Abstract
Fungi can colonize most of the substrata on Earth. Honey, a sugary food produced by bees (and other insects) has been studied little in terms of its fungal diversity. We have surveyed and evaluated the presence of xerotolerant and xerophilic fungi in a set of honey bee samples collected from across Spain. From 84 samples, a total of 104 fungal strains were isolated, and morphologically and phylogenetically characterized. We identified 32 species distributed across 16 genera, most of them belonging to the ascomycetous genera Aspergillus, Bettsia, Candida, Eremascus, Monascus, Oidiodendron, Penicillium, Skoua, Talaromyces and Zygosaccharomyces. As a result of this survey, eight new taxa are proposed: i.e. the new family Helicoarthrosporaceae, two new genera, Helicoarthrosporum and Strongyloarthrosporum in Onygenales; three new species of Eurotiales, Talaromyces affinitatimellis, T. basipetosporus, and T. brunneosporus; and two new species of Myxotrichaceae, Oidiodendron mellicola, and Skoua asexualis.
Keywords: Eurotiales, Fungi, Honey, New taxa, Onygenales, Osmophiles, Xerophiles
INTRODUCTION
Honey is a natural sweetener produced by honey bees (insects of the genus Apis of the order Hymenoptera) from nectar (blossom honey or nectar honey) or from carbohydrate-rich secretions of living green parts of plants or excretions of plant-sucking phytophagous aphids (insects of the family Aphidida, order Hemiptera) (honeydew honey) after combination with the bee’s specific substances, placement, dehydration, and storage in the honey comb to ripen and mature. Honey is mostly composed of monosaccharides (dextrose and fructose), at a concentration of not lower than 60% and a much lesser amount of oligosaccharides, organic acids, enzymes (amylases and α-glucosidase) and solid particles. Due to its particular physicochemical nature and biological origin, honey should be an ideal substratum for the development of xerotolerant and xerophilic fungi. However, little information has been gathered about these fungi and their relationships with honey and honey products. Nonetheless, most of the fungal species from honey had been reported as new for science. Representative ascomycetous yeasts found in honey are Blastobotrys meliponae, Candida lundiana, C. magnoliae, C. sorbosivorans, C. suthepensis, Schizosaccharomyces octosporus, Trichosporon mucoides, Zygosaccharomyces favi, Z. mellis, Z. richteri, Z. rouxii, and Z. siamensis (Lochhead & Farrell 1931; Ruiz-Argueso & Rodriguez-Navarro 1975; Carvalho et al. 2010; Saksinchai et al. 2012a, b; Čadež et al. 2015; Crous et al. 2016). The obligate xerophiles Ascosphaera apis and Bettsia alvei have been reported in honey, as well as several xerotolerant species of Alternaria, Aspergillus, Cladosporium and Penicillium and a few mucoralean fungi (Snowdon & Cliver 1996; Kačániová et al. 2009; Pettersson & Leong 2011; Kačániová et al. 2012; Sinacori et al. 2014; Grabowski & Klein 2015). Recently, Monascus mellicola, Penicillium apimei, P. meliponae, P. mellis, and Talaromyces brasiliensis were reported from honey produced by stingless bees (Melipona scutellaris, family Apidae, order Hymenoptera) inhabiting Brazilian forests (Barbosa et al. 2017, 2018). Common environmental and plant pathogenic species of fungi have been reported in samples of honey collected in Spain (Pérez-Sánchez et al. 1997; Seijo et al. 2011; Magyar et al. 2016; Terrab et al. 2019) and Portugal (Martíns et al. 2003). In another study, the yeast Metschnikowia reukaufii was, surprisingly, the only fungus reported for floral honey from Portugal and Spain (Magyar et al. 2005). Although honey should be a substratum amenable for the development of xerotolerant and xerophilic fungi, few studies have intentionally targeted these fungi. Therefore, the main objective of this study was to assess the diversity of honey-associated fungi, by employing a selective culture medium to a set of samples collected predominantly in Spain, and to characterize the morphology, physiology and phylogeny of new isolates and those considered of taxonomic interest.
MATERIALS AND METHODS
Fungal isolation
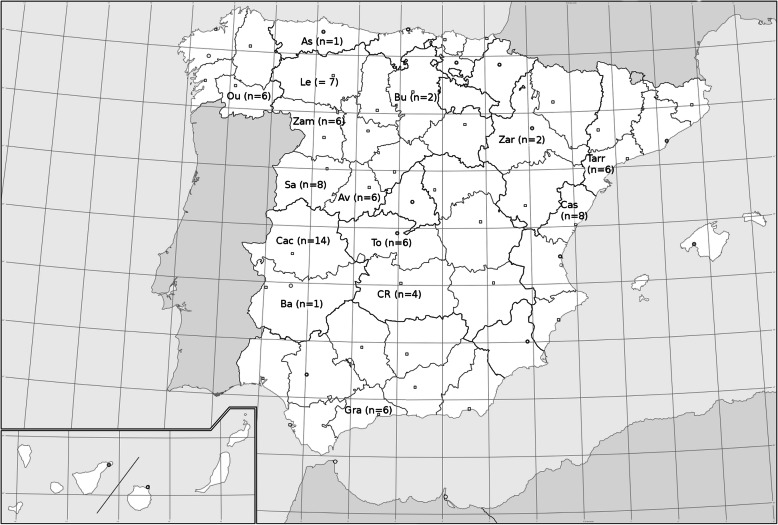
A total of 83 samples of honeydew and blossom (nectar) honey from different locations in Spain (Fig. 1), and one from Argentina (San Martín, Buenos Aires province), have been processed. All samples were of the harvest in 2014, stored in settling tanks, and after a variable period of time clarified by filtration (with one exception, which was by centrifugation). Seventy-two of the Spanish samples corresponded to honeydew honeys, 45 from trading companies and 27 collected and processed by beekeepers. A few of the samples provided by commercial companies were categorized (according to the nature of the honeydew) as oak, holm oak and forest honey. The 11 samples of blossom honey were provided by beekeepers, and these were classified as multifloral. All samples provided by commercial companies were subjected to a thermal treatment, subjecting the honey at 45–55 °C for a few hours up to 2 days, or pasteurized (2 min at 80 °C). The samples provided by beekeepers have not undergone any heat treatment. For each sample, 10 g of honey was dissolved into 90 mL of sterile water in a sterile disposable plastic container, and 1 mL of such dilution (1:10) was aseptically plated onto two 90 mm diam. plastic Petri dishes and mixed with 15 mL of molten (at 50–55 °C) 18% glycerol agar (G18; DG18 [Hocking & Pitt 1980] without dichloran: 5 g peptone, 10 g dextrose, 1 g KH2PO4, 0.5 g MgSO4·7H2O, 15 g agar-agar, 110 g glycerol, 1 L tap water, and supplemented with 250 mg/L of L-chloramphenicol). Once the medium had solidified, one of the Petri dishes was incubated in darkness at 15 °C and the other at 25 °C for up to 2 months. The colonies developed were examined under a stereomicroscope. Fungal structures from selected (representative of all morphological variety) colonies were transferred to 50 mm diam. Petri dishes containing G18 by using a sterile insulin-type needle and incubated in the same conditions to obtain pure cultures.
Fig. 1.
Locations of Spain where the samples were collected. Asturias (As), Ávila (Av), Badajoz (Ba), Burgos (Bu), Cáceres (Cac), Castellón (Cas), Ciudad Real (CR), Granada (Gra), León (Le), Ourense (Ou), Salamanca (Sa), Tarragona (Tarr), Toledo (To), Zamora (Zam), and Zaragoza (Zar)
Phenotypic study
For cultural characterization, suspensions of spores from the isolates were prepared in a semi-solid medium (0.2% agar; 0.05% Tween 80), and 0.5 μL of such suspension was inoculated onto malt extract agar (MEA; Difco, Detroit, USA; Samson et al. 2010), oatmeal agar (OA; 30 g of filtered oat flakes, 15 g agar-agar, 1 L tap water; Samson et al. 2010), Czapek yeast extract agar (CYA; 30 g sucrose, 3 g NaNO3, 5 g yeast extract, 1 g K2HPO4, 0.5 g KCl, 0.5 g MgSO4·7H2O, 0.01 g FeSO4, 15 g agar-agar, 1 L tap water; Pitt 1979), yeast extract sucrose agar (YES; 20 g yeast extract, 150 g sucrose, 0.5 g MgSO4·7H2O, 20 g agar-agar, 1 L tap water; Frisvad 1981), creatine sucrose agar (CREA; 3 g creatine, 30 g sucrose, 1.6 g K3PO4·7H2O, 0.5 g MgSO4·7H2O, 0.5 g KCl, 0.01 g FeSO4·7H2O, 0.05 g bromocresol purple, 20 g agar-agar, 1 L tap water; Frisvad 1981), G18, potato dextrose agar (PDA; Pronadisa, Madrid, Spain; Hawksworth et al. 1995), 25% glycerol nitrate agar (G25 N; 7.5 g Czapek concentrate, 0.75 g K2HPO4, 3.7 g yeast extract, 250 mL glycerol, 12 g agar-agar, 1 L tap water; Pitt 1979), bromocresol purple milk solids glucose agar (BCP-MS-G; 80 g skim milk powder, 40 g glucose, 10 mL of 1.6% of bromocresol purple in 95% ethanol, 30 g agar-agar,1 L tap water; Kane & Smitka 1978), test opacity tween medium (TOTM; 10 g bacteriological peptone, 5 g NaCl, 1 g CaCl2, 5 mL Tween 80, 15 g agar-agar, 1 L tap water; Slifkin 2000), phytone yeast extract agar (PYE; Becton, Dickinson & Co., Sparks, MD, USA; Carmichael & Kraus 1959), malt extract yeast extract 70% fructose-glucose (MY70FG; 6 g malt extract, 6 g yeast extract, 10 g peptone, 350 g fructose, 350 g glucose, 12 g agar-agar, 1 L tap water; Beuchat & Hocking 1990), and blood agar (Becton, Dickinson & Co., Sparks, MD, USA). Colonies were characterized after three wk. at 25 °C in darkness. G18 medium was used to determine the minimum, optimal and maximum temperatures of growth. Christensen’s urea agar (EMD Millipore, Darmstadt, Germany; Christensen 1946) was inoculated and incubated during 4–7 days at 25 °C in darkness to detect the production of urease. Cycloheximide tolerance of the fungal strains was tested on Sabouraud dextrose agar (SDA; Pronadisa, Spain) supplemented with 0.2% of cycloheximide (Sigma, USA) after incubation at 30 °C for two wk. Fungal tolerance to NaCl was evaluated on SDA adding 3, 10 and 20% w/v NaCl, with the same incubation conditions as in the previous test. Colour notations were according to Kornerup & Wanscher (1978). The microscopic structures were characterized and measured from wet mountings of slide cultures, using water and 60% lactic acid. Photo micrographs were taken using a Zeiss Axio-Imager M1 light microscope (Oberkochen, Germany) with a DeltaPix Infinity X digital camera, using Nomarski differential interference contrast. The samples for scanning electron microscopy (SEM) were processed according to Figueras & Guarro (1988), and SEM micrographs were taken at 15 keV with a JEOL JSM 840 microscope.
DNA extraction, amplification and sequencing
Total deoxyribonucleic acid (DNA) was extracted according to Marimon et al. (2006), and a fragment of the 28S nrRNA gene (LSU) was amplified and sequenced using the primer pair LR0R (Rehner & Samuels 1994) and LR5 (Vilgalys & Hester 1990). For some isolates the following markers were amplified and sequenced: ribosomal internal transcribed spacers (ITS) (ITS5/ITS4; White et al. 1990); and fragments of the beta-tubulin (BenA) (Bt2a/Bt2b; Glass & Donaldson 1995), calmodulin (CaM) (Cmd5/Cmd6; Hong et al. 2005) and RNA polymerase II subunit 2 (rpb2) (RPB2-5F/RPB2-7cR; Liu et al. 1999) genes. Amplicons were sequenced at Macrogen Europe (Macrogen, Amsterdam, The Netherlands). Consensus sequences were obtained using the SeqMan software v. 7 (DNAStar Lasergene, Madison, WI, USA). Sequences we generated were deposited in GenBank (Table 1).
Table 1.
Fungal taxa recovered with their nucleotide sequence accession number, and the geographic origin of the honey samples processed
| Taxon | Culture collection accession number | EMBL/GenBank nucleotide sequence accession number | Geographic origin (province, community) | ||||
|---|---|---|---|---|---|---|---|
| BenA | CaM | rpb2 | ITS | LSU | |||
| Alternaria multiformis | FMR 16018 | – | – | – | LT963545 | LT963546 | Salamanca, Castilla y León |
| Ascosphaera atra | FMR 16318 | – | – | – | LT964944 | LT984552 | Cáceres, Extremadura |
| Aspergillus asperescens | FMR 16310 | LT963510 | – | – | LT986672 | Zamora, Castilla y León | |
| Aspergillus montevidensis | FMR 15994 | LR027804 | – | – | LT963466 | LT984537 | Castellón, Valencia |
| Aspergillus pseudoglaucus | FMR 9392 | LT963512 | – | – | – | LT984695 | Castellón, Valencia |
| Aspergillus pseudoglaucus | FMR 15992 | LT963513 | – | – | – | LT984696 | Castellón, Valencia |
| Aspergillus pseudoglaucus | FMR 15993 | LT963514 | – | – | – | LT984697 | Castellón, Valencia |
| Aspergillus pseudoglaucus | FMR 16011 | LT963518 | – | – | – | LT984701 | Salamanca, Castilla y León |
| Aspergillus pseudoglaucus | FMR 16112 | LT963515 | – | – | – | LT984698 | Ciudad Real, Castilla-La Mancha |
| Aspergillus pseudoglaucus | FMR 16281 | LT963516 | – | – | – | LT984699 | Ciudad Real, Castilla-La Mancha |
| Aspergillus pseudoglaucus | FMR 16317 | LT963517 | – | – | – | LT984700 | Zamora, Castilla y León |
| Bettsia alvei | FMR 15670 | – | – | – | – | LT963566 | Castellón, Valencia |
| Bettsia alvei | FMR 15672 | – | – | – | – | LT963567 | Castellón, Valencia |
| Bettsia alvei | FMR 15678 | – | – | – | – | LT963568 | Castellón, Valencia |
| Bettsia alvei | FMR 15681 | – | – | – | – | LT963569 | Castellón, Valencia |
| Bettsia alvei | FMR 15685 | – | – | – | – | LT963570 | Castellón, Valencia |
| Bettsia alvei | FMR 16111 | – | – | – | – | LT963571 | Cáceres, Extremadura |
| Bettsia alvei | FMR 16115 | – | – | – | – | LT963572 | Toledo, Castilla-La Mancha |
