30%) | 213 (46.30%) | 153 (58.62%) |
| Nodular goiter | 2745 | 71 | 166 | 127 |
| Adenomatous goiter | 551 | 11 | 45 | 26 |
| Thyroid granuloma | 518 | |||
| Follicular adenoma | 182 | 2 | ||
| Malignant nodules | 6997 (63.65%) | 69 (45.70%) | 247 (53.70%) | 108 (41.38%) |
| PTC | 6910 | 69 | 246 | 108 |
| MTC | 65 | 1 | ||
| FTC | 20 | |||
| ATC | 2 | |||
| Total images | 23504 | 530 | 1475 | 1032 |
| Benign | 9084 | 289 | 881 | 502 |
| Malignant | 14420 | 241 | 594 | 530 |
| Types of machine | Philips EPIQ 5 | Philips IU22 | Philips EPIQ 7 | GE LOGIQ E9 |
| Philips IU Elite | Philips IU22 | Mindray DC-8 | ||
| Philips IU22 | Siemens Acuson Qxana 2 | Siemens Acuson S2000 | ||
| Philips HD11 | Siemens Acuson S2000 | |||
| TOSHIBA Aplio 500 | ||||
| TOSHIBA Aplio 400 | ||||
PTC – papillary thyroid carcinoma; MTC – medullary thyroid carcinoma; FTC – follicular thyroid carcinoma; ATC – anaplastic thyroid carcinoma.
Classification by EDLC-TN
In this paper, accuracy, specificity and sensitivity were the main evaluation criteria for classification. Two models were similar in structure, so we analyzed the experimental results of one them, Classifier1, as the main model. The results of ensemble learning using different combination strategies are shown in Table 3. Among them, the method of voting requires at least three weak models, so two instances of weak classifier 1 are used.
Table 3.
Comparison of the diagnostic performance of EDLC-TN with radiologists.
| Accuracy | Sensitivity | Specificity | |
|---|---|---|---|
| EDLC-TN | 93.70% | 93.19% | 94.01% |
| Radiologist 1 | 91.55% | 91.45% | 91.71% |
| Radiologist 2 | 87.26% | 96.34% | 72.19% |
| Radiologist 3 | 93.07% | 92.56% | 93.92% |
| Average of radiologists | 90.63% | 93.45% | 85.94% |
| Radiologists and EDLC-TN | 96.54% | 97.11% | 95.58% |
EDLC-TN – ensemble deep learning classification model of thyroid nodules.
The accuracy rate of the two weak models was already high. Strong classifier 1 and strong classifier 2 both were obtained by combining two models. Of the three methods, the averaging method calculates the arithmetic mean of the results obtained from the two models, the competition method takes the higher confidence level of two results as the predicted value, and the voting method combines the results of multiple (more than 3) models. All the models vote for benignancy and malignancy, with the majority of votes serving as the final result. Therefore, we found that the strong classifiers had higher accuracy than each weak classifier. The test results for weak and strong classifiers in diagnosis of thyroid nodules are shown in Supplementary Table 4.
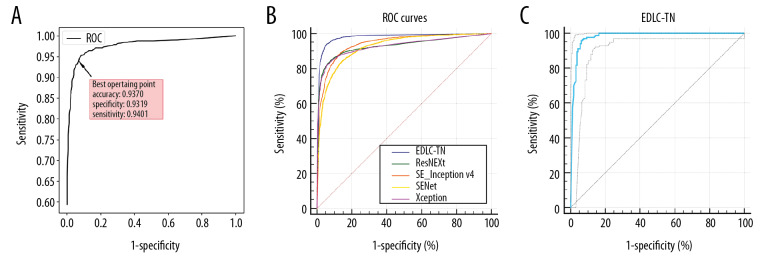
The model proposed in this paper is the structure of “classification after segmentation”. The performance of ensemble learning is shown in Figure 3A. With the changing threshold, accuracy, specificity, and sensitivity continue to change. When the threshold is around 0.54, the accuracy, sensitivity, and specificity were all at the high level (93.70%, 93.19% and 94.01%, respectively).
Figure 3.
