Abstract
Barnyard millet (Echinochloa species) has become one of the most important minor millet crops in Asia, showing a firm upsurge in world production. The genus Echinochloa comprises of two major species, Echinochloa esculenta and Echinochloa frumentacea, which are predominantly cultivated for human consumption and livestock feed. They are less susceptible to biotic and abiotic stresses. Barnyard millet grain is a good source of protein, carbohydrate, fiber, and, most notably, contains more micronutrients (iron and zinc) than other major cereals. Despite its nutritional and agronomic benefits, barnyard millet has remained an underutilized crop. Over the past decades, very limited attempts have been made to study the features of this crop. Hence, more concerted research efforts are required to characterize germplasm resources, identify trait-specific donors, develop mapping population, and discover QTL/gene (s). The recent release of genome and transcriptome sequences of wild and cultivated Echinochloa species, respectively has facilitated in understanding the genetic architecture and decoding the rapport between genotype and phenotype of micronutrients and agronomic traits in this crop. In this review, we highlight the importance of barnyard millet in the current scenario and discuss the up-to-date status of genetic and genomics research and the research gaps to be worked upon by suggesting directions for future research to make barnyard millet a potential crop in contributing to food and nutritional security.
Keywords: barnyard millet, Echinochloa species, micronutrients, small millets, genetic and genomic resources, value addition
Introduction
Barnyard millet (Echinochloa species) is an ancient millet crop grown in warm and temperate regions of the world and widely cultivated in Asia, particularly India, China, Japan, and Korea. It is the fourth most produced minor millet, providing food security to many poor people across the world. Globally, India is the biggest producer of barnyard millet, both in terms of area (0.146 m ha–1) and production (0.147 mt) with average productivity of 1034 kg/ha during the last 3 years (IIMR, 2018). The details on major areas of cultivation and worldwide production are presented in Figures 1, 2. Barnyard millet is primarily cultivated for human consumption, though it is also used as a livestock feed. Among many cultivated and wild species of barnyard millet, two of the most popular species are Echinochloa frumentacea (Indian barnyard millet) and Echinochloa esculenta (Japanese barnyard millet) (Sood et al., 2015). Barnyard millet is a short duration crop that can grow in adverse environmental conditions with almost no input and can withstand various biotic and abiotic stresses. In addition to these agronomic advantages, the grains are valued for their high nutritional value and lower expense as compared to major cereals like rice, wheat, and maize. It contains a rich source of protein, carbohydrates, fiber, and, most notably, micronutrients like iron (Fe) and zinc (Zn) (Singh et al., 2010; Saleh et al., 2013; Chandel et al., 2014) that are related to numerous health benefits (Saleh et al., 2013; Ugare et al., 2014). All these features make barnyard millet an ideal supplementary crop for subsistence farmers and also as an alternate crop during the failure of monsoons in rice/major crop cultivating areas (Gupta et al., 2009).
FIGURE 1.
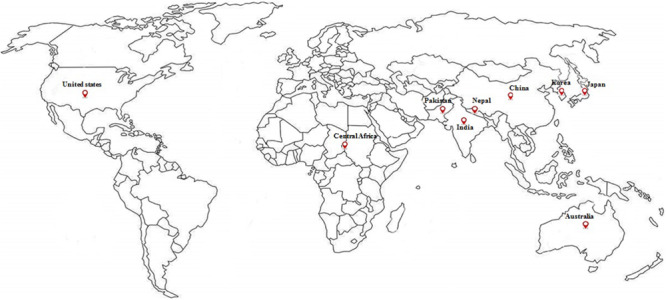
Major cultivation and production of barnyard millet around the world (from IIMR, 2018).
FIGURE 2.
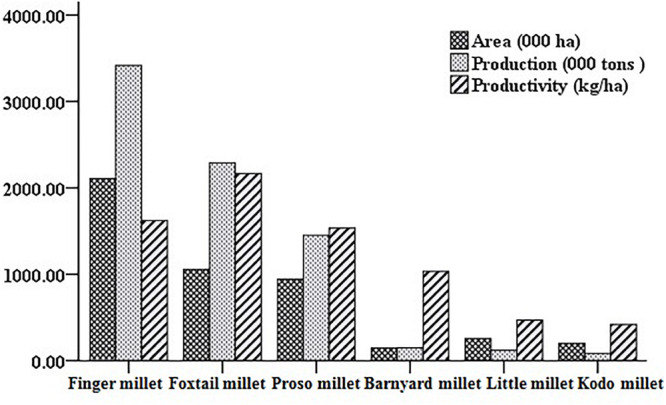
World area, production, and productivity scenario of small millets (from IIMR, 2018).
Despite barnyard millet’s excellent nutritional and agronomic value, the lack of awareness has led this crop to be considered as a neglected and underutilized crop. Over the past decades, efforts made to study the features of barnyard millet are limited compared to other minor millets. So far, most of the studies have been aimed at exploring the knowledge of diversity within the germplasm through morphological (Mehta et al., 2005; Gupta et al., 2009; Nirmalakumari and Vetriventhan, 2009; Sood et al., 2015; Renganathan et al., 2017) and molecular markers (Nozawa et al., 2006; Altop and Mennan, 2011; Prabha et al., 2012; Wallace et al., 2015; Manimekalai et al., 2018; Murukarthick et al., 2019). Also, several studies disclosed the nutritional profile of barnyard millet, particularly the high Fe and Zn content in the grains (Veena et al., 2005; Saleh et al., 2013; Chandel et al., 2014; Renganathan et al., 2017). However, comprehensive research is needed to understand the accurate details of germplasm accessions, identify the trait-specific donors, develop mapping population, and discover the quantitative trait locus (QTLs) and gene. Genomic resources are helpful for the progress of any crop species and they assist effective characterization of germplasm resources and their subsequent use in the discovery of QTL/gene(s) for the crop improvement program. However, genome research in barnyard millet is still in the early stage and far behind the other minor millets. This is mainly due to the complex nature of the genome (2n = 6x = 54, hexaploid). Recently, second and third-generation sequencing technologies unlocked several genome sequencing issues and facilitated to identify the genome sequence of wild and transcriptome sequences in cultivated Echinochloa species (Li et al., 2013a, b; Yang et al., 2013; Nah et al., 2015; Xu et al., 2015; Guo et al., 2017; Murukarthick et al., 2019). These genome resources facilitated the chance for better genotyping studies such as genetic diversity analysis, development of highly dense linkage maps and accurate physical maps, and detection of QTLs associated with micronutrients and agronomic traits. For instance, Wallace et al. (2015) developed the genome through single nucleotide polymorphism (SNP) markers and analyzed the genetic diversity in the barnyard millet core collection. Murukarthick et al. (2019) investigated the transcriptional changes between E. frumentacea and E. cru-galli and discovered genes related to drought and micronutrient content.
Accumulating evidence suggests that the interest in barnyard millet research has increased markedly over recent years; since 2010, more than 350 publications on barnyard millet have been available in the National Center for Biotechnology Information (NCBI) PubMed database1 (last accessed on December 2019) (Figure 3A). This review discusses the origin and taxonomy, nutritional value and health benefits, stress adaptation as well as the current status of genetic and genomics research in barnyard millet. The final section highlights the research gap and future research directions needed to promote barnyard millet as a potential crop for food and nutritional security.
FIGURE 3.
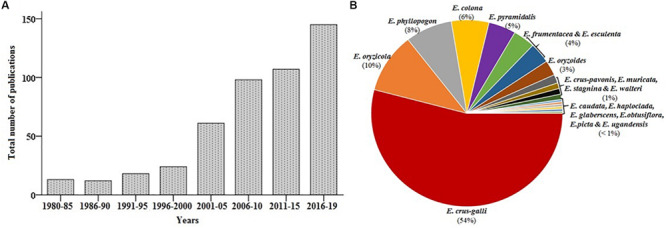
Graphical representations of literature and nucleotide sequence availability. (A) PubMed (https://www.ncbi.nlm.nih.gov/pubmed) (B) nucleotide sequence (https://www.ncbi.nlm.nih.gov/). (Note: Data verified till December 2019).
Origin, Taxonomy and Genomic Relationship of Echinochloa Species
Barnyard millet belongs to the genus Echinochloa, the family Poaceae, and the sub-family Panicoideae (Clayton and Renvoize, 2006). The genus Echinochloa consists of approximately 250 annual and perennial species that are widely distributed in the warmer and temperate parts of the world (Bajwa et al., 2015). However, the lack of clarity over the Echinochloa species makes it hard to differentiate themselves via the morphological markers due to low interspecific and intraspecific variations in nature and their phenotype plasticity (Chauhan and Johnson, 2011). Despite this challenge, 35 species have been identified to date for their taxa and phylogenetic relationship through morphological, cytological, and molecular marker studies (Yabuno, 1966, 1987; Yuichiro et al., 1999; Yamaguchi et al., 2005). Among them, most of the Echinochloa species, including E. crus-galli (allohexaploid, 2n = 6x = 54), E. colona (allohexaploid, 2n = 6x = 54), E. oryzicola (allotetraploid, 2n = 4x = 36), and others, have been designated as problematic weeds in major crop fields (Yabuno, 1966, 1987; Wanous, 1990; Yuichiro et al., 1999; Yamaguchi et al., 2005; Kraehmer et al., 2015). E. crus-galli is a predominant weed in rice fields in more than 60 countries, due to its quick germination (even in hypoxic conditions, up to 100 mm deep), rapid growth, mimicking character of rice, broad ecological tolerance, and profuse seed production (Barrett, 1983).
Echinochloa species have very few cultivatable forms and thereby are cultivated as minor millet by marginal farmers in warmer and temperate regions of the world. E. frumentacea (Roxb.) Link; syn. E. colona var. frumentacea (allohexaploid, 2n = 6x = 54), commonly known as Indian barnyard millet, originated from wild E. colona (L.) (Jungle rice), and exhibits a parallel line of evolution in India and Africa. E. frumentacea species has four races, namely stolonifera, intermedia, robusta, and laxa, that are widely cultivated in Central Africa, India, Malawi, Nepal, Pakistan, and Tanzania (Doggett, 1989; Upadhyaya et al., 2014). Another cultivated allohexaploid species, E. esculenta (A. Braun) H. Scholz; syn. E. utilis var. esculenta; known as Japanese barnyard millet, originated from wild E. crus-galli (L.) (Barnyard grass) was domesticated some 4,000 years ago in the temperate regions of Japan (De Wet et al., 1983; Doggett, 1989). Utilis and intermedia are two races of E. crus-galli, widely cultivated in Japan, Korea, China, Russia, and Germany (De Wet et al., 1983; Yabuno, 1987; Upadhyaya et al., 2014). Both wild and cultivated Echinochloa species are different from each other in terms of growth habitat, general morphology, and other characteristics (Table 1). The interspecific relationship between Echinochloa species was unclear till a series of prominent taxonomic reports by Yabuno, (1962, 1984, 1996, 2001). The interspecific hybrids between wild species and its progenitor, i.e., E. crus-galli × E. esculenta and E. colona × E. frumentacea produce normal meiotic division (27 bivalents) i.e., fertile. But, interspecific hybrids between two cultivated species and their respective wild counterparts, E. esculenta × E. frumentacea and E. crus-galli × E. colona, showed irregular meiotic division that leads to sterility (Yabuno, 1966, 1984). Collectively, all the cytological studies reveal the poor genomic affinity among species of Echinochloa. Besides, Yamaguchi et al. (2005) confirmed three cross compatible groups identified by Yabuno (1966) using chloroplast DNA sequence analysis, and separated these Echinochloa complexes into E. oryzicola complex, E. crus-galli complex, and E. colona-frumentacea complex. The E. oryzicola complex consists of two weedy species, E. oryzicola and E. phyllopogon, and one rare cultivated Mosuo barnyard millet. The E. crus-galli complex includes four wild species, E. crus-galli var. crus-galli, E. crus-galli var. praticola, E. crus-galli var. formosensis, and E. crus-galli var. oryzoides, and one major cultivated species, E. esculenta (Japanese barnyard millet). The third, E. colona-frumentacea complex, consists of one wild species, E. colona, and one major cultivated species, E. frumentacea (Indian barnyard millet). Further, through molecular analysis, Aoki and Yamaguchi (2008) reported that, though all these three groups exhibit different cytoplasmic lineages, the nuclear lineage between E. oryzicola complex and E. crus-galli complex have a higher affinity than E. colona-frumentacea complexes proving Yabuno’s hypothesis that E. oryzicola is the probable paternal parent of E. crus-galli (Aoki and Yamaguchi, 2008). However, information regarding ancestors of E. colona and their cultivated E. frumentacea remains almost unknown. Therefore, analyses the meiotic behavior of inter or intra specific hybrid combinations through advanced cytogenetic techniques like genomic in situ hybridization (GISH)/fluorescent in situ hybridization (FISH) would not only provides their ancestral information but also differentiate many unresolved genomes of Echinochloa species.
TABLE 1.
Morphological differences among wild and cultivated species of Echinochloa species.
| Traits | Echinochloa colonaa | Echinochloa crus-gallia | Echinochloa frumentacea | Echinochloa esculenta |
| Common name | Jungle rice | Barnyard grass | Indian barnyard millet/sawa millet | Japanese barnyard millet |
| Synonyms | Echinochloa colonum, Echinochloa crus-galli subsp. colona, Panicum colonum, Panicum cumingianum, Panicum zonale, and Milium colonum, Oplismenus colonus | Panicum crus-galli, Panicum hispidulum, Milium crus-galli, and Pennisetum crus-galli | Billion Dollar grass, sawa millet, sama millet | Japanese millet, marsh millet, Siberian millet, and white millet |
| Origin | China and Japan | China, Japan, and Korea | India, Pakistan, and Nepal | Eastern Asia, Japan, China, and Korea |
| Distribution | Widely distributed in South and Southeast Asia, Australia, Africa, Europe, and America | Widely distributed in South and Southeast Asia, Africa, Europe, and America | Widely distributed in Central Africa, Africa, Temperate Asia, Tropical Asia, Australasia, and South America. | Widely distributed in India, Temperate Asia, Australasia, and Pacific. |
| Habitat | Annual, warmer region | Annual, temperate region | Annual, warmer region | Annual, warm-season |
| Cultivable range | – | – | Altitude below 1,900 m | Altitude up to 2,500 m |
| General morphology | Erect to decumbent, 60 cm tall, short leaf length 10-15 cm, red tinges at the basal portion of leaf, leaf blade surface smooth, leaf blades 3–30 cm long and 2–8 mm wide | Erect, 200 cm tall, leaf 10-40 cm long, leaf blade surface smooth, leaf blades 0.5–35 cm long and 6–20 mm wide | Erect, 242 cm tall, leaf length 15–40 cm long and 1–2.5 cm wide, plants mostly green, however, purple tinges also found in vegetative and reproductive parts, leaf blades are smooth and glabrous, culms slender to robust | Robust, 60–122 cm tall, leaf sheath smooth 10–50 cm long and 7–25 mm wide, plants green, however, light to dark purple pigmentation in various plant parts, thicker stem |
| Inflorescence morphology | Green to purple, inflorescence length 5-15 cm, simple ascending racemes 5–15 numbers, 0.5–3 cm long raceme | Green to purple, inflorescence length 10-25 cm, compound ascending racemes 5–15 numbers, 2–10 cm long, slightly hairy, green to purplish awns 2-5 mm long | Green to purple, usually erect and compact, inflorescence length 1-28 cm long, racemes numerous 20–70, 1–3 cm long, rarely drooping, awnless | Brown to purple, compact inflorescence 12–15 cm long, racemes 5–15 numbers, arcuate to flexuous, 0.5–3 cm long, rarely awned |
| Spikelets on panicle | Spikelets arranged in 4 uniform rows on the primary rachis, spikelets 1-3 mm long | Open, branched spikelets on the rachis, setae on the primary branches | Compact, non-branched spikelets on the rachis, tightly clustered, 2–4 mm long, acute and awnless. | Compact, branched spikelets, larger spikelets and longer primary branches, dense clustered, 3–4 mm long, shortly cuspidate and rarely awned. |
| Caryopsis | Straw white, brown | Dark gray, brown | Turgid, whitish | Dull, pale yellow to light brown |
| Seed dormancy | Present | Absent | Present | Absent |
| Seed shattering | High | Low | High | Low |
Plant Architecture, Floral Biology and Seed Traits
The cultivated barnyard millet is an annual, robust, and tall crop that grows up to a height of 220 cm (Denton, 1987; Padulosi et al., 2009). The inflorescence is a terminal panicle with varying shapes (cylindrical, pyramidal, and globose to elliptic), colors (green, light purple, and dark purple) and compactness (compact, intermediate, and open) (Gupta et al., 2009; Prakash and Vanniarajan, 2013; Sood et al., 2014; Renganathan et al., 2017; Kuraloviya et al., 2019) (Figure 4). Racemes are present in one side, two sides, or around the axis of rachis and vary from 22 to 64 numbers per inflorescence (Renganathan et al., 2017). The arrangement of spikelets is either on one side or around the rachis of the raceme. Each spikelet contains two florets in which the lower floret is sterile and consists of lemma with small palea, while the upper floret is bisexual with a shiny lemma that partially encloses palea. Fertile lemma and palea have three stamens varying from a white color to a dark purplish color with stigma plumose and bifid, ranging from white to dark purple (Figure 4). The two unequal glumes further enclose the seed kernel (Gupta et al., 2010b; Singh et al., 2010). Anthesis and pollination progress in the direction from top to bottom of the inflorescence in the early morning (5 am) and reaches a maximum during 6 am -7 am, while it closes at 10 am (Sundararaj and Thulasidas, 1976; Jayaraman et al., 1997). Sundararaj and Thulasidas (1976) further reported on the requirement of 10–14 days of duration for the flowering process. Though self-pollination is a strict rule, the reception of stigmatic branches before dehiscence of anther provides some chances for cross-pollination (Seetharam et al., 2003).