| Bettsia alvei | FMR 16305 | – | – | – | – | LT963574 | Ourense, Galicia |
| Bettsia alvei | FMR 16313 | – | – | – | – | LT963575 | Ourense, Galicia |
| Bettsia alvei | FMR 16568 | – | – | – | – | LT963573 | Cáceres, Extremadura |
| Bettsia alvei | FMR 16570 | – | – | – | – | LT963576 | Ourense, Galicia |
| Candida magnoliae | FMR 16311 | – | – | – | – | LT963487 | Ourense, Galicia |
| Candida magnoliae | FMR 16314 | – | – | – | – | LT963488 | Ourense, Galicia |
| Candida magnoliae | FMR 16496 | – | – | – | – | LT963486 | Ourense, Galicia |
| Candida sorbosivorans | FMR 16278 | – | – | – | – | LT963489 | Ourense, Galicia |
| Cunninghamella bertholletiae | FMR 16008 | – | – | – | LT963490 | LR215930 | Salamanca, Castilla y León |
| Eremascus albus | FMR 16116 | – | – | – | – | LT964975 | Cáceres, Extremadura |
| Eremascus albus | FMR 16118 | – | – | – | – | LT964976 | Cáceres, Extremadura |
| Eremascus albus | FMR 16119 | – | – | – | – | LT964977 | Toledo, Castilla-La Mancha |
| Eremascus albus | FMR 16493 | – | – | – | – | LT964978 | Cáceres, Extremadura |
| Helicoarthrosporum mellicola | FMR 15673 | – | – | – | – | LT978462 | Castellón, Valencia |
| Helicoarthrosporum mellicola T | FMR 15679 = CBS 143838 | – | – | – | – | LT906535 | Castellón, Valencia |
| Helicoarthrosporum mellicola | FMR 16307 | – | – | – | – | LT978463 | León, castilla y León |
| Helicoarthrosporum mellicola | FMR 16308 | – | – | – | – | LT906536 | Zamora, Castilla y León |
| Helicoarthrosporum mellicola | FMR 16315 | – | – | – | – | LT906537 | Cáceres, Extremadura |
| Monascus pilosus | FMR 16306 | – | – | – | LT963491 | LT984551 | Zamora, Castilla y León |
| Monascus purpureus | FMR 16283 | – | – | – | LT963492 | LR215932 | Ávila, Castilla y León |
| Monascus purpureus | FMR 16316 | – | – | – | LT963493 | LT984550 | Cáceres, Extremadura |
| Monascus purpureus | FMR 16321 | – | – | – | LT963494 | LR215933 | Cáceres, Extremadura |
| Monascus ruber | FMR 16284 | – | – | – | LT963495 | LT986673 | Zamora, Castilla y León |
| Mucor plumbeus | FMR 16012 | – | – | – | LT963539 | LR215934 | Ciudad Real, Castilla-La mancha |
| Mucor plumbeus | FMR 16013 | – | – | – | LT963540 | LT984540 | Salamanca, Castilla y León |
| Mucor plumbeus | FMR 16017 | – | – | – | LT963541 | LT984548 | Salamanca, Castilla y León |
| Oidiodendron mellicola | FMR 15680 | – | – | – | LT906540 | LT978465 | Tarragona, Catalonia |
| Oidiodendron mellicola T | FMR 15683 = CBS 143839 | – | – | – | LT906544 | LT978464 | Castellón, Valencia |
| Oidiodendron mellicola | FMR 16023 | – | – | – | LT978506 | LT978470 | Salamanca, Castilla y León |
| Oidiodendron mellicola | FMR 16031 | – | – | – | LT906541 | LT978466 | Ciudad Real, Castilla-La mancha |
| Oidiodendron mellicola | FMR 16117 | – | – | – | LT978503 | LT978467 | Ciudad Real, Castilla-La Mancha |
| Oidiodendron mellicola | FMR 16120 | – | – | – | LT978507 | LT978471 | Toledo, Castilla-La Mancha |
| Oidiodendron mellicola | FMR 16274 | – | – | – | LT978509 | LT978473 | Burgos, Castilla y León |
| Oidiodendron mellicola | FMR 16282 | – | – | – | LT978508 | LT978472 | Toledo, Castilla-La Mancha |
| Oidiodendron mellicola | FMR 16503 | – | – | – | LT978504 | LT978468 | Ciudad Real, Castilla-La Mancha |
| Oidiodendron mellicola | FMR 16504 | – | – | – | LT978505 | LT978469 | Ourense, Galicia |
| Penicillium camemberti | FMR 16016 | LR027805 | – | – | LT963578 | LT984541 | Salamanca, Castilla y León |
| Penicillium citrinum | FMR 16028 | LT963451 | – | – | – | LT984702 | Salamanca, Castilla y León |
| Penicillium corylophilum | FMR 16010 | LR027808 | – | – | LT963581 | LT984538 | Asturias |
| Penicillium corylophilum | FMR 16027 | LT963452 | – | – | – | LT986674 | Asturias |
| Penicillium corylophilum | FMR 16030 | LR027809 | – | – | LT963582 | LT984547 | Cáceres, Extremadura |
| Penicillium cravenianum | FMR 16019 | LR027807 | – | – | LT963580 | LT984542 | Salamanca, Castilla y León |
| Penicillium cravenianum | FMR 16020 | LR027806 | – | – | LT963579 | LT984549 | Cáceres, Extremadura |
| Rhizopus oryzae | FMR 16022 | – | – | – | LT963543 | LR215931 | Cáceres, Extremadura |
| Schizosaccharomyces octosporus | FMR 16279 | – | – | – | – | LT963544 | Ourense, Galicia |
| Skoua asexualis | FMR 16497 | – | – | – | LT964664 | LT964665 | Cáceres, Extremadura |
| Skoua asexualis | FMR 16567 | – | – | – | LT964666 | LT964667 | Cáceres, Extremadura |
| Skoua asexualis T | FMR 16572 = CBS 144072 | – | – | – | LT964668 | LT964669 | León, castilla y León |
| Skoua fertilis | FMR 10812 | LR585993 | – | LR586005 | LR585979 | LT965019 | Castellón, Valencia |
| Skoua fertilis | FMR 10813 | LR585994 | – | LR586006 | LR585980 | LT965023 | Castellón, Valencia |
| Skoua fertilis | FMR 10814 | LR585995 | – | – | LR585981 | LT965016 | Castellón, Valencia |
| Skoua fertilis | FMR 10815 | – | – | LR586007 | LR585982 | LT965015 | Castellón, Valencia |
| Skoua fertilis | FMR 15671 | LR585996 | – | LR586008 | LR585983 | LT965014 | Castellón, Valencia |
| Skoua fertilis | FMR 15676 | LR585997 | – | LR586009 | LR585984 | LT965017 | Castellón, Valencia |
| Skoua fertilis | FMR 15682 | LR585998 | – | LR586010 | LR585985 | LT965018 | Castellón, Valencia |
| Skoua fertilis | FMR 15686 | LR585999 | – | LR586011 | LR585986 | LT965020 | Castellón, Valencia |
| Skoua fertilis | FMR 15687 | LR586000 | – | LR586012 | LR585987 | LT965021 | Castellón, Valencia |
| Skoua fertilis | FMR 15689 | LR586001 | – | – | LR585988 | LT965022 | Castellón, Valencia |
| Skoua fertilis | FMR 16032 | – | – | – | LR585989 | LT965024 | Asturias |
| Skoua fertilis | FMR 16320 | – | – | – | LR585990 | LT965025 | Zamora, Castilla y León |
| Skoua fertilis | FMR 16492 | – | – | – | LR585991 | LT965026 | Cáceres, Extremadura |
| Skoua fertilis | FMR 16571 | LR586002 | – | LR586013 | LR585992 | LT965027 | Badajoz, Extremadura |
| Strongyloarthrosporum catenulatum T | FMR 16121 = CBS 143841 | – | – | – | – | LT906534 | Toledo, Castilla-La Mancha |
| Talaromyces affinitatimellis | FMR 15674 | LT965001 | – | – | – | LT968852 | Tarragona, Catalonia |
| Talaromyces affinitatimellis | FMR 15675 | LT965002 | – | – | – | LT968853 | Tarragona, Catalonia |
| Talaromyces affinitatimellis | FMR 15677 | LT965003 | – | – | – | LT968854 | Tarragona, Catalonia |
| Talaromyces affinitatimellis | FMR 15684 | LT965004 | – | – | – | LT968855 | Castellón, Valencia |
| Talaromyces affinitatimellis | FMR 15688 | LT906553 | LT906550 | LT906547 | LT906538 | LT964941 | Castellón, Valencia |
| Talaromyces affinitatimellis T | FMR 15690 = CBS 143840 | LT906552 | LT906549 | LT906546 | LT906543 | LT964939 | Castellón, Valencia |
| Talaromyces affinitatimellis | FMR 16029 | LT965005 | – | – | – | LT968856 | Cáceres, Extremadura |
| Talaromyces affinitatimellis | FMR 16033 | LT906554 | LT906551 | LT906548 | LT906539 | LT964942 | Salamanca, Castilla y León |
| Talaromyces affinitatimellis | FMR 16114 | LT965006 | – | – | – | LT968857 | Salamanca, Castilla y León |
| Talaromyces affinitatimellis | FMR 16125 | LT965009 | – | – | – | LT968860 | Zamora, Castilla y León |
| Talaromyces affinitatimellis | FMR 16126 | LT965012 | – | – | – | LT968861 | Zamora, Castilla y León |
| Talaromyces affinitatimellis | FMR 16276 | LT965010 | – | – | – | LT968862 | Zamora, Castilla y León |
| Talaromyces affinitatimellis | FMR 16494 | LT965011 | – | – | – | LT968863 | Zamora, Castilla y León |
| Talaromyces affinitatimellis | FMR 16499 | LT965007 | – | – | – | LT968858 | Cáceres, Extremadura |
| Talaromyces affinitatimellis | FMR 16501 | LT965008 | – | – | – | LT968859 | Cáceres, Extremadura |
| Talaromyces basipetosporus T | FMR 9720 = CBS 143836 | LT906563 | – | LT906545 | LT906542 | LT964940 | Buenos Aires, Argentina |
| Talaromyces brunneosporus T | FMR 16566 = CBS 144320 | LT962483 | LT962488 | LT962485 | LT962487 | LT964943 | Salamanca, Castilla y León |
| Xerochrysium xerophilum | FMR 15669 | – | – | – | LT986724 | LT986675 | Castellón, Valencia |
| Zygosaccharomyces gambellarensis | FMR 16277 | – | – | – | – | LT963549 | Salamanca, Castilla y León |
| Zygosaccharomyces gambellarensis | FMR 16569 | – | – | – | – | LT963548 | Cáceres, Extremadura |
| Zygosaccharomyces mellis | FMR 16280 | – | – | – | – | LT963550 | Ourense, Galicia |
| Zygosaccharomyces mellis | FMR 16312 | – | – | – | – | LT963551 | Ourense, Galicia |
| Zygosaccharomyces siamensis | FMR 16034 | – | – | – | LT963547 | LT984543 | Salamanca, Castilla y León |
FMR = Faculty of Medicine of Reus culture collection; CBS = Westerdijk Fungal Biodiversity Institute (ex Centraalbureau voor Schimmelcultures). T = ex type
Phylogenetic analysis
A preliminary molecular identification of the isolates was carried out with LSU sequences using Basic Local Alignment Search Tool (BLAST; https://blast.ncbi.nlm.nih.gov/Blast.cgi) and only the type sequences or reliable reference strains from GenBank were considered for identification, and a maximum level of identity (MLI) of ≥98% was used for identification at the rank of species and < 98% at the rank of genus. BenA for to the genera Aspergillus, Penicillium, and Talaromyces, and ITS for the genera Monascus, Oidiodendron and Skoua were used for identification at the rank of species. An LSU tree was built to determine the phylogenetic relationships of all our isolates. Phylogenetic trees of ITS and a combination of ITS-BenA-CaM-rpb2 were also built to distinguish the members of Myxotrichaceae and the genus Talaromyces, respectively. Cunninghamella bertholletiae (CBS 693.68), Mucor plumbeus (DAOM 220743), Mucor racemosus (ATCC 42647), and Rhizopus oryzae (CBS 112.07 and CBS 130146) were used as outgroup for the LSU tree; Aphanoascus keratinophilus (IMI 319010) for the Myxotrichaceae taxa tree; and Trichocoma paradoxa (CBS 247.57) for the Talaromyces tree. The sequence alignments and the maximum-likelihood (ML) and Bayesian-inference (BI) phylogenetic analyses were performed as described previously (Valenzuela-Lopez et al. 2018). The final matrices used for the phylogenetic analysis were deposited in TreeBASE (www.treebase.org; accession number: S23122).
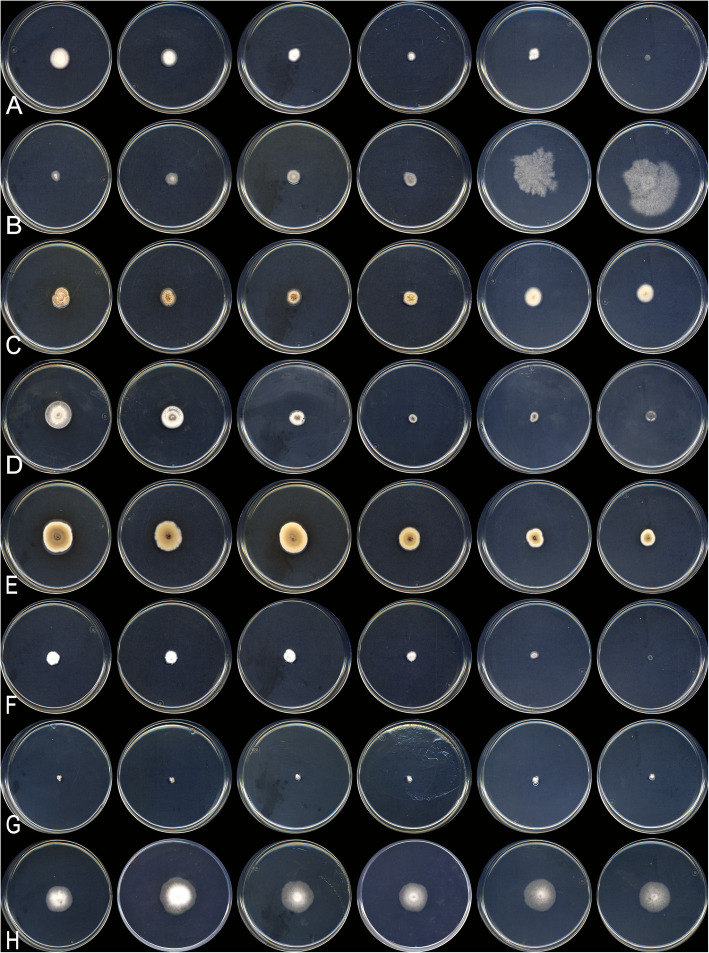
Growth at different water activities (aw)
To test the capacity of growth in different water activities, media containing malt extract (1% w/w), yeast extract (0.25% w/w) and agar-agar (1% w/w) at pH 5.3 were adjusted at six different aw (0.97, 0.95, 0.93, 0.92, 0.88 and 0.82) by adding equal weights of fructose and glucose (corresponding to 22, 30, 40, 44, 48, and 55% w/w of sugars, respectively) (Pitt & Hocking 1977). Water activity was measured in duplicate by a water activity meter (Aqualab, Decagon Devices CX3 02734) with an accuracy of ±0.002 at 25 °C. Triplicate plates were inoculated at their centre with 5 μL of spore suspension of selected fungi, and incubated at 25 °C in darkness, with the exception of FMR 15880, FMR 15883 and FMR 16031, which were at 15 °C (because of their poor growth at 25 °C). The colony diam. was measured after 21 days.
RESULTS
Fungal diversity
All honey samples produced fungal colonies on G18 at 15 °C as well as at 25 °C. Table 1 summarizes the fungal strains identified phenotypically and molecularly. With the exception of a few ascomycetous yeasts and of Mucorales, most of the fungi were filamentous Ascomycota. From the latter, the highest number of strains corresponded to Skoua (syn. Eremascus) fertilis, Bettsia alvei, and Oidiodendron sp., followed by an unknown arthrosporic fungus, Eremascus albus and Skoua sp. Alternaria multiformis, Ascosphaera atra, another unknown arthrospored fungus and Xerochrysium xerophilum were isolated only once. Obligate xerophilic species of Aspergillus were not found, but the xerotolerant A. pseudoglaucus, A. asperescens and A. montevidensis were isolated. Three species of Monascus were identified, i.e. M. pilosus, M. purpureus, and M. ruber. The isolates of Penicillium were classified as P. camemberti, P. citrinum, P. corylophilum, and P. cravenianum. Members of Talaromyces were classified at the rank of section, i.e. section Trachyspermi and section Purpurei. We only identified three species of Mucoromycota, viz. Cunninghamella bertholletiae, Mucor plumbeus, and Rhizopus oryzae. Regardless of their geographical origin, type of honey (nectar or honeydew) and if honey was or not thermally treated, S. fertilis and B. alvei were present in all honey samples.