Performance of the EDLC-TN in identification of thyroid cancer in different datasets. (A) Performance of the EDLC-TN on the training dataset. The accuracy, sensitivity and specificity were 93.70%, 93.19%, and 94.01%, respectively. (B) Diagnostic performance of the EDLC-TN and four other state-of-the-art machine learning algorithms. The EDLC-TN demonstrated the highest value for AUC (0.941, 95% CI: 0.935–0.946), sensitivity (93.77%), specificity (94.44%), and accuracy (98.51%). (C) The performance of EDLC-TN on the external validation dataset. The EDLC-TN achieved an accuracy of 95.76%, with a sensitivity of 95.88%, a specificity of 93.75% and an AUC of 0.979 (95% CI: 0.958–0.992).
EDLC-TN vs. radiologists
In this experiment, three thyroid disease radiologists in the hospital were randomly selected to independently evaluate benign and malignant thyroid ultrasound images (the same test data set used for deep learning) and annotate them. The accuracy of each doctor and their average values are shown in Table 3. Those results indicate that the deep learning model proposed in this paper is more accurate than that of individual radiologists.
In addition, we also carried out relevant experiments with multi-expert cooperating diagnosis, that is, the three radiologists simultaneously performed benign and malignant judgments and voted on one ultrasound image, and the majority of the votes were the final results. After comparing the results of a single model and a single radiologist, the highest accuracy of the model was 93.70%. However, compared with the accuracy of the model, the result of the medical consultation of three radiologists was more accurate, with a rate of 95.43%. Finally, the accuracy was 96.54% with analyses of the model and radiologist combined, which was higher than that for independent diagnosis by either (Table 3).
The ICC and Kappa value were used to assess test-retest reliability and inter-reader agreement for three radiologists. As a result, the ICC of diagnosing results from three radiologists was 0.7052 (95%IC: 0.6836–0.7260). The Kappa values for Radiologist 1 vs. 2, Radiologist 2 vs. 3 and Radiologist 1 vs. 3 were 0.649 (95%IC: 0.609–0.689), 0.656 (95%IC: 0.616–0.696), 0.774 (95%IC: 0.741–0.808), separately.
EDLC-TN vs. other four AI models
The diagnostic performance of the four machine learning algorithms is shown in Table 4 and Figure 3B. The EDLC-TN model demonstrated the highest value for AUC (0.941, 95% CI: 0.935–0.946), which was significantly higher than the other four models (P<0.0001). Also, the EDLC-TN model performed had the highest values for sensitivity (93.77%), specificity (94.44%), and accuracy (98.51%).
Table 4.
Comparison of the diagnostic performance of EDLC-TN with other four state-of-the-art algorithms.
| AUC | Sensitivity (%) | Specificity (%) | Accuracy (%) | |
|---|---|---|---|---|
| EDLC-TN | 0.941 (0.936–0.946) | 93.77 | 94.44 | 98.51 |
| ResNeXt | 0.882 (0.875–0.889)* | 85.53 | 90.86 | 82.83 |
| SE_Inception_v4 | 0.874 (0.866–0.881)* | 90.33 | 84.38 | 97.12 |
| SE_Net | 0.840 (0.832–0.848)* | 88.64 | 79.35 | 96.52 |
| Xception | 0.880 (0.872–0.887)* | 84.68 | 91.26 | 93.84 |
EDLC-TN – ensemble deep learning classification model of thyroid nodules; AUC – area under the ROC curve; AUCs of EDLC-TN and other three models were calculated by the method of DeLong et al. P – The difference of AUCs between the EDLC-TN and other four models was compared by Z-test,
P<0.05.
Generalizability of EDLC-TN
To investigate the generalizability of EDLC-TN in diagnosis of thyroid cancer, we applied the same deep learning framework to ultrasound images from Peking University BinHai Hospital (Center 4), which were not contained in the training set (Table 1). In this test, the EDLC-TN achieved an accuracy of 95.76%, with a sensitivity of 95.88% and a specificity of 93.75% in differentiating between benign and malignant thyroid nodules. The ROC curve is shown in Figure 3C and the area under the ROC curve of EDLC-TN for diagnosing thyroid cancer was 0.979 (95% CI: 0.958–0.992).