FIGURE 4.

Genetic diversity of various morphological traits of barnyard millet. (Panel 1) Ear head diversity of different barnyard millet accessions. (Panel 2) Variation in ear head compactness (a) compact, (b) intermediate, and (c) open. (Panel 3) Variation in ear head shape (a) cylindrical, (b) pyramidal, and (c) globose to elliptic. (Panel 4) Grain variation in color and size of different barnyard millet accessions (a) straw white, medium (b) light gray, bold (c) gray, medium, and (d) dark gray, narrow. (Panel 5) variation in ear head color (a) green, (b) medium purple, and (c) dark purple. (Panel 6) Anther color variation (a) white, (b) light purple, and (c) dark purple. (Panel 7) Variation in tillering ability and pigmentation (a) high tillers, green, (b) medium tillers, light pigmentation, and (c) low tillers, dark pigmentation (from Prakash and Vanniarajan, 2013; Renganathan et al., 2017; Kuraloviya et al., 2019).
Compared to other minor millets like kodo and foxtail millet, barnyard millet grains are less hard. The mature pericarp of the seed consists of two epidermal layers with cells of the inner epidermis completely compressed over the outer epidermis (Singh et al., 2010). The cell wall of the aleuronic layer cutinized (Zee and O’brien, 1971), and also contains a maximum amount of carbohydrate (57–66%), followed by fiber (6.4–12.2%), protein (5–8.5%), fat (3.5–4.6%), and ash (2.5–4.0%) content. Starch granules are simple and are spherical to polygonal shapes with a diameter of 1.2–10 μm, which is larger than other small millets (Kumari and Thayumanavan, 1998). The pericarp color of grain differs among genotypes from straw white to light gray and dark gray (Renganathan et al., 2017; Kuraloviya et al., 2019). The seeds usually germinate easily under proper storage conditions at 12°C and are able to retain their viability for up to 13 months (Kannan et al., 2013) and beyond, although improper or poor storage may lead to loss of viability in both species of barnyard millet. The seed dormancy, a major limiting factor in the cultivation of small millets, has not been studied yet in detail. However, in barnyard millet, both wild and freshly harvested seeds of cultivated species reported to have seed dormancy (Maun and Barrett, 1986; Sung et al., 1987; Manidool, 1992). Although the deep physiological dormancy in E. crus-galli grain was the most probable feature for its prolonged existence (Song et al., 2015), the innate dormancy present in cultivated Echinochloa species further hinders the evaluation or multiplication of seeds in germplasm conservation centers (Kovach et al., 2010). Despite this, the dormancy breaking treatments in Echinochloa also varies with species; some accessions may require light or dark and cold or heat or a combination of both (Kovach et al., 2010). Seed application of 100 ppm of IAA (Indoleacetic acid) improved germination percentage (18%), speed of germination (5.58 days earlier), and increased the seed length (11%), dry matter (3.80%), and vigor index (21%) (Sujatha et al., 2013). In another study, barnyard millet seeds treated with Pseudomonas fluorescens enhanced the seed germination and seedling growth attributes in barnyard millet (Sridevi and Manonmani, 2016). This is mainly due to the direct suppression of deleterious pathogens or the indirect production of growth hormones that ultimately increases the uptake, solubilization, and translocation of less available minerals (Olanrewaju et al., 2017).
Echinochloa species are generally considered to be a short-day plant (Muldoon, 1985) exhibiting photoperiodism and perform as per the different ranges of photoperiods from short days (8–13 h) to long days (16 h) (Maun and Barrett, 1986; Mitich, 1990). For instance, the variety CO (Kv) 2 reported having variable flowering times in temperate and hot regimes within regions of southern parts of India, with hindered uniform grain yield across the state (Vanniarajan et al., 2018). To alleviate that, the latest variety MDU 1, released in Tamil Nadu, India, has been found to have a short duration along with stable grain yield across the state (Vanniarajan et al., 2018).
Relevance of Barnyard Millet in Climate Change and Nutritional Security
Responses to Biotic and Abiotic Stresses
The Echinochloa species generally has potential resistance against various biotic and abiotic stresses. However, cultivated species such as E. esculenta and E. frumentacea are widely threatened by pest and diseases (i.e., shoot fly, stem borer, grain smut, and loose smut) at different growth stages of the crop (Jain et al., 1997; Jagadish et al., 2008). Aphid’s infection at the vegetative stage causes considerable yield reduction to E. frumentacea. So far, DHBM 996 and TNEF-204 were found to be resistant genotypes for shoot fly and stem borer (Rawat et al., 2019). Meanwhile, Kim et al. (2008) reported that some E. frumentacea accessions have the potential for antifeeding activity against brown plant hopper, which is among the major pests that affect rice production. On the other hand, loose smut (Ustilago tritici) and grain smut (Ustilago panici frumentacea) are major fungal diseases that affect the grain formation in both the cultivated species of Echinochloa (Jain et al., 1997; Gupta et al., 2010a). A heavy infestation of smuts during head formation leads to a significant reduction in grain yield and quality (Gupta et al., 2010a). However, Nagaraja and Mantur (2008) and Gupta et al. (2010a) showed that some of the E. esculenta accessions had the immunity against both smut diseases and further provide the chance to breed the resistance lines.
Abiotic stresses are a major threat to important food crops such as rice, wheat, and maize, and cause serious yield loss across the world. However, Echinochloa species have a high degree of tolerance to various abiotic stresses (Gupta et al., 2010b; Singh et al., 2010). For instance, a recent investigation from Arthi et al. (2019) showed that among the 89 Echinochloa accessions, CO (Kv) 2, MDU 1, PRJ1, TNEf 301, TNEf 204, TNEf 361, TNEf 364, and VL 29 exhibited better germination as compared to the rice variety, White Ponni, at 200 mM NaCl concentration. Similarly, Echinochloa species are also the preferable choice of farmers for cultivation in various adverse environments such as those prone to drought or flooding. These features showed that the Echinochloa species might have some specialized rhizosphere organizations that can facilitate the uptake and release of oxygen (O2) from their roots at stressful conditions. Zegada-Lizarazu and Iijima (2005) reported higher water uptake efficiency (deep root) of barnyard millet (E. utilis) over other minor millets, including pearl millet, and found that barnyard millet sustained and increased the water use efficiency, leaf area index, and dry matter production in both drought and flooding conditions. Therefore, it is also worth investigating the Echinochloa species mechanism behind the tolerance to drought and flooding stress. Further, identification and characterization of genes and pathways associated with resistance to saline, drought, and flooding stress in Echinochloa species may not only be useful to develop superior cultivars but also assist in improving the tolerance in a major cereal crop. It is also well known for its excellent nitrogen-use efficiency over cereal crops (Goron and Raizada, 2015) and has been recommended as a natural phyto-extractor in heavy metal (lead, cadmium, and chromium) contaminated soils and sodic soils due to hyper accumulation nature. Since heavy metals are currently of much environmental concern, phyto-based soil reclamation is an alternative, cost-effective, and eco-friendly approach (Subhashini and Swamy, 2014) that needs to be imparted in soil health restoration programs.
Nutritional Significance and Health Benefits
In terms of nutritive value, barnyard millet is superior to major and minor millets. Barnyard millet grains are a rich source of dietary fiber, iron, zinc, calcium, protein, magnesium, fat, vitamins, and some essential amino acids (Singh et al., 2010; Saleh et al., 2013; Chandel et al., 2014). The nutritional composition of barnyard millet is presented in Supplementary Table S1. The average carbohydrate content of barnyard millet varies between 51.5 and 62.0 g/100 g (Saleh et al., 2013), which is lower than that of other major and minor millets. Ugare et al. (2014) reported that the crude fiber of barnyard millet is higher than any other cereal, ranging between 8.1 and 16.3%. The high ratio of carbohydrate to crude fiber ensures the slower release of sugars in the blood, and so thus aids in maintaining blood sugar level. The resistant starch in barnyard millet has shown to lower blood glucose, serum cholesterol, and triglycerides in rats (Kumari and Thayumanavan, 1998). In a clinical study with human volunteers, Ugare et al. (2014), confirmed a lower glycemic index (GI) in type 2 diabetic groups during regular consumption of barnyard millet meal. Existing evidence showed that the protein content (11.2–12.7%) in barnyard millet was reasonably higher than other major cereals and millets. Although the total minerals, ash, fat, and amino acid content in barnyard millet were although comparable with other cereals and millets, the iron content in the grain was significantly higher than others. For instance, the iron content in barnyard millet grain is about 15.6–18.6 mg/100 g (Saleh et al., 2013; Renganathan et al., 2017; Vanniarajan et al., 2018), which is rationally higher than major cereals and millets. In addition, a lower phytate (3.30–3.70 mg/100 g) content in grains (Panwar et al., 2016) followed by the dehulling process has also decreased phytic acids significantly, favoring the bioavailability of minerals. This makes barnyard millet an ideal food not only for people with lifestyle diseases, but also for anemic patients and especially women in developing countries. The polyphenols and carotenoids are known to have several potential benefits to humans, and are twofold higher in barnyard millet than finger millet (Panwar et al., 2016). Similarly, alkaloids, steroids, carbohydrates, glycosides, tannins, phenols, and flavonoids present in barnyard millet have various ethno-medical properties like being antioxidant, anti-carcinogenic, anti-inflammatory, antimicrobial, having a wound healing capacity, biliousness, and alleviating constipation-associated diseases (Kim et al., 2011; Ajaib et al., 2013; Moreno Amador et al., 2014; Borkar et al., 2016; Nguyen et al., 2016; Sharma et al., 2016; Sayani and Chatterjee, 2017). Collectively, all these features make the barnyard millet a suitable and secured food for present-day consumers in their overall physical and nutritional well-being.
Physico-Chemical Properties and Relationship With Cooking and Value Addition
The studies on the physical and mechanical properties of grains are an important criterion to design any processing instruments like dehullers, polishers, sorters, storage, and other processing machineries (Singh et al., 2010). In barnyard millet, grain moisture is the prime criteria playing a key role not only in storage but also in the development of processing machineries. The moisture level of barnyard millet grain highly influences the quality as well as the time of milling and polishing (Lohani et al., 2012). For instance, the 8% moisture content of the grain is better for polishing than at 14% moisture. However, at 14% moisture, the degree of polishing increases grain recovery and decreases the loss of protein, fat, ash, and fiber. Based on this, Lohani et al. (2012) suggested 10% as the optimum moisture level for polishing. Similarly, physical (grain diameter, grain surface area, 1,000 grain mass, true density, dynamic angle of repose, coefficient of internal friction, and coefficient of static friction), aerodynamic (terminal velocity), and mechanical (specific deformation and rupture energy) properties are other major parameters influenced by the moisture content of the grains. All these factors ultimately influence the processing of grains in the machines (Singh et al., 2010). Therefore, a study in detail on these properties should be conducted in order to design and develop better milling, polishing, grading, and sorting machineries for barnyard millet.
The cooking and flour quality of the grain are primary standards to assess consumer acceptability. At the same time, different processing techniques aim to increase the storage time of grain/flour as well as increase the physicochemical accessibility of nutrients with reduced anti-nutrient losses during consumption. Barnyard millet grains are usually parboiled-dehulled-cooked and consumed in a similar way to rice (Surekha et al., 2013). It requires about 12 min to cook. The grain can also transform into flour for the preparation of various food formulation by processing techniques. Nevertheless, different processing methods cause the variations in the functional, nutritional, anti-nutritional, and pasting properties of barnyard millet flour (Nazni and Shobana, 2016). In addition, physical parameters, such as bulk density and porosity are important criteria in flour storage and oxidation related problems. Nazni and Shobana (2016) compared the flour of raw and germinated rice for setting up different processing methods for storage and transportation. The study found that the germinated flour exhibited decreased bulk density and porosity (air spacing) than raw rice flour. Therefore, the germinated flours are comparatively less prone to autoxidation than raw rice due to reduced air space between the flour molecules, and this could prevent the spoilage of flour and facilitate easy packaging and enable long-distance transportation. Apart from that, the germinated flours also have an increased oil absorption capacity that makes the flour suitable for enrichment in flavor and mouth feel. Similarly, value-added food products are not only free of anti-nutritional factors but also increase nutritional compounds, making barnyard millet a good base ingredient for infant food formulas. Besides, the flour is highly amenable for various food preparations such as baby foods, snacks, and dietary foods (Vijayakumar et al., 2009; Anju and Sarita, 2010; Surekha et al., 2013). The flour is also highly compatible to blend with other food flours for making novel or any value-added products without affecting the flavor and taste (Veena et al., 2004; Surekha et al., 2013). For instance, a ready-to-eat snack food can successfully be prepared with barnyard millet, potato mash, and tapioca powder in the ratio of 60:37:3 (Jaybhaye and Srivastav, 2015). However, tannin content (0.21–0.36%) in the grain affects the in vitro protein digestibility (IVPD). Although, compared to other small millets (kodo millet and finger millet), it was very low. Therefore, it is suggested that there are many chances for the application of several processing techniques to improve barnyard millet flour quality and nutritional properties, especially in the value-addition strategy of food industries.