Molecular phylogeny
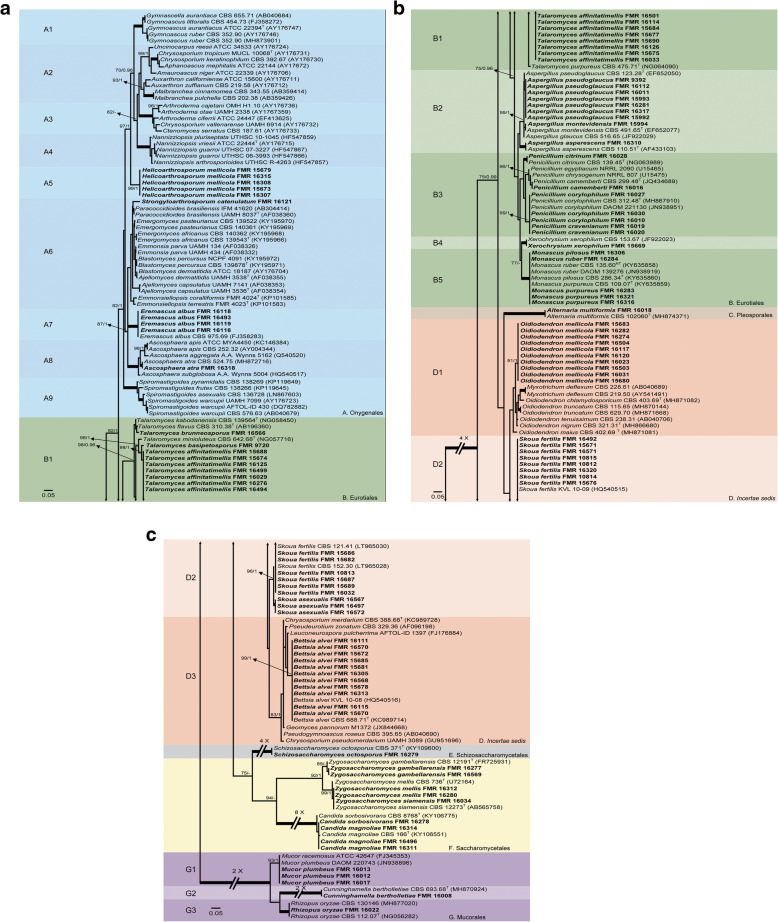
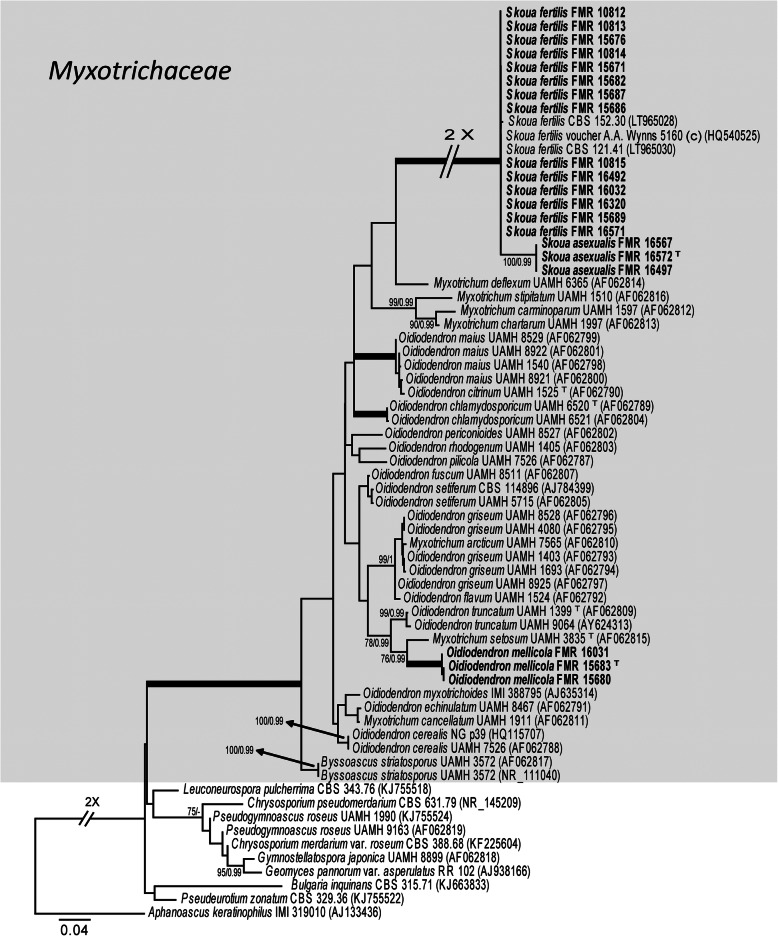
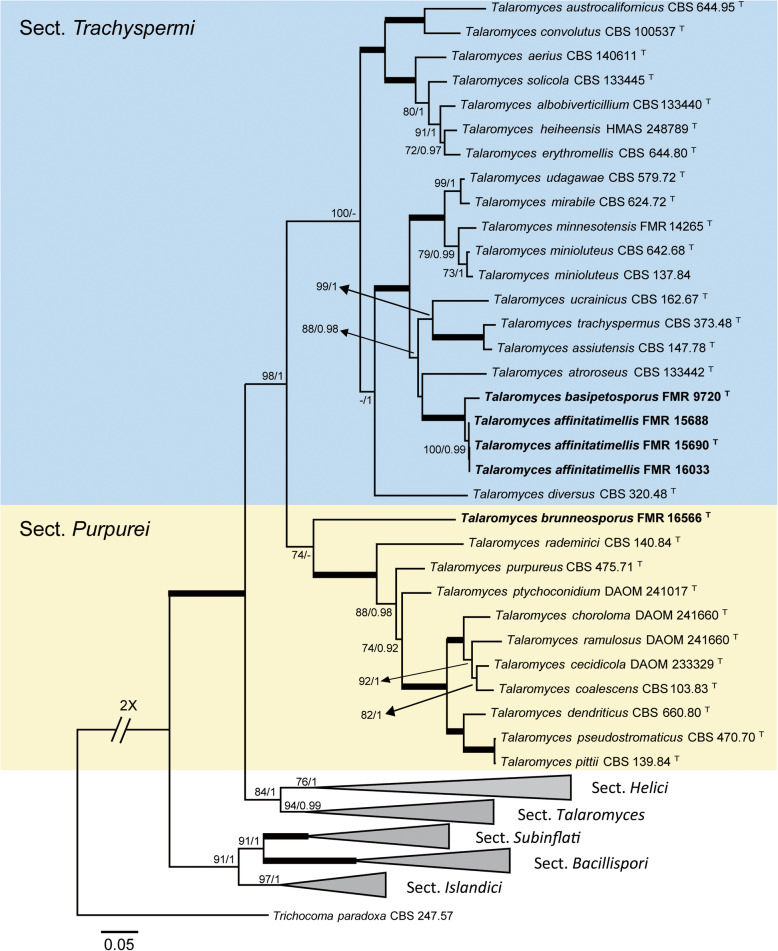
Our first phylogenetic study included 206 LSU sequences with a total of 606 characters, including gaps, 352 of them being parsimony informative. The ML analysis was congruent with that obtained in the BI analysis, both displaying trees with similar topologies. The isolates were distributed across two main clades (Fig. 2a-c), the first (100% BS / 1 PP) corresponding to the Ascomycota and including 99 isolates, and the second (100% BS / 1 PP) involving the rest of the isolates and pertaining to the Mucoromycota. The first main clade was divided into six subclades: A (82% BS / 1 PP), which represents Onygenales; B (75% BS / 0.96 PP), Eurotiales; C (100% BS / 1 PP); Pleosporales, D (unssuported) as incertae sedis; E (100% BS / 1 PP), Schizosaccharomycetales, and F (94% BS / - PP), Saccharomycetales. Subclade A contains seven well-supported groups, six of which represent the known families of Onygenales, i.e. Gymnoascaceae (A1), Arthrodermataceae (A3), Nannizziopsiaceae (A4), Eremascaceae (A7), Ascosphaeriaceae (A8), and Spiromastigaceae (A9), and a seventh group (A5) composed of five of our strains probably representing a new family. The groups representing Ajellomycetaceae (A6) and Onygenaceae (A2) were unsupported. Strains in subclade A were distributed as follows: the five mentioned above into A5, FMR 16121 into a separate branch of the Ajellomycetaceae (A6), four strains conspecific with Eremascus albus (A7), and one (FMR 16318) identified as Ascosphaera atra (A8). Thirty-nine strains were placed in Eurotiales (Subclade B). One (FMR 16566) was placed together with Talaromyces flavus and T. kabodanensis in an unsupported branch, and 16 strains near to T. minioluteus into a well-supported sister clade (B1). Into B2 (unsupported), which includes species of Aspergillus, eight of the strains were placed in a branch (99% BS / 1 PP) together with A. glaucus, A. montevidensis and A. pseudoglaucus (sect. Aspergillus). For the final identification of these eight strains, we used BenA sequence comparison, which were found to be A. montevidensis (one strain) and A. pseudoglaucus (seven strains). FMR 16310 was placed in a branch together with the ex-type sequence of A. asperescens (sect. Nidulantes). Seven strains grouped into the sister clade B3 (unsupported), representing five species of Penicillium. FMR 15669 was identified as Xerochrysium xerophilum (B4), and five strains were initially identified as Monascus spp. Based on the comparison of ITS sequences, these five strains were finally identified as M. pilosus (one strain), M. purpureus (three strains), and M. ruber (one strain). Strain FMR 16018 was located together with Alternaria multiformis (Subclade C, Pleosporales). Subclade D (unsupported) was divided into three groups: D1, representing the Myxotrichaceae; D2, the genus Skoua; and D3, the Pseudeurotiaceae. This group had 38 strains, 10 among the genera Oidiodendron and Myxotrichum (D1), 17 together with Skoua fertilis (D2), and 11 within Bettsia alvei (D3). Subclade E (Schizosaccharomycetales), grouped FMR 16279 together with the ex-type sequence of Schizosaccharomyces octosporus. Subclade F (Saccharomycetales), had nine strains belonging to Zygosaccharomyces spp. (five strains) and Candida spp. (four strains). Clade G had 5 strains, Mucorales, divided into three groups that comprised Mucor spp. (three strains), Cunninghamella bertholletiae (FMR 16008) and Rhizopus oryzae (FMR 16022), respectively. Figures 3, 4 show the trees resulting from the phylogenetic analyses of Myxotrichaceae and Talaromyces, respectively. The phylogenetic tree based on the analysis of the ITS (Fig. 3), included 67 sequences belonging to Myxotrichaceae and Pseudeurotiaceae, whose alignments encompassed a total of 547 characters, including gaps, 204 of which were parsimony informative. The ML and BI analyses showed a similar tree topology. It comprised a main clade of Myxotrichaceae, where 20 strains were located, 17 of Skoua (14 identified as S. fertilis), and the remaining three in a separate branch that might represent a new species of the genus. Finally, three strains phylogenetically distant from the others appeared in a separate branch close to Myxotrichum setosum and Oidiodendron truncatum. The tree based on four concatenated loci (BenA, CaM, rpb2 and ITS; Table 2; Fig. 4) was built to resolve the phylogenetic relationships of the Talaromyces strains. The dataset contained 123 sequences with a total of 2265 characters, including gaps, (520 of them for ITS, 377 for BenA, 516 for CaM and 852 for rpb2), of which 1069 were parsimony informative (195 for ITS, 217 for BenA, 308 for CaM and 349 for rpb2). The sequence datasets did not show conflict in the tree topologies for the 70% reciprocal bootstrap trees, which allowed the multi-locus analysis. The ML analysis showed similar tree topology and was congruent with the Bayesian analysis. In this tree (Fig. 4), the five Talaromyces strains we obtained were located in two different clades: one corresponding to the section Trachyspermi (100% BS / - PP), with four strains phylogenetically distant from T. atroroseus, one of them (FMR 9720) in a separate branch; and the second corresponding to the section Purpurei (74% BS / - PP), where the fifth strain (FMR 16566) was located in a distant branch.
Fig. 2.
a-c ML phylogenetic tree based on the analysis of LSU nucleotide sequences for all fungi isolated from honey. Members of Mucoromycota were chosen as out-group. Support in nodes is indicated above thick branches and is represented by posterior probabilities (BI analysis) of 0.95 and higher and/or bootstrap values (ML analysis) of 70% and higher. Fully supported branched (100% BS /1 PP) are indicated in bold. T = ex type. Alignment length 606 bp. The sequences generated by us are in Table 1
Fig. 3.
ML phylogenetic tree based on the analysis of ITS nucleotide sequences of representative taxa of the families Myxotrichaceae (in grey balckground) and Pseudeurotiaceae. Aphanoascus keratinophilus IMI 319010 was chosen as out-group. Support in nodes is indicated above thick branches and is represented by posterior probabilities (BI analysis) of 0.95 and higher and/or bootstrap values (ML analysis) of 70% and higher. Fully supported branched (100% BS /1 PP) are indicated in bold. T = ex type. Alignment length 544 bp
Fig. 4.
ML phylogenetic tree built using the ITS, BenA, CaM and rpb2 concatenated dataset for species of the genus Talaromyces. Species of the section Trachyspermi are indicated in a blue background and those of the section Purpurei in yellow. Trichocoma paradoxa CBS 247.57 was chosen as out-group. Support in nodes is indicated above thick branches and is represented by posterior probabilities (BI analysis) of 0.95 and higher and/or bootstrap values (ML analysis) of 70% and higher. Fully supported branched (100% BS /1 PP) are indicated in bold. T = ex-type strain. Alignment length 2265 bp
Table 2.
Talaromyces spp. nucleotide sequences employed to build a phylogram to locate phylogenetically our strains from honey
| Species name | Section | Strain no. | GenBank accession # | |||
|---|---|---|---|---|---|---|
| BenA | CaM | rpb2 | ITS | |||
| Talaromyces aculeatus | Talaromyces | CBS 289.48 = IMI 040588 = NRRL 2129 | KF741929 | KF741975 | KM023271 | KF741995 |
| Talaromyces adpressus | Talaromyces | CBS 140620 = CGMCC3.18211 = DTO 317-G4 | KU866844 | KU866741 | KU867001 | KU866657 |
| Talaromyces alveolaris | Talaromyces | UTHSC DI16–146 | LT559085 | LT795594 | LT795595 | LT558968 |
| Talaromyces amazonensis | Talaromyces | CBS 140373 = IBT 23215 = DTO 093-F9 | KX011490 | KX011502 | – | KX011509 |
| Talaromyces amestolkiae | Talaromyces | CBS 132696 = DTO 179-F5 | JX315623 | KF741937 | JX315698 | JX315660 |
| Talaromyces angelicae | Talaromyces | KACC 46611 | KF183640 | KJ885259 | – | KF183638 |
| Talaromyces apiculatus | Talaromyces | CBS 312.59 = FRR 635 = IMI 068239 | KF741916 | KF741950 | KM023287 | JN899375 |
| Talaromyces aurantiacus | Talaromyces | CBS 314.59 = IMI 099722 = NRRL 3398 | KF741917 | KF741951 | – | JN899380 |
| Talaromyces beijingensis | Talaromyces | CBS 140617 = CGMCC3.18200 = DTO 317-D8 | KU866837 | KU866733 | KU866993 | KU866649 |
| Talaromyces calidicanius | Talaromyces | CBS 112002 | HQ156944 | KF741934 | KM023311 | JN899319 |
| Talaromyces cnidii | Talaromyces | KACC 46617 | KF183641 | KJ885266 | KM023299 | KF183639 |
| Talaromyces derxii | Talaromyces | CBS 412.89 | JX494305 | KF741959 | KM023282 | JN899327 |
| Talaromyces duclauxii | Talaromyces | CBS 322.48 = IMI 040044 = MUCL 28672 = NRRL 1030 | JX091384 | KF741955 | JN121491 | JN899342 |
| Talaromyces euchlorocarpius | Talaromyces | PF 1203 = DTO 176-I3 = DTO 176-I4 | KJ865733 | KJ885271 | KM023303 | AB176617 |
| Talaromyces flavovirens | Talaromyces | CBS 102801 = IBT 27044 | JX091376 | KF741933 | – | JN899392 |
| Talaromyces flavus | Talaromyces | CBS 310.38 = IMI 197477 = NRRL 2098 | JX494302 | KF741949 | JF417426 | JN899360 |
| Talaromyces francoae | Talaromyces | CBS 113134 = IBT 23221 = DTO 056-D9 | KX011489 | KX011501 | – | KX011510 |
| Talaromyces funiculosus | Talaromyces | CBS 272.86 = IMI 193019 | JX091383 | KF741945 | KM023293 | JN899377 |
| Talaromyces fusiformis | Talaromyces | CBS 140637 = CGMCC3.18210 = DTO 317-F4 | KU866843 | KU866740 | KU867000 | KU866656 |
| Talaromyces galapagensis | Talaromyces | CBS 751.74 = IFO 31796 | JX091388 | KF741966 | – | JN899358 |
| Talaromyces indigoticus | Talaromyces | CBS 100534 = IBT 17590 | JX494308 | KF741931 | – | JN899331 |
| Talaromyces intermedius | Talaromyces | CBS 152.65 = BDUN 267 = IFO 31752 = IMI 100874 | JX091387 | KJ885290 | – | JN899332 |
| Talaromyces kabodanensis | Talaromyces | DI16–149 | LT559088 | LT795598 | LT795599 | LT558971 |
| Talaromyces liani | Talaromyces | CBS 225.66 = IMI 098480 = NRRL 3380 = VKM F-301 | JX091380 | KJ885257 | – | JN899395 |
| Talaromyces macrosporus | Talaromyces | CBS 317.63 = FRR 404 = IMI 197478 | JX091382 | KF741952 | KM023292 | JN899333 |
| Talaromyces mangshanicus | Talaromyces | CGMCC 3.18013 | KX447530 | KX447528 | KX447527 | KX447531 |
| Talaromyces marneffei | Talaromyces | CBS 388.87 | JX091389 | KF741958 | KM023283 | JN899344 |
| Talaromyces muroii | Talaromyces | CBS 756.96 = PF 1153 | KJ865727 | KJ885274 | – | JN899351 |
| Talaromyces neofusisporus | Talaromyces | AS3.15415 = CBS 139516 | KP765381 | KP765383 | – | KP765385 |
| Talaromyces oumae-annae | Talaromyces | CBS 138208 = DTO 269-E8 | KJ775213 | KJ775425 | – | KJ775720 |
| Talaromyces panamensis | Talaromyces | CBS 128.89 = IMI 297546 | HQ156948 | KF741936 | KM023284 | JN899362 |
| Talaromyces paucisporus | Talaromyces | PF 1150 = IFM 53616 | – | – | – | AB176603 |
| Talaromyces pinophilus | Talaromyces | CBS 631.66 = CECT 2809 = DSM 1944 = IAM 7013 = IMI 114933 | JX091381 | KF741964 | KM023291 | JN899382 |
| Talaromyces primulinus | Talaromyces | CBS 321.48 = CBS 439.88 = FRR 1074 = IMI 040031 = MUCL 31321 = NRRL 1074 | JX494305 | KF741954 | KM023294 | JN899317 |
| Talaromyces purgamentorum | Talaromyces | CBS 113145 = IBT 23220 = DTO 056-E1 | KX011487 | KX011500 | – | KX011504 |
| Talaromyces purpurogenus | Talaromyces | CBS 286.36 = IMI 091926 | JX315639 | KF741947 | JX315709 | JN899372 |
| Talaromyces qii | Talaromyces | AS3.15414 = CBS 139515 | KP765380 | KP765382 | – | KP765384 |
| Talaromyces rapidus | Talaromyces | UTHSC DI16–148 = CBS 142382 T | LT559087 | LT795600 | LT795601 | LT558970 |
| Talaromyces ruber | Talaromyces | CBS 132704 = DTO 193-H6 = IBT 10703 = CBS 113137 | JX315629 | KF741938 | JX315700 | JX315662 |
| Talaromyces rubicundus | Talaromyces | CBS 342.59 = IMI 099723 = NRRL 3400 | JX494309 | KF741956 | KM023296 | JN899384 |
| Talaromyces sayulitensis | Talaromyces | CBS 138204 = DTO 245-H1 | KJ775206 | KJ775422 | – | KJ775713 |
| Talaromyces siamensis | Talaromyces | CBS 475.88 = IMI 323204 | JX091379 | KF741960 | KM023279 | JN899385 |
| Talaromyces stipitatus | Talaromyces | CBS 375.48 = NRRL 1006 = IMI 39805 | KM111288 | KF741957 | KM022380 | JN899348 |
| Talaromyces stollii | Talaromyces | CBS 408.93 | – | JX315646 | JX315712 | JX315674 |
| Talaromyces thailandensis | Talaromyces | CBS 133147 = KUFC 3399 | JX494294 | KF741940 | KM023307 | JX898041 |
| Talaromyces verruculosus | Talaromyces | CBS 388.48 = DSM 2263 = IMI 040039 = NRRL 1050 | KF741928 | KF741944 | KM023306 | KF741994 |