Discussion
Many researchers have made significant contributions to the field of deep learning models for differentiating between benign and malignant thyroid lesions. Xia J et al. [20] proposed an extreme learning machine (ELM) based on ultrasound features, such as composition, echogenicity, margin, shape, and calcification, to classify malignant and benign thyroid nodules and it achieved 87.72% diagnostic accuracy. Liu T et al. [21] used the CNN model learned from ImageNet as a pretrained feature extractor for an ultrasound image dataset. Their experimental results with 1 037 images demonstrated an accuracy of 93.1%. Li et al. [6] also structured an ensemble model for diagnosis of thyroid cancer based on ResNet 50 and Darknet 19. However, the diagnostic accuracy was only 85.7% to 88.9% because the types of two sub-models were similar.
In this study, we proposed a new ensemble deep learning classification model called EDLC-TN for classifying benign and malignant thyroid nodules by ultrasound with evidence from multiple centers. The strengths of EDLC-TN model are fourfold. The core of this method is performing deep learning model training on the basis of segmenting the ROI, which is the area where the thyroid nodule is located. The accuracy of this model is the highest among the state-of-the-art algorithms and other models mentioned above. The accuracy of our model in diagnosing benign and malignant thyroid nodules was higher than that of a single radiologist and the model could help improve the diagnostic accuracy of radiologists. This model represents a generalized platform that can be universally applied to ultrasound images from different medical centers. Moreover, remarkable progress has been made with deep learning in the field of image processing, resulting in mature models of segmentation, localization, and classification for natural images. We used ensemble learning methods to connect the results of multiple models of deep learning. With that method, it was possible to distinguish between malignant and benign nodules with the highest accuracy, in contrast to other advanced deep learning models. The diagnostic performance of the radiologists in diagnosing thyroid cancer can be significantly improved if combined with EDLC-TN. Therefore, it could benefit radiologists in diagnosis to a large extent.
Furthermore, our network is a general platform that can be universally applied to ultrasound images from different medical centers. When applying the EDLC-TN model to ultrasound images from a hospital with totally different types of ultrasound equipment, the EDLC-TN achieved excellent accuracy, sensitivity, and specificity. Even compared to a radiologist’s performance, our model also has advantages. The high accuracy with model in our study suggests that the EDLC-TN model has the potential to effectively learn from different types of medical images with a high degree of generalization. This could benefit screening programs and produce more efficient referral systems in all medical fields, particularly in low-resource or remote areas. The result might a wide-ranging impact on both clinical care and public health.
There are several limitations to this study. Our benign datasets contained a high percentage of malignant nodules and nodular goiters, which may have introduced bias. Only three senior radiologists were chosen as the matched group, contributing to study bias. This model did not analyze extensive pathological types of thyroid nodules; they will be assessed in future studies. Our algorithm only gives a classification result and not provide a classification standard or texture analysis. In medicine, a good predictive algorithm often is insufficient. What is needed is the ability to explain an algorithm’s decisions and increase the credibility of diagnostic results [22]. We did not know whether this model can be applied to other types of medical images. These limitations will be overcome by expanding the ultrasound images datasets with various image types.
Conclusions
In this work, we proposed an ensemble deep learning classification model called EDLC-TN for distinguishing between benign and malignant thyroid nodules in ultrasound images. In addition, our network represents a generalized platform that can potentially be applied to different medical centers to assist radiologists.
Supplementary Data
Supplementary Table 1.
The algorithm for finding the upper and lower boundaries of a nodule.
| Algorithm 1. Detector for the upper and lower boundaries of a given nodule. | |
|---|---|
| Input: mask: Distinguish whether a pixel belongs to the nodule with 0 or 1 label. | |
|
Output: up_bound: Upper boundary of the nodule; low_bound: Lower boundary of the nodule. | |
| 1: | RS = ∑ (mask, axis=1) // The sum of each line of the mask. |
| 2: | RS.append(0) // In order to simplify the calculation process. |
| 3: | start, maxLen, curLen = 0, 0, 0 |
| 4: | for i, v in enumerate(RS) do |
| 5: | if v > threshold then |
| 6: | curLen +=1 |
| 7: | else |
| 8: | if curLen > maxLen then |
| 9: | start = i – curLen |
| 10: | maxLen = curLen |
| 11: | end if |
| 12: | end if |
| 13: | end for |
| 14: | up_bound = start, low_bound = start + maxLen |
| 15: | return up_bound, low_bound |
Supplementary Table 2.