Versatile Research and Industrial Applications
The amylose-rich barnyard millet starch has now attracted attention in biodegradable film making industries as an antioxidant packaging material (Cao et al., 2017). The incorporation of borage seed oil in barnyard millet starch increases the elongation range and decreases the tensile strength, water permeability, and moisture content properties of the starch, which makes it suitable for biofilm production. These biofilms are found to be resistant against various microbes and block light transparency and free radical formation in food industries (Cao et al., 2017). Research on Nanoparticle by Kumar et al. (2016) suggested the use of an aqueous extract from aerial parts of E. colona plant in the synthesis of silver nanoparticles (AgNPs) as a new eco-friendly approach in bio-synthesizing nanoparticle in plants. Such synthesis of AgNPs from plant extracts could be a safe and eco-friendly approach with possibilities for application at large-scale in the near future in the field of medicine, engineering, and agriculture. Mosovska et al. (2010) reported that E. esculenta extract showed antimutagenicity against 3-(5-nitro-2-furyl) acrylic acid in strains of Salmonella typhimurium due to its higher polyphenolic content, thereby playing a major role as antioxidants in scavenging H2O2 radicals. Similarly, a novel antifungal peptide, EcAMP1, was identified in the seeds of E. crus-galli, a unique antimicrobial peptide with a wide spectrum of antifungal activity against phytopathogens such as Alternaria, Botrytis, Fusarium, and Trichoderma. This peptide has a unique disulfide stabilized α-helical hairpin structure that intensively binds to the surface of fungal conidia, accumulates in the cytoplasm, and finally inhibits the elongation of hyphae without lysis of the cytoplasmic membrane (Nolde et al., 2011). This property could be exploited in future protein engineering technologies for the synthesis of novel antimicrobials in the agriculture and pharmaceutical industries. Besides all these, Barnyard millet has a higher straw yield and fodder value even at multiple cuttings (Bandyopadhyay, 2009). The fodder yield is about 6.3 tons/ha (Vanniarajan et al., 2018). Fodder contains a good amount of protein (7.6%), digestible fiber (23%), ash (12%), and fat (2.0%). Besides its superior feed quality, higher digestibility and nitrogen concentrations have meant barnyard millet is used as a potential livestock feed crop in the dry areas of the Deccan plateau to the extreme hills of the temperate sub-Himalayan region (Singh and Singh, 2005; Bandyopadhyay, 2009; Yadav and Yadav, 2013; Sood et al., 2015).
Germplasm Resources and Utilization
The major collections of barnyard millet germplasm accessions are housed in India and Japan. Vivekananda Parvatiya Krishi Anusandhan Sansthan (VPKAS), India; Indian Institute of Millets Research (IIMR), India; National Institute of Agrobiological Sciences (NIAS), Japan, and Consultative Group on International Agricultural Research like International Crop Research Institute for the Semi-Arid Tropics (ICRISAT), India are actively working on germplasm evaluation for various agronomic, biotic, and abiotic stresses, grain, and nutritional content traits in barnyard millet. India made a series of collaborative exploration missions (i.e., Indo-Australian missions, Indo-Japanese missions, and Indo-Soviet protocol) for the improvement of barnyard millet and other millets with different countries across the world. So far, six hundred and one exotic barnyard millet accessions have been introduced into India between the period of 1976 and 2007 to increase the food and fodder production (Gomashe, 2017). The major source of introduction was from Australia, Canada, France, Germany, Ghana, Italy, Japan, Kenya, Malawi, the Philippines, Russian Federation, South Africa, Spain, United States of America, and Yugoslavia. During this period, Indian barnyard millet accessions were also introduced in the United States, Canada, and Australia for feed and forage purposes (Gomashe, 2017). At present, 8,000 barnyard millet germplasms have been conserved at different centers throughout the world (Table 2).
TABLE 2.
Major organizations across the globe conserving the Echinochloa species (till December 2019).
| Country | Number of accessions | Organization |
| India | 1888 | National Bureau of Plant Genetic Resources, New Delhi |
| 985 | University of Agricultural Sciences, Bangalore | |
| 749 | International Crop Research Institute for the Semi-Arid Tropics, Patancheru | |
| 300 | Vivekananda Parvatiya Krishi Anusandhan Sansthan, Almora | |
| 1561 | Indian Institute of Millets Research, Hyderabad | |
| Japan | 3671 | National Institute of Agrobiological Sciences, Kannondai |
| 65 | Plant Germplasm Institute, Kyoto University | |
| United States | 232 | USDA Agricultural Research Service, Washington |
| 306 | National Centre for Genetic Resources Conservation, Collins | |
| 304 | North Central Regional Plant Introduction Station, Ames | |
| China | 717 | Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing |
| Kenya | 208 | National Gene bank of Kenya, Crop Plant Genetic Resources Centre, Muguga |
| Ethiopia | 92 | International Livestock Research Institute, Addis Ababa |
| Australia | 66 | Australian Plant Genetic Resource Information Service, Queensland |
| Pakistan | 50 | Plant Genetic Resources Program, Islamabad |
| Norway | 44 | Svalbard Gene Bank, Spitsbergen |
| United Kingdom | 44 | Millennium Seed Bank Project, Seed Conservation Department, Royal Botanic Gardens, London |
| Germany | 36 | Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben |
To obtain basic knowledge about germplasm, morphological characterization is the preliminary step for characterizing and classifying any collected/introduced materials. This not only provides the heritability of traits but also increases the utility of promising materials in breeding programs. In a study, Gupta et al. (2009) evaluated barnyard millet germplasm at the Himalayan regions of Uttarakhand, India, and identified some promising donors for plant height (<120 and >200 cm), productive tillers (>4), inflorescence length (>28 cm), raceme number (>50), raceme length (>3.1 cm), and grain yield (>16 g). In another study, to enable efficient use of genetic resources and to increase its access for breeders, barnyard millet core collection comprising of 89 accessions had also been established based on phenotypic and genotypic characterization (Upadhyaya et al., 2014). It was revealed that the mean difference between 89 core accessions and 736 whole accessions for most of the agronomic traits were not significant, indicating that the entire genetic variation had been sufficiently preserved in this core collection (Upadhyaya et al., 2014). The coefficient of variation among core germplasm varied from 0.79 to 36.43% for days to maturity and basal tiller number, while the heritability (broad sense) varied between 70.14 and 99.87% for inflorescence length and days to maturity, respectively.
Further, the multidimensional principal component analysis (PCA)-based phenotypic characterization of these 89 accessions resulted in three different groups for agronomic and other phenotypic traits based on its origin (Sood et al., 2015). The study has also identified some promising genotypes, which could be efficiently used in a breeding program for the improvement of early maturity, grain yield, and yield contributing traits. Similarly, the IIMR, Hyderabad, evaluated the 146 barnyard millet accessions and found a larger variation for grain yield and yield contributing traits, which led to the identification of 18 promising accessions for barnyard millet breeding programs (IIMR, 2016). A comparison of agronomic traits from various trials conducted across India is given in Supplementary Table S2, which revealed barnyard millet genotypes to have considerable variation for yield and yield-related traits. For instance, the genotypes with higher grain yield and yield contributing traits (panicle length, number of raceme, and grain yield) were identified in the Southern States of India viz., Telangana and Tamil Nadu. In contrast, early maturing (58–90 days) genotypes were mostly found in the Northern States of India.
On the other hand, registration of trait-specific germplasms in the National Gene Banks (NGB) not only protects the natural resources from Intellectual Property Rights (IPRs) but also facilitates the breeders to access important/valuable genotypes for any crop improvement programs. In crop plants, 60% of the registered traits of germplasms belong to cereals, oilseeds, and legumes for resistance against various biotic and abiotic stresses. With regard to cereals, the maximum number of germplasms were registered in paddy and wheat mainly for biotic stress related traits (Radhamani et al., 2011). However, in millets except for sorghum and pearl millet, most of them were registered with limited traits only, mainly, cytoplasmic male sterile (CMS) line in foxtail millet (Radhamani et al., 2011), waxy trait in proso millet (Santra et al., 2015), and easy de-hulling in barnyard millet. The registered barnyard millet genotype B29 by VPKAS, Almora, showed a 42–146.4% faster de-hulling percentage over other check varieties (Gupta et al., 2014). Therefore, despite the focus on higher grain yields alone, barnyard millet breeding programs should also include the strategy of registration of unique traits that might be conserved in the landraces, germplasms, or rejected entries from the evaluation trials.
The successful utilization of barnyard millet genetic resources resulted in the release of more than 20 varieties and cultures across India (Gomashe, 2017). The first variety of K1 was developed by the pureline selection method from local landraces of Tenkasi, Tamil Nadu, India, released during 1970, which possesses an average state yield of 1,000 kg ha–1. Later, several varieties were released against various pests and diseases across India through pureline selection from local landraces or exotic germplasm accessions. Among these, the notable variety PRJ 1, a direct selection from exotic collections of ICRISAT was released during 2003, by Vivekananda Institute of Hill Agriculture, Almora, Uttarakhand, India, possess a higher grain yield (2,500 kg ha–1) with resistance against various smuts (Upadhyaya et al., 2008). Recently, MDU 1, a variety developed by Agricultural College and Research Institute, Tamil Nadu Agricultural University, Madurai, India, through pureline selection of local landrace of Tamil Nadu possesses the characteristic features of short duration (<100 days) and higher grain yield (2,500 kg ha–1) (Vanniarajan et al., 2018). Besides, this variety also possesses a higher amount of iron content (16 mg/100 g) in the grains with good milling and cooking quality. In Japan, the “Noge-Hie,” a low amylose grain-containing cultivar was identified from a local landrace possessing natural deletion in one of three waxy genes (Hoshino et al., 2010). At the same time, “Chojuromochi,” a mutant developed through artificial γ-radiation, was completely devoid of Waxy (Wx) protein synthesis. In addition, the waxy protein trait was found to be stably inherited. Such glutinous variety in barnyard millet is in huge demand from Japanese consumers and industries for various food preparations similar to rice from paddy.
Hybridization is a difficult task in small millets, however, the hot water-based method followed by the contact method of crossing was found to be effective in finger millet (Raj et al., 1964; Nandini and Fakrudin, 1999) and foxtail millet (Siles et al., 2001). The same method was also applied in barnyard millet hybridization programs (Renganathan et al., 2015; Sood et al., 2015). Prior to pollination (early morning), the panicles which started flowering were selected for emasculation (Renganathan et al., 2015; Sood et al., 2015). The selected panicle was trimmed for 1/3rd portion by removing the opened and immature flowers in the respective upper and lower portions of the racemes, and then the remaining middle portion was immersed in hot water at 52°C in a thermos flask for 1 min. The emasculated panicles were then covered with butter paper bags to avoid contamination. The panicles in which flowering had already commenced were chosen as a pollinator source and panicle to panicle contacts were made by tying them together with a thread. The male and female panicles thus secured together were covered by a butter paper bag to avoid contamination with foreign pollen. However, priority should be given for the development of CMS line in barnyard millet for the better exploitation of variability as followed in foxtail millet.
Genomic Resources and Utilization
Whole genome sequence (WGS) is fundamental to understand the genome composition and gene repertoire of a crop. It helps to identify important genes and pathways related to economically important traits in crops. Recent advances in second and third-generation sequencing technologies have facilitated simple and cost-effective sequencing platforms to generate genome and transcriptome sequences. Among millets, the whole genome sequencing was completed in sorghum, pearl millet, foxtail millet, finger millet, and proso millet by various researchers (Zhang et al., 2012; Mace et al., 2013; Hittalmani et al., 2017; Varshney et al., 2017; Zou et al., 2019). Genomic resources are also considerably well-defined in sorghum, pearl millet, foxtail millet, and finger millet due to the presence of genetic linkage maps, physical maps, cytogenetic stocks, and large-insert libraries (as reviewed by Varshney et al., 2006; Gomashe, 2017). However, in barnyard millet, very limited attempts have been made to discover the genomic structure and associated downstream processes due to its genome complexity and lack of research funding on this orphan crop.
Chloroplast Genomes and Phylogeny Analysis
The chloroplast genome of Echinochloa is highly conserved for its genome structure, organization, and gene order (Ye et al., 2014). So far, chloroplast genomes of seven Echinochloa species including E. crus-galli, E. ugandensis, E. stagnina, E. colona, E. esculenta, E. frumentacea, and E. oryzicola (Ye et al., 2014; Perumal et al., 2016; Lee et al., 2017; Sebastin et al., 2019) have been sequenced. It revealed their genome structure (quadripartite), identity (99.5%), and size (139,592–139,851 bp) of the chloroplast genomes. The quadripartite structure generally confers a major impact on the evolution of plastome sequences of an organism (Yang et al., 2013). Such a quadripartite genome comprises a pair of inverted repeats (IR) separated by a small single-copy region (SSC) and a large-single copy region (LSC). The comparison of chloroplast genomes of wild and cultivated Echinochloa is given in Table 3. The size of IR, LSC, and SSC regions varied from 22,618 to 22,748, 81,837 to 82,053, and 12,518 to 12,519 bp, respectively. Similar to other angiosperms, the chloroplast genome of Echinochloa species comprises of 38.6% GC regions and 61% AT regions (Sebastin et al., 2019). In contrast, the number of genes varied from 111 to 131 among the species of Echinochloa, with the cultivated species exhibiting minimum. This could be mainly due to the reorganization of gene copy number and structure during the course of evolution or speciation. The divergence in copy number of any gene further creates the genetic polymorphism between the species, which contributes a major variation in their genome size and phenotype (Suryawanshi et al., 2016). As reported in angiosperms by Wendel (2015), the high morphological variation among the wild and its cultivated species occurs due to the consequences of genome reorganization during the evolutionary process. The comparative analysis of Echinochloa chloroplast genomes revealed that they are closer to the Panicum virgatum than other grasses (Ye et al., 2014). Further, molecular divergence clock analysis of grass species revealed that Echinochloa species had diverged 21.6 Mya than others. The wild species, such as E. oryzicola and E. crus-galli, diverged around 3.3 Mya while another wild species, E. colona, diverged from E. oryzicola and E. crus-galli between 2.65 and 3.18 Mya, respectively (Lee et al., 2017). Later, the cultivated species (E. frumentacea) diverged from E. oryzicola and E. crus-galli in and around 1.9–2.7 Mya (Perumal et al., 2016) (Figure 5). However, both wild and cultivated Echinochloa species have a high sequence identity with P. virgatum and Sorghum bicolor and low sequence identity with Triticum aestivum and Oryza sativa (Ye et al., 2014), which further concludes that Echinochloa species are more closely related to the P. virgatum and S. bicolor than Triticum and Oryza.
TABLE 3.
Summary statistics of chloroplast genomes for wild and cultivated barnyard millet.
| Genome composition | E. crus-gallia | E. colonab | E. esculentac | E. frumentacead |
| Genome size (bp) | 139,851 | 139,592 | 139,851 | 139,593 |
| Inverted Repeat size (bp) | 22,640 | 22,618 | 22,748 | 22,618 |
| Large-single copy region size (bp) | 82,053 | 81,837 | 81,837 | 81,839 |
| Small-single copy region size (bp) | 12,518 | 12,519 | 12,518 | 12,518 |
| Number of genes | 131 | 131 | 111 | 112 |
| Number of Protein coding genes | 88 | 88 | 76 | 77 |
| Number of tRNA | 40 | 40 | 30 | 30 |
| Number of rRNA | 4 | 4 | 4 | 4 |
| GC contents (%) | 38.6 | 38.6 | 38.6 | 38.6 |
FIGURE 5.
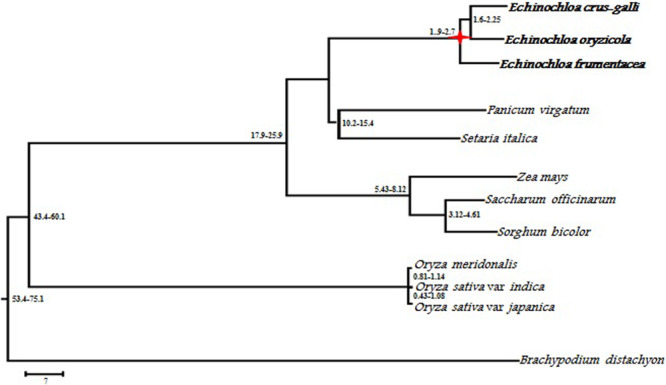
Phylogenetic tree showing relationships among Echinochloa species and grass species (from Perumal et al., 2016).