| Talaromyces viridis | Talaromyces | CBS 114.72 = ATCC 22467 = NRRL 5575 | JX494310 | KF741935 | JN121430 | AF285782 |
| Talaromyces viridulus | Talaromyces | CBS 252.87 = FRR 1863 = IMI 288716 | JX091385 | KF741943 | JF417422 | JN899314 |
| Talaromyces aerugineus | Helici | CBS 350.66 = BDUN 276 = IMI 105412 | KJ865736 | KJ885285 | JN121502 | AY753346 |
| Talaromyces bohemicus | Helici | CBS 545.86 = CCF 2330 = IAM 14789 | KJ865719 | KJ885286 | JN121532 | JN899400 |
| Talaromyces boninensis | Helici | CBS 650.95 = IBT 17516 | KJ865721 | KJ885263 | KM023276 | JN899356 |
| Talaromyces cinnabarinus | Helici | CBS 267.72 = NHL 2673 | AY753377 | KJ885256 | JN121477 | JN899376 |
| Talaromyces diversiformis | Helici | CBS 141931 = CGMCC3.18204 = DTO 317-E3 | KX961216 | KX961259 | KX961274 | KX961215 |
| Talaromyces georgiensis | Helici | UTHSC DI16–145 = CBS 142380 | LT559084 | – | LT795606 | LT558967 |
| Talaromyces helicus | Helici | CBS 335.48 = DSM 3705 = IMI 040593 = NRRL 2106 | KJ865725 | KJ885289 | KM023273 | JN899359 |
| Talaromyces reverso-olivaceus | Helici | CBS 140672 = CGMCC3.18195 = DTO 317-C3 | KU866834 | KU866730 | KU866990 | KU866646 |
| Talaromyces ryukyuensis | Helici | NHL 2917 = DTO 176-I6 | – | – | – | AB176628 |
| Talaromyces varians | Helici | CBS 386.48 = IMI 040586 = NRRL 2096 | KJ865731 | KJ885284 | KM023274 | JN899368 |
| Talaromyces cecidicola | Purpurei | CBS 101419 = DAOM 233329 | FJ753295 | KJ885287 | KM023309 | AY787844 |
| Talaromyces chlorolomus | Purpurei | DAOM 241016 = CV 2802 | GU385736 | KJ885265 | KM023304 | FJ160273 |
| Talaromyces coalescens | Purpurei | CBS 103.83 | JX091390 | KJ885267 | KM023277 | JN899366 |
| Talaromyces dendriticus | Purpurei | CBS 660.80 = IMI 216897 | JX091391 | KF741965 | KM023286 | JN899339 |
| Talaromyces pittii | Purpurei | CBS 139.84 = IMI 327871 | KJ865728 | KJ885275 | KM023297 | JN899325 |
| Talaromyces pseudostromaticus | Purpurei | CBS 470.70 = FRR 2039 | HQ156950 | KJ885277 | KM023298 | JN899371 |
| Talaromyces ptychoconidium | Purpurei | DAOM 241017 = CV 2808 = DTO 180-E7 | GU385733 | JX140701 | KM023278 | FJ160266 |
| Talaromyces purpureus | Purpurei | CBS 475.71 = FRR 1731 = IMI 181546 | GU385739 | KJ885292 | JN121522 | JN899328 |
| Talaromyces rademirici | Purpurei | CBS 140.84 = CECT 2771 = IMI 282406 | KJ865734 | – | KM023302 | JN899386 |
| Talaromyces ramulosus | Purpurei | DAOM 241660 = CV 2837 = DTO 184-B8 | FJ753290 | JX140711 | KM023281 | EU795706 |
| Talaromyces aerius | Trachyspermi | CBS 140611 = CGMCC3.18197 = DTO 317-C7 | KU866835 | KU866731 | KU866991 | KU866647 |
| Talaromyces albobiverticillius | Trachyspermi | CBS 133440 T = DTO 166-E5 = YMJ 1292 | KF114778 | KJ885258 | KM023310 | HQ605705 |
| Talaromyces assiutesis | Trachyspermi | CBS 147.78 T | KJ865720 | KJ885260 | KM023305 | N899323 |
| Talaromyces atroroseus | Trachyspermi | CBS 133442 T = IBT 32470 = DTO 178-A4 | KF114789 | KJ775418 | KM023288 | KF114747 |
| Talaromyces austrocalifornicus | Trachyspermi | CBS 644.95 T = IBT 17522 | KJ865732 | KJ885261 | – | JN899357 |
| Talaromyces convolutus | Trachyspermi | CBS 100537 T = IBT 14989 | KF114773 | – | JN121414 | JN899330 |
| Talaromyces diversus | Trachyspermi | CBS 320.48 T = DSM 2212 = IMI 040579 = NRRL 2121 | KJ865723 | KJ885268 | KM023285 | KJ865740 |
| Talaromyces erythromellis | Trachyspermi | CBS 644.80 T = FRR 1868 = IMI 216899 | HQ156945 | KJ885270 | KM023290 | JN899383 |
| Talaromyces heiheensis | Trachyspermi | CGMCC 3.18012 | KX447525 | KX447532 | KX447529 | KX447526 |
| Talaromyces minioluteus | Trachyspermi | CBS 137.84 | KF114798 | – | – | NR138301 |
| Talaromyces minioluteus | Trachyspermi | CBS 642.68 = IMI 089377 = MUCL 28666 | KF114799 | KJ885273 | JF417443 | JN899346 |
| Talaromyces minnesotensis | Trachyspermi | FMR 14265 T = CBS 142381 | LT559083 | LT795604 | LT795605 | LT558966 |
| Talaromyces mirabile | Trachyspermi | CBS 624.72 | KF114797 | – | – | NR138300 |
| Talaromyces solicola | Trachyspermi | DAOM 241015 T = CV 2800 = DTO 180-D4 | GU385731 | KJ885279 | KM023295 | FJ160264 |
| Talaromyces trachyspermus | Trachyspermi | CBS 373.48 T = IMI 040043 | KF114803 | KJ885281 | JF417432 | JN899354 |
| Talaromyces ucrainicus | Trachyspermi | CBS 162.67 T = FRR 3462 = NHL 6086 | KF114771 | KJ885282 | KM023289 | JN899394 |
| Talaromyces udagawae | Trachyspermi | CBS 579.72 T = FRR 1727 = IMI 197482 | KF114796 | – | – | JN899350 |
| Talaromyces bacillisporus | Bacillispori | CBS 296.48 = IMI 040045 = NRRL 1025 | AY753368 | KJ885262 | JF417425 | KM066182 |
| Talaromyces columbiensis | Bacillispori | CBS 113151 = IBT 23206 = DTO 058-F3 | KX011488 | KX011499 | – | KX011503 |
| Talaromyces emodensis | Bacillispori | CBS 100536 = IBT 14990 | KJ865724 | KJ885269 | JN121552 | JN899337 |
| Talaromyces hachijoensis | Bacillispori | PF 1174 = IFM 53624 | – | – | – | AB176620 |
| Talaromyces mimosinus | Bacillispori | CBS 659.80 = FRR 1875 = IMI 223991 | KJ865726 | KJ885272 | – | JN899338 |
| Talaromyces proteolyticus | Bacillispori | CBS303.67 = NRRL 3378 | KJ865729 | KJ885276 | KM023301 | JN899387 |
| Talaromyces unicus | Bacillispori | CBS 100535 = CCRC 32703 = IBT 18385 | KJ865735 | KJ885283 | – | JN899336 |
| Talaromyces palmae | Subinflati | CBS 442.88 = IMI 343640 | HQ156947 | KJ885291 | KM023300 | JN899396 |
| Talaromyces subinflatus | Subinflati | CBS 652.95 = IBT 17520 | KJ865737 | KJ885280 | KM023308 | JN899397 |
| Talaromyces acaricola | Islandici | CBS 137386 = DTO 183-B3 = DAOM 241025 = IBT 32387 | JX091610 | JX140729 | KF984956 | JX091476 |
| Talaromyces allahabadensis | Islandici | CBS 304.63 | KF984614 | KF984768 | KF985006 | KF984873 |
| Talaromyces atricola | Islandici | CBS 255.31 = NRRL 1052 = FRR 1052 = Thom 4640.439 | KF984566 | KF984719 | KF984948 | KF984859 |
| Talaromyces brunneus | Islandici | CBS 227.60 = FRR 646 = IFO 6438 = IHEM 3907 = IMI 078259 = MUCL 31318 | KJ865722 | KJ885264 | KM023272 | JN899365 |
| Talaromyces cerinus | Islandici | CBS 140622 = CGMCC3.18212 = DTO 318-A2 | KU866845 | KU866742 | KU867002 | KU866658 |
| Talaromyces chlamydosporus | Islandici | CBS 140635 = CGMCC3.18199 = DTO 317-D5 | KU866836 | KU866732 | KU866992 | KU866648 |
| Talaromyces columbinus | Islandici | NRRL 58811 | KF196843 | KJ885288 | KM023270 | KJ865739 |
| Talaromyces crassus | Islandici | CBS 137381 = DTO 181-C5 = DAOM 241027 = IBT 32814 | JX091608 | JX140727 | KF984914 | JX091472 |
| Talaromyces infraolivaceus | Islandici | CBS 137385 = DTO 182-I2 = DAOM 241024 = IBT 32487 | JX091615 | JX140734 | KF984949 | JX091481 |
| Talaromyces islandicus | Islandici | CBS 338.48 = IMI 040042 = MUCL 31324 = NRRL 1036 | KF984655 | KF984780 | KF985018 | KF984885 |
| Talaromyces loliensis | Islandici | CBS 643.80 = FRR 1798 = IMI 216901 = MUCL 31325 | KF984658 | KF984783 | KF985021 | KF984888 |
| Talaromyces neorugulosus | Islandici | CBS 140623 = CGMCC3.18215 = DTO 318-A8 | KU866846 | KU866743 | KU867003 | KU866659 |
| Talaromyces piceus | Islandici | CBS 361.48 = IMI 040038 = NRRL 1051 | KF984668 | KF984680 | KF984899 | KF984792 |
| Talaromyces radicus | Islandici | CBS 100489 = FRR 4718 | KF984599 | KF984773 | KF985013 | KF984878 |
| Talaromyces rotundus | Islandici | CBS 369.48 = IMI 040589 = NRRL 2107 | KJ865730 | KJ885278 | KM023275 | JN899353 |
| Talaromyces rugulosus | Islandici | CBS 371.48 = IMI 040041 = MUCL 31201 = NRRL 1045 | KF984575 | KF984702 | KF984925 | KF984834 |
| Talaromyces scorteus | Islandici | CBS 340.34 = NRRL 1129 = FRR 1129 | KF984565 | KF984684 | KF984916 | KF984892 |
| Talaromyces subaurantiacus | Islandici | CBS 137383 = DTO 181-I2 = DAOM 241020 = IBT 32838 | JX091609 | JX140728 | KF984960 | LT558965 |
| Talaromyces tardifaciens | Islandici | CBS 250.94 | KC202954 | KF984682 | KF984908 | JN899361 |
| Talaromyces tratensis | Islandici | CBS 133146 = KUFC 3383 | KF984559 | KF984690 | KF984911 | KF984891 |
| Talaromyces wortmannii | Islandici | CBS 391.48 = IMI 040047 = NRRL 1017 | KF984648 | KF984756 | KF984977 | KF984829 |
| Talaromyces yelensis | Islandici | DTO 268E5 | KJ775210 | – | – | KJ775717 |
| Trichocoma paradoxa | – | CBS 247.57 | JF417468 | JF417505 | JF417421 | JF417485 |
TAXONOMY
Subclade A: Onygenales
Based on the above phylogenetic analyses, we suggest the following novel taxonomic arrangements: Helicoarthrosporaceae fam. nov. (Fig. 2; sister clade A5), phylogenetically close to the family Gymnoascaceae, with Helicoarthrosporum gen. nov. as type genus and H. mellicola sp. nov. as the type species; based on the strain FMR 16121, we introduce Strongyloarthrosporum gen. nov. with S. catenulatum sp. nov. as its type species. These new taxa are described and illustrated below.
Helicoarthrosporaceae Stchigel, Rodr.-Andr. & Cano, fam. nov. MycoBank MB 832226.
Diagnosis: Differing from other families of Onygenales by the production of long, sinuous to helical chains of arthroconidia (which are shorter, right, curved or contorted in other taxa).
Type genus: Helicoarthrosporum Stchigel et al. 2019.
Description: Hyphae hyaline, septate. Asexual morph reduced to sinuous, helical or zig-zag lateral branches, terminal part becoming fertile, disarticulating into conidia. Conidia hyaline, prismatic to cuboid, holo- and enteroarthric conidia. Sexual morph not observed.
Helicoarthrosporum Stchigel, Cano & Rodr.-Andr., gen. nov. MycoBank MB 823584.
Etymology. From Greek έλικα-, helix, −άρθρωση-, joint, and -σπορά, spore, referring to the morphology of the conidiophores.
Diagnosis: Distinguished from other phylogenetically related genera by its long, sinuous to helical chains of prismatic to cuboid arthroconidia, and by its extreme xerotolerance.
Type species: Helicoarthrosporum mellicola Stchigel et al. 2019.
Description: Mycelium composed by hyaline, septate hyphae. Conidiophores consisting in fertile lateral branches and terminal part of the hyphae, sinuous, helical or zig-zag, disarticulating in hyaline, mostly prismatic to cuboid, holo- and enteroarthric conidia.
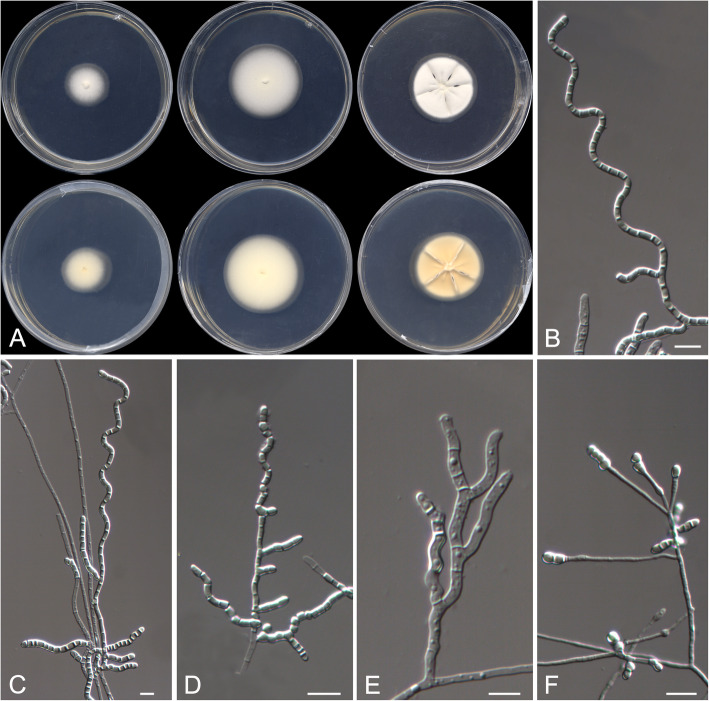
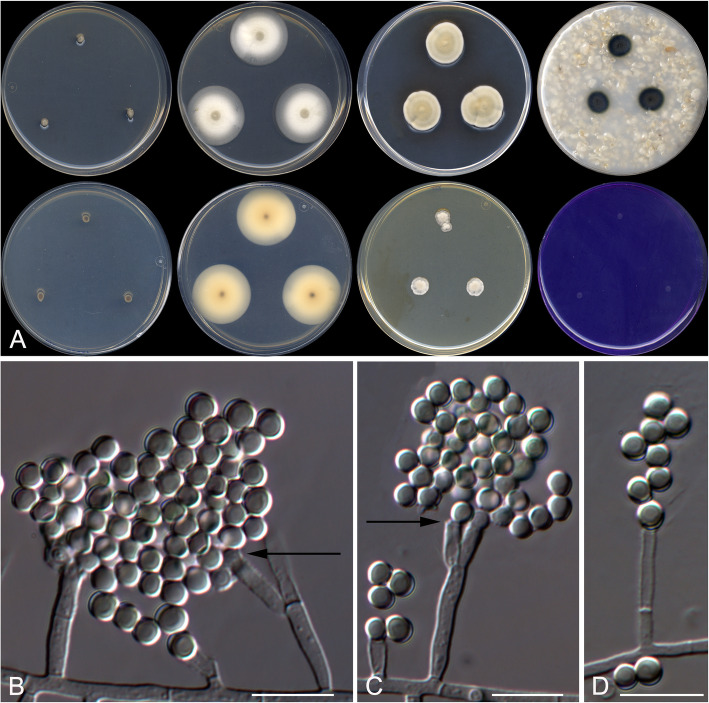
Helicoarthrosporum mellicola Stchigel, Cano & Rodr.-Andr., sp. nov. Fig. 5. MycoBank MB 823585.
Fig. 5.
Helicoarthrosporum mellicola CBS 143838 T. a Colonies on G18 at 15 °C and at 25 °C, and on PDA at 25 °C (from left to right), surface and reverse (from top to bottom). b–e Conidiophores. d Chlamydospores on OA. Scale bar = 10 μm
Etymology: From Latin mellis-, honey, and -cola, to reside, referring to the habitat of the fungus.
Diagnosis: Helicoarthrosporum mellicola morphologically resembles Scytalidium cuboideum (syn. Arthrographis cuboidea), S. ganodermophthorum, and S. sphaerosporum in producing long chains of cuboid arthroconidia (Kang et al. 2010). Helicoarthrosporum mellicola grows slowly on PDA and shows a high xerotolerance, whereas Scytalidium spp. grow fast on PDA and do not show a xerotrophic habit; also, S. ganodermophthorum and S. sphaerosporum produce both asexual and sexual morphs, while H. mellicola only displays an asexual one.
Type: Spain: Valencia community: Castellón province, from decanted and filtered honey, 10 May 2014, A. Gómez Pajuelo (CBS H-23368 – holotype; CBS 143838 = FMR 15679 – ex-type cultures; LSU sequence GenBank LT906535).
Description: Colonies on G18 reaching 38–41 mm diam after 3 wk. at 25 °C, flattened, velvety, yellowish white (4A2) at the centre, margins regular, sporulation sparse; exudate absent; reverse pale yellow (4A3), diffusible pigment absent. Mycelium composed of hyaline to subhyaline, septate, smooth- and thin-walled hyphae, 1.5–4 μm wide; racquet hyphae present. Conidiophores reduced (mostly) to fertile side branches and to the terminal part of a vegetative hyphae, sinuous to helical or in zig-zag, mostly simple, sometimes branched, 15–180 μm long, hyaline, disarticulating in conidia. Conidia mostly 1-celled, sometimes up to 4-celled, mostly holoarthric, occasionally enteroarthric, in chains of up to 30, mostly barrel-shaped, prismatic or cuboid, sometimes triangular and “Y”-shaped, smooth-walled, thicker than the hyphae, thickener at the ends, 2–8 × 2–5 μm, hyaline, disarticulating by schizolysis or rhexolysis from the conidiogenous hyphae. Chlamydospores produced on OA, terminally on or intercalary in the fertile hyphae hyaline, one to multicellular, smooth- and thick-walled, globose, ovoid, pyriform, clavate or irregularly-shaped, truncate at the base or at both ends, to 10 μm long and 3–5 μm wide.