ROI extraction algorithm structure.
| Processing | Layer | Output size | Activation |
|---|---|---|---|
| Down-sampling | conv1_1 | 224×224 | Relu |
| conv1_2 | 224×224 | Relu | |
| pool1 | 112×112 | ||
| conv2_1 | 112×112 | Relu | |
| conv2_2 | 112×112 | Relu | |
| pool2 | 56×56 | ||
| conv3_1 | 56×56 | Relu | |
| conv3_2 | 56×56 | Relu | |
| conv3_3 | 56×56 | Relu | |
| conv3_4 | 56×56 | Relu | |
| pool3 | 28×28 | ||
| conv4_1 | 28×28 | Relu | |
| conv4_2 | 28×28 | Relu | |
| conv4_3 | 28×28 | Relu | |
| conv4_4 | 28×28 | Relu | |
| pool4 | 14×14 | ||
| conv5_1 | 14×14 | Relu | |
| conv5_2 | 14×14 | Relu | |
| Down-sampling [continued] | conv5_3 | 14×14 | Relu |
| conv5_4 | 14×14 | Relu | |
| pool5 | 7×7 | ||
| conv6 | 7×7 | Relu | |
| conv7 | 7×7 | Relu | |
| conv8 | 7×7 | Relu | |
| Up-sampling | deconv1 | 14×14 | |
| deconv2 | 28×28 | ||
| deconv3 | 224×224 |
Supplementary Table 3.
Classification algorithm structure.
| Layer | Detail | Output size |
|---|---|---|
| Convolution | 3×3 conv | 64×64×16 |
| Dense Block1 | {3×3 conv }×17 | 64×64×220 |
| Transition Layer1 | 1×1 conv | 32×32×220 |
| 2×2 avg pool | ||
| Dense Block2 | {3×3 conv }×17 | 32×32×424 |
| Transition Layer2 | 1×1 conv | 16×16×424 |
| 2×2 avg pool | ||
| Dense Block3 | {3×3 conv }×17 | 16×16×628 |
| Transition Layer3 | 1×1 conv | 8×8×628 |
| 2×2 avg pool | ||
| Dense Block4 | {3×3 conv }×17 | 8×8×832 |
| Transition Layer4 | 1×1 conv | 4×4×832 |
| 2×2 avg pool | ||
| Dense Block5 | {3×3 conv }×17 | 4×4×1036 |
| Batch Normalization | 4×4×1036 | |
| Relu | 4×4×1036 | |
| Pooling | 4×4 avg pool | 1×1×1036 |
| Fully Connection | 1036 | |
| Fully Connection | 2 | |
| Softmax | 2 |
Supplementary Table 4.
Test results of weak and strong classifiers in the diagnosis of thyroid nodules.
| Model | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| Weak Model 1 | 92.24% | 95.22% | 87.29% |
| Weak model 2 | 92.31% | 91.89% | 93.00% |
| Weak Model 3 | 91.89% | 91.00% | 92.26% |
| Strong Model | 93.70% | 93.19% | 94.01% |
Footnotes
Conflict of interest
None.