Transcriptome Analysis
With the advent of next-generation sequencing technologies (NGS), RNA-seq (RNA-sequencing) has now superseded the previous microarray technologies and a huge number of genomic resources are being generated in a cost and time effective manner (Weber, 2015). It not only generates differential genes, but also the functional molecular markers like simple sequence repeats (SSRs) and SNPs in various minor millet species. Enormous transcript profiles have been developed in weedy Echinochloa species till date for various traits associated with invasiveness and adaptations such as herbicide resistance, photosynthesis, flooding response, and other homeobox genes (Li et al., 2013a, b; Yang et al., 2013; Nah et al., 2015; Xu et al., 2015; Guo et al., 2017; Gao et al., 2018). Recently, transcriptome sequences developed from cultivated E. frumentacea variety CO (Kv) 2, yielded 97,065 transcripts with an average length of 94 Mbp (Murukarthick et al., 2019). Further de novo assembly, functional annotation, and comparison to E. crus-galli transcripts identified some key genes regulating Fe and Zn accumulation and drought tolerance. In addition, the study also generated 300 SSR primer pairs from 10,881 SSR loci targeting major repeats of trinucleotide (122) followed by dinucleotide (121), tetra-nucleotide (35), penta-nucleotide (20), and hexa-nucleotide (2).
A review of transcriptomes data in the NCBI database has revealed the presence of 952 gene sequences to date, generated from E. crus-galli (170), E. oryzicola (132), E. frumentacea (130), E. esculenta (130), E. colona (130), E. ugandensis (131), and E. stagnina (129). The details of transcriptome sequences published in the NCBI database were consolidated and presented in Table 4. Most of the genes were related to photosynthesis (PS I, PS II, NADH-plastoquinone oxidoreductase, ATP synthase), C4 pathways (phosphoenolpyruvate carboxylase, aldolase, maturase K, kinase), micronutrient transportation [Fe2+ transport protein 2-like protein (IRT2) gene, nicotianamine synthase 1 (NAS1), nicotianamine synthase 2 (NAS2), polymerases (RNA, DNA)], herbicide resistances (1-aminocyclopropane-1-carboxylic acid synthase 3, acetolactate synthase, calcineurin, cyclophilin 2, cytochrome P450, GH31, glutathione S-transferase), flooding tolerances (enolase, alcohol dehydrogenase), waxy grains (granule-bound starch synthase), non-shattering grains (sh4), ribosomal RNA, and transfer RNAs, etc. The proteomics exploration also revealed that a total of 540 proteins are found to be commonly expressed in Echinochloa species, of which most of the annotated protein sequences are tRNA, ribosomal, and other photosystem proteins. In addition, most of the proteins found in the Echinochloa species showed orthologs among themselves for proteins of C4 pathways, calcium binding protein, photosynthesis, bZIP transcription factor 1, translational initiation factors, transporters, and hypothetical proteins, etc. (Yang et al., 2013). However, some uniquely expressing proteins were also identified in the Echinochloa species. For instance, the maximum expression of quinclorac-resistant proteins, Cu/Zn superoxide dismutase, defensin, cadmium tolerant, viral nucleoprotein, and antimicrobial peptides was observed in E. crus-galli (Odintsova et al., 2008), multiple-herbicide-resistant proteins in E. phyllopogan (Iwakami et al., 2014), and granule-bound starch synthase in E. esculenta (Ishikawa et al., 2013). However, we require more research/data to draw a valid conclusion on the species-specific expression.
TABLE 4.
Details of transcriptome sequences of Echinochloa species.
| Species | Parts | Data generated (Gb) | Platform | Accession number |
| E. frumentacea | Leaf | 18.3 | ILLUMINA (Illumina HiSeq 2500) | SRX5210765 |
| E. frumentacea | Leaf | 48.6 | ILLUMINA (NextSeq 500) | SRX3029505 |
| E. esculenta | NA | 11.6 | ILLUMINA (Illumina HiSeq 2000) | SRX2698648 |
| E. crus-galli var. zelayensis | Leaf | 07.3 | ILLUMINA (Illumina HiSeq 2000) | SRX3574154 |
| E. crus-galli | NA | 32.8 | PacBio | SRX3081138 |
| E. colona* | Leaf | 11.6 | ILLUMINA (Illumina HiSeq 2500) | SRX2588690 |
| E. stagnina | Leaf | 02.4 | ILLUMINA (Illumina HiSeq 2500) | SRX3330321 |
| E. stagnina | Roots | 01.5 | ILLUMINA (Illumina HiSeq 2500) | SRX3330365 |
| E. glabrescens | NA | 05.0 | ILLUMINA (NextSeq 2000) | ERX990971 |
| Barnyard millet core collection | NA | 69.0 | ILLUMINA (Illumina HiSeq 2500) | SRX734221 |
∗Imazamox (herbicide) treated; NA, not available. Source: https://www.ncbi.nlm.nih.gov/search/all/?term=echinochloa..
Genome Sequence
Research conducted in China released the whole genome sequence of weedy E. crus-galli during 2017 and was annotated successfully for its unique nature of invasiveness and adaptation in the fields of crop plants (Guo et al., 2017). The total sequence length of the genome at a depth of 171× was estimated to the size of 1.27 Gb, representing around 90.7% of the predicted genome size. The genomic libraries range between2 160 bp and 20 Kb with a total number of contigs of 4534 with minimum and maximum contigs size of 1 kb and 11.7 Mb, respectively. The gene annotation of E. crus-galli further revealed 108,771 protein-coding genes, 785 miRNAs, 514 Mb repetitive elements, and non-coding RNAs. As of 2019, the genomic resources available in the NCBI domain include 1,246 nucleotide sequences, 822 gene sequences, 2,468 protein sequences, 105 short read archive (SRA) sequences, 74 Expressed Sequence Tags (ESTs), and one Gene Expression Omnibus (GEO) dataset related to various species of Echinochloa. Among the species E. crus-galli (652), E. oryzicola (126), E. phyllopogon (96), E. colona (76), E. pyramidalis (46), E. esculenta (44), E. frumentacea (43), and E. oryzoides (32) hold the maximum number of sequences (Figure 3B). To date, 54% of nucleotide sequences are available for E. crus-galli, while cultivated barnyard millet E. frumentacea and E. esculenta have only 4%. The comparative scenario of genomic resources among small millets is presented in Supplementary Table S3, which further emphasized the need to enrich the cultivated barnyard millet genome in the future.
Molecular Markers and Its Application
Genetic Diversity Analysis
Molecular markers are nucleotide sequences that are widely used for genetic diversity, linkage map construction, and marker assisted selection of crop plants (Muthamilarasan and Prasad, 2014). Early in the period of molecular marker research, Random Amplified Polymorphic DNA (RAPD) markers were utilized to access the genetic diversity and phylogeny among Echinochloa species (Hilu, 1994). Hilu (1994) proving that RAPD markers are effective in distinguishing both the cultivated and wild progenitors of the Echinochloa species at the genomic level. The genetic diversity among E. frumentacea was also found to be more diverse than E. utilis populations. However, isozyme marker analysis between these two species revealed that the accessions within the same species formed two different clusters and accessions from different species grouped into the same cluster, creating the possibility of the existence of intergrades and overlaps between the species (Prabha et al., 2010). Previously, Rutledge et al. (2000) obtained 90 polymorphic bands using 21 primer pairs with an average of 4.3 alleles per primer and Ruiz-Santaella et al. (2006) obtained 75 polymorphic bands using 13 primer pairs with an average of 5.8 alleles per primer. This suggests a low exhibition of polymorphism in the germplasm by the RAPD markers. The low level of polymorphism using RAPD markers has also been previously reported in finger millet diversity studies (Muza et al., 1995). Notwithstanding, the Amplified fragment length polymorphism (AFLP) marker system later developed a higher ability in revealing the genetic diversity in Echinochloa species compared to RAPD markers (Danquah et al., 2002; Tabacchi et al., 2009), since it generates more alleles per primer. For instance, a total of 166 polymorphic bands were produced in four primer pairs with an average of 41.5 per primer pairs in 28 genotypes. Whereas, the polymorphism information content (PIC) value of markers ranged between 0.44 and 0.52 (Kaya et al., 2014). This was in accordance with the previous report of Tabacchi et al. (2009), where seven primer pairs produced 156 polymorphic bands with an average of 22.3 alleles per primer in 80 genotypes. Similarly, polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) techniques were successfully applied for species identification in E. oryzicola and E. crus-galli (Yamaguchi et al., 2005; Yasuda et al., 2006). Recently, InDel markers for rbcL, matK, and ITS genes identified in E. colona have been widely used as a cost effective approach in the DNA barcoding of E. colona, E. oryzicola, and E. crus-galli (Lee et al., 2014). All these studies emphasized that with insufficient sequence information, the RAPD and isozymes markers are the only choice, not only helpful in differentiating Echinochloa species, but also in laying the foundation for molecular breeding in barnyard millet.
Later on, a significant development in the sequencing technologies further eliminated the limitations present in the RAPD, RFLP, and AFLP techniques through sequence-based markers such as SSRs, EST-SSRs (Expressed sequence tags-simple sequence repeats), and SNPs. The sequenced-based markers are more desirable in genetic diversity studies due to their co-dominant, reproducible, highly polymorphic, and effective utilization in many crop plants (Lin et al., 2011). The information available on sequence-based markers in barnyard millet is still in its infancy, despite the reports of microsatellite markers related to genetic diversity studies in germplasm accessions gaining attention today. For instance, utilizing five SSR markers, 155 accessions of Echinochloa species including E. esculenta (49), E. crus-galli (94), and E. esculenta var. formosensis (12) were grouped into three separate clusters (Nozawa et al., 2006). The same study reported that the accessions belonging to E. esculenta were less diverse than those of E. crus-galli or E. esculenta var. formosensis. More recently, the ESTs markers also been proven to be a very informative and effective tool for the analysis of genetic diversity in many small millets. Extensive transcriptomics and annotation studies previously performed on herbicide resistant varieties of E. crus-galli resulted in 74 ESTs (Li et al., 2013b; Yang et al., 2013). However, those ESTs were limitedly used in the marker development and diversity studies in barnyard millet, since they are weedy ancestors. For instance, the in silico mining of E. crus-galli ESTs resulted in the identification of 22 pairs of EST-SSR primers (Babu and Chauhan, 2017). The study also reported that frequent SSR repeats were found to be tetra-nucleotide repeat followed by the penta- and hexa- nucleotide repeats. Among the repeats, GT (dimer), AGG and AGA (trimer), CAAA (tetra), TGTTT (penta), and AGACGA (hexa) were the most common type of repeat motifs in barnyard millet. On the other hand, a restriction-site associated DNA (RAD) approach combined with Illumina DNA sequencing strategy was performed in E. phyllopogon for the rapid and mass discovery of SSR and SNP markers by Chen et al. (2017). This study yields sequencing reads of 4132 contigs, of which 4710 are annotated to be putative SSRs and 49,179 are probable SNPs. Out of 4710 putative SSR markers, 78 were potentially polymorphic. Besides, the most frequent motif was AT and maximum motif length was dinucleotide type (>82%) followed by tri, tetra, penta, and hexa. The further validation of eight SSRs in four E. phyllopogon population resulted in 66 alleles with an average of 3.1–4.8 alleles from locus per population. Moreover, the study also identified a higher percentage of GC (48.9%) content in the genome, proving their successful nature of adaptation against freezing and desiccation, with GC% indicating more stability in an organism. Hence, SSRs and SNPs markers developed from E. phyllopogon may be very useful in studying not only the diversity, origin, and distribution of herbicides-resistant population (Osuna et al., 2011; Okada et al., 2013), but also for predicting gene location and molecular breeding in cultivated types. Recently, Manimekalai et al. (2018) and Murukarthick et al. (2019) used EST-SSR markers developed from the cultivated, E. frumentacea transcriptome sequence to analyze the genetic diversity of Indian barnyard millet germplasm. Manimekalai et al. (2018) used 51 EST-SSR markers to study the genetic diversity of 61 barnyard millet germplasms. Among 51 EST-SSR markers, 14 were polymorphic and produced 29 alleles with the PIC value ranging between 0.276 and 0.652. Similarly, Murukarthick et al. (2019) identified 10 polymorphic markers from 30 EST-SSRs and showed clear polymorphism in the 30 Indian barnyard millet germplasms.
Apart from SSR markers, a total of 21,000 SNPs were identified and characterized through the whole-genome genotyping-by-sequencing (GBS) method using core germplasm comprising of 95 barnyard millet accession (Wallace et al., 2015). About 10,816 out of 21,000 SNPs were spread across 65 biotypes of E. colona, and 8,217 SNPs across 22 biotypes of E. crus-galli. The SNPs discriminating among E. colona and E. crus-galli biotypes were 1,299 and 1,444, respectively. Further, population structure analysis with SNPs strongly separated these two species with four clusters in E. colona and three clusters in E. crus-galli (Wallace et al., 2015).
Gene/QTL Mapping
The use of molecular markers such as SSRs and SNPs provide opportunities for breeders to identify the QTL/gene(s) for important micronutrient and agronomical traits in barnyard millet. So far, many SSR and SNP markers (Wallace et al., 2015; Chen et al., 2017; Manimekalai et al., 2018; Murukarthick et al., 2019) have been developed to speed up the linkage map construction and QTL mapping in barnyard millet, but no genetic linkage map or QTLs published yet compares to other millets such as foxtail millet and finger millet (Supplementary Table S4). To date, two mapping studies only have been published on barnyard millet (Table 5). Ishikawa et al. (2013) identified functional SNP markers for waxy traits and found that these waxy traits are controlled by three loci, namely EeWx1, EeWx1, and EeWx3. The plants with functional alleles in all three loci exhibited normal amylose content (wild), while any one of the alleles (natural mutant) and/or completely homozygous mutant alleles (artificial mutant) exhibited low amylose and very low amylose content (waxy), respectively. Besides, the phylogenetic analysis also revealed that the waxy gene sequences are highly conserved among grass species. In another study, bulk segregant analysis (BSA) and 51 EST-SSR markers were used to analyze the F2 individuals of ACM 331 × MA 10, contrast parents for anthocyanin pigments, and the results showed that the SSR marker, BMESSR 39, was linked with anthocyanin pigments in barnyard millet (Renganathan et al., 2019). Conclusively, progress in barnyard millet genome mapping remains slow and is still in its initial stage. Moreover, these published two reports contain preliminary results only; further experimental investigation is required to apply for marker-assisted selection (MAS).
TABLE 5.
Molecular markers associated with waxy and anthocyanin pigment traits in barnyard millet.
| Markers | Trait | F2 Mapping population | Segregation ratio (χ2 analysis) | References |
| SNP | Waxy | Wild × Mutant (γ) and Natural mutant × Mutant (γ) | 3:1 and 63:1 | Ishikawa et al., 2013 |
| ESTSSR | Anthocyanin pigment | Pigmented × Green | 1:2:1 | Renganathan et al., 2019 |
SNP, single nucleotide polymorphism; ESTSSR, EST-derived simple sequence repeat, EMS, ethyl methane sulfonate.
Comparative Genomics and Synteny Analyses
Comparative genomics studies brought considerable benefit to barnyard millet crop. Mainly, SSR markers obtained from the cereals and millets were successfully utilized to characterize the barnyard millet germplasm. The summary of cross transferable molecular markers developed in other cereals and millet are presented in Table 6. Due to the non-availability of whole genome sequencing in barnyard millet, the genomes of rice, maize, finger millet, and foxtail millet have served as essential models to study the marker-based syntenic relationships. The genomic SSR (gSSR) markers developed through in silico mining of the foxtail millet sequence showed a high degree of cross-transferability in barnyard millet and other related small millet species. Among the 159 gSSRs, 58 were found to show consistent amplification in barnyard millet, that is, 91.3% cross-species amplification ability (Pandey et al., 2013). Similarly, 106 eSSR (EST-derived simple sequence repeats) markers from Setaria showed consistent amplification in millet and non-millet species and also exhibited high cross species transferability in barnyard millet (90.6%) (Kumari et al., 2013). Muthamilarasan and Prasad (2014) reported that 100 out of 5,000 intron-length polymorphic markers (ILP) mined from the foxtail millet genome showed amplification in various small millets with 94 percentage cross-transferability in barnyard millet. Yadav et al. (2014), also found that the rice genic SSR primers from calcium transporters and calcium kinases group showed 100% and 72.2% cross transferability, respectively, in barnyard millet.
TABLE 6.