Colonies on G18 reaching 22–27 mm diam after 3 wk. at 15 °C, flat, velvety, yellowish-white (4A2), margins regular, sporulation sparse, exudate absent; reverse pale yellow (4A3), diffusible pigment absent; no growth on G18 over 35 °C; on PDA reaching 31–35 mm diam after 3 wk. at 25 °C, slightly elevated, velvety, slightly sulcate, yellowish (3A2) at the centre and white (3A1) at the edge, exudate absent; reverse reddish yellow (4A6) at the centre and pale orange (5A3) at the edge, diffusible pigment absent; on OA at 25 °C after 3 wk. very small, 7–8 mm diam, velvety, white (4A1), sporulation sparse, exudate absent; reverse pale orange (5A3), diffusible pigment absent.
Minimum, optimal and maximum temperature of growth on G18 are 15 °C, 25 °C, and 30 °C, respectively; no hemolysis observed on blood agar at 25 °C, and on BCP-MS-G casein hydrolyzed without pH changes. Lipase negative, urease positive. Inhibited by cycloheximide and 20% NaCl, but tolerant to 3% and to 10% NaCl on Sabouraud dextrose agar.
Other specimens examined: Spain: Valencia community: Castellón province, from decanted and filtered honey, 10 May 2014, A. Gómez Pajuelo (FMR 15673). Castilla y León community: León province, from decanted, filtered and thermally treated honey, 20 May 2014, A. Terrab (FMR 16307). Castilla y León community: Zamora province, from decanted and filtered honey, 5 Oct. 2014, A. Gómez Pajuelo (FMR 16308). Extremadura community: Cáceres province, from decanted, filtered and thermally treated honey, 16 May 2014, A. Terrab (FMR 16315).
Strongyloarthrosporum Rodr.-Andr., Cano & Stchigel, gen. nov. MycoBank MB 823587.
Etymology: From Greek στρογγυλός-, globose, −άρθρωση-, joint, and -σπορά, spore, referring to the morphology of the conidia.
Diagnosis: Distinguished from other genera of Onygenales by the production of thick-walled globose arthroconidia, and because this fungus is an obligate xerophile.
Type species: Strongyloarthrosporum catenulatum Rodr.-Andr. et al. 2019.
Description: Mycelium of hyaline, septate hyphae. Conidiophores fertile lateral branches and part of the vegetative hyphae, disarticulating. Conidia enteroarthic, hyaline, mostly globose.
Strongyloarthrosporum catenulatum Rodr.-Andr., Cano & Stchigel, sp. nov. Fig. 6. MycoBank MB 823588.
Fig. 6.
Strongyloarthrosporum catenulatum CBS 143841 T. a Colonies on G18, G25 N and MY70FG at 25 °C (from left to right), surface and reverse (from top to bottom). b–e Conidiophores and conidia. Scale bar = 10 μm
Etymology: From Latin catenulatus, in chains, referring to the disposition of the conidia.
Diagnosis: Strongyloarthrosporum catenulatum is phylogenetically close to the Ajellomycetaceae, a family of non-xerophilic fungi characterized by their thermally dimorphic nature and, consequently, pathogenic for animals. By contrast, S. catenulatum is an obligate xerophilic fungus with globose conidia sometimes disposed in chains.
Type: Spain: Castilla-La Mancha community: Toledo province, from decanted, filtered and thermally treated honey, 12 May 2014, A. Terrab (CBS H- 23371 – holotype; CBS 143841 = FMR 16121 – ex-type cultures; LSU sequence GenBank LT906534).
Description: Colonies on G18 reaching 20–21 mm diam after 3 wk. at 25 °C, elevated, velvety, sulcate, sporulation sparse, exudate absent, yellowish white (4A2) at the centre and white (3A1) at the edge; reverse orange-grey (5B2), diffusible pigment absent. Mycelium composed of hyaline, septate, smooth, thin- to thick-walled, anastomosing hyphae, 1.5–4 μm wide. Conidiophores reduced mostly to single fertile side branches and to the terminal part of the vegetative hyphae, 5–60 μm long, hyaline, disarticulating in conidia. Conidia hyaline, mostly one-celled, occasionally two-celled, holo- and enteroarthric, solitary, disposed terminally, intercalary or sessile on the fertile hyphae, or produced in basipetal chains of up to ten conidia, smooth-walled, thicker than the hyphae, thickener at the ends, mostly globose, 3–6 μm diam, flattened or not at one or both ends, disarticulating by rhexolytic secession from the conidiogenous hyphae. Chlamydospores and racquet hyphae absent.
Colonies on G25 N reaching 19–20 mm diam after 3 wk. at 25 °C, elevated, velvety, sulcate, exudate absent, sporulation sparse, light orange (5A4) at the centre and grey (5B1) at the edge; reverse greyish orange (5B5), diffusible pigment absent; on MY70FG reaching 29–30 mm diam after 3 wk. at 25 °C, flat, floccose, margins entire, sporulation sparse, white; reverse light yellow (4A4), diffusible pigments absent.
Minimum, optimal and maximum temperature of growth on G18 are 15 °C, 25 °C, and 35 °C, respectively, does not grow on blood agar, BCP-MS-G, Sabouraud dextrose agar with different NaCl concentrations, TOTM, OA, PYE nor on Christensen’s urea agar.
Subclade B: Eurotiales
Due to both LSU-based (Fig. 2; sister clade B1) and ITS-BenA-CaM-rpb2-based (Fig. 4) phylogenetetic trees, four of our Talaromyces strains were placed in section Trachyspermi in a well-supported subclade divided in two branches, and one more strain was placed into the section Purpurei in a basal position (Fig. 4), phylogenetically distant and phenotypically different from other species of Talaromyces in this section, consequenly, we propose the recognition of three new species of the genus.
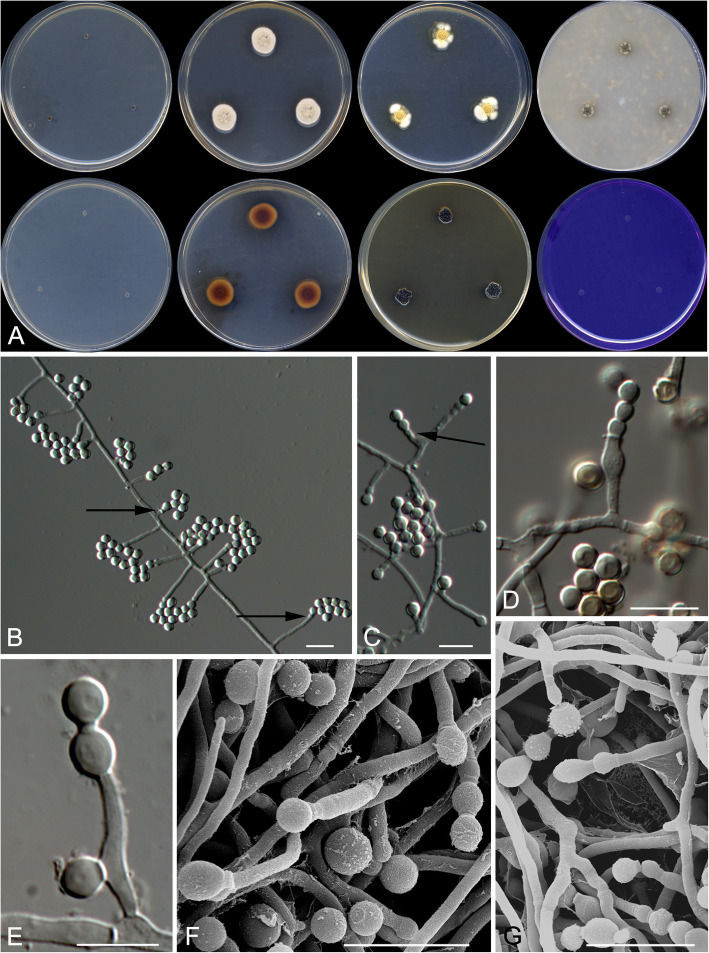
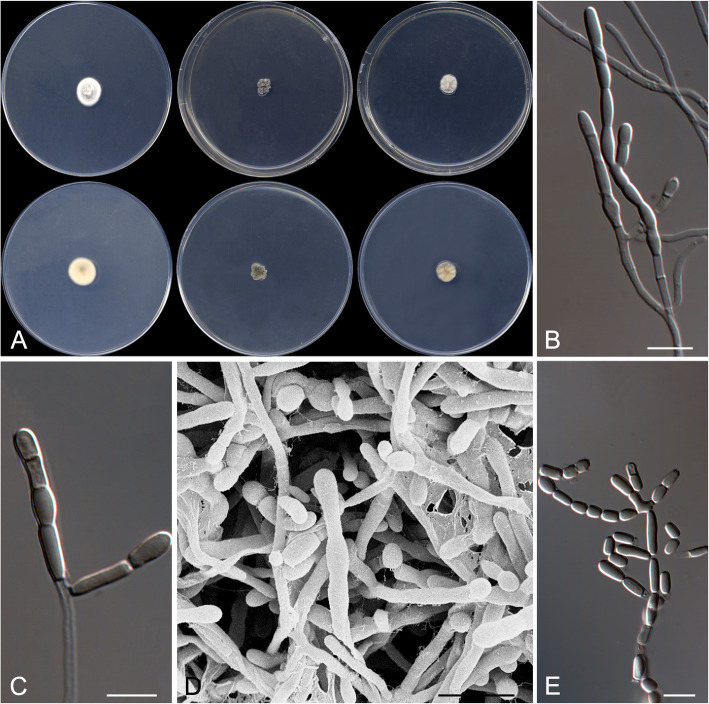
Talaromyces basipetosporus Stchigel, Cano & Rodr.-Andr., sp. nov. Fig. 7. MycoBank MB 823589.
Fig. 7.
Talaromyces basipetosporus CBS 143836 T. a Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES and CREA. b–g Conidiophores and conidia; the arrows shows the conidia formed basipetally. Scale bar = 10 μm
Etymology: After the morphological similarity to the asexual morph of Basipetospora (formerly applied to the asexual morph of Monascus).
Diagnosis: Differs from other species in sect. Trachyspermi in that the conidiogenesis is very similar to that of Monascus (syn. Basipetospora), characterized by retrogressively produced conidia, which have not been previously described in Talaromyces (see diagnosis of Talaromyces affinitatimellis).
Type: Argentina: Buenos Aires province: San Martín, from decanted, filtered and thermally treated honey, 1 Oct. 2007, M. A. Álvarez (CBS H-23365 – holotype; CBS 143836 = FMR 9720 – ex-type cultures; LSU sequence GenBank LT964940).
Description: Colonies on MEA reaching 10–11 mm diam after 3 wk. at 25 °C, slightly elevated, velvety to floccose, margins entire, yellowish grey (4B2) at the centre and white (4A1) at the edge, exudate absent, sporulation sparse; reverse brownish red (8C8) at the centre and greyish orange (5B6) at the edge, diffusible pigments absent. Mycelium abundant, composed of subhyaline to pale brown, smooth to echinulate, thin-walled, septate, anastomosing hyphae, of 2–3 μm wide. Conidiophores mostly reduced to a single conidiogenous cell, sometimes slender and with an additional conidiogenous locus near the base, arising alternately or oppositely at both sides of the vegetative hyphae, mostly separate from the vegetative hyphae by a basal septum. Conidiogenous cells smooth-walled to echinulate, mostly cylindrical and occasionally slightly slender towards the apex, sometimes broadening below the apex, but also flask- or barrel-shaped, very variable in length, 3–20(− 45) × 1–2.5 μm, conidiogenesis retrogressive. Conidia one-celled, hyaline and echinulate when young, becoming brown to dark brown and nearly smooth-walled with the age, formed basipetally, in false chains of up to ten conidia, mostly globose, 3.0–5.0 μm diam. Sexual morph not observed.
Colonies on DG18 reaching 13–14 mm diam after 3 wk. at 25 °C, colonies moderately elevated, texture floccose, yellowish orange (4B7) with mycelium white (5A1) at edge, sporulation dense, exudate absent, diffusible pigments absent, reverse reddish golden (6C7) at centre and pale yellow (3A4) at edge; on G18 reaching 10–11 mm diam after 3 wk. at 25 °C, slightly elevated, velvety to floccose, margins regular, yellowish white (3A2), exudates uncolored, diffusible pigment absent, reverse pale orange (5A3) at the centre and white at the edge; on OA reaching 5–6 mm diam. After 3 wk. at 25 °C, flat, margins entire, mycelium grey, texture velvety to floccose, sporulation dense, diffusible pigments absent, exudate absent, colonies dark brown (5D4) at centre and grey with olive-brown (6B1-4E6) patches at edge; on PDA reaching 10–11 mm diam. After 3 wk. at 25 °C, elevated, velvety, brown (7E7) at the centre and brownish grey (4D2) at the edge, sporulation abundant, exudate absent, diffusible blackish olive (2G6) pigment present, reverse dark brown (7F4) at centre and brown (7E8) at the edge; on YES reaching 7–8 mm diam after 3 wk. at 25 °C, moderately elevated, sulcate, rough, sporulation strong, blackish brown (6G8), diffusible pigments absent, exudates absent, reverse yellowish brown (5E8).
Minimum, optimal and maximum temperature of growth on G18 are 15, 25, and 30 °C, respectively; does not grow on CYA, Czapek 20% sucrose, CREA, Starch agar, or MY70FG.
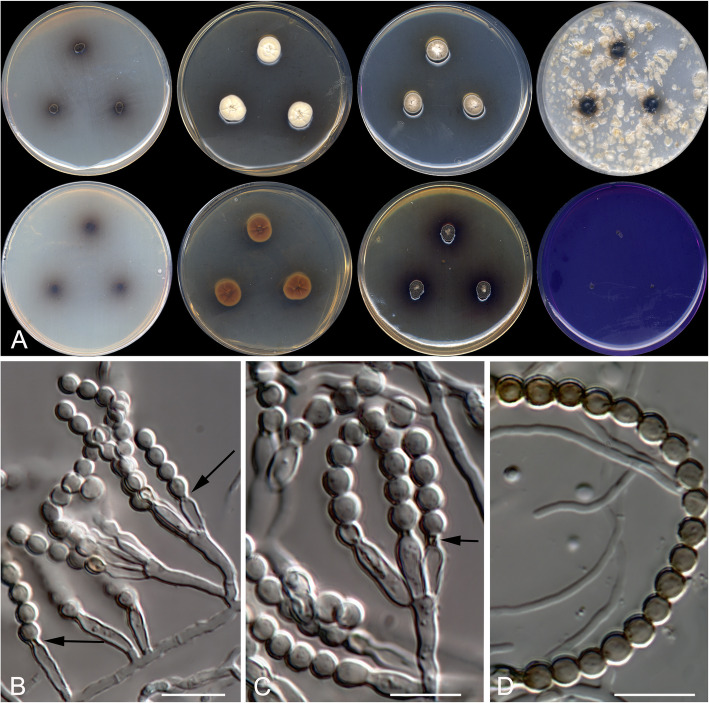
Talaromyces brunneosporus Rodr.-Andr., Cano & Stchigel, sp. nov.
Figure 8. MycoBank MB 823590.
Fig. 8.
Talaromyces brunneosporus CBS 144320T. a Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES and CREA. b, c Poorly-developed (single phialide) and well-developed (monoverticillate) conidiophores; the arrows indicate the conspicuous collarette at the top of the phialides. d A chain of globose, dark brown, verrucose conidia. Scale bar = 10 μm
Etymology: From Latin brunneus-, brown, and -sporarum, spore, in reference to the colour of the conidia.
Diagnosis: Distinguished from other species in sect. Purpurei, with the exception of T. purpurei (the type species of the section), by the production of solitary phialides and monoverticillate conidiophores (biverticillate conidiophores in the other species of the section). However, T. brunneosporus can be differentiated from T. purpureus because lack of a sexual morph (present in the latter species), and produces penicillate conidiophores (having an aspergillate look in T. purpureus) and verrucose conidia (ornamented with spiral ridges in T. purpureus).
Type: Spain: Castilla y León community: Salamanca province, from decanted, filtered and thermally treated honey, 1 Oct. 2014, A. Terrab (CBS H-23375 – holotype; CBS 144320 = FMR 16566 – ex-type cultures; LSU sequence GenBank LT964943).
Description: Colonies on MEA reaching 13–14 mm diam after 3 wk. at 25 °C, slightly elevated, velvety to floccose, margins irregular, yellowish white (4A3), exudate absent, sporulation sparse, reverse light brown (6D8) at the centre and yellowish brown (5D6) at the edge, diffusible yellowish brown (5E6) pigment present. Mycelium abundant, composed of subhyaline, smooth- and thin-walled, septate, anastomosing hyphae 2–3 μm wide. Conidiophores mostly stalked, monoverticillate, smooth- and thin-walled, bearing one to four conidiogenous cells at the top, frequently arising oppositely at both sides of the vegetative hyphae, sometimes reduced to a single conidiogenous cell, sessile or integrated to the vegetative hyphae (= adelophialides). Conidiogenous cells phialidic, smooth-walled, mostly slender towards the apex, flask-shaped, 8–12 × 2.5–3.5 μm, with a darkened apical area when the conidiogenous cells have produced several conidia, conidiogenesis enteroblastic. Conidia one-celled, globose, hyaline and smooth-walled when young, becoming brownish-green to dark brown and verrucose with the age, 3–4 μm diam, in long false chains of up to 25 conidia. Sexual morph not observed.
Colonies on CYA reaching 4–5 mm diam after 3 wk. at 25 °C, elevated, velvety, dark brown (8F4) at the centre and greyish-brown (7E3) at the edge, exudate absent, sporulation abundant, reverse dark brown (8F6) at the centre and reddish brown (8E5) at the edge, diffusible brown (6E7) pigment present; on DG18 reaching 10–11 mm diam after 3 wk. at 25 °C, moderately elevated, floccose, margins irregular, yellowish white (4A2) at the centre and olive-brown (4D6) at the edge, exudate absent, sporulation strong, reverse light brown (5D7), diffusible yellowish brown (5D5) soluble pigment present; on OA reaching 9–10 mm diam after 3 wk. at 25 °C, flat, floccose, margins entire, exudate absent, sporulation strong, colonies blackish olive (2G6) at the centre and brown (6E6) at the edge, diffusible olive brown (4E8) pigment present; on YES reaching 8–9 mm diam after 3 wk. at 25 °C, flat, floccose, black at the centre and yellowish-brown (5E6) at the edge, exudate absent, sporulation sparse, reverse dark violet (8E8), diffusible blackish brown (6G8) pigment present.