Source of support: This study was supported by the National Natural Science Foundation of China (81771852); Major Scientific and Technological Projects for a New Generation of Artificial Intelligence of Tianjin (Grant No. 18ZXZNSY00300)
References
- 1.Tessler FN, Middleton WD, Grant EG, et al. ACR thyroid imaging, reporting and data system (TI-RADS): White paper of the ACR TI-RADS Committee. J Am Coll Radiol. 2017;14:587–95. doi: 10.1016/j.jacr.2017.01.046. [DOI] [PubMed] [Google Scholar]
- 2.Haugen BR, Alexander EK, Bible KC, et al. 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association Guidelines Task Force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoang JK, Middleton WD, Farjat AE, et al. Reduction in thyroid nodule biopsies and improved accuracy with American College of Radiology Thyroid Imaging Reporting and Data System. Radiology. 2018;287:185–93. doi: 10.1148/radiol.2018172572. [DOI] [PubMed] [Google Scholar]
- 4.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–18. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zech JR, Badgeley MA, Liu M, et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Zhang S, Zhang Q, et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: A retrospective, multicohort, diagnostic study. Lancet Oncol. 2019;20:193–201. doi: 10.1016/S1470-2045(18)30762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, Tian J, Pei S, et al. Machine learning-assisted system for thyroid nodule diagnosis. Thyroid. 2019;29:858–67. doi: 10.1089/thy.2018.0380. [DOI] [PubMed] [Google Scholar]
- 8.Jeong EY, Kim HL, Ha EJ, et al. Computer-aided diagnosis system for thyroid nodules on ultrasonography: Diagnostic performance and reproducibility based on the experience level of operators. Eur Radiol. 2019;29:1978–85. doi: 10.1007/s00330-018-5772-9. [DOI] [PubMed] [Google Scholar]
- 9.Buda M, Wildman-Tobriner B, Hoang JK, et al. Management of thyroid nodules seen on us images: Deep learning may match performance of radiologists. Radiology. 2019;292:695–701. doi: 10.1148/radiol.2019181343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim KJ, Choi CS, Yoon DY, et al. Computer-aided diagnosis for the differentiation of malignant from benign thyroid nodules on ultrasonography. Acad Radiol. 2008;15:853–58. doi: 10.1016/j.acra.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 11.Ma J, Wu F, Zhu J, et al. A pre-trained convolutional neural network based method for thyroid nodule diagnosis. Ultrasonics. 2017;73:221–30. doi: 10.1016/j.ultras.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Igelnik B, Pao YH, LeClair SR, Shen CY. The ensemble approach to neural-network learning and generalization. IEEE Trans Neural Netw. 1999;10:19–30. doi: 10.1109/72.737490. [DOI] [PubMed] [Google Scholar]
- 13.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017; pp. 4700–8. [Google Scholar]
- 14.Xie S, Girshick R, Dollár P, et al. Aggregated residual transformations for deep neural networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Honolulu, HI, USA. 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- 15.Szegedy C, Loffe S, Vanhoucke V, Alemi AA. Inception-v4, inception-resnet and the impact of residual connections on learning. AAAI. 2017;4:12. [Google Scholar]
- 16.Hu J, Shen L, Sun G. Squeeze-and-excitation networks; Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2018; pp. 7132–41. [Google Scholar]
- 17.Chollet F. Xception: Deep learning with depthwise separable convolutions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017; pp. 1251–58. [Google Scholar]
- 18.Lee JH, Ha EJ, Kim D, et al. Application of deep learning to the diagnosis of cervical lymph node metastasis from thyroid cancer with CT: External validation and clinical utility for resident training. Eur Radiol. 2020;30:3066–72. doi: 10.1007/s00330-019-06652-4. [DOI] [PubMed] [Google Scholar]
- 19.Wu P, Cui Z, Gan Z, Liu F. Three-dimensional resnext network using feature fusion and label smoothing for hyperspectral image classification. Sensors (Basel) 2020;20:1652. doi: 10.3390/s20061652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia J, Chen H, Li Q, et al. Ultrasound-based differentiation of malignant and benign thyroid Nodules: An extreme learning machine approach. Comput Methods Programs Biomed. 2017;147:37–49. doi: 10.1016/j.cmpb.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Liu T, Xie S, Yu J, et al. Book classification of thyroid nodules in ultrasound images using deep model based transfer learning and hybrid features. 2017. Classification of thyroid nodules in ultrasound images using deep model based transfer learning and hybrid features; pp. 919–23. [Google Scholar]
- 22.Sollini M, Cozzi L, Chiti A, Kirienko M. Texture analysis and machine learning to characterize suspected thyroid nodules and differentiated thyroid cancer: Where do we stand? Eur J Radiol. 2018;99:1–8. doi: 10.1016/j.ejrad.2017.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
The algorithm for finding the upper and lower boundaries of a nodule.