Transferability details of cross cereal species markers to barnyard millet.
| Species | Marker | Number of amplified markers | PIC | Polymorphic marker | Cross-transferability (%) | References |
| Finger millet | eSSR | 15/104 | NA | NA | 14.2 | Arya et al., 2014 |
| gSSR | 20/132 | NA | NA | 15.5 | Arya et al., 2014 | |
| eSSR, gSSR | 33/101 | NA | NA | 55.4 | Krishna et al., 2018 | |
| SSR | 07/18 | 0.16–0.53 | 06 | 85.7 | Babu et al., 2018a | |
| SSR | 32/32 | 0.27–0.73 | 29 | 90.6 | Babu et al., 2018b | |
| Pearl millet | SSR | 10/32 | NA | NA | 31.2 | Arya et al., 2014 |
| Foxtail | SSR | 53/58 | NA | NA | 91.3 | Pandey et al., 2013 |
| eSSR | 106 | 0.00–0.48 | NA | 90.6 | Kumari et al., 2013 | |
| ILP | 94/100 | 0.03–0.47 | NA | 94.1 | Muthamilarasan and Prasad, 2014 | |
| SSR | 46/64 | 0.02–0.66 | NA | 73.4 | Gupta et al., 2013 | |
| SSR | 7/26 | NA | 2 | 65.38 | Krishna et al., 2018 | |
| Sorghum | eSSR | 42 | NA | NA | 80.9 | Yadav et al., 2014 |
| Rice | eSSR | 102 | NA | NA | 72.1 | Yadav et al., 2014 |
| SSR | 85/120 | 0.15–0.67 | 41 | 48.2 | Babu et al., 2018b | |
| Maize | SSR | 32/46 | 0.25–0.73 | 26 | 70.0 | Babu et al., 2018a |
SSR, simple sequence repeat; eSSR, EST-derived simple sequence repeat; gSSR, genomic simple sequence repeat; ILP, intron length polymorphism; PIC, polymorphic information content; NA, not available.
The orthologs and paralogs analyses of the genome E. crus-galli against some of the grass family revealed that the approximate divergence times of Oryza – Sorghum and Sorghum – Echinochloa were estimated to be 48.5 and 28.5 Mya, respectively, followed by the polyploidization and speciation events by 7.8 Mya (Guo et al., 2017). Three copies of gene clusters related to the biosynthesis of DIMBOA (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one), a unique allelopathic compound reported in maize (Frey et al., 2009), were found in the E. crus-galli genome and each of them showed a perfect synteny with segments of BX1-5 and BX8 of the maize genome. Similarly, E. crus-galli also exhibited synteny with rice for momilactones, a phytoalexin compound expressed to protect against blast pathogens (Guo et al., 2017). Babu and Chauhan (2017) also found homology of some barnyard millet ESTs against the chromosomal regions of 2, 5, 6, 8, 9, and 12 of rice, the waxy gene of maize, granule-bound starch synthase I (GBSSI-S) gene of Panicum repens, Setaria italica, Panicum miliaceum, and super oxide dismutase (SOD) gene of Colletotrichum eremochloae. On the other hand, Babu et al. (2018a;b) compared rice, maize, and finger millet gSSRs for cross species amplification in barnyard millet and reported that maize and finger millet SSRs exhibited higher PIC values, efficient cross species amplification, and polymorphism percentage than rice SSRs. However, the comparative genetic mapping between rice and barnyard millet showed several putative syntenic regions across the genome that regulated the traits including seed dormancy, plant height, panicle length, spikelet characters, leaf senescence, seed weight/yield-related traits, shattering character, root traits, blast resistance, brown plant hopper (BPH) resistance, and amylose content (Babu et al., 2018b). Eventually, using the available literature in the published reports, we concluded that EST-derived SSR markers had higher cross-genome amplification than genomic SSR markers, indicating higher conservation of the former than the latter across the species of the grass family. Therefore, cross transferability mechanisms could be exploited in barnyard millet for trait-based marker identification.
Conclusion and Future Prospects
Despite its nutritional and agronomic benefits, barnyard millet has remained an underutilized crop and has received very little attention from researchers as well as farmers across the globe. Barnyard millet breeding programs have stagnated due to limited funding from various funding agencies and research organizations. Therefore, considerable efforts are needed to develop varieties or hybrids with farmer/consumer preferred traits. More breeding programs have to be designed in the future for harnessing the genetic variability for high yield potential, yield stability, improved salinity tolerance, pest and disease resistance, as well as enhanced nutritional quality, especially micronutrient composition. However, the progress of barnyard millet breeding programs is very slow due to the lack of genetic and genomic resources. With respect to genetic resources, the size of the core collection in barnyard millet is comparatively less than that of other minor millets (foxtail and finger millet) and so far, breeding populations have not been developed. Therefore, core and mini-core collections representing maximum diversity as well as bi-parental and multiparent populations have to be established and evaluated for various nutritional and agronomic traits. These resources will be useful to track the genomic regions associated with targeted traits by the linkage-based QTL mapping, genome-wide association study (GWAS), and genomic selection (GS), as well as for the detection of candidate genes.
Despite genome research in barnyard millet being at its infancy and far behind other minor millets, transcriptome sequencing has allowed researchers to develop several genomic resources, including EST-SSRs and SNPs, that could be useful for marker-assisted breeding. However, extensive efforts are needed in the future to develop the reference genome, genome-wide SSR and SNP markers, construction of genetic linkage maps, and physical maps. The recent release of the genome sequence of a weedy ancestor (E. crus-galli), together with the genomic resources from major and minor millet crops, offers an initial framework for enriching genomic research in an orphan crop like barnyard millet by comparative genomic approaches. It is also fruitful to use the E. crus-galli genome as a reference genome for cultivated barnyard millet species similar to the case in bread wheat. It helps not only to understand the genome composition of cultivated barnyard millet species and increases mapping accuracy, but also helps us to know the effect of variants on protein function.
Barnyard millet is a potential crop for the biofortification of micronutrients. The grains of barnyard millet are rich in micronutrients (Fe and Zn) and hence, the identification of potential genes related to the accumulation of micronutrients (Fe and Zn) will be helpful to transfer these genes to high yielding barnyard millet cultivars or even to other major crops like rice, wheat, maize, etc. Strategies for the improvement of micronutrients in barnyard millet are presented in Figure 6, which are also applicable to other agronomic traits. Furthermore, barnyard millet is well adapted to both warm and temperate regions and it is a rich source of genes responsible for stress tolerance. Therefore, understanding the molecular mechanism of plant responses to stress in inherently stress-tolerant crops such as barnyard millet will be useful in developing highly stress-tolerant cultivars. So far, several stress tolerance genes were identified in barnyard millet, but the function of these genes has not been tested by overexpression studies, mainly due to the lack of a genetic transformation system. To date, very limited reports have been published on genetic transformation in barnyard millet. Therefore, there is an immense need to develop an efficient transformation system for barnyard millet in the future so that it also paves the way for functional genomics studies related to tolerance against biotic and abiotic stresses as well as micronutrient traits.
FIGURE 6.
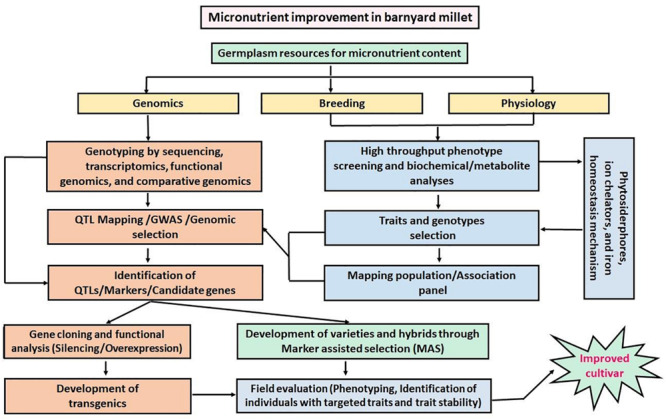
Schematic representation of the proposed strategy for micronutrient improvements in barnyard millet.
Besides these research gaps, the farming community is still unaware of the true potential of barnyard millet cultivation in terms of nutritional value and productivity. Farmers generally cultivate this crop under marginal areas, but they still depend on low yielding local landraces. Therefore, support from non-government organizations (NGOs) can help in increasing awareness among the farmers, stakeholders, nutritionists, and consumers to adopt and promote barnyard millet cultivation as well as consumption. Moreover, being a polyploid, ratooning (or) multi-cutting practices have to be standardized, like in sorghum, for better utilization of the growing season for grain and green fodder production. There is also an urgent need for advancements in post-harvest technologies for better processing and value-addition in the barnyard and other minor millets. At the same time, a change in consumer preference toward small millets with simultaneous development of suitable food products, along with an increase in market price, would fetch better returns for farmers and healthier choices for consumers. Finally, when these challenges are overcome, barnyard millet, being nutritionally sound and environmentally hardy, is going to be a promising crop for sustainable food and nutritional security in future climate scenarios.
Author Contributions
VR built the layout of the manuscript, collected literature, and wrote the manuscript. CV, JR, and AK provided suggestions. VR and AK revised and prepared the final draft of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Centre of Excellence – Post Doctoral program, Tamil Nadu Agricultural University to VR. All the authors would like to extend their sincere appreciation to Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu and India. We are also thankful to editors and reviewers for their detailed suggestions in improving the manuscript.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00500/full#supplementary-material
References
- Ajaib M., Khan K. M., Perveen S., Shah S. (2013). Antimicrobial and antioxidant activities of Echinochloa colona (Linn.) link and Sporobolus coromandelianus (Retz.) kunth. J. Chem. Soc. Pakistan. 35 1384–1398. [Google Scholar]
- Altop K. E., Mennan H. (2011). Genetic and morphologic diversity of Echinochloa crus-galli populations from different origins. Phytoparasitica 39 93–102. 10.1007/s12600-010-0135-3 [DOI] [Google Scholar]
- Anju T., Sarita S. (2010). Suitability of foxtail millet (Setaria italica) and barnyard millet (Echinochloa frumentacea) for development of low glycemic index biscuits. Malays. J. Nutr. 16 361–368. [PubMed] [Google Scholar]
- Aoki D., Yamaguchi H. (2008). Genetic relationship between Echinochloa crus-galli and Echinochloa oryzicola accessions inferred from internal transcribed spacer and chloroplast DNA sequences. Weed Biol. Manag. 8 233–242. 10.1111/j.1445-6664.2008.00303.x [DOI] [Google Scholar]
- Arthi N., Rajagopal B., Geethanjali S., Nirmalakumari A., Natesan S. (2019). Screening of barnyard millet (Echinochloa frumentacea) germplasm for salinity tolerance. Electron. J. Plant Breed. 10 659–666. 10.5958/0975-928X.2019.00083.8 [DOI] [Google Scholar]
- Arya L., Chauhan D., Yadav Y., Verma M. (2014). “Transferability of simple sequence repeat (SSR) markers developed in Finger Millet, and Pearl Millet to Kodo Millet and barnyard Millet,” in Innovative Approach in Stem Cell Research, Cancer Biology and Applied Biotechnology, eds Johri A. K., Mishra G. C., (New Delhi: Excellent Publishing House; )60–64. [Google Scholar]
- Babu B. K., Agrawal P. K., Pandey D., Jaiswal J. P., Kumar A. (2014a). Association mapping of agro-morphological characters among the global collection of finger millet genotypes using genomic SSR markers. Mol. Biol. Rep. 41 5287–5297. 10.1007/s11033-014-3400-6 [DOI] [PubMed] [Google Scholar]
- Babu B. K., Agrawal P., Pandey D., Kumar A. (2014b). Comparative genomics and association mapping approaches for opaque2 modifier genes in finger millet accessions using genic, genomic and candidate gene-based simple sequence repeat markers. Mol. Breed. 34 1261–1279. 10.1007/s11032-014-0115-2 [DOI] [Google Scholar]
- Babu B. K., Dinesh P., Agrawal P. K., Sood S., Chandrashekara C., Bhatt J. C., et al. (2014c). Comparative genomics and association mapping approaches for blast resistant genes in finger millet using SSRs. PLoS One 9:e99182. 10.1371/journal.pone.0099182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu B. K., Pandey D., Agrawal P. K., Sood S., Kumar A. (2014d). In-silico mining, type and frequency analysis of genic microsatellites of finger millet (Eleusine coracana (L.) Gaertn.): a comparative genomic analysis of NBS–LRR regions of finger millet with rice. Mol. Biol. Rep. 41 3081–3090. 10.1007/s11033-014-3168-8 [DOI] [PubMed] [Google Scholar]
- Babu B., Chauhan R. (2017). In-Silico Identification of EST Based Microsatellite Markers and SNPs, and Comparative Genomic Analysis of ESTs in Barnyard Millet for their Omics Applications. Curr. Agric. Res. J. 5 279–287. 10.12944/CARJ.5.3.03 [DOI] [Google Scholar]
- Babu B., Rashmi C., Sood S. (2018a). Cross transferability of finger millet and maize genomic SSR markers for genetic diversity and population structure analysis of barnyard millet. Indian J. Genet. Plant Breed. 78 364–372. 10.31742/IJGPB.78.3.5 [DOI] [Google Scholar]
- Babu B., Sood S., Kumar D., Joshi A., Pattanayak A., Kant L., et al. (2018b). Cross-genera transferability of rice and finger millet genomic SSRs to barnyard millet (Echinochloa spp.). 3 Biotech 8:95. 10.1007/s13205-018-1118-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu B. K., Joshi A., Sood S., Agrawal P. K. (2017). Identification of microsatellite markers for finger millet genomics application through cross transferability of rice genomic SSR markers. Indian J. Genet. 77 92–98. [Google Scholar]
- Bai H., Cao Y., Quan J., Dong L., Li Z., Zhu Y., et al. (2013). Identifying the genome-wide sequence variations and developing new molecular markers for genetics research by re-sequencing a landrace cultivar of foxtail millet. PLoS One 8:e73514. 10.1371/journal.pone.0073514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa A., Jabran K., Shahid M., Ali H. H., Chauhan B., Ehsanullah (2015). Eco-biology and management of Echinochloa crus-galli. Crop Prot. 75 151–162. 10.1016/j.cropro.2015.06.001 [DOI] [Google Scholar]
- Bandyopadhyay B. B. (1999). Genotypic differences in relation to climatic adaptation of two cultivated barnyard millet at Garhwal hills. Indian J. Genet. 59 105–108. [Google Scholar]
- Bandyopadhyay B. B. (2009). Evaluation of barnyard millet cultivars for fodder yield under single and double cut treatments at higher elevation of hills. Agric. Sci. Dig. 29 66–68. [Google Scholar]