Minimum, optimal and maximum temperature of growth on G18 are 15, 25, and 30 °C, respectively; no growth on CYA at 37 °C nor on CREA at 25 °C.
Notes: Talaromyces brunneosporus and T. purpureus grow more slowly on CYA and MEA than other species of the section. However, T. brunneosporus produces dark brown colonies with a dark brown diffusible pigment on CYA, while the colonies of T. purpureus are pale beige and without diffusible pigments. Also, the colonies on OA and MEA are purplish in T. purpureus and pale coloured and dark brown in T. brunneosporus.
Talaromyces affinitatimellis Rodr.-Andr., Stchigel & Cano, sp. nov. Fig. 9. MycoBank MB 823591.
Fig. 9.
Talaromyces affinitatimellis CBS 143840 T. a Colonies from left to right (top row) CYA, MEA, DG18 and OA; (bottom row) CYA reverse, MEA reverse, YES and CREA. b–d Conidiophores and conidia; the arrows shows the conidia formed basipetally. Scale bar = 10 μm
Etymology: From Latin affinitatis-, affinity, and -mellis, honey, after the substrate from which the fungus was isolated.
Diagnosis: Differing from all other species in sect. Trachyspermi (with the exception of T. basipetosporus) by the production of conidia by retrogressive conidiogenesis. Talaromyces affinitatimellis differs from T. basipetosporus by the production cylindrical, smooth-walled to echinulate conidiogenous cells ending in a greenish brown, broad collarette-like structure (conidiogenous cells irregularly-shaped, smooth-walled, and without such apical structure in T. basipetosporus).
Type: Spain: Valencia community: Castellón province, from decanted and filtered blossom honey, 10 May 2014, A. Gómez Pajuelo (CBS H- 23370 – holotype; CBS 143840 = FMR 15690 – ex-type cultures; LSU sequence GenBank LT964939).
Description: Colonies on MEA reaching 29–30 mm diam. After 3 wk. at 25 °C, flat, floccose, not sulcate, margins entire, olive (3D3) at the centre and white (4A1) at edge, exudate absent, sporulation sparse; reverse pale orange (5A3) at centre and pale yellow (4A3) at edge, diffusible pigment absent. Mycelium abundant, composed of subhyaline to pale brown, smooth- and thin-walled, septate, anastomosing hyphae, of 2–4 μm wide. Conidiophores hyaline to pale brown, reduced to a single conidiogenous cell, occasionally with an additional conidiogenous locus near the base or lateraly disposed, or short-stalked and bearing two conidiogenous cells, sometimes with an additional lateral conidiogenous cell arising alternately at both sides of the vegetative hyphae, separate from them by a basal septum. Conidiogenous cells hyaline to pale brown, smooth-walled, mostly cylindrical and occasionally slightly slender towards the apex, sometimes ending in a greenish-brown, broad collarette-like structure, 3–20 × 1.5–3 μm, conidiogenesis retrogressive but enteroblastic. Conidia one-celled, hyaline and echinulate, becoming brown to dark brown and nearly smooth-walled with the age, produced basipetally in false chains of up to ten in number, mostly globose, 3.0–5.0 μm diam. Sexual morph not observed.
Colonies on DG18 reaching 13–14 mm diam after 3 wk. at 25 °C, moderately elevated, floccose, yellowish orange (4B7) with white (5A1) margins, exudates absent, sporulation strong; reverse reddish golden (6C7) at the centre and pale yellow (A4) at the edge, diffusible pigment absent; on G18 reaching 21–24 mm diam at 25 °C, slightly elevated, velvety to floccose, margins regular, yellowish white (4A4), exudates absent, sporulation abundant, reverse greyish orange (5B6), diffusible pigment absent; on OA reaching 12–13 mm diam after 3 wk. at 25 °C, flat, velvety to floccose, margins entire, black, exudates absent, sporulation abundant; colonies grey (7F1) at the centre and dark brown (6F4) to black at the edge, diffusible pigment absent; on PDA reaching 39–43 mm diam after 3 wk. at 25 °C, flat, velvety, margins slightly irregular, yellowish-brown (5F6) at the centre, grey (7F1) and yellowish brown (5E4) at the middle part, and light grey (5B1) at the edge, exudate absent, sporulation scarce, reverse dark brown (7F7) at the centre and brownish yellow (5C7) at the edge, diffusible pigment absent; on YES reaching 10–11 mm diam after 3 wk. at 25 °C, moderately elevated, floccose, white (4A1), exudate absent, sporulation sparse, reverse greyish orange (5B6), diffusible pigment absent.
Minimum, optimal and maximum temperature of growth on G18 are 15, 25, and 35 °C, respectively; no growth on CYA, Czapek 20% or CREA, or at 40 °C on all tested media.
Other specimens examined: Spain: Catalonia community: Tarragona province, from decanted and filtered blossom honey, 10 May 2014, A. Gómez Pajuelo (FMR 15674, FMR 15675, and FMR 15677); Valencia community: Castellón province, from decanted and filtered blossom honey, 10 May 2014, A. Gómez Pajuelo (FMR 15684 and FMR 15688); Extremadura community: Cáceres province, from decanted, filtered and thermally treated honeydew honey, 16 May 2014, A. Terrab (FMR 16029, FMR 16499, and FMR 16501); Castilla y León community: Salamanca province, from decanted, filtered and thermally treated honeydew honey, 01 Oct. 2014, A. Terrab (FMR 16033 and FMR 16114); Zamora province, from decanted, filtered and thermally treated honeydew honey, 05 Oct 2014, A. Terrab (FMR 16125, FMR 16126, FMR 16276, and FMR 16494).
Subclade D: Incertae sedis
Based on both LSU-based (Fig. 2; sister clade D1) and ITS-based (Fig. 3) phylogenetic trees, ten of our strains were located in a well-supported and separated branch related to species of the genera Oidiodendron and Myxotrichum, and phylogenetically distant from the most similar taxa included in the study, M. setosum and O. truncatum (Fig. 3). Recognition of all of these distinct strains was also supported by unique phenotypic characteristics; therefore, we propose the recognition of the new species Oidiodendron mellicola. Furthermore, because three of our strains were placed near Skoua fertilis in both LSU-based (Fig. 2; sister clade D2) and ITS-based (Fig. 3) phylogenies and because they showed different phenotypic features and enough phylogenetic distance relative to S. fertilis, we also propose the introduction of a further new species, Skoua asexualis.
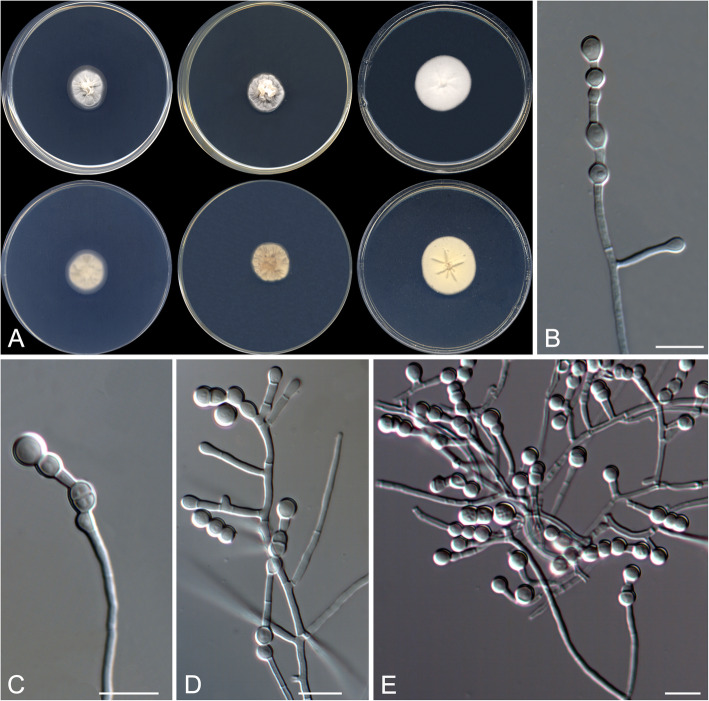
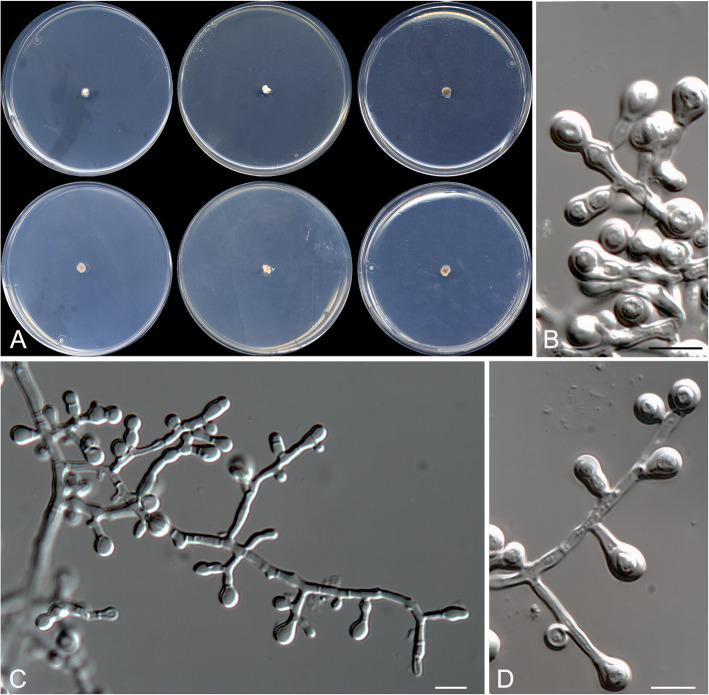
Oidiodendron mellicola Rodr.-Andr., Cano & Stchigel, sp. nov. Fig. 10. MycoBank MB 823586.
Fig. 10.
Oidiodendron mellicola CBS 143839 T. a Colonies on PDA at 15 °C and at 25 °C, and on G18 at 25 °C (left to right), surface and reverse (from top to bottom). b–d Conidiophores. e Disarticulating chains of conidia. Scale bar = 10 μm
Etymology: From Latin mellis-, honey, and -cola dwelling on, referring to the habitat.
Diagnosis: Forming a terminal clade together with O. truncatum and M. setosum at a significant phylogenetic distance (5.3% from the other two species), and differing morphologically from other known species of Oidiodendron and the asexual morphs of Myxotrichum in the absence of well-differentiated conidiophores, and the slow growth.
Type: Spain: Valencia community: Castellón province, from decanted and filtered blossom honey, 10 May 2014, A. Gómez Pajuelo (CBS H-23369 – holotype; CBS 143839 = FMR 15683 – ex-type cultures; ITS sequence GenBank LT906544).
Description: Colonies on PDA at 15 °C reaching 15–16 mm diam after 3 wk., white (5A1), sporulation sparse (seen after 6 wk. of incubation), exudate absent, reverse orange-white (6A2) at the centre and orange-grey (6B2) at the edge, diffusible pigment absent. Mycelium composed of hyaline, septate, smooth- and thin-walled hyphae, 1–3 μm wide. Conidiophores reduced to fertile side branches and the terminal part of a vegetative hyphae, mostly simple or once branched near or at the base, 10–40 μm long, pale olive, disarticulating in conidia. Conidia one-celled, mostly holoarthric, sometimes enteroarthric, mostly in chains of up to ten, occasionally solitary and sessile, mostly barrel-shaped, sometimes cylindrical, conical or “Y”-shaped, 5–14 × 2.5–5 μm, pale olive, disarticulating by schizolytic or rhexolytic secession from the hyphae. Chlamydospores absent. Sexual morph absent.
Colonies on PDA reaching 10–11 mm diam. After 3 wk. at 25 °C, elevated, compact, velvety, margins irregular, olive brown (4E3), exudates absent, sporulation abundant; reverse olive brown (4E5) at the center, grey (5D1) at the edge, diffusible pigment absent. Colonies on G18 reaching 11–12 mm diam after 3 wk. at 25 °C, elevated, velvety to floccose, yellowish white (4A2) at the centre and white (4A1) at the edge, margins regular, sporulation absent, reverse pale yellow (4A3), diffusible pigment absent; on G18 at 15 °C reaching 12–15 mm diam after 3 wk., similar in aspect than at 25 °C; on MY70FG and MEA 2% at 25 °C after 3 wk. reaching 1–3 mm diam.
Minimum, optimal and maximum temperature of growth on G18 are 5, 15, and 25 °C, respectively; no growth on OA or PCA at 25 °C.
Other specimens examined: Spain: Catalonia community: Tarragona province, from decanted and filtered blossom honey, 10 May 2014, A. Gómez Pajuelo (FMR 15680); Castilla-La Mancha community, Ciudad Real province, from decanted, filtered and thermally treated honeydew honey, 10 May 2014, A. Terrab (FMR 16031, FMR 16117, and FMR 16503); Toledo province, from decanted, filtered and thermally treated honeydew honey, 12 May 2014, A. Terrab (FMR 16120 and FMR 16282); Galicia community: Ourense province, from decanted, filtered and thermally treated honeydew honey, 03 May 2014, A. Terrab (FMR 16504); Castilla y León community: Salamanca province, from decanted, filtered and thermally treated honeydew honey, 01 Oct. 2014, A. Terrab (FMR 16023); Burgos province, from decanted, filtered and thermally treated honeydew honey, 23 May 2014, A. Terrab (FMR 16274).
Skoua asexualis Rodr.-Andr., Cano & Stchigel, sp. nov. Fig. 11. MycoBank MB 824092.
Fig. 11.
Skoua asexualis CBS 144072 T. a Colonies on G18, MEA and PDA at 25 °C (left to right), surface and reverse (from top to bottom). b–d Conidiophores and conidia. Scale bar = 10 μm
Etymology: From Latin asexualis, without sex, because of lack of a known sexual morph.
Diagnosis: Differing from the other known species of the genus, S. fertilis, in asexual reproduction, as the latter only produces ascospores within globose asci arising from the mycelium.
Type: Spain: Castilla y León community: León province, from decanted, filtered and thermally treated honeydew honey, 1 Oct. 2014, A. Terrab (CBS H-23397 – holotype; CBS 144072 = FMR 16572 – ex-type cultures; ITS sequence GenBank LT964668).
Description: Colonies on PDA reaching 6–7 mm diam after 3 wk. at 25 °C, elevated, velvety, sporulation abundant, exudates absent, diffusible pigment absent, colonies brown (7E6) at the centre and whitish at the edge, reverse brownish orange (6C5) at the centre and greyish orange (5B3) at the edge. Mycelium composed of hyaline, repeatedly septate, smooth- and thin-walled hyphae, 2–6 μm wide. Conidiophores absent. Conidia mostly one-celled, occasionally two- to three-celled, hyaline, solitary or in short chains, smooth- and thick-walled, mostly globose, occasionally broadly ellipsoidal, pyriform, or irregular-shaped, truncate at one or both ends, 3–7 μm diam, conidiogenesis holoblastic when sessile or terminal, and holothallic when intercalary, disarticulating by rhexolytic secession; the holoblastic and holothallic conidia produce a succession of secondary holoblastic conidia, forming a big, radiating mass of cells of up to 50 μm diam, which eventually detach as complex asexual propagules from the fertile hyphae. Chlamydospores similar to the conidia but thicker, mostly non- or occasionally one-septate, intercalary or terminal. Sexual morph unknown.
Colonies on MEA reaching 3–4 mm diam after 3 wk. at 25 °C, colonies elevated, velvety to floccose, margins irregular, sporulation abundant, diffusible pigment absent, mycelium yellowish white (4A2), reverse pale yellow (4A3); on G18 reaching 4–5 mm diam after 3 wk. at 25 °C, elevated, floccose, margins irregular, sporulation sparse, diffusible pigment absent, exudates absent, colonies pale yellow (4A3) at the centre, reverse orange-grey (5B2).
Minimum, optimal and maximum temperature of growth on G18 are 15, 25, and 30 °C, respectively; no growth on CYA, CREA, OA, or YES at 25 °C.
Other specimens examined: Spain: Extremadura community: Cáceres province, from decanted, filtered and thermally treated honeydew honey, 16 May 2014, A. Terrab (FMR 16497 and FMR 16567).