| Algorithm 1. Detector for the upper and lower boundaries of a given nodule. | |
|---|---|
| Input: mask: Distinguish whether a pixel belongs to the nodule with 0 or 1 label. | |
|
Output: up_bound: Upper boundary of the nodule; low_bound: Lower boundary of the nodule. | |
| 1: | RS = ∑ (mask, axis=1) // The sum of each line of the mask. |
| 2: | RS.append(0) // In order to simplify the calculation process. |
| 3: | start, maxLen, curLen = 0, 0, 0 |
| 4: | for i, v in enumerate(RS) do |
| 5: | if v > threshold then |
| 6: | curLen +=1 |
| 7: | else |
| 8: | if curLen > maxLen then |
| 9: | start = i – curLen |
| 10: | maxLen = curLen |
| 11: | end if |
| 12: | end if |
| 13: | end for |
| 14: | up_bound = start, low_bound = start + maxLen |
| 15: | return up_bound, low_bound |
Supplementary Table 2.
ROI extraction algorithm structure.
| Processing | Layer | Output size | Activation |
|---|---|---|---|
| Down-sampling | conv1_1 | 224×224 | Relu |
| conv1_2 | 224×224 | Relu | |
| pool1 | 112×112 | ||
| conv2_1 | 112×112 | Relu | |
| conv2_2 | 112×112 | Relu | |
| pool2 | 56×56 | ||
| conv3_1 | 56×56 | Relu | |
| conv3_2 | 56×56 | Relu | |
| conv3_3 | 56×56 | Relu | |
| conv3_4 | 56×56 | Relu | |
| pool3 | 28×28 | ||
| conv4_1 | 28×28 | Relu | |
| conv4_2 | 28×28 | Relu | |
| conv4_3 | 28×28 | Relu | |
| conv4_4 | 28×28 | Relu | |
| pool4 | 14×14 | ||
| conv5_1 | 14×14 | Relu | |
| conv5_2 | 14×14 | Relu | |
| Down-sampling [continued] | conv5_3 | 14×14 | Relu |
| conv5_4 | 14×14 | Relu | |
| pool5 | 7×7 | ||
| conv6 | 7×7 | Relu | |
| conv7 | 7×7 | Relu | |
| conv8 | 7×7 | Relu | |
| Up-sampling | deconv1 | 14×14 | |
| deconv2 | 28×28 | ||
| deconv3 | 224×224 |
Supplementary Table 3.
Classification algorithm structure.
| Layer | Detail | Output size |
|---|---|---|
| Convolution | 3×3 conv | 64×64×16 |
| Dense Block1 | {3×3 conv }×17 | 64×64×220 |
| Transition Layer1 | 1×1 conv | 32×32×220 |
| 2×2 avg pool | ||
| Dense Block2 | {3×3 conv }×17 | 32×32×424 |
| Transition Layer2 | 1×1 conv | 16×16×424 |
| 2×2 avg pool | ||
| Dense Block3 | {3×3 conv }×17 | 16×16×628 |
| Transition Layer3 | 1×1 conv | 8×8×628 |
| 2×2 avg pool | ||
| Dense Block4 | {3×3 conv }×17 | 8×8×832 |
| Transition Layer4 | 1×1 conv | 4×4×832 |
| 2×2 avg pool | ||
| Dense Block5 | {3×3 conv }×17 | 4×4×1036 |
| Batch Normalization | 4×4×1036 | |
| Relu | 4×4×1036 | |
| Pooling | 4×4 avg pool | 1×1×1036 |
| Fully Connection | 1036 | |
| Fully Connection | 2 | |
| Softmax | 2 |
Supplementary Table 4.
Test results of weak and strong classifiers in the diagnosis of thyroid nodules.
| Model | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| Weak Model 1 | 92.24% | 95.22% | 87.29% |
| Weak model 2 | 92.31% | 91.89% | 93.00% |
| Weak Model 3 | 91.89% | 91.00% | 92.26% |
| Strong Model | 93.70% | 93.19% | 94.01% |