- Barrett S. H. (1983). Crop mimicry in weeds. Econ. Bot. 37 255–282. 10.1007/bf02858881 [DOI] [Google Scholar]
- Bennetzen J. L., Schmutz J., Wang H., Percifield R., Hawkins J., Pontaroli A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30 555–561. 10.1038/nbt.2196 [DOI] [PubMed] [Google Scholar]
- Borkar V. S., Senthil Kumaran K., Senthil Kumar K. L., Gangurde H. H., Chordiya M. A. (2016). Ethno medical properties of Echinochloa colona and Hydrolea zeylanica: a review. World J. Pharmaceut. Res. 5 354–360. [Google Scholar]
- Cao T. L., Yang S. Y., Song K. B. (2017). Characterization of barnyard millet starch films containing borage seed oil. Coatings 7:183 10.3390/coatings7110183 [DOI] [Google Scholar]
- Chandel G., Meena R., Dubey M., Kumar M. (2014). Nutritional properties of minor millets: neglected cereals with potentials to combat malnutrition. Curr. Sci. 107 1109–1111. [Google Scholar]
- Chauhan B., Johnson D. E. (2011). Ecological studies on Echinochloa crus-galli and the implications for weed management in direct-seeded rice. Crop Prot. 30 1385–1391. 10.1016/j.cropro.2011.07.013 [DOI] [Google Scholar]
- Chen G., Zhang W., Fang J., Dong L. (2017). Identification of massive molecular markers in Echinochloa phyllopogon using a restriction-site associated DNA approach. Plant Divers. 39 287–293. 10.1016/j.pld.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. I., Chung J. W., Lee G. A., Ma K. H., Dixit A., Gwag J. G., et al. (2010). Development and characterization of twenty-five new polymorphic microsatellite markers in proso millet (Panicum miliaceum L.). Genes Genomics 32 267–273. 10.1007/s13258-010-0007-8 [DOI] [Google Scholar]
- Cidade F. W., de Souza-Chies T. T., Batista L. A. R., Dall’agnol M., Zucchi M. I., Jungmann L., et al. (2009). Isolation and characterization of microsatellite loci in Paspalum notatum Flüggé (Poaceae). Conserv. Genet. 10 1977–1980. 10.1007/s10592-009-9872-6 [DOI] [Google Scholar]
- Cidade F. W., de Souza-Chies T. T., Souza F. H. D., Batista L. A. R., Agnol M. D., Valls J. F. M., et al. (2010). Microsatellite loci for Paspalum atratum (Poaceae) and cross-amplification in other species. Am. J. Bot. 97 107–110. 10.3732/ajb.1000207 [DOI] [PubMed] [Google Scholar]
- Cidade F. W., Vigna B. B. Z., de Souza F. H. D., Valls J. F. M., Dall’Agnol M., Zucchi M. I., et al. (2013). Genetic variation in polyploid forage grass: assessing the molecular genetic variability in the Paspalum genus. BMC Genet. 14:50. 10.1186/1471-2156-14-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton W. D., Renvoize S. A. (2006). Genera Graminum: Grasses of the world in Kew Bulletin Additional Series XIII, Royal Botanical Gardens Kew. Chicago, IL: University of Chicago Press. [Google Scholar]
- Danquah E. Y., Hanley S. J., Brookes R. C., Aldam C., Karp A. (2002). Isolation and characterization of microsatellites in Echinochloa (L.) Beauv. spp. Mol. Ecol. Notes 2 54–56. 10.1046/j.1471-8286.2002.00144.x [DOI] [Google Scholar]
- De Wet J., Prasada Rao K., Mengesha M., Brink D. (1983). Domestication of mawa millet (Echinochloa colona). Econ. Bot. 37 283–291. 10.1007/BF02858883 [DOI] [Google Scholar]
- Denton D. C. (1987). “Food crops for waterfowl,” in Fireside Waterfowler: Fundamentals of Duck and Goose Ecology, eds Wesley D. E., Leitch W. G., (Mechanicsburg, PA: Stackpole Books; ), 352. [Google Scholar]
- Dida M. M., Srinivasachary, Ramakrishnan S., Bennetzen J. L., Gale M. D., Devos K. M. (2007). The genetic map of finger millet, Eleusine coracana. Theor. Appl. Genet. 114 321–332. 10.1007/s00122-006-0435-7 [DOI] [PubMed] [Google Scholar]
- Doggett H. (1989). “Small millets-a selective overview,” in Small Millets in Global Agriculture, eds Seetharam A., Riley K. W., Harinarayana G., (Oxford: Oxford; ), 3–18. [Google Scholar]
- Fang X., Dong K., Wang X., Liu T., He J., Ren R., et al. (2016). A high density genetic map and QTL for agronomic and yield traits in Foxtail millet [Setaria italica (L.) P. Beauv.]. BMC Genomics 17:336. 10.1186/s12864-016-2628-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M., Schullehner K., Dick R., Fiesselmann A., Gierl A. (2009). Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70 1645–1651. 10.1016/j.phytochem.2009.05.012 [DOI] [PubMed] [Google Scholar]
- Gao Y., Li J., Pan X., Liu D., Napier R., Dong L. (2018). Quinclorac resistance induced by the suppression of the expression of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase genes in Echinochloa crus-galli var. zelayensis. Pestic. Biochem. Physiol. 146 25–32. 10.1016/j.pestbp.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Gimode D., Odeny D. A., de Villiers E. P., Wanyonyi S., Dida M. M., Mneney E. E., et al. (2016). Identification of SNP and SSR markers in finger millet using next generation sequencing technologies. PLoS One 11:e0159437. 10.1371/journal.pone.0159437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomashe S. S. (2017). Barnyard millet: present status and future thrust areas. Millets Sorghum Biol. Genet. Improv. 134 184–198. 10.1002/9781119130765.ch7 [DOI] [Google Scholar]
- Goron T. L., Raizada M. N. (2015). Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution. Front. Plant Sci. 6:157. 10.3389/fpls.2015.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Qiu J., Ye C.-Y., Jin G., Lingfeng M., Zhang H., et al. (2017). Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 8:1031. 10.1038/s41467-017-01067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Joshi D., Mahajan V., Gupta H. (2010a). Screening barnyard millet germplasm against grain smut (Ustilago panici-frumentacei Brefeld). Plant Genet. Resour. 8 52–54. 10.1017/S1479262109990141 [DOI] [Google Scholar]
- Gupta A., Mahajan V., Gupta H. S. (2010b). “Genetic resources and varietal improvement of small millets for Indian Himalaya,” in Biodiversity Potentials of the Himalaya, eds Tewari L. M., Pangtey Y. P. S., Tewari G., (Gorakhpur: Gyanodaya Prakashan press; ), 305–316. [Google Scholar]
- Gupta A., Mahajan V., Kumar M., Gupta H. (2009). Biodiversity in the barnyard millet (Echinochloa frumentacea Link. Poaceae) germplasm in India. Genet. Resour. Crop Evol. 56 883–889. 10.1007/s10722-009-9462-y [DOI] [Google Scholar]
- Gupta A., Sood S., Agrawal P. K., Bhatt J. C. (2014). B 29: an Easy Dehulling Barnyard Millet (Echinochloa frumentacea Link) Genotype. Natl. Acad. Sci. Lett. 38 21–24. 10.1007/s40009-014-0295-7 [DOI] [Google Scholar]
- Gupta P., Raghuvanshi S., Tyagi A. (2001). Assessment of the efficiency of various gene promoters via biolistics in leaf and regenerating seed callus of millets, eleusine coracana and Echinochloa crusgalli. Plant Biotechnol. 18 275–282. 10.5511/plantbiotechnology.18.275 [DOI] [Google Scholar]
- Gupta S., Kumari K., Muthamilarasan M., Alagesan S., Prasad M. (2013). Development and utilization of novel SSRs in foxtail millet [Setaria italica (L.) P. Beauv.]. Plant Breed. 132 367–374. 10.1111/pbr.12070 [DOI] [Google Scholar]
- Hilu K. (1994). Evidence from RAPD markers in the evolution of Echinochloa millets (Poaceae). Plant Syst. Evol. 189 247–257. 10.1007/BF00939730 [DOI] [Google Scholar]
- Hittalmani S., Mahesh H. B., Shirke M. D., Biradar H., Uday G., Aruna Y. R., et al. (2017). Genome and Transcriptome sequence of Finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics 18:465. 10.1186/s12864-017-3850-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., Nakamura T., Seimiya Y., Kamada T., Ishikawa G., Ogasawara A., et al. (2010). Production of a fully waxy line and analysis of waxy genes in the allohexaploid crop, Japanese barnyard millet. Plant Breed. 129 349–355. [Google Scholar]
- Hou S., Sun Z., Li Y., Wang Y., Ling H., Xing G., et al. (2017). Transcriptomic analysis, genic ssr development, and genetic diversity of proso millet (Panicum miliaceum;Poaceae). Appl. Plant Sci. 5:1600137. 10.3732/apps.1600137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt H. V., Denyer K., Packman L. C., Jones M. K., Howe C. J. (2010). Molecular basis of the waxy endosperm starch phenotype in broomcorn millet (Panicum miliaceum L.). Mol. Biol. Evol. 27 1478–1494. 10.1093/molbev/msq040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IIMR, (2016). Annual Report 2016-17. Hyderabad: Indian Institute of Millets Research. [Google Scholar]
- IIMR, (2018). Annual Report 2017-18. Hyderabad: Indian Institute of Millets Research. [Google Scholar]
- Ishikawa G., Seimiya Y., Saito M., Nakamura T., Hoshino T. (2013). Molecular characterization of spontaneous and induced mutations in the three homoeologous waxy genes of Japanese barnyard millet [Echinochloa esculenta (A. Braun) H. Scholz]. Mol. Breed. 31 69–78. 10.1007/s11032-012-9769-9 [DOI] [Google Scholar]
- Iwakami S., Uchino A., Kataoka Y., Shibaike H., Watanabe H., Inamura T. (2014). Cytochrome P450 genes induced by bispyribac-sodium treatment in a multiple-herbicide-resistant biotype of Echinochloa phyllopogon. Pest Manag. Sci. 70 549–558. 10.1002/ps.3572 [DOI] [PubMed] [Google Scholar]
- Jagadish P. S., Mohapatra H. K., Chakravarthy M. K., Srivastava N., Nangia N. (2008). A Compendium of Insect Pests of Finger Millet and Other Small Millets. Available online at: http://www.aicrpsm.res.in/Downloads/Publications//A%20compendium%20of%20Insect%20pests%20of%20FM%20&%20other%20SM.pdf [Google Scholar]
- Jain A. K., Jain S. K., Yadava H. S. (1997). Assessment of yield losses due to grain smut in barnyard millet. Indian Phytopathol. 50 49–52. [Google Scholar]
- Jaiswal V., Gupta S., Gahlaut V., Muthamilarasan M., Bandyopadhyay T., Ramchiary N., et al. (2019). Genome-wide association study of major agronomic traits in foxtail millet (Setaria italica L.) Using ddRAD sequencing. Sci. Rep. 9:5020. 10.1038/s41598-019-41602-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman N., Suresh S., Nirmala Ganeshan N. M. (1997). “Genetic enhancement and breeding strategies in small millets,” in Proceedings of the National Seminar on Small Millets: Current Research Trends and Future Priorities as Food Feed and in Processing for Value Addition, Bangalore, 23–24. [Google Scholar]
- Jaybhaye R. V., Srivastav P. P. (2015). Development of barnyard millet ready-to-eat snack food: Part II. Food Sci. Res. J. 6 285–291. 10.15740/HAS/FSRJ/6.2/285-291 [DOI] [Google Scholar]
- Jia G., Huang X., Zhi H., Zhao Y., Zhao Q., Li W., et al. (2013). A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45 957–961. 10.1038/ng.2673 [DOI] [PubMed] [Google Scholar]
- Jia X., Zhang Z., Liu Y., Zhang C., Shi Y., Song Y., et al. (2009). Development and genetic mapping of SSR markers in foxtail millet [Setaria italica (L.) P. Beauv.]. Theor. Appl. Genet. 118 821–829. 10.1007/s00122-008-0942-9 [DOI] [PubMed] [Google Scholar]
- Johnson M., Deshpande S., Vetriventhan M., Upadhyaya H. D., Wallace J. G. (2019). Genome-Wide population structure analyses of three minor millets: kodo millet, little millet, and proso millet. Plant Genome 12:190021 10.3835/plantgenome2019.03.0021 [DOI] [PubMed] [Google Scholar]
- Joshi R. P., Jain A. K., Chauhan S. S., Singh G. (2015). Characterization of barnyard millet (Echinochloa frumentacea (Roxb.) Link.) landraces for agro-morphological traits and disease resistance. Electron. J. Plant Breed. 6 888–898. [Google Scholar]
- Joshi V. (2013). Assessment of genetic variability and identification of genotypes for different traits in Barnyard millet (Echinochola spp.). Int. J. Agric. Food Sci. Technol. 4 65–67. [Google Scholar]
- Kannan S. M., Thooyavathy R. A., Kariyapa R. T., Subramanian K., Vijayalakshmi K. (2013). Data from: Seed Production Techniques for Cereals and Millets. Seed node of the Revitalizing Rainfed Agriculture Network Centre for Indian Knowledge Systems (CIICS). Available at: http://www.ciks.org/downloads/seeds/5.%20Seed%20Production%20Techniques%20for%20Cereals%20and%20Millets.pdf (Accessed August 09, 2019). [Google Scholar]
- Kaya H., Demirci M., Tanyolac B. (2014). Genetic structure and diversity analysis revealed by AFLP on different Echinochloa spp. from northwest Turkey. Plant Syst. Evol. 300 1–11. 10.1007/s00606-013-0965-9 [DOI] [Google Scholar]
- Kim C. S., Alamgir K. M., Matsumoto S., Tebayashi S. I., Koh H. S. (2008). Antifeedants of Indian barnyard millet, Echinochloa frumentacea link, against brown planthopper, Nilaparvata lugens (Stål). Sect. C J. Biosci. 63 755–760. 10.1515/znc-2008-9-1022 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Chang J. K., Park B.-R., Han S.-I., Choi K.-J., Kim S.-Y., et al. (2011). Physicochemical and antioxidative properties of selected barnyard millet (Echinochloa utilis) species in Korea. Food Sci. Biotechnol. 20 461–469. 10.1007/s10068-011-0064-z [DOI] [Google Scholar]
- Kovach D. A., Widrlechner M., Brenner D. (2010). Variation in seed dormancy in Echinochloa and the development of a standard protocol for germination testing. Seed Sci. Technol. 38 559–571. 10.15258/sst.2010.38.3.04 [DOI] [Google Scholar]
- Kraehmer H., Jabran K., Mennan H., Chauhan B. (2015). Global distribution of rice weeds - A review. Crop Prot. 80 73–86. 10.1016/j.cropro.2015.10.027 [DOI] [Google Scholar]
- Krishna T. P. A., Maharajan T., David R. H. A., Ramakrishnan M. S., Ceasar A., Duraipandiyan V., et al. (2018). Microsatellite markers of finger millet (Eleusine coracana (L.) Gaertn) and foxtail millet (Setaria italica (L.) Beauv) provide resources for cross-genome transferability and genetic diversity analyses in other millets. Biocat. Agrl. Biotechnol. 16 493–450. [Google Scholar]
- Kumar A., Yadav S., Panwar P., Gaur V., Sood S. (2015). Identification of anchored simple sequence repeat markers associated with calcium content in finger millet (Eleusine coracana). Proc. Natl. Acad. Sci. India B Biol. Sci. 85 311–317. 10.1007/s40011-013-0296-1 [DOI] [Google Scholar]
- Kumar L. D., Siva Sankar S., Venkatesh P., Hepcy Kalarani D. (2016). Green synthesis of silver nanoparticles using aerial parts extract of Echinochloa colona and their characterization. Eur. J. Pharm. Med. Res. 3 325–328. [Google Scholar]
- Kumari K., Muthamilarasan M., Misra G., Gupta S., Alagesan S., Parida S., et al. (2013). Development of eSSR-Markers in Setaria italica and their applicability in studying genetic diversity, cross-transferability and comparative mapping in millet and non-millet species. PLoS One 8:e67742. 10.1371/journal.pone.0067742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari K. S., Thayumanavan B. (1998). Characterization of starches of proso, foxtail, barnyard, kodo, and little millets. Plant Foods Hum. Nutr. 53:47 10.1023/A:1008083020810 [DOI] [PubMed] [Google Scholar]