DISCUSSION
This is the most comprehensive assessment of the diversity of the xerotolerant and xerophilic fungi of honey intended for human consumption to date. We have isolated selectively and identified, by a polyphasic approach, six species of ascomycetous yeasts and 27 of filamentous ascomycetes, some representing new taxa, from honey samples. The yeasts, Candida magnoliae, C. sorbosivorans, Schizosaccharomyces octosporus, Zygosaccharomyces barkeri, Z. mellis, and Z. gambellarensis, had been reported from honey before, and C. magnoliae has also been associated with living honeybees (Gilliam et al. 1974b). All these yeasts have been described as osmophilic and able to grow at aw of 0.80 or lower (Tilbury 1967; van Eck et al. 1993; Ganthala et al. 1994; Erickson & McKenna 1999; Torriani et al. 2011). We found C. magnoliae and C. sorbosivorans were phylogenetically closely related (see Fig. 2), and it was reported that both differ only in a few physiological characteristics (James et al. 2001). To our knowledge, none of the species of Aspergillus that we isolated (A. asperescens, A. montevidensis, and A. pseudoglaucus) have previously been reported from honey. Aspergillus asperescens was originally isolated from soil and bat dung (Stolk 1954), but also from rotten wood and soybean seeds; however, most of the isolates were from cave soil (probably linked to bat dung). Aspergillus montevidensis and A. pseudoglaucus have been reported as the most important food-spoilage species of the genus (Pitt & Hocking 1977; Kozakiewicz 1989), but are known from extreme environments such as salterns (Butinar et al. 2005). Aspergillus montevidensis has been reported from various environmental samples (air, soil, etc.), and even on honeybees and bee larvae (http://gcm.wfcc.info/; Talice & Mackinnon 1931; Gilliam et al. 1974a); A. pseudoglaucus has been reported in air, paper and soil (http://gcm.wfcc.info/; Blochwitz 1929). Aspergillus montevidensis and A. pseudoglaucus are able to grow at aw values of 0.80 (Snow 1949; Armolik & Dickson 1956; Guynot et al. 2003). Monascus is a well-known genus with species (especially M. purpureus and M. ruber) of economic importance due to their use in production of foodstuffs, bioactive compounds, pigments and enzymes. Currently, Monascus is placed in Aspergillaceae (syn. Trichocomaceae) based on phylogenetic studies, and closely related to Leiothecium ellipsoideum and Xeromyces bisporus (Houbraken & Samson 2011; Pettersson et al. 2011). Recently, three new species were added, all of them associated with stingless bees: M. flavipigmentosus, M. mellicola, and M. recifensis (Barbosa et al. 2017). We found a small number of isolates, including M. pilosus, M. purpureus, and M. ruber. These species have been frequently reported in fermented and spoiled foods (van Tieghem 1884; Hesseltine 1965; Lin 1975; Hawksworth & Pitt 1983). Monascus ruber has also been found in soil and human clinical specimens (Hawksworth & Pitt 1983). Species of Monascus have been previously reported in honey by Snowdon & Cliver (1996) and by Barbosa et al. (2017). Monascus pilosus, M. purpureus, and M. ruber were reported previously (Hawksworth & Pitt 1983) as able to grow well on G25 N (aw = 0.93). The species of Penicillium we found in honey included P. camemberti, P. citrinum, P. corylophilum, and P. cravenianum. The most common source of isolation of P. camemberti is blue cheeses, but it can also be found on a wide variety of substrata (Thom 1906; http://gcm.wfcc.info/). Penicillium citrinum was originally reported in milk and bread in the USA (Thom 1910), but it is found globally and easy to recover from spoiled foods and diverse environmental sources (www.cabri.org/collections.html) including honey, pollen and bee nests (Barbosa et al. 2018). Penicillium corylophilum (Dierckx 1901) mostly occurs in damp buildings in North America and Western Europe, but also in foods and mosquitoes (Da Costa & De Oliveira 1998; McMullin et al. 2014), and honey (Sinacori et al. 2014). The minimum aw reported for the growth of P. camemberti, P. citrinum and P. corylophilum was around 0.80 (Abellana et al. 2001; Fontana 2008; Kalai et al. 2017). Penicillium cravenianum, a species moderately xerotolerant (grows on G25 N), has only been reported in soil (Visagie et al. 2016). Notably, all the isolates of Talaromyces that we found in honey belonged to three unrecognized species. Talaromyces basipetosporus was recovered from a honey sample in Buenos Aires province, Argentina, and is characterized by simple conidiophores that mimic those of the asexual morph of Monascus (syn. Basipetospora), which develops conidia by a retrogressive mode of conidiogenesis, a feature not previously reported in Talaromyces. Talaromyces affinitatimellis displays a similar conidiogenesis to T. basipetosporus and both species are phylogenetically closely related but phenotypically differentiated as T. affinitatimellis grows faster and produces more complex conidiophores. Talaromyces brunneosporus differs from the other species of sect. Purpurei, apart from T. purpureus, in having monophialidic and monoverticillate conidiophores (they are biverticillate in the other species). However, both species are distinguishable because T. brunneosporus produces penicillate conidiophores (not aspergillate as in T. purpureus), longer phialides, and verrucose conidia with a flattened base (T. purpureus conidia are ornamented by spiral ridges). Talaromyces basipetosporus has a high xerotolerance, with similar growth rates on MEA with sugars up to aw 0.82. Despite the decreasing growth rates of T. brunneosporus and T. affinitatimellis when sugar concentration increases, both fungi are able to grow at aw 0.82 (Fig. 12). Xerochrysium xerophilum (Pitt et al. 2013; syn. Chrysosporium xerophilum, Pitt 1966), is an extreme xerophile with a minimum aw for growth of 0.66 (Gock et al. 2003; Leong et al. 2011). This fungus, previously reported from chocolate, coconut, dried prunes, and stored corn (Pitt & Hocking 2009; Pitt et al. 2013), has not been found in honey until now. This species is phylogenetically close to Monascus (Pitt et al. 2013). Among the species of Onygenales, Ascosphaera atra and Eremascus albus were recovered once and four times, respectively. Ascosphaera atra (Skou & Hackett 1979) was originally reported from dead larvae of the alfalfa leafcutter bee covered in cysts of Ascosphaera aggregata (Skou 1975), and from pollen in the gut of healthy leafcutter larvae. This fungus was subsequently reported from grass silage (Skou 1986). Ascosphaera atra is homothallic and saprobic, probably being a common contaminant of pollen (Skou & Hackett 1979), which would explain its presence in honey samples. Eremascus albus is a well-known xerophilic fungus, with spores that can germinate at aw as low as 0.70 (Pitt 1965). This fungus has been reported to spoil malt extract (Eidam 1883), chocolate cake, dried fruits, and mustard powder (Harrold 1950), but never previously from honey. We identified several isolates belonging to the newly described family Helicoarthrosporaceae, which only includes the new monotypic genus Helicoarthrosporum, and a single strain belonging to the new monotypic genus Strongyloarthrosporum (Ajellomycetaceae). The morphology of Helicoarthrosporum mellicola resembles species of Scytalidium (S. cuboideum, S. ganodermophthorum, and S. sphaerosporum) because of the production of cuboid arthroconidia in long chains. However, Helicoarthrosporum is phylogenetically distant from Scytalidium, as the latter is related to Myxotrichaceae. Strongyloarthrosporum catenulatum was found to be phylogenetically close to Ajellomycetaceae, whose members are thermally dimorphic and pathogenic to animals (including the humans), and has never been reported as xerotolerant. However, having features not seen in that family, S. catenulatum is unequivocally a xerophilic fungus, only growing on G18, G25 N and MY70FG, and producing globose arthroconidia, either singly or in chains. The sole xerophilic fungus phylogenetically close to S. catenulatum is Eremascus albus (Eremascaceae), but it only develops a sexual morph. Regarding the family Myxotrichaceae, Skoua fertilis, which was detected in all honey samples, resembles Eremascus albus (Eidam 1883) in having naked asci arising directly out of the mycelium and formed by the fusion of two equal cells borne on short entwined hyphae. Both taxa can be only morphologically differentiated by the shape of the ascospores and by sexual reproductive details. While S. fertilis (syn. E. fertilis) belongs to Leotiomycetes, closely related to Myxotrichaceae (Wynns 2015), E. albus is located in Eurotiomycetes, closely related to Onygenales (Cai et al. 1996; Berbee 2001; Wynns 2015). Skoua was introduced for E. fertilis (i.e. Skoua fertilis) and has been reported on bee bread, honeycomb, dried prunes and spoiled moist prunes, green compost, and shortcake (www.cabri.org/collections.html; http://gcm.wfcc.info/; Harrold 1950), but not so far on honey. The minimum aw for growth and sporulation reported for S. fertilis was 0.77 (Pitt 1965; Wynns 2015), a similar value observed in all our strains (0.82). We isolated three strains of Skoua phylogenetically different from S. fertilis, and named them as Skoua asexualis because they form asexual spores instead of the sexual spores as observed in the type species of the genus. Bettsia alvei (Skou 1972, 1975), the other fungus identified in all honey samples, belongs to Pseudeurotiaceae and is characterized by dark, closed ascomata (usually called “spore cysts”) and hyaline globose ascospores, forming a sticky mass. Bettsia alvei has been isolated from hives in Europe as well as the USA (Burnside 1929), and from bakery products, spoiled chocolate, desiccated coconut, honeycomb, concentrated jelly, dried and spoiled prunes, pollen, table jelly, bee wax, and wine starters (www.cabri.org/collections.html; http://gcm.wfcc.info/). It was also isolated from chocolate in Austria (aw less than 0.3), but thus far had not been recorded from honey. The lowest aw tested for growth of this species was 0.88 (Beuchat & Pitt 1990) and 0.89 (Udagawa & Toyazaki 2000), similar values to those we found. All our isolates of B. alvei developed the chrysosporium-like asexual morph but failed in the production of the sexual morph. Among the most frequent species we isolated was an undescribed species of Oidiodendron, O. mellicola. Species of this genus are mostly recovered from soil and other substrata rich in cellulose, and are found worldwide (Domsch et al. 1980; Calduch et al. 2004; Rice & Currah 2005). Oidiodendron mellicola is phylogenetically related to O. truncatum and M. setosum, the former characterized by well-differentiated dark conidiophores and barrel-shaped conidia with a dark scar at one or both ends (typical features of Oidiodendron), and the latter by hyaline conidiophores and conidia, and by dark brown to black, spinose, gymnothecial ascomata (typical of the genus Myxotrichum). Interestingly, M. setosum is reported as a common hive fungus in Europe (Burnside 1929). Oidiodendron mellicola is the only species of the genus reported from honey, and it can be distinguished morphologically from other species of the genus by its absence of stipitate conidiophores, and the production of long chains of conidia, which are pale, smooth, ellipsoidal to cylindrical, truncated (but not darkened, as in O. truncatum) at one or both ends, and by the slow growing colonies. Like most of the species of the genus, O. mellicola grows better at 15 °C than 25 °C. Other fungi rarely found in our study were Alternaria multiformis, previously only reported from soil (Simmons 1998), and the mucoralean Cunninghamella bertholletiae, Mucor plumbeus, and Rhizopus oryzae, all found worldwide. These probably represent environmental contaminants. Although all the new taxa that we propose displayed a high xerotolerance, only Strongyloarthrosporum catenulatum can be considered an obligate xerophile, because it was able to grow faster at the lowest aw tested (Fig. 12).
Fig. 12.
Relatedness between the growth of the new fungal taxa and the decreasing water activity (aw) of the culture medium. Surface of the colonies grown on MEA with aw values of 0.97, 0.95, 0.93, 0.92, 0.88 and 0.82 (from left to right, respectively). a Helicoarthrosporum mellicola. b Strongyloarthrosporum catenulatum. c Talaromyces basipetosporus. d Talaromyces brunneosporus. e Talaromyces affinitatimellis. f Oidiodendron mellicola. g Skoua asexualis. h Skoua fertilis (as reference, highly xerotolerant fungus)
CONCLUSION
The application of G18 as a selective culture medium for isolation of xerotolerant/xerophilic fungi from honey samples enabled the recovery and identification of 13 genera and 29 species of Ascomycota, and three genera (one species for each) of Mucoromycota. Many of these fungi have never reported from honey before. Among them, we proposed a new family (Helicoarthrosporaceae), two new genera (Strongyloarthrosporum and Helicoarthrosporum) and seven new species (Strongyloarthrosporum catenulatum, Helicoarthrosporum mellicola, Oidiodendron mellicola, Skoua asexualis Talaromyces basipetosporus, T. brunneosporus, and T. affinitatimellis). All fungal taxa that we isolated from honey were able to grow at low water activity (up to 0.82), but only Ascosphaera atra, Bettsia alvei (two fungi strongly associated to honeybees and their life-style), Eremascus albus, Strongyloarthrosporum catenulatum (one of the new taxa we described) and Xerochrysium xerophylum can be considered obligate xerophiles. Also, because several of the honey samples were thermally treated, these fungi can be considered as hot-resistant. Honey is evidently a reservoir of xerotolerant and xerophilic fungi, which survives to the thermal treatment used to make honey non-crystallisable. Some of these fungi are related to the honeybee life-style; however, as is in the case of the new taxa described here, the origin in nature remains unknown. In the latter case, flowers and aphids could play an important role as a source of such fungi. During the course of the study, the most important pathogenic fungi for honeybees, Aspergillus flavus and Ascosphaera apis, were not found. Several of the fungi found in honey samples (Aspergillus and Pencillium spp.) are potential producers of mycotoxins, but this does not mean that the honey may represent a risk to the health of the consumer, because (in general) the production of mycotoxins or the fungal growth are suppressed at water activities lower than 0.70 (Mannaa & Kim 2017), as is the case of honey (aw of 0.60 or less). Honey should be considered as a “living food” and, consequently, its “normal” mycobiota merits more extensive study. It is expected that such “normal” mycobiota may vary qualitatively and quantitatively, depending on the geographic origin, the botanical type and water activity of the honey, among other physicochemical and biological parameters. Honey is clearly one of the relatively unexplored habitats for the missing fungal diversity, especially as the new taxa we found came from samples from just two countries.
Acknowledgements
The authors are indebted to Antonio Gómez-Pajuelo (Spain) and Marcela Álvarez (Argentina) for providing honey samples and their useful suggestions regarding the preparation of this article. We also thank to Stella Maris Romero by measuring the water activity in the media used.
Adherence to national and international regulations
The authors confirm that this manuscript respects the Nagoya Protocol to the Convention on Biological Diversity.
Abbrevations
- aw
water activity
- BCP-MS-G
Bromcresol purple milk solids glucose agar
- BEA
Bile esculin agar
- BenA
fragment of the beta-tubulin gene
- BI
Bayesian-inference
- BLAST
Basic Local Alignment Search Tool
- CaM
fragment of the calmodulin gene
- CREA
Creatine sucrose agar
- CYA
Czapek yeast extract agar
- DG18
Dichloran 18% glycerol agar
- DNA
Deoxyribonucleic acid
- G18 DG18
without dichloran
- G25 N 25%
glycerol nitrate agar
- ITS
Ribosomal internal transcribed spacers
- LSU
Large sub unit of the ribosomal genes
- MEA
Malt extract agar
- ML
Maximum-likelihood
- MLI
Maximum level of identity
- MY70FG
Malt extract yeast extract 70% fructose-glucose
- nrRNA
Nuclear ribosomal ribonucleic acid
- OA
Oatmeal agar
- PDA
Potato dextrose agar
- PYE
Phytone yeast extract agar
- rpb2
fragment of the RNA polymerase II subunit 2 gene
- SEM
Scanning electron microscopy
- TOTM
Test opacity tween medium
- TreeBASE
a repository of user-submitted phylogenetic trees and data used to build them
- YES
Yeast extract sucrose agar
Authors’ contributions
ER-A performed all the experimental work, culturing the samples, isolating in pure culture the fungi and performing their phenotypic characterization, as well as the DNA extraction and purification, gene sequencing and data processing for phylogenetic analysis, being one of the major contributors of this manuscript. AMS, because their experience on fungi from honey, supervised all steps of the experimental work by ER-A, collaborating in the description of the novel fungi and in the writing of chapters “Introduction” and “Discussion”, reviewing of the draft several times. AT provided most of the samples analyzed in this work, gave useful suggestions to write the manuscript and reviewed the draft several times. JG contributed actively in the identification and taxonomy of the fungal strains, and reviewed the draft several times. JFC-L supervised the nucleotide sequence alignment and phylogenetic reconstruction, took the pictures that appear in the figures, contributed actively in the identification and taxonomy of the fungal strains, gave useful suggestions to write the manuscript and reviewed several times the draft. All authors read and approved the final manuscript.
Funding
The authors are indebted to the Instituto de Ciencia, Tecnología e Innovación (Mexico) and the Consejo Nacional de Ciencia y Tecnología (Mexico) for the scholarship 440135 with scholar 277137. This work was supported by the Spanish Ministerio de Economía y Competitividad, grant CGL2017–88094-P.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abellana M, Sanchis V, Ramos AJ. Effect of water activity and temperature on growth of three Penicillium species and Aspergillus flavus on a sponge cake analogue. International Journal of Food Microbiology. 2001;71:151–157. doi: 10.1016/S0168-1605(01)00596-7. [DOI] [PubMed] [Google Scholar]
- Armolik N, Dickson J. Minimum humidity requirements for germination of conidia of fungi associated with storage of grain. Phytopathology. 1956;46:462–465. [Google Scholar]
- Barbosa RN, Bezerra JDP, Souza-Motta CM, Frisvad JC, Samson RA, et al. New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek. 2018;111:1883–1912. doi: 10.1007/s10482-018-1081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa R.N., Leong S.L., Vinnere-Pettersson O., Chen A.J., Souza-Motta C.M., Frisvad J.C., Samson R.A., Oliveira N.T., Houbraken J. Phylogenetic analysis of Monascus and new species from honey, pollen and nests of stingless bees. Studies in Mycology. 2017;86:29–51. doi: 10.1016/j.simyco.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee ML. The phylogeny of plant and animal pathogens in the Ascomycota. Physiological and Molecular Plant Pathology. 2001;59:165–187. doi: 10.1006/pmpp.2001.0355. [DOI] [Google Scholar]
- Beuchat LR, Hocking AD. Some considerations when analyzing foods for the presence of xerophilic fungi. Journal of Food Protection. 1990;53:984–989. doi: 10.4315/0362-028X-53.11.984. [DOI] [PubMed] [Google Scholar]
- Beuchat LR, Pitt JI. Influence of water activity and temperature on survival of and colony formation by heat-stressed Chrysosporium farinicola aleuriospores. Applied and Environmental Microbiology. 1990;56:2951–2956. doi: 10.1128/aem.56.10.2951-2956.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochwitz A. Die Gattung Aspergillus. Neue Spezies. Diagnosen. Synonyme. Annales Mycologici. 1929;27:205–240. [Google Scholar]
- Burnside CE. Saprophytic fungi associated with the honey bee. Bee World. 1929;10:42. doi: 10.1080/0005772X.1929.11092782. [DOI] [Google Scholar]
- Butinar L, Zalar P, Frisvad JC, Gunde-Cimerman N. The genus Eurotium - members of indigenous fungal community in hypersaline waters of salterns. FEMS Microbiology Ecology. 2005;51:155–166. doi: 10.1016/j.femsec.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Čadež N, Fülöp L, Dlauchy D, Péter G. Zygosaccharomyces favi sp. nov., an obligate osmophilic yeast species from bee bread and honey. Antonie Van Leeuwenhoek. 2015;107:645–654. doi: 10.1007/s10482-014-0359-1. [DOI] [PubMed] [Google Scholar]
- Cai J, Roberts IN, Collins MD. Phylogenetic relationships among members of the ascomycetous yeast genera Brettanomyces, Debaryomyces, Dekkera, and Kluyveromyces deduced by small-subunit rRNA gene sequences. International Journal of Systematic Bacteriology. 1996;46:542–549. doi: 10.1099/00207713-46-2-542. [DOI] [PubMed] [Google Scholar]
- Calduch M, Gené J, Cano J, Stchigel AM, Guarro J. Three new species of Oidiodendron Robak from Spain. Studies in Mycology. 2004;50:159–170. [Google Scholar]
- Carmichael JW, Kraus HJ. The cattle ringworm fungus, Trichophyton verrucosum, in Alberta. Alberta Medical Bulletin. 1959;24:201–203. [Google Scholar]
- Carvalho CM, Meirinho S, Estevinho ML, Choupina A. Yeast species associated with honey: different identification methods. Archivos de Zootecnia. 2010;59:103–113. doi: 10.21071/az.v59i225.4896. [DOI] [Google Scholar]
- Christensen WB. Urea decomposition as means of differentiating Proteus and Paracolon cultures from each other and from Salmonella and Shigella types. Journal of Bacteriology. 1946;52:461–466. doi: 10.1128/jb.52.4.461-466.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, Le Roux JJ, Strasberg D, et al. Fungal Planet description sheets: 400–468. Persoonia. 2016;36:316–458. doi: 10.3767/003158516X692185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa GL, De Oliveira PC. Penicillium species in mosquitoes from two Brazilian regions. Journal of Basic Microbiology. 1998;38:343–347. doi: 10.1002/(SICI)1521-4028(199811)38:5/6<343::AID-JOBM343>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Dierckx RP (1901) Un essai de revision du genre Penicillium Link. Note preliminaire. Annales de la Societe Scientifique de Bruxelles 25:83–89
- Domsch KH, Gams W, Anderson TH. Compendium of soil fungi. London: Academic Press; 1980. [Google Scholar]
- Eidam E. Zur kenntniss der entwickelung bei den Ascomyceten. Beiträge zur Biologie der Pflanzen. 1883;3:377–425. [Google Scholar]
- Erickson JP, McKenna DN. Zygosaccharomyces. In: Robinson RK, Batt CA, Patel PD, editors. Encyclopedia of food microbiology. 3. San Diego: Academic Press; 1999. pp. 2359–2365. [Google Scholar]
- Figueras MJ, Guarro J. A scanning electron microscopic study of ascoma development in Chaetomium malaysiense. Mycologia. 1988;80:298–306. doi: 10.2307/3807625. [DOI] [Google Scholar]
- Fontana AJ (2008) Appendix D: minimum water activity limits for growth of microorganisms. In: Barbosa-Cánovas GV, Fontana AJ, Schmidt SJ, Labuza TP (eds) Water activity in foods: fundamentals and applications. Blackwell Publishing Ltd.