- Kuraloviya M., Vanniarajan C., Vetriventhan M., Babu C., Kanchana S., Sudhagar R. (2019). Qualitative characterization and clustering of early-maturing barnyard millet (Echinochloa spp.) germplasm. Electron. J. Plant Breed. 10:535 10.5958/0975-928x.2019.00067.x [DOI] [Google Scholar]
- Lata C., Prasad M. (2012). Validation of an allele-specific marker associated with dehydration stress tolerance in a core set of foxtail millet accessions. Plant Breed. 132 496–499. 10.1111/j.1439-0523.2012.01983.x [DOI] [Google Scholar]
- Lee J., Kim C.-S., Lee I.-Y. (2017). Discrimination of Echinochloa colona (L.) Link from other Echinochloa Species using DNA Barcode. Weed Turfgrass Sci. 4 225–229. 10.5660/WTS.2015.4.3.225 [DOI] [Google Scholar]
- Lee J., Park K., Lee I., Kim C., Kown O., Park T. (2014). Simple sequence repeat analysis of genetic diversity among Acetyl-CoA carboxylase inhibitor-resistant and -susceptible Echinochloa crus-galli and E. oryzicola populations in Korea. Weed Res. 55 90–100. 10.1111/wre.12119 [DOI] [Google Scholar]
- Li G., Wu S., Cai L., Wang Q., Zhao X., Wu C. (2013a). Identification and mRNA expression profile of glutamate receptor-like gene in quinclorac-resistant and susceptible Echinochloa crus-galli. Gene 531 489–495. 10.1016/j.gene.2013.09.013 [DOI] [PubMed] [Google Scholar]
- Li G., Wu S. G., Yu R. X., Cang T., Chen L. P., Zhao X. P., et al. (2013b). Identification and expression pattern of a glutathione S-transferase in Echinochloa crus-galli. Weed Res. 53 314–321. 10.1111/wre.12031 [DOI] [Google Scholar]
- Lin H.-S., Chiang C. Y., Chang S.-B., Kuoh C.-S. (2011). Development of simple sequence repeats (SSR) Markers in Setaria italica (Poaceae) and cross-amplification in related species. Int. J. Mol. Sci. 12 7835–7845. 10.3390/ijms12117835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohani U., Pandey J., Shahi N. (2012). Effect of degree of polishing on milling characteristics and proximate compositions of barnyard millet (Echinochloa frumentacea). Food Bioprocess Technol. 5 1113–1119. 10.1007/s11947-011-0518-6 [DOI] [Google Scholar]
- Lule D., de Villiers S., Fetene M., Odeny D. A., Rathore A., Das R. R., et al. (2018). Genetic diversity and association mapping of Ethiopian and exotic finger millet accessions. Crop Pasture Sci. 69 879–891. 10.1071/CP18175 [DOI] [Google Scholar]
- Mace E. S., Tai S., Gilding E. K., Li Y., Prentis P. J., Bian L., et al. (2013). Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 4:2320. 10.1038/ncomms3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manidool C. (1992). “Echinochloa crus-galli (L.) P. Beauv,” in Plant Resources of South-East Asia, eds t’ Mannetje, Jones R. M., (Jodhpur: Pudoc Scientific Publishers; ), 303. [Google Scholar]
- Manimekalai M., Dhasarathan M., Karthikeyan A., Murukarthick J., Renganathan V. G., Thangaraj K., et al. (2018). Genetic diversity in the barnyard millet (Echinochola frumentacea) germplasms revealed by morphological traits and simple sequence repeat markers. Curr. Plant Biol. 14 71–78. 10.1016/j.cpb.2018.09.006 [DOI] [Google Scholar]
- Maun M. A., Barrett S. C. (1986). The biology of Canadian weeds, Echinochloa crus- galli (L.) Beauv. Can. J. Plant Sci. 66 739–759. 10.4141/cjps86-093 [DOI] [Google Scholar]
- Mauro-Herrera M., Wang X., Barbier H., Brutnell T. P., Devos K. M., Doust A. N. (2013). Genetic Control and Comparative Genomic Analysis of Flowering Time in Setaria (Poaceae). G3 3 283L–295L. 10.1534/g3.112.005207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta H., Tyagi P. C., Mohapatra K. P. (2005). Genetic diversity in Barnyard millet (Echinochloa frumentacea Roxb.). Indian J. Genet. 65 293–295. [Google Scholar]
- Mishra A. K., Puranik S., Bahadur R. P., Prasad M. (2012). The DNA-binding activity of an AP2 protein is involved in transcriptional regulation of a stress-responsive gene, SiWD40, in foxtail millet. Genomics 100 252–263. 10.1016/j.ygeno.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Mitich L. W. (1990). Barnyardgrass. Weed Technol. 4 918–920. [Google Scholar]
- Monteiro P. V., Sudharshana L., Ramachandra G. (1988). Japanese barnyard millet (Echinochloa frumentacea): protein content, quality and SDS-PAGE of protein fractions. J. Sci. Food Agric. 43:17e25. [Google Scholar]
- Moreno Amador M., de Comino I., Sousa C. (2014). Alternative grains as potential raw material for gluten-free food development in the diet of celiac and gluten-sensitive patients. Austin J. Nutr. Food Sci. 2:9. [Google Scholar]
- Mosovska S., Mikulasova M., Brindzova L., Valik L., Mikusova L. (2010). Genotoxic and antimutagenic activities of extracts from pseudocereals in the Salmonella mutagenicity assay. Food Chem. Toxicol. 48 1483–1487. 10.1016/j.fct.2010.03.015 [DOI] [PubMed] [Google Scholar]
- Muldoon D. K. (1985). The effect of photoperiod on the growth and development of Echinochloa spp. Millets. Aust. J. Exp. Agric. 25, 428–433. 10.1071/EA9850428 [DOI] [Google Scholar]
- Muldoon K. D., Pearson C. J., Wheeler J. (1982). The effect of temperature on growth and development of Echinochloa millets. Ann. Bot. 50 665–672. 10.1093/oxfordjournals.aob.a086408 [DOI] [Google Scholar]
- Murukarthick J., Manimekalai M., Karthikeyan A., Perumal S., Dhasarathan M., Kandasamy T., et al. (2019). Transcriptomes of Indian barnyard millet and barnyardgrass reveal putative genes involved in drought adaptation and micronutrient accumulation. Acta Physiol. Plant. 41:66 10.1007/s11738-019-2855-4 [DOI] [Google Scholar]
- Muthamilarasan M., Prasad M. (2014). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128 1–14. 10.1007/s00122-014-2399-3 [DOI] [PubMed] [Google Scholar]
- Muza F. R., Lee D. J., Andrews D. J., Gupta S. C. (1995). Mitochondrial DNA variation in finger millet [Eleusine coracana (L.) Gaertn.]. Euphytica 81 199–205. 10.1007/BF00025434 [DOI] [Google Scholar]
- Nagaraja A., Mantur S. G. (2008). Evaluation of barnyard millet entries for grain smut resistance and yield. Mysore J. Agric. Sci. 42 375–377. [Google Scholar]
- Nah G., Im J.-H., Kim J.-W., Park H.-R., Yook M.-J., Yang T.-J., et al. (2015). Uncovering the differential molecular basis of adaptive diversity in three echinochloa leaf transcriptomes. PLoS One 10:e0134419. 10.1371/journal.pone.0134419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandini R., Fakrudin B. (1999). Emasculation in finger millet using hot water. Mysore J. Agric. Sci. 33 6–10. [Google Scholar]
- Nazni P., Shobana D. R. (2016). Effect of processing on the characteristics changes in barnyard and foxtail millet. J. Food Process. Technol. 7:566 10.4172/2157-7110.1000566 [DOI] [Google Scholar]
- Nguyen H. D., Bingtian Z., Le D. A. T., Yoon Y. H., Ko J. Y., Woo K. S., et al. (2016). Isolation of lignan and fatty acid derivatives from the grains of Echinochloa utilis and Their Inhibition of Lipopolysaccharide-Induced Nitric Oxide Production in RAW 264.7 cells. J. Agric. Food Chem. 64 425–432. 10.1021/acs.jafc.5b05638 [DOI] [PubMed] [Google Scholar]
- Nirgude M., Babu B. K., Shambhavi Y., Singh U. M., Upadhyaya H. D., Kumar A. (2014). Development and molecular characterization of genic molecular markers for grain protein and calcium content in finger millet (Eleusine coracana (L.) Gaertn.). Mol. Biol. Rep. 41 1189–1200. 10.1007/s11033-013-2825-7 [DOI] [PubMed] [Google Scholar]
- Nirmalakumari A., Vetriventhan M. (2009). Phenotypic analysis of anther and pollen in diversified genotype of barnyard millet (Echinochloa frumentaceae) floral characters. ICFAI Univ. J. Genet. Evol. 2 12–16. [Google Scholar]
- Nolde S., Vassilevski A. A., Rogozhin E. A., Barinov N. A., Balashova T. A., Samsonova O. V., et al. (2011). Disulfide-stabilized helical hairpin structure and activity of a novel antifungal peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus-galli). J. Biol. Chem. 286 25145–25153. 10.1074/jbc.M110.200378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa S., Takahashi M., Nakai H., Sato Y.-I. (2006). Difference in SSR Variations between Japanese Barnyard Millet (Echinochloa esculenta) and its Wild Relative E. crus-galli. Breed. Sci. 56 335–340. 10.1270/jsbbs.56.335 26081539 [DOI] [Google Scholar]
- Obidiegwu O. N., Obidiegwu J. E., Parzies H. (2013). Development of SSR for foxtail millet (Setaria italica (L.) P. Beauv.) and its utility in genetic discrimination of a core set. Genes Genomics 35 609–615. 10.1007/s13258-013-0110-8 [DOI] [Google Scholar]
- Odintsova T. I., Rogozhin E. A., Baranov Y., Musolyamov A. K., Yalpani N., Egorov T. A., et al. (2008). Seed defensins of barnyard grass Echinochloa crusgalli (L.) Beauv. Biochimie 90 1667–1673. 10.1016/j.biochi.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Okada M., Hanson B. D., Hembree K. J., Peng Y., Shrestha A., Wright D., et al. (2013). Evolution and spread of glyphosate resistance in Conyza canadensis in California. Evol. Appl. 6 761–777. 10.1111/eva.12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanrewaju O. S., Glick B. R., Babalola O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33:197. 10.1007/s11274-017-2364-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna M., Okada M., Ahmad R., Fischer A., Jasieniuk M. (2011). Genetic Diversity and spread of thiobencarb resistant early watergrass (Echinochloa oryzoides) in California. Weed Sci. 59 195–201. 10.1614/WS-D-10-00124.1 [DOI] [Google Scholar]
- Padulosi S., Bhag M. C., Bala R. S., Gowda J. V., Gowda K. T. K., Shanthakumar G. C., et al. (2009). Food security and climate change: role of plant genetic resources of minor millets. Indian J. Plant Genet. Resour. 22 1–16. [Google Scholar]
- Pandey G., Misra G., Kumari K., Gupta S., Kumar Parida S., Chattopadhyay D., et al. (2013). Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res. 20 197–207. 10.1093/dnares/dst002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar P., Dubey A., Verma A. K. (2016). Evaluation of nutraceutical and antinutritional properties in barnyard and finger millet varieties grown in Himalayan region. J. Food Sci. Technol. 53 2779e2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal S., Jayakodi M., Kim D.-S., Yang T.-J., Natesan S. (2016). The complete chloroplast genome sequence of Indian barnyard millet, Echinochloa frumentacea (Poaceae). Mitochondrial DNA 1 79–80. 10.1080/23802359.2015.1137832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabha D., Negi Y. K., Khanna V. K. (2010). Morphological and isozyme diversity in the accessions of two cultivated species of barnyard millet. Nat. Sci. 8 71–76. [Google Scholar]
- Prabha D., Negi Y. K., Khanna V. K. (2012). Assessment of genetic diversity of barnyard millet accessions using molecular markers. Indian J. Plant Genet. Resour. 25 174–179. [Google Scholar]
- Prakash R., Vanniarajan C. (2013). Genetic variability for panicle characters in indigenous and exotic barnyard millet (Echinochloa frumentacea (Roxb.) Link) germplasm over environment. Veg. An Int. J. Plant Res. 26:297 10.5958/j.2229-4473.26.2.088 [DOI] [Google Scholar]
- Qie L., Jia G., Zhang W., Schnable J., Shang Z., Li W., et al. (2014). Mapping of Quantitative Trait Locus (QTLs) that contribute to germination and early seedling drought tolerance in the interspecific cross Setaria italica×Setaria viridis. PLoS One 9:e101868. 10.1371/journal.pone.0101868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhamani J., Pandey A., Srinivasan K., Tyagi V. (2011). Conserving millet genetic resources in India. Proc. Indian Natl. Sci. Acad. 77 295–304. [Google Scholar]
- Raj S. M., Mahudeswaran K., Shanmugasundaran A. (1964). Observation on the hot water technique of emasculation of ragi flowers (Eleusine coracana (L.) Gaertn.). Madras Agric. J. 51 71–75. [Google Scholar]
- Rajput S. G., Santra D. K. (2016). Evaluation of genetic diversity of proso millet germplasm available in the United States using simple-sequence repeat markers. Crop Sci. 56 2401–2409. 10.2135/cropsci2015.10.0644 [DOI] [Google Scholar]
- Rajput S. G., Santra D. K., Schnable J. (2016). Mapping QTLs for morpho-agronomic traits in proso millet (Panicum miliaceum L.). Mol. Breed. 36:37 10.1007/s11032-016-0460-4 [DOI] [Google Scholar]
- Ramakrishnan M., Antony Ceasar S., Duraipandiyan V., Al-Dhabi N. A., Ignacimuthu S. (2016). Assessment of genetic diversity, population structure and relationships in Indian and non-Indian genotypes of finger millet (Eleusine coracana (L.) Gaertn) using genomic SSR markers. Springerplus 5 1–11. 10.1186/s40064-015-1626-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan M., Ceasar S. A., Vinod K. K., Duraipandiyan V., Ajeesh Krishna T. P., Upadhyaya H. D., et al. (2017). Identification of putative QTLs for seedling stage phosphorus starvation response in finger millet (Eleusine coracana L. Gaertn.) by association mapping and cross species synteny analysis. PLoS One 12:e0183261. 10.1371/journal.pone.0183261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat L., Nautiyal A., Bisht T. S., Prasad S., Naithani D., Makhloga K., et al. (2019). Screening of barnyard millet germplasm against shoot fly and stem borer damage under field conditions. Int. J. Curr. Microbiol. Appl. Sci. 8 1221–1226. 10.20546/ijcmas.2019.802.142 [DOI] [Google Scholar]
- Renganathan V. G., Ibrahim S. M., Vanniarajan C. (2015). Combining ability for quantitative traits in Barnyard millet (Echinochloa frumentaceae (Roxb.) Link). Electron. J. Plant Breed. 6 389–394. [Google Scholar]
- Renganathan V. G., Vanniarajan C., Nirmalakumari A., Raveendran M., Thiyageshwari S. (2017). Cluster analyses for qualitative and quantitative traits in barnyard millet Echinochloa frumentacea (Roxb.Link) germplasm. Bioscan 12 1927–1931. [Google Scholar]
- Renganathan V. G., Vanniarajan C., Ramalingam J. (2019). “Genetic analysis and identification of molecular markers linked to the anthocyanin pigmentation in barnyard millet [Echinochloa frumentacea Roxb (Link)],” in Proceedings of the Neglected and Underutilized crop species for Food, Nutrition, Energy and Environment, (New Delhi: NIPGR; ), 43. [Google Scholar]
- Ruiz-Santaella P. J., Bastida F., Franco A., Prado R. (2006). Morphological and molecular characterization of different Echinochloa spp. and Oryza sativa populations. J. Agric. Food Chem. 54 1166–1172. 10.1021/jf0520746 [DOI] [PubMed] [Google Scholar]
- Rutledge J., Talbert R. E., Sneller C. H. (2000). RAPD analysis of genetic variation among propanil-resistant and -susceptible Echinochloa crus-galli populations in Arkansas. Weed Sci. 48 669–674. 10.1614/0043-1745(2000)048[0669:raogva]2.0.co;2 [DOI] [Google Scholar]
- Saleh A., Zhang Q., Chen J., Shen Q. (2013). Millet grains: nutritional quality, processing, and potential health benefits. Compr. Rev. Food Sci. Food Saf. 12 281–295. 10.1111/1541-4337.12012 [DOI] [Google Scholar]
- Santra D., Heyduck R., Baltensperger D., Graybosch R., Nelson L., Frickel G., et al. (2015). Registration of ‘Plateau’ Waxy (Amylose-Free) proso millet. J. Plant Regist. 9:41 10.3198/jpr2013.11.0067crc [DOI] [Google Scholar]
- Sato K., Mukainari Y., Naito K., Fukunaga K. (2013). Construction of a foxtail millet linkage map and mapping of spikelet-tipped bristles1 (stb1) by using transposon display markers and simple sequence repeat markers with genome sequence information. Mol. Breed. 31 675–684. 10.1007/s11032-012-9825-5 [DOI] [Google Scholar]
- Sayani R., Chatterjee A. (2017). Nutritional and biological importance of the weed Echinochloa colona: a review. Int. J. of Food Sci. Biotechnol. 2 31–37. 10.11648/j.ijfsb.20170202.13 [DOI] [Google Scholar]
- Sebastin R., Lee K. J., Cho G. T., Lee J. R., Shin M. J., Kim S. H., et al. (2019). The complete chloroplast genome sequence of Japanese Millet Echinochloa esculenta (A. braun) H. scholz (Poaceae). Mitochondrial DNA Part B Resour. 4 1392–1393. 10.1080/23802359.2019.1598787 [DOI] [Google Scholar]
- Seetharam A., Gowda J., Halaswamy J. H. (2003). “Small millets,” in Nucleus and Breeder Seed Production Manual, eds Chowdhury S. K., Lal S. K., (New Delhi: Indian Agricultural Research Institute; ), 54–67. [Google Scholar]
- Sharma A., Sood S., Agrawal P. K., Kant L., Bhatt J. C., Pattanayak A. (2016). Detection and assessment of nutraceuticals in methanolic extract of finger (Eleusine coracana) and Barnyard Millet (Echinochloa frumentacea). Asian J. Chem. 28 1633–1637. 10.14233/ajchem.2016.19790 [DOI] [Google Scholar]
- Sharma D., Tiwari A., Sood S., Jamra G., Singh N. K., Meher P. K., et al. (2018). Genome wide association mapping of agro-morphological traits among a diverse collection of finger millet (Eleusine coracana L.) genotypes using SNP markers. PLoS One 13:e19444. 10.1371/journal.pone.0199444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siles M., Baltensperger D., Nelson L. A. (2001). Technique for Artificial Hybridization of Foxtail Millet [(L.) Beauv.]. Crop Sci. 41 1408–1412. 10.2135/cropsci2001.4151408x [DOI] [Google Scholar]
- Singh H. S., Singh K. (2005). “Status and needs of pasture and fodder management in Uttaranchal,” in Road Map for Pasture and Fodder Development in NWHR for Livestock Sustenance, eds Bisht J. K., Srivastava A. K., (Almora: Vivekananda Parvatiya Krishi Anusandhan Sansthan; ), 39–64. [Google Scholar]
- Singh K. P., Mishra H. N., Saha S. (2010). Moisture-dependent properties of barnyard millet grain and kernel. J. Food Eng. 96 598–606. 10.1016/j.jfoodeng.2009.09.007 [DOI] [Google Scholar]
- Song B. Y., Shi J. X., Song S. Q. (2015). Dormancy release and germination of Echinochloa crus-galli grains in relation to galactomannan-hydrolysing enzyme activity. J. Integr. Agric. 14 1627–1636. 10.1016/S2095-3119(14)60940-0 [DOI] [Google Scholar]
- Sood S., Khulbe R., Kumar R. A., Agrawal P. K., Upadhyaya H. (2015). Barnyard millet global core collection evaluation in the sub mountain Himalayan region of India using multivariate analysis. Crop J. 3 517–525. 10.1016/j.cj.2015.07.005 [DOI] [Google Scholar]
- Sood S., Khulbe R., Saini N., Gupta A., Agrawal P. K. (2014). Research Note Interspecific Hybrid between Echinochloa esculenta (Japanese barnyard millet) and E. frumentacea (Indian barnyard millet) – A New Avenue for Genetic Enhancement of Barnyard Millet. Electron. J. Plant Breed. 5 248–253. [Google Scholar]
- Sridevi R., Manonmani V. (2016). Seed priming effect on physiological traits of kodo millet and barnyard millet. Int. J. Agric. Sci. Res. 6 187–194. [Google Scholar]
- Sridhar R., Lakshminarayana G. (1992). Lipid class contents and fatty acid composition of small millets: barnyard little (Panicum sumatrense), kodo (Paspalum scrobiculatum), and barnyard (Echinochloa colona). J. Agric. Food Chem. 40:2131e2134. [Google Scholar]
- Subhashini V., Swamy A. V. V. S. (2014). Screening Potential of three native grass species for phyto remediation of heavy metals. Int. J. Eur. Acad. Res. 4 5887–5903. [Google Scholar]
- Sujatha K., Selvarani K., Vijayalakshmi V., Vanniarajan C., Sivasubramaniam K. (2013). Seed fortification studies in barnyard millet (Echinochloa frumentacea) cv. CO1. IOSR J. Agric. Vet. Sci. 5 22–24. 10.9790/2380-0542224 [DOI] [Google Scholar]
- Sun J., Luu N. S., Chen Z., Chen B., Cui X., Wu J., et al. (2019). Generation and Characterization of a Foxtail Millet (Setaria italica) Mutant Library. Front. Plant Sci. 10:369. 10.3389/fpls.2019.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaj D. P., Thulasidas G. (1976). Botany of Field Crops. New Delhi: Macmillan Publisher. [Google Scholar]
- Sung S.-J., Leather G. R., Hale M. G. (1987). Development and germination of barnyardgrass (Echinochloa crus-galli) seeds. Weed Sci. 35 211–215. 10.1017/S0043174500079078 [DOI] [Google Scholar]
- Surekha N., Ravikumar S. N., Mythri S., Rohini D. (2013). Barnyard Millet (Echinochloa frumentacea Link) cookies: development, value addition, consumer acceptability, nutritional and shelf life evaluation. IOSR J. Environ. Sci. Toxicol. Food Technol. 7 01–10. 10.9790/2402-0730110 [DOI] [Google Scholar]
- Suryawanshi V., Talke I. N., Weber M., Eils R., Brors B., Clemens S., et al. (2016). Between-species differences in gene copy number are enriched among functions critical for adaptive evolution in Arabidopsis halleri. BMC Genomics 17:1034. 10.1186/s12864-016-3319-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabacchi M., Mantegazza R., Spada A., Ferrero A. (2009). Morphological traits and molecular markers for classification of Echinochloa species from Italian rice fields. Weed Sci. 54 1086–1093. 10.1614/WS-06-018R1.1 [DOI] [Google Scholar]
- Trivedi A. K., Arya L., Verma S., Tyagi R., Hemantaranjan A. (2017). Evaluation of barnyard millet diversity in central Himalayan region for environmental stress tolerance. J. Agric. Sci. 155 1–11. 10.1017/S0021859617000545 [DOI] [Google Scholar]
- Ugare R., Chimmad B., Naik R., Bharati P., Itagi S. (2014). Glycemic index and significance of barnyard millet (Echinochloa frumentacae) in type II diabetics. J. Food Sci. Technol. 51 392–395. 10.1007/s13197-011-0516-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya H., Dwivedi S. L., Singh S. K., Sube S., Vetriventhan M., Sharma S. (2014). Forming core collections in Barnyard, Kodo, and little millets using morphoagronomic descriptors. Crop Sci. 54 2673–2682. 10.2135/cropsci2014.03.0221 [DOI] [Google Scholar]
- Upadhyaya H., Gowda C., Sastry D. (2008). Plant genetic resources management: collection, characterization, conservation and utilization. J. SAT Agric. Res. 6:16. [Google Scholar]
- Vanniarajan C., Anand G., Kanchana S., Arun Giridhari V., Renganathan V. G. (2018). A short duration high yielding culture - Barnyard millet ACM 10145. Agric. Sci. Dig. A Res. J. 8 123–126. 10.18805/ag.D-4574 [DOI] [Google Scholar]
- Varshney R. K., Hoisington D. A., Tyagi A. K. (2006). Advances in cereal genomics and applications in crop breeding. Trends Biotechnol. 24 490–499. 10.1016/j.tibtech.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Varshney R. K., Shi C., Thudi M., Mariac C., Wallace J., Qi P., et al. (2017). Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat. Biotechnol. 35 969–976. 10.1038/nbt.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veena B., Chimmad B. V., Naik R. K., Shantakumar G. (2004). Development of barnyard millet based traditional foods. Karnataka J. Agric. Sci. 17, 522–527. [Google Scholar]
- Veena S., Chimmad B. V., Naik R. K., Shanthakumar G. (2005). Physico-chemical and nutritional studies in Barnyard millet. Karnataka J. Agric. Sci. 18:101e105. [Google Scholar]
- Veeranagamallaiah G., Puli C., Jyothsnakumari G., Sudhakar C. (2007). Glutamine synthetase expression and pyrroline-5-carboxylate reductase activity influence proline accumulation in two cultivars of foxtail millet (Setaria italica L.) with differential salt sensitivity. Environ. Exp. Bot. 60 239–244. 10.1016/j.envexpbot.2006.10.012 [DOI] [Google Scholar]
- Vijayakumar T. P., Mohankumar J. B., Jaganmohan R. (2009). Quality evaluation of chapati from millet flour blend incorporated composite flour. Indian J. Nutr. Diet. 46 144–155. [Google Scholar]
- Wallace G. J., Upadhyaya H., Vetriventhan M., Buckler E., Hash Charles J., Ramu P. (2015). The genetic makeup of a global barnyard millet germplasm collection. Plant Genome 08 01–07. 10.3835/plantgenome2014.10.0067 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Z., Du X., Yang H., Han F., Han Y., et al. (2017). A high-density genetic map and QTL analysis of agronomic traits in foxtail millet [Setaria italica (L.) P. Beauv.] using RAD-seq. PLoS One 12:e0179717. 10.1371/journal.pone.0179717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang Z. L., Yang H. Q., Yuan F., Guo E. H., Tian G., et al. (2013). Genetic analysis and preliminary mapping of a highly male-sterile gene in foxtail millet (Setaria italica L. Beauv.) using SSR markers. J. Integr. Agric. 12 2143–2148. 10.1016/S2095-3119(13)60392-5 [DOI] [Google Scholar]
- Wanous M. K. (1990). Origin, taxonomy and ploidy of the millets and minor cereals. Plant Var. Seeds 3 99–112. [Google Scholar]
- Weber A. (2015). Discovering new biology through RNA-Seq. Plant Physiol. 169:01081.2015. 10.1104/pp.15.01081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F. (2015). The wondrous cycles of polyploidy in plants. Am. J. Bot. 102 1753–1756. 10.3732/ajb.1500320 [DOI] [PubMed] [Google Scholar]
- Xu W., Di C., Zhou S., Liu J., Li L., Liu F., et al. (2015). Rice transcriptome analysis to identify possible herbicide quinclorac detoxification genes. Front. Genet. 6:306. 10.3389/fgene.2015.00306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuno T. (1962). Cytotaxonomic studies on the two cultivated species and the wild relatives in the genus Echinochloa. Cytologia 27 296–305. 10.1508/cytologia.27.296 25058746 [DOI] [Google Scholar]
- Yabuno T. (1966). Biosystematic study of the genus Echinochloa. Jpn. J. Bot. 19 277–323. [Google Scholar]
- Yabuno T. (1984). A biosystematic study on Echinochloa oryzoides (Ard.) Fritsch. Cytologia 49 673–678. 10.1508/cytologia.49.673 25058746 [DOI] [Google Scholar]
- Yabuno T. (1987). Japanese barnyard millet (Echinochloa utilis. Poaceae) in Japan. Econ. Bot. 41 484–493. 10.1007/BF02908141 [DOI] [Google Scholar]
- Yabuno T. (1996). “Taxonomy and phylogeny of the genus Echinochloa,” in Natural History of Genus Echinochloa, eds Yabuno T., Yamaguchi H., (Tokyo: Daw Elanco/Dow Chemical Company; ), 16–28. [Google Scholar]
- Yabuno T. (2001). “Taxonomy and phylogeny of the genus Echinochloa,” in Natural History of Genus Echinochloa, eds Yabuno T., Yamaguchi H., (Tokyo: Zennokyo Shuppan; ), 15–30. [Google Scholar]
- Yadav R., Yadav V. (2013). Comparative performance of Indian and Japanese barnyard millet cultivars under varied fertility conditions for dual use in Indian Central Himalaya. Range Manag. Agrofor. 34 175–178. [Google Scholar]
- Yadav S., Gaur V., Jaiswal J. P., Kumar A. (2014). Simple sequence repeat (SSR) analysis in relation to calcium transport and signaling genes reveals transferability among grasses and a conserved behavior within finger millet genotypes. Plant Syst. Evol. 300 1–8. 10.1007/s00606-014-0982-3 [DOI] [Google Scholar]
- Yamaguchi H., Utano A. Y. A., Yasuda K., Yano A., Soejima A. (2005). A molecular phylogeny of wild and cultivated Echinochloa in East Asia inferred from non-coding region sequences of trnT-L-F. Weed Biol. Manag. 5 210–218. 10.1111/j.1445-6664.2005.00185.x [DOI] [Google Scholar]
- Yang X., Yu X.-Y., Li Y. F. (2013). De novo assembly and characterization of the barnyardgrass (Echinochloa crus-galli) transcriptome using next-generation pyrosequencing. PLoS One 8:e69168. 10.1371/journal.pone.0069168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K., Yano A., Nakayama Y., Yamaguchi H. (2006). Molecular identification of Echinochloa oryzicola Vasing. and E. crus-galli (L.) Beauv. using a polymerase chain reaction–restriction fragment length polymorphism technique. Weed Biol. Manag. 2 11–17. 10.1046/j.1445-6664.2002.00041.x [DOI] [Google Scholar]
- Ye C.-Y., Lin Z., Li G., Wang Y., Qiu J., Fu F., et al. (2014). Echinochloa chloroplast genomes: insights into the evolution and taxonomic identification of two weedy species. PLoS One 9:e113657. 10.1371/journal.pone.0113657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitsu Y., Takakusagi M., Abe A., Takagi H., Uemura A., Yaegashi H., et al. (2017). QTL-seq analysis identifies two genomic regions determining the heading date of foxtail millet. Setaria italica (L.) P.Beauv. Breed. Sci. 67 518–527. 10.1270/jsbbs.17061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuichiro M., Shinya U., Yamaguchi H. (1999). Identification of polyploid groups in the genus Echinochloa by isozyme analysis. J. Weed Sci. Technol. 44 205–217. 10.3719/weed.44.205 [DOI] [Google Scholar]
- Zee S. Y., O’brien T. P. (1971). Aleurone Transfer Cells and other Structural Features of the Spikelet of Millet. Aust. J. Biol. Sci. 24 391–396. [Google Scholar]
- Zegada-Lizarazu W., Iijima M. (2005). Deep root water uptake ability and water use efficiency of pearl millet in comparison to other millet species. Plant Prod. Sci. 8 454–460. 10.1626/pps.8.454 [DOI] [Google Scholar]
- Zhang G., Liu X., Quan Z., Cheng S., Xu X., Pan S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30 549–554. 10.1038/nbt.2195 [DOI] [PubMed] [Google Scholar]
- Zhang K., Fan G., Zhang X., Zhao F., Wei W., Du G., et al. (2017). Identification of QTLs for 14 agronomically important traits in Setaria italica based on SNPs generated from high-throughput sequencing. G3 7 1587–1594. 10.1534/g3.117.041517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Tang C., Zhao Q., Li J., Yang L., Qie L., et al. (2014). Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in Foxtail millet [Setaria italica (L.) P. Beauv.]. BMC Genomics 15:78. 10.1186/1471-2164-15-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Lee G. A., Kwon S. W., Ma K. H., Lee M. C., Park Y. J. (2012). Development and use of novel SSR markers for molecular genetic diversity in Italian millet (Setaria italica L.). Genes Genom. 34 51–57. 10.1007/s13258-011-0102-5 [DOI] [Google Scholar]
- Zou C., Li L., Miki D., Li D., Tang Q., Xiao L., et al. (2019). The genome of broomcorn millet. Nat. Commun. 10:436. 10.1038/s41467-019-08409-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