- Frisvad JC. Physiological criteria and mycotoxin production as AIDS in identification of common asymmetric penicillia. Applied and Environmental Microbiology. 1981;41:568–579. doi: 10.1128/aem.41.3.568-579.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganthala BP, Marshall JH, May JW. Xerotolerance in fission yeasts and the role of glycerol as compatible solute. Archives of Microbiology. 1994;162:108–113. doi: 10.1007/BF00264382. [DOI] [Google Scholar]
- Gilliam M, Prest DB, Morton HL. Fungi isolated from honey bees, Apis mellifera, fed 2,4-D and antibiotics. Journal of Invertebrate Pathology. 1974;24:213–217. doi: 10.1016/0022-2011(74)90013-5. [DOI] [PubMed] [Google Scholar]
- Gilliam M, Wickerham LJ, Morton HL, Martin RD. Yeasts isolated from honey bees, Apis mellifera, fed 2,4-D and antibiotics. Journal of Invertebrate Pathology. 1974;24:349–356. doi: 10.1016/0022-2011(74)90143-8. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gock MA, Hocking AD, Pitt JI, Poulos PG. Influence of temperature, water activity and pH on growth of some xerophilic fungi. International Journal of Food Microbiology. 2003;81:11–19. doi: 10.1016/S0168-1605(02)00166-6. [DOI] [PubMed] [Google Scholar]
- Grabowski NT, Klein G. Microbiology and foodborne pathogens in honey. Critical Reviews in Food Science and Nutrition. 2015;57:1852–1862. doi: 10.1080/10408398.2015.1029041. [DOI] [PubMed] [Google Scholar]
- Guynot ME, Marin S, Sanchis V, Ramos AJ. Modified atmosphere packaging for prevention of mold spoilage of bakery products with different pH and water activity levels. Journal of Food Protection. 2003;66:1864–1872. doi: 10.4315/0362-028X-66.10.1864. [DOI] [PubMed] [Google Scholar]
- Harrold CE. Studies in the genus Eremascus I The rediscovery of Eremascus albus Eidam and some new observations concerning its life-history and cytology. Annals of Botany. 1950;14:127–148. doi: 10.1093/oxfordjournals.aob.a083239. [DOI] [Google Scholar]
- Hawksworth DL, Kirk PM, Sutton BC, Pegler DN. Ainsworth & Bisby’s dictionary of the Fungi. 8. Wallingford: CAB International; 1995. [Google Scholar]
- Hawksworth DL, Pitt JI. A new taxonomy for Monascus species based on cultural and microscopical characters. Australian Journal of Botany. 1983;31:51–61. doi: 10.1071/BT9830051. [DOI] [Google Scholar]
- Hesseltine CW. A millennium of fungi, food, and fermentation. Mycologia. 1965;57:149–197. doi: 10.1080/00275514.1965.12018201. [DOI] [PubMed] [Google Scholar]
- Hocking AD, Pitt JI. Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Applied and Environmental Microbiology. 1980;39:488–492. doi: 10.1128/aem.39.3.488-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97:1316–1329. doi: 10.3852/mycologia.97.6.1316. [DOI] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SA, Bond CJ, Roberts IN. Candida sorbosivorans sp. nov., a new member of the genus Candida Berkhout. International Journal of Systematic and Evolutionary Microbiology. 2001;51:1215–1219. doi: 10.1099/00207713-51-3-1215. [DOI] [PubMed] [Google Scholar]
- Kačániová M, Kňazovická V, Felšöciová S, Rovná K. Microscopic fungi recovered from honey and their toxinogenity. Journal of Environmental Science and Health. 2012;47:1659–1664. doi: 10.1080/10934529.2012.687242. [DOI] [PubMed] [Google Scholar]
- Kačániová M, Pavlicová S, Hascík P, Kociubinski G, Kňazovická V, et al. Microbial communities in bees, pollen and honey from Slovakia. Acta Microbiologica et Immunologica Hungarica. 2009;56:285–295. doi: 10.1556/AMicr.56.2009.3.7. [DOI] [PubMed] [Google Scholar]
- Kalai S, Anzala L, Bensoussan M, Dantigny P. Modelling the effect of temperature, pH, water activity, and organic acids on the germination time of Penicillium camemberti and Penicillium roqueforti conidia. International Journal of Food Microbiology. 2017;240:124–130. doi: 10.1016/j.ijfoodmicro.2016.03.024. [DOI] [PubMed] [Google Scholar]
- Kane J, Smitka C. Early detection and identification of Trichophyton verrucosum. Journal of Clinical Microbiology. 1978;8:740–747. doi: 10.1128/jcm.8.6.740-747.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Sigler L, Lee J, Gibas CF, Yun SH, et al. Xylogone ganodermophthora sp. nov., an ascomycetous pathogen causing yellow rot on cultivated mushroom Ganoderma lucidum in Korea. Mycologia. 2010;102:1167–1184. doi: 10.3852/09-304. [DOI] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. Methuen handbook of colour. 3. London: Methuen; 1978. [Google Scholar]
- Kozakiewicz Z. Aspergillus species on the stored products. Mycological Papers. 1989;161:1–188. [Google Scholar]
- Leong SL, Pettersson OV, Rice T, Hocking AD, Schnurer J. The extreme xerophilic mould Xeromyces bisporus-growth and competition at various water activities. International Journal of Food Microbiology. 2011;145:57–63. doi: 10.1016/j.ijfoodmicro.2010.11.025. [DOI] [PubMed] [Google Scholar]
- Lin CF. Studies on the Monascus isolated from the startor of kaoliang brandy. Chinese Journal of Microbiology. 1975;8:152–160. [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Lochhead AG, Farrell L. The types of osmophilic yeasts found in normal honey and their relation to fermentation. Canadian Journal of Research. 1931;5:665–672. doi: 10.1139/cjr31-105. [DOI] [Google Scholar]
- Magyar D, Gönczöl J, Révay Á, Grillenzoni F, Seijo-Coello MD. Stauro-and scolecoconidia in floral and honeydew honeys. Fungal Diversity. 2005;20:103–120. [Google Scholar]
- Magyar D, Mura-Mészáros A, Grillenzoni F. Fungal diversity in floral and honeydew honeys. Acta Botanica Hungarica. 2016;58:145–166. doi: 10.1556/034.58.2016.1-2.6. [DOI] [Google Scholar]
- Mannaa M, Kim KD. Influence of temperature and water activity on deleterious fungi and mycotoxin production during grain storage. Mycobiology. 2017;45:240–254. doi: 10.5941/MYCO.2017.45.4.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R, Gene J, Cano J, Trilles L, Dos Santos LM, et al. Molecular phylogeny of Sporothrix schenckii. Journal of Clinical Microbiology. 2006;44:3251–3256. doi: 10.1128/JCM.00081-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martíns HM, Martíns ML, Bernardo FM. Bacillaceae spores, fungi and aflatoxins determination in honey. Revista Portuguesa de Ciências Veterinárias. 2003;98:85–88. [Google Scholar]
- McMullin DR, Nsiama TK, Miller JD. Secondary metabolites from Penicillium corylophilum isolated from damp buildings. Mycologia. 2014;106:621–628. doi: 10.3852/13-265. [DOI] [PubMed] [Google Scholar]
- Pérez-Sánchez MC, Del Baño-Breis F, Candela-Castillo ME, Egea-Gilabet C. Microbial flora in honey from the región of Murcia, Spain. Anales de Biologia. 1997;22:155–164. [Google Scholar]
- Pettersson OV, Leong SL (2011) Fungal xerophiles (Osmophiles) in: encyclopedia of life sciences. Chichester. 10.1002/9780470015902.a0000376.pub2
- Pettersson OV, Leong SL, Lantz H, Rice T, Dijksterhuis J, et al. Phylogeny and intraspecific variation of the extreme xerophile, Xeromyces bisporus. Fungal Biology. 2011;115:1100–1111. doi: 10.1016/j.funbio.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Pitt JI. Microbiological problems in prune preservation. University of new South Wales. Australia: Sydney; 1965. [Google Scholar]
- Pitt JI. Two new species of Chrysosporium. Transactions of the British Mycological Society. 1966;49:467–470. doi: 10.1016/S0007-1536(66)80092-X. [DOI] [Google Scholar]
- Pitt JI. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. London: Academic Press; 1979. [Google Scholar]
- Pitt JI, Hocking AD. Influence of solute and hydrogen ion concentration on the water relations of some xerophilic fungi. Journal of General Microbiology. 1977;101:35–40. doi: 10.1099/00221287-101-1-35. [DOI] [PubMed] [Google Scholar]
- Pitt JI, Hocking AD. Fungi and food spoilage. 3. New York: Springer; 2009. [Google Scholar]
- Pitt JI, Lantz H, Pettersson OV, Leong SL. Xerochrysium gen. Nov. and Bettsia, genera encompassing xerophilic species of Chrysosporium. IMA Fungus. 2013;4:229–241. doi: 10.5598/imafungus.2013.04.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research. 1994;98:625–634. doi: 10.1016/S0953-7562(09)80409-7. [DOI] [Google Scholar]
- Rice AV, Currah RS. Oidiodendron: a survey of the named species and related anamorphs of Myxotrichum. Studies in Mycology. 2005;53:83–120. doi: 10.3114/sim.53.1.83. [DOI] [Google Scholar]
- Ruiz-Argueso T, Rodriguez-Navarro A. Microbiology of ripening honey. Applied Microbiology. 1975;30:893–896. doi: 10.1128/am.30.6.893-896.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksinchai S, Suzuki M, Chantawannakul P, Ohkuma M, Lumyong S. A novel ascosporogenous yeast species, Zygosaccharomyces siamensis, and the sugar tolerant yeasts associated with raw honey collected in Thailand. Fungal Diversity. 2012;52:123–139. doi: 10.1007/s13225-011-0115-z. [DOI] [Google Scholar]
- Saksinchai S, Suzuki M, Lumyong S, Ohkuma M, Chantawannakul P. Two new species of the genus Candida in the Zygoascus clade, Candida lundiana sp. nov. and Candida suthepensis sp. nov., isolated from raw honey in Thailand. Antonie Van Leeuwenhoek. 2012;101:633–640. doi: 10.1007/s10482-011-9677-8. [DOI] [PubMed] [Google Scholar]
- Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. Food and indoor fungi. Utrecht: CBS-KNAW Fungal Biodiversity Centre; 2010. [Google Scholar]
- Seijo MC, Escudero O, Fernández-González M. Fungal diversity in honeys from Northwest Spain and their relationship to the ecological origin of the product. Grana. 2011;50:55–62. doi: 10.1080/00173134.2011.559555. [DOI] [Google Scholar]
- Simmons EG. Multiplex conidium morphology in species of the Ulocladium atrum group. Canadian Journal of Botany. 1998;76:1533–1539. doi: 10.1139/b98-091. [DOI] [Google Scholar]
- Sinacori M, Francesca N, Alfonzo A, Cruciata M, Sannino C, et al. Cultivable microorganisms associated with honeys of different geographical and botanical origin. Food Microbiology. 2014;38:284–294. doi: 10.1016/j.fm.2013.07.013. [DOI] [PubMed] [Google Scholar]
- Skou JP. Ascosphaerales. Friesia. 1972;10:1–24. [Google Scholar]
- Skou JP. Two new species of Ascosphaera and notes on the conidial state of Bettsia alvei. Friesia Nordisk Mykologisk Tidsskrift. 1975;11:62–74. [Google Scholar]
- Skou JP. Notes on the habitats, morphology and taxonomy of spores cyst fungi. Apimondia. 1986;30:260–264. [Google Scholar]
- Skou JP, Hackett K. A new, homothallic species of Ascosphaera. Friesia Nordisk Mykologisk Tidsskrift. 1979;11:265–272. [Google Scholar]
- Slifkin M. Tween 80 opacity test responses of various Candida species. Journal of Clinical Microbiology. 2000;38:4626–4628. doi: 10.1177/147323000203000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow D. Germination of mould spores at controlled humidities. Annals of Applied Biology. 1949;36:1–13. doi: 10.1111/j.1744-7348.1949.tb06395.x. [DOI] [PubMed] [Google Scholar]
- Snowdon JA, Cliver DO. Microorganisms in honey. International Journal of Food Microbiology. 1996;31:1–26. doi: 10.1016/0168-1605(96)00970-1. [DOI] [PubMed] [Google Scholar]
- Stolk AC. Aspergillus asperescens n. sp. Antonie Van Leeuwenhoek. 1954;20:299–304. doi: 10.1007/BF02543732. [DOI] [PubMed] [Google Scholar]
- Talice RV, Mackinnon J-E (1931) Aspergillus (Eurotium) montevidensis, n. sp. isolé d'un cas d'otomycose chez l'homme. Société de Biologie de Montevideo 108:1007–1009
- Terrab A, Berjano R, Sanchez JA, Pajuelo A, Díez MJ. Palynological and geographical characterisation of Spanish oak honeydew honeys. Grana. 2019;58:63–77. doi: 10.1080/00173134.2018.1509124. [DOI] [Google Scholar]
- Thom C. Fungi in cheese ripening; camembert and Roquefort. Bureau of Animal Industry Bulletin. 1906;82:1–39. [Google Scholar]
- Thom C. Cultural studies of species of Penicillium. Bureau of Animal Industry Bulletin. 1910;118:1–107. [Google Scholar]
- Tilbury RH (1967) Studies on the microbiological deterioration of raw cane sugar, with special reference to osmophilic yeasts and the preferential utilization of laevulose in invert. M. S., thesis. University of Bristol, Bristol, England.
- Torriani S, Lorenzini M, Salvetti E, Felis GE. Zygosaccharomyces gambellarensis sp. nov., an ascosporogenous yeast isolated from an Italian 'passito' style wine. International Journal of Systematic and Evolutionary Microbiology. 2011;61:3084–3088. doi: 10.1099/ijs.0.031146-0. [DOI] [PubMed] [Google Scholar]
- Udagawa SI, Toyazaki N. Growth characteristics of Chrysosporium farinicola isolated from spoiled imported Austrian chocolate. Japan Journal of Food Microbiology. 2000;17:163–169. doi: 10.5803/jsfm.17.163. [DOI] [Google Scholar]
- Valenzuela-Lopez N, Cano-Lira JF, Guarro J, Sutton DA, Wiederhold N, et al. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Studies in Mycology. 2018;90:1–69. doi: 10.1016/j.simyco.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eck JH, Prior BA, Brandt EV. The water relations of growth and polyhydroxy alcohol production by ascomycetous yeasts. Journal of General Microbiology. 1993;139:1047–1054. doi: 10.1099/00221287-139-5-1047. [DOI] [Google Scholar]
- van Tieghem P. Monascus genre nouveau de I'ordre des ascomyctes. Bulletin de la Societe Botanique de France. 1884;31:226–231. doi: 10.1080/00378941.1884.10828230. [DOI] [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J, Samson RA, Jacobs K. A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus. 2016;7:75–117. doi: 10.5598/imafungus.2016.07.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Shinsky J, White T, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. [Google Scholar]
- Wynns AA. Convergent evolution of highly reduced fruiting bodies in Pezizomycotina suggests key adaptations to the bee habitat. BMC Evolutionary Biology. 2015;15:145–156. doi: 10.1186/s12862-015-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